In this study, we investigate a role for p8 in autophagy in vitro and in vivo, by using p8 −/− mice. In both settings, silencing of p8 is associated with basal up-regulation of autophagy and apoptosis. In vivo, the hearts of p8 knockout mice develop features that provoke a decreased left ventricular functionality.
Abstract
Autophagy is a cytoprotective pathway used to degrade and recycle cytoplasmic content. Dysfunctional autophagy has been linked to both cancer and cardiomyopathies. Here, we show a role for the transcriptional regulator p8 in autophagy. p8 RNA interference (RNAi) increases basal autophagy markers in primary cardiomyocytes, in H9C2 and U2OS cells, and decreases cellular viability after autophagy induction. This autophagy is associated with caspase activation and is blocked by atg5 silencing and by pharmacological inhibitors. FoxO3 transcription factor was reported to activate autophagy by enhancing the expression of autophagy-related genes. P8 expression represses FoxO3 transcriptional activity, and p8 knockdown affects FoxO3 nuclear localization. Thus, p8 RNAi increases FoxO3 association with bnip3 promoter, a known proautophagic FoxO3 target, resulting in higher bnip3 RNA and protein levels. Accordingly, bnip3 knockdown restores cell viability and blocks apoptosis of p8-deficient cells. In vivo, p8 −/− mice have higher autophagy and express higher cardiac bnip3 levels. These mice develop left ventricular wall thinning and chamber dilation, with consequent impaired cardiac function. Our studies provide evidence of a p8-dependent mechanism regulating autophagy by acting as FoxO3 corepressor, which may be relevant for diseases associated with dysregulated autophagy, as cardiovascular pathologies and cancer.
INTRODUCTION
Autophagy is a cellular response to insufficient nutrient availability and stress, which involves the sequestration of cytosol and organelles within autophagosomes for their delivery to lysosomes (Xie and Klionsky, 2007; Klionsky et al., 2008). Through autophagy, cells recycle and generate new metabolic substrates and adapt to reduced nutrient availability (Kundu and Thompson, 2008; Meijer and Codogno, 2008). Autophagy eliminates toxic proteins, damaged and aged cells or organelles, and can eventually elicit a self-destructive system leading to cell death (Baehrecke, 2005; Maiuri et al., 2007). Accordingly, the disruption of this process is associated with multiple pathophysiological conditions, including cancer, diabetes, and cardiovascular diseases (Levine and Kroemer, 2008; Mizushima et al., 2008).
In unstressed cells, autophagy is inhibited by the mammalian target of rapamycin complex 1 (mTORC1). Nutrient depletion, serum starvation, rapamycin inhibition of mTORC1, or activation of 5′AMP-activated protein kinase (AMPK) by 5-aminoimidazole-4-carboxamide-1-d-ribofuranoside (AICAR) promotes the activation of a set of autophagy-regulating proteins (Atg). In turn, some Atgs contribute to the formation of the autophagosomes, which contain the cellular components destined for degradation (Klionsky et al., 2008; Kundu and Thompson, 2008).
Autophagy requires two ubiquitin-like systems, one leading to the conjugation of Atg12 to Atg5 and the second converting the microtubule-associated protein 1 light chain 3 (LC3 or Atg8) soluble form (LC3-I) to the autophagic vesicle-associated form (LC3-II) (Klionsky et al., 2008).
Aside from cellular relocalization and posttranslational modifications of Atgs, the autophagic process includes the activation of a transcriptional program (Attaix and Bechet, 2007). Up-regulation of lysosomal enzymes, such as cathepsin L, and of multiple atg genes has been reported in several cell systems and organisms (Attaix and Bechet, 2007; Meijer and Codogno, 2008). Indeed, the transcription factor E2F was shown to drive lc3, atg1, and dram expression in U2OS cells, whereas the transcription factor FoxO3 was shown to control the expression of bnip3, beclin1, and lc3 among others, in cultured myocytes and in vivo (Mammucari et al., 2007; Zhao et al., 2007; Polager et al., 2008). However, the mechanisms involved in the fine-tuning, repression, and termination of this cellular program are still poorly investigated.
In the heart, autophagy is important for the turnover of organelles at low basal levels under normal conditions (Nakai et al., 2007). Cardiac basal autophagy is altered by stress triggered by cardiovascular diseases, including ischemic injury, cardiac hypertrophy, cardiac remodeling, and heart failure (De Meyer and Martinet, 2008; Rothermel and Hill, 2008a). Basal autophagy seems to play a protective role, because atg7 silencing in rat neonatal cardiomyocytes reduces cell viability, and adult cardiomyocytes from cardiac-specific atg5-deficient hearts are more susceptible to isoproterenol (Nakai et al., 2007). Yet, its role in the heart is still poorly understood.
p8 (nupr1 or com1) is a nuclear basic helix-loop-helix protein strongly induced in response to stress (Chowdhury et al., 2009; Goruppi and Iovanna, 2009). p8 has been implicated in several diverse context-dependent functions, including transcriptional regulation, cell cycle control, diabetic renal and cardiomyocyte hypertrophy, as well as apoptotic regulation (Goruppi et al., 2002, 2007; Vasseur et al., 2002b; Quirk et al., 2003; Carracedo et al., 2006; Malicet et al., 2006; Salazar et al., 2009; Sambasivan et al., 2009). Accordingly, p8 acts as a transcriptional coactivator and interacts with members of the transcriptional machinery, including AP1 complex, p53, and p300, among others (Hoffmeister et al., 2002; Goruppi et al., 2007; Clark et al., 2008).
We have shown that p8 is induced in failing human hearts. p8 is a transcriptional regulator required for endothelin-1– and phenylephrine-induced hypertrophy in rat cardiomyocytes and for tumor necrosis factor (TNF)-induced activation of matrix metalloprotease (MMP) 9 in rat cardiac fibroblasts (Goruppi et al., 2007). Accordingly, p8 associates with and activates the MMP9 promoter in primary fibroblasts and cancer cells.
In this study, we investigate a role for p8 in autophagy in vitro and in vivo, by using the p8 −/− mice. In both settings, silencing of p8 is associated with basal up-regulation of autophagy and apoptosis. In vivo, the hearts of p8 knockout mice develop features that provoke a decreased left ventricular functionality.
MATERIALS AND METHODS
Cell Culture and Treatments
U2OS, 293, and H9C2 cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS). Primary rat cardiomyocytes, human cardiac fibroblasts, and human aortic endothelial cells were from ScienCell Research Laboratories (Carlsbad, CA). Rapamycin, AICAR, 2-deoxyglucose (2DG), 3-methyladenine, methyl pyruvate, TNF, zVADfmk, E64D, pepstatin, puromycin, and G418 were all from Calbiochem (San Diego, CA). Survival assays and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT; Calbiochem) labeling of the cells were performed as reported previously (Goruppi et al., 2001). In brief, the U2OS cells were seeded in 12-well tissue culture dishes and were transfected the following day, when 60–70% confluent, with either crtl or p8 small interfering RNA (siRNA) by using the Transit Plus transfection reagent (Mirus, Madison, WI), as indicated by the manufacturer. After 48 h, the cells were treated as indicated for further 48 h; the culture medium was then changed with fresh medium containing 10% FBS and 10 mg/ml MTT, and the cultures labeled for 4 h. All experiments were carried out in triplicate. Formazan salt products were solubilized in 10% SDS and 0.1 M HCl for the spectrophotometric quantification.
Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
Total RNA was isolated and RT-PCRs were performed as described in Goruppi et al. (2002). β-actin or gapdh served as controls. Primer sequences were as follows: for human p8, ATGGCCACCTTCCCACCAGCAA sense and GGCTGCCGTGCGTGTCTATTTA antisense; for mouse lc3, CGTCCTGGACAAGACCAAGT sense and ATGTGGGTGCCTACGTTCTC antisense; and for mouse bnip3, GGGTTTTCCCCAAAGGAATA sense and GACCACCCAAGGTAATGGTG antisense.
siRNA Interference, Cellular Fractionation, and Chromatin Immunoprecipitation (ChIP)
For human p8 siRNA, we used two pairs of oligonucleotides (OLs), which silenced both at a similar extent the endogenous p8. Rat p8 siRNA has been described in Goruppi et al. (2002). sc-37007, sc-41445, sc-37451 (Santa Cruz Biotechnology, Santa Cruz, CA) were used for control (ctrl), atg5, and bnip3 RNAi, respectively. In some experiments, control oligonucleotides were a mix of two nontargeting sequences. For ChIPs, 24 h after p8 or crtl RNAi U2OS cells were kept in 10% FBS or serum starved for 20 h, as indicated. ChIPs were performed using SimpleChIP Enzymatic Chromatin IP kit (Cell Signaling Technology, Danvers, MA). Human p8, OL1: GGAGGACCCAGGACAGGATTT sense and AAATCCTGTCCTGGGTCCTCC antisense, and OL2 were from Carracedo et al. (2006). Annealed siRNAs were introduced in the cells using Transit Plus transfection reagent (Mirus), as indicated by the manufacturer. In each experiment, the effective gene silencing was monitored by RT-PCR. p8 and control lentiviruses, which contain three to five target-specific hairpin constructs to knockdown gene expression (or a nontargeting mix control), were from Santa Cruz Biotechnology. U2OS cells were infected as indicated by the manufacturer and selected for one week with 3 μg/ml puromycin.
Nuclear/Cytosol (N/C) Separation.
U2OS cells either overexpressing or having p8 silenced or infected with crtl virus were grown to confluence in 10-cm cell culture dishes and then either serum starved for 24 h or left in complete culture medium (10% FBS). After this time, the cells were collected by scraping in 1 ml of ice-cold phosphate-buffered saline (PBS) containing protease inhibitors (BIOMOL Research Laboratories, Plymouth Meeting, PA), and the nuclear fractions were separated from cytosolic fractions using a Nuclear/Cytosol fractionation kit (BioVision, Mountain View, CA), as indicated by the manufacturer. The cellular lysates were quantified by Bradford assays (Bio-Rad Laboratories, Hercules, CA) and 30 μg of proteins for each sample was separated on SDS-polyacrylamide gel electrophoresis (PAGE) for Western blot analysis with the reported antibodies.
ChIP.
Endogenous FoxO3 or acetylated histone H3 (Lys9) were immunoprecipitated from soluble chromatin as described previously (Goruppi et al., 2007). Two sets of primers, amplifying the proximal and a distal FoxO binding regions (A and B, respectively), were used to amplify segments of the human bnip3 promoter from the copurified DNA: Oligo A, GACACGACTGTCTGGGAAGC sense and CGTGGAGGCACTTTTCAGAG antisense; and oligo B, TGCGCCAGCATGTAATACTC sense and GAGGATGGGGCTTTGGTACT antisense. Parallel ChIPs were carried out using same amounts of affinity purified nonimmune rabbit antibodies (Cell Signaling Technology), and the associated DNA was amplified as a background control. An aliquot of total chromatin was amplified to determine the equal amount of chromatin used in each assay.
Immunoprecipitations and Immunoblotting
U2OS cells stable transfected with either green fluorescent protein (GFP) or GFPp8 were grown to confluence in 15-cm Petri dishes and either serum starved for 24 h or left untreated (10% FBS). The Western blots were performed as described previously (Goruppi et al., 2007). Expression levels were normalized to loading controls by using ImageJ software (http://rsb.info.nih.gov/ij/). Immunoprecipitation: Cells were recovered by gentle scraping in 1 ml of ice-cold PBS containing protease inhibitors (Calbiochem), and total cellular lysates were prepared as described in Goruppi et al. (2007). Immunoprecipitations were carried out using a mix of two anti-p8 antibodies (5 μg each) (Goruppi and Kyriakis, 2004) together with 10 μl of Ultralink Plus Protein A/G (Pierce Chemical, Rockford, IL), and the immunocomplexes washed three times in lysis buffer before SDS-PAGE and Western blot analysis with the indicated antibodies. Total proteins were normalized before immunoprecipitation.
Western Blots.
Primary antibodies used include anti-LC3B, ATG12-5, caspase-3, cleaved caspase-3, cleaved rat caspase-9, poly(ADP-ribose) polymerase (PARP), cleaved PARP, Survivin, cIAP1, Sgk1, FoxO3, phosphorylated (p)-AMPKα, p-AMPKβ, p-S6, p-c-Jun NH2-terminal kinase, and p-Akt from Cell Signaling Technology, and antibodies against p8, Bnip3, FoxO3, GFP, glutathione transferase (GST), Lamin A, tubulin, and actin were from Santa Cruz Biotechnology and Goruppi and Kyriakis (2004). Appropriate peroxidase-conjugated secondary were from Santa Cruz Biotechnology or Jackson ImmunoResearch laboratories (West Grove, PA), whereas enhanced chemiluminescence reagents were from Boston BioProducts (Ashland, MA).
Stable Red-LC3 Cell Line and Autophagy Detection
U2OS were transfected with pHcRed1-LC3, encoding for the far-red fluorescent protein (Clontech, Mountain View, CA) fused to LC3 and for neomycin resistance (kindly provided by Alessia Di Nardo, Harvard Medical School, Boston MA). The percentage of cells having Red-LC3 translocation into dots (a minimum of 100 cells/sample, in three separate experiments) was counted in the Red-LC3–expressing U2OS cells fixed at room temperature with 3.7% paraformaldehyde (Sigma-Aldrich, St. Louis, MO) in PBS, containing 10 μM Hoechst 33342 for nuclear staining. Cells with only nuclear Red-LC3 were considered negatives, whereas cells with more than five dots were considered as positive (Klionsky et al., 2008; Herrero-Martin et al., 2009). Cells with only cytosolic dots were considered positive (if >5).
U2OS Red-LC3 Stable Cells.
Two days after transfection, the cells were divided and the following day they were selected using 0.5 mg/ml G418 (Calbiochem) for 2 weeks. Several clones were expanded and selected by immunofluorescence for homogeneous recombinant protein expression. The autophagosome induction in the cell line was confirmed by rapamycin and Hanks' balanced salt solution treatment for 3 h. In the untreated U2OS cells, the Red-LC3 was nuclear or diffusely distributed in the cytosol, and only 2% of the cells presented dotted staining. The percentage was slightly increased (4–5%) in the cells transfected with control (crtl) siRNA (data not shown).
For immunocytochemical imaging, images were obtained using an E800 microscope (Nikon, Tokyo, Japan) coupled with a digital camera (Hamamatsu, Bridgewater, NJ). The software used was OpenLab 5 (PerkinElmer, Waltham, MA) on a Mac OS 10.5.6 platform. Mounting medium used for the images was Advantage permanent mounting media (Axell, Westbury, NY). Objectives used for image capture were 40× and 63× (Nikon). For the preparation of the figures, images were processed using Photoshop CS (Adobe Systems, Mountain View, CA).
Transient Transfection, DNA Constructs, and Luciferase Assays
Transfection methods, pEGFP, pEGFPp8, and pcDNA His5 p8 have been described previously (Goruppi and Kyriakis, 2004). The luciferase reporter containing six copies of FoxO binding site and wild-type FoxO3 have been described previously (Brunet et al., 1999; Furuyama et al., 2000). For analysis of p8 and FoxO3 transactivation activity, U2OS cells were transfected in triplicate using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA), and the luciferase assays were carried out as described previously (Goruppi et al., 2007). A plasmid encoding for β-galactosidase, pCMV βgal (5 ng/well), was coexpressed to normalize the transfection efficiencies.
