Abstract
Studies of the phosphodiesterase PDE7 family are impeded by there being only one commercially-available PDE7 inhibitor, BRL50481. We have employed a high throughput screen of commercial chemical libraries, using a fission yeast-based assay, to identify PDE7 inhibitors that include steroids, podocarpanes, and an unusual heterocyclic compound, BC30. In vitro enzyme assays measuring the potency of BC30 and two podocarpanes, in comparison with BRL50481, produce data consistent with those from yeast-based assays. In other enzyme assays, BC30 stimulates the PDE4D catalytic domain, but not full-length PDE4D2, suggesting an allosteric site of action. BC30 significantly enhances the anti-inflammatory effect of the PDE4 inhibitor rolipram as measured by release of TNFα from activated monocytes. These studies introduce several new PDE7 inhibitors that may be excellent candidates for medicinal chemistry due to the requirements for drug-like characteristics placed on them by the nature of the yeast-based screen.
Keywords: Schizosaccharomyces pombe, cAMP, phosphodiesterase, high throughput, inhibitors, PDE7
Introduction
Cyclic nucleotide phosphodiesterases (PDEs) convert cyclic AMP (cAMP) to 5′AMP and cyclic GMP (cGMP) to 5′GMP, to regulate levels of these important second messengers. Mammals possess 21 genes encoding 11 PDE families that are distinguished by their substrate specificity, their sensitivity to various small molecules, and the presence of conserved domains outside of the catalytic domains (as reviewed by (1-3)). Differences in tissue-specific expression as well as subcellular localization of individual proteins (3, 4) suggest that the various PDE families might serve as therapeutic targets for a wide range of indications.
Enzymes of the PDE4, PDE7, and PDE8 families are cAMP-specific and are expressed by four, two, and two genes, respectively. Rolipram, the first PDE4-specific inhibitor, has been used to demonstrate that inhibition of PDE4 can reduce inflammation, enhance cognition, inhibit HIV infection and replication, and induce apoptosis in chronic lymphocytic leukemia (CLL) cells, thus demonstrating a wide range of biological roles for this enzyme family (1-3, 5). Much less is known about the more recently-discovered PDE7 and PDE8 enzymes, in part due to the limited availability of family-specific inhibitors. While PDE7 is a potential target for treatment of inflammation (6), only one PDE7 inhibitor is commercially-available, BRL50481 (7), and this compound is significantly less active against PDE7B than it is against PDE7A (see Results). Since most PDE inhibitor discovery is performed through the structural characterization of the active site, combined with the novel synthesis and testing of compounds from a particular structural class, the compounds identified in these studies are generally not readily available to the research community (8, 9). Even fewer tools are available to study the post-genomically-identified PDE8A and PDE8B enzymes, with the broad-spectrum PDE inhibitor dipyridamole being used for PDE8 inhibition (1).
The fission yeast Schizosaccharomyces pombe monitors extracellular glucose by a cAMP signaling pathway (10). High glucose levels detected by a putative G protein coupled receptor, Git3, lead to adenylate cyclase activation via the Gpa2-Git5-Git11 heterotrimeric G protein. Adenylate cyclase produces a cAMP signal that activates protein kinase A (PKA), which negatively regulates transcription of genes involved in gluconeogenesis and sexual development. Most of the genes of the PKA pathway have been identified in genetic screens that utilize a fusion of the PKA-repressed fbp1+ gene to the ura4+ gene of the uracil biosynthetic pathway. Defects in cAMP signaling lead to constitutive fbp1-ura4 expression, allowing growth on glucose-rich medium lacking uracil, while conferring sensitivity to the pyrimidine analog 5-fluoro orotic acid (5FOA) (11). Screens for suppressors of this low PKA phenotype, which restore 5FOA-resistant growth, identified mutations in the cgs1+ and cgs2+ genes that encode the regulatory subunit of PKA and the cAMP PDE, respectively (12, 13).
The growth phenotypes conferred by the fbp1-ura4 fusion were further exploited to develop a high throughput screening platform for PDE inhibitors by replacing the cgs2+ PDE gene with mammalian PDE genes and screening for compounds that confer 5FOA-resistant growth to strains that possess defects in glucose-signaling (14). Our initial screens, which focused on the PDE4 family, showed that the furanocoumarin imperatorin is a PDE4B inhibitor, thus explaining a prior study that demonstrated its ability to inhibit HIV replication (15), an outcome also observed with PDE4 inhibition by rolipram (5). Additional PDE4 inhibitors will be described elsewhere.
This current study describes the development and deployment of a PDE7A inhibitor screen and the identification of compounds that inhibit PDE7A and PDE7B activity. Foremost among these is BC30, which displays potent synergy with rolipram in inhibiting TNFα release by LPS-treated U937 cells. While the mechanism of PDE7 inhibition by BC30 remains to be determined, it appears to act at a site other than the cAMP-binding site, as suggested by its unusual ability to stimulate cAMP hydrolysis in an in vitro reaction that contains the catalytic domain of the PDE4D enzyme, which displays ∼60% similarity to PDE7A and PDE7B catalytic domains. These studies demonstrate the utility of our screening platform for the discovery of novel PDE inhibitors.
Materials and Methods
Yeast strains, media and growth conditions
Strains CHP1189 (h+ fbp1-ura4+ ura4∷fbp1-lacZ leu1-32 gpa2Δ pap1Δ cgs2∷PDE7A1), CHP1209 (h- fbp1-ura4+ ura4∷fbp1-lacZ leu1-32 git3Δ pap1Δ cgs2∷PDE7B1), CHP1262 (h- fbp1-ura4+ ura4∷fbp1-lacZ leu1-32 gpa2Δ pap1Δ cgs2∷PDE4A1), CHP1268 (h- fbp1-ura4+ ura4∷fbp1-lacZ leu1-32 git3Δ pap1Δ cgs2∷PDE4B2), and CHP1186 (h+ fbp1-ura4+ ura4∷fbp1-lacZ leu1-32 git11Δ pap1Δ cgs2∷PDE4D3) were grown in YEA rich or EMM defined media as previously described (13).
