Abstract
The major mood disorders, which include bipolar disorder (BD) and major depressive disorder (MDD), are substantially heritable, but few risk loci have been identified. We performed a meta-analysis of 5 major mood disorder case-control samples, including over 13,600 unique individuals genotyped with approximately 500,000 to 1 million single nucleotide polymorphism (SNP) markers on high-density arrays. Allele-wise association results were meta-analyzed with a method that weights results by sample size. We found genome-wide significant evidence that SNPs in a region of chromosome 3p21.1were associated with major mood disorders. The SNP rs2251219 returned the smallest meta-analysis p-value, 3.63 × 10−8, with a pooled odds ratio of 0.87. Supportive results were observed in 2 out of 3 independent samples tested in a replication study. These results implicate one or more genes in this region in the etiology of major mood disorders and suggest that BD and MDD share genetic risk factors.
Keywords: bipolar, depression, GAIN, STAB1, NT5DC2, PBRM1, NEK4, SPCS1, GNL3, GLT8D1, ITIH
The major mood disorders have a total lifetime prevalence up to 20%, and may soon become the leading cause of morbidity worldwide1. Similarities in symptoms and treatment response, twin concordance, and shared familial risk (reviewed in ref. 2), have long fed the suspicion that the major mood disorders share genetic risk factors, but molecular evidence remains scarce. Genome-wide association study (GWAS) data offer the opportunity to take a fresh look at genetic factors involved in these common disorders.
Samples (see Table 1) consisted of cases with a major mood disorder and controls, all of European ancestry, ascertained and genotyped as previously described(see Methods). One published GWAS of MDD 3 was not included in the meta-analysis, because complete results were not available, but key SNPs were tested later in a replication study. SNP genotype data were obtained from dbGaP (National Institute of Mental Health BD [NIMH-BP] and Genetic Association Information Network [GAIN-MDD] samples), the Wellcome Trust Case Control Consortium (WTCCC) sample, and through collaborators (German sample). Data from the Systematic Treatment Enhancement Program for Bipolar Disorder (STEP-BD) sample were obtained from the authors’ website. Data from the NIMH-BP, GAIN-MDD, and German samples were used to impute genotypes for about two million HapMap Phase 2 markers. The WTCCC and STEP-BD samples were both genotyped on the same platform, so only the observed data were used.
Table 1.
Descriptive statistics for the samples analyzed. WTCCC: Wellcome-Trust Case Control Consortium bipolar disorder sample; GAIN Depression: Genetic Association Information Network major depressive disorder sample; STEP-BD: STEP-BD bipolar disorder sample; NIMH Bipolar: NIMH bipolar disorder sample; German: German bipolar disorder sample. Counts refer to subjects who passed all quality control filters (see Methods).
| Sample | Cases | Case Diagnosis | Controls | Platform |
|---|---|---|---|---|
| WTCCC | 1854 | Bipolar I, bipolar II, schizoaffective bipolar | 2943 | Affymetrix 500K |
| GAIN MDD | 1722 | Major depressive disorder | 1774 | Perlegen |
| STEP-BD | 1461 | Bipolar I, bipolar II, schizoaffective bipolar | 2008 | Affymetrix 500K |
| NIMH Bipolar | 1001 | Bipolar I, schizoaffective bipolar | 1033 | Affymetrix 6.0 |
| German | 645 | Bipolar I | 1310 | Illumina HumanHap 550 |
| Total | 6683 | 9068 | ||
A total of 317,889 markers could be reliably scored across 4 or 5 samples (Figure 1a). The final results revealed no evidence of residual bias, with a mean genome-wide Z-score of 0.005, close to the theoretical null value of 0 (Supplementary Fig 1). Complete meta-analysis results are presented in Supplementary Table 1.