Animal Experiments
p8 −/− mice and wild-type, p8 +/+, littermate controls in C57BL/6J background have been described previously (Vasseur et al., 2002b). Ten- to 12-week-old age-matched animals were analyzed in the study, and the hearts were removed under deep anesthesia. For echocardiography, mice were imaged in the left lateral decubitus position by using a Phillips-Sonos 7500 echocardiography system equipped with 11-MHz sector array and 10- to 15-MHz linear array transducers. Digital echocardiographic images were analyzed with Phillips-based analysis software (qLAB) on XP-based platform coupled with the echocardiography system. Left ventricular end-diastolic diameter (LVEDD) and left ventricular end-systolic diameter (LVESD) were measured and fractional shortening (FS) was calculated as (LVEDD − LVESD/LVEDD) × 100%. All studies and analysis were made by investigators blinded to mice genotype. All procedures were performed according to the animal protocol approved by Tufts University Animal Care and Use Committee.
Statistical Analysis
Independent experiments were pooled when the coefficient of variance could be assumed to be equal. Statistical significance was evaluated with Prism 5.0 software (GraphPad Software, San Diego, CA) by using nonpaired, two-tailed student's t test (n is number of experiments or animals analyzed). p values below 0.05 were considered significant and if lower are indicated.
RESULTS
p8 Silencing Increases Autophagy
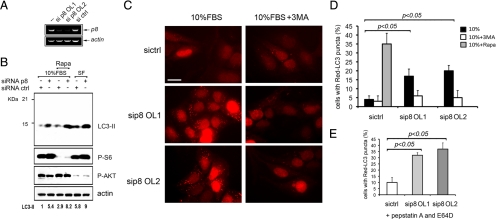
To determine whether p8 plays a role in autophagy, we used siRNA-mediated silencing to knock down U2OS cell p8 expression. Two sequences targeting human p8 were used, OL1 and OL2, with both oligonucleotides completely silencing p8 expression to a similar extent after 24 h, as detected by RT-PCR (Figure 1A). Serum starvation and inhibition of mTOR by rapamycin are known inducers of autophagy (Klionsky et al., 2008); we thus asked whether p8 could affect autophagy induced by these stimuli. LC3 is a key component of autophagic complex, and LC3-I to LC3-II processing is a prominent marker for autophagy (Klionsky et al., 2008). After p8 or ctrl RNAi, U2OS cells were either maintained for 48 h in complete medium (10% FBS) or autophagy was induced by addition of 100 nM rapamycin (Rapa) or by serum starvation (SF) for 16 h.
Figure 1.
p8 silencing causes an increase of autophagy. (A) p8 RNAi in U2OS cells. Cells were transfected with two different p8 siRNAs (OL1 and OL2) or with a mix of two nontargeting siRNA as control (ctrl). After 24 h total RNA was analyzed by RT-PCR, by using p8-specific and actin-specific oligonucleotides. (B) p8 RNAi causes LC3-II processing in U2OS cells. Western blot analysis of total cellular lysates from U2OS cells after RNAi as described in A, and either left untreated or treated with 100 nM rapamycin (Rapa) or serum starved (SF) for additional 24 h. Membranes were developed using anti LC3B, p-S6, p-Akt, and actin antibodies. In this experiment, p-S6, p-Akt antibodies were used as treatment controls for serum starvation and Rapa addition. The LC3B antibody has a stronger reactivity for LC3B-II form. Film was quantitated by densitometry and fold induction of LC3-II, normalized to total actin, is shown. (C) p8 silencing induces autophagosome formation in U2OS cells. Red-LC3–expressing U2OS cells after either OL1 or OL2 or crtl RNAi. The cells were fixed for the analysis after 48 h. When indicated, 10 mM 3MA or 100 nM rapamycin were added for 24 h. (C and D), Representative immunofluorescent images (63×; bar, 20 μm), and the percentage of cells with LC3 puncta is shown. (E) Percentage of cells with LC3 puncta in experiments as described in C and D having pepstatin A and E64D added for 4 h before analysis. The values represent the mean ± SEM for three experiments. Samples were subjected to unpaired Student's t test.
Silencing of p8 strongly stimulated LC3-II formation, even in medium containing 10% serum—a condition that should not provoke autophagy (Figure 1B and Supplemental Figure 1, A and B). Rapamycin or serum starvation each induced a strong up-regulation of LC3-II, but this induction was consistently higher after p8 RNAi (Figure 1B). Thus, p8 silencing induces LC3-II processing, indicating activation of autophagy.
The formation of LC3-II is associated with the aggregation of LC3-I in the autophagosomes. This aggregated LC3-II appears as puncta detectable by cytochemical methods. A fluorescent protein fused to LC3 is commonly used as an alternative approach to analyze autophagy (Klionsky et al., 2008). We thus established a U2OS cell line expressing the LC3 fused to a far-red fluorescent protein, Red-LC3 (Figure 1C). U2OS Red-LC3 cells were subjected to p8 RNAi by using either OL1 or OL2 (Figure 1A), and 3-methyl adenine (3MA), an inhibitor of autophagy was used in parallel experiments (Figure 1D). As positive control, after ctrl RNAi, Red-LC3 cells were treated with 100 nM rapamycin. In all the experiments, cells were visualized by nuclear Hoechst staining, and the percentage of cells bearing at least five puncta was quantitated, as described in Klionsky et al. (20080 and Herrero-Martin et al. (2009). Consistent with Figure 1B, p8 silencing leads to autophagosomes accumulation with at least 20% of the cells manifesting autophagic puncta. The appearance of puncta was inhibited by 3MA addition (Figure 1, C and D). When E64D and pepstatin were added for 4 h before the analysis, we found a similar higher percentage of Red-LC3–positive cells after p8 silencing, strongly suggesting an increase of the “on rate” of the autophagic process (Figure 1E). Thus, our data indicate that silencing of p8 is sufficient to increase the basal autophagosome formation in U2OS cells and is blocked by autophagy inhibitors incubation.
p8 Protects from Autophagy-induced Apoptosis
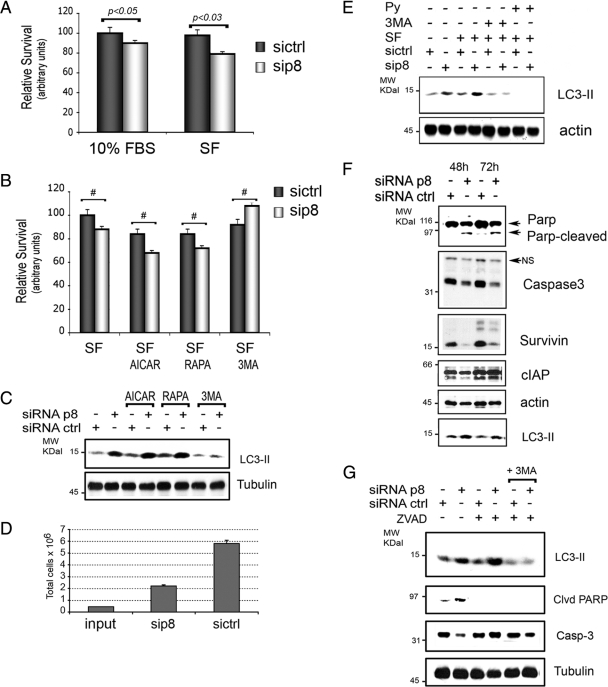
Autophagy can be either protective or deleterious on cellular fate (Maiuri et al., 2007). We thus set out to determine the effects of p8 RNAi on cellular viability and consequent to autophagy activation by serum starvation, AMPK stimulation with AICAR, or by mTOR inhibition with rapamycin. Thus, after p8 RNAi we applied autophagic stimuli and assessed cell viability by MTT assays. Silencing of p8 decreased the survival of cells cultured in the presence of 10% FBS (Figure 2A) and enhanced cell death to an even greater degree upon activation of autophagy by serum starvation, AICAR, or rapamycin (Figure 2, A and B). The effects of p8 RNAi on cell death provoked by serum depletion were reversed when cells were simultaneously treated with autophagy inhibitor 3MA (Figure 2B). These findings were confirmed in assays with U87MG cells, a glioblastoma cell line (Supplemental Figure 1, B and C). In parallel experiments, we monitored autophagy levels in the same conditions as in Figure 2B (Figure 2C). A further increase of LC3-II upon pharmacological activation of autophagy in serum starved cells was detected and was blocked by 3MA addition (Figure 2C). On line with this evidence, 96 h of p8 RNAi resulted in a decrease in total U2OS cell number, as quantified by direct cell count and consistent with an effect on cellular viability (Figure 2D).