Construction of S. pombe strains that express human PDE4 and PDE7 enzymes
The human PDE4A1 (Genbank accession number U68532), PDE4B2 (Genbank accession number L20971), PDE4D3 (Genbank accession number U50159.1), PDE7A1 (Genbank accession number NM_002603) and PDE7B1 (Genbank accession number NM_018945) open reading frames were PCR amplified using oligonucleotides that provide targeting sequences to the S. pombe cgs2 PDE gene locus. As previously described (14), the PCR products were integrated by homologous recombination into the cgs2/pde1 locus, which had been disrupted with ura4+ to allow detection by 5FOA counterselection (16). Subsequent crosses were carried out to introduce fbp1-lacZ and fbp1-ura4 reporters, along with mutations in the glucose/cAMP pathway and a deletion of the pap1+ transcriptional activator gene to enhance compound sensitivity (17).
HTS screening and 5FOA growth assays
High throughput screens (HTSs) were performed at the Broad Institute's Chemical Biology Program screening facility. Prior to screening, each HTS was optimized as follows. First, various mutations with respect to cAMP production were introduced into the strain background to determine whether they confer significant 5FOA sensitivity, a reflection of low basal cAMP levels (10). Secondly, we determined the optimal concentration of cAMP that must be present in the culture prior to the HTS in order to repress the fbp1-ura4+ reporter that controls the 5FOA growth phenotype. If the reporter is not repressed prior to the screen, cells will be 5FOA-sensitive due to the pre-existing Ura4 enzyme. Finally, we determined the optimal concentration of cells to inoculate into the microtiter dishes in order to produce an HTS with a high Z factor (see below). For the PDE7A HTS, strain CHP1189 (gpa2Δ PDE7A) was cultured to exponential phase in EMM complete medium containing 2.5 mM cAMP to repress fbp1-ura4 transcription. Cells were collected by centrifugation and resuspended in 5FOA medium (11), and 25 μl was transferred to 384-well microtiter dishes (untreated, with flat clear bottoms) that contained 25 μl 5FOA medium plus 100 nl of compounds (stock solutions were generally 10mM). The starting cell concentration was 1 × 105 cells/ml. Positive control wells contained 5mM cAMP in the 5FOA medium. Cultures were grown for 48 hours at 30°C while sealed in an airtight container with moist paper towels to prevent evaporation. Optical densities (OD600) of the cultures were determined using a microplate reader to measure growth. Bioinformatic analysis of the results to determine composite Z scores was performed as previously described (18). The Z factor of an assay is determined by multiplying the sum of the standard deviations of the positive and negative controls by three, dividing by the absolute difference in the means of the positive and negative controls, and subtracting from the number one. An assay with a Z factor of greater than 0.5 is considered sufficiently robust for high throughput screening. Within a screen, individual wells are assigned a Z score, which represents the number of standard deviations above or below the mean of the negative control wells in that same assay plate. Duplicate Z scores for each compound are plotted onto a grid (Figure 1) and projected perpendicularly to the diagonal that represents identity between duplicate Z scores. The composite Z score is the distance from this point on the diagonal to the origin. 5FOA growth assays for strains expressing PDE4 subtypes and PDE7B were carried out under similar conditions as in the PDE7A screen.
Figure 1.
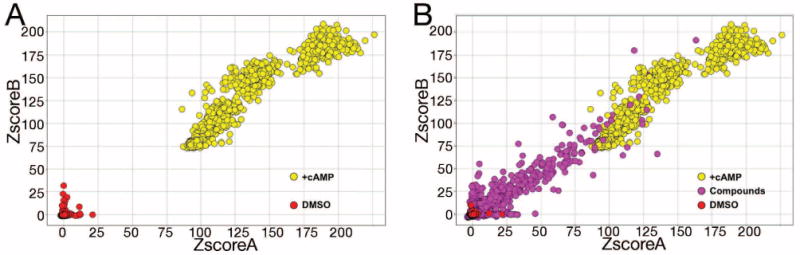
High throughput screening data summary. A) Z scores for duplicate wells including 10,578 DMSO-pinned (negative control, red circles) and 1,920 cAMP-supplemented wells (positive control, yellow circles) are presented. B) Z scores for duplicate wells from 48,176 compounds are superimposed on the negative and positive control data from panel A. Images were created using the Spotfire software package (TIBCO Software Inc.).
In vitro enzyme assays
In vitro PDE assays were carried out as described by Wang et al. (19), in which the amount of cAMP remaining in a reaction is measured by scintillation counting after differential Ba(OH)2 precipitation of the 3H-AMP produced by hydrolysis from the unreacted 3H-cAMP. PDE4D (both catalytic domain and full-length PDE4D2) and PDE7B enzymes were generously provided by Drs. H. Wang and H. Ke; PDE7A enzyme was purchased from BioMol International. Assays were conducted under conditions that resulted in less than 25% hydrolysis of cAMP in the absence of inhibitor. The rate of hydrolysis, which was constant over the 15 minutes' duration of the reaction, was proportional to enzyme concentration. The initial cAMP concentration, 25nM, was slightly below the published Kms for cAMP of 100-200 nM for PDE7A and 30-70 nM for PDE7B (1), as well as our observed Km for PDE7B of ∼50 nM, which was obtained using Michaelis-Menten kinetics analyzed by Lineweaver-Burke and Eadie-Hofstee plots (not shown). The IC50 is the concentration of inhibitor that reduces the rate of cAMP hydrolysis to half of that of the vehicle-treated control reactions.
TNFα assays
Myelomonocytic U937 cultures growing in fresh medium (RPMI 1640 + 10% fetal bovine serum) at 106 cells/ml were pre-incubated for 17 hr. with 300 ng/ml PMA. Aliquots (105 cells) were diluted in equal volume of fresh medium and treated with compounds (10 μM final concentration) for 1 hr in 96-well plates. Control cultures received vehicle only (ethanol containing < 0.1% DMSO). Cultures were activated with 25 μg/ml LPS to stimulate TNFα release (20), and culture supernatants were collected after 3 or 20 hrs of LPS stimulation. TNFα was measured using a human TNF ELISA kit (BD Biosciences, Franklin Lakes, NJ) according to the manufacturer's instructions. Statistical significance was determined using the Excel one-tailed Student's t Test function, with the differences being considered significant at p < 0.05.
