Abstract
Genomic instability and alterations in gene expression are hallmarks of eukaryotic aging. The yeast histone deacetylase Sir2 silences transcription and stabilizes repetitive DNA, but during aging or in response to a DNA break, the Sir complex relocalizes to sites of genomic instability, resulting in the desilencing of genes that cause sterility, a characteristic of yeast aging. Using embryonic stem cells, we show that mammalian Sir2, SIRT1, represses repetitive DNA and a functionally diverse set of genes across the mouse genome. In response to DNA damage, SIRT1 dissociates from these loci and relocalizes to DNA breaks to promote repair, resulting in transcriptional changes that parallel those in the aging mouse brain. Increased SIRT1 expression promotes survival in a mouse model of genomic instability and suppresses age-dependent transcriptional changes. Thus, DNA damage-induced redistribution of SIRT1 and other chromatin modifying proteins may be a conserved mechanism of aging in eukaryotes.
Keywords: epigenetics, sirtuin, chromatin, histone, DNA repair
Introduction
Chromosomes are arguably the most difficult structure for an organism to maintain over a lifetime. Chromosomes break, mutations accumulate, and youthful gene expression patterns are progressively lost. Some changes in gene expression have been interpreted as beneficial responses to cellular damage (Narita et al., 2006; Niedernhofer et al., 2006). However, there are numerous stochastic changes in gene expression that have no apparent long-term benefit to the organism and may in fact be detrimental (Bahar et al., 2006). The "Heterochromatin Island Hypothesis" and related hypotheses propose that alterations in chromatin and the resulting gene expression changes can drive the aging process, but evidence is lacking (Cutler, 1995; Imai and Kitano, 1998; Vijg, 2004; Villeponteau, 1997).
Potential clues about the relationship between epigenetic changes and aging come from studies in Saccharomyces cerevisiae, where epigenetic changes are a primary cause of the aged phenotype. The SIR2 gene, encoding a nutrient-responsive NAD+-dependent histone deacetylase, has emerged as a key regulator of health and lifespan in yeast and other organisms (Haigis and Guarente, 2006). Two major functions of Sir2 are to repress gene expression at the silent mating type loci HML and HMR (Klar et al., 1979; Rine et al., 1979) and to suppress recombination at the ribosomal DNA (rDNA) locus, which gives rise to toxic rDNA circles (ERCs) (Sinclair and Guarente, 1997). As yeast cells age, the Sir protein complex dissociates from HM loci and moves to the nucleolus in response to ERC accumulation causing sterility, a hallmark of yeast aging (Kennedy et al., 1997; Sinclair and Guarente, 1997; Smeal et al., 1996). Thus, a redistribution of chromatin modifying factors and the resulting changes in transcription are the cause of primary yeast aging phenotypes.
Aging is not the only stimulus that causes yeast Sir proteins to relocalize. DNA breakage causes Sir proteins to dissociate from HM loci and relocate to DNA breaks, in a DNA damage checkpoint dependent manner (Martin et al., 1999; McAinsh et al., 1999; Mills et al., 1999). The effect of relocalization appears to be two fold: (i) expression of HM genes promotes DNA repair and (ii) Sir proteins directly modify chromatin surrounding the break site, possibly to facilitate DNA repair (Lee et al., 1999; Tamburini and Tyler, 2005).
There is some evidence that related processes occur in mammals. First, cells damaged by oxidative stress in vitro undergo stochastic transcriptional changes that parallel those in aged heart tissue (Bahar et al., 2006). Second, a deficiency in the DNA repair factor ERCC1 accelerates aging phenotypes and generates gene expression profiles reminiscent of aged animals (Niedernhofer et al., 2006). Third, cells that senesce due to replicative aging in vitro or in aged tissues in vivo exhibit alterations in heterochromatin (Herbig et al., 2006; Narita et al., 2006) and secrete growth factors that can drive tumorigenesis (Campisi, 2005). Finally, oxidative DNA damage at promoters correlates with gene repression in the aging human brain (Lu et al., 2004) and has been linked to both transcriptional and epigenetic changes that may contribute to Alzheimer disease (Wu et al., 2008).
To date, no study has tested whether Sir2-mediated alterations in chromatin contribute to aging in mammals. Several observations, however, are consistent with this possibility. The mammalian ortholog of Sir2, SIRT1, regulates both the expression of individual genes (Picard et al., 2004; Pruitt et al., 2006; Vaquero et al., 2004) and the formation of facultative heterochromatin (Vaquero et al., 2007). SIRT1 has also been linked to the DNA damage response via regulation of p53 (Luo et al., 2001; Vaziri et al., 2001) and its interaction with Nbs1, a component of the DNA damage sensor complex MRN (Mre11-Rad50-Nbs1) (Yuan et al., 2007). Furthermore, SIRT1 has recently been implicated in the regulation of DNA methylation patterns at damaged CpG-rich DNA (O'Hagan et al., 2008). Deletion of another Sir2 homolog, SIRT6, reduces base excision DNA repair and causes an accelerated aging phenotype in mice (Mostoslavsky et al., 2006). In this study, we map the interaction between SIRT1 and the mouse genome and identify an evolutionarily conserved DNA damage response that may drive changes in gene expression during aging.
Results
Loss of Sir2 and SIRT1-dependent silencing in response to oxidative stress
Our previous studies on the relocalization of yeast Sir2 utilized highly artificial means to induce DNA damage, such as EcoRI and the yeast HO endonuclease (Mills et al., 1999). To test whether a stress more relevant to aging results in desilencing of mating-type loci, a yeast strain carrying a GFP reporter at the HMR locus (HMR::GFP) was exposed to oxidative stress (i.e. H2O2). There was a tight correlation between H2O2 levels and HMR derepression (Figure 1A and Figure S1A).
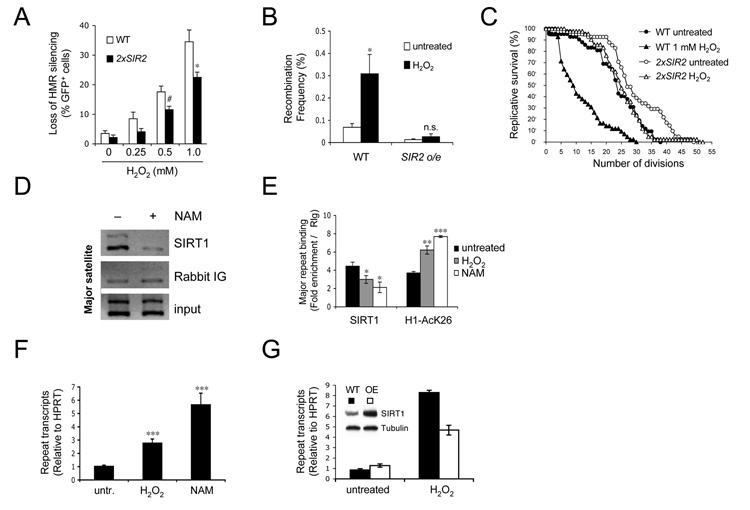
Figure 1.
Oxidative stress reduces Sir2/SIRT1-mediated repetitive DNA silencing in yeast and mammalian cells. (A–C). Effects of H2O2 on yeast aging. (A) H2O2 causes loss of silencing at the mating type loci. Shown is the fraction of GFP+ cells in 2xSIR2 and WT cells. Unless otherwise noted, P values are based on Student’s two-tailed t test with #P≤ 0.1, *P≤ 0.05, **P≤ 0.01, ***P≤ 0.001. (B) Frequency of H2O2-induced rDNA recombination in the absence presence of extra Sir2 (WT and Sir2 o/e). (C) Replicative lifespan of WT and 2xSIR2 cells grown in the absence or presence of 1 mM H2O2. (D) Mouse SIRT1 binds to repetitive genomic DNA. ChIP for SIRT1 or control immunoglobulin (Ig) at major satellite repeats in the absence or presence of NAM (25 mM) (E) q-PCR analysis of ChIP experiments using antibodies specific for SIRT1 or H1AcK26. ES cells were left untreated or treated with H2O2 (2 mM) or NAM for 1 h. (F) Oxidative stress increases transcription of satellite repeat DNA. ES cells were treated with NAM for 24 h or with H2O2 for 1 h, followed by 23 h recovery. (G) Cells with a targeted extra copy of SIRT1 and control cells were treated with H2O2 and analyzed as in (F). The inset shows a Western blot of SIRT1 in WT (black) and SIRT1 overexpressing ES cells (white). Data are represented as mean +/− SEM.
An additional copy of SIR2 (2xSIR2) extends replicative lifespan (Kaeberlein et al., 1999), but its effect on oxidative stress resistance and loss of silencing is unknown. When treated with H2O2, strains with increased Sir2 levels maintained HMR silencing and had more stable rDNA compared to wild type cells (Figures 1A and B). At non-lethal concentrations of H2O2, the 2xSIR2 strain had a significantly greater replicative lifespan (Figure 1C and Figure S1B). Thus, additional SIR2 suppresses the toxicity, genomic instability, and desilencing caused by genotoxic stress.