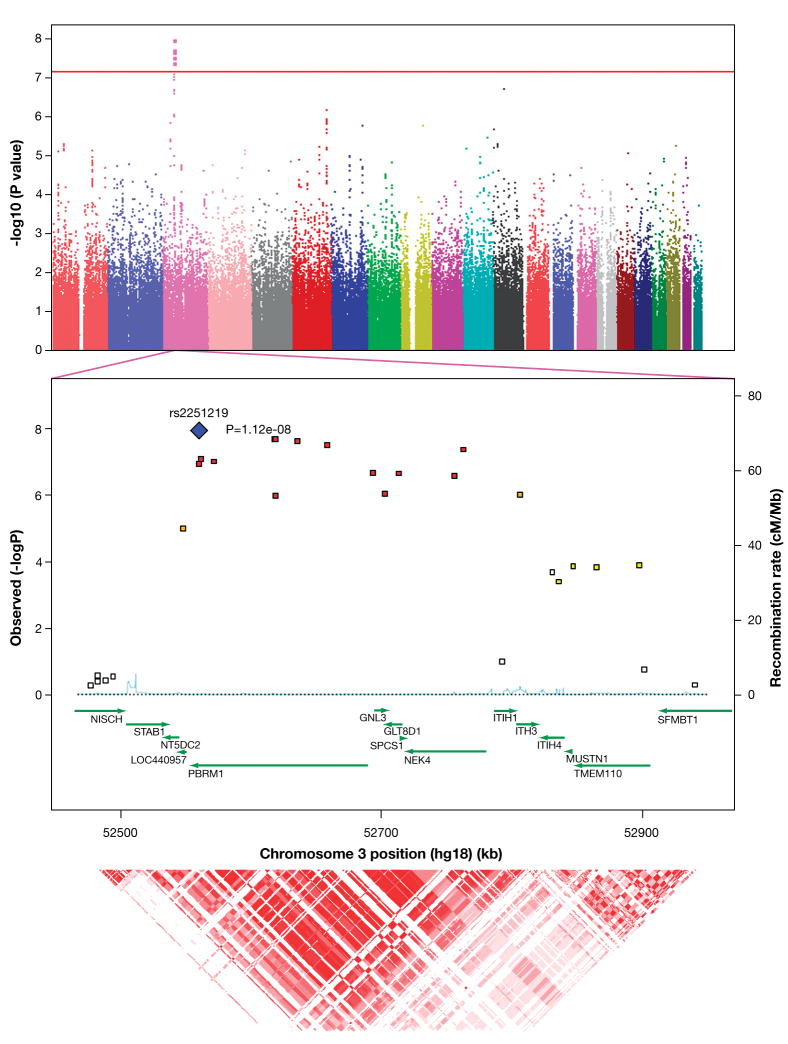
Figure 1.
a. Manhattan plot of the meta-analysis results, generated by Haploview 4.0. Physical position is shown along the x-axis, -log (meta-p-value) is shown along the y-axis, and each chromosome is shown in a distinct color. The red guideline indicates the threshold of genome-wide significance (7.2 ×10−8). b. Detail of the associated region, generated by SNAP 2.0. Physical position and gene annotations (HapMap release 22) are shown along the x-axis, -log (meta-p-value) is shown on the left y-axis, recombination rate (CEU) on the right y-axis. c. Linkage disequilibrium (r2) as estimated from HapMap 3 phased genotypes, generated by UCSC Genome Browser. Darker red indicates higher values.
Six SNPs on chromosome 3p21 were associated with major mood disorder at the p < 7.2 × 10−8 level (Figure 1b; Supplementary Table 2). This corresponds to a genome-wide corrected p<0.05 in samples of European ancestry4. At the most significant marker, rs2251219, the C-allele was consistently less common in cases than controls (p-value = 1.12 × 10−8; Table 2).
Table 2.
Association results for rs2251219 in each sample and in the meta-analysis.
