Abstract
Purpose
The three main treatment options for primary prostate cancer are surgery, radiation, and active surveillance. Surgical and radiation intervention for prostate cancer can be associated with significant morbidity. Therefore, accurate stratification predictive of outcome for prostate cancer patients is essential for appropriate treatment decisions. Nomograms that use clinical and pathologic variables are often used for risk prediction. Favorable outcomes exist even among men classified by nomograms as being at high risk of recurrence.
Experimental Design
Previously, we identified a set of DNA-based biomarkers termed Genomic Evaluators of Metastatic Prostate Cancer (GEMCaP) and have shown that they can predict risk of recurrence with 80% accuracy. Here, we examined the risk prediction ability of GEMCaP in a high-risk cohort and compared it to a Kattan nomogram.
Results
We determined that the GEMCaP genotype alone is comparable with the nomogram, and that for a subset of cases with negative lymph nodes improves upon it.
Conclusion
Thus, GEMCaP shows promise for predicting unfavorable outcomes for negative lymph node high-risk cases, where the nomogram falls short, and suggests that addition of GEMCaP to nomograms may be warranted.
Prostate-specific antigen (PSA) remains the only well-validated biomarker for stratification by risk of recurrence in routine clinical use for prostate cancer. The absence of additional biomarkers predicting recurrence has prompted researchers to develop predictive tools based on statistical models using disease features. Among the strategies for risk stratification is the use of nomograms. Nomograms are models that predict outcomes using specific clinical, pathologic, and patient information for each individual patient (2). Our working hypothesis is that genome copy number profiles can define genotypes that predict a patient's risk of postoperative disease recurrence and metastasis and that these genotypes can be incorporated into nomograms thus increasing their accuracy.
Using BAC-based array comparative genomic hybridization (aCGH), we discovered a suite of DNA-based biomarkers that seem to predict prostate cancer recurrence and metastasis (1). These map to 39 loci termed GEMCaP for Genomic Evaluators of Metastatic Prostate Cancer. The GEMCaP loci were identified through an application of evolutionary theory and computational analysis comparing the frequency of copy number changes in primary tumors from patients who did not recur following radical prostatectomy (RP; median follow-up, 11 y; 8 years minimum) to two independent cohorts with bone metastasis recurrence or organ metastases (1). We then tested whether the GEMCaP genotypes could predict recurrence in an independent cohort of primary prostate tumors from 27 patients for which clinical and pathologic parameters were known. The risk of postoperative recurrence, defined as two consecutive PSA measurements of ≥0.2 ng/mL and/or local or distant disease, was assessed using both GEMCaP and the Kattan nomogram. The overall accuracy of the Kattan postoperative nomogram was 75%. Analysis of copy number changes at the GEMCaP loci accurately classified recurrence for 78% of the patients (3). The Kattan nomogram predicts outcome for higher risk patients better than other existing nomograms (4). Therefore, in the current study, we aimed to assess GEMCaP in a larger cohort of high-risk tumors and to then compare the GEMCaP biomarkers to a Kattan nomogram in predicting outcome.
Materials and Methods
Patient selection
This is a retrospective case-control study of high-risk patients whose primary initial treatment for localized prostate cancer at the University of California at San Francisco (between 1989 and 2004) was restricted to RP. All study patients had pT2C or pT3 stage disease. All available high-risk cases were identified from our urological database and included patients who experienced biochemical failure (two consecutive PSA measurements of >0.2 ng/mL) within 1 y of RP and/or had positive lymph nodes identified at the time of surgery. Controls were randomly selected from all patients with similar high-risk disease features who had a minimum disease-free follow-up of 24 mo. None of the controls received any other treatment for their prostate cancer and none had recurrent disease at last follow-up, with a median follow-up of 64 mo. Other disease features considered when identifying appropriate controls to reduce the possibility of confounding are listed in Table 1 (please see Supplementary Table S1 for detailed clinical information). By design, this resulted in a fairly uniform patient sample, with the number evaluated not based on a test hypothesis.
Table 1.