Figure 2.
p8 presence protects from autophagy-induced apoptosis. (A–C) p8 RNAi decreases cellular viability in response to autophagy stimuli. U2OS were cultured for 48 h after p8 or ctrl RNAi, and then the cells were cultured for further 48 h with 10% FBS or without FBS (SF) before cell viability assays. When indicated, 1 mM AICAR (AICAR) or 100 nM rapamycin (RAPA) or 10 mM 3MA was added. (B) Values were normalized to ctrl-silenced cells in serum-free conditions. Values represent the mean ± SEM for five experiments in triplicate. Samples were subjected to unpaired Student's t test. #p < 0.05. (C) Western blot analysis of the levels of LC3-II in total cellular lysates of U2OS treated as described in B. (D) Decrease of total cell number following p8 RNAi. U2OS were cultured for 96 h after p8 (sip8) or ctrl (sictrl) RNAi, and cell number was evaluated using a hemocytometer. The values represent the mean ± SEM for three experiments. (F) p8 RNAi-induced LC3 processing is blocked by autophagy inhibitors. Western blot analysis after p8 or ctrl RNAi as described in Figure 1A and serum starvation (SF) or treatment with 10 mM 3MA or 10 mM methylpyruvate (Py) for 24 h. Western blots were performed with anti-LC3B and actin antibodies. (E) p8 RNAi induces autophagy and concomitant apoptosis in U2OS cells. U2OS were cultured in 10% FBS for 48 and 72 h after p8 (sip8) or ctrl (sictrl) RNAi. Total cellular lysates were analyzed with antibodies to autophagy, survival, and apoptosis markers, as indicated. Actin was used to normalize the loading. Representative Western blots are shown. (G) p8 RNAi-induced LC3 processing is not blocked by apoptosis inhibitors. Analysis after p8 or ctrl RNAi as in Figure 1A and treatment with 20 μM zVADfmk (ZVAD) caspase inhibitor alone or with ZVAD and 10 mM 3MA combined for 16 h in 10% FBS. The Western blots were carried out with LC3B, caspase-3, cleaved PARP, and tubulin antibodies.
We further analyzed the mechanism responsible for autophagy induction by p8 RNAi. As for cellular viability (Figure 2B), the addition of 3MA to the cell medium suppressed the increase in LC3-II upon p8 RNAi (Figure 2E). The enhancement, by p8 RNAi, of serum starvation-induced cell death may be due to the disruption of intracellular energy balance as methylpyruvate, a growth factor-independent energy source, reversed the p8-dependent increase of LC3-II processing (Figure 2E). Thus, p8 seems to suppress autophagy and cell death associated with autophagic stimuli, rendering cells less sensitive to energy stressors.
We further assessed whether the decrease in cell viability observed after p8 silencing was due to apoptotic cell death. To this end, we analyzed targets of caspase machinery and found that p8 siRNA triggered both nuclear PARP cleavage and a decrease in total caspase-3, indicative of its processing. Accordingly, we observed a decrease in the caspase-3 inhibitor survivin, whereas no significant changes for c-IAP1 were found (Figure 2F).
To determine whether blocking apoptosis had an effect on autophagy, we performed an experiment where ctrl- and p8-silenced U2OS cells were treated with the caspase inhibitor zVADfmk (ZVAD) and, as positive control, simultaneously with ZVAD and autophagy inhibitor 3MA. The effects on autophagy and apoptosis were monitored with LC3B, cleaved PARP and total caspase-3 antibodies. We found that ZVAD treatment did not affect p8 silencing-induced LC3 processing, but, as expected, it blocked PARP cleavage. Addition of both ZVAD and 3MA abolished both autophagy and apoptosis (Figure 2G). Together, the results in Figure 2 indicate that the presence of p8 protects cells from autophagy and that p8 silencing provokes autophagy and cell death associated with caspase activation.
p8 Represses FoxO3 Transactivation
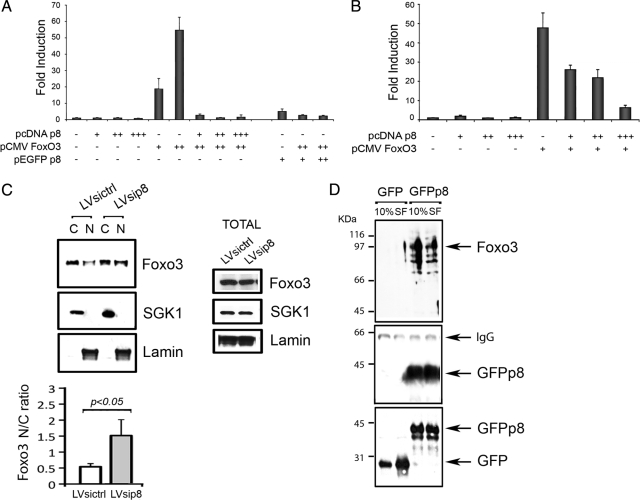
We next sought to identify the mechanisms by which p8 regulates autophagy. We have shown that p8 is a cotranscriptional regulator in cardiovascular and cancer cells (Goruppi et al., 2007). In many instances, the induction of autophagy is accompanied by the expression of a number of genes (Attaix and Bechet, 2007; Klionsky et al., 2008). Thus, we asked whether p8 might regulate the induction of autophagy via a transcriptional mechanism. The transcription factor FoxO3 has been reported to control autophagy (Mammucari et al., 2007; Zhao et al., 2007). We used a FoxO luciferase reporter to test the effects of p8 expression on FoxO3 transactivation. We found that p8 coexpression triggered a complete repression of the FoxO3-dependent reporter transactivation (p < 001) (Figure 3A). Likewise, expression of a GFP-tagged p8 had no significant reporter activity and, when coexpressed, inhibited the FoxO3 reporter transactivation (Figure 3A). When FoxO3 was expressed with decreasing concentrations of p8 a dose dependent derepression was detected, ruling out the possibility of a toxic effect due to protein overexpression (Figure 3B).
Figure 3.
p8 interacts with and represses Foxo3 transactivation. (A and B) p8 represses FoxO3 transactivation in a dose dependent manner. U2OS cells were transfected with a FoxO transcription factors reporter and a combination of pcDNAp8, pEGFPp8, and pCMVFoxO3 wild type (wt). The next day, cells were serum starved (0.1% FBS) for 16 h. Luciferase activity was measured in triplicate and normalized to β-galactosidase activity. The FoxO3 concentration having maximal transactivation was cotransfected with increasing amounts of pcDNAp8, in B. Mean ± SEM for four experiments is shown. (C) U2OS cells were infected either with a control silencing lentivirus (LV sicrtl) or a lentivirus silencing human p8 (LV sip8). Confluent cells were subjected to nuclear/cytosolic separation (N and C). The Western blots were carried out with the indicated antibodies, quantitated by densitometry, and the normalized values are indicated as nuclear/cytosol ratio in the histograms (mean ± SEM for three experiments; p < 0.05). Separate cultures were analyzed for total protein expression (right). (D) p8 binds to endogenous FoxO3. U2OS cells expressing enhanced green fluorescent protein (EGFP) or EGFPp8 were either left untreated (10%) or serum starved (SF) for 24 h before immunoprecipitation with anti-p8 antibodies. The western blots were carried out with anti-FoxO3 and p8, whereas total lysates were stained with GFP.
