Abstract
PBPA from Mycobacterium tuberculosis (Mtb) is a class B-like penicillin-binding protein (PBP) that is not essential for cell growth in Mtb, but is important for proper cell division in M. smegmatis. We have determined the crystal structure of PBPA at 2.05 Å resolution, the first published structure of a PBP from this important pathogen. Compared to other PBPs, PBPA has a relatively small N-terminal domain and conservation of a cluster of charged residues within this domain suggests that PBPA is more related to Class B PBPs than previously inferred from sequence analysis. The C-terminal domain is a typical transpeptidase fold and contains the three conserved active site motifs that characterize penicillin-interacting enzymes. Whilst the arrangement of the SxxK and KTG motifs is similar to that observed in other PBPs, the SxN motif is markedly displaced away from the active site, such that its serine (Ser281) is not involved in hydrogen bonding with residues of the other two motifs. A disulphide bridge between Cys282 (the “x” of the SxN motif) and Cys266, which resides on an adjacent loop, may be responsible for this unusual conformation. Another interesting feature of the structure is a relatively long connection between β5 and α11, which restricts the space available in the active site of PBPA, and suggests that conformational changes would be required to accommodate peptide substrate or β-lactam antibiotics during acylation. Finally, the structure shows that one of the two threonines postulated to be targets for phosphorylation is inaccessible (Thr362), whereas the other (Thr437) is well placed on a surface loop near the active site.
Keywords: penicillin-binding proteins, tuberculosis, peptidoglycan, X-ray crystallography
Introduction
Penicillin-binding proteins (PBPs) are transpeptidases (TPases) responsible for the final stages of cell-wall synthesis in bacteria and the molecular targets for β-lactam antibiotics (for reviews see 1,2). Penicillin and other β-lactam antibiotics inhibit TPases by forming a long-lived covalent complex that blocks the active site from reaction with the natural peptide substrate. Although β-lactams have been remarkably successful for treating many bacterial diseases, historically they have been considered ineffective against tuberculosis (TB). One reason for this natural resistance is that Mycobacterium tuberculosis (Mtb) expresses a β-lactamase called BlaC 3 that hydrolyzes the β-lactam before it can reach its killing target. Another is that the complicated cell envelope of Mtb, comprised of layers of complex lipids outside the peptidoglycan layer 4, is believed to create a permeability barrier against β-lactam and other compounds 5. Nevertheless, when combined with β-lactamase inhibitors, β-lactams show good antimycobacterial activity in vitro 6–9 10 11 and have been used for the treatment of tuberculosis patients 12,13 14. Thus, PBPs remain potential clinical targets in Mtb and are therefore important to study.
Mtb strain H37Ra expresses at least four PBPs with apparent molecular masses of 94, 82, 52 and 37 kDa 15. The three high-molecular mass (HMM) PBPs were shown to bind most β-lactam antibiotics with high affinity and this binding was associated with antibacterial activity 15, suggesting these are the in vivo targets for β-lactams. The 94 and 83 kDa PBPs have been tentatively assigned as PBP1 (Rv0050) and PBP3 (Rv2163), respectively 16, the latter based on its homology with PBP3 of E. coli, a Class B PBP that localizes to the septum and may be involved in protein-protein interactions associated with the so-called ‘divisome’ 17 18.
The 52 kDa protein is encoded by the pbpA gene (Rv0016c) and has been classified as a Class B-like PBP 16. Although PBPA does not appear to be essential for growth of M. tuberculosis 19, deletion of the pbpA gene in M. smegmatis leads to reduced cell growth and defective cell septation, suggesting it has an important role in cell division 20. Interestingly, pbpA is part of a cluster of genes that also encode two serine/threonine protein kinases (PknA and PknB), and a phosphatase (PstP). This, apparently, is not a coincidence as PBPA can be phosphorylated by PknB at threonines 362 and 437 20, suggesting that the activity and/or cellular localization of PBPA may be controlled by a two-component signaling system. Together, these data suggest that PBPA serves an important biological function in Mtb.
As a step toward understanding the biological function of PBPA in M. tuberculosis, we have determined its crystal structure from Mtb strain H37Rv, the first published structure of a PBP from this organism. Although the protein contains a typical transpeptidase fold, it manifests some interesting structural features that may relate to its specific function in Mtb. These include a relatively small N-terminal domain, a displaced SxN motif and a long connecting loop that appears to restrict access to the active site in the unacylated enzyme. Finally, the structure is used to assess the likelihood of two threonines as targets for phosphorylation by a protein kinase.
Results and Discussion
Structure Determination
The crystal structure of a soluble construct of PBPA from M. tuberculosis H37Rv was solved at 2.05 Ǻ resolution and refined to an R factor of 21.7 % (Rfree = 25.2 %) with excellent stereochemistry (Tables 1 and 2). This construct comprises residues 35–491 of the protein and lacks 34 residues at the N-terminus that are predicted to encode the signal sequence and membrane anchor. Initial attempts at phasing were hampered by the presence of pseudo-translational symmetry and merohedral twinning that was overcome by collecting data from small crystals that had been excised from larger crystals. There are two molecules in the asymmetric unit and these are structurally very similar, with a root mean square deviation (RMSD) between all common main chain atoms of 0.47 Å. There is no evidence of a biological dimer and the protein elutes as a monomer by size-exclusion chromatography (data not shown). For both molecules, weak electron density occurs in four regions: the N terminus, the connecting loop between α3n and β6n, β2c and its connecting loops, and the C terminus (Fig. 1 and Supplementary Table 1) Additional regions of weak or absent density occur in molecule B for residues 371–385 (β2h-β2i-β2j) and 411–418 (α9-β3 loop). In all cases, these residues were not included in the final model. Since molecule A exhibits more order than molecule B and has a lower mean B factor (45.5 Å2 for all protein atoms compared to 59.3 Å2 for molecule B), it will be the focus of the following description.
Table 1.
Data collection statistics for diffraction data used to determine the crystal structure of PBPA from M. tuberculosis by multiwavelength anomalous dispersion.
| Data set | Peak | Inflection | Remote |
|---|---|---|---|
| Wavelength (Å) | 0.97932 | 0.97946 | 0.97180 |
| Resolution range (Å) | 48.11-2.4 (2.49-2.4) |
48.06-2.2 (2.28-2.2) |
48.11-2.4 (2.49-2.4) |
| No. of observations | 357,544 | 463,056 | 357,499 |
| No. of unique reflections | 31,689 | 41,092 | 31,716 |
| Rmerge* (%) | 9.7 (39.4) | 10.7 (38.7) | 9.8 (44.0) |
| Completeness (%) | 100 (100) | 100 (100) | 100 (100) |
| Redundancy | 11.2 (11.0) | 11.18 (10.9) | 11.2 (11.0) |
| <I>/<σI> | 11.6 (4.3) | 10.2 (4.3) | 11.0 (3.6) |
Data were collected in three wavelengths around the absorption edge of selenium. Numbers in parentheses are for the outer shell of data.
Table 2.
X-ray diffraction data and model refinement statistics for the crystal structure of PBPA
| Data collection | |
| Wavelength (Å) | 1.0 |
| Space group | P61 |
| Cell dimensions (Å) | a=b=120.6, c=92.2 |
| Resolution range (Å) | 36.3 - 2.05 (2.12-2.05) |
| Rmerge* (%) | 7.6 (54.8) |
| Completeness (%) | 99.6 (99.9) |
| Redundancy | 11.2 (9.8) |
| <I>/<σI> | 45.9 (2.6) |
| Refinement | |
| Resolution range (Å) | 36.30 - 2.05 |
| No. of unique reflections | 47,630 |
| No. reflections used to calculate Rfree | 2,388 |
| No. of non-hydrogen protein atoms | 5908 |
| No. of water molecules | 76 |
| R cryst (%) | 21.7 |
| R free (%) | 25.2 |
| RMS deviations from ideal stereochemistry:- | |
| bond lengths (Å) | 0.011 |
| bond angles (°) | 1.30 |
| B factors: | |
| Mean B factor (main chain) (Å2) | 50.3 |
| RMS. deviation in main chain B factors (Å2) | 1.22 |
| Mean B factor (side chains & waters) (Å2) | 53.3 |
| RMS deviation in side chain B factors (Å2) | 1.75 |
| Ramachandran plot: | |
| Residues in most favored region (%) | 90.6 |
| Residues is disallowed region (%) | 8.4 |
| Residues in generously allowed regions | 0.8 |
| Residues in disallowed regions | 0.2 |
| PDB code | 3LO7 |
where Rmerge = Σ|Ii−Im|/ Σ Ii, where Ii is the intensity of the measured reflections and Im is the mean intensity of all symmetry-related reflections. Figures within brackets are for the outer resolution shells.
