Abstract
Engineering squid rhodopsin with modified retinal analogues is essential for understanding the conserved steric and electrostatic interaction networks that govern the architecture of the Schiff base binding site. Depriving the retinal of its steric and electrostatic contacts affects the positioning of the Schiff-base relative to the key residues Asn87, Tyr111, and Glu180. Displacement of the W1 and W2 positions and the impact on the structural rearrangements near the Schiff base binding region reiterates the need for the presence of internal water molecules and the accessibility of binding sites to them. Also, the dominant role of the Glu180 counterion in inducing the S1/S2 state reversal for SBR is shown for the first time in squid rhodopsin.
Squid rhodopsin is an invertebrate α-helical Gq protein-coupled receptor (GPCR) that consists of 448 amino acids. Its chromophore, the 11-cis-retinal, is bound to Lys305 as a protonated Schiff base (PSBR).1 Recently, two X-ray crystal structures of squid rhodopsin have been reported at 3.72 and 2.5 Å resolutions, and they showed Tyr111 and Asn87 to be the prospective H-bonding partners for PSBR.1 Subsequent QM/MM evaluation of the structure and spectral tuning mechanism in squid rhodopsin identified a negatively charged Glu180 residue situated at 4.29 Å from SBN+ that acts as the non-H-bonded counterion to the PSBR.3
Generally, the remarkable stability and spectral sensitivity of visual pigments is attributed to specific bonded and non-bonded retinal–protein interactions.4 In the case of squid rhodopsin, due to the absence of high-resolution X-ray, NMR, and long-range MD simulation data, details of the retinal architecture, especially the positioning of the Schiff base relative to the key residues Asn87, Tyr111, and Glu180, remain unclear. Therefore, in order to examine this issue, in the present paper, the wildtype (WT) retinal is drawn out of its comfort zone and replaced by demethyl (DM) and deprotonated Schiff base (SBR) retinal analogues. In other words, (i) the methyl groups attached to C1, C5, C9 and C13 carbon atoms and (ii) the hydrogen atom at the SB terminal of the 11-cis-retinal, are removed. In the absence of experimental measurements of these retinal analogues, theoretical predictions derived from this study may help in clarifying some of the uncertainties associated with the structure of the active site in squid rhodopsin.
To begin with, the QM/MM-optimized structure of wildtype squid rhodopsin is prepared starting from chain-A of the 2.5 Å X-ray structure. The details of the model building and computational setup have been described elsewhere.3 In short, the overall charge of the system after adding the missing hydrogen atoms is +1/0 due to PSB/SB linkage between 11-cis-retinal and Lys305. The resulting coordinates are fully optimized with a hybrid QM/MM (QM = B3LYP/6-31G*; MM = AMBER) method in ONIOM (Our own N-layer Integrated molecular Orbital + molecular Mechanics) electronic embedding scheme implemented in Gaussian03.5,6
The corresponding retinal analogue structures were prepared using the WS2 system setup of ref 3 as a template (see Supporting Information). Herein, the wildtype PSBR (WT) was replaced by demethyl (DM) and then by deprotonated SB chromophore in the presence of the protonated (SBRcip) and deprotonated (SBRcidp) Glu180 counterion. The resulting structures were then reoptimized without any constraints. The QM model part always consists of the retinal plus side-chain N–H moiety of Lys305 along with a hydrogen link atom at the QM/MM frontier region.
Ab initio multireference QM/MM calculations on the resulting structures were carried out using the ORCA 2.6.19 program package.7 Spectroscopy oriented configuration interaction (SORCI+Q) method was applied on top of three-root state-averaged complete active space self-consistent field (CASSCF) wave functions to calculate the absorption and circular dichroism (CD) spectra. The active space consisted of six electrons in six orbitals, and the 6-31G* basis set was used in all calculations. The first (S1) and second (S2) excited states and oscillator and rotatory strengths were calculated for all of the structures discussed in this study. We estimate the accuracy of this computational setup to be within ±15 nm, and the setup has been shown to yield results in excellent agreement with experimental measurements.3,8
It looks like binding to the protein reduces the differences between the WT, DM, SBRcip, and SBRcidp models significantly, making them all structurally alike (Figure 1 and Table 1). However, a closer look reveals a different picture. Compared to the WT (Figure 1A), removal of the bulky methyl substituents deprives the DM of its natural steric contacts and thus causes a significant contraction of the retinal (Figure 1B). As a consequence, the length of the chromophore conjugation (from C5 to SBN+) is drastically reduced from 10.61 Å in WT to 10.23 Å in DM. In particular, the H-bonded network between SB and Asn87 involving a bridged water molecule (W1) is disrupted. This in turn causes Asn87 to draw closer by almost 1.0 Å (from 3.93 Å in WT to 2.78 Å in DM) to form a strong H-bond with SBN+. In contrast, contraction induces both Tyr 111 and Glu180 to move farther away by 0.06 Å (3.41 Å in WT to 3.47 Å in DM) and by 0.26 Å (4.29 Å in WT to 4.55 Å in DM).