We next sought to examine whether these findings are relevant to mammals. We first tested if SIRT1 associates with highly repetitive DNA such as pericentromeric major satellite repeats in mouse ES cells, a cell type that has been previously used to study satellite repeat silencing (Kanellopoulou et al., 2005). Histone H1 acetylation on lysine 26 (H1AcK26) served as a read-out for SIRT1 deacetylase activity (Vaquero et al., 2004). Using chromatin immunoprecipitation (ChIP) and quantitative PCR (q-PCR), we detected an association between SIRT1 and major satellite repeats that was disrupted by the pansirtuin inhibitor nicotinamide (NAM) (Figure 1D and 1E). This coincided with an increase in repeat transcripts (Figure 1F) and H1AcK26 (Figure 1E). Consistent with known redundancy among histone deacetylases (Zhu et al., 2004), trichostatin A (TSA), NAM and sirtinol, pan-class I/II and pan-class III (sirtuin) inhibitors, respectively, caused desilencing of major satellite repeats (Figure 1F and Figures S1D and E, (Kanellopoulou et al., 2005)), whereas no significant increase in repeat transcripts was seen after stable knock-down of SIRT1 (Figure S1C and data not shown). Ongoing work is aimed at identifying the role of other HDACs that may contribute to satellite repeat silencing.
In accordance with our observations in yeast, treatment of cells with non-cytotoxic levels of H2O2 greatly decreased the amount of SIRT1 bound to repeats, coinciding with an increase in H1AcK26 (Figure 1E, Figure S2 and S3). Similar to the effect of NAM and TSA, oxidative stress increased the transcription of these loci (Figure 1F), an effect that was counteracted by overexpression of SIRT1 (Figure 1G). Together, these data indicate that SIRT1 binds and can contribute to the silencing of major satellite repeats.
Global changes in promoter-associated SIRT1 in response to oxidative stress
Given the role of yeast Sir2 in silencing HM loci, we hypothesized that mammalian SIRT1 might regulate a number of protein-coding genes. To test this, we used ChIP in combination with a genome-wide promoter tiling array (ChIP on chip) to identify putative SIRT1 target genes. Immunoprecipitated DNA from either untreated or H2O2-treated cells was hybridized to a NimbleGen MM5 array to detect SIRT1- and H1AcK26-associated promoter segments (Figure 2A). Based on gene ontology (GO), overrepresented groups included genes for chromatin assembly and transcriptional repression (e.g. methyl-CpG binding proteins 2, -3 and -4, SIRT7, and several histone genes), ubiquitin-regulated protein degradation, and cell cycle regulation (Figure 2B, S4A and Table S1).
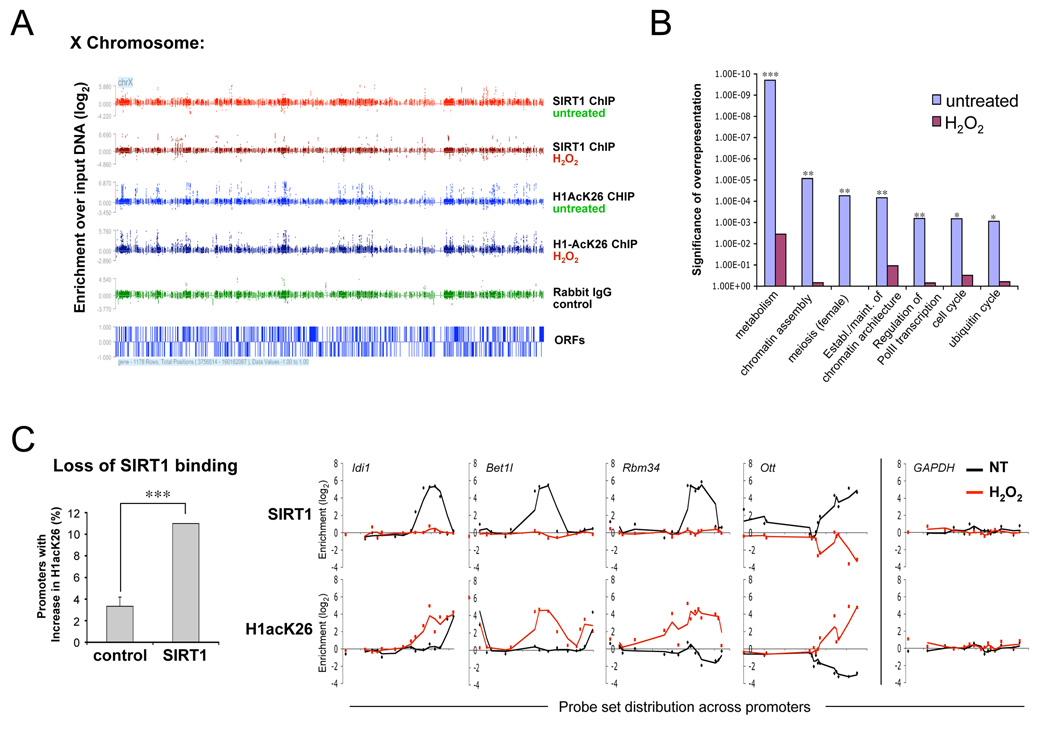
Figure 2.
Oxidative stress causes a major redistribution of SIRT1. (A) Distribution of SIRT1 and H1AcK26 along a representative chromosome. ES cells were either untreated or treated with H2O2 for 1 h, followed by ChIP for SIRT1, H1AcK26 or control Ig. The x-axis depicts probe sets spanning the promoters of annotated ORFs, signals across the y-axis reflect the log2 change of IP DNA over input DNA. (B) Significance of overrepresentation of selected gene ontology (GO) groups amongst SIRT1-associated genes before and after H2O2 treatment. Asterisks indicate significance after Benjamini- Hochberg false discovery rate correction. (C) Loss of SIRT1 binding correlates with increased H1K26 acetylation. Promoters with ≥ 2log2 enrichment of SIRT1 prior to H2O2 treatment, but no signal thereafter were compared to non-SIRT1 associated promoters. The y-axis shows the fraction of promoters with a ≥ 2-fold increase in H1AcK26 enrichment after exposure to H2O2. SIRT1 and H1AcK26 probe set binding is shown for four of these promoters. GAPDH served as a negative control.
Next, we determined if SIRT1-binding and H1 acetylation patterns change in response to oxidative stress. Paralleling the yeast response, oxidative stress caused a redistribution of SIRT1 at the chromatin level (Figure 2A), such that less than 10% of SIRT1-associated promoters overlapped between untreated and H2O2-treated cells (Figure S4B), and the resulting binding pattern did not cluster into functional GO groups, indicating a shift to random SIRT1 distribution (Figure 2B). There was a significant negative correlation between the loss of SIRT1 binding and H1K26 acetylation (χ2=12.12, P<0.001, Figure 2C), supporting the notion that SIRT1 regulates these genes, at least in part, through H1 deacetylation. Together, these observations indicate that SIRT1 associates with considerably more genes than currently known and that oxidative stress causes a major change in SIRT1 distribution across the genome.
Oxidative stress causes SIRT1 dependent transcriptional deregulation
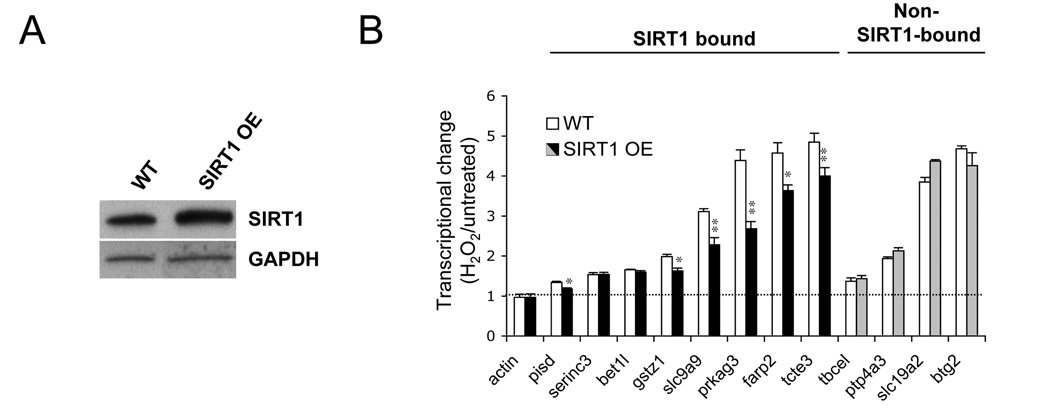
To determine how the redistribution of promoter-associated SIRT1 affects transcription, we performed a combination of microarray-based transcriptional profiling (Table S2), and q-RT-PCR analysis of SIRT1-associated and non-associated genes, comparing standard growth conditions with H2O2 treatment, with and without SIRT1 overexpression. We identified a diverse set of SIRT1-associated genes that significantly increased in expression upon H2O2 treatment, coincident with SIRT1 release, including regulators of metabolism (pisd, prkag3, gstz1), ion transport (slc9a9), cell motility (tcte3) and G-protein signaling (farp2). Of these genes, 75% were repressed by modest overexpression of SIRT1 (Figure 3) and H1K26 acetylation inversely correlated with SIRT1 binding (Figure S5A). Demonstrating specificity, SIRT1 overexpression did not repress the induction of genes that were not associated with SIRT1 (Figure 3B). The reduction in SIRT1 binding was confirmed for a random selection of promoters by q-PCR using two distinct types of genotoxic stress: H2O2 and methyl-methane-sulfonate (MMS) (Figure S5B). Inactivation of SIRT1 resulted in transcriptional deregulation of six of the eight SIRT1-associated loci, further corroborating a regulatory role for SIRT1 at these sites (Figure S5C). SIRT1 was recently shown to negatively regulate HDAC1 and can, thus, function both as transcriptional activator and repressor (Binda et al., 2008), which may explain why knockdown of SIRT1 does not always alter gene expression in the same direction as oxidative stress. Together, our results indicate that DNA damage induces a change in SIRT1 distribution that affects the expression of individual genes, and that this effect can be suppressed by increasing SIRT1 levels.