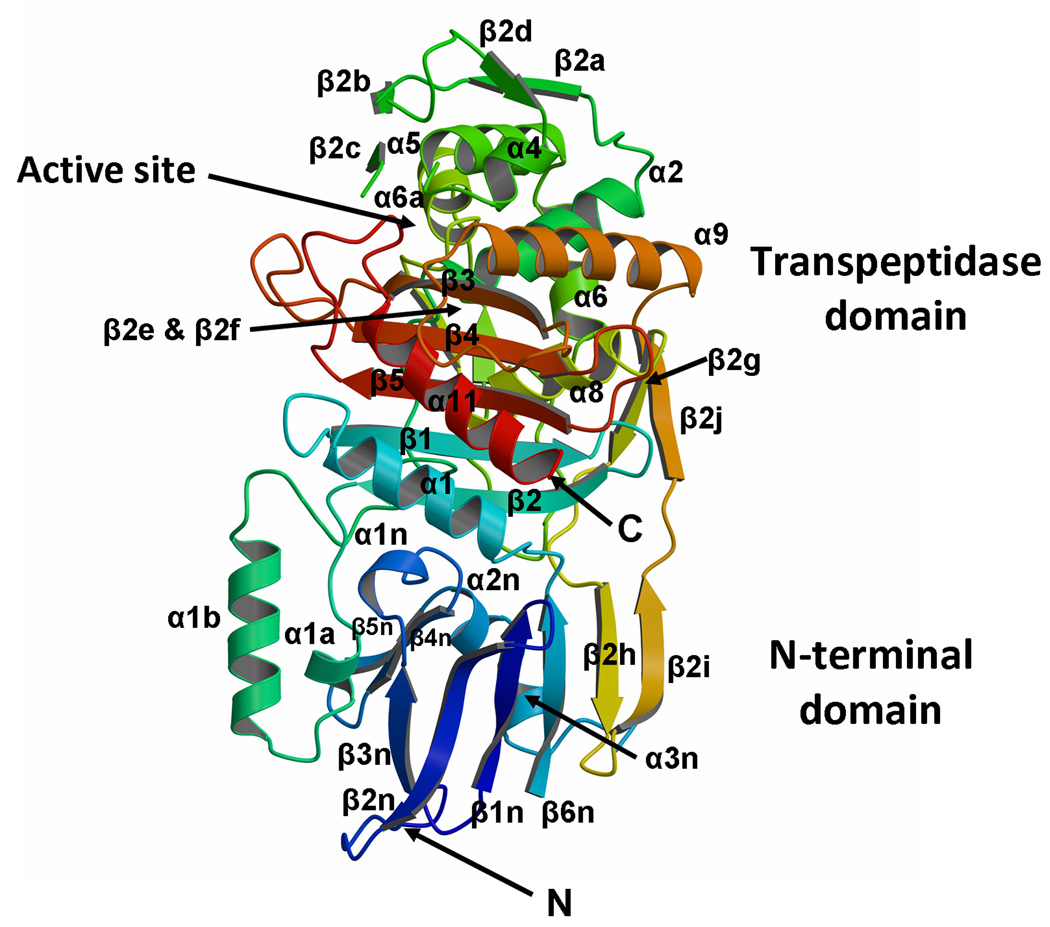
Fig. 1.
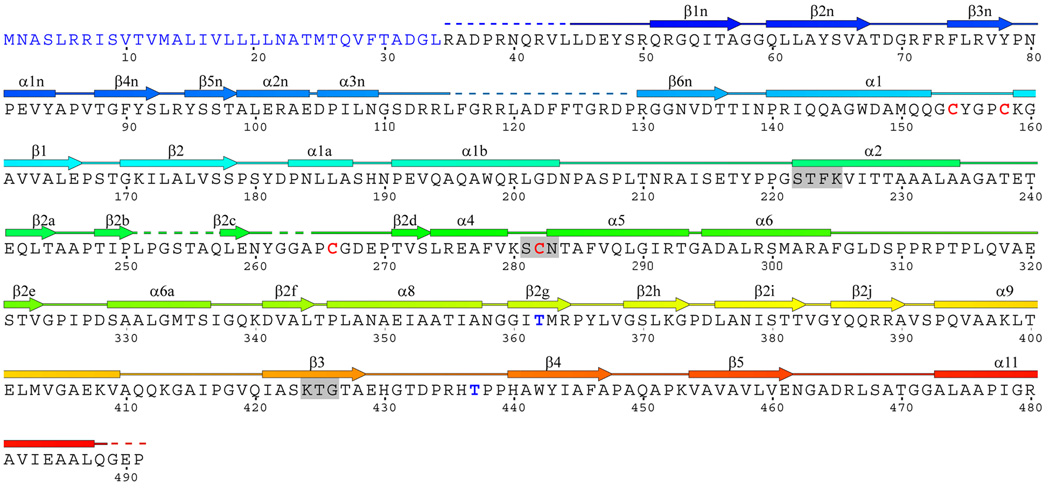
The structure of PBPA from M. tuberculosis. A. Ribbon representation of the molecule in which the structure is color-ramped blue-to-red in the N-terminal to C-terminal direction. Elements of secondary structure are labeled according to Pares et al 24 and Lobowsky et al. 25. Accordingly, some elements of secondary structure are absent, i.e. α3, α7 and α10. In this view, the transpeptidase domain is at the top and the N terminal domain, comprising the interdomain linker, is at the bottom. In the native protein, the membrane anchor is at the N-terminal end of the protein (at the bottom). (B) The amino acid sequence and secondary structure of PBPA. Regions that could not be modeled due to apparent disorder (in molecule A of the asymmetric unit) are shown as dashed lines. The three conserved motifs of the active site are highlighted by grey boxes. The cysteines involved in disulphide bridges and the threonines that are reported to be phosphorylated in vivo 20 are shown in red and blue, respectively. Figure produced in part by SecSeq (D. E. Brodersen, unpublished software, http://xray.imsb.au.dk/~deb/secs).
N-terminal domain
PBPA contains two domains: an N-terminal mostly β domain (NTD) and a C-terminal transpeptidase (TPase or penicillin-binding) domain that is found in all non-metallo PBPs (Fig. 1). Compared to other Class B HMM PBPs of known structure, the NTD is relatively small because it contains only the so-called interdomain linker region 1 and lacks any other domains that found in other PBPs. Except for three conserved motifs at the heart of the interdomain linker region (described below), there is no detectable sequence similarity with the NTDs of Class B PBPs of known structure, such as PBP2x, PBP1a and PBP1b of S. pneumoniae or with PBP2 of N. gonorrhoeae. The core of the NTD in PBPA is a six-stranded anti-parallel β-sheet, in which two of the strands (β2h and β2i) are contributed by the TPase domain via an extension of the connecting loop between β2g and β2j. A similar addition is also observed in the S. pneumoniae PBPs: PBP2X, PBP1a and PBP1b 1. The NTD β sheet of PBPA is exposed to solvent on one side, but is packed on the other by the connection between β3n and β6n, which comprises three short helices and β hairpin in the order α1n, β4n, β5n, α2n and α3n.
The size and domain structures of the NTDs of HMM PBPs vary considerably and these differences may be related to their functional roles in the respective organisms. One possible function ascribed to these domains is to mediate protein-protein interactions that may be involved in the assembly of multiprotein complexes 21. The best example of this is the divisome in E. coli, which is a complex of ~12 proteins that localizes to the septum during cell division 22. One important player in this complex is E. coli PBP3 23, a class B PBP. The classification of PBPA as a class B-like PBP 16 and its apparent requirement for cell separation in M. smegmatis 20 suggests that PBPA is also required for cell division in M. tuberculosis. It is interesting to note, however, that the relatively small size of the NTD presents fewer opportunities for protein-protein interactions compared to other PBPs.
A more likely function for the NTD of PBPA is to project the TPase away from the membrane and toward its peptidoglycan substrate in the periplasm. If so, given the relatively small size of the NTD and even allowing for the ~10 residues absent at the N terminus due to disorder, the peptidoglycan in M. tuberculosis (at the least in the vicinity of this particular PBP) may be relatively closer to the cytoplasmic membrane than in other bacteria.