Figure 1.
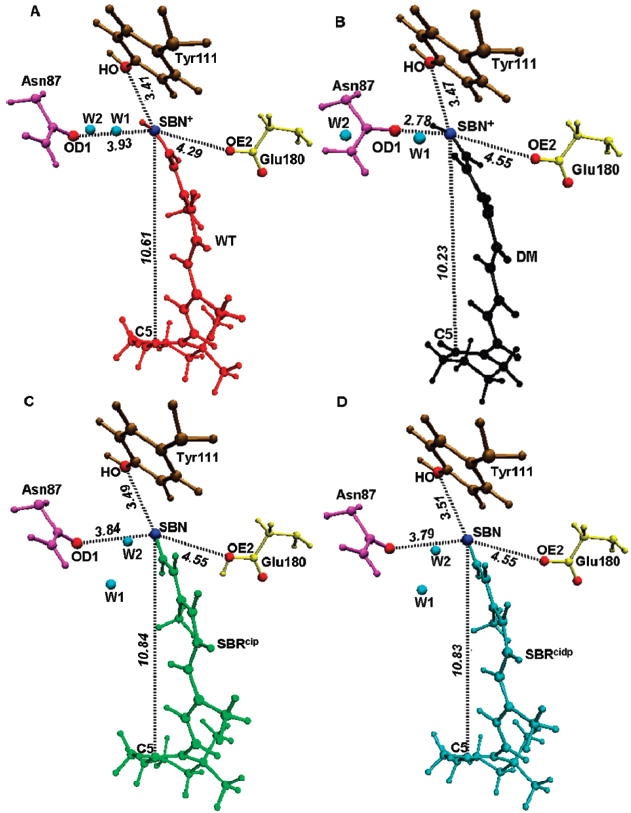
Optimized QM/MM geometries of (A) wildtype (WT) in red, (B) demethyl (DM) in black, (C) deprotonated (SBRcip) in the presence of protonated counterion Glu180 in green, and (D) deprotonated (SBRcidp) in the presence of deprotonated counterion Glu180 in cyan. Also shown are the distances (black dashed lines) between SBN+ or the SBN atom to the OH atom of Tyr111 (ochre), the OD1 atom of Asn87 (magenta), the OE2 atom of Glu180 (yellow), and the C5 atom of the retinal. Also shown are the two internal water molecules W1 and W2 (blue circles) near the SB.
Table 1.
Comparison of Distances from the PSB Nitrogen Atom to the Side Chain Oxygen Atom of the Asn87, Tyr111, and Glu180 Residues between the Optimized QM/MM Wildtype (WT), Demethyl (DM), and Deprotonated (SBRcip, SBRcidp) Retinal Geometriesa
| distance (Ǻ) | QM/MM-optimized structures |
|||
|---|---|---|---|---|
| WT | DM | SBRcip | SBRcidp | |
| OH (Tyr111) ↔ N (PSBR) | 3.41 | 3.47 | 3.49 | 3.51 |
| OD1 (Asn87) ↔ N (PSBR) | 3.93 | 2.78 | 3.84 | 3.79 |
| OE2 (Glu180) ↔ N (PSBR) | 4.29 | 4.55 | 4.55 | 4.55 |
| C5 ↔ SBN+/SBN | 10.61 | 10.23 | 10.84 | 10.83 |
| A-DBL | 1.365 | 1.359 | 1.348 | 1.347 |
| A-SBL | 1.432 | 1.427 | 1.454 | 1.457 |
| BLA | 0.067 | 0.068 | 0.106 | 0.100 |
A-DBL refers to the average double bond length. A-SBL refers to the average single bond length. BLA refers to the bond length alternation, which is A-SBL minus A-DBL. Details of the geometric parameters are given in the Supporting Information.