| Sample | Allele | Frequency (cases) | Frequency (controls) | Allelic Chi-square | Allelic p-value | OR | N |
|---|---|---|---|---|---|---|---|
| Discovery Samples | |||||||
| NIMH BP | C | 0.39 | 0.41 | 1.43 | 0.2312 | 0.93 | 2034 |
| German BP | C | 0.38 | 0.44 | 9.51 | 0.0020 | 0.81 | 1955 |
| STEP-BD | C | 0.38 | 0.40 | 5.67 | 0.0172 | 0.89 | 1461 |
| WTCCC | C | 0.36 | 0.40 | 13.68 | 0.0002 | 0.85 | 4797 |
| GAIN MDD | C | 0.39 | 0.42 | 4.98 | 0.0256 | 0.90 | 3496 |
| Pooled discovery | 0.38 | 0.41 | 1.1 × 10−8 | 0.87 | 13743 | ||
| Replication Samples | |||||||
| GSK-BP | C | not available | 0.0023 | 0.57 | 1536 | ||
| GSK-Lausanne | C | 0.3219 | 1.21 | 1349 | |||
| GSK-Munich | C | 0.1227 | 0.79 | 1792 | |||
| Pooled Replication | 0.0012 | 0.84 | 4677 | ||||
| Pooled Discovery and Replication | 1.7 × 10−9 | 18420 | |||||
Observed genotypes at rs2251219 were available in all samples except for GAIN-MDD. For that sample, we used imputed genotypes at rs2251219, but the imputation is expected to be highly reliable. Imputation methods perform well in European-ancestry samples5, and there was strong linkage disequilibrium (LD) among the markers flanking rs2251219 (r2 ≥ 0.98). One nearby SNP that was genotyped in the GAIN-MDD sample, rs2289247, returned a meta-analysis p-value of 8.96 × 10−7, similar to rs2251219. Random masking of 5% of genotypes had little effect on the imputation results (data not shown). Imputation statistics are presented in Supplementary Table 2.
The analysis of rs2251219 was repeated under a random effects model, with similar results (p=3.62 × 10−8; Supplementary Table 3). The pooled OR was 0.87 (95% CI: 0.83–0.92), with no evidence of heterogeneity (Q=2.93, df=3, p=ns; I2 = 0). GAIN-MDD returned a similar OR of 0.90 (95% CI: 0.82–0.99), consistent with a similar contribution of this locus to both BP and MDD.
To assess robustness, we repeated the analysis 5 times, removing one sample each time. Random effects p-values at rs2251219 ranged from 10−6 – 10−9, reflecting varying sample sizes, but the ORs remained stable. (Supplementary Table 3). Thus it appears that the results are not driven by any single sample.
We subsequently obtained association results at rs2251219 in an independent BP case-control sample (n=1536) genotyped by GlaxoSmithKline (GSK)6. The C-allele of rs2251219 was significantly under-represented in BP cases compared to controls (p=0.002, OR = 0.57; Table 2), replicating our main meta-analysis result. Data from two independent MDD case-control samples were also obtained from GSK3: A clinical sample from Munich (n=1792) and a population-based sample from Lausanne (n=1349). No significant association with rs2251219 was found in either MDD sample, although in the larger one the 95% CI of the OR (0.83–1.77) substantially overlapped with that of the present study (0.83–0.92, Table 2). Additional, nearby markers also showed evidence of association in one or more samples (data not shown). When all 3 samples were combined with our original results, the evidence of association at rs2251219 increased (fixed effects p = 1.67 × 10−9; random effects p=4.99 × 10−9). These data provide support for our findings in independent samples, but the findings are more robust in BP than in MDD.
The association signals on chromosome 3p span a ≥246 kb region containing several annotated transcripts (Figure 1b). SNP rs2251219 is a synonymous variant in the gene PBRM1, which encodes polybromo-1, important in chromatin remodeling7. The nearby SNP rs2289247 is a non-synonymous (V → M) variant in the gene GNL3, encoding the GTP-ase nucleostemin, involved in proliferation of stem cells, especially in the central nervous system8. These genes are good biological candidates, but the LD across the locus is very strong (r2 > 0.9; Figure 1c), complicating efforts to localize functional marker(s) in individual genes by association mapping alone.
To help prioritize genes for further study, we examined gene expression in brain tissue. PBRM1 was over-expressed in the dorsolateral prefrontal cortex of patients with BP (p=0.018; Supplementary Figure 2), compared to healthy controls. This finding, not confounded by linkage disequilibrium in the region, independently implicates PBRM1, at least in BP. We also mined 2 brain expression datasets9,10 to test 22 disease-associated SNPs for cis-association with expression of all known transcripts in the 3p region. Apparent associations between rs2251219 and expression of PBRM1 and GLT8D1 involved probes that overlap common SNPs, an important source of artifact11. Neither were confirmed by qPCR in the Stanley Brain Foundation Samples, and the GLT8D1 finding could not be confirmed in cDNA from the original study9 using probes that did not overlap known SNPs (data not shown).