Results
Identification of PDE7 inhibitors using a yeast growth assay
To identify PDE7 inhibitors, we constructed a fission yeast strain whose only PDE activity comes from the human PDE7A gene and whose growth behavior reflects its intracellular cAMP level. Using homologous recombination, we replaced the fission yeast cgs2+ PDE gene with PDE7A as described for other mammalian PDE genes (14). The CHP1189 screening strain also contains fbp1-ura4 and fbp1-lacZ reporter genes (11), a deletion of the pap1+ transcriptional activator (to enhance compound sensitivity) (21), and a gpa2+ deletion to reduce cAMP synthesis so as to confer 5FOA-sensitive growth (22). After determining growth conditions that optimize the detection of PDE7A inhibitors, 48,176 compounds (most at 20μM, Figure 1B, purple circles) were screened in duplicate for their ability to stimulate 5FOA-resistant growth in 384 well microtiter dishes. An additional 10,578 DMSO-only negative control wells (Figure 1A, red circles) and 1920 positive control wells supplemented with 5 mM cAMP (Figure 1A, yellow circles) were included in the screen. The OD600 (detecting the turbidity of the cultures) of the positive controls after 48 hours growth was 1.27 ± 0.068, while the OD600 of the negative controls was 0.062 ± 0.011. The Z factor for this screen (an assessment of the robustness of an HTS) is 0.804, well above the value 0.5 that serves as the benchmark for a high-quality screen. Sixty-one compounds stimulated growth to an average OD600 > 0.6, or approximately half the OD600 of the saturated positive controls. Thus, ∼ 0.1% of the screened compounds display the potential for being effective PDE7A inhibitors.
In comparing the structures of the 24 compounds that conferred the highest average OD600 values (>0.821), two related compound families were immediately obvious. Two compounds are the steroids androsterone acetate (Microsource 00107113) and canrenone (Microsource 01505248), while another three are podocarpanes, which possess the same arrangement of three six-carbon rings as found in steroids (Figure 2A). Previously, Lee and coworkers described the PDE7A inhibitor IC242 as being a steroid-like compound (23); thus these compounds are related in structure to at least one previously-identified PDE7 inhibitor. On the other hand, they bear no resemblance to a variety of purine-based (24), pyrimidine-based (25), or thioxoquinazoline-based (26) PDE7 inhibitors described in the literature.
Figure 2.
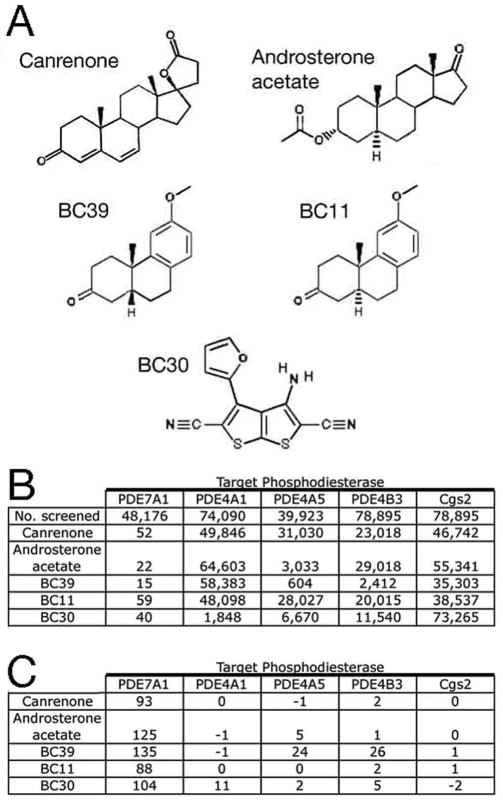
Structure and HTS data for some PDE7A inhibitors. A) Structures of five of the most effective PDE7A inhibitors. B) HTS screening data for compounds shown in panel A. Strains expressing human PDE7A1, mouse PDE4A1, rat PDE4A5, mouse PDE4B3, and S. pombe PDE Cgs2 were grown in the presence of compounds at 20 uM. The first row indicates the number of compounds screened, while subsequent rows indicate the rank of each compound with respect to Composite Z score from the initial HTSs. C) Composite Z scores from the initial 20μM HTSs for the compounds shown in panel A.
In addition to steroids and podocarpanes, an unusual heterocyclic compound was identified in this screen, 3-amino-4-(2-furyl) thieno[2,3- b]thiophene-2,5- dicarbonitrile (Maybridge KM03472SC), which we refer to as BC30 (Figure 2A), and which is also structurally distinct from previously described PDE7 inhibitors. While BC30 conferred only the 40th highest Composite Z score in the 20μM screen (Figure 2B, 2C), it was one of the most effective PDE7A inhibitors upon rescreening at 2μM (data not shown). The five compounds shown in Figure 2A were also examined in separate HTSs using strains that express the following genes: mouse PDE4A1 (Genbank accession number AF208021), rat PDE4A5 (Genbank accession number L27057), mouse PDE4B3 (Genbank accession number AF208023), and S. pombe PDE cgs2+/pde1+ (Genbank accession number S64907). The podocarpane BC39 showed modest activity against PDE4A5 and PDE4B3, while BC30 showed very weak activity against PDE4A1 when tested at 20μM. Based on this degree of selectivity, we chose to carry out further studies of BC30, the podocarpanes BC11 (Microsource 00100646) and BC39 (Microsource 00100254), and the commercially-available PDE7 inhibitor BRL50481 (7).