However, removal of the SB proton causes the retinal to expand, resulting in the sharp increase of the length of conjugation from 10.61 Å in WT to 10.84 Å in SBRcip (Figure 1C) and to 10.83 Å in SBRcidp (Figure 1D). Even though W1 is also displaced in this case, due to the loss of positive charge on the SB terminal, the effect of expansion has only a negligible effect on the distance of Asn87, which decreases from 3.93 Å in WT to 3.84 Å in SBRcip and to 3.79 Å in SBRcidp, and on the distance of Tyr111, which increases from 3.41 Å in WT to 3.47 Å in SBRcip and to 3.51 Å in SBRcidp. However, as expected, the loss of the SB proton repels Glu180 from the retinal by 0.26 Å, irrespective of its protonation state (4.29 Å in WT to 4.55 Å in both SBRcip and SBRcidp).
Geometric parameters of the QM/MM-optimized WT, DM, and SBR models show significant single/double bond length alternation (BLA) along the retinal backbone (Figure 2, bottom). In both WT and DM cases, BLA reduction starts to set in from C8 and extends up to the SB terminal, regardless of whether they pertain to formal single or double bonds. Compared to the WT, the average double bond lengths decrease by ~0.017 Å, and the average single bond lengths increase by ~0.025 Å in SBR. The calculated average BLA evaluates to ~0.07 Å for both WT and DM and ~0.10 Å for both SBR models.
Figure 2.
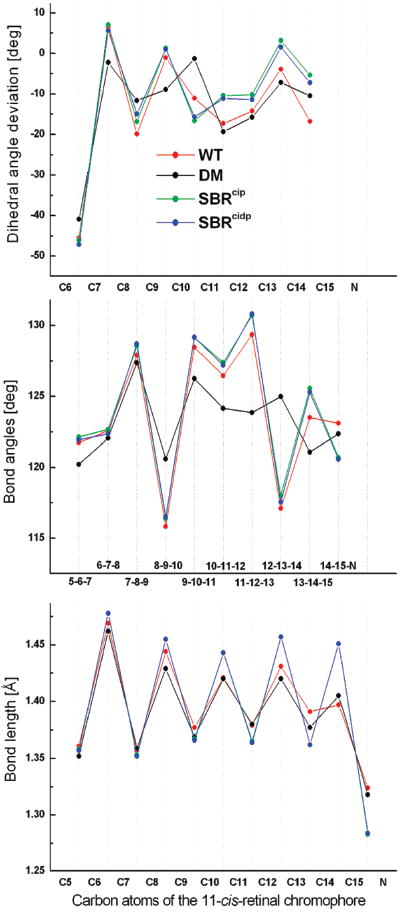
Comparison of the bond length alternation (bottom), bond angles (middle), and dihedral angles (top) along the conjugated carbon chain of the optimized QM/MM wildtype PSBR (red), demethyl PSBR (black), deprotonated SBRcip (green), and SBRcidp (blue) geometries. The dihedral angle deviations are from either the cis (0°) or the trans (180°) configuration.
In the case of DM, the smallest bond angles (Figure 2, middle) at C9 and C13 become larger, which indicates the absence of spacious methyl substituents at those positions. Removal of the SB proton (SBRcip and SBRcidp) causes the bond angles near the photoisomerization region to widen by ~2°. Also, it does not seem to affect the orientation of the β-ionone ring toward the polyene chain. However, loss of methyl groups reduces the C6–C7 twist angle (Figure 2, top) by 4° (−45° in WT; −41 ° in DM).
The C11–C12 double bond has been shown to be almost planar in vacuo.9,10 Therefore, the origin of both the sign and magnitude of the negative pretwist (− 17°) should arise from the protein environment. However, this property is often attributed to the specific nonbonded steric interaction between C13-methyl and C10-hydrogen atoms in the protein. Surprisingly, the present QM/MM calculations rule out this effect as in the absence of methyl groups, the twist angle increases by ~2° (from −17° in WT to −19° in DM). Despite this finding, one cannot rule out the possibility of the involvement of steric interaction between C13-methyl and C10-hydrogen in accelerating the photoisomerization pathway.11 On the contrary, removal of the SB proton (SBRcip and SBRcidp) reduces the C11 – C12 twist angle by 10°. Therefore, half of the negative pretwist on the WT should arise from the weak BLA due to localization of the positive charge at the SB terminal.8,12
The calculated excited-state properties of the QM/MM-optimized structures of squid rhodopsin are given in Table 2. In the gas phase, the WT, DM, SBRcip, and SBRcidp models absorb at 604,3 577, 326, and 333 nm. Inclusion of the electrostatic and polarization effects from the protein shifts the S1 absorbance to 490,3,13,14 471, 314, and 313 nm, respectively.