The interdomain linker region of PBPA
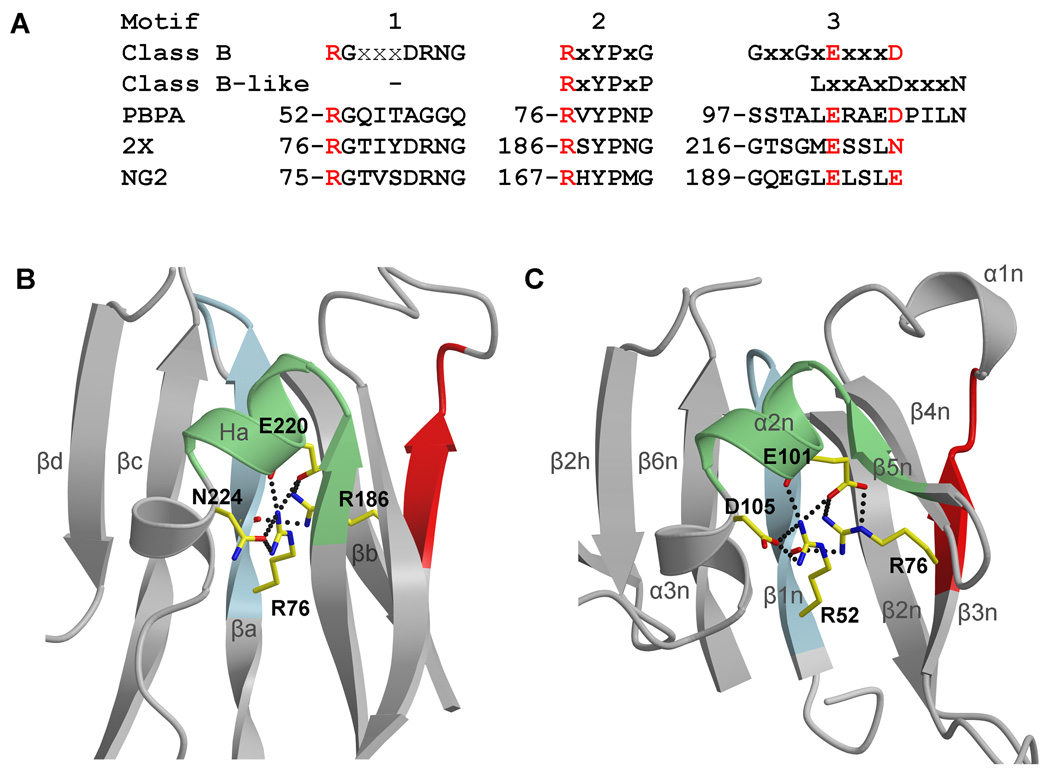
The functional importance of the NTD of PBPA is reinforced by the presence of three conserved motifs at the heart of the interdomain linker region 16. For Class B PBPs, these are defined as: motif 1 - RGx3DRN/SG, motif 2 - RxYPxG and motif 3 Gx2GxEx3D (where bold residues are invariant and x is any residue) 16 (Fig. 2A). As observed in the crystal structure of PBP2X from S. pneumoniae, the first Arg of motifs 1 and 2, as well as the Glu of motif 3, cluster together three-dimensionally to form a dense electrostatic network that probably stabilizes the interdomain linker region of the molecule 1 (Fig. 2B). According to the existing classification by Goffin and Ghuysen, Class B-like PBPs are a subclass of Class B PBPs that lack motif 1 and contain a different motif 3 (Lx2AxDx3T) 16. Examination of this same region in PBPA, which is classified by this scheme as a class B-like PBP, however, shows that two arginines and a Glu are also present in equivalent positions as those observed in PBP2X and form similar interactions (Fig. 2C). The primary difference is that PBPA contains an Asp at position 105 instead of Asn224 in PBP2X. Hence, the presence of Arg52 indicates that PBPA does contain a motif 1 that is similar to that of Class B PBPs, but, aside from the Arg and Gly at the N-terminal end, lacks its conserved residues. Furthermore, the observation of Glu101 within the cluster and not an Asp also suggests that motif 3 belongs to the consensus for Class B PBPs rather than the one defined by Goffin & Ghuysen for Class B-like PBPs. With the benefit of structural alignment afforded by the crystal structure of PBPA, it is clear that motif 3 of Class B-like PBPs, as defined, is simply misaligned by four residues (Fig. 2A). The presence of a motif 1 and a motif 3 that aligns with that of Class B PBPs suggests that, at the structural level, PBPA is more related to Class B PBPs than could be inferred from sequence analysis alone, and the same could be true for PBPs within this sub-class.
Fig. 2.
A comparison of the interdomain linker regions of PBPA, N. gonorrhoeae PBP2 and S. pneumoniae PBP2X. A. The three consensus sequence regions of the interdomain region, as defined by Goffin and Ghuysen 16, including those for motif 3 in Class B and Class B-like PBPs. Aligned with these are the sequences from PBPA, S. pneumoniae PBP2X (“2X”) and N. gonorrhoeae PBP2 (“NG2”). Note that motif 1 is present in PBPA and that motif 3 of PBPA aligns with the consensus sequence for motif 3 of a Class B PBPA, whereas the Class B-like motif 3 is simply four residues downstream. B. The structure of PBP2X in the interdomain linker region showing the clustering of four conserved residues contributed by the three motifs, which form an electrostatic network at the core of the domain and are colored red in (A). C. The same view in M. tuberculosis PBPA showing a similar cluster. Both structures are shown in ribbon format, where the three motifs that comprise the interdomain linker are colored light blue, green and red, respectively, for motifs 1 to 3. The secondary structure for PBP2X is labeled according to Macheboeuf et al 1. Dashed lines represent potential hydrogen bonds..
Transpeptidase domain
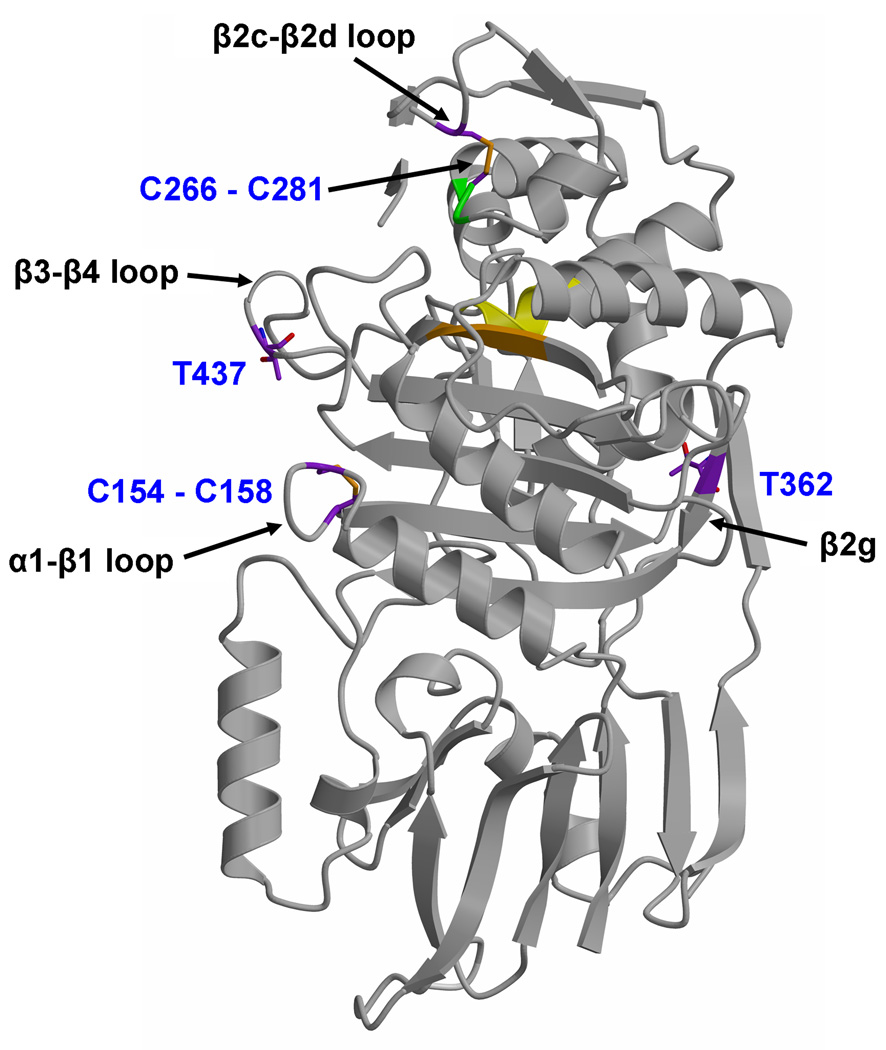
The TPase domain of PBPA shares the same fold as other (non-metallo) penicillin-interacting enzymes, including PBPs and most classes of β-lactamase, and contains the active site. It can be considered as two sub-domains, one α/β and the other predominantly α helical. The α/β sub-domain comprises a five-stranded anti-parallel sheet (β1-β5) packed on one side by α helices α1 and α11, and on the other by α8, whereas α9 lies across the top of the sheet (the nomenclature adopted for the TPase domain (Fig. 1B) follows that of Pares et al 24, which itself is adapted from 25). As viewed in Fig. 1A, the α domain lies above the α/β sub-domain and contains the helices α2, α4-α6 and α6a. A β-hairpin extension between α2 and α4, comprising four short β strands β2a-β2d, is located at the top of the molecule above the active site. This region exhibits apparent disorder in both molecules of the asymmetric unit as judged by weak electron density. Within the residue range 251–264, only residues 258–261 can be modeled in molecule A (and residues 259–260 for molecule B), comprising β2c. Just these residues are visible because of the interactions between this stand and β2b, whereas its connecting loops appear to be flexible. An interesting feature of the TPase domain is the presence of two disulphide bridges: one between Cys282 of the SxN motif (i.e. SCN) and Cys266 of the β2c-β2d loop (described below), and the other between Cys154 at the C-terminal end of α1 and Cys158 in the following loop that connects with β1 (Fig. 3). All of these cysteines are conserved in PBPA orthologues in different mycobacterial species 20.