In the presence of serum, FoxO3 is kept inactive and sequestered in the cytosol through a phosphatidylinositol 3-kinase/Akt-dependent mechanism. On serum withdrawal, FoxO3 enters the nucleus and becomes active (Brunet et al., 1999). To determine whether p8 expression might interfere with FoxO3 intracellular localization, we generated a U2OS cell line in which p8 was silenced by lentiviral short hairpin RNA (shRNA) (LV sip8), and control cells were generated by infecting with empty shRNA lentiviruses (LV sictrl) (Supplemental Figure 2B). Nuclear and cytosolic extracts were prepared and FoxO3 detected on immunoblots. SGK1 and Lamin A provided cytosolic and nuclear markers, respectively (Figure 3C). The levels of FoxO3 were quantified and expressed as the nuclear (N) to cytoplasmic (C) ratio. We found that cells in which p8 expression was silenced had significantly higher levels of nuclear FoxO3 even in presence of serum (0.5 ± 2 vs. 1.7 ± 5, respectively; p < 0.05; n = 3), indicative of a possible functional interaction between FoxO3 and p8 (Figure 3C). No change in total FoxO3 protein expression was detected in total cellular lysates made in SDS-loading buffer (Figure 3C, middle).
To investigate whether the observed increase in nuclear FoxO3 localization in p8 knockdown cells might be a consequence of a direct interaction with p8, we performed immunoprecipitations from U2OS expressing either GFP or GFPp8 (Supplemental Figure 2A) by using anti-p8 antibodies. FoxO3 was clearly detected in cells expressing p8 and the interaction was not apparently influenced if the cells were serum starved for 24 h (Figure 3D). Altogether, our results indicate that p8 expression is able to repress FoxO3-dependent transactivation in U2OS cells and that p8 knockdown increases nuclear FoxO3.
p8 Suppresses FoxO3 Association with the bnip3 Promoter and Modulates bnip3 RNA and Protein Levels in U2OS Cells
The above-mentioned studies suggested that p8 might regulate FoxO3-dependent induction of autophagic genes. The promoters of several autophagy genes (lc3, bnip3, and atg12) contain multiple FoxO consensus cis-acting sequences (Mammucari et al., 2007; Zhao et al., 2007). We focused on Bnip3, a protein whose inhibition blocks autophagy in cardiomyocytes and hypoxic tumors, whereas if overexpressed it can induce autophagy and repress mTORC1 (Hamacher-Brady et al., 2006; Hamacher-Brady et al., 2007; Li et al., 2007). Several studies suggest a major role for Bnip3 in FoxO3-induced autophagy in vitro and in vivo (Mammucari et al., 2007; Zhang and Ney, 2009).
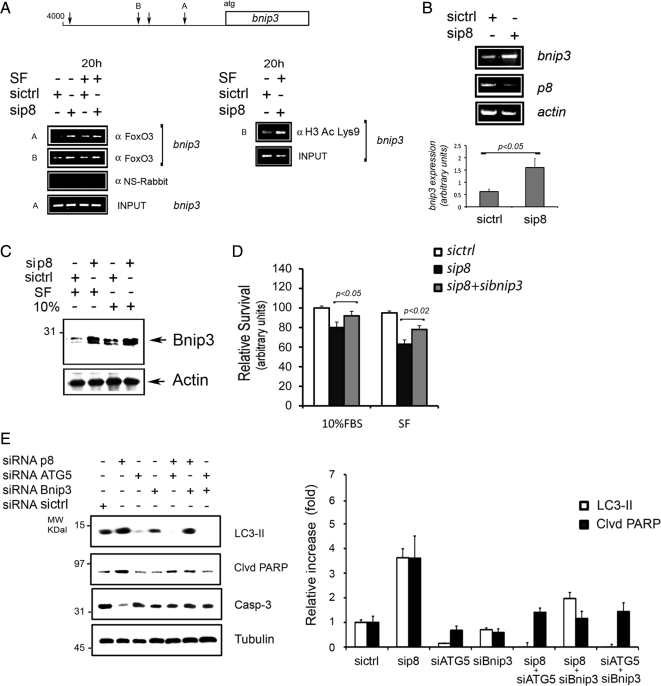
We tested the effects of p8 RNAi on FoxO3's association with two different regions on the bnip3 promoter (A and B in Figure 4A). After p8 or control RNAi treatment, U2OS cells were either left in presence of serum or serum starved for 20 h before performing ChIPs. DNA Copurified with FoxO3 and NRS ChIPs were amplified by PCR using the oligonucleotide surrounding sequences A and B (Figure 4A). We found that silencing of p8 increases the association of FoxO3 with chromatin containing the bnip3 promoter. Along with the results in Figure 3A, these data suggest that p8 negatively regulates FoxO3 transactivation of bnip3. Accordingly, p8 silencing also increased the levels of Bnip3 mRNA, as detected by RT-PCR (Figure 4B). Moreover, when separate ChIPs were performed using acetylated-histone H3 (Lys9) antibodies, which recognize transcriptionally active chromatin, we found that p8 RNAi increases the levels of chromatin containing acetylated histone H3. These data suggest that in situ p8 reduces the level of active chromatin (Figure 4A). In accordance with our RNA and ChIP studies, we found that Bnip3 protein levels are increased upon p8 silencing (Figure 4C).
Figure 4.
p8 RNAi increases FoxO3 association to bnip3 promoter resulting in higher bnip3 RNA and protein levels. (A) Increased FoxO3 association to endogenous bnip3 promoter after p8 RNAi. U2OS 24 h after p8 or crtl RNAi were either left untreated or serum starved (SF) for additional 20 h before harvesting. Chromatin immunoprecipitations of endogenous FoxO3: either anti FoxO3 (αFoxO3) or a nonimmune serum (αNS-Rabbit) were used. Total cellular chromatin (INPUT). Two FoxO-responsive elements in the bnip3 promoter (A and B in the bnip3 promoter scheme) were analyzed. p8 RNAi increases histone H3 acetylation on bnip3 promoter. ChIPs were performed from U2OS chromatin by using anti-lysine 9-acetylated histone H3 antibodies. PCR reactions were made using B oligonucleotides. (B) p8 RNAi enhances bnip3 RNA expression. U2OS were treated as described in Figure 2A, and RT-PCRs were run using bnip3, p8, and actin oligonucleotides. The histograms show the mean values ± SEM for three experiments. p < 0.05. (C) p8 RNAi increases Bnip3 protein levels. Western blot analysis of total cellular lysates from U2OS cells treated as described in Figure 5A, using either anti Bnip3 or actin antibodies, as loading control. (D) Knockdown of bnip3 decreases cell death associated with p8 silencing. U2OS were cultured for 48 h after ctrl, p8 alone, or p8 and bnip3 RNAi. Cellular viability assays (MTT) were performed after additional 48 h. The values represent the mean ± SEM for three experiments in triplicate. Samples were subjected to unpaired Student's t test. (E) p8 RNAi induces atg5-dependent autophagy and bnip3-dependent apoptosis. U2OS were cultured for 48 h in serum-free after ctrl, atg5, p8, and bnip3 alone or after combined p8+atg5, p8+bnip3, and atg5+bnip3 RNAi. Western blot analysis of total cellular lysates using LC3B, anti-cleaved PARP, caspase-3, and tubulin antibodies. The efficiency of atg5 and bnip3 knockdown is shown in Supplemental Figure 2C. The quantification of two separate Western blots is shown on the right.