Table 2.
Calculated SORCI+Q Vertical Excitation Energies (λ) in nm, Oscillator (f) and Rotatory (R) Strengths in au of the WT, DM, SBRcip, and SBRcidp Chromophores in Gas-Phase (gp) and Protein Environmentsa,b
| structure | excited states | Excited-State Properties of Squid Rhodopsin |
|||||
|---|---|---|---|---|---|---|---|
| gas phase |
protein |
||||||
| λ | f | R | λ | f | R | ||
| WT | S1 | 604 | 0.93 | 0.16 | 490 | 1.14 | 0.32 |
| S2 | 429 | 0.30 | 0.48 | 393 | 0.14 | 0.16 | |
| N87‡ | S1 | 510 | 1.10 | 0.32 | |||
| S2 | 397 | 0.17 | 0.20 | ||||
| Y111‡ | S1 | 501 | 1.12 | 0.31 | |||
| S2 | 395 | 0.15 | 0.18 | ||||
| E180‡ | S1 | 591 | 0.98 | 0.17 | |||
| S2 | 422 | 0.29 | 0.42 | ||||
| DM | S1 | 577 | 1.06 | -0.08 | 471 | 1.22 | 0.07 |
| S2 | 425 | 0.16 | 0.16 | 392 | 0.05 | 0.04 | |
| N87┴ | S1 | 504 | 1.15 | 0.00 | |||
| S2 | 402 | 0.11 | 0.09 | ||||
| Y111┴ | S1 | 478 | 1.19 | 0.03 | |||
| S2 | 394 | 0.09 | 0.07 | ||||
| E180┴ | S1 | 551 | 1.10 | -0.01 | |||
| S2 | 419 | 0.14 | 0.11 | ||||
| SBRcip | S1 | 318 | 1.50 | 0.22 | 335 | 0.00 | 0.00 |
| S2 | 326 | 0.05 | 0.02 | 314 | 1.59 | 0.31 | |
| N87¶ | S1 | 335 | 0.02 | 0.01 | |||
| S2 | 319 | 1.56 | 0.32 | ||||
| Y111¶ | S1 | 334 | 0.01 | 0.01 | |||
| S2 | 315 | 1.58 | 0.32 | ||||
| E180¶ | S1 | 323 | 1.51 | 0.29 | |||
| S2 | 334 | 0.05 | 0.02 | ||||
| SBRcidp | S1 | 322 | 1.52 | 0.32 | 333 | 0.00 | 0.00 |
| S2 | 333 | 0.05 | 0.03 | 313 | 1.58 | 0.34 | |
| N87# | S1 | 332 | 0.00 | 0.01 | |||
| S2 | 313 | 1.53 | 0.28 | ||||
| Y111# | S1 | 333 | 0.02 | 0.00 | |||
| S2 | 313 | 1.57 | 0.32 | ||||
| E180# | S1 | 327 | 1.47 | 0.29 | |||
| S2 | 332 | 0.07 | 0.03 | ||||
Charges of N87, Y111, and E180 are turned off, one at a time, in the corresponding ‡WT, ┴DM, ¶SBRcip, and #SBRcidp QM/MM calculations.
cipCounterion Glu180 is protonated (charge = 0). cidpCounterion Glu180 is deprotonated (charge = −1). The properties of the optically allowed transition are given in bold.
To disentangle the origin of this blue shift, the charges of Asn87, Tyr 111, and Glu180 near the SB binding site are turned off, one at a time (see Table 2). The results indicate that Asn87 contributes 20 nm in the WT and 33 nm in DM. This increase in the spectral shift is attributed to the 1.15 Å decrease in the distance of Asn87 to SBN+ in DM. In contrast, a slight increase of 0.06 Å in the position of Tyr111 causes a decrease of 4 nm (11 nm in WT to 7 nm in DM). However, turning off the charges of the counterion (Glu180) induces a significant spectral shift of 101 nm (490 to 591 nm) in WT and 80 nm (471 to 551 nm) in DM. Therefore, the retinal–protein interaction causes an increase in the excitation energy and thus blue shifts the absorbance from gas phase by ~100 nm, an aspect also seen in the studies of bovine rhodopsin.3,8,15,16
Removal of the methyl groups induces a blue shift of 27 nm in the gas phase (604 to 577 nm) and 19 nm (490 to 471 nm) in the protein environment. The 27 nm spectral shift in the gas phase correlates well with the 43 nm blue shift calculated at the CC2/def2-TZVPP level using the MP2/TZVP molecular structure, 17 whereas removal of the SB proton induces a blue shift of almost ~300 nm in the gas phase (from 604 to ~320 nm) and ~175 nm in protein (from 490 to ~315 nm). Irrespective of the protonation state of the counterion, the role of the protein environment on SBR is minimal as the 114 nm spectral shift (604 to 490 nm) in the WT is reduced to less than 10 nm (318 to 314 nm in SBRcip and 322 to 313 nm SBRcidp).8
Generally, the electronic spectrum of the deprotonated SBR in proteins as well as that in solutions is characterized by the peculiar excited-state (S1/S2) level ordering reversal.18,19 This property is of significant diagnostic value for characterizing the counterion environment around the protein–bound retinal. In the case of squid rhodopsin, the calculated state reversal in the protein compared to that in the gas phase is in good agreement with the experimental findings.18 Surprisingly, when the charges of Glu180 is turned off, state reversal does not occur, and S1 is above S2.