Distribution of patient features
| Cases (n = 30) | Controls (n = 24) | |
|---|---|---|
| Preoperative PSA | ||
| Median | 10.9 ng/mL | 6.6 ng/mL |
| Range | 4.7-66.7 | 3.2-24.1 |
| % >15 | 30% | 21% |
| Pathologic Gleason sum | ||
| 5-6 | 7% | 17% |
| 7 | 70% | 54% |
| 3+4 | 43% | 42% |
| 4+3 | 27% | 12% |
| 8-9 | 23% | 29% |
| Percent positive margins | 47% | 25% |
| Percent seminal vesicle involvement | 38% | X |
| Percent extracapsular extension | 70% | 46% |
| Percent positive lymph nodes | 50% | X |
| Year of RP | ||
| Range | 1989-2004 | 1994-2002 |
| <2000 | 40% | 58% |
| ≥2000 | 60% | 42% |
| Median follow-up | X | 63.5 mo |
| Range | X | 27-148 |
| % >5 y | X | 63% |
NOTE: X, determined by study design.
Translational Relevance.
We describe the use of 39 BAC-based markers of metastasis to assess recurrence risk in silico for high-risk radical prostatectomy cases. This set of biomarkers, the Genomic Evaluators of Metastatic Prostate Cancer (GEMCaP), were previously identified through array comparative genomic hybridization–based experiments of both primary and metastatic prostate tumors (1). Herein, we determined that the GEMCaP genotype alone is comparable with a Kattan nomogram, the risk assessment tool commonly used by urologists. Moreover, for a subset of cases with negative lymph nodes, GEMCaP improves upon the Kattan nomogram. If our findings are replicated, then it will be possible to identify patients who are good candidates for postoperative surveillance and immediate adjuvant therapy.
All investigators involved with the sample processing or genotype analysis for this study were blinded about the patient clinical information and treatment outcome.
Archived tissue processing for aCGH
Fifteen 15-μm slices were cut for each patient from formalin fixed, paraffin-embedded RP prostate tissue blocks. H&E stains were performed on 5-μm slices representative of the beginning and the end of the cut section. A single pathologist outlined areas of >80% tumor for macrodissection with a scalpel. DNA was extracted using the Puregene DNA isolation kit (Gentra Systems) as per the manufacturer's instructions. Phenol/chloroform extraction was done after the Gentra kit's final elution step. This kit has yielded good quality DNA from formalin fixed, paraffin-embedded material for aCGH in our laboratory (1, 5).
Array comparative genomic hybridization
The human BAC arrays were purchased from the University of California at San Francisco Array Core. Each array consists of 2,464 BAC clones spotted in triplicate on chromium slides (6). The resolution is ∼1.4 Mb. The imaging set up and custom software are described elsewhere (6). We followed our published hybridization protocol (5), but with a 72-h hybridization. Imaging processing was done with the University of California at San Francisco SPOT version 2.1 and SPROC version 2.0 software packages (7).
aCGH analysis
The tumor/reference fluorescence intensity ratios were converted to the log2 domain and the replicate spots were averaged. The observed log2 ratios were not included if there were fewer than two replicate spots (out of 3) or if the SD of the replicates was >0.2. Each array was normalized to have a median log2 ratio of 0 and denoised using in-house software. To identify copy number changes in individual samples, we explored three thresholding approaches, which are termed floating, fixed, and integrated.
Floating threshold approach
aCGH data were analyzed using circular binary segmentation (8) with default parameters, as implemented in the DNA copy package in R/Bioconductor (9), to translate intensity measurements into regions of equal copy number. Missing values were imputed using the maximum value of two flanking segments, producing smoothed values. The Merge Level procedure (10) was applied to the smoothed values to further merge the segments. For each sample, gain/loss status for each probe was assigned by considering the merged values closest to zero as the level of no change, whereas those above or below it as having a gain or loss. Experimental variation for each sample was estimated by calculating the median absolute deviation of the difference between the observed and smoothed values.
Fixed threshold approach
A fixed threshold of 2.5 times the sample median absolute deviation, as defined above, was applied to the log2 ratio values to determine gain/loss status of each probe (11).
Integrated threshold approach
Compared with known clinical status, only a decreased sensitivity with increased specificity was achieved when the fixed threshold was applied, and this mainly reflected copy number losses. The opposite was observed with the floating thresholding so that individually neither was found to be informative to characterize the entire patient sample. Therefore, the two thresholding methods were combined to use the strengths of each. Our integrated approach involved using a fixed threshold for calling copy number losses (subset of 23 loci) and the floating threshold method for the copy number gains (subset of 16 loci).