Characterization of PDE7A and PDE7B inhibition via yeast-growth assays
As an initial test of compounds BC11, BC30, BC39, and BRL50481, we repeated the 5FOA growth assay on strains that express PDE7A or PDE7B, using a range of compound concentrations (Figure 3). For these assays, newly-purchased compounds were used to ensure that activity detected in the HTS represented the original compound and not a breakdown product. All four compounds confer dose-dependent stimulation of growth in 5FOA medium, which is highly responsive to small changes in compound concentration. These steep response curves are consistent with the fact that growth in 5FOA medium is not proportional to the level of fbp1-ura4 transcription; while mutations can produce a >200-fold increase in transcription, 5FOA-sensitivity is conferred by as little as a 30-fold increase (11). BRL50481 is the most effective compound for stimulation of 5FOA-resistant growth of the PDE7A-expressing strain; compound BC30 is also very potent at low micromolar concentrations (Figure 3A). The plateau in the OD600 seen for BC30 may represent a problem with solubility at higher concentrations or a lack of stability whereby the PDE is not inhibited long enough to permit growth to saturation. Higher concentrations of compounds BC11 and BC39 are required for stimulating 5FOA-resistance, although these two compounds are fairly effective compared with most other compounds tested. Most remarkably, according to our 5FOA growth assays, BRL50481 is a rather poor inhibitor of PDE7B, equal to BC11 and BC39, and far less effective than BC30 (Figure 3B).
Figure 3.
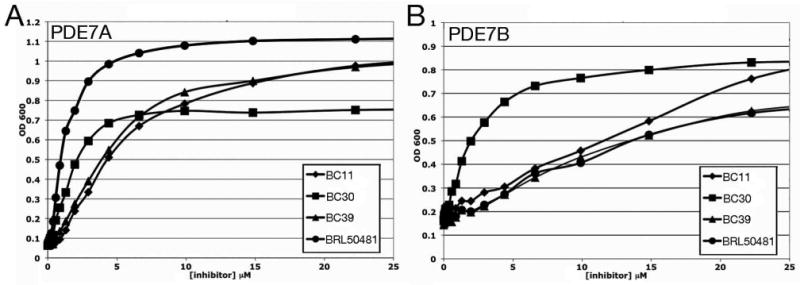
5FOA growth response curves of candidate PDE7A and PDE7B inhibitors. Optical density of cultures of a (A) PDE7A-expressing strain and a (B) PDE7B-expressing strain grown in 384 well microtiter wells were measured after 48 hours incubation at 30°C. This is plotted against the concentration of compound (in μM) in the wells. Each value represents the average of four wells. DMSO levels are kept below 0.2% as growth inhibition is seen when DMSO levels reach 1% or higher.
In vitro enzyme assays of PDE7A and PDE7B inhibition
Assays of purified PDE7A and PDE7B were carried out in order to compare the inhibitory potency of compounds BC11, BC30 BC39, and BRL50481 (Table 1). We observed a similar IC50 (∼150 nM) for BRL50481 against PDE7A to that of the previously published value (7). Consistent with our cell-based assay, BRL50481 proved to be a much weaker inhibitor of PDE7B than of PDE7A, displaying an 80-fold higher IC50 (Table 1). The podocarpanes BC11 and BC39, which were similar to each other in 5FOA growth assays (Figure 3), likewise displayed nearly identical IC50 values (Table 1); in both cases having more potency against PDE7A than PDE7B. Results with BC30 in vitro did not precisely recapitulate the growth assay in the in vitro assays: while BC30 is equally effective in the cell-based assay with strains expressing PDE7A and PDE7B, it has a four-fold lower IC50 on PDE7A than PDE7B. In summary, nevertheless, for both the cell-based assay and the in vitro assay, BC30 is found to be intermediate between BRL50481 and the podocarpanes in efficacy against PDE7A, but the best inhibitor of PDE7B.
Table 1.
IC50 values for PDE inhibitors against PDE7A and PDE7B
| Inhibitor IC50 (μM) | ||||
|---|---|---|---|---|
| Enzyme | BRL50481 | BC30 | BC11 | BC39 |
| PDE7A | 0.15 | 1.1 | 3.2 | 3.8 |
| PDE7B | 12.1 | 3.7 | 26.3 | 26.6 |
In vitro enzyme assays were carried out as described in the Materials and Methods. IC50 values were determined by linear interpolation using the three concentrations that resulted in the closest to 50% inhibition from a series of assays testing 8-9 inhibitor concentrations.
BC30 activity against PDE4 in yeast-based and in vitro enzyme assays
Because PDE4 and PDE7 enzymes possess closely-related catalytic domains, and because BC30 weakly stimulates growth of a strain that expresses mouse PDE4A1 (Figure 2B, 2C), we inquired whether compound BC30 inhibits human PDE4 enzymes. BC30 showed no effect on the growth of strains that express human PDE4A1, PDE4B2, or PDE4D3, while displaying equal potency on strains expressing PDE7A and PDE7B (Figure 4). These assays were extended to strains that express genes from other mammalian PDE families. For cGMP-specific PDEs, the assay was modified by using strains that lack adenylate cyclase and included the addition of 30μM to 1mM cGMP to the 5FOA growth medium, a concentration sufficient to activate PKA once the cGMP PDE is inhibited. As with the PDE4-expressing strains (Figure 4), BC30 had no effect on growth of strains expressing PDE1B, PDE1C, PDE2A, PDE3B, PDE5A, PDE8A, PDE9A, or PDE11A (data not shown). Each assay included a positive control of growth stimulation by either a known PDE inhibitor or a cyclic nucleotide added to the medium. These results suggest that BC30 is primarily a PDE7-specific inhibitor. However, a strain expressing PDE10A shows a modest response, reaching OD600 = 0.53 in the presence of 40μM BC30 (data not shown). Thus, BC30 appears to be a weak PDE10A inhibitor.
Figure 4.