We next examined whether bnip3 expression was responsible for the decrease in viability after p8 silencing. To this end, we either silenced p8 or both bnip3 and p8, before inducing autophagy for 48 h, as in Figure 2. We found that although p8 silencing alone impaired cellular viability, concomitant silencing of p8 and bnip3 restored the cellular viability both in the presence or the absence of serum (Figure 4D).
We further investigated the effects on cell survival of simultaneous p8 and autophagy knockdown by silencing the essential component of the autophagic machinery atg5 (Klionsky et al., 2008). To this end, we either silenced p8, atg5, and bnip3 alone or in combination before serum starvation for 48 h (Figure 4E and Supplemental Figure 2C). We found that simultaneous silencing of atg5 and p8 blocked LC3 processing and decreased apoptosis, as detected by PARP cleavage and total caspase3 levels (Figure 4E). Bnip3 and p8 silencing blocked apoptosis, but a higher basal LC3 processing was still detectable. From Figure 4, A–E, we can conclude that p8 silencing up-regulates Bnip3 and causes ATG5-dependent autophagy and Bnip3-dependent apoptosis. Interestingly, in absence of apoptosis, higher levels of LC3-II were detected in bnip3 and p8 cosilenced cells (Figure 4E). Together, our results in Figures 3, A–C and 4, A–E, indicate that p8 suppresses the association of FoxO3 with the bnip3 promoter and that p8 absence increases bnip3 transcription and protein levels. More importantly, Bnip3 seems to be a key target mediating cell death induced by p8 silencing.
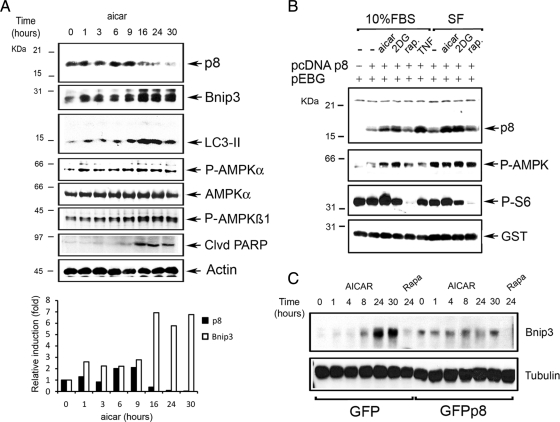
Cellular Energy Stress Acts on p8 Expression and Stability
Metabolic stress results in a decrease in ATP and an accumulation of AMP, which activates AMPK (Hardie, 2007). Likewise, amino acid depletion deactivates mTORC1, resulting in reduced protein synthesis (Meijer and Codogno, 2008). To determine whether metabolic stresses could act on p8 expression, we induced autophagy in U2OS cells with AICAR and followed p8 protein levels over time. The expression of endogenous Bnip3 and the induction of LC3-II were monitored in parallel. As controls, we followed the levels of AMPK activation and the induction of apoptosis. We found that endogenous p8 protein was transiently induced between 1 and 9 h after AICAR addition. Induction of autophagy by AICAR was associated by a slight increase of total Bnip3 protein (Figure 5A). After 16 h, endogenous p8 levels were down-regulated and concomitantly the induction of Bnip3 was up-regulated, as were LC3-II levels. Endogenous p8 stabilization by AICAR was transient. Accordingly, both amino acid and glucose starvation stabilized p8 in primary rat cardiomyocytes (Supplemental Figure 3, A and B). AMPK phosphorylation was first detected at 1 h from AICAR addition and remained higher than basal at later time points, when PARP cleavage became evident (Figure 5A). Thus, although autophagic stimuli such as AICAR induce p8 transiently, markers of autophagy are fully up-regulated in AICAR-stimulated cells when p8 levels decline back to basal. These findings are consistent with our observations indicating that p8 suppresses autophagy by reducing the transcription of Bnip3.
Figure 5.
Cellular energy stress acts on p8 expression and stability. (A) Endogenous p8 is down-regulated by AMPK-signaling in U2OS cells. Analysis of U2OS cells stimulated for the indicated times with 1 mM AICAR. The Western blots were carried out with anti-p8, Bnip3, LC3B, p-AMPKα, p-AMPKβ, AMPK-cleaved PARP, and actin antibodies. For p8 and Bnip3, values were quantified as fold of induction relative to the untreated cells levels. (B) Ectopic expressed p8 is stabilized by 2DG and AICAR. 293 cells were cotransfected with pcDNAp8 and a vector expressing GST protein (pEGB). After 2 d, the cells were treated for 8 h with 20 mM 2DG, 1 mM AICAR, 20 ng/ml rapamycin, and 20 ng/ml TNF (positive control) either in the presence (10% FBS) or absence (SF) of serum in the culture medium. The Western blot analysis of total lysates was performed using anti-p8, anti-p-AMPK, p-S6, and GST as treatments and efficiency of transfection controls, respectively. (C) p8 overexpression blocks endogenous Bnip3 up-regulation by AICAR. Analysis of EGFP- or EGFPp8-expressing cells stimulated for the indicated times with AICAR as described in A. The Western blots were made with anti-Bnip3 and tubulin antibodies.
We and others have reported that p8 polypeptide undergoes multiple posttranslational modifications, some of them required for its stabilization. Endothelin-1 and β-adrenergic agonists act at transcriptional level, whereas others, such as TNF, modify both expression and protein stability. In addition, p8 turnover is regulated by Akt-glycogen synthase kinase 3 signaling, which is sensitive to serum withdrawal (Goruppi and Kyriakis, 2004; Goruppi et al., 2007; Chowdhury et al., 2009; Goruppi and Iovanna, 2009). To determine whether the transient increase of endogenous p8 polypeptide is determined by signaling regulating the autophagic process, we transfected 293 cells with pcDNA p8 and treated them with either AICAR, 2DG, rapamycin, or TNF as positive control, both in presence of serum (10%) or under serum-free conditions. Immunoblotting with anti p8 antibodies showed that serum depletion increased p8 polypeptide levels modestly, whereas AICAR and 2DG stimulated robust increases in p8 protein, both under normal serum conditions and in serum-free conditions. Rapamycin did not affect p8 levels (Figure 5B). Thus p8 polypeptide stability is a target of AMPK-activated signaling but not of mTOR or serum withdrawal.
Because AICAR-induced autophagy is associated with endogenous Bnip3 up-regulation and a decrease in endogenous p8 levels (Figure 5A), we explored the effects of p8 overexpression on AICAR-induced Bnip3 up-regulation. To this end, we took advantage of U2OS cells expressing either GFP or GFPp8 (Figure 4D and Supplemental Figure 2A) and analyzed over time the levels of endogenous Bnip3 after AICAR addition. As in parental U2OS cells (Figure 5A), in GFP-expressing U2OS cells the basal Bnip3 levels were strongly up-regulated after 24 h from AICAR addition (Figure 5C). By contrast, and consistent with a p8 corepressor function, no significant endogenous Bnip3 up-regulation was detected in GFPp8-expressing U2OS cells (Figure 5C). Together these data suggest that, after a transient stabilization, the induction of autophagy by stimuli mimicking energy stress is associated with the down-regulation of endogenous p8, which leads to a concomitant up-regulation of endogenous Bnip3.