Figure 3.
Transcriptional deregulation of SIRT1-associated genes in response to oxidative stress is repressed by increasing SIRT1 levels. (A) SIRT1 expression in wild-type (WT) and SIRT1 overexpressing (OE) ES cells. (B) q-RT-PCR analysis of putative SIRT1 target genes (SIRT1-bound), non-SIRT1-bound control genes and β-actin in wild-type (open bars) and SIRT1 overexpressing ES cells (closed bars). Shown is the fold change of expression compared to untreated samples. Data are represented as mean +/− SEM.
SIRT1 is recruited to DNA double strand breaks
Given that the yeast Sir complex redistributes from silent loci to sites of DNA repair (Martin et al., 1999; Mills et al., 1999), we wondered if SIRT1 also behaves in this way. When isolating chromatin-bound and non-chromatin-bound protein fractions from untreated or H2O2-treated cells, we observed a substantial, transient, and dose-dependent increase in chromatin-associated SIRT1. Treatment with MMS produced a similar effect, supporting the idea that SIRT1 is recruited to damaged DNA from promoters and from the soluble nuclear pool (Figure 4A, Figures S6A and S6B).
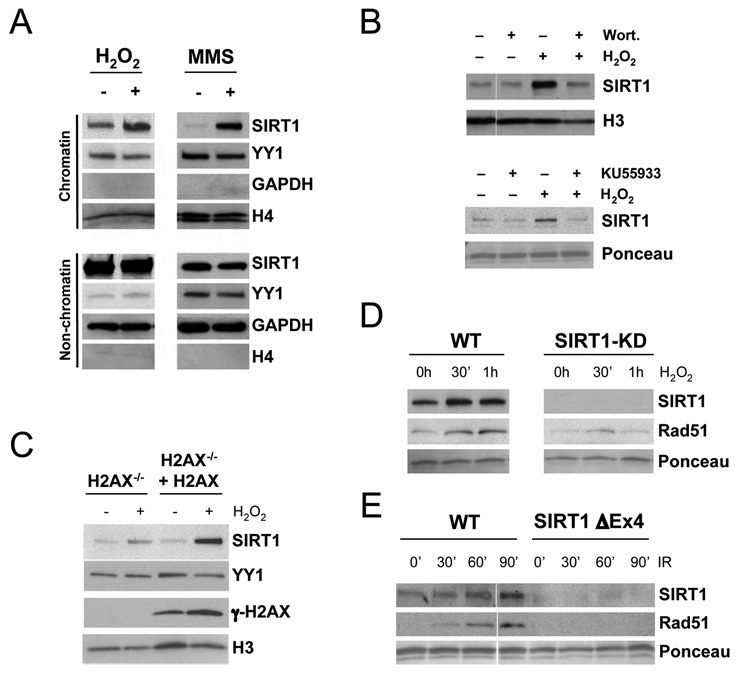
Figure 4.
SIRT1 is recruited to chromatin upon DNA damage in an ATM-dependent manner. (A) Western blot analysis of SIRT1 and indicated control proteins in both chromatin-bound and non-chromatin-bound protein fractions from ES cells that were either left untreated or treated with H2O2 or MMS for 1 h. (B and C) Western blot analysis of chromatin-bound SIRT1 in response to H2O2 with or without pretreatment with wortmannin or KU55933 (B), and in the presence or absence of H2AX (C). (D and E) Recruitment of SIRT1 and Rad51 to chromatin in WT or SIRT1-deficient (SIRT1-Δex4) cells in response to H2O2 or IR.
In yeast, the recruitment of Sir proteins to a DNA double-strand break (DSB) requires DNA damage signaling through MEC1, an ortholog of the mammalian PI3-kinases ATR/ATM (Martin et al., 1999; McAinsh et al., 1999; Mills et al., 1999). To test whether DNA damage signaling is required for SIRT1 redistribution, cells were treated with the PI3 kinase inhibitor wortmannin or the ATM inhibitor KU55933 prior to H2O2 exposure. The increase in chromatin-associated SIRT1 was strongly reduced by both compounds (Figure 4B). We then investigated whether SIRT1 recruitment is dependent on histone H2AX, one of the immediate targets of ATM (Burma et al., 2001). In H2AX-deficient ES cells there was a 2–3 fold reduction in the amount of chromatin-bound SIRT1 in response to H2O2 or MMS compared to cells reconstituted with wild-type H2AX (Figure 4C and Figure S6C, (Xie et al., 2004)). Thus, efficient recruitment of SIRT1 to damaged DNA requires DNA damage signaling through ATM and H2AX.
Rad51, a critical component of the homologous DSB repair (HR) process, was recruited to chromatin concomitant with SIRT1 in response to both H2O2 and exposure to DSB-inducing ionizing irradiation (IR). Importantly, Rad51 recruitment was impaired in the absence of SIRT1 (Figure 4D–E). These data indicated that SIRT1 might physically associate with DSBs and perform a key step in the DNA repair process.
To test this, we employed a cell-based system in which a DSB is induced by transient transfection with a vector encoding the endonuclease I-SceI (Weinstock et al., 2006). This system has been used previously to identify the DSB-binding patterns of a number of DNA repair enzymes (Rodrigue et al., 2006). Nbs1, a component of the MRN complex served a positive control for binding (Berkovich et al., 2007). Concomitant with recruitment of Nbs1, SIRT1 binding was detected 24 h after transfection with I-SceI at the DSB site. Similar kinetics were recently shown for the interaction between SIRT1 and DSBs in CpG-rich DNA (O'Hagan et al., 2008).
Interestingly, the association of Nbs1 with the break site was delayed and strongly reduced in the absence of SIRT1 (Figures 5A and B). A similar effect was observed for recruitment of Rad51 (Figure 5C). These data show that SIRT1 physically associates with sites of DNA damage and agree with recent studies indicating that chromatin-modifying enzymes are recruited to the DSB to prepare the site for incoming DNA repair factors (Botuyan et al., 2006; Tamburini and Tyler, 2005).
Figure 5.