Fig. 3.
Location of two disulphide bridges, and two threonines proposed to be targets of phosphorylation by PknB 20, in PBPA. Set against the fold of PBPA (displayed in grey ribbon) are the cysteines that form disulphide bridges and threonines 362 and 437. In both cases, carbon bonds are colored purple. Nearby elements of secondary structure to each site are marked by arrows. The active site is indicated by the cluster of conserved motifs, with the ribbon colored yellow for the SxxK motif, green for the SxN motif and orange for the KTG motif.
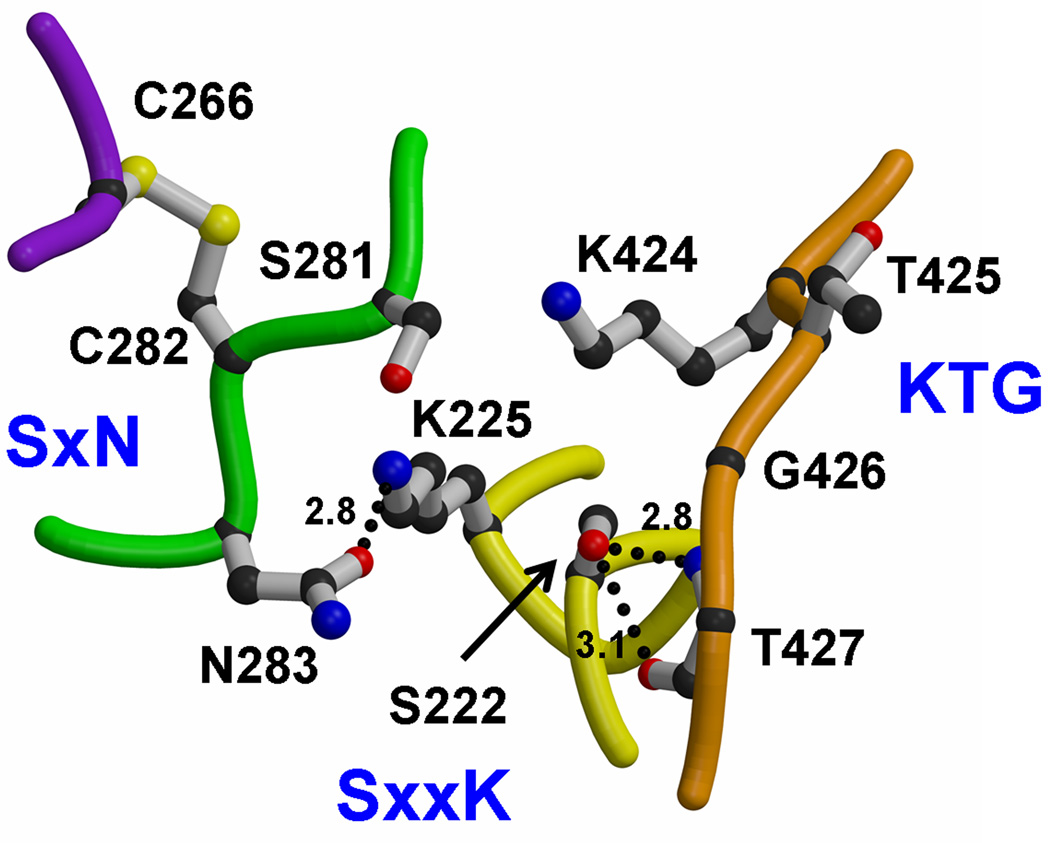
Active site
The active site of PBPA contains the three conserved sequence motifs that are the hallmark of most penicillin-interacting enzymes 26(Fig. 4). The SxxK motif is located at the N-terminal end of the helix α2 and contains two residues known to be important for catalysis in PBPs: Ser222, the serine nucleophile that is acylated by the peptide substrate and β-lactam antibiotics, and Lys225, whose equivalent in other PBPs is believed to enhance the nucleophilicity of the serine 27. The SxN motif is situated on the loop connecting α4 and α5, and comprises Ser281, Cys282 and Asn283. Lastly, the KTG motif, comprising Lys424, Thr425 and Gly426, lies in the middle of β3.
Fig. 4.
Active site of M. tuberculosis PBPA. Shown are the three conserved motifs, where the SxxK motif is yellow, the SxN motif is green and the KTG motif is orange. Residues are shown in bond format. Potential hydrogen bonds are shown as dashed lines and distances are given in Å. The fourth loop, colored purple, is the connecting loop between β2c and β2d that is tethered to the SxN motif by a disulphide bridge between Cys282 and Cys266.
In keeping with its function as a nucleophile, Ser222 occupies a central location in the active site. The rotamer position of its side chain varies in known structures of unacylated PBPs and in PBPA it is hydrogen bonded to Thr427, which immediately follows the KTG motif. For this reason, Lys225 is not hydrogen bonded to Ser222, as observed in many PBP structures, but is within hydrogen bonding distance of Asn283. In contrast to other PBP structures, Lys225 is not hydrogen bonded to Ser281 due to the unusual conformation of the SxN motif (see below); in fact, the Oγ atom of Ser281 and Nζ atom of Lys225 are separated by 5.6 Å. Finally, Lys424 of the KTG motif is hydrogen bonded with the main chain carbonyl of Phe278 at the C-terminal end of α4 (not shown), whereas in some PBPs, such as E. coli PBP5 28 and N. gonorrhoeae PBP2 29, this residue forms hydrogen bonds with the equivalent of Ser281 in the SxN motif.
Comparison with structures of other Class B PBPs
The TPase domain of PBPA was compared with that of two other Class B HMM PBPs: PBP2x from S. pneumoniae 30 and PBP2 from N. gonorrhoeae (NgPBP2) 29. Structure-based sequence alignment reveals very low sequence identities between the TPase domain of PBPA and those from the other ClassB PBPs (23 and 28%, respectively). The structures were superimposed using 11 residues of the three conserved motifs (i.e. residues 222–225, 281–283 and 424–427 of PBPA) with a root mean square deviations (RMSD) of 1.3 Å for PBP2X and 1.4 Å for NgPBP2.
At the broad level, the three structures are very similar, but there are interesting differences in the interconnecting loops, some of which may impact the shape and accessibility of the active site. One notable difference is the connection between β2 and α2, a region that forms a protrusion that lies away from the base of the TPase domain (see Fig. 1A). PBPA is marked by a long α helix within this region (α1b), whereas in both PBP2X and NgPBP2, this loop is shorter and there is proportionally more irregular structure present. Interestingly, α1a, which is a short segment of α helix that precedes α1b, is relatively more conserved than α1b and this may be because it interacts with β3n and β4n of the interdomain linker region. One key interaction is made by Asn184, whose side chains forms hydrogen bonds with both the carbonyl and the amide nitrogen of Tyr78.