This is the first psychiatric study to find genome-wide significant association on chromosome 3p. Previous studies have detected suggestive evidence of association in this region6,12. A meta-analysis of BP GWAS in samples that overlap with those in the present study found suggestive signals6; the most significant SNP in that study was nominally significant in the present study (rs1042779; p=0.001). Considering the strong local LD, all of these studies may have detected signals arising from the same risk allele(s).
Previous major mood disorder GWAS have highlighted several genes elsewhere in the genome We detected association with most of these genes (Supplementary Table 4), but our results are not independent replications since the samples we used overlap with those in the previous studies. No signal was significant after genome-wide correction for multiple testing. If many genes play a role in risk, signals may vary from study to study, reflecting small sample differences and other unmeasured factors Thus, even this large sample may be too small or heterogeneous to detect all important risk alleles at genome-wide significance. A complementary analysis is expected in the future, when the Psychiatric GWAS Consortium13 completes its study of fully-imputed data in these and other samples.
We report molecular support for the prior epidemiologic evidence of genetic overlap between BP and MDD2. These data do not explain why, among all those carrying risk allele(s), some develop BP, others develop MDD, and still others remain apparently well. This phenomenon may reflect a large number of risk alleles, few of which have been detected to date, environmental influences and -- perhaps -- epigenetic factors.
The genetic association findings to date seem to account for little of the inherited risk for mood disorders. Since GWA studies usually omit SNPs with minor allele frequencies below 3–5%, we can say nothing about alleles in that frequency range, even if they have relatively strong effects. If rare alleles of large effect exist, each can account for only a small proportion of cases. More common autosomal alleles (frequency >20%) conferring a heterozygote relative risk of 1.3 would have been detected with >90% power in this sample. Since we found no such alleles, they probably do not exist in this sample, the largest mood disorder sample studied to date. Many loci may add together to confer risk, as some studies suggest14,15, or fewer loci may interact, but to our knowledge, strong epistasis has not yet been demonstrated in complex human traits.
The genetic architecture of the major mood disorders appears to be multi-genic and/or highly heterogeneous. As robust findings accumulate and sample sizes grow, the identified genes may triangulate pathways of etiologic relevance. The GWAS remains an important means to that end, even though each individual finding may represent only a small step forward.
METHODS
Study Samples
The samples used in the meta-analysis have been described previously16–19. Details, including quality control procedures applied to the genotyping data and a description of the replication samples, is provided as a supplement to the published report (Supplementary Note).
Imputation
Genotype data from the NIMH-BP, GAIN-MDD, and German samples were used to impute data on 2.1 million HapMap Phase 2 SNPs by use of the program Markov Chain Haplotyping (MACH), version 1.0 20. MACH uses Markov chain haplotyping to resolve haplotypes, and thereby missing genotypes, from observed genotypes in unrelated individuals. We used the “greedy” algorithm, as recommended by the authors. SNPs that were flagged as having different alleles than in HapMap CEU or as monomorphic were reviewed, after which they were either recoded for the reverse strand (flipped) or dropped. SNPs that were flagged for allele frequencies that were markedly different from HapMap CEU were also reviewed. Palindromic SNPs whose allele frequencies were consistent with reversed coding were flipped. Other SNPs with unexpected allele frequencies were dropped. PLINK21 (vers. 1.4) was used to flip and drop SNPs as necessary. After all allele-coding, monomorphism, and palindrome issues were resolved, imputation was run again. SNPs in the results files were dropped if the MAF in cases or controls was <0.05 or if the error rate (as reported in the .erate output file) was >0.01. Finally, the imputed data were formatted into PLINK binaries for analysis.
Meta-analysis
PLINK output (.assoc) files were modified with columns for direction of association, sample size, and strand. For most samples, sample size equaled the sum of cases and controls included in the final analysis, after quality control. For the STEP-BD sample, sample size was set to equal the number of cases only. This was done to avoid over-weighting the results from the NIMH control sample, largely overlapping portions of which were included in both the NIMH bipolar disorder and STEP-BD samples. Modified files were loaded into Metal (July, 2008 version), then processed using the GENOMICCONTROL option, which applies a genomic control22 correction in samples where the genomic inflation factor is greater than 1.0. Metal weights each sample based on the square root of the sample size.