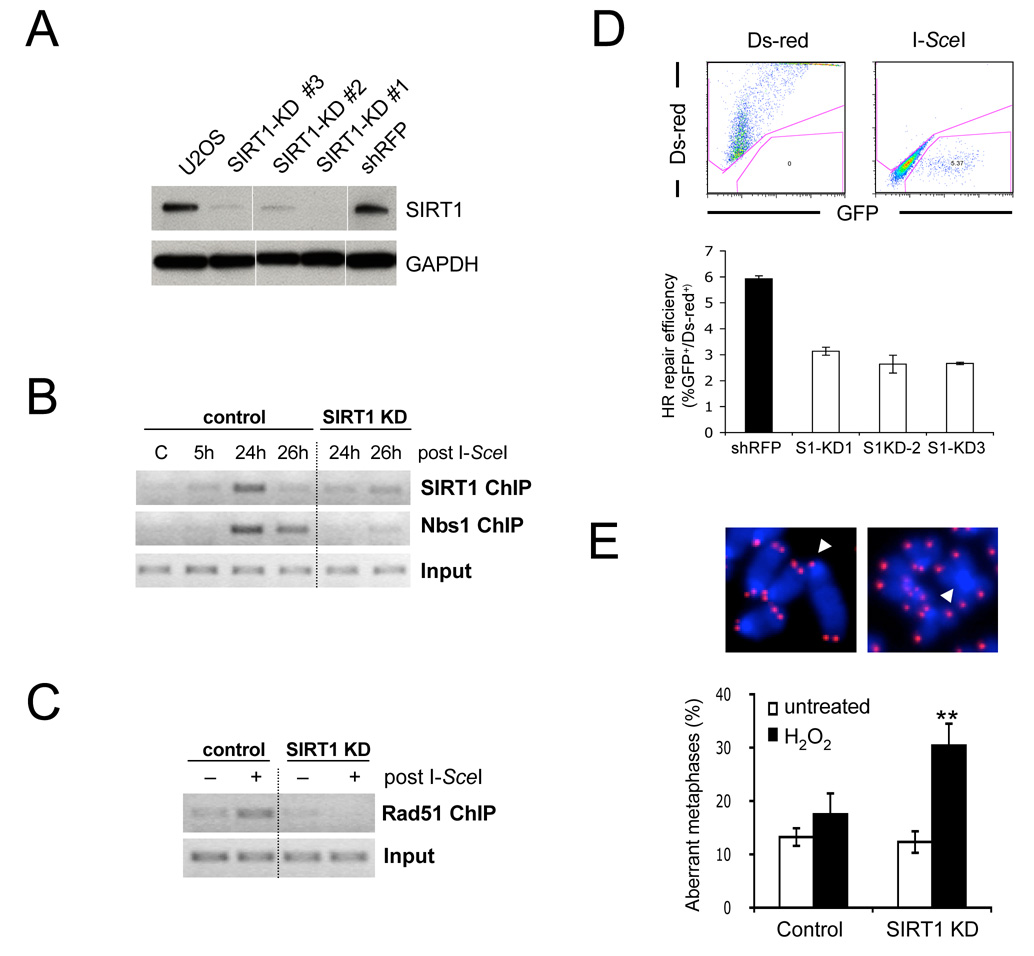
SIRT1 is recruited to DNA breaks and is required for efficient DSB repair. (A) SIRT1 expression in parental U2OS-DRGFP cells, three independent shSIRT1 expressing cell lines (SIRT1-KD) and shRFP expressing controls. (B) ChIP analysis of SIRT1 and Nbs1 binding to a DSB in shRFP (control) and SIRT1 KD cells from (A). (C) DSB ChIP analysis of Rad51 recruitment using Ds-red-transfected (−) or I-SceI transfected cells (+) from (B) 24h after transfection. (D) SIRT1 knock-down results in reduced DSB repair as measured by GFP rexpression (see Figure S7A). Control and SIRT1-KD cell lines from (A) were transfected as in (C) and analyzed by FACS. (E) Loss of SIRT1 causes increased genomic instability upon oxidative stress. Untreated or H2O2 treated shSIRT1- or control shRNA-expressing ES cells were subjected to Q-FISH analysis. Telomeres are shown in red. Arrows indicate a chromatid break (left) and a fused centromere (right). The fraction of aberrant metaphases is shown. Data are represented as mean +/− SEM.
SIRT1 is required for efficient DSB repair and genomic stability
DSB repair occurs through two major pathways: HR and non-homologous end-joining (NHEJ). Our Rad51 data suggested a role for SIRT1 in HR, which can be measured by determining the repair frequency with which a defective GFP gene is restored to wild-type upon I-SceI transfection (see Figure S7A). Inhibition of SIRT1 activity with NAM, the specific SIRT1 inhibitor S91211 (Solomon et al., 2006), or stable knockdown of SIRT1 resulted in a 25% to 50% reduction in GFP+ cells, indicating that SIRT1 is necessary for efficient HR-mediated repair (Figure 5D, Figure S7B). Repair by NHEJ, the prominent DSB repair pathway in G1 and post-mitotic cells, was also reduced in this assay system, but to a lesser extent (Figure S7C).
To test whether SIRT1 is necessary for the maintenance of genomic stability, SIRT1 knock-down ES cell lines were transiently exposed to H2O2 and analyzed for metaphase aberrations. In the absence of genotoxic stress, there was no significant difference in chromosomal stability between SIRT1-deficient and control cells, consistent with previous findings (Chua et al., 2005). Strikingly, H2O2 treatment caused a significant increase in chromosomal aberrations specifically in SIRT1 deficient cells (Figure 5E). The frequency of chromatid breaks was comparable between knock-down and control ES cells, but the number of chromosomal fusions, in particular dicentric chromosomes and Robertsonian translocations, was significantly higher in the absence of SIRT1 (Table S3). Chromosome fusions are generally a result of aberrantly repaired DNA breaks, further supporting a role for SIRT1 in DSB repair. Although a general DSB defect should also increase chromatid breaks, we predominantly detected stably inherited aberrations such as fusions as metaphases were analyzed 48 h (approx. two divisions) after exposure to H2O2.
Increased SIRT1 levels protect from irradiation-induced cancer in mice
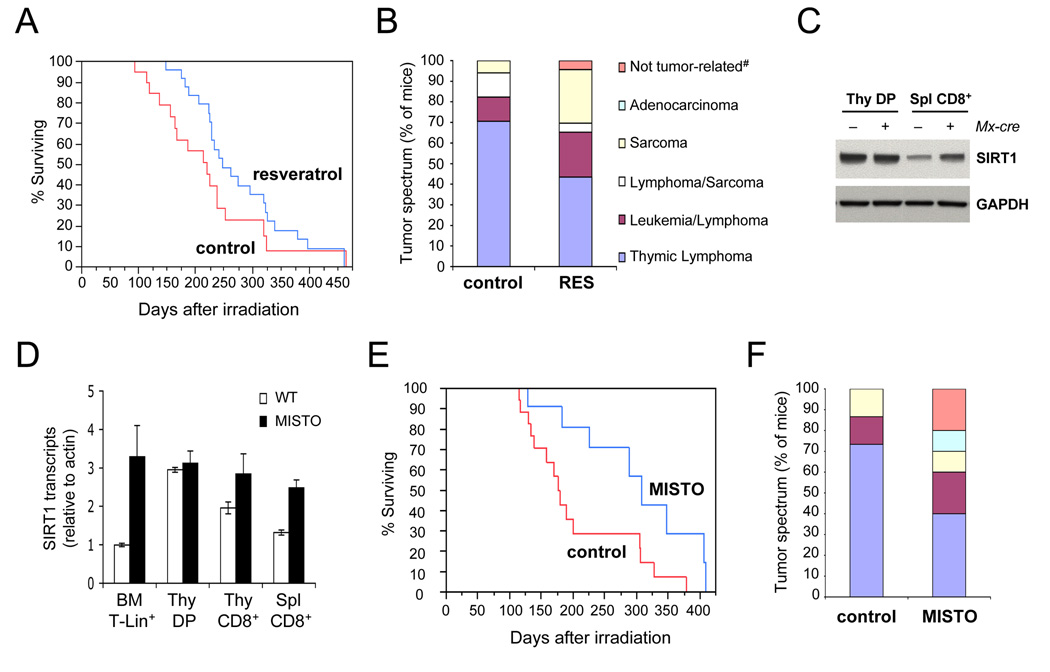
To test if SIRT1 promotes genomic stability in vivo, we used p53+/− mice (Jacks et al., 1994), which, when exposed to IR, show a high incidence of malignant thymic lymphoma arising from a loss of heterozygosity (LOH) at the p53 locus (Kemp et al., 1994). To explore the role of SIRT1 in damage-induced LOH, p53+/− mice were fed the SIRT1 activator resveratrol (Baur and Sinclair, 2006) three weeks prior to irradiation. Resveratrol-treated animals showed a 24% increase in survival (χ2=5.0, P = 0.025, Wilcoxon rank sum test) and a 45% reduction in the frequency of fatal thymic lymphomas (Figures 6A and B), resulting in a tumor spectrum highly reminiscent of non-irradiated p53+/− mice (Donehower et al., 1992; Jacks et al., 1994),
Figure 6.
Increased SIRT1 activity alters tumor spectrum and increases survival of irradiated p53+/− mice. (A) Survival of irradiated p53+/− mice fed normal or resveratrol-supplemented chow (n=19 and 25, respectively). Tumor-related deaths were recorded in days after irradiation. (B) Tumor spectrum from mice in (A), legend lists dominant tumor at time of death; #no tumors detected in necropsy. (C) Western blot analysis of MACS-purified thymic CD4/CD8 double-positive (DP) T cells and splenic CD8+ T cells from MISTO mice (+ Mx-cre) and littermate controls (− Mx-cre). Mice were analyzed 14 days after Mx-cre induction; Thy: Thymus, Spl: Spleen. (D) SIRT1 mRNA expression in lymphocyte subsets from MISTO mice (closed bars) and littermate controls (open bars). BM: Bone marrow, Lin+: Lineage-positive. (E) Survival of MISTO and control p53+/− mice (n=12 and 16, respectively) in response to a single dose of 4 Gy γ-irradiation. Tumor-related deaths were recorded in days after irradiation. (F) Tumor spectrum in mice from (E), legend as in (B).
Given that the effects of resveratrol may not be limited to SIRT1 activation, we generated a SIRT1 transgenic mouse strain (SIRT1STOP), that allows for Cre-mediated temporal and tissue-specific overexpression of SIRT1 via the deletion of a transcriptional STOP cassette (Firestein et al., 2008). The SIRT1STOP strain was crossed to p53+/− mice carrying the interferon (IFN) type I inducible Mx-cre transgene (Kuhn et al., 1995). Mice with Mx-cre-dependent, IFN-inducible SIRT1 overexpression are referred to as "MISTO mice". Injection with the IFN inducer poly(I)poly(C), MISTO mice increased expression of SIRT1 in bone marrow lymphocyte progenitors (3–4 fold) as well as mature B and T cells (Figures 6C and 6D). Thymocytes had intrinsically high levels of SIRT1. Two weeks after SIRT1 induction, p53+/− MISTO mice p53+/− controls were exposed to 4 Gy of γ-irradiation, and monitored for tumor-related deaths. Deletion of the STOP cassette in tumor tissues was examined by Southern blot or q-PCR (Figure S8A and B).
The mean survival of MISTO mice was ~46% greater than in control animals (Figure 6E, χ2=5.68, P=0.017, Log-Rank test; χ2=4.9, P=0.027, Wilcoxon rank sum test). Furthermore, the frequency of fatal thymic lymphoma was reduced by 45% in MISTO mice, consistent with our finding in resveratrol-treated animals (Figure 6F). Tumor cells exhibited LOH at the p53 locus both in control and MISTO mice (Figure S8C and D). Together, our data indicate that increasing SIRT1 activity or quantity can increase genomic stability in vivo and suppress tumorigenesis.
SIRT1-associated genes are deregulated in the aged brain
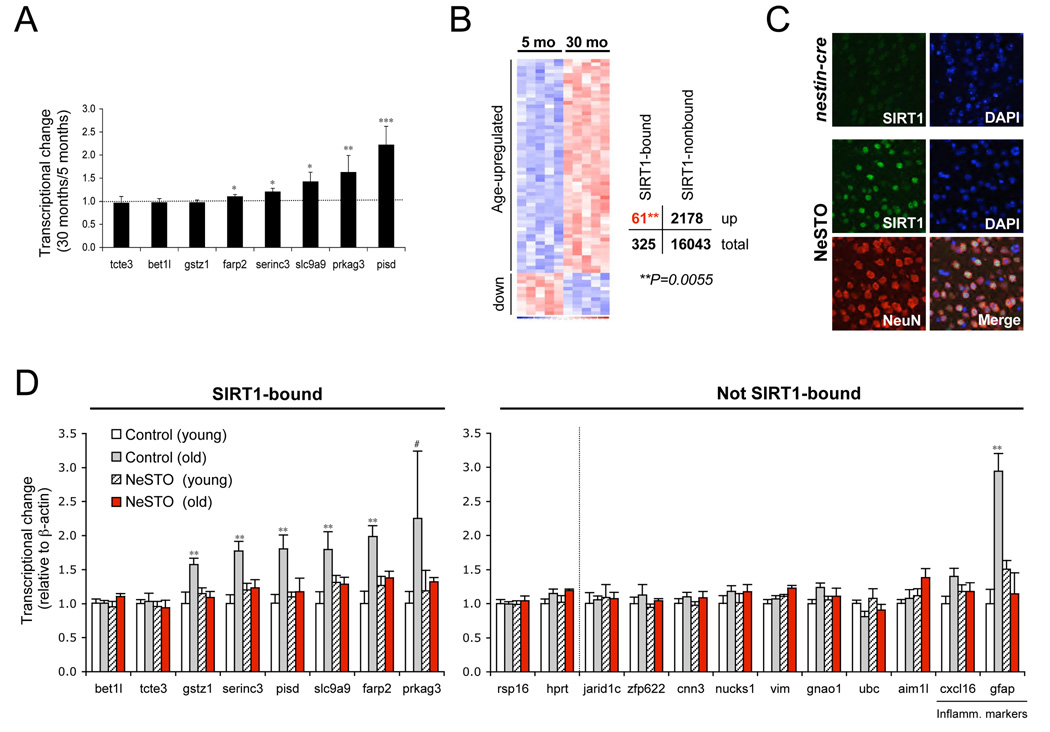
Our observations indicated that SIRT1, like its yeast counterpart, is recruited to DSBs in response to genotoxic stress, resulting in a loss of silencing at repetitive DNA elements and SIRT1-regulated genes. To test if these transcriptional changes were relevant to aging, we compared the transcriptional changes caused by oxidative stress with those associated with aging using q-RT-PCR and microarrays. We chose to examine neocortex because age-related gene expression changes have been well characterized in this tissue (Lee et al., 2000; Lu et al., 2004). Interestingly, two thirds of SIRT1-bound genes that were derepressed by oxidative stress in vitro (Figure 3) were also derepressed during aging (Figures 7A and D). Moreover, SIRT1-target genes identified by ChIP on chip in vitro (see Figure 2) were significantly overrepresented amongst age-upregulated genes (χ2=7.3, P=0.0055; Figure 7B and Table S4). The abundance of major satellite repeat transcripts also increased significantly with age (Figure S9A).
Figure 7.
Transcriptional deregulation of SIRT1 target genes occurs during normal aging. (A) Expression of SIRT1-associated gene from Figure 3 in the neocortex from young (5 months) and old (30 months) B6C3F1 mice (n ≥ 6 per group), analyzed by q-PCR. Shown is the fold change in expression in old relative to young mice. P values are based on student’s one-tailed t-test. (B) Microarray expression analysis of SIRT1 target genes in the neocortex of young and old B6C3F1 mice (n = 5 per group). SIRT1 target genes are significantly overrepresented amongst age-upregulated genes (χ2 = 7.28, P = 0.0055). (C) Immunofluorescence analysis of SIRT1 and NeuN expression in the neocortex of NeSTO mice and Nestin-cre littermate controls. (D) q-RT-PCR analysis of SIRT1-bound genes from (A) and non-SIRT1-bound genes including housekeeping genes (rps16, hprt) and genes upregulated in 30 months old mice by microarray. NeSTO mice and Nestin-cre littermate controls (Control) were analyzed at 8–10 months (young, n = 3–5 and 6, respectively) or 18–19 months of age (old, n = 4 and 5, respectively). P values are based on student’s one-tailed t-test. Data are represented as mean +/− SEM.
To gain mechanistic insights, we tested whether overexpression of SIRT1 in the brain could delay these transcriptional changes, paralleling the ability of SIR2 to suppress the expression of yeast mating type genes. The SIRT1STOP transgenic mouse was crossed to a brain-specific Cre-driver (Nestin-cre) to generate Nestin-cre; SIRT1STOP mice, referred to as "NeSTO mice." Transcript levels of SIRT1-bound genes as well as age-upregulated non-SIRT1 bound (control) genes were examined in NeSTO mice with comparable SIRT1 overexpression (~10 fold, Figure 7C, Figure S9B and C) at 8–10 or 18–19 months. Strikingly, transcriptional derepression was almost exclusively observed for SIRT1-associated genes and was completely suppressed in aged NeSTO mice (Figure 7D). The only non-SIRT1 associated genes to change with ~19 months of age were inflammatory markers associated with gliosis, a characteristic of brain aging (Nichols et al., 1995). This upregulation was also repressed in NeSTO mice, indicating that there are secondary, beneficial effects of SIRT1 overexpression. The effect of SIRT1 overexpression in mice older than 19 months of age is not yet known. Together, our results confirm that SIRT1-bound genes are derepressed in the aging brain and that SIRT1 overexpression can suppress these age-related changes.
Discussion
The discovery that the yeast Sir complex relocalizes during aging and in response to DNA damage (Kennedy et al., 1995; Kennedy et al., 1997; Martin et al., 1999; McAinsh et al., 1999; Mills et al., 1999) led us to propose a model whereby aging is caused, in part, by the DNA-damage induced reorganization of chromatin, a phenomenon we have termed the "RCM response," for redistribution of chromatin modifiers (Imai and Kitano, 1998; Oberdoerffer and Sinclair, 2007; Villeponteau, 1997). Here, we present evidence that the RCM response exists in mammals, and that it may contribute to age-related changes in gene expression.
SIRT1 is recruited to DSBs and is required for efficient DNA repair
A role for chromatin modifiers in DNA repair has been convincingly shown in yeast (Tamburini and Tyler, 2005) and mammals (Bassing et al., 2002; Bhaskara et al., 2008; Botuyan et al., 2006; Celeste et al., 2002; Xie et al., 2004). Here, we show that SIRT1 binds to hundreds of promoters in the mouse genome, and that this binding pattern is altered in response to genotoxic stress, coincident with the relocalization of SIRT1 to damaged DNA.
The recruitment of SIRT1 to DSBs is reminiscent of yeast Sir2 and other histone modifying enzymes that bind to a DSB, resulting in epigenetic changes surrounding the break site (Chen et al., 2008; Tamburini and Tyler, 2005). As shown for the methylation of H4 at lysine 20, chromatin alterations surrounding a DSB can promote the recruitment of DNA repair factors such as 53BP1 (Botuyan et al., 2006). DSB associated SIRT1 may serve to deacetylate histones or DNA repair factors. Consistent with the latter, SIRT1 was shown to directly interact with Nbs1 (Yuan et al., 2007) and we show here that both proteins coexist at the break site (Figure 5B). How SIRT1 recruitment to DSBs is initiated at the molecular level will require further study, but our data indicate that ATM-mediated signaling through H2AX phosphorylation is important (Figure 4B and 4C). We further demonstrate SIRT1 is required for efficient DSB repair and genome maintenance in response to oxidative stress. The direct association with DSBs as well as the finding that SIRT1-deficient cells are checkpoint proficient suggest a DNA repair defect rather than a checkpoint defect (Cheng et al., 2003). Consistent with a role for SIRT1 in DNA repair, high doses of IR have been reported to induce a cell cycle delay in SIRT1- deficient fibroblasts (Yuan et al., 2007), reminiscent of SIRT6-deficient cells, which show a DNA repair defect and prolonged S phase but no checkpoint defect (Mostoslavsky et al., 2006).
Based on studies showing that SIRT1 can deacetylate and inactivate p53, some researchers predicted that SIRT1 will promote tumorigenesis in vivo (Lim, 2006). In contrast, p53+/− mice with increased SIRT1 activity are less susceptible to irradiation-induced thymic lymphoma with a 20–46% greater mean lifespan (Figure 6). It is appealing to speculate that increased SIRT1 levels protect from irradiation-induced LOH by increasing DSB repair efficiency. Our finding that MISTO transgenic mice overexpressed SIRT1 in early lymphocyte precursors but not at later stages during thymic T cell development (Figures 6C and 6D) is consistent with this hypothesis and argues against a protective role for SIRT1 at later stages of tumor progression. We cannot rule out, however, that additional protective mechanisms contributed to the protection from tumorigenesis.
Derepression of SIRT1-associated loci in response to oxidative stress and aging
SIRT1 has previously been reported to contribute to the formation of facultative heterochromatin (Vaquero et al., 2007). Our data show that SIRT1-mediated repression can also occur at constitutive heterochromatic regions such as pericentromeric DNA, as well as a number of specific genes, most prominently regulators of chromatin assembly and transcription. Oxidative stress reduces the association of SIRT1 with both repetitive loci (Figure 1E) and individual genes (Figure 2). Not surprisingly, this change in SIRT1 localization is associated with functional consequences on silencing of heterochromatic repetitive DNA (Figure 1F) as well as the expression of individual genes (Figure 3). In cells and in the aging brain, the majority of these changes were counteracted by overexpressing SIRT1 (Figure 3 and Figure 7D). Loss of SIRT1 binding, however, did not always lead to transcriptional derepression, indicating that chromatin alterations may be necessary but not always sufficient to cause transcriptional deregulation. Further work will be required to identify other chromatin modifiers or transcription factors that are involved in the RCM response. HDAC1 and HDAC2 are candidates for the RCM response given their increased binding to chromatin after DNA damage (see Figure S10).
Although a wealth of data on changes in gene expression with age has been catalogued in recent years, there is still much debate about their physiological relevance (Oberdoerffer and Sinclair, 2007; Vijg, 2004). Transcriptional changes in the elderly may be a beneficial defense response to cellular damage (Niedernhofer et al., 2006). Conversely, age-related changes in gene expression may be deleterious, yet exist because of the weak forces of natural selection at older ages. Thus, while a transient RCM response in young individuals is likely beneficial, constitutive triggering of RCM may actually contribute to aging. This duality is clearly evidenced in yeast, where the transient derepression of silent HM loci increases HR and resistance to DNA damaging agents (Lee et al., 1999) yet the constitutive derepression of HM loci in old cells is a cause of sterility (Smeal et al., 1996). In mammals, defective DNA repair is often associated with premature aging (Lombard et al., 2005). Conversely, the lack of a DNA damage response can be beneficial in situations of chronic DNA damage due to telomere dysfunction (Choudhury et al., 2007; Schaetzlein et al., 2007). Furthermore, exposure to genotoxic stress early in life seems to accelerate changes in gene expression that have been associate with age-related diseases such as amyloidogenesis (Wu et al., 2008). Interestingly, we found that constitutive overexpression of a set of age-deregulated SIRT1 target genes promotes apoptosis in primary neurons (Figure S11), however more work is needed to determine the physiological relevance of this observation.
Perspective
We have identified SIRT1 as participant in a stress response that may provide a direct link between DNA damage and gene expression changes that occur during aging. While DNA damage has been previously suggested to directly inhibit gene repression (Lu et al., 2004), our data explains how ostensibly undamaged genes may become deregulated over time. We speculate that the RCM response may also cause permanent changes to the chromatin structure at sites of repair, leading to stable transcriptional changes that accumulate over a lifetime (Oberdoerffer and Sinclair, 2007). Indeed, a recent report showed that SIRT1 recruitment to a DNA break in CpG islands can result in DNA methylation changes and heritable gene silencing (O'Hagan et al., 2008).
Because age-related transcriptional changes are not limited to SIRT1-regulated loci (Botuyan et al., 2006; Tamburini and Tyler, 2005), multiple mechanisms involving other chromatin modifiers are likely to be involved. What sets SIRT1 apart is its link to calorie restriction (CR), a dietary regimen that slows aging in mammals (Sinclair, 2005). Given that increased SIR2/SIRT1 expression can suppress genomic instability and gene expression alterations, perhaps CR promotes genomic stability and delays aging in mammals via a similar mechanism.
Experimental Procedures
Yeast experiments
All experiments were on log phase yeast growing in YPD (2% glucose). HMR::GFP cells or HMR::GFP 2xSIR2 cells (Park et al., 1999) were exposed to H2O2 for 30 min, followed by a 4 h recovery period, then analyzed FACS. Replicative lifespans and rDNA recombination analyses were performed as described (Lamming et al., 2005). For rDNA recombination, WT or Sir2o/e W303AR cells were treated for 30 min with H2O2 (1.5 mM) prior to plating. For lifespan analysis, WT or 2xSIR2 cells were plated on regular YPD agar or agar supplemented with H2O2 (1 mM).
Cell culture and treatments
Mouse ES cells were cultured on gelatinized tissue culture dishes as described (Kanellopoulou et al., 2005). Stable SIRT1 overexpressing V6.5–C10 ES cells were obtained from SIRT1STOP ES cells (Firestein et al., 2008) by Cre-mediated deletion of a loxP-flanked STOP cassette. SIRT1 knock-down ES cells were generated by lentiviral infection of V6.5–C10 ES cells (Beard et al., 2006). Cells were infected with either a luciferase-specific or a SIRT1-specific shRNA lentiviral vector (Araki et al., 2004). Generation of SIRT1 knock-out ES cells and respective wild-type cells is described (Chua et al., 2005). Cells were γ-irradiated (4 Gy, 137Cs irradiator, Shepherd and Associates) or treated with H2O2 or MMS for 1 h at 37°C. Treatment with 50 µM wortmannin (Sigma) or 25 µM KU55933 (AstraZeneca) was started 2–3 h prior to other treatments. NAM (Sigma, 25 mM), TSA (Sigma, 0.1 µM) or sirtinol (100 µM) were added for the indicated times.
DRGFP-transgenic U2OS cells and I-SceI- or Ds-red-encoding plasmids pCBASceI and pCAGGS-Dsred are described (Weinstock et al., 2006). Stable SIRT1 knock-down and control lines were generated by lentiviral gene transfer using shRNA vectors from Open Biosystems. Transfection with pCBASce or pCAGGS-Dsred was performed using Fugene 6 transfection reagent (Roche). When indicated, cells were treated with NAM (10 mM) or S91211/EX-527 (50 µM, (Solomon et al., 2006)) starting 2 h prior to transfection. After 48 h, cells were analyzed by FACS.
Chromatin immunoprecipitation (ChIP)
Approximately 107 cells were cross-linked with 1% formaldehyde for 15–20 minutes at 37°C and quenched with glycine. Cell lysates were sonicated (Branson sonifier) and incubated overnight with rabbit α-Sir2α (Upstate), α-Nbs1 (Novus), α-Rad51 (Calbiochem) or α-H1AcK26 (Vaquero et al., 2004). Immunoprecipitation was performed as described (Upstate) and eluates were purified using QIAgen PCR purification, followed by (q-) PCR analysis.
RNA isolation, reverse transcription, PCR analysis
Total RNA was isolated using Trizol reagent (Invitrogen), followed by DNase treatment (Turbo Dnase free, Ambion). RNA was reverse transcribed with Invitrogen’s Thermoscript RT PCR system using a combination of random hexamers and oligo dT primers. PCR was performed using Taq Platinum (Roche). q-PCR was performed using SYBR green-based detection on a Roche Light Cycler or Roche LC480. See Table S5 for primers and PCR conditions.
Microarray analysis and statistics
For Nimblegen promoter tiling array analysis (Nimblegen-Roche), ChIP DNA was amplified by ligation-mediated PCR. IP and input DNA samples were labeled using 9mer Cy3 and Cy5-labeled primers. IP and total DNAs were co-hybridized to the NimbleGen MM5 minimal promoter tiling array and analyzed using NimbleScan software (NimbleGen). Peak data files were generated by searching for 4 or more probes with significant enrichment using a 500 bp sliding window. Each peak was assigned a false discovery rate (FDR) score based on randomization. Gene ontology cluster analysis of SIRT1-associated promoters (FDR ≤ 0.1) was performed using the BiNGO plug-in in Cytoscape v2.5.
For Affymetrix expression analysis, total RNA was hybridized to the mouse genome 430 2.0 array. CEL files were analyzed for significance and fold changes between experimental groups using DChip software. For the comparison of Nimblegen and Affymetrix array data, Gene IDs of both arrays were matched using DAVID. Analysis was limited to genes with highly significant SIRT1 promoter enrichment (FDR < 0.005) and transcriptional increase (a ≥ 10%, P ≤ 0.005). χ2-based P values were calculated using Pearson's Chi-squared test with Yates' continuity correction. Microarray data have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE13121 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE13121).
Cellular fractionation and Western blotting
Chromatin-bound protein was purified as described (Cha et al., 2005). Primary antibodies were rabbit α-Sir2α, α-histone H3, α-Histone H4 (Upstate), rabbit α-YY1 (Santa Cruz), mouse α-GAPDH (Chemicon) and rabbit α-Rad51 (Dr. R. Scully); α-rabbit and α-mouse HRP-coupled secondary antibodies were from GE Healthcare.
Metaphase analysis
Metaphase spreads were performed as previously described (Mostoslavsky et al., 2006). At least 80 metaphases of each genotype were scored per experiment.
Mouse breeding and treatments
All mice were housed pathogen-free. SIRT1STOP (Firestein et al., 2008), Mx-cre (gift from Dr. K. Rajewsky), Nestin-cre and p53−/− mice (Jackson Laboratory) were crossed as to obtain the indicated genotypes. Experimental animals were on C57BL/6 × 129/Sv mixed genetic background. For Mx-cre induction, mice were injected with 400 µg poly(I)poly(C) (Amersham) at 6–10 weeks of age (Kuhn et al., 1995). For tumor studies, mice were γ-irradiated 10–14 days thereafter with a single dose of 4 Gy. Animals were sacrificed when moribund. When not obvious, mice were submitted to necropsy to identify the cause of death. Deaths not related to tumors and mice too decomposed for analysis were censored. Kaplan-Meier survival curves were generated from two separate cohorts of irradiated animals using JMP7 software. Resveratrol was fed at 2.4 mg/kg food as previously described (Baur et al., 2006). Two cohorts were pooled for survival analysis.
Supplementary Material
Acknowledgements
D.A.S. wishes to thank Bruce Stillman for advice and scientific discussions. We are also grateful to K. Rajewsky, D. Reinberg, D. Lamming, L. Guarente, A. Vaquero , and M. Jasin for advice and reagents. We thank E. Vollmann for assistance with MACS sorting, R. Bronson for mouse necropsy. We are especially grateful to P. Glenn, M. Collins, and L. Ellison for their support of aging research. P.O. was supported by a fellowship from the National Space Biomedical Research Institute (grant PF00903); the Sinclair Lab by grants RO1GM068072 and R01AG19719 (NIH) and the Glenn Foundation for Medical Research. D.A.S. and F. W. A. are Ellison Medical Foundation Senior Scholars. R. M. is a V and Kimmel Foundation Scholar. A. B. supported by NIH grant NS047188, R.S. by GM 07394 and J.V., S.P. and T.A.P. by R01AG 020681. F.W.A. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Araki T, Sasaki Y, Milbrandt J. Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science. 2004;305:1010–1013. doi: 10.1126/science.1098014. [DOI] [PubMed] [Google Scholar]
- Bahar R, Hartmann CH, Rodriguez KA, Denny AD, Busuttil RA, Dolle ME, Calder RB, Chisholm GB, Pollock BH, Klein CA, Vijg J. Increased cell-to-cell variation in gene expression in ageing mouse heart. Nature. 2006;441:1011–1014. doi: 10.1038/nature04844. [DOI] [PubMed] [Google Scholar]
- Bassing CH, Chua KF, Sekiguchi J, Suh H, Whitlow SR, Fleming JC, Monroe BC, Ciccone DN, Yan C, Vlasakova K, et al. Increased ionizing radiation sensitivity and genomic instability in the absence of histone H2AX. Proc Natl Acad Sci U S A. 2002;99:8173–8178. doi: 10.1073/pnas.122228699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baur JA, Sinclair DA. Therapeutic potential of resveratrol: the in vivo evidence. Nat Rev Drug Discov. 2006;5:493–506. doi: 10.1038/nrd2060. [DOI] [PubMed] [Google Scholar]
- Beard C, Hochedlinger K, Plath K, Wutz A, Jaenisch R. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis. 2006;44:23–28. doi: 10.1002/gene.20180. [DOI] [PubMed] [Google Scholar]
- Berkovich E, Monnat RJ, Jr, Kastan MB. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat Cell Biol. 2007;9:683–690. doi: 10.1038/ncb1599. [DOI] [PubMed] [Google Scholar]
- Bhaskara S, Chyla BJ, Amann JM, Knutson SK, Cortez D, Sun ZW, Hiebert SW. Deletion of histone deacetylase 3 reveals critical roles in S phase progression and DNA damage control. Mol Cell. 2008;30:61–72. doi: 10.1016/j.molcel.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binda O, Nassif C, Branton PE. SIRT1 negatively regulates HDAC1-dependent transcriptional repression by the RBP1 family of proteins. Oncogene. 2008 doi: 10.1038/sj.onc.1211014. [DOI] [PubMed] [Google Scholar]
- Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, Mer G. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burma S, Chen BP, Murphy M, Kurimasa A, Chen DJ. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J Biol Chem. 2001;276:42462–42467. doi: 10.1074/jbc.C100466200. [DOI] [PubMed] [Google Scholar]
- Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 2005;120:513–522. doi: 10.1016/j.cell.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Celeste A, Petersen S, Romanienko PJ, Fernandez-Capetillo O, Chen HT, Sedelnikova OA, Reina-San-Martin B, Coppola V, Meffre E, Difilippantonio MJ, et al. Genomic instability in mice lacking histone H2AX. Science. 2002;296:922–927. doi: 10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha TL, Zhou BP, Xia W, Wu Y, Yang CC, Chen CT, Ping B, Otte AP, Hung MC. Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science. 2005;310:306–310. doi: 10.1126/science.1118947. [DOI] [PubMed] [Google Scholar]
- Chen CC, Carson JJ, Feser J, Tamburini B, Zabaronick S, Linger J, Tyler JK. Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell. 2008;134:231–243. doi: 10.1016/j.cell.2008.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HL, Mostoslavsky R, Saito S, Manis JP, Gu Y, Patel P, Bronson R, Appella E, Alt FW, Chua KF. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci U S A. 2003;100:10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury AR, Ju Z, Djojosubroto MW, Schienke A, Lechel A, Schaetzlein S, Jiang H, Stepczynska A, Wang C, Buer J, et al. Cdkn1a deletion improves stem cell function and lifespan of mice with dysfunctional telomeres without accelerating cancer formation. Nat Genet. 2007;39:99–105. doi: 10.1038/ng1937. [DOI] [PubMed] [Google Scholar]
- Chua KF, Mostoslavsky R, Lombard DB, Pang WW, Saito S, Franco S, Kaushal D, Cheng HL, Fischer MR, Stokes N, et al. Mammalian SIRT1 limits replicative life span in response to chronic genotoxic stress. Cell Metab. 2005;2:67–76. doi: 10.1016/j.cmet.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Cutler RG. Longevity determinant genes, cellular dysdifferentiation and oxidative stress. In: Cutler RG, Packer L, Bertram J, Mori A, editors. Oxidative stress and aging. Birkhauser Press; 1995. pp. 15–19. [Google Scholar]
- Donehower LA, Harvey M, Slaglex BL, McArthur MJ, Montgomery CA, Jr, Butel JS, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- Firestein R, Blander G, Michan S, Oberdoerffer P, Ogino S, Campbell J, Bhimavarapu A, Luikenhuis S, de Cabo R, Fuchs C, et al. The SIRT1 deacetylase suppresses intestinal tumorigenesis and colon cancer growth. PLoS ONE. 2008;3:e2020. doi: 10.1371/journal.pone.0002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigis MC, Guarente LP. Mammalian sirtuins--emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- Herbig U, Ferreira M, Condel L, Carey D, Sedivy JM. Cellular senescence in aging primates. Science. 2006;311:1257. doi: 10.1126/science.1122446. [DOI] [PubMed] [Google Scholar]
- Imai S, Kitano H. Heterochromatin islands and their dynamic reorganization: a hypothesis for three distinctive features of cellular aging. Exp Gerontol. 1998;33:555–570. doi: 10.1016/s0531-5565(98)00037-0. [DOI] [PubMed] [Google Scholar]
- Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp CJ, Wheldon T, Balmain A. p53-deficient mice are extremely susceptible to radiation-induced tumorigenesis. Nat Genet. 1994;8:66–69. doi: 10.1038/ng0994-66. [DOI] [PubMed] [Google Scholar]
- Kennedy BK, Austriaco NR, Jr, Zhang J, Guarente L. Mutation in the silencing gene SIR4 can delay aging in S. cerevisiae. Cell. 1995;80:485–496. doi: 10.1016/0092-8674(95)90499-9. [DOI] [PubMed] [Google Scholar]
- Kennedy BK, Gotta M, Sinclair DA, Mills K, McNabb DS, Murthy M, Pak SM, Laroche T, Gasser SM, Guarente L. Redistribution of silencing proteins from telomeres to the nucleolus is associated with extension of life span in S. cerevisiae. Cell. 1997;89:381–391. doi: 10.1016/s0092-8674(00)80219-6. [DOI] [PubMed] [Google Scholar]
- Klar AJ, Fogel S, Macleod K. MAR1-a Regulator of the HMa and HMalpha Loci in SACCHAROMYCES CEREVISIAE. Genetics. 1979;93:37–50. doi: 10.1093/genetics/93.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- Lamming DW, Latorre-Esteves M, Medvedik O, Wong SN, Tsang FA, Wang C, Lin SJ, Sinclair DA. HST2 mediates SIR2-independent life-span extension by calorie restriction. Science. 2005;309:1861–1864. doi: 10.1126/science.1113611. [DOI] [PubMed] [Google Scholar]
- Lee CK, Weindruch R, Prolla TA. Gene-expression profile of the ageing brain in mice. Nat Genet. 2000;25:294–297. doi: 10.1038/77046. [DOI] [PubMed] [Google Scholar]
- Lee SE, Paques F, Sylvan J, Haber JE. Role of yeast SIR genes and mating type in directing DNA double-strand breaks to homologous and non-homologous repair paths. Curr Biol. 1999;9:767–770. doi: 10.1016/s0960-9822(99)80339-x. [DOI] [PubMed] [Google Scholar]
- Lim CS. SIRT1: tumor promoter or tumor suppressor? Med Hypotheses. 2006;67:341–344. doi: 10.1016/j.mehy.2006.01.050. [DOI] [PubMed] [Google Scholar]
- Lombard DB, Chua KF, Mostoslavsky R, Franco S, Gostissa M, Alt FW. DNA repair, genome stability, and aging. Cell. 2005;120:497–512. doi: 10.1016/j.cell.2005.01.028. [DOI] [PubMed] [Google Scholar]
- Lu T, Pan Y, Kao SY, Li C, Kohane I, Chan J, Yankner BA. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429:883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- Martin SG, Laroche T, Suka N, Grunstein M, Gasser SM. Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell. 1999;97:621–633. doi: 10.1016/s0092-8674(00)80773-4. [DOI] [PubMed] [Google Scholar]
- McAinsh AD, Scott-Drew S, Murray JA, Jackson SP. DNA damage triggers disruption of telomeric silencing and Mec1p-dependent relocation of Sir3p. Curr Biol. 1999;9:963–966. doi: 10.1016/s0960-9822(99)80424-2. [DOI] [PubMed] [Google Scholar]
- Mills KD, Sinclair DA, Guarente L. MEC1-dependent redistribution of the Sir3 silencing protein from telomeres to DNA double-strand breaks. Cell. 1999;97:609–620. doi: 10.1016/s0092-8674(00)80772-2. [DOI] [PubMed] [Google Scholar]
- Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- Narita M, Krizhanovsky V, Nunez S, Chicas A, Hearn SA, Myers MP, Lowe SW. A novel role for high-mobility group a proteins in cellular senescence and heterochromatin formation. Cell. 2006;126:503–514. doi: 10.1016/j.cell.2006.05.052. [DOI] [PubMed] [Google Scholar]
- Nichols NR, Finch CE, Nelson JF. Food restriction delays the age-related increase in GFAP mRNA in rat hypothalamus. Neurobiol Aging. 1995;16:105–110. doi: 10.1016/0197-4580(95)80013-h. [DOI] [PubMed] [Google Scholar]
- Niedernhofer LJ, Garinis GA, Raams A, Lalai AS, Robinson AR, Appeldoorn E, Odijk H, Oostendorp R, Ahmad A, van Leeuwen W, et al. A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature. 2006;444:1038–1043. doi: 10.1038/nature05456. [DOI] [PubMed] [Google Scholar]
- O'Hagan HM, Mohammad HP, Baylin SB. Double Strand Breaks Can Initiate Gene Silencing and SIRT1-Dependent Onset of DNA Methylation in an Exogenous Promoter CpG Island. PLoS Genet. 2008;4:e1000155. doi: 10.1371/journal.pgen.1000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberdoerffer P, Sinclair DA. The role of nuclear architecture in genomic instability and ageing. Nat Rev Mol Cell Biol. 2007;8:692–702. doi: 10.1038/nrm2238. [DOI] [PubMed] [Google Scholar]
- Park PU, Defossez PA, Guarente L. Effects of mutations in DNA repair genes on formation of ribosomal DNA circles and life span in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:3848–3856. doi: 10.1128/mcb.19.5.3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW, Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt K, Zinn RL, Ohm JE, McGarvey KM, Kang SH, Watkins DN, Herman JG, Baylin SB. Inhibition of SIRT1 reactivates silenced cancer genes without loss of promoter DNA hypermethylation. PLoS Genet. 2006;2:e40. doi: 10.1371/journal.pgen.0020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rine J, Strathern JN, Hicks JB, Herskowitz I. A suppressor of mating-type locus mutations in Saccharomyces cerevisiae: evidence for and identification of cryptic mating-type loci. Genetics. 1979;93:877–901. doi: 10.1093/genetics/93.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigue A, Lafrance M, Gauthier MC, McDonald D, Hendzel M, West SC, Jasin M, Masson JY. Interplay between human DNA repair proteins at a unique double-strand break in vivo. Embo J. 2006;25:222–231. doi: 10.1038/sj.emboj.7600914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaetzlein S, Kodandaramireddy NR, Ju Z, Lechel A, Stepczynska A, Lilli DR, Clark AB, Rudolph C, Kuhnel F, Wei K, et al. Exonuclease-1 deletion impairs DNA damage signaling and prolongs lifespan of telomere-dysfunctional mice. Cell. 2007;130:863–877. doi: 10.1016/j.cell.2007.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair DA. Toward a unified theory of caloric restriction and longevity regulation. Mech Ageing Dev. 2005;126:987–1002. doi: 10.1016/j.mad.2005.03.019. [DOI] [PubMed] [Google Scholar]
- Sinclair DA, Guarente L. Extrachromosomal rDNA circles--a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- Smeal T, Claus J, Kennedy B, Cole F, Guarente L. Loss of transcriptional silencing causes sterility in old mother cells of S. cerevisiae. Cell. 1996;84:633–642. doi: 10.1016/s0092-8674(00)81038-7. [DOI] [PubMed] [Google Scholar]
- Solomon JM, Pasupuleti R, Xu L, McDonagh T, Curtis R, DiStefano PS, Huber LJ. Inhibition of SIRT1 catalytic activity increases p53 acetylation but does not alter cell survival following DNA damage. Mol Cell Biol. 2006;26:28–38. doi: 10.1128/MCB.26.1.28-38.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamburini BA, Tyler JK. Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Mol Cell Biol. 2005;25:4903–4913. doi: 10.1128/MCB.25.12.4903-4913.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquero A, Scher M, Erdjument-Bromage H, Tempst P, Serrano L, Reinberg D. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature. 2007;450:440–444. doi: 10.1038/nature06268. [DOI] [PubMed] [Google Scholar]
- Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- Vijg J. Impact of genome instability on transcription regulation of aging and senescence. Mech Ageing Dev. 2004;125:747–753. doi: 10.1016/j.mad.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Villeponteau B. The heterochromatin loss model of aging. Exp Gerontol. 1997;32:383–394. doi: 10.1016/s0531-5565(96)00155-6. [DOI] [PubMed] [Google Scholar]
- Weinstock DM, Nakanishi K, Helgadottir HR, Jasin M. Assaying double-strand break repair pathway choice in mammalian cells using a targeted endonuclease or the RAG recombinase. Methods Enzymol. 2006;409:524–540. doi: 10.1016/S0076-6879(05)09031-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Basha MR, Zawia NH. The environment, epigenetics and amyloidogenesis. J Mol Neurosci. 2008;34:1–7. doi: 10.1007/s12031-007-0009-4. [DOI] [PubMed] [Google Scholar]
- Xie A, Puget N, Shim I, Odate S, Jarzyna I, Bassing CH, Alt FW, Scully R. Control of sister chromatid recombination by histone H2AX. Mol Cell. 2004;16:1017–1025. doi: 10.1016/j.molcel.2004.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z, Zhang X, Sengupta N, Lane WS, Seto E. SIRT1 regulates the function of the Nijmegen breakage syndrome protein. Mol Cell. 2007;27:149–162. doi: 10.1016/j.molcel.2007.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P, Huber E, Kiefer F, Gottlicher M. Specific and redundant functions of histone deacetylases in regulation of cell cycle and apoptosis. Cell Cycle. 2004;3:1240–1242. doi: 10.4161/cc.3.10.1195. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.