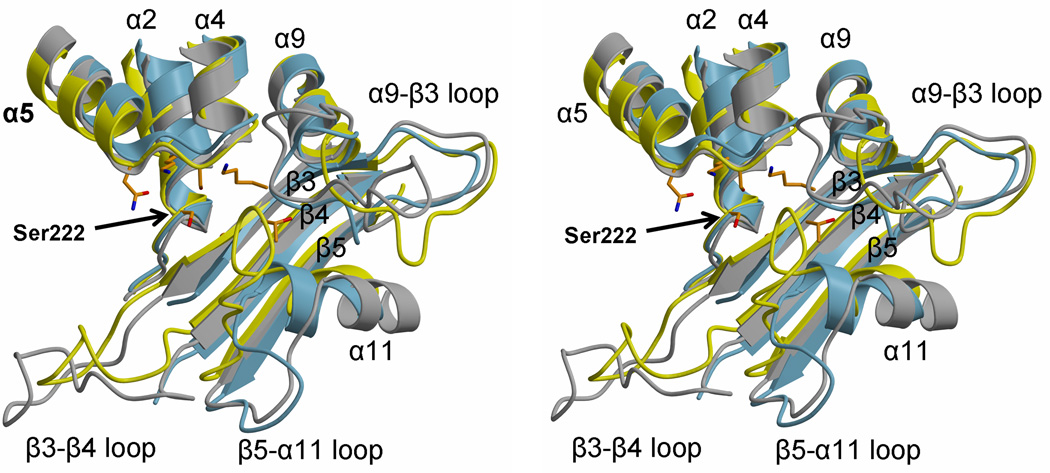
Several significant differences between the three enzymes occur near the active site involving the connecting loops β3-β4, α9-β3 and β5-α11 (Fig. 5A). The β3-β4 loop in PBPA lies relatively closer to the active site and is significantly shorter than its equivalent in PBP2X. Interestingly, the same loop in NgPBP2 is disordered and contains mutations associated with penicillin resistance of N. gonorrhoeae 29.
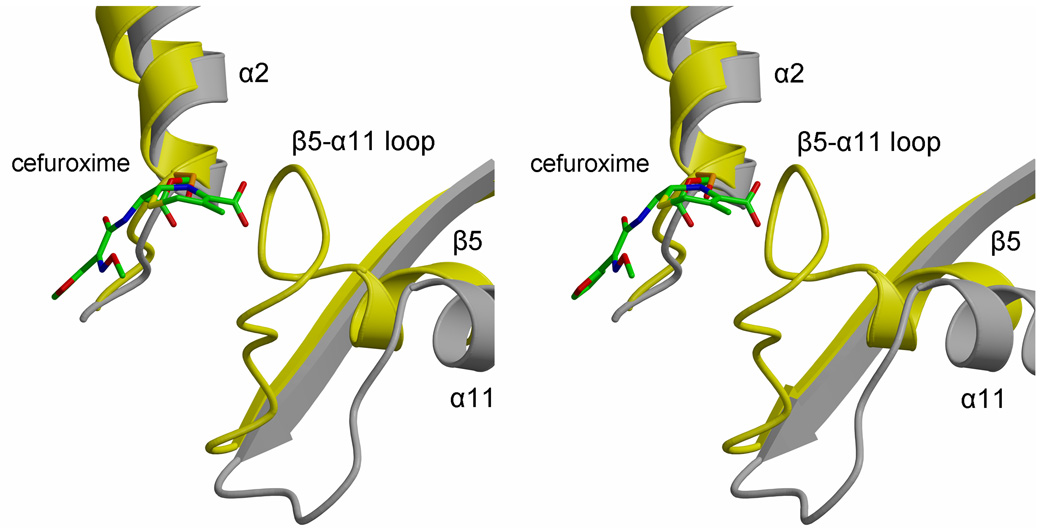
Fig. 5.
Varying lengths and conformations of connecting loops in the active site region of class B PBPs. A. In this stereoview of the superimposition, PBPA is colored yellow, PBP2X is grey and NgPBP2 is blue. Each structure is shown in ribbon format and the active site residues of PBPA are shown as orange bonds. Elements of secondary structure are labeled, as is the serine nucleophile. B. A close-up stereoview showing PBPA and PBP2X only, the latter in complex with cefuroxime, displayed with green bonds 30. This shows the close proximity of the β5-α11 loop of PBPA with the carboxylate of the cephalosporin in acylated PBP2X.
Compared to both PBP2x and NgPBP2, there is a much more direct route between α9 and β3 in PBPA, which means this loop is relatively further away from the active site compared to the equivalent loop in PBP2X and NgPBP2, which in these PBPs lies near the SxN motif. By contrast, the β5-α11 loop, which is adjacent to the α9-β3 loop, is longer and is significantly nearer the active site in PBPA than its respective counterpart in PBP2X and NgPBP2. This restricts the space available in the active site of PBPA and may impact its recognition of β-lactams. This is illustrated by a superimposition of PBPA with that of PBP2X in complex with cefuroxime (Fig. 5B) in which the β5-α11 loop of PBPA lies very close to the carboxylate of the cepaholosporin and suggests that conformational changes in this loop are required for the binding of substrate or β-lactams to PBPA. It is interesting to note that the latter two differences near the active site may be correlated. A shorter and more direct α9-β3 loop in PBPA is coupled with a longer β5-α11 loop, whereas the opposite occurs in PBP2X and NgPBP2. Overall, these differences in connecting loops in the immediate vicinity of the active site may relate to the respective substrate specificities of the three enzymes, which, in turn, is probably dictated by the specific peptidoglycan structures present in these organisms.
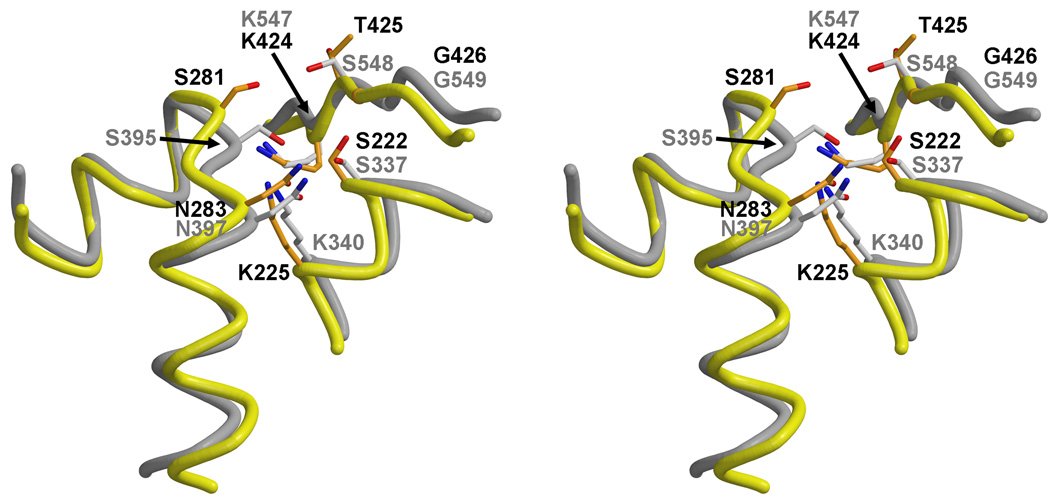
Unusual conformation of the SxN motif
The superimposition with class B PBPs also shows that the SxN motif of PBPA occupies a position that is further from the core of the active site than is observed in other PBPs (Fig. 6), which places Ser281 beyond hydrogen bonding distance with residues of the SxxK and KTG motifs. This difference is slightly more pronounced in molecule B of the asymmetric unit compared to molecule A. One possible reason for the unusual conformation of the SxN motif in PBPA may be the disulphide bridge between Cys282 and Cys266 of the β2c-β2d loop. The latter belongs to the β-hairpin extension that lies across the top of the molecule (as viewed in Fig. 1). These region appears to be important for function in PBPs because, in some PBPs, e.g. NgPBP2 29, E. faecium PBP5fm 31,32 and PBP2X 33, the β-hairpin extension harbors mutations associated with penicillin resistance. In NgPBP2, insertion of an aspartate in the β-hairpin extension is believed to disrupt the interaction between this region and the SxN motif 29.
Fig. 6.
The unusual conformation of the SxN motif in PBPA. A. Superimposition of the SxN motifs of PBPA (yellow) and PBP2X (grey) shows the relative displacement of this motif away from the active site in PBPA. Residues of PBPA are colored orange and labeled in black, whereas those of PBP2X are displayed and labeled in grey.