CD spectroscopy is a useful tool to distinguish between different retinal isomers.20 Turning to the CD spectra of squid rhodopsin, the positive helicity for the retinal arises from the negatively and positively twisted C11–C12 and C12–C13 bonds.3,21 The strong contribution of methyl groups on the calculated rotatory strength (R) is evident as R is reduced from +0.32 au in WT to +0.07 au in DM. Also, the chiral discrimination exerted by the electrostatic influence of the protein environment on DM is evident as the sign of R changes from −0.08 au in the gas phase to +0.07 au in protein. However, removal of the SB proton does not seem to exert any significant effect on the magnitude of R.
Engineering squid rhodopsin with modified retinal analogues is essential for understanding the conserved steric and electrostatic interaction networks that govern the architecture of the Schiff base binding site. Depriving the retinal of its steric and electrostatic contacts affects the positioning of the Schiff base relative to the key residues Asn87, Tyr111, and Glu180. Although the use of the SBRcidp model is purely hypothetical, theoretical curiosity on modeling such an active site allows us to study the dominant role of Glu180 in inducing the S1/S2 state reversal in squid rhodopsin. Displacement of the W1 and W2 positions and its impact on the structural rearrangements near the binding site reiterates the need for the presence of internal water molecules and the accessibility of binding sites to them. Although we recognize the individual role of methyl groups and theoretical evaluation of the origin of the protonation state of Glu180 in squid compared to that of Glu181 in bovine rhodopsin to be missing in this study, detailed calculations in this direction are in progress.
Supplementary Material
Acknowledgments
The authors thank the anonymous referees for their critical comments. The authors also thank Dr. A. Altun for valuable discussions during the course of this work. The work at Emory is supported, in part, by a grant from the National Institutes of Health (R01EY016400-04), and that at Kyoto is supported by a Core Research for Evolutional Science and Technology (CREST) grant in the Area of High Performance Computing from JST.
Footnotes
SUPPORTING INFORMATION AVAILABLE ONIOM (QM/MM)-optimized Cartesian coordinates and geometric parameters of the models discussed in this study. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Murakami M, Kouyama T. Crystal Structure of Squid Rhodopsin. Nature. 2008;453:363–367. doi: 10.1038/nature06925. [DOI] [PubMed] [Google Scholar]
- 2.Shimamura T, Hiraki N, Takahashi T, Hori H, Ago K, Masuda K, Takio M, Ishiguro M, Miyano M. Crystal Structure of Squid Rhodopsin with Intracellularly Extended Cytoplasmic Region. J Biol Chem. 2008;283:17753–17756. doi: 10.1074/jbc.C800040200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sekharan S, Altun A, Morokuma K. Photochemistry of Visual Pigment in a Gq Protein-Coupled Receptor (GPCR)—Insights from Structural and Spectral Tuning Studies on Squid Rhodopsin. Chem—Eur J. 2010 doi: 10.1002/chem.200903194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birge RR. Photophysics and Molecular Electronic Applications of the Rhodopsins. Annu Rev Phys Chem. 1990;41:683–733. doi: 10.1146/annurev.pc.41.100190.003343. [DOI] [PubMed] [Google Scholar]
- 5.Vreven T, Byun KS, Komáromi I, Dapprich S, Montgomery JA, Jr, Morokuma K, Frisch MJ. Combining Quantum Mechanics Methods with Molecular Mechanics Methods in ONIOM. J Chem Theory Comput. 2006;2:815–826. doi: 10.1021/ct050289g. [DOI] [PubMed] [Google Scholar]