Great care was taken in combining results from different samples and platforms to avoid confusing alleles, especially at palindromic SNPs. To check this, we inspected all SNPs whose range of allele frequencies in the meta-analysis was greater than 0.2, a commonly-used threshold. Most of these SNPs were palindromes and, as expected, their minimum and maximum allele frequencies across the study samples added to approximately 1. Using the “STRANDLABEL” and “USESTRAND ON” commands in METAL, these SNPs were recoded to ensure consistent allele coding across the samples analyzed. Because the German sample was genotyped on the Illumina platform that contains no palindromic SNPs, we used that sample as the gold standard. A final check identified 36 SNPs whose allele frequency ranges still exceeded 0.2. Most of these were not palindromes, and those that were did not show complementary allele frequencies. We concluded these were unreliable SNPs and dropped them from the analysis.
Selected results were confirmed, and heterogeneity statistics were calculated, using Comprehensive Meta-analysis version 2.0. This performs a random-effects meta-analysis that is robust to sample heterogeneity, as well as explicit tests of heterogeneity using Q and I statistics.23 In order to account for overlap in control samples, we grouped cases from the NIMH-BP and STEP-BD samples (which do not overlap), and compared them, as a whole, with the controls used in the STEP-BD report, as recommended by some authorities24,25.
We used a threshold of genome-wide significance (7.2 × 10−8) derived from a published, genome-wide simulation of common variants in samples of European ancestry4. This threshold is more conservative than the value of 1.6 × 10−7 that would represent a Bonferroni-corrected p=0.05 for the approx. 318,000 markers tested, but is consistent with accepted thresholds for genome-wide significance26. Although we set out to find variants shared in common between BP and MDD, we have considered only BP in other studies16. The results at rs2251219 would remain significant even if multiplied by 2 to account for this.
Power Analysis was done with Genetic Power Calculator27. We assumed a trait prevalence of 2%, minor allele frequency of 20%, an alpha of 7.2 × 10−8, and a marker-allele D’ value of 0.8.
Gene Expression Analysis
Brain RNA and genomic DNA samples were obtained from the Stanley Medical Research Institute (SMRI). This collection comprises 3 diagnostic groups, each with 35 samples: healthy control, bipolar disorder and schizophrenia. All experiments were done after the specimen code was broken and they were thus unblinded. RNA samples originated from the dorsolateral prefrontal cortex. Of these, 101 samples provided sufficient RNA for reverse transcription (Transcriptor First Strand cDNA Synthesis kit with oligo-dT priming, Roche Applied Science, Indianapolis, IN) and subsequent real-time PCR analysis. We amplified PBRM1 mRNA with a primer pair common to all known RefSeq transcript variants (Eurofins MWG operon, Huntsville, AL) and a FAM-labeled probe (Roche Applied Science Universal Probe Libraray probe # 41). No known SNPs overlapped with primer or probe sequences (Supplementary Figure 2). For normalization, we used a pre-designed endogenous control assay interrogating PGK1 (Applied Biosystems, Foster City, CA; catalog number 4333765F, FAM-labeled). Reactions were carried out in triplicate in a 384-well LightCycler 480 (Roche Applied Science) in reaction volumes of 8 uL, with 5 ng of reverse-transcribed RNA, 1x final concentration of Roche LightCycler 480 Probes master mix, 450 nM of each primer, and 125 nM of fluorescent probe. Assay efficiencies were determined using two-fold serial dilutions of pooled cDNA. Relative expression levels for PBRM1 were calculated using the efficiency-corrected comparative threshold method 28. We used the sample with the median PBRM1 expression level as calibrator and log2-transformed expression levels. One hundred samples were successfully assayed. (As recommended by SMRI, one case sample was omitted due to a known degenerative neurological disorder). Of the remaining 99 samples (31 with bipolar disorder, 34 with schizophrenia, and 33 healthy controls), 97 were from donors of European ancestry. Ninety-six of these samples were successfully genotyped at rs2251219 with standard exonuclease methods (TaqMan, Applied Biosystems, Boston, MA). Data were analyzed by ANOVA (Xlstat 6.0), with diagnosis as the independent, and relative expression level as the dependent, variable. Specific comparisons were performed with the Tukey HSD test. The entire data set has been uploaded to the SMRI data bank.