Interestingly, flexibility in the SxN motif of PBPA is suggested by the absence of clear electron density for the side chain of Ser281 (in both molecules) and the presence of some positive peaks of electron density (in molecule A of the asymmetric unit) corresponding to a position for Ser281 that is closer toward the active site. This is important because the SxN motif appears to play a central role in the mechanism of deacylation of PBPs 34,35. Different positions of the SxN motif have been observed in crystal structures of PBP5 as a result of covalent modification of a nearby cysteine by a mercurial compound 36, and disordering of the region immediately prior to the SxN motif has been observed in PBP2X derived from penicillin-resistant strains of S. pneumoniae 33. Given that flexibility apparently occurs in both the β-hairpin extension (near β2c) and the SxN motif, one function of the disulphide bridge in PBPA may be to tether these regions together.
Phosphorylation sites
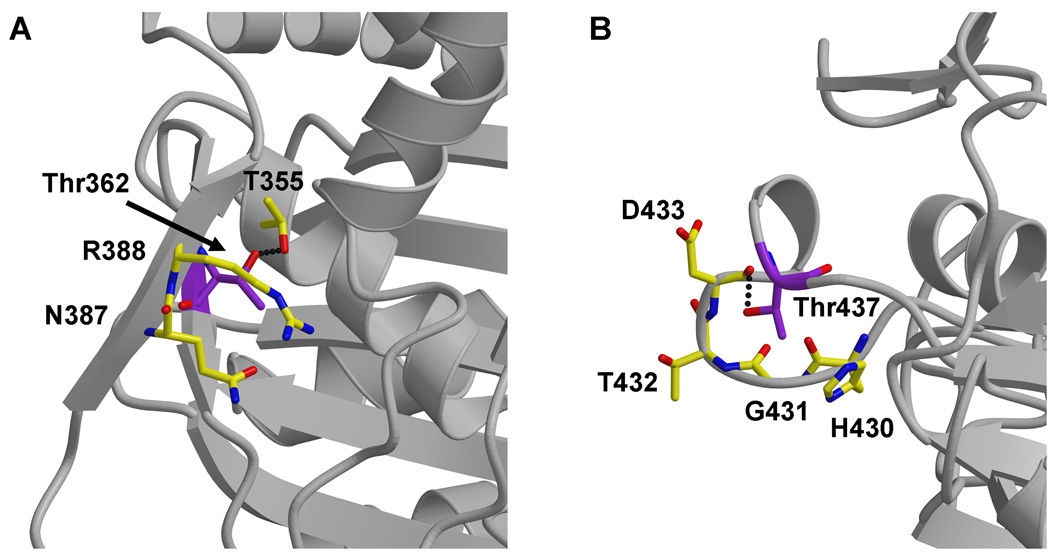
Another intriguing feature of PBPA is evidence that suggests it is phosphorylated in vivo 20. In mycobacteria, the gene encoding PBPA resides within an operon that also includes two Ser/Thr protein kinases (PknA and PknB) and a Ser/Thr phosphatase (PstP). Co-expression of PknB with PBPA (in E. coli) leads to phosphorylation of PBPA at two threonines, Thr362 and Thr437 20. Moreover, T437A mutants of PBPA fail to localize to the septum during cell division in M. tuberculosis, which suggests that phosphorylation may regulate the cellular localization of PBPA 20. With the crystal structure of PBPA, the likelihood of these two residues being targets for phosphorylation can be examined. Thr362 is located on β2g and its side chain points toward the interior of the protein, where it forms a putative hydrogen bond with Thr355 (Fig. 7A). Without a conformational change in this region, this residue does not appear to be sufficiently exposed for phosphorylation by the PknB kinase.
Fig. 7.
Two Thr residues in PBPA reported to be targets for phosphorylation 20. A. Thr362 lies on the inward face of β2g and appears too buried for phosphorylation. B. Thr437 resides on the β3-β4 loop near the active site and appears sufficiently exposed for phosphorylation. In both cases, the threonines are colored with purple bonds, whereas surrounding residues are colored yellow. The backbone of the surrounding structure is displayed in ribbon format.
By contrast, Thr437, which is part of the connecting loop between β3 and β4 that protrudes away from the main body of the TPase domain, appears well placed for phosphorylation (Fig. 7B). The electron density indicates the loop is well ordered in both molecules of the asymmetric unit, with the exception of the side chains of Lys435 and His436. Interestingly, this loop is located sufficiently close to the active site of the enzyme such that phosphorylation of Thr437 could influence enzymatic activity. In agreement with this idea, the equivalent loop in S. pneumoniae PBP1a and NgPBP2 contains mutations that are associated with resistance to β-lactams 29,37,38.
Conclusion
The crystal structure of PBPA from Mtb shows an unusual architecture in the active site region in which in the SxN motif lies in a position where it appears to be less than optimal for catalysis. In addition, the active site appears partially blocked to peptide substrate and β-lactams by a relatively long connection between β5 and α11. Both observations suggest that conformational changes in the active site region are likely to occur prior to or during acylation to allow access to the active site and for the SxN motif to perform its presumed role in deacylation. Apparent flexibility in the SxN motif and in the adjacent β-hairpin extension is consistent with conformational changes in the active site region. Structures of PBPA in acylated form will show whether this is indeed the case. Another goal is to examine the impact of phosphorylation of Thr437 on both the structure of PBPA and its acylation rate to determine whether this modification is of biological relevance for the function of the enzyme. Of equal interest is the nature of the putative interaction between PknB and PBPA.
Materials and methods
Cloning
The gene encoding PBPA, but excluding codons 1–34 (the putative signal sequence and membrane anchor), was amplified from the genomic DNA of M. tuberculosis strain H37Rv using the following primers: forward, 5’-AGAGGATCCCGTGCCGATCCCCGCAAC-3’ (BamHI restriction site underlined) and reverse, 5’-AGAGAATTCTCATGGTTCCCCCTGCAGTG-3’ (EcoRI site underlined and stop codon in boldface). The PCR product was cloned into the pT7HTb vector, which incorporates a hexa-histidine tag and an intervening cleavage site for tobacco etch virus (TEV) protease at the N-terminus of the protein, and transformed into E. coli BL21(DE3) cells.
Protein expression and recovery of inclusion bodies
One liter of E. coli BL21(DE3) cells in Luria-Bertani broth containing 50 mg/L kanamycin was grown at 37 °C until the OD600nm reached 0.6–0.8, at which point protein expression was induced by addition of 0.5 mM isopropyl β-D-thiogalactoside (IPTG), followed by overnight incubation at 20 °C. After centrifugation at 4,500 g for 15 min., the cells were resuspended in 10 ml of lysis buffer (20 mM Tris-HCl pH 8, 500 mM NaCl, 2 mM EDTA, 10% glycerol and 1 mM PMSF). Cells were lysed by three freeze/thaw cycles, followed by incubation with 20 µg/ml lysozyme at 4 °C for 30 min., and then 90 s of sonication (3 × 30 s. with 60 s. interval between each cycle). Inclusion bodies (IB) were recovered by centrifugation at 10,000 g at 4 °C. The pellet was washed twice with 10 mM Tris-HCl, 1 mM EDTA, pH 8, first with, and then without, 1% Triton X-100. The pellet was then dissolved in 10 ml of 8 M urea, and remaining particulates were removed by centrifugation at 15,000 g for 30 min.
Protein refolding and purification
Solubilized and clarified inclusion bodies were diluted slowly into 200 ml of ice-cold refolding buffer (50 mM KH2PO4, pH 10.7, 500 mM NaCl and 10% glycerol). After stirring for 2 hr at 4 °C, the pH was reduced to 8 with HCl and stirred overnight at 4 °C. After centrifugation at 10,000 g for 10 min. to remove particulates, the volume of the refolded protein was increased 30 % by the addition of buffer A (20 mM Tris-HCl pH 8, 0.5M NaCl, 10 % glycerol and 20 mM imidazole) and the solution passed over a 10 ml Ni2+-affinity resin (Chelating Sepharose Fast Flow, GE Healthcare) equilibrated with buffer A. After washing with 10 column volumes of buffer A, and then 10 column volumes of buffer A containing 100 mM imidazole, PBPA was eluted with buffer A containing 500 mM imidazole. Purified His-tagged PBPA was concentrated to 11 mg/ml in buffer A with 500 mM imidazole and stored at −80 °C. In the absence of imidazole, the protein showed a tendency to precipitate.