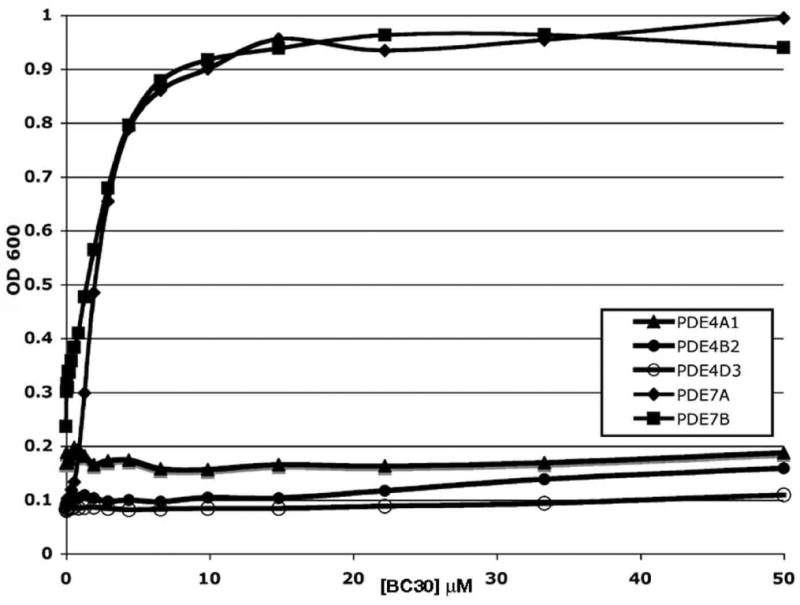
5FOA growth response curves for BC30 on strains expressing PDE4 or PDE7 enzymes. Assays were carried out as described in Figure 3 using strains that express human PDE4A1, PDE4B2, PDE4D3, PDE7A1, or PDE7B1.
BC30 was also examined as part of a panel of compounds in in vitro enzyme assays using 2μM compound and the catalytic domains from PDE4A, PDE4B, and PDE4D expressed in and purified from E. coli (19). Surprisingly, BC30 and three unrelated PDE7A inhibitors significantly stimulate cAMP hydrolysis by the PDE4D catalytic domain (residues 86-413 of PDE4D2; data not shown). BC30 acts at a remarkably low concentration; 25 nM produced the maximum increase in activity under conditions where the concentration of enzyme was 16 nM, doubling the Vmax of the enzyme (Fig. 5). However, neither BC30 nor the other compounds that stimulate the catalytic domain of PDE4D affect the activity of the full-length PDE4D2 (residues 1-507; data not shown). In addition, there are no data to suggest that PDE4D is stimulated either in our yeast strains or in mammalian cells by any of these compounds. Therefore, while we view this effect on the PDE4D catalytic domain to be an artifact unique to the in vitro enzyme assay, it represents a highly specific interaction between compound and enzyme. This activation phenomenon is specific to a minority of compounds tested and suggests that they bind somewhere other than the cAMP-binding site.
Figure 5.
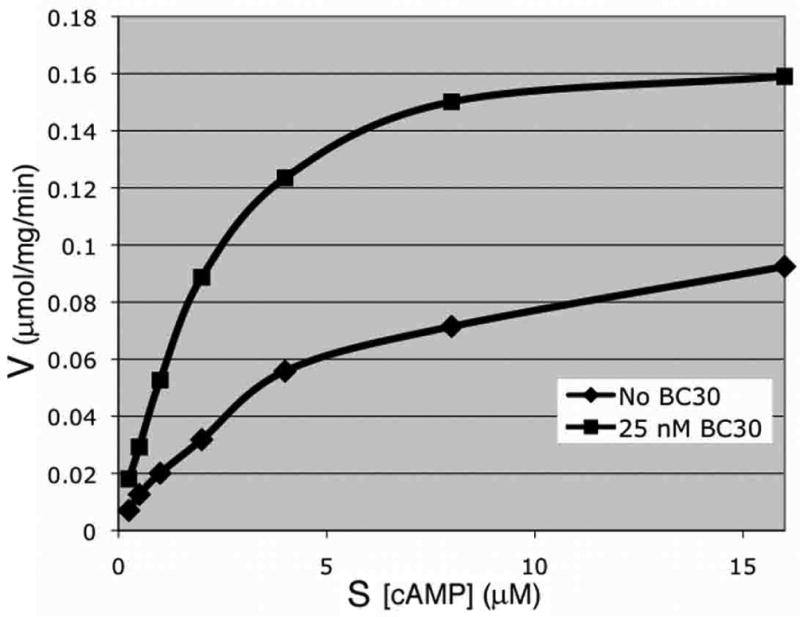
BC30 stimulation of cAMP hydrolysis by the PDE4D catalytic domain. In vitro enzyme assays were carried out using the PDE4D catalytic domain (PDE4D2 residues 86-413) in the absence or presence of 25 nM BC30 at varying concentrations of cAMP.
TNFα release assays assess anti-inflammatory potential of PDE7 inhibitors
BRL50481 was previously shown to enhance the anti-inflammatory effect of PDE4 inhibitors (7), consistent with studies showing that PDE7A and PDE7B are significantly expressed in cells of the immune system (27, 28). We therefore examined the effect of BRL50481, BC11, BC30, and BC39 on TNFα release by LPS-treated U937 cells, alone or in combination with the PDE4 inhibitor rolipram. In vehicle-treated samples, we observe a nearly 50-fold increase in TNFα release as a result of LPS treatment after three hours, and a nearly 100-fold increase after 20 hours (data not shown). Of the five compounds tested, only BC30 at both time points and rolipram at the 20 hour time point conferred a significant reduction in TNFα release (Figure 6A; p < 0.05 as determined by a Student's t Test). When combined with rolipram, 10μM BRL50481 modestly enhances the rolipram-mediated reduction of TNFα release at both time points, although the difference between rolipram-treated cells and rolipram+BRL50481-treated cells is statistically significant for only the 20 hour time point (Figure 6; note that this is a concentration that would only partially inhibit PDE7B based on both the yeast-based and enzyme assays). In contrast, the combination of BC30 and rolipram produces a dramatic reduction of TNFα release at both time points. We observe a similar synergistic effect between BC30 and four of our PDE4-specific inhibitors (to be described elsewhere). Therefore, while we cannot exclude the possibility that partial inhibition of PDE10A by BC30 is responsible for the synergistic effect with PDE4 inhibition, these results could signify that either PDE7B plays a greater role than PDE7A in mediating the TNFα release in response to LPS treatment of activated U937 cells, or that inhibition of both PDE7A and PDE7B is required for significant synergy with PDE4 inhibition.