- 6.Bakowies D, Thiel W. Hybrid Models for Combined Quanum Mechanical and Molecular Mechanical Approaches. J Phys Chem. 1996;100:10580–10594. [Google Scholar]
- 7.Neese F. A Spectroscopy Oriented Configuration Interaction Procedure. J Chem Phys. 2003;119:9428–9443. [Google Scholar]
- 8.Altun A, Yokoyama S, Morokuma K. Mechanism of Spectral Tuning Going from Retinal in Vacuo to Bovine Rhodopsin and its Mutants: Multireference Ab Initio and Quantum Mechanics/Molecular Mechanics Studies. J Phys Chem B. 2008;112:16883–16890. doi: 10.1021/jp807172h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Röhrig UF, Guidoni L, Rothilsberger U. Solvent and Protein Effects on the Structure and Dynamics of the Rhodopsin Chromophore. ChemPhysChem. 2005;6:1836–1847. doi: 10.1002/cphc.200500066. [DOI] [PubMed] [Google Scholar]
- 10.Bravaya K, Bochenkova A, Granovsky A, Nemukhin A. An Opsin Shift in Rhodopsin: Retinal S0–S1 Excitation in Protein, in Solution and in the Gas Phase. J Am Chem Soc. 2007;129:13035–13042. doi: 10.1021/ja0732126. [DOI] [PubMed] [Google Scholar]
- 11.Wang Q, Kochendoerfer GG, Schoenlein RW, Verdegem PJE, Lugtenburg J, Mathies RA, Shank CV. Femtosecond Spectroscopy of 13-Demethylrhodopsin Visual Pigment Analogue: The Role of Nonbonded Interactions in the Isomerization Process. J Phys Chem. 1996;100:17388–17394. [Google Scholar]
- 12.Sugihara M, Hufen J, Buss V. Origin and Consequences of Steric Strain in the Rhodopsin Binding Pocket. Biochemistry. 2006;45:801–810. doi: 10.1021/bi0515624. [DOI] [PubMed] [Google Scholar]
- 13.Hubbard R, St. George RCC. The Rhodopsin Systemof the Squid. J Gen Physiol. 1958;41:501–528. doi: 10.1085/jgp.41.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hara T, Hara R, Takeuchi J. Vision in Octopus and Squid. Nature. 1967;214:572–573. doi: 10.1038/214572a0. [DOI] [PubMed] [Google Scholar]
- 15.Gascon JA, Sproviero EM, Batista V. Computational Studies of the Primary Phototransduction Event in Visual Rhodopsin. Acc Chem Res. 2006;39:184–193. doi: 10.1021/ar050027t. [DOI] [PubMed] [Google Scholar]
- 16.Sekharan S, Sugihara M, Weingart O, Okada T, Buss V. Protein Assistance in the Photoisomerization of Rhodopsin and 9-cis-Rhodopsin—Insights from Experiment and Theory. J Am Chem Soc. 2007;129:1052–105. doi: 10.1021/ja066970p. [DOI] [PubMed] [Google Scholar]
- 17.Send R, Sundholm D. Coupled Cluster Studies of the Lowest Excited States of the 11-cis-Retinal Chromophore. Phys Chem Chem Phys. 2007;9:2862–2867. doi: 10.1039/b616137e. [DOI] [PubMed] [Google Scholar]
- 18.Birge RR, Murray LP, Pierce BM, Akita H, Balogh-Nair V, Findsen LA, Nakanishi K. Two-Photon Spectroscopy of Locked-11-cis-Rhodopsin: Evidence for a Protonated Schiff Base in the Neutral Protein Binding Site. Proc Natl Acad Sci U S A. 1985;82:4117–4121. doi: 10.1073/pnas.82.12.4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bachilo SM, Bondarev SL, Gillbro T. Fluorescent Properties of Protonated and Unprotonated Schiff Bases of Retinal at Room Temperature. J Photochem Photobiol B. 1996;34:39–46. [Google Scholar]
- 20.Send R, Sundholm D. The Molecular Structure of a Curl-Shaped Retinal Isomer. J Mol Model. 2008;14:717–726. doi: 10.1007/s00894-008-0284-1. [DOI] [PubMed] [Google Scholar]
- 21.Shichida S, Tokunaga F, Yoshizawa T. Circular Dichroism of Squid Rhodopsin and its Intermediates. Biochim Biophys Acta. 1978;504:413–430. doi: 10.1016/0005-2728(78)90064-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