Site-directed mutagenesis and incorporation of selenomethionine
There are 5 methionine residues in the PBPA construct, but initial attempts to solve the structure by the multiwavelength anomalous diffraction (MAD) method using selenomethionine were unsuccessful. An additional methionine was introduced by site-directed mutagenesis of Leu141, which was predicted to lie within a hydrophobic core region of the protein, using the QuikChange protocol (Stratagene, Inc). The selenomethionine-derivative of this mutant (SeMet-PBPA-L141M) was prepared by growing transformed BL21 (DE3) cells under conditions where methionine biosynthesis is inhibited by high concentrations of hydrophobic amino acids 39. Cells were grown at 37 °C in 1L of LB medium containing 50 mg/L kanamycin until the OD600nm reached 0.4. Cells were then harvested by centrifugation for 15 min. at 4000 rpm and resuspended in 1L of M9 minimal medium containing 4 g/L glucose and 50 mg/L kanamycin. After further incubation to an OD600nm of 0.6, 100 mg/L each of lysine, phenylalanine, and threonine, 50 mg/L isoleucine and valine, and 60 mg/L selenomethionine were added. The culture was then shaken for an additional 15 min. at 37 °C, cooled to 20 °C, and protein expression was induced by addition of 0.5 mM IPTG. SeMet-PBPA-L141M was then purified as described for the wild-type protein except that 2 mM β-mercaptoethanol was included in all buffers.
In contrast to wild-type PBPA, the His6 tag was then removed from the purified fusion protein. SeMet-PBPA-L141M was digested with TEV protease (50:1 weight ratio), followed by dialysis against buffer A with 10 mM β-mercaptoethanol overnight at 4°C. The cleaved product was separated from the digestion mixture by second passage over a Ni2+-affinity column (HiTrap HP, GE Healthcare) pre-equilibrated with buffer A containing 10 mM β-mercaptoethanol. SeMet-PBPA-L141M passed straight through the column, whereupon it was concentrated to 8.4 mg/ml (in the same buffer). Successful incorporation of SeMet was established by mass spectrometry (data not shown).
Crystallization and data collection
Wild-type PBPA was subjected to a search for crystallization conditions, initially beginning with Crystal Screens 1 and 2 (Hampton Research, Aliso Viejo, CA, USA) and the JCSG+ Suite (Qiagen, Inc.). Crystal trays were set up using a hanging-drop vapor diffusion method in which 1 µl of protein was mixed with 1 µl of well solution. After optimization, the best crystals of PBPA were obtained at 18 °C using a 9 mg/ml protein solution over wells containing 35 % PEG 1000, 0.2 M ammonium acetate, and 0.1 M sodium acetate pH 4.6. By a similar approach, crystals of SeMet-PBPA-L141M were obtained over wells containing 22–25% PEG 3350, 0.1 M Bis-Tris pH 5.5. Both wild-type and L141M mutant crystals were cryoprotected by passage through a solution containing 35 % PEG 8000, 16 % PEG 1000, 0.2 M ammonium acetate, and 0.1M sodium acetate, pH 4.6.
X-ray diffraction data extending to 2.05 Å resolution were collected at the SER-CAT ID-22 beamline of the Advanced Photon Source (Argonne National Laboratory, Chicago, IL) from crystals of wild-type PBPA. The crystals belong to space group P61 or P65 with cell dimensions a=120.5 and c=92.2 Å. There are two molecules in the asymmetric unit. For crystals of SeMet-PBPA-L141M mutant of PBPA, data extending to 2.2 Å resolution were collected at three wavelengths corresponding to the peak, high-energy remote and inflection points of the K absorption edge of selenium (Table 1). All data were processed with HKL2000 40. The structure was solved in space group P61 by SOLVE and RESOLVE 41 using all three data sets. Electron density maps were displayed in O 42 and used to construct an initial model. This was refined by automated refinement using REFMAC5 43 and manual adjustments using O. After several such rounds, higher resolution data collected from the wild-type crystals were introduced for refinement (containing the same assignments for calculation of R free 44). In later rounds, waters were added to the model using Phenix 45. The stereochemistry of the model was monitored by PROCHECK 46. The numbering of the model corresponds to the gene sequence for PBPA, such that the first molecule of the construct is Arg35.
Supplementary Material
Acknowledgements
This work was supported by the National Institutes of Health grants GM66861 to C.D. and AI36901 to R.A.N. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-ENG-38. Data were collected at Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. Supporting institutions may be found at www.ser-cat.org/members.html. The authors would like to thank Miriam Braunstein of the University of North Carolina-Chapel Hill for kindly providing the M. tuberculosis genomic DNA from which pbpA was amplified. Figures were prepared using MOLSCRIPT 47 and RASTER3D 48 except where stated.
Abbreviations used
- HMM
high molecular weight
- LMM
low molecular weight
- Mtb
Mycobacterium tuberculosis
- NTD
N terminal domain
- PBP
penicillin-binding protein
- RMSD
root mean square deviation
- TPase
transpeptidase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Acession Code:
The coordinates and structure factors have been deposited with the Protein Data Bank with accession code 3LO7.
References
- 1.Macheboeuf P, Contreras-Martel C, Job V, Dideberg O, Dessen A. Penicillin binding proteins: key players in bacterial cell cycle and drug resistance processes. FEMS Microbiol. Rev. 2006;30:673–691. doi: 10.1111/j.1574-6976.2006.00024.x. [DOI] [PubMed] [Google Scholar]
- 2.Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev. 2008;32:234–258. doi: 10.1111/j.1574-6976.2008.00105.x. [DOI] [PubMed] [Google Scholar]
- 3.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, 3rd, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 4.Dover LG, Cerdeno-Tarraga AM, Pallen MJ, Parkhill J, Besra GS. Comparative cell wall core biosynthesis in the mycolated pathogens, Mycobacterium tuberculosis and Corynebacterium diphtheriae. FEMS Microbiol. Rev. 2004;28:225–250. doi: 10.1016/j.femsre.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Jarlier V, Gutmann L, Nikaido H. Interplay of cell wall barrier and beta-lactamase activity determines high resistance to beta-lactam antibiotics in Mycobacterium chelonae. Antimicrob. Agents Chemother. 1991;35:1937–1939. doi: 10.1128/aac.35.9.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cynamon MH, Palmer GS. In vitro activity of amoxicillin in combination with clavulanic acid against Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 1983;24:429–431. doi: 10.1128/aac.24.3.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sorg TB, Cynamon MH. Comparison of four beta-lactamase inhibitors in combination with ampicillin against Mycobacterium tuberculosis. J. Antimicrob. Chemother. 1987;19:59–64. doi: 10.1093/jac/19.1.59. [DOI] [PubMed] [Google Scholar]
- 8.Segura C, Salvado M, Collado I, Chaves J, Coira A. Contribution of beta-lactamases to beta-lactam susceptibilities of susceptible and multidrug-resistant Mycobacterium tuberculosis clinical isolates. Antimicrob. Agents Chemother. 1998;42:1524–1526. doi: 10.1128/aac.42.6.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dincer I, Ergin A, Kocagoz T. The vitro efficacy of beta-lactam and beta-lactamase inhibitors against multidrug resistant clinical strains of Mycobacterium tuberculosis. Int. J. Antimicrob. Agents. 2004;23:408–411. doi: 10.1016/j.ijantimicag.2003.09.023. [DOI] [PubMed] [Google Scholar]
- 10.Hugonnet JE, Tremblay LW, Boshoff HI, Barry CE, 3rd, Blanchard JS. Meropenem-clavulanate is effective against extensively drug-resistant Mycobacterium tuberculosis. Science. 2009;323:1215–1218. doi: 10.1126/science.1167498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watt B, Edwards JR, Rayner A, Grindey AJ, Harris G. In vitro activity of meropenem and imipenem against mycobacteria: development of a daily antibiotic dosing schedule. Tuber. Lung Dis. 1992;73:134–136. doi: 10.1016/0962-8479(92)90145-A. [DOI] [PubMed] [Google Scholar]
- 12.Chambers HF, Kocagoz T, Sipit T, Turner J, Hopewell PC. Activity of amoxicillin/clavulanate in patients with tuberculosis. Clin. Infect. Dis. 1998;26:874–877. doi: 10.1086/513945. [DOI] [PubMed] [Google Scholar]