The overall GEMCaP score is the proportion of aberrant (gain or loss) loci, calculated from the aCGH data and determined by the threshold technique, among the total number of evaluable loci (maximum of 39). As determined in our prior studies, if the total GEMCaP score was ≥20%, the patient was considered to be at high risk of recurrence, and if the score was <20%, he was a low-risk patient (1, 3). Although statistically different, the GEMCaP distributions for the cases and controls using the integrated threshold overlap so that the difference between subsets is not as clear. Therefore, we retained the cutoff used in our prior studies to determine whether our initial results with this cut-point were generalizable (1, 3). The distribution of GEMCaP scores was compared across thresholding approaches.
Nomograms to predict progression-free survival
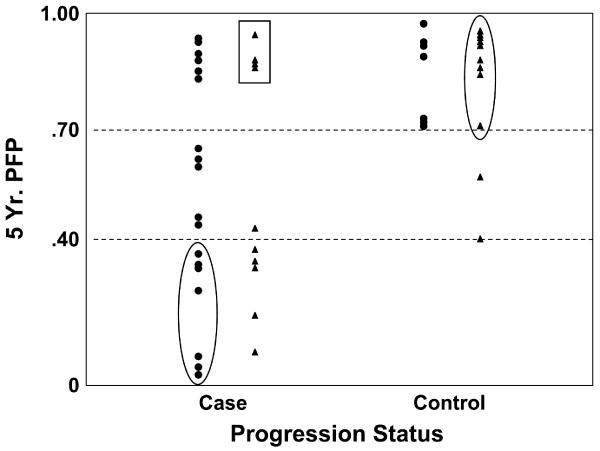
Kattan's postoperative nomogram4 was used to obtain 5-y estimates of progression-free probability (PFP; ref. 12). This nomogram was selected because the tumor genotype assessed by GEMCaP is determined using the surgical specimen. The PFP estimate is a function of pathologic Gleason score, surgical margin status, seminal vesicle or lymph node involvement, extracapsular extension, and preoperative PSA. As shown in Fig. 1, the minimum predicted 5-y PFP among controls was 40%, suggesting a cut-point between the two patient subsets to be used in these analyses.
Fig. 1.
Agreement with GEMCaP and nomogram predictions. Horizontal dotted lines, 40% and 70% nomogram cut-points. ▲, favorable GEMCaP predictions; ●, unfavorable GEMCaP predictions. Data points within the ovals are where the nomogram and GEMCaP classification agree. Cases misclassified by both approaches are within the rectangle. Patients with intermediate nomogram scores (i.e., between the 40% and 70% cut-points) are those that might benefit the most from genomic analysis.
Statistical methods
Pearson's correlation coefficient was calculated to evaluate the relationship between the genomic and nomogram scores. To compare scores between subsets of patients (e.g., cases and controls), either the t statistic or the Mann-Whitney statistic was used. The three genomic and nomogram distributions were each dichotomized with a GEMCaP score of ≥20% and a nomogram probability estimate of <40%, indicating an increased risk of recurrence. Using these binary random variables, agreement in risk classification (favorable or unfavorable) was analyzed using McNemar's test. Agreement between the known recurrence status and each of the alternative classifications was summarized by the point estimates of sensitivity, specificity, and accuracy. Although this is a case-control study, positive and negative predictive values were included for reference for future studies.
A logistic regression model was used to explore how each of the three thresholding GEMCaP models and the 5-y nomogram PFP could predict known recurrence status. The GEMCaP scores were considered as continuous and binary variables (using the 20% cut-point) individually and in combination with the nomogram probability. Statistical significance defined as a probability of <0.05 was determined using the likelihood ratio test. For each logistic model, the receiver operating characteristic curve was calculated and the area under the curve (AUC) estimated the fit. Analyses were done using Statistica software (StatSoft, Inc. version 6.0).
Results
GEMCaP and Kattan scores
To evaluate the role of GEMCaP in predicting clinical status, confounding by known disease factors were avoided by selecting cases and controls with comparable baseline features (Supplementary Table S1; Table 1). The risk classifications according to the fixed, floating, and integrated GEMCaP scores are shown in Supplementary Table S1 and include the 5-year postoperative PFP using the Kattan historical nomogram (13). All aCGH log2 ratios, along with probe information, are provided in Supplementary Table S2.
Summary of predictive model scores
The summary features for each of the four prediction models are displayed in Table 2A. As would be expected, the three GEMCaP scores are highly correlated (P < 0.001 for each pairwise comparison), but none were correlated with the 5-year nomogram prediction of PFP (P > 0.35 for each comparison). A significant difference between cases and controls was observed in the nomogram distributions (P = 0.0001), and a borderline difference between clinical subsets was observed using the floating and the integrated GEMCaP scores (P = 0.08 and 0.09, respectively).