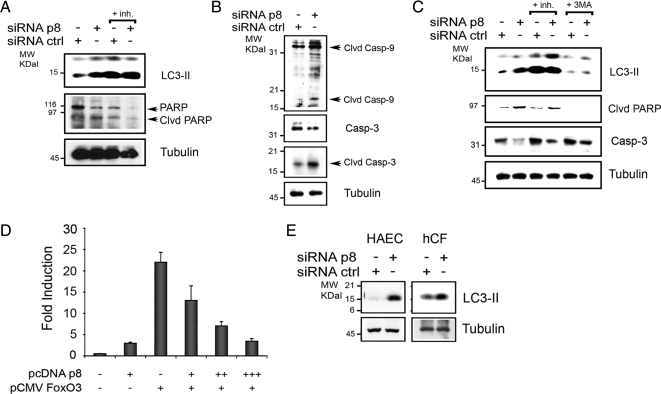
p8 Silencing Increases Autophagy and Apoptosis in Cardiac Cells
Our results in Figure 1 and 2 demonstrate that p8 silencing in U2OS cells is followed by an increase in basal autophagy levels and apoptosis. We have reported previously that p8 is expressed in cardiac cells (Goruppi et al., 2007); we thus explored the consequences of p8 silencing in primary neonatal rat cardiomyocytes and in rat H9C2 cardiomyoblast cells. We monitored the autophagy levels and apoptosis 72 h after either ctrl or p8 RNAi (Goruppi et al., 2007). In both cardiomyocytes and H9C2 cells, we studied the effects of p8 silencing in the presence or absence of lysosome inhibitors (E64D and pepstatin) and in H9C2 cells also in presence/absence of the autophagy inhibitor (3MA). The induction of autophagy was detected with LC3B antibodies and the activation of apoptosis was monitored with caspase-3, caspase-9, and PARP antibodies by Western blot. We found that silencing of p8 resulted in LC3 processing in both cardiomyocytes and H9C2 cells, which was dramatically increased by the lysosomal inhibitors, thus ruling out LC3-II flux as an explanation for increased LC3-II levels after p8 RNAi (Figure 6, A–C). This was correlated with a significant apoptosis in both cardiac cells (Figure 6, A–C) and was blocked by 3MA addition in H9C2 cells (Figure 6C). Three results are consistent with our evidence in U2OS cells and show an increase of 3MA-dependent autophagy after p8 silencing in cardiomyocytes, which is associated with caspase activation.
Figure 6.
p8 silencing in cardiovascular cells. (A–C) p8 silencing increases autophagy and apoptosis in cardiac cells. p8 RNAi in primary neonatal rat cardiomyocytes (A and B) and in rat H9C2 cardiac cells (C). Cells were cultured for 72 h in 10% FBS after (rat) p8 or ctrl RNAi. When indicated, both pepstatin A and E64D were added for 4 h (+inh) or 10 mM 3-methyladenine (+3MA) was added for 16 h before analysis. The Western blots were performed with anti-LC3B, PARP, rat-specific cleaved (clvd) caspase-9, and total and cleaved (clvd) caspase-3 antibodies for cardiomyocytes and LC3B and cleaved (clvd) PARP and caspase-3 antibodies for H9C2 cells. Tubulin was used as loading control. (D) p8 represses FoxO3 transactivation in a dose-dependent manner in H9C2 cells. H9C2 cells were transfected in triplicate with a FoxO luciferase reporter and pcDNAp8 and pCMVFoxO3 wt as described for Figure 3A). In separate experiments, FoxO3 was cotransfected with increasing amounts of pcDNAp8. Luciferase activity was measured in triplicate and normalized to coexpressed β-galactosidase activity. Means ± SEM for two experiments are shown. (E) p8 silencing increases autophagy in primary human cells. p8 RNAi in primary hCFs and in primary HAECs. Cells were cultured for 72 h in 10% FBS after p8 or ctrl RNAi, and the Western blot analysis was performed as described in A.
To further support our findings in the cardiovascular system, we investigated the effects of p8 expression on FoxO3-dependent transactivation. As for the U2OS cells (Figure 3, A and B), we found that p8 coexpression in the H9C2 cardiac cell line resulted in a repression of the FoxO3-dependent reporter transactivation (Figure 6D). Likewise, when FoxO3 was expressed with increasing concentrations of p8, a dose-dependent repression was detected (Figure 6D).
In addition to cardiomyocytes, several different cell types can be found in the human heart (Rothermel and Hill, 2008b). We thus explored whether a similar increase in basal autophagy levels after p8 silencing, as detected in cardiomyocytes, occurred in primary human cardiac fibroblasts (hCFs) and in primary human aortic endothelial cells (HAECs). In both primary human cell types, p8 silencing for 72 h was associated with an increase of basal LC3 processing (Figure 5D). These studies strengthen our findings in U2OS cells and extend their relevance to the cardiovascular system.
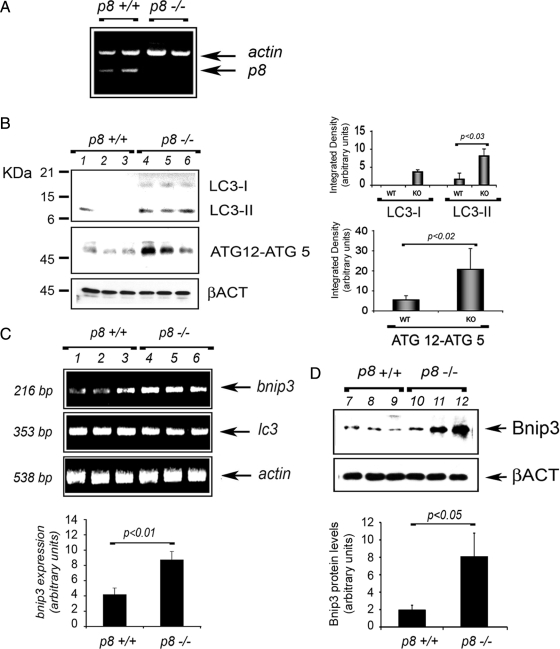
Cardiac Tissue from p8 −/− Mice Exhibits Higher Basal Autophagy
We have shown that p8 plays a crucial role in regulation of cardiomyocyte hypertrophy and cardiac fibroblast production of MMPs, suggesting a function for p8 in the heart (Goruppi et al., 2007). We took advantage of p8 −/− mice to investigate whether disruption of p8 is associated with increased autophagy in the heart. RT-PCR confirmed the expression of p8 in the left ventricles (LVs) of wild-type (p8 +/+) mice, and, as expected, its absence in p8 −/− mice (Figure 7A). We analyzed by Western blot the levels of LC3-II and Atg12–5 in the LV total lysates of age-matched p8 +/+ and p8 −/− mice. We found that disruption of p8 led to significantly elevated levels of LC3-I and LC3-II, and of Atg12–5 (p < 0.03, n = 5 and p < 0.02, n = 5, respectively), indicating a greater basal autophagy (Figure 7B). The increase in LC3-I and LC3-II observed was not apparently due to a direct transcriptional effect, because we detected no significant difference in lc3 mRNA between p8 −/− and p8 +/+ mice (Figure 7C), by RT-PCR. Consistent with our RNAi and overexpression results, we found that both RNA and protein expression of bnip3 were higher in p8 −/− (p < 0.01, n = 6 and p < 0.05, n = 4, respectively) (Figure 7, C and D). These findings indicate that, as with silencing of p8 in cultured cells, the disruption of p8 is associated with an increase in the expression of autophagy markers also in vivo.
Figure 7.
Cardiac tissue from p8 −/− mice exhibits higher basal autophagy and present increased levels of both Bnip3 RNA and protein. (A) p8 is expressed in the heart. RT-PCR analysis of total RNA from LVs of wilt type (p8 +/+) and knockout mice (p8 −/−). p8 and actin were coamplified, and PCR products were separated on 1.8% agarose gel. (B) p8 −/− mice have higher LC3-II and ATG12–5 levels. Western blot analysis of LV extracts from p8 +/+ and p8 −/− mice by using anti-LC3B, anti-ATG12-5, and β-actin antibodies. The quantifications are shown in the histograms. The values represent the mean ± SEM. Samples were subjected to unpaired Student's t test (p < 0.03 and p < 0.02, respectively; n = 5). (C) p8 −/− mice have higher bnip3 expression. bnip3, lc3, and actin were amplified from LV RNA of p8 +/+ and p8 −/− mice. PCR products were quantified and subjected to unpaired Student's t test (p < 0.01; n = 6). (D) p8 −/−mice have higher Bnip3 protein levels. Western blot analysis of LV extracts from p8 +/+ and p8 −/− mice by using anti Bnip3 and β-actin antibodies, as indicated. The quantifications are shown in the histograms (p < 0.05; n = 4). The values represent the means ± SEM.