Figure 6.
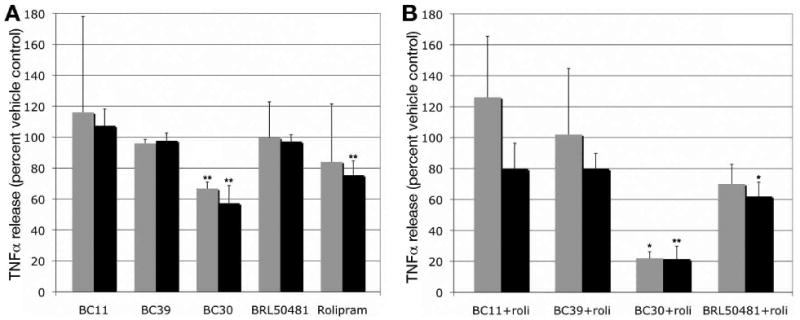
Effect of PDE inhibitors on LPS-stimulated TNFα release in activated U937 cells. U937 cells were activated by PMA and then treated with LPS to stimulate TNFα production and release. The effect of a one hour treatment with individual (panel A) and combinations (panel B) of PDE inhibitors (at 10μM) prior to LPS addition was determined by measuring TNFα in the growth medium at 3 hour (gray bars) and 20 hour (black bars) time points. Data is presented as the percent of the release observed in vehicle-treated cells that were subjected to LPS stimulation, and represent the average and standard deviation from two to four independent experiments for the three hour time point and from three to six experiments for the 20 hour time point. The statistical significance, P ≤ 0.05 (*) or P ≤ 0.01 (**), of the reduction of TNFα release was calculated using a paired one-tailed t-test. Values in panel A were compared to the vehicle-treated control, while the values in panel B were compared to the rolipram-treated values from panel A. The compounds tested do not confer significant cell injury or cytotoxicity as judged by LDH release assays and trypan blue staining (not shown).
Discussion
The work presented here demonstrates that we have developed an effective fission yeast-based HTS for PDE7 inhibitors. Among the most potent inhibitors identified from over 48,000 compounds screened were two steroids and three podocarpanes, which are structurally related to steroids. This is noteworthy as ICOS Corporation previously identified a steroid-like compound, IC242, as a PDE7-specific inhibitor (23); however, as with most compounds under development by pharmaceutical companies, this compound is not readily available nor has its structure been revealed. As BRL50481 is the only commercially-available PDE7 inhibitor, we set out to compare it to some compounds identified in our screen, all of which are commercially-available from either Microsource or Maybridge.
BRL50481 was developed based on its ability to inhibit PDE7A as judged by in vitro enzyme assays, and in both the 5FOA cell growth assay and in vitro enzyme assays of PDE7A inhibition, it is superior to our PDE7 inhibitors. In contrast, BRL50481 is not nearly as effective for inhibition of PDE7B as judged by our cell-based assay and by in vitro enzyme assays (Figure 3, Table 1). While we were initially surprised by this degree of subtype-selectivity, it should be noted that the PDE7A and PDE7B catalytic domains are only 66% identical, compared with PDE4A, PDE4B, PDE4C, and PDE4D, where the catalytic domains are ∼85% identical. We have identified several other PDE7 and PDE4/7 inhibitors that produce a growth response in our PDE7B strain at lower concentrations than required for BRL50481 (Figure 3, data not shown). Further work with BC30 or these other compounds could lead to the development of more potent PDE7 inhibitors, including ones based on a compound in our collection that inhibits PDE7B better than PDE7A (data not shown).
An unusual observation from the in vitro enzyme assays is that several compounds originally identified as inhibitors of PDE7, including BC30, stimulate the activity of the PDE4D catalytic domain (Figure 5). Although we see no evidence that such stimulation occurs in mammalian cells or in our yeast strains that express full-length PDE4D2 or PDE4D3, this finding is quite intriguing. Because BC30 increases the Vmax of the PDE4D catalytic domain, and because it does not interfere with substrate binding, BC30 appears to bind PDE4D somewhere other than at the substrate-binding site. Given that the PDE4D catalytic domain is ∼33% identical and 60% similar to those of the PDE7 enzymes, it is possible that a similar allosteric interaction occurs to inhibit the PDE7 enzymes. BC30 appears to increase the Km of PDE7B (data not shown), however, such a response is not incompatible with allosteric binding. Much remains to be learned about the mechanism of BC30 action, including why the stimulatory effect is not observed with the full-length PDE4D2 enzyme. It may simply be that the N-terminal or C-terminal ends of the protein interfere with either the site or the outcome of binding. We would like to emphasize that it is unlikely that compounds that act through allosteric sites will be found via structure-based drug design approaches that target PDE active sites or through medicinal chemistry programs that are based on previously-identified PDE inhibitors. In contrast, our screening platform identifies PDE inhibitors regardless of the site of action.
Finally, we show that BC30 reduces the inflammatory effect of LPS on activated U937 cells, as judged by TNFα release assays. In particular, we observe a potent synergistic effect upon treatment with BC30 and rolipram (Figure 6B). While we anticipated that combining PDE4 and PDE7 inhibition would produce such synergy, the degree to which TNFα release was inhibited was unexpected, as BRL50481 plus rolipram produces only a modest improvement over rolipram alone. In both our growth assays and in enzyme assays, BRL50481 is superior to BC30 as a PDE7A inhibitor, and is fully effective at 10μM, the concentration used in the TNFα assay. On the other hand, 10μM BRL50481 only partially inhibits PDE7B as judged by either assay, while BC30 is a fully effective PDE7B inhibitor. Therefore, it appears that PDE7B may be more important than PDE7A in this assay, or that dual inhibition of both subtypes is required for the synergistic effect seen with rolipram-mediated inhibition of PDE4. Alternatively, the anti-inflammatory effect of BC30 may be due to a target other than PDE7. 5FOA growth assays using strains expressing members of ten of the eleven mammalian PDE families show BC30 to be a potent PDE7 inhibitor and a weak PDE10A inhibitor. As transcription of PDE10A is induced in rat peritoneal macrophages upon LPS treatment (29), it is possible that PDE10A inhibition could have an effect on TNFα release.
The yeast growth-based screening method used to discover BC30 as a PDE7A/PDE7B inhibitor with potent anti-inflammatory properties requires that the compounds identified have drug-like characteristics in addition to being effective PDE inhibitors. These compounds must show little toxicity to fission yeast, which should exclude compounds that bind a large number of proteins, as such promiscuous binding would likely reduce cell growth. In addition, these compounds must be cell permeable to fission yeast, and remain active for most or all of the 48 hour incubation period of the assay. Therefore, compounds identified by this method may represent superior starting material with regard to drug development relative to compounds identified by in vitro enzyme assays that prioritize candidates solely on the basis of binding affinity. (At the same time, the relative potencies of the compounds discovered by this method are confirmed by in vitro enzyme assays (Figure 3, Table 1).) In addition to BC30, podocarpanes could serve as starting material to develop PDE7 inhibitors that might avoid the side-effects associated with treatment with steroids, which also inhibit PDE7. Thus, this work opens up new avenues for drug development in the PDE7 arena.
Acknowledgments
This work was supported by a grant from Boston College and by NIH grant R21 GM079662 to C.S.H. We thank Joe Beavo and Miles Houslay for clones and for advice and encouragement during the course of this project, Nicola Tolliday, Lynn VerPlank, Jason Burbank, and Stephanie Norton for guidance on high throughput screening, and Chip Stewart for assistance with statistical analyses. The project has been funded in part with Federal funds from the National Cancer Institute's Initiative for Chemical Genetics, National Institutes of Health, under Contract No. N01-CO-12400 and has been performed with the assistance of the Chemical Biology Platform of the Broad Institute of Harvard and MIT.
Abbreviations
- PDE
cyclic nucleotide phosphodiesterase
- cAMP
adenosine 3′,5′-cyclic monophosphate
- PMA
phorbol 12-myristate 13-acetate
- LPS
lipopolysaccharide
- HTS
high throughput screen
Footnotes
The final, definitive version of the article is available at http://online.sagepub.com/.
References
- 1.Bender AT, Beavo JA. Cyclic nucleotide phosphodiesterases: molecular regulation to clinical use. Pharmacol Rev. 2006 Sep;58(3):488–520. doi: 10.1124/pr.58.3.5. [DOI] [PubMed] [Google Scholar]
- 2.Lerner A, Epstein PM. Cyclic nucleotide phosphodiesterases as targets for treatment of haematological malignancies. Biochem J. 2006 Jan 1;393(Pt 1):21–41. doi: 10.1042/BJ20051368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conti M, Beavo J. Biochemistry and physiology of cyclic nucleotide phosphodiesterases: essential components in cyclic nucleotide signaling. Annu Rev Biochem. 2007;76:481–511. doi: 10.1146/annurev.biochem.76.060305.150444. [DOI] [PubMed] [Google Scholar]
- 4.Baillie GS, Scott JD, Houslay MD. Compartmentalisation of phosphodiesterases and protein kinase A: opposites attract. FEBS Lett. 2005 Jun 13;579(15):3264–70. doi: 10.1016/j.febslet.2005.03.089. [DOI] [PubMed] [Google Scholar]
- 5.Angel JB, Saget BM, Walsh SP, Greten TF, Dinarello CA, Skolnik PR, et al. Rolipram, a specific type IV phosphodiesterase inhibitor, is a potent inhibitor of HIV-1 replication. Aids. 1995 Oct;9(10):1137–44. doi: 10.1097/00002030-199510000-00004. [DOI] [PubMed] [Google Scholar]
- 6.Giembycz MA, Smith SJ. Phosphodiesterase 7A: a new therapeutic target for alleviating chronic inflammation? Curr Pharm Des. 2006;12(25):3207–20. doi: 10.2174/138161206778194123. [DOI] [PubMed] [Google Scholar]
- 7.Smith SJ, Cieslinski LB, Newton R, Donnelly LE, Fenwick PS, Nicholson AG, et al. Discovery of BRL 50481 [3-(N,N-dimethylsulfonamido)-4-methyl-nitrobenzene], a selective inhibitor of phosphodiesterase 7: in vitro studies in human monocytes, lung macrophages, and CD8+ T-lymphocytes. Mol Pharmacol. 2004 Dec;66(6):1679–89. doi: 10.1124/mol.104.002246. [DOI] [PubMed] [Google Scholar]
- 8.Card GL, Blasdel L, England BP, Zhang C, Suzuki Y, Gillette S, et al. A family of phosphodiesterase inhibitors discovered by cocrystallography and scaffold-based drug design. Nat Biotechnol. 2005 Feb;23(2):201–7. doi: 10.1038/nbt1059. [DOI] [PubMed] [Google Scholar]
- 9.Huai Q, Wang H, Sun Y, Kim HY, Liu Y, Ke H. Three-dimensional structures of PDE4D in complex with roliprams and implication on inhibitor selectivity. Structure. 2003 Jul;11(7):865–73. doi: 10.1016/s0969-2126(03)00123-0. [DOI] [PubMed] [Google Scholar]
- 10.Hoffman CS. Glucose sensing via the protein kinase A pathway in Schizosaccharomyces pombe. Biochem Soc Trans. 2005 Feb;33(Pt 1):257–60. doi: 10.1042/BST0330257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoffman CS, Winston F. Isolation and characterization of mutants constitutive for expression of the fbp1 gene of Schizosaccharomyces pombe. Genetics. 1990;124(4):807–16. doi: 10.1093/genetics/124.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stiefel J, Wang L, Kelly DA, Janoo RTK, Seitz J, Whitehall SK, et al. Suppressors of an adenylate cyclase deletion in the fission yeast Schizosaccharomyces pombe. Eukaryotic Cell. 2004;3(3):610–9. doi: 10.1128/EC.3.3.610-619.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang L, Griffiths K, Zhang YH, Ivey FD, Hoffman CS. Schizosaccharomyces pombe adenylate cyclase suppressor mutations suggest a role for cAMP phosphodiesterase regulation in feedback control of glucose/cAMP signaling. Genetics. 2005;171:1523–33. doi: 10.1534/genetics.105.047233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ivey FD, Wang L, Demirbas D, Allain C, Hoffman CS. Development of a fission yeast-based high-throughput screen to identify chemical regulators of cAMP phosphodiesterases. J Biomol Screen. 2008 Jan;13(1):62–71. doi: 10.1177/1087057107312127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sancho R, Marquez N, Gomez-Gonzalo M, Calzado MA, Bettoni G, Coiras MT, et al. Imperatorin inhibits HIV-1 replication through an Sp1-dependent pathway. J Biol Chem. 2004 Sep 3;279(36):37349–59. doi: 10.1074/jbc.M401993200. [DOI] [PubMed] [Google Scholar]
- 16.Mochizuki N, Yamamoto M. Reduction in the intracellular cAMP level triggers initiation of sexual development in fission yeast. Mol Gen Genet. 1992;233(1-2):17–24. doi: 10.1007/BF00587556. [DOI] [PubMed] [Google Scholar]
- 17.Toda T, Shimanuki M, Yanagida M. Fission yeast genes that confer resistance to staurosporine encode an AP-1-like transcription factor and a protein kinase related to the mammalian ERK1/MAP2 and budding yeast FUS3 and KSS1 kinases. Genes Dev. 1991 Jan;5(1):60–73. doi: 10.1101/gad.5.1.60. [DOI] [PubMed] [Google Scholar]
- 18.Franz AK, Dreyfuss PD, Schreiber SL. Synthesis and cellular profiling of diverse organosilicon small molecules. J Am Chem Soc. 2007 Feb 7;129(5):1020–1. doi: 10.1021/ja067552n. [DOI] [PubMed] [Google Scholar]
- 19.Wang H, Liu Y, Chen Y, Robinson H, Ke H. Multiple elements jointly determine inhibitor selectivity of cyclic nucleotide phosphodiesterases 4 and 7. J Biol Chem. 2005 Sep 2;280(35):30949–55. doi: 10.1074/jbc.M504398200. [DOI] [PubMed] [Google Scholar]
- 20.Mander T, Hill S, Hughes A, Rawlins P, Clark C, Gammon G, et al. Differential effects on TNF alpha production by pharmacological agents with varying molecular sites of action. Int J Immunopharmacol. 1997 Aug;19(8):451–62. doi: 10.1016/s0192-0561(97)00088-x. [DOI] [PubMed] [Google Scholar]
- 21.Toone WM, Kuge S, Samuels M, Morgan BA, Toda T, Jones N. Regulation of the fission yeast transcription factor Pap1 by oxidative stress: requirement for the nuclear export factor Crm1 (Exportin) and the stress-activated MAP kinase Sty1/Spc1. Genes Dev. 1998 May 15;12(10):1453–63. doi: 10.1101/gad.12.10.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nocero M, Isshiki T, Yamamoto M, Hoffman CS. Glucose repression of fbp1 transcription of Schizosaccharomyces pombe is partially regulated by adenylate cyclase activation by a G protein α subunit encoded by gpa2 (git8) Genetics. 1994;138(1):39–45. doi: 10.1093/genetics/138.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee R, Wolda S, Moon E, Esselstyn J, Hertel C, Lerner A. PDE7A is expressed in human B-lymphocytes and is up-regulated by elevation of intracellular cAMP. Cell Signal. 2002 Mar;14(3):277–84. doi: 10.1016/s0898-6568(01)00250-9. [DOI] [PubMed] [Google Scholar]
- 24.Pitts WJ, Vaccaro W, Huynh T, Leftheris K, Roberge JY, Barbosa J, et al. Identification of purine inhibitors of phosphodiesterase 7 (PDE7) Bioorg Med Chem Lett. 2004 Jun 7;14(11):2955–8. doi: 10.1016/j.bmcl.2004.03.021. [DOI] [PubMed] [Google Scholar]
- 25.Kempson J, Pitts WJ, Barbosa J, Guo J, Omotoso O, Watson A, et al. Fused pyrimidine based inhibitors of phosphodiesterase 7 (PDE7): synthesis and initial structure-activity relationships. Bioorg Med Chem Lett. 2005 Apr 1;15(7):1829–33. doi: 10.1016/j.bmcl.2005.02.025. [DOI] [PubMed] [Google Scholar]
- 26.Castano T, Wang H, Campillo NE, Ballester S, Gonzalez-Garcia C, Hernandez J, et al. Synthesis, structural analysis, and biological evaluation of thioxoquinazoline derivatives as phosphodiesterase 7 inhibitors. ChemMedChem. 2009 May;4(5):866–76. doi: 10.1002/cmdc.200900043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith SJ, Brookes-Fazakerley S, Donnelly LE, Barnes PJ, Barnette MS, Giembycz MA. Ubiquitous expression of phosphodiesterase 7A in human proinflammatory and immune cells. Am J Physiol Lung Cell Mol Physiol. 2003 Feb;284(2):L279–89. doi: 10.1152/ajplung.00170.2002. [DOI] [PubMed] [Google Scholar]
- 28.Zhang L, Murray F, Zahno A, Kanter JR, Chou D, Suda R, et al. Cyclic nucleotide phosphodiesterase profiling reveals increased expression of phosphodiesterase 7B in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2008 Dec 9;105(49):19532–7. doi: 10.1073/pnas.0806152105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Witwicka H, Kobialka M, Siednienko J, Mitkiewicz M, Gorczyca WA. Expression and activity of cGMP-dependent phosphodiesterases is up-regulated by lipopolysaccharide (LPS) in rat peritoneal macrophages. Biochim Biophys Acta. 2007 Feb;1773(2):209–18. doi: 10.1016/j.bbamcr.2006.10.008. [DOI] [PubMed] [Google Scholar]