Table 2.
| A. Summary of four prediction models (n = 54) | ||||
|---|---|---|---|---|
| Median (range) | Kattan nomogram | |||
| GEMCaP score | ||||
| Fixed | Floating | Integrated | ||
| Controls | 14% (0%-50%) | 21% (0%-74%) | 16% (3%-66%) | 93% (40%-99%) |
| Cases | 18% (3%-46%) | 29% (0%-77%) | 23% (3%-64%) | 53% (3%-96%) |
| B. Agreement in classification between GEMCaP and the nomogram | ||||
| Agreement in classification* | ||||
| %Favorable | % Unfavorable | % Total | ||
| Nomogram and: | ||||
| Fixed threshold | 52% | 9% | 61% | |
| Floating threshold | 28% | 17% | 45% | |
| Integrated threshold | 37% | 13% | 50% | |
| All three GEMCaP models | 26% | 9% | 35% | |
Favorable is defined as a GEMCaP score of <20% and as a nomogram probability of ≥40%.
Agreement in risk classification among the models
The overall agreement between each of the GEMCaP models and the nomogram score was investigated. Note that this is not an agreement with known disease recurrence status, but a summary of concurrence among the four methods. All three GEMCaP methods classified 31% of the patients as having a favorable risk and 31% as having an unfavorable risk of recurrence. Differences in classification occurred among the remaining third of the study sample. The classification of patients significantly differed between the integrated threshold method compared with both the fixed and floating approaches (McNemar's test: P = 0.02 for each comparison).
Overall, the Kattan nomogram classified 35% of the patients identically as all three GEMCaP methods, 26% favorable and 9% unfavorable (Table 2B). Using the GEMCaP fixed method, agreement with the nomogram occurred for 61% of the patients, but both groupings only identified 9% of the entire sample as being at increased risk of progression. A difference in classification between the nomogram and both the floating and integrated methods was observed (McNemar's test: P < 0.0001 and 0.002, respectively). Because of the differing classification, we investigated the agreement between individual models and the combination of GEMCaP scores with the Kattan nomogram.
Agreement between predicted and known recurrence status
The known postoperative recurrence status was used as the reference to evaluate the ability of the four proposed methods to predict outcome. The floating method had the highest sensitivity (80%), whereas the fixed method had the highest specificity (75%; Table 3A). The fixed threshold approach did not sufficiently identify cases displaying a sensitivity of 43%, and the floating threshold method resulted in a specificity of 50% for identifying controls. Integration of the floating and fixed GEMCaP models achieved a sensitivity, specificity, and accuracy of ∼65%. Changing the GEMCaP cut-point did not improve the accuracy of any of the three GEMCaP models. Due to the selection of the classification cut-point for the predicted 5-year PFP from the nomogram for this analysis, all control patients were correctly identified. With this cut-point, the sensitivity of the nomogram was only 40%, which is similar to the fixed thresholding results.
Table 3.
| A. Predictability of GEMCaP score with clinical outcome (n = 54) | |||
|---|---|---|---|
| Threshold method | |||
| Fixed | Floating | Integrated | |
| Sensitivity | 43% | 80% | 67% |
| Specificity | 75% | 50% | 63% |
| Positive predictive value | 68% | 67% | 60% |
| Negative predictive value | 51% | 67% | 69% |
| Accuracy | 57% | 67% | 65% |
| B. Frequency of identifying cases based on lymph node status | |||
| No. of unfavorable cases identified | |||
| Lymph node negative (n = 15) | Lymph node positive (n = 15) | ||
| Nomogram | 2 | 10 | |
| GEMCaP: | |||
| Fixed | 6 | 7 | |
| Floating | 12 | 12 | |
| Integrated | 10 | 10 | |
NOTE: Sensitivity is the proportion of cases who are correctly classified based on the GEMCaP score (≥20%) as being at high risk of recurrence; specificity is the proportion of controls who are correctly classified as being at low risk of recurrence. The positive predictive value is the proportion of patients classified by the GEMCaP score to be at high risk of recurrence who are cases, and the negative predictive value is the proportion of patients classified to be at low risk of recurrence who are controls. The limitations of these last two estimates for a case-control study are known, but they are included only as a reference for future studies. Accuracy is the overall proportion of correctly classified cases and controls. Cases were identified by values >20% for GEMCaP and <40% for the nomogram.