Supplementary Material
Acknowledgments
Funded by the National Institute of Mental Health (NIMH) Intramural Research Program, Deutsche Forschungsgemeinschaft (DFG), the National Genome Research Network (NGFN) of the Federal German Ministry of Education and Research, NARSAD (Independent Investigator Award to FJM and Young Investigator Award to TGS), the Alfried Krupp von Bohlen und Halbach-Stiftung, the DFG-Graduate College 793, University of Heidelberg, AND grants from the NIMH and National Human Genome Research Institute (NHGRI) to JRK (MH078151, MH081804, MH059567 supplement). This research was also supported, in part, by the Intramural Research Program of the National Library of Medicine, NIH. The replication samples were supported by the Swiss National Science Foundation (#3200B0 105993, #32003B-118308 and #33CSCO-122661) and GlaxoSmithKline (Psychiatry Center of Excellence for Drug Discovery and Genetics Division, Verona).
Genotyping of the GAIN major depression and NIMH bipolar disorder samples was provided through the Genetic Association Information Network (GAIN), Foundation for NIH. The datasets used for the analyses described in this manuscript were obtained from the database of Genotypes and Phenotypes. Samples and associated phenotype data were provided by the contributing studies.
We thank the Wellcome Trust Case Control Consortium, the STEP-BD group, the Netherlands Study of Depression and Anxiety, and the Netherlands Twin Registry for making data/results available for analysis.
Unless noted otherwise, gene annotations reflect the UCSC Genes track in UCSC Genome Browser, March 2006 Build. This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, MD. Postmortem brain tissue was supplied by the Stanley Medical Research Institute.
Additional acknowledgements are included in the Supplementary Material.
Footnotes
DATABASE ACCESSION NUMBERS
NIMH bipolar disorder sample: dbGAP phs000017.v1.p1.c1-c3
GAIN major depression sample: dbGAP phs000020.v1.p1
Author Contributions
F.J.M. designed the study, led the analysis, and wrote the manuscript. N.A., T.G.S., and C.J.M.S. contributed to the data management and analysis. S.D.-W., T.G.S., P.M., W.M., F.H., M.R., J.I.N., and H.J.E. edited the manuscript. J.R.W. performed the gene expression experiments. P.M., F.T., R.B., J.S., M.M., T.W.M., W.M, M.M.N., S.C., A.F., J.B.V., F.H., M.P., M.R., and BiGS collected samples and/or shared genetic association results. All authors reviewed the manuscript.
URLs
MACH, http://www.sph.umich.edu/csg/abecasis/MACH/; METAL, http://www.sph.umich.edu/csg/abecasis/Metal/; Haploview, http://www.broad.mit.edu/mpg/haploview; SNAP, http://www.broad.mit.edu/mpg/snap; UCSC Genome Browser, http://genome.ucsc.edu; HapMap, http://www.hapmap.org; PLINK, http://pngu.mgh.harvard.edu/~purcell/plink/; Comprehensive Meta-analysis, http://www.meta-analysis.com; Genetic Power Calculator, http://pngu.mgh.harvard.edu/~purcell/gpc/; Xlstat, http://www.xlstat.com; Stanley Medical Research Institute, http://www.stanleyresearch.org; dbGAP, http://www.ncbi.nlm.nih.gov/gap
References
- 1.Ustun TB, Ayuso-Mateos JL, Chatterji S, Mathers C, Murray CJ. Global burden of depressive disorders in the year 2000. Br J Psychiatry. 2004;184:386–92. doi: 10.1192/bjp.184.5.386. [DOI] [PubMed] [Google Scholar]
- 2.Goodwin FK, Jamison K. Manic-Depressive Illness. New York: Oxford Univ Press; pp. 8–26. [Google Scholar]
- 3.Muglia P, et al. Genome-wide association study of recurrent major depressive disorder in two European case-control cohorts. Mol Psychiatry. 2008 doi: 10.1038/mp.2008.131. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 4.Dudbridge F, Gusnanto A. Estimation of significance thresholds for genomewide association scans. Genet Epidemiol. 2008;32:227–34. doi: 10.1002/gepi.20297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang L, et al. Genotype-imputation accuracy across worldwide human populations. Am J Hum Genet. 2009;84:235–50. doi: 10.1016/j.ajhg.