- 13.Nadler JP, Berger J, Nord JA, Cofsky R, Saxena M. Amoxicillin-clavulanic acid for treating drug-resistant Mycobacterium tuberculosis. Chest. 1991;99:1025–1026. doi: 10.1378/chest.99.4.1025. [DOI] [PubMed] [Google Scholar]
- 14.Chambers HF, Turner J, Schecter GF, Kawamura M, Hopewell PC. Imipenem for treatment of tuberculosis in mice and humans. Antimicrob. Agents Chemother. 2005;49:2816–2821. doi: 10.1128/AAC.49.7.2816-2821.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chambers HF, Moreau D, Yajko D, Miick C, Wagner C, Hackbarth C, Kocagoz S, Rosenberg E, Hadley WK, Nikaido H. Can penicillins and other beta-lactam antibiotics be used to treat tuberculosis? Antimicrob. Agents Chemother. 1995;39:2620–2624. doi: 10.1128/aac.39.12.2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goffin C, Ghuysen JM. Biochemistry and comparative genomics of SxxK superfamily acyltransferases offer a clue to the mycobacterial paradox: presence of penicillin-susceptible target proteins versus lack of efficiency of penicillin as therapeutic agent. Microbiol. Mol. Biol. Rev. 2002;66:702–738. doi: 10.1128/MMBR.66.4.702-738.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nguyen-Disteche M, Fraipont C, Buddelmeijer N, Nanninga N. The structure and function of Escherichia coli penicillin-binding protein 3. Cell Mol. Life Sci. 1998;54:309–316. doi: 10.1007/s000180050157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weiss DS, Chen JC, Ghigo JM, Boyd D, Beckwith J. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J. Bacteriol. 1999;181:508–520. doi: 10.1128/jb.181.2.508-520.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 20.Dasgupta A, Datta P, Kundu M, Basu J. The serine/threonine kinase PknB of Mycobacterium tuberculosis phosphorylates PBPA, a penicillin-binding protein required for cell division. Microbiology. 2006;152:493–504. doi: 10.1099/mic.0.28630-0. [DOI] [PubMed] [Google Scholar]
- 21.Holtje JV. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev. 1998;62:181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weiss DS. Bacterial cell division and the septal ring. Mol. Microbiol. 2004;54:588–597. doi: 10.1111/j.1365-2958.2004.04283.x. [DOI] [PubMed] [Google Scholar]
- 23.Wissel MC, Weiss DS. Genetic analysis of the cell division protein FtsI (PBP3): amino acid substitutions that impair septal localization of FtsI and recruitment of FtsN. J. Bacteriol. 2004;186:490–502. doi: 10.1128/JB.186.2.490-502.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pares S, Mouz N, Petillot Y, Hakenbeck R, Dideberg O. X-ray structure of Streptococcus pneumoniae PBP2x, a primary penicillin target enzyme. Nat. Struct. Biol. 1996;3:284–289. doi: 10.1038/nsb0396-284. [DOI] [PubMed] [Google Scholar]
- 25.Lobkovsky E, Moews PC, Liu H, Zhao H, Frere JM, Knox JR. Evolution of an enzyme activity: crystallographic structure at 2-A resolution of cephalosporinase from the ampC gene of Enterobacter cloacae P99 and comparison with a class A penicillinase. Proc. Natl. Acad. Sci. U S A. 1993;90:11257–11261. doi: 10.1073/pnas.90.23.11257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goffin C, Ghuysen JM. Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol. Mol. Biol. Rev. 1998;62:1079–1093. doi: 10.1128/mmbr.62.4.1079-1093.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davies C, White SW, Nicholas RA. Crystal structure of a deacylation-defective mutant of penicillin-binding protein 5 at 2.3 Å resolution. J. Biol. Chem. 2001;276:616–623. doi: 10.1074/jbc.M004471200. [DOI] [PubMed] [Google Scholar]
- 28.Nicholas RA, Krings S, Tomberg J, Nicola G, Davies C. Crystal structure of wild-type penicillin-binding protein 5 from E. coli: Implications for deacylation of the acyl-enzyme complex. J. Biol. Chem. 2003;278:52826–52833. doi: 10.1074/jbc.M310177200. [DOI] [PubMed] [Google Scholar]
- 29.Powell AJ, Tomberg J, Deacon AM, Nicholas RA, Davies C. Crystal structures of penicillin-binding protein 2 from penicillin-susceptible and - resistant strains of Neisseria gonorrhoeae reveal an unexpectedly subtle mechanism for antibiotic resistance. J. Biol. Chem. 2009;284:1202–1212. doi: 10.1074/jbc.M805761200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gordon E, Mouz N, Duee E, Dideberg O. The crystal structure of the penicillin-binding protein 2x from Streptococcus pneumoniae and its acylenzyme form: implication in drug resistance. J. Mol. Biol. 2000;299:477–485. doi: 10.1006/jmbi.2000.3740. [DOI] [PubMed] [Google Scholar]
- 31.Rybkine T, Mainardi JL, Sougakoff W, Collatz E, Gutmann L. Penicillin-binding protein 5 sequence alterations in clinical isolates of Enterococcus faecium with different levels of beta-lactam resistance. J. Infect. Dis. 1998;178:159–163. doi: 10.1086/515605. [DOI] [PubMed] [Google Scholar]
- 32.Sauvage E, Kerff F, Fonze E, Herman R, Schoot B, Marquette JP, Taburet Y, Prevost D, Dumas J, Leonard G, Stefanic P, Coyette J, Charlier P. The 2.4 Å crystal structure of the penicillin-resistant penicillin-binding protein PBP5fm from Enterococcus faecium in complex with benzylpenicillin. Cell. Mol. Life Sci. 2002;59:1223–1232. doi: 10.1007/s00018-002-8500-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dessen A, Mouz N, Gordon E, Hopkins J, Dideberg O. Crystal structure of PBP2x from a highly penicillin-resistant Streptococcus pneumoniae clinical isolate: a mosaic framework containing 83 mutations. J. Biol. Chem. 2001;276:45106–45112. doi: 10.1074/jbc.M107608200. [DOI] [PubMed] [Google Scholar]
- 34.Silvaggi NR, Anderson JW, Brinsmade SR, Pratt RF, Kelly JA. The crystal structure of phosphonate-inhibited D-Ala-D-Ala peptidase reveals an analogue of a tetrahedral transition state. Biochemistry. 2003;42:1199–1208. doi: 10.1021/bi0268955. [DOI] [PubMed] [Google Scholar]
- 35.Nicola G, Peddi S, Stefanova ME, Nicholas RA, Gutheil WG, Davies C. Crystal structure of Escherichia coli penicillin-binding protein 5 bound to a tripeptide boronic acid inhibitor: a role for Ser-110 in deacylation. Biochemistry. 2005;44:8207–8217. doi: 10.1021/bi0473004. [DOI] [PubMed] [Google Scholar]
- 36.Nicola G, Fedarovich A, Nicholas RA, Davies C. A large displacement of the SXN motif of Cys115-modified penicillin-binding protein 5 from Escherichia coli. Biochem. J. 2005;392:55–63. doi: 10.1042/BJ20050449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Contreras-Martel C, Job V, Di Guilmi AM, Vernet T, Dideberg O, Dessen A. Crystal structure of penicillin-binding protein 1a (PBP1a) reveals a mutational hotspot implicated in beta-lactam resistance in Streptococcus pneumoniae. J. Mol. Biol. 2006;355:684–696. doi: 10.1016/j.jmb.2005.10.030. [DOI] [PubMed] [Google Scholar]
- 38.Job V, Carapito R, Vernet T, Dessen A, Zapun A. Common alterations in PBP1a from resistant Streptococcus pneumoniae decrease its reactivity toward beta-lactams: structural insights. J. Biol. Chem. 2008;283:4886–4894. doi: 10.1074/jbc.M706181200. [DOI] [PubMed] [Google Scholar]
- 39.Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol. 1993;229:105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- 40.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Meths. Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 41.Terwilliger TC. Automated structure solution, density modification and model building. Acta Cryst. 2002;D58:1937–1940. doi: 10.1107/s0907444902016438. [DOI] [PubMed] [Google Scholar]
- 42.Jones TA, Zou J-Y, Cowan SW, Kjeldgaard M. Improved methods for building protein structures in electron-density maps and the location of errors in these models. Acta Cryst. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 43.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Cryst. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 44.Brünger AT. Free R value: a novel statistical quantity for assessing the accuracq gf crystal structures. Nature. 1992;355:472–474. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 45.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 46.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- 47.Kraulis PJ. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Cryst. 1991;24:946–950. [Google Scholar]
- 48.Merritt EA, Murphy MEP. Raster3D version 2.0. A program for photorealistic molecular graphics. Acta Cryst. 1994;D50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.