Among the 17 patients classified as favorable by all three GEMCaP models, there were five mismatches with the clinical status. The nomogram also misclassified two of these five patients. Similarly, among the 17 classified as unfavorable with all three GEMCaP thresholding approaches, 5 were mismatches with known status. The nomogram prediction also misclassified these five genomic mismatches, but incorrectly classified seven others in this unfavorable subset.
The nomogram score is a continuous variable with no accepted standard cut-points to indicate increased risk of recurrence. Because it is a validated and well-used method by clinicians to estimate outcome, we defined the nomogram cut-point of above 40% to identify all control patients based on this study sample as displayed in Fig. 1. Data points within ovals are where the nomogram and GEMCaP classification agree above 70% and below 40%. Both scoring systems misclassified cases (see data points within rectangle) and a similar number of cases and controls were misclassified by each approach (circles with values, >70%). All but one of those patients with intermediate nomogram scores (i.e., between 40% and 70%) had accurate GEMCaP classifications.
Detailed evaluation of agreement for cases
The difference between the nomogram and integrated classifications in identifying cases was explored further. For this study, patients were selected to be a case if they had positive lymph nodes determined at the time of RP or recurred within 1 year of surgery. The postoperative nomogram PFP score decreases when a patient has positive lymph nodes, whereas those cases who recurred within 1 year with negative lymph nodes would have a similar PFP estimate to the high-risk controls. Therefore, the nomogram had a low sensitivity when detecting true cases.
For all three GEMCaP methods, the distribution of the GEMCaP signature was consistent for all cases. In contrast with this, a significant difference was observed in the nomogram distributions between lymph node–positive cases and cases who recurred within 1 year of surgery (P = 0.0006). Even if the cut-point for the nomogram was increased, this difference would still be observed. There were 15 lymph node–negative cases in this study. GEMCaP identified 10 such cases, whereas the nomogram identified only 2 (1 sample overlapped). Descriptive data are shown in Table 3B.
Summary predictive model
To combine these observations, a multivariate analysis was done, assuming a logistic regression model to predict the observed disease recurrence status. Individually, only the nomogram was predictive of disease recurrence, which is consistent with the previous results indicating a difference in distributions of the PFP between cases and controls (Table 4A). The three GEMCaP approaches using the actual scores all resulted in AUCs for the receiver operating characteristic curves in the range of 0.60 to 0.64 whereas the AUC for the nomogram was 0.81. When the GEMCaP scores were dichotomized, the binary outcomes using the integrated and floating threshold classifications were each significant predictors of disease status using the logistic model, but an increase in the AUC was not achieved. These significant results reflect the ∼65% accuracy with either of these two approaches for a binary GEMCaP score (Table 3A).
Table 4.
| A. Prediction of disease recurrence status assuming a logistic regression model | |||
|---|---|---|---|
| Continuous/binary coding | |||
| Probability for model: likelihood ratio test | AUC | ||
| Nomogram | <0.0001/− | 0.81/− | |
| GEMCaP: | |||
| Fixed | 0.34/0.16 | 0.60/0.59 | |
| Floating | 0.18/0.02 | 0.64/0.65 | |
| Integrated | 0.19/0.03 | 0.63/0.65 | |
| Probability of GEMCaP as an independent predictor of status in addition to the nomogram: | |||
| GEMCaP (Binary): | |||
| Fixed | Not significant | 0.80 | |
| Floating | P = 0.02 | 0.85 | |
| Integrated | P = 0.055 | 0.84 | |
| B. Prediction of the added benefit of GEMCaP | |||
| Model | |||
| Nomogram score | Nomogram + floating | Nomogram + integrated | |
| Sensitivity | 60% | 77% | 77% |
| Specificity | 71% | 79% | 75% |
| Positive predictive value | 72% | 82% | 79% |
| Negative predictive value | 59% | 73% | 72% |
| Accuracy | 65% | 78% | 76% |
NOTE: Due to the selection of the cut-point for the nomogram, a binary classification to predict disease status is not possible.
Importantly, the integrated and floating GEMCaP signatures were able to detect the cases with negative lymph nodes who recurred within 1 year of surgery more often than the nomogram (Table 3B). Thus, the addition of a binary GEMCaP classification to the nomogram probability in predicting the known disease status was tested. For both the integrated and floating methods, in addition to the nomogram PFP, the GEMCaP classification was a significant, independent predictor of recurrence status (likelihood ratio tests: nomogram P = 0.0001: plus integrated P = 0.055; plus floating P = 0.02). This resulted in a simultaneous increase in sensitivity, specificity, and accuracy compared with the nomogram prediction alone as well as an increase in the AUC for the receiver operating characteristic curve to 0.84 and 0.85, respectively (Table 4B). Thus, this indicates the additional benefit of the GEMCaP signature in predicting disease progression.
Discussion
Men diagnosed with clinically similar prostate cancer often exhibit widely varying outcomes following local therapy, even for those classified at high risk of recurrence using nomograms. Because surgical and/or radiation intervention can be associated with morbidity that impacts quality of life, methods for stratifying patients into risk groups independent of, or in combination with, existing tools are needed for improved patient management. Previously, we identified a group of 39 DNA-based biomarkers termed GEMCaP (1) and showed that they can predict risk of recurrence with 80% accuracy (3). Moreover, it was hypothesized that a group of widely distributed genome biomarkers might be better suited for analyzing tumors that are inherently heterogeneous. Here, we examined the risk prediction ability of GEMCaP in a high-risk cohort.
A GEMCaP score is based on aCGH copy number measurements at each GEMCaP locus and a calculation of the percent that are aberrant. There is debate in the field as to how to best “threshold” for aCGH copy number. In addition, typical prostate tumor genomes have relatively low-level copy number changes, possibly due to heterogeneity, and this complicates thresholding. We chose to explore multiple methods (fixed, floating, and integrated) because each method may behave differently for copy number gains versus deletions and for aggressive versus indolent tumors. When considered by known disease status, 50% of the control patients had a low GEMCaP score (<20%) and 80% of the cases had a high score when using the floating threshold approach. Similarly, 67% of the cases were classified to be at high risk and 63% of the controls were classified to be at low risk with the integrated approach. The results are comparable, but the integrated approach is also presented here because the increased specificity among controls would aid in identifying those patients able to avoid more aggressive therapy. When the GEMCaP score is categorized as a binary random variable (<20% versus ≥20%), it is a significant predictor of clinical status using the floating and integrated threshold methods. Performing this type of study in a high-risk cohort is complicated by the fact that all the tumors come from patients who are by definition at high risk of recurrence. In an effort to insure the accuracy of recurrence status, multiple clinical updates were done on this study sample. Nonetheless, it is probable that some of the controls will recur, affecting statistical comparisons between the clinical outcome with the GEMCaP classification and the Kattan nomogram probability.
There were cases where the Kattan nomogram predicted recurrence risk better than GEMCaP. This especially applies to those patients with PFP estimates of <40%. This cut-point might not be appropriate for all patient sets. One explanation for this result is that the Kattan postoperative nomogram was developed using cases representing all recurrence risk levels, whereas the GEMCaP algorithm was determined using intermediate to high-risk patients. An alternate explanation is that a subset of tumors has genotypes dominated by either copy number gains or losses that may confound the GEMCaP algorithm. Manual inspection of cases (i.e., patients who recurred after RP in <1 year) where GEMCaP failed to predict outcome did reveal that a subset of these cases was dominated by either GEMCaP gains or losses, resulting in a low (<20%; i.e., favorable) overall GEMCaP score. This asymmetry is observed in ∼10% of our cases to date5 and may represent subtypes of prostate cancer.
Importantly, there were patients where GEMCaP and the nomogram differed in their risk predictions. Therefore, GEMCaP has the potential of adding information to the nomogram and improving risk prediction. As shown in Fig. 1, the GEMCaP classification can identify patients with an unfavorable outcome despite a high predicted 5-year progression-free nomogram estimate (see circles for cases above 70% cut-point). The benefit of GEMCaP in predicted recurrence risk was also observed for patients with nomogram probabilities in the mid-range (40-70%) for cases and controls. Significantly, this reflects the ability of GEMCaP to uniformly identify high-risk cases especially including those cases with negative lymph nodes. This is consistent with the concept that GEMCaP is composed of metastatic genotypes (1). The ability to identify aggressive cancer despite negative lymph nodes could be very important in the clinical setting. Together, these observations support our hypothesis that pathologic features alone can be misleading and that the underlying tumor genotype can complement these for identification of aggressive tumors. Thus, it may be possible to use GEMCaP to help identify patients at high-risk of recurrence who may benefit from adjuvant treatment. These encouraging results are similar to the efforts by Kattan et al. (14) to add biomarkers, specifically interleukin-6 soluble receptor (IL6SR) and transforming growth factor β1, to a nomogram's standard clinical predictors. Including the biology of the tumor in the form of interleukin-6 soluble receptor and transforming growth factor β1, plasma levels were found to improve the ability of a nomogram to predict biochemical progression after RP. In this study, we have evaluated GEMCaP in predicting risk of recurrence in a high-risk cohort. The results suggest that incorporation of GEMCaP into standard clinical tools such as the Kattan nomogram may improve predictive accuracy.
Supplementary Material
Acknowledgments
Grant Support: This work was supported by an RO1 (CA115484) and a UCSF Prostate Cancer SPORE (P50CA89520) from the National Institutes of Health.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
P.L. Paris, unpublished data.
Disclosure of Potential Conflicts of Interest: C. Collins: scientific advisory board, Combimatrix Molecular Diagnostics. P.L. Paris and C. Collins are inventors on a patent issued to UCSF. The other authors disclosed no potential conflicts of interest.
References
- 1.Paris PL, Andaya A, Fridlyand J, et al. Whole genome scanning identifies genotypes associated with recurrence and metastasis in prostate tumors. Hum Mol Genet. 2004;13:1303–13. doi: 10.1093/hmg/ddh155. [DOI] [PubMed] [Google Scholar]
- 2.Ross PL, Scardino PT, Kattan MW. A catalog of prostate cancer nomograms. J Urol. 2001;165:1562–8. [PubMed] [Google Scholar]
- 3.Paris PL, Weinberg V, Simko J, et al. Preliminary evaluation of prostate cancer metastatic risk biomarkers. Int J Biol Markers. 2005;20:141–5. doi: 10.1177/172460080502000301. [DOI] [PubMed] [Google Scholar]
- 4.Kattan MW, Eastham JA, Stapleton AM, Wheeler TM, Scardino PT. A preoperative nomogram for disease recurrence following radical prostatectomy for prostate cancer. J Natl Cancer Inst. 1998;90:766–71. doi: 10.1093/jnci/90.10.766. [DOI] [PubMed] [Google Scholar]
- 5.Paris PL, Albertson DG, Alers JC, et al. High-resolution analysis of paraffin-embedded and formalin-fixed prostate tumors using comparative genomic hybridization to genomic microarrays. Am J Pathol. 2003;162:763–70. doi: 10.1016/S0002-9440(10)63873-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pinkel D, Segraves R, Sudar D, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998;20:207–11. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- 7.Jain AN, Tokuyasu TA, Snijders AM, Segraves R, Albertson DG, Pinkel D. Fully automatic quantification of microarray image data. Genome Res. 2002;12:325–32. doi: 10.1101/gr.210902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–72. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 9.Team RDC. R: A language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing; 2005. [Google Scholar]
- 10.Willenbrock H, Fridlyand J. A comparison study: applying segmentation to array CGH data for downstream analyses. Bioinformatics. 2005;21:4084–91. doi: 10.1093/bioinformatics/bti677. [DOI] [PubMed] [Google Scholar]
- 11.Fridlyand J, Snijders AM, Pinkel D, Albertson DG, Jain A. Application of Hidden Markov Models to the analysis of the array CGH data. Journal of Multivariate Analysis (Special Genomic Issue) 2004;90:132–53. [Google Scholar]
- 12.Kattan MW, Wheeler TM, Scardino PT. Postoperative nomogram for disease recurrence after radical prostatectomy for prostate cancer. J Clin Oncol. 1999;17:1499–507. doi: 10.1200/JCO.1999.17.5.1499. [DOI] [PubMed] [Google Scholar]
- 13.Stephenson AJ, Scardino PT, Eastham JA, et al. Postoperative nomogram predicting the 10-year probability of prostate cancer recurrence after radical prostatectomy. J Clin Oncol. 2005;23:7005–12. doi: 10.1200/JCO.2005.01.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kattan MW, Shariat SF, Andrews B, et al. The addition of interleukin-6 soluble receptor and transforming growth factor β1 improves a preoperative nomogram for predicting biochemical progression in patients with clinically localized prostate cancer. J Clin Oncol. 2003;21:3573–9. doi: 10.1200/JCO.2003.12.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.