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scott LJ, et al. Genome-wide association and meta-analysis of bipolar disorder in individuals of European ancestry. Proc Natl Acad Sci U S A. 2009;106:7501–6. doi: 10.1073/pnas.0813386106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thompson M. Polybromo-1: the chromatin targeting subunit of the PBAF complex. Biochimie. 2009;91:309–19. doi: 10.1016/j.biochi.2008.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsai RYL, McKay RDG. A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes Dev. 2002;16:2991–3003. doi: 10.1101/gad.55671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Myers AJ, et al. A survey of genetic human cortical gene expression. Nat Genet. 2007;39:1494–9. doi: 10.1038/ng.2007.16. [DOI] [PubMed] [Google Scholar]
- 10.Heinzen EL, Ge D, Cronin KD, Maia JM, Shianna KV, Gabriel WN, Welsh-Bohmer KA, Hulette CM, Denny TN, Goldstein DB. Tissue-specific genetic control of splicing: Implications for the study of complex traits. PLoS Biol. 2008;6:e1. doi: 10.1371/journal.pbio.1000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doss S, Schadt EE, Drake TA, Lusis AJ. Cis-acting expression quantitative trait loci in mice. Genome Res. 2005;15:681–91. doi: 10.1101/gr.3216905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moskvina V, et al. Gene-wide analyses of genome-wide association data sets: Evidence for multiple common risk alleles for schizophrenia and bipolar disorder and for overlap in genetic risk. Mol Psychiatry. 2009;14:252–60. doi: 10.1038/mp.2008.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Psychiatric GWAS Consortium. A framework for interpreting genome-wide association studies of psychiatric disorders. Mol Psychiatry. 2009;14:10–7. doi: 10.1038/mp.2008.126. [DOI] [PubMed] [Google Scholar]
- 14.Baum AE, et al. A genome-wide association study implicates diacylglycerol kinase eta (DGKH) and several other genes in the etiology of bipolar disorder. Mol Psychiatry. 2008;13:197–207. doi: 10.1038/sj.mp.4002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.International Schizophrenia Consortium. Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–52. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith EN, et al. Genome-wide association study of bipolar disorder in European American and African American individuals. Mol Psychiatry. 2009;14:755–63. doi: 10.1038/mp.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boomsma DI, et al. Genome-wide association of major depression: description of samples for the GAIN Major Depressive Disorder Study: NTR and NESDA biobank projects. Eur J Hum Genet. 2008;16:335–42. doi: 10.1038/sj.ejhg.5201979. [DOI] [PubMed] [Google Scholar]
- 18.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sklar P, et al. Whole-genome association study of bipolar disorder. Mol Psychiatry. 2008;13:558–69. doi: 10.1038/sj.mp.4002151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, Abecasis GR. MACH 1.0: Rapid haplotype reconstruction and missing genotype inference. Am J Hum Genet. 2006;S79:2290. [Google Scholar]
- 21.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Devlin B, Roeder K. Genomic Control for Association Studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 23.Ioannidis JP. Interpretation of tests of heterogeneity and bias in meta-analysis. J Eval Clin Pract. 2008;14:951–7. doi: 10.1111/j.1365-2753.2008.00986.x. [DOI] [PubMed] [Google Scholar]
- 24.Sagoo GS, Tatt I, Salanti G, Butterworth AS, Sarwar N, van Maarle M, Jukema JW, Wiman B, Kastelein JJ, Bennet AM, de Faire U, Danesh J, Higgins JP. Seven lipoprotein lipase gene polymorphisms, lipid fractions, and coronary disease: A HuGE association review and meta-analysis. Am J Epidemiol. 2008;168:1233–46. doi: 10.1093/aje/kwn235. [DOI] [PubMed] [Google Scholar]
- 25.Littell JH, Corcoran J, Pillai VK. Systematic reviews and meta-analysis. Vol. 62. New York: Oxford Univ. Press; 2008. [Google Scholar]
- 26.Manolio TA, Brooks LD, Collins FS. A HapMap harvest of insights into the genetics of common disease. J Clin Invest. 2008;118:1590–605. doi: 10.1172/JCI34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19:149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- 28.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.