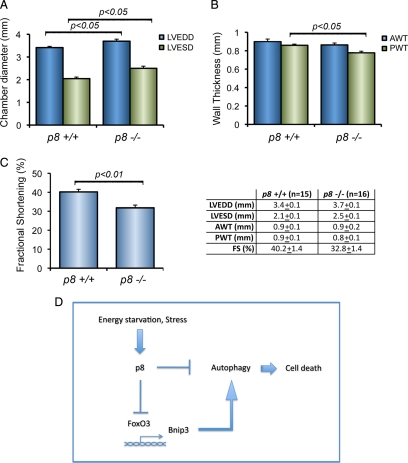
p8 Genetic Deletion Is Associated with a Cardiac Phenotype
Autophagy in the heart under baseline conditions is a homeostatic mechanism for the maintenance of normal cardiac function and morphology. However, unrestrained or excessive autophagic activity can result in cardiac cellular loss and cardiac dysfunction (De Meyer and Martinet, 2008; Kundu and Thompson, 2008; Rothermel and Hill, 2008a). We have shown previously that p8 is induced in human failing hearts and by stimuli associated with cardiac remodeling (Goruppi et al., 2007). To determine whether p8 genetic deletion was associated with a change in heart function, we performed echocardiographic recordings of p8 +/+ (n = 15) and p8 −/− (n = 16) mice and measured their left ventricle wall dimensions and ventricle performance. We found that compared with their p8 +/+ littermates, the p8 −/− mice present a small but significant LV dilation (p < 0.05), as detected by end-diastolic and end-systolic dimensions (Figure 8A), which result in a lower fractional shortening (p < 0.01) (Figure 8C). In addition, compared with p8 +/+ mice, p8 −/− mice develop a LV posterior wall thinning (p < 0.05), whereas no significant differences were found for the LV anterior wall (Figure 8B). Our findings indicate that in vivo p8 absence causes an increase in bnip3 proautophagic gene expression associated with an increase of autophagic markers. In the heart, in vivo p8 deletion results in a decreased cardiac functionality.
Figure 8.
p8 genetic deletion is associated with a cardiac phenotype. (A) p8 −/− hearts display chamber dilation. Echocardiographic data from LV of p8 +/+ and p8 −/− mice (*p < 0.05); n = 15 for each group). LVEDD and LVESD are measured in millimeters. (B) p8 −/− hearts display LV wall thinning. AWT and PW, anterior and posterior wall thickness (in millimeters) (*p < 0.05 for PWT +/+ vs. −/−; n = 15 for each group). (C) p8 −/− mice have lower fractional shortening when compared with p8 +/+ mice. FS, fractional shortening, calculated from A as (LVEDD − LVESD/LVEDD) × 100% (*p < 0.01 +/+ vs. −/−; n = 15). Numeric values for echocardiographic data are shown in the right panel. (D) Scheme of the mechanism regulating autophagy by p8.
DISCUSSION
Autophagy has been implicated in the pathogenesis of various diseases, including cancer, neurodegeneration, and cardiomyopathies. Central to the discussion of autophagy in disease pathogenesis is its role in adapting to cellular stress versus its role in contributing to cell death (Maiuri et al., 2007; Mizushima et al., 2008; Rothermel and Hill, 2008a). Multiple stimuli induce autophagy in mammalian cells; and although there is evidence for a transcriptional regulation, the mechanisms involved in this process are still poorly understood. The present study unveils a role for p8 in autophagy control and provides new insights into the mechanisms by which FoxO3 regulates autophagy. In particular, we show that in vitro and in vivo p8 depletion is associated with the up-regulation of autophagy and with the transcription of a known FoxO3 target gene involved in the autophagic process.
Silencing of p8 Causes Autophagy and Apoptosis
Under basal conditions the low level of autophagy is tightly regulated and is required to maintain cellular homeostasis (Codogno and Meijer, 2005; Maiuri et al., 2007). The results we present show that basal p8 expression represses autophagy and protects cells from autophagy-induced cell death. p8 RNAi increases puncta formation, indicative of autophagosome accumulation, albeit to a lower level than rapamycin or serum starvation, two known inducers of autophagy. Autophagosomes accumulation can reflect either an increase of formation, due to an increase in autophagic activity, or a reduction in their turnover. In our system, addition of autophagy inhibitors further increased both puncta formation and LC3 processing, strongly suggesting that p8 silencing increases the on rate of the autophagic process. In addition, such LC3 processing was inhibited by atg5 silencing, 3MA, and methyl pyruvate addition, pointing thus to a canonical autophagic pathway activation (Klionsky et al., 2008).
Although enhanced autophagy may confer a growing advantage, a constant increase in self-cannibalism may reduce survival rates when environmental stresses appear. We show here that the inability to regulate p8 renders the cells more sensitive to metabolic stressors. Accordingly, the mouse embryonic fibroblasts derived from p8 knockout mice have been shown to proliferate faster, but they fail to produce colonies when challenged in the absence of adhesion (Vasseur et al., 2002a).
p8 Represses FoxO3 Transactivation of bnip3
p8 is activated by various stressors, including agents affecting DNA structure, cytokines, and growth conditions. This induction by stress involves multiple mechanisms that often lead to stabilization of the protein. Once activated, p8 mediates several effects ranging from genomic stability, apoptosis, cellular growth, and cell cycle arrest. Some of these effects depend on p8 ability to modify transcriptional activity (Goruppi et al., 2007; Chowdhury et al., 2009). Whether p8 induces adaptive responses (cell survival) or cell death has been linked so far to distinct transcriptional programs and might depend on the stimuli or cellular microenvironment (Chowdhury et al., 2009; Goruppi and Iovanna, 2009). Our evidences indicate that p8 represses FoxO transcription and that p8 absence is associated with increased FoxO3-dependent expression of bnip3, in vitro and in vivo.
FoxO3 controls the activation of autophagy, as overexpression of an activated FoxO3 induces autophagy in a muscle cell line and regulates the expression of atgs in the skeletal muscle (Mammucari et al., 2007; Zhao et al., 2007). In this setting, a key role is played by FoxO3-induced bnip3, because its overexpression stimulates autophagosome formation, whereas bnip3 silencing decreases FoxO3-induced autophagy. Accordingly, we show that p8 represses Bnip3 up-regulation by AICAR that bnip3 knockdown restores cell viability of p8 deficient cells and blocks p8 RNAi-induced apoptosis, thus reinforcing p8 corepressor role toward FoxO3. Nevertheless, bnip3 RNAi does not completely restore cell viability of p8 deficient cells, and these cells have yet higher autophagy levels, strongly pointing to the existence of additional proautophagic targets repressed by p8.
The precise mechanism of Bnip3 effects on autophagy remains to be determined. Bnip3 might induce autophagy (or apoptosis) by competing with Bcl2 and BclXL for Beclin1 interaction (Pattingre et al., 2005; Zhang and Ney, 2009). Accordingly, dissociation of Beclin1/BclXL complex by the BH3-mimetic peptide or after Dap-kinase phosphorylation promotes autophagy (Oltersdorf et al., 2005; Zalckvar et al., 2009). Alternatively, deregulated Bnip3 expression might cause mitochondrial damage, cytochrome c release, and reactive oxygen species increase (Zhang and Ney, 2009). Finally, Bