Abstract
MicroRNAs (miRNAs) are small non-coding RNAs that are expressed in higher eukaryoates and have even been found in viral genomes. They usually act as endogenous repressors of target genes by either inhibiting translation, causing mRNA degradation, or by a combination of both mechanisms. More than 850 mature miRNA sequences have been identified in humans, and although this accounts for less than 2% of human genes, it is predicted that 30% of mRNAs are targeted by miRNAs. miRNAs play critical roles in most cellular processes including development, differentiation, and the homeostasis of both a cell and an organism. Moreover, many disease states, including cancer, occur or are sustained by miRNA dysregulation. Here we will review the latest reports of miRNA involvement and aberrant expression in human disease with an emphasis on cancer.
Keywords: cancer, epigenetics, microRNA, disease, development
Introduction
MicroRNAs (miRNAs) are ~22 nucleotide long small RNA molecules that usually act as endogenous repressors of gene activity. Although miRNAs were initially discovered in the nematode C. elegans, they have subsequently been found in various organisms and are thought to be expressed in all metazoan eukaryotes [1]. The importance of miRNAs in humans continues to become apparent and it is clear that faithful miRNA expression is critical for a myriad of biological processes ranging from differentiation and development to physiological homeostasis.
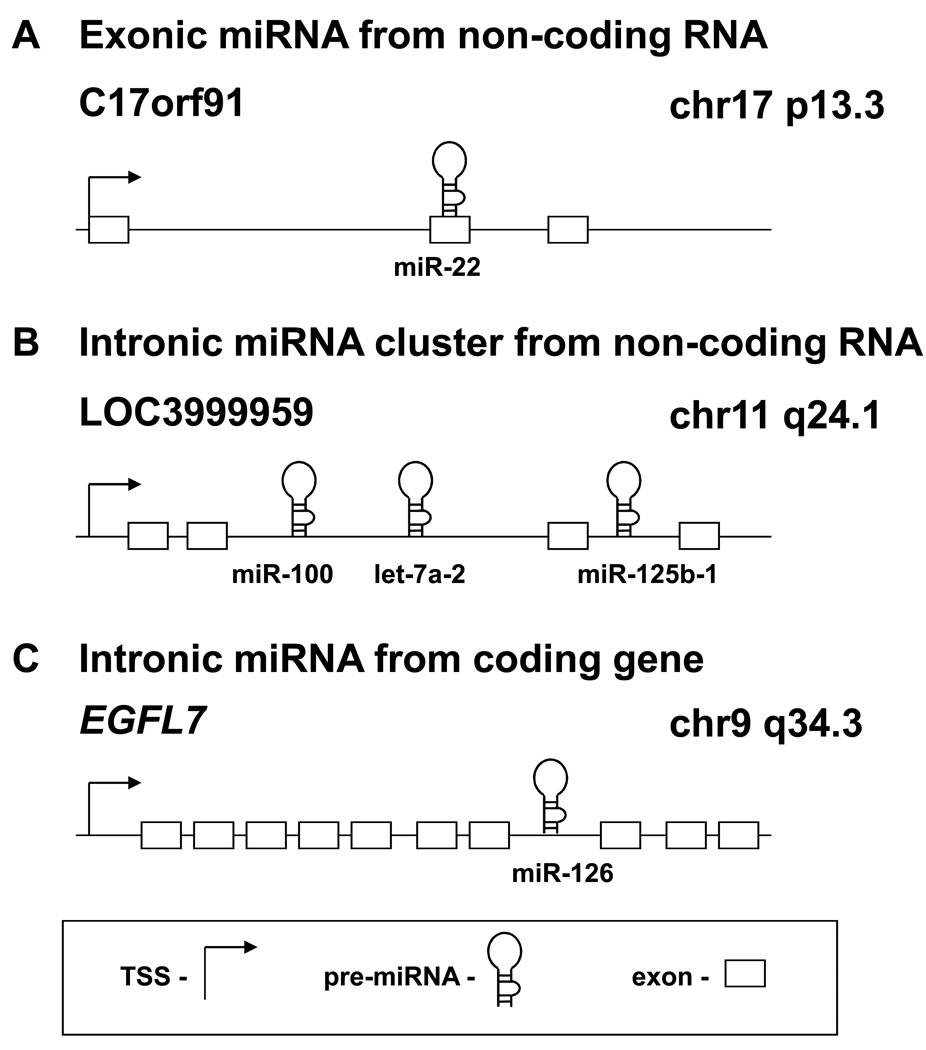
There are currently 851 mature human miRNA sequences in the Sanger database version 13.0, many of which are highly conserved in other organisms [2]. miRNAs are either intronic or exonic, and may be located in non-coding regions between annotated genes (intergenic) or in coding regions (Fig. 1)[3, 4]. The intergenic miRNAs are likely transcribed from their own promoters and many are located in miRNA clusters that generate long polycistronic transcipts [4, 5]. The intronic miRNAs located in protein coding genes are usually coordinately expressed with their host genes and are therefore regulated by the same factors as the host gene [1]. In addition, many miRNAs are expressed in a tissue-specific or developmental stage-specific manner, indicating the importance of coordinated regulation and function of miRNAs [6–8].
Figure 1.
Genomic locations of miRNAs. miRNAs may be located in almost any region of the genome including repeats. They may be grouped into clusters or situated alone, and may be exonic or intronic. A, a miRNA may be part of the exon of a non-coding RNA transcript, such as miR-22. B, polycistronic miRNAs may be grouped into a cluster and transcribed as a pri-miRNA, which yields several miRNAs. This occurs with the miR-100/let-7a-2/miR-125b-1 cluster, which is transcribed as an intron of a non-coding RNA. C, miRNAs may be located in the introns of canonical genes. The mature miRNA may be processed from the intron of the host gene transcript. This occurs when miR-126 is processed from the EGFL7 transcript. Arrow, transcription start site; hairpin, pre-miRNA; rectangle, exon.
miRNA biogenesis and basic function
Most miRNAs are transcribed by RNA polymerase II as long (>1 kb) primary miRNAs (pri-miRNA) that contain a 5′ 7-methyl guanosine cap and a 3′ poly adenosine tail, similar to mRNAs [9]. However, miRNAs embedded in repetitive elements such as Alus can be transcribed by RNA polymerase III [10]. The hairpin structure in the pri-miRNA is normally recognized and cleaved by the nuclear RNase III enzyme Drosha and its cofactor DiGeorge syndrome critical region 8 (DGCR8), although there are exceptions that bypass Drosha processing [11, 12]. Drosha/DGCR8 mediated pri-miRNA cleavage yields a precursor miRNA (pre-miRNA) of ~70 nt that forms a hairpin, which is exported to the cytoplasm via the nuclear transport receptor exportin-5 and the cofactor RanGTP [12]. Once in the cytoplasm, the pre-miRNA is cleaved by the RNase III enzyme Dicer into a double stranded RNA of ~22 nt. The strand with the less stable 5′ hydrogen bonding is usually selected as the mature miRNA, and is then incorporated into the RNA-induced silencing complex (RISC) [1].
RISC directs the miRNA to binding sites in the target mRNAs, which usually leads to gene repression, although there have been some reports of gene upregulation [13, 14]. The miRNA binding sites of the target mRNA are often perfectly complementary to the “seed” sequence (5′ nt 2–7) of the miRNA and are located in the 3′ untranslated region (UTR), but many important targets do not follow these rules [13, 15, 16]. The exact mechanisms of gene repression are still being elucidated, but there is evidence for translational initiation inhibition, translational elongation inhibition, premature translational termination, and cotranslational protein degradation [17]. In addition, several studies suggest that miRNA binding enhances mRNA degradation, such that transcript levels decrease along with protein levels [17]. Transcriptomics and proteomics approaches indicate that there may be >200 targets per miRNA [18] and a miRNA can directly affect the translation of hundreds of genes [19, 20]. These widespread effects of miRNAs are not surprising considering the promiscuous nature of miRNA:mRNA interactions.
miRNAs regulate important biological processes
The importance of faithful miRNA expression has been implicated in numerous biological and cellular events. The miRNA let-7 is critical for developmental timing [21], a developmentally regulated miRNA (bantam) controls cell proliferation via regulation of apoptosis [22], and miRNAs control ES cell differentiation [16] and stem cell division [23]. Other examples are miR-196, which is involved in hindlimb development [24], and the brain-specific miR-134, which contributes to the spatiotemporal control of mRNA translation that is necessary for synaptic development and plasticity [25]. Skin differentiation is promoted by miR-203, which represses p63 in stratified epithelial tissues [26], while precise levels of miR-1 are critical in cardiogenesis [27]. Normal immune function is dependent on miR-155 [28] and B-cell differentiation is controlled by miR-150 mediated repression of the transcription factor c-Myb [29]. In addition, the pancreatic islet cell-specific miR-375 regulates insulin secretion by inhibiting Myotrophin, a component of the exocytosis pathway [30].
Recent studies have shown that miRNAs may play causative roles in several diseases. In neurological diseases, the loss of the miR-20a/b-1 cluster has been implicated in Alzheimer’s disease [31] and the loss of miR-133b may contribute to the decrease in dopaminergic neurons seen in Parkinson’s disease [32]. In heart disease, the expression of miR-21 in cardiac fibroblasts contributes to interstitial fibrosis and cardiac hypertrophy [33], while miR-1 and miR-133 in cardiomyocytes protect against hypertrophy [34]. As a defense against viral infection, interferon-β upregulates miRNAs that target hepatitis C virus RNA and decrease replication and infection [35]. On the other hand, during latent infection herpes simplex virus 1 expresses miRNAs that target viral transcripts [36], so miRNAs have evolved to play roles in both aiding viruses and defending against them.
miRNAs involved in cancer
miRNA misexpression also has been well documented in cancer and some of these are listed in Table 1. The first high-throughput study using 334 patient samples of various malignancies showed that miRNA profiles can distinguish the developmental lineage and differentiation state of the tumors [37]. Another report was even able to identify the tissue of origin of metastatic tumors with unknown primary origin based on the miRNA profiles [38]. Profiling experiments have established miRNA deregulation in various cancers including pancreatic cancer [39, 40], liver cancer [41], breast cancer [42], colorectal cancer [43, 44], neuroblastoma [45], prostate cancer [46, 47], bladder cancer [48], cervical cancer [49, 50], leukemias [51, 52], lung cancer [53, 54], esophageal cancer [55], ovarian cancer [56], thyroid cancer [57], and sarcomas [58]. Intriguingly, many of these studies show that miRNA signatures have diagnostic and prognostic value, and may become valuable clinical tools in cancer therapy [59].
Table 1.
miRNAs that are implicated in human cancer.
| miRNA | Target | Function in Cancer | References |
|---|---|---|---|
| miR-15a/16-1 cluster |
BCL2, DDND1, WNT3A |
Tumor suppressor | [60–62] |
| Let-7 family | HMGA2, RAS | Tumor suppressor | [64–67] |
| miR-34 family | CCNE2, CDK4, MET |
Tumor suppressor | [68–70, 90] |
| BIC/miR-155 | TP53INP1 | Oncogene | [72–74] |
| miR-17~92 cluster |
HIF-1α, Tsp1, CTGF |
Oncogene | [75–79] |
| miR-221/222 | p27(CDKN1B), p57(CDKN1C) |
Oncogene | [80, 81] |
| miR-127 | BCL6 | Tumor suppressor | [84] |
| miR-124a | CDK6 | Tumor suppressor | [85] |
| miR-223 | NFI-A | Tumor suppressor | [86, 87] |
| miR-203 | ABL1, BCR- ABL1 |
Tumor suppressor | [88] |
| miR-1 | FoxP1, HDAC4, MET |
Tumor suppressor | [89] |
| miR-29 family | YYI, DNMT3A/B |
Tumor suppressor | [49, 92] |
| miR-101 | EZH2 | Tumor suppressor | [48, 95] |
| miR-128 | BMI1 | Tumor suppressor | [96] |
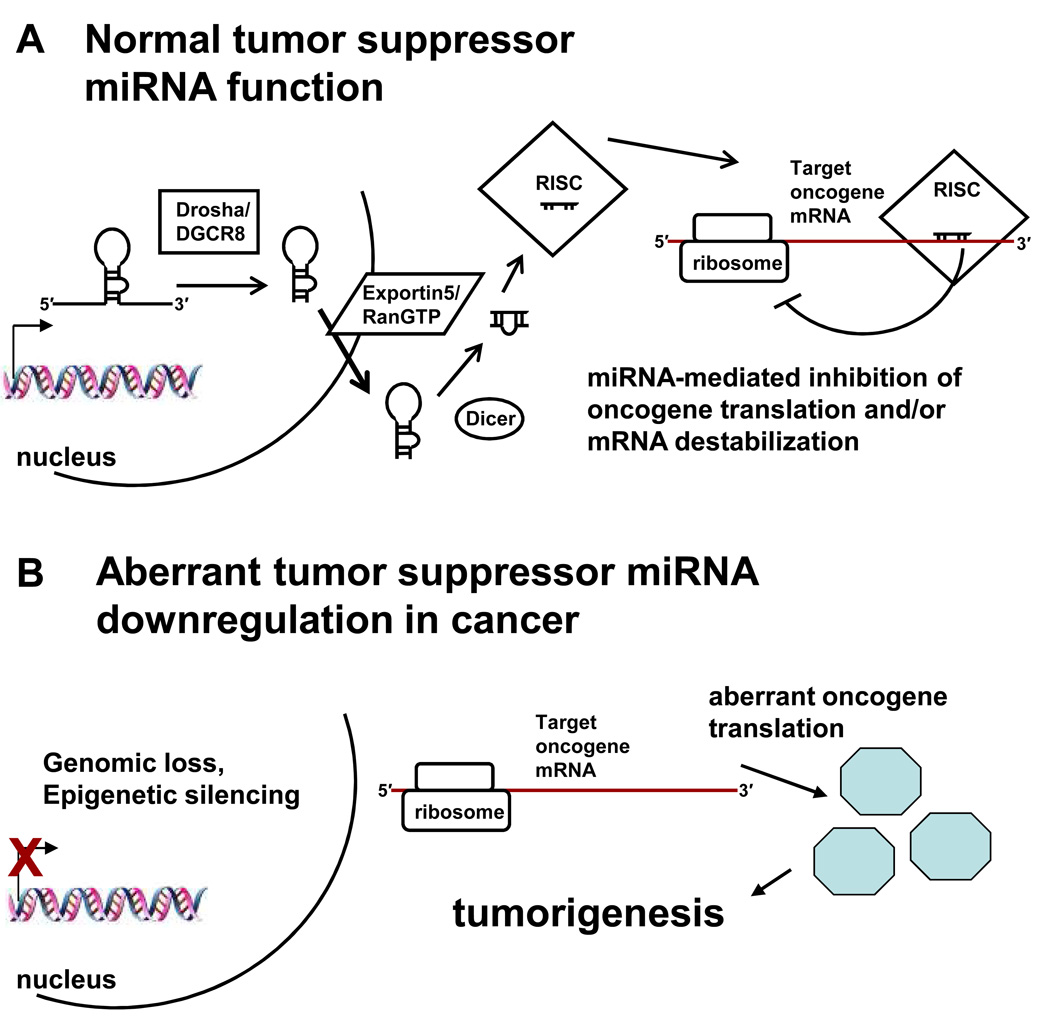
Thorough elucidation of the impact each specific miRNA can have on neoplastic processes will take many years. However, several examples of important oncogenic and tumor suppressor miRNAs have been reported. The first was the frequent down-regulation and deletion of the miR15a/16-1 cluster at 13q14 in chronic lymphocytic leukemia (CLL) [60]. Subsequent work showed that both miR-15a and miR-16-1 likely serve a tumor suppressor function by targeting the anti-apoptotic protein Bcl2 (Fig. 2) [61]. In addition, a recent report using a prostate cancer model revealed that the miR-15a/16-1 cluster also regulates tumorigenic activities such as survival, proliferation and invasion by targeting DDND1 and WNT3A [62].
Figure 2.
The function of tumor suppressor miRNAs. A, normally, a tumor suppressor miRNA is transcribed in the nucleus as a pri-miRNA, processed by Drosha/DGCR8 to a pre-miRNA, exported to the cytoplasm by Exportin-5/RanGTP, processed by Dicer and incorporated into RISC, which mediates translational repression and/or mRNA degradation of the target oncogene. B, decreased levels of a tumor suppressor miRNA, whether by genomic loss or epigenetic silencing as shown (processing defects and transcriptional blocks via trans acting factors may also occur), de-represses the target oncogene, leading to aberrant oncogene translation and tumorigenesis. In the case of miR-101, genomic loss decreases miR-101, causing an increase in its target EZH2, which promotes tumorigenesis.
Another well studied tumor suppressor miRNA family is the 11 member let-7 family, which is downregulated in lung cancer, and lower levels correlated with poor prognosis (Table 1) [63]. Let-7 targets two important oncogenes, HMGA2 [64] and RAS [65], and a common 12q15 translocation in cancer replaced the let-7 binding site in the HMGA2 mRNA, leading to a loss of repression [66]. A recent study extended the realm of let-7 to breast cancer by showing that let-7 decreased proliferation by targeting RAS and increased differentiation by targeting HMGA2 in breast tumor-initiating cells [67]. In 2007, three separate reports showed that miRNAs were intimately involved with the tumor suppressor p53 [68–70]. These groups demonstrated that p53 transactivates the miR-34 family, which causes apoptosis and decreases progression through the cell cycle by targeting CCNE2, CDK4, and MET [71].
The first oncogenic miRNA to be identified was miR-155, which is generated from the B-cell integration cluster (BIC) non-coding RNA [72]. miR-155 levels were increased in lymphomas and overexpression of miR-155 promoted B-cell proliferation and eventual malignancy in a transgenic mouse model [73]. miR-155 may cause these effects by targeting TP53INP1, a p53 target gene that mediates apoptosis and cell cycle arrest [74].
Another well-studied oncogenic non-coding RNA is the miR-17-92 cluster, which codes for 7 miRNAs. The miR-17-92 cluster is frequently amplified in B-cell lymphoma and its overexpression, along with c-myc, promoted tumor development in a mouse model [75]. In addition, the miR-17-92 cluster is amplified and upregulated in lung cancer and enforced expression of the miR-17-92 cluster enhanced lung cancer cell growth [76], while administration of antisense oligonucleotides promoted apoptosis [77]. This group later identified hypoxia inducible factor (HIF) 1 alpha as a target of the miR-17-92 cluster, elucidating a mechanism for c-myc induced downregulation of HIF-1 alpha [78]. Using mouse colonocytes, another study showed that c-Myc induced expression of the miR-17-92 cluster, which mediated tumor angiogenesis by targeting Tsp1 and CTGF [79].
miR-221 and miR-222 are transcribed together and have also been identified as oncogenic miRNAs. Thyroid cancer miRNA profiling revealed that miR-221/222 were upregulated in tumors and this correlated with a loss of KIT [57]. A forward genetics approach showed that miR-221/222 target the tumor suppressor and cell cycle regulator p27(Kip1), while miR-221/222 levels inversely correlated with p27(Kip1) expression in glioblastoma [80]. A recent study expanded the scope of miR-221 by showing that it targeted another cell cycle regulator p57(CDKN1C) and that miR-221 levels inversely correlated with p27(CDKN1B) and p57(CDKN1C) in hepatocellular carcinoma [81].
Epigenetics and miRNAs
The causes of miRNA misexpression in cancer may be due to DNA copy number amplification or deletion [59], inappropriate transactivation [79], genetic mutation [82], altered post transcriptional processing [83], or epigenetic mechanisms [84]. Epigenetic silencing of miRNAs could contribute to carcinogenesis by leading to the permanent upregulation of the miRNA target genes. The first example of an epigenetically regulated miRNA was the putative tumor suppressor miR-127, which is located in a CpG island and is normally expressed as part of a miRNA cluster. In cancer cells this cluster is silent and miR-127 is methylated, but treatment with a DNA demethylating agent and a histone deacetylase inhibitor induced miR-127 expression from its own promoter [84]. Further analysis showed that miR-127 may have a tumor suppressor function by repressing the oncogene BCL6 [84].
Subsequently, several reports have demonstrated that other tumor suppressor miRNAs are epigenetically silenced in various cancers. miR-124a was silenced by DNA methylation in cancer and targeted the oncogene cyclin D kinase 6, while leading to the phosphorylation of the tumor suppressor retinoblastoma [85]. The most common acute myeloid leukemia-associated fusion protein AML/ETO aberrantly epigenetically silences miR-223, which controls myelopoiesis by targeting NFI-A [86, 87]. Ectopic expression of miR-223, knockdown of AML/ETO, or treatment with DNA demethylating drugs caused leukemia cells to differentiate, revealing another mechanism by which AML/ETO promotes leukemia [87].
Another example of a miRNA that targets a fusion protein in hematopoietic malignancies is miR-203. miR-203 is either genetically lost or epigenetically silenced in leukemias and lymphomas and its re-expression directly repressed both ABL1 and BCR-ABL1 fusion protein (Philadelphia chromosome), causing an inhibition of cancer cell proliferation [88]. A study showed that miR-1 is silenced by hypermethylation in hepatocellular carcinoma and functions as a tumor suppressor by targeting the oncogenes FoxP1, MET and HDAC4 [89]. Moreover, the p53 regulated miR-34b/c was frequently methylated in colorectal cancer and DNA demethylating treatment re-expressed miR-34b/c in colon cancer cell lines [90].
In addition to DNA hypermethylation, repressive histone modifications may epigenetically silence miRNAs in cancer as well. During skeletal myogenesis low NF-κB and YY1 levels increase miR-29 expression, which targets its repressor YY1 in a positive feedback loop [91]. The elevated NF-κB levels in rhabdomyosarcoma (RMS) led to aberrant miR-29 silencing through YY1. The expression of miR-29 inhibited RMS tumor growth and promoted differentiation, implying that miR-29 acts as a tumor suppressor by promoting myogenesis [91].
miRNAs also control epigenetic mechanisms by targeting key chromatin modifying proteins. In lung cancer, the miR-29 family (miR-29a, -29b, -29c) was downregulated and its expression inversely correlated with DNA methyltransferase (DNMT) 3A and 3B levels [92]. Enforced expression of miR-29s in lung cancer cell lines inhibited tumorigenicity, decreased DNMT3A and DNMT3B levels, restored normal DNA methylation patterns, and re-expressed hypermethylation-silenced tumor suppressor genes [92].
The Polycomb group protein EZH2 is a histone methyltransferase that epigenetically silences genes by trimethylating histone H3 lysine 27 (H3K27me3) [93]. EZH2 acts as an oncogene in various malignancies, but the mechanism for EZH2 overexpression had not been elucidated [94]. Two recent studies showed that miR-101, which is frequently downregulated in cancer, targets EZH2 [48, 95]. Ectopic expression of miR-101 decreased EZH2 levels, decreased global H3K27me3, re-expressed aberrantly silenced Polycomb target genes, and inhibited cancer cell proliferation (Fig. 2) [48, 95].
Another Polycomb group protein BMI1 is involved in epigenetic gene silencing and promotes tumorigenesis [96]. miRNA profiling of glioblastoma revealed that miR-128 is downregulated in the tumor tissues, which inversely correlated with BMI1 expresssion [96]. Inducing miR-128 expression inhibited glioma cell proliferation and self-renewal, decreased H3K27me3, and increased p21(CIP1), all of which is consistent with BMI1 downregulation [96].
The future of miRNAs in medicine
Because miRNAs play such important roles in disease, the development of miRNA-based diagnoses and therapies is gaining traction. Techniques have been established to isolate miRNAs from cell-free bodily fluids such as serum and urine [97]. The miRNA profiles generated from these samples are robust and could discriminate between pregnant and non-pregnant women [97]. A recent report demonstrated that miRNAs are present and very stable in plasma, and miRNAs originating from prostate tumor xenografts are easily measured and discriminate tumor-bearing mice from controls [98]. In addition, the detection of miR-141 in plasma could distinguish prostate cancer patients from healthy controls, clearly showing the potential for miRNA-based diagnosis [98].
miRNA-based therapies will be difficult to develop because of the inherent instability of administered RNA. However, engineered oligonucleotides complementary to endogenous miRNAs termed “antagomirs” were developed and are stable when intravenously administered to mice [99]. The antagomir to the liver-specific miR-122 caused a robust decrease in miR-122 levels and an increase in transcripts with miR-122 binding sites [99]. Another group used miR-122 as the target to validate systemic delivery of locked-nucleic-acid-modified oligonucleotides (LNA-antimiR) in non-human primates [100]. A LNA contains at least one monomer with a modified sugar that is locked in a RNA-like conformation and shows very high hybridization affinity to complementary oligonucleotides [101]. The intravenously administered LNA-antimiR was taken up into hepatocytes to form a stable duplex with miR-122. This lead to a dose dependent depletion of mature miR-122 and a decrease in plasma cholesterol, a marker of miR-122 downregulation [100]. Moreover, there was no evidence of toxicity or histopathological changes from the treatment, indicating that LNA-antimiRs could be valuable tools to both elucidate miRNA function in animals and to treat disease in humans [100].
Summary
miRNAs are a fundamental part of coordinated gene regulation in eukaryotic cells and the recent explosion of reports on miRNA involvement in various biological processes continues unabated. It now seems that miRNA involvement in a cellular pathway or function is the rule rather than the exception, although the specific and intricate roles of each miRNA will take some time to determine. Several human diseases, from neurological disease to heart disease to cancer, are caused or propagated by miRNA misexpression, which has generated great interest in therapies, diagnoses and prognoses based on disease-specific miRNAs. Recent work in this regard has shown tremendous promise and the successful translation of miRNA research from novel bench work to medical practice and patients may open up a new avenue to treat disease in humans.
References
- 1.Cullen BR. Transcription and processing of human microRNA precursors. Mol Cell. 2004;16(6):861–865. doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 2.Berezikov E, et al. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120(1):21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 3.Rodriguez A, et al. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14(10A):1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129(7):1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farh KK, et al. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310(5755):1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 7.Stark A, et al. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123(6):1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 8.Zeng Y. Principles of micro-RNA production and maturation. Oncogene. 2006;25(46):6156–6162. doi: 10.1038/sj.onc.1209908. [DOI] [PubMed] [Google Scholar]
- 9.Lee Y, et al. MicroRNA genes are transcribed by RNA polymerase II. Embo J. 2004;23(20):4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13(12):1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 11.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448(7149):83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6(5):376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 13.Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30(4):460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 14.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318(5858):1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 15.Brodersen P, Voinnet O. Revisiting the principles of microRNA target recognition and mode of action. Nat Rev Mol Cell Biol. 2009 doi: 10.1038/nrm2619. [DOI] [PubMed] [Google Scholar]
- 16.Tay Y, et al. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455(7216):1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 17.Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132(1):9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 18.Lim LP, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433(7027):769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 19.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455(7209):64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Selbach M, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455(7209):58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 21.Reinhart BJ, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403(6772):901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 22.Brennecke J, et al. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113(1):25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 23.Hatfield SD, et al. Stem cell division is regulated by the microRNA pathway. Nature. 2005;435(7044):974–978. doi: 10.1038/nature03816. [DOI] [PubMed] [Google Scholar]
- 24.Hornstein E, et al. The microRNA miR-196 acts upstream of Hoxb8 and Shh in limb development. Nature. 2005;438(7068):671–674. doi: 10.1038/nature04138. [DOI] [PubMed] [Google Scholar]
- 25.Schratt GM, et al. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439(7074):283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 26.Yi R, et al. A skin microRNA promotes differentiation by repressing ‘stemness’. Nature. 2008;452(7184):225–229. doi: 10.1038/nature06642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436(7048):214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 28.Rodriguez A, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316(5824):608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao C, et al. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131(1):146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 30.Poy MN, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432(7014):226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 31.Hebert SS, et al. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased BACE1/beta-secretase expression. Proc Natl Acad Sci U S A. 2008;105(17):6415–6420. doi: 10.1073/pnas.0710263105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim J, et al. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317(5842):1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thum T, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456(7224):980–984. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 34.Care A, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13(5):613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 35.Pedersen IM, et al. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449(7164):919–922. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Umbach JL, et al. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature. 2008;454(7205):780–783. doi: 10.1038/nature07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 38.Rosenfeld N, et al. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol. 2008;26(4):462–469. doi: 10.1038/nbt1392. [DOI] [PubMed] [Google Scholar]
- 39.Roldo C, et al. MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J Clin Oncol. 2006;24(29):4677–4684. doi: 10.1200/JCO.2005.05.5194. [DOI] [PubMed] [Google Scholar]
- 40.Szafranska AE, et al. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. 2007;26(30):4442–4452. doi: 10.1038/sj.onc.1210228. [DOI] [PubMed] [Google Scholar]
- 41.Murakami Y, et al. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25(17):2537–2545. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- 42.Iorio MV, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65(16):7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 43.Cummins JM, et al. The colorectal microRNAome. Proc Natl Acad Sci U S A. 2006;103(10):3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schepeler T, et al. Diagnostic and prognostic microRNAs in stage II colon cancer. Cancer Res. 2008;68(15):6416–6424. doi: 10.1158/0008-5472.CAN-07-6110. [DOI] [PubMed] [Google Scholar]
- 45.Chen Y, Stallings RL. Differential patterns of microRNA expression in neuroblastoma are correlated with prognosis, differentiation, and apoptosis. Cancer Res. 2007;67(3):976–983. doi: 10.1158/0008-5472.CAN-06-3667. [DOI] [PubMed] [Google Scholar]
- 46.Porkka KP, et al. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007;67(13):6130–6135. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
- 47.Ambs S, et al. Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res. 2008;68(15):6162–6170. doi: 10.1158/0008-5472.CAN-08-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Friedman JM, et al. The putative tumor suppressor microRNA-101 modulates the cancer epigenome by repressing the polycomb group protein EZH2. Cancer Res. 2009;69(6):2623–2629. doi: 10.1158/0008-5472.CAN-08-3114. [DOI] [PubMed] [Google Scholar]
- 49.Wang X, et al. Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. PLoS ONE. 2008;3(7):e2557. doi: 10.1371/journal.pone.0002557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lui WO, et al. Patterns of known and novel small RNAs in human cervical cancer. Cancer Res. 2007;67(13):6031–6043. doi: 10.1158/0008-5472.CAN-06-0561. [DOI] [PubMed] [Google Scholar]
- 51.Calin GA, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353(17):1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 52.Calin GA, et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci U S A. 2004;101(32):11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu SL, et al. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell. 2008;13(1):48–57. doi: 10.1016/j.ccr.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 54.Yanaihara N, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9(3):189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 55.Guo Y, et al. Distinctive microRNA profiles relating to patient survival in esophageal squamous cell carcinoma. Cancer Res. 2008;68(1):26–33. doi: 10.1158/0008-5472.CAN-06-4418. [DOI] [PubMed] [Google Scholar]
- 56.Zhang L, et al. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc Natl Acad Sci U S A. 2008;105(19):7004–7009. doi: 10.1073/pnas.0801615105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.He H, et al. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci U S A. 2005;102(52):19075–19080. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Subramanian S, et al. MicroRNA expression signature of human sarcomas. Oncogene. 2008;27(14):2015–2026. doi: 10.1038/sj.onc.1210836. [DOI] [PubMed] [Google Scholar]
- 59.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 60.Calin GA, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99(24):15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cimmino A, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102(39):13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bonci D, et al. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14(11):1271–1277. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- 63.Takamizawa J, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64(11):3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 64.Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21(9):1025–1030. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Johnson SM, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120(5):635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 66.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315(5818):1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu F, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131(6):1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 68.He L, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447(7148):1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chang TC, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26(5):745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Raver-Shapira N, et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26(5):731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 71.He L, et al. microRNAs join the p53 network--another piece in the tumour-suppression puzzle. Nat Rev Cancer. 2007;7(11):819–822. doi: 10.1038/nrc2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Eis PS, et al. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci U S A. 2005;102(10):3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Costinean S, et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci U S A. 2006;103(18):7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gironella M, et al. Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proc Natl Acad Sci U S A. 2007;104(41):16170–16175. doi: 10.1073/pnas.0703942104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.He L, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435(7043):828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hayashita Y, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65(21):9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 77.Matsubara H, et al. Apoptosis induction by antisense oligonucleotides against miR-17-5p and miR-20a in lung cancers overexpressing miR-17-92. Oncogene. 2007;26(41):6099–6105. doi: 10.1038/sj.onc.1210425. [DOI] [PubMed] [Google Scholar]
- 78.Taguchi A, et al. Identification of hypoxia-inducible factor-1 alpha as a novel target for miR-17-92 microRNA cluster. Cancer Res. 2008;68(14):5540–5545. doi: 10.1158/0008-5472.CAN-07-6460. [DOI] [PubMed] [Google Scholar]
- 79.Dews M, et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet. 2006;38(9):1060–1065. doi: 10.1038/ng1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.le Sage C, et al. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. Embo J. 2007;26(15):3699–3708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fornari F, et al. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene. 2008;27(43):5651–5661. doi: 10.1038/onc.2008.178. [DOI] [PubMed] [Google Scholar]
- 82.Jazdzewski K, et al. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc Natl Acad Sci U S A. 2008;105(20):7269–7274. doi: 10.1073/pnas.0802682105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Melo SA, et al. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat Genet. 2009 doi: 10.1038/ng.317. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 84.Saito Y, et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9(6):435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 85.Lujambio A, et al. Genetic Unmasking of an Epigenetically Silenced microRNA in Human Cancer Cells. Cancer Res. 2007;67(4):1424–1429. doi: 10.1158/0008-5472.CAN-06-4218. [DOI] [PubMed] [Google Scholar]
- 86.Fazi F, et al. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123(5):819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 87.Fazi F, et al. Epigenetic silencing of the myelopoiesis regulator microRNA-223 by the AML1/ETO oncoprotein. Cancer Cell. 2007;12(5):457–466. doi: 10.1016/j.ccr.2007.09.020. [DOI] [PubMed] [Google Scholar]
- 88.Bueno MJ, et al. Genetic and epigenetic silencing of microRNA-203 enhances ABL1 and BCR-ABL1 oncogene expression. Cancer Cell. 2008;13(6):496–506. doi: 10.1016/j.ccr.2008.04.018. [DOI] [PubMed] [Google Scholar]
- 89.Datta J, et al. Methylation mediated silencing of MicroRNA-1 gene and its role in hepatocellular carcinogenesis. Cancer Res. 2008;68(13):5049–5058. doi: 10.1158/0008-5472.CAN-07-6655. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 90.Toyota M, et al. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res. 2008;68(11):4123–4132. doi: 10.1158/0008-5472.CAN-08-0325. [DOI] [PubMed] [Google Scholar]
- 91.Wang H, et al. NF-kappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell. 2008;14(5):369–381. doi: 10.1016/j.ccr.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fabbri M, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A. 2007;104(40):15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6(11):846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 94.Varambally S, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419(6907):624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 95.Varambally S, et al. Genomic Loss of microRNA-101 Leads to Overexpression of Histone Methyltransferase EZH2 in Cancer. Science. 2008 doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Godlewski J, et al. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 2008;68(22):9125–9130. doi: 10.1158/0008-5472.CAN-08-2629. [DOI] [PubMed] [Google Scholar]
- 97.Gilad S, et al. Serum microRNAs are promising novel biomarkers. PLoS ONE. 2008;3(9):e3148. doi: 10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mitchell PS, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105(30):10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Krutzfeldt J, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438(7068):685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 100.Elmen J, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452(7189):896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 101.Vester B, Wengel J. LNA (locked nucleic acid): high-affinity targeting of complementary RNA and DNA. Biochemistry. 2004;43(42):13233–13241. doi: 10.1021/bi0485732. [DOI] [PubMed] [Google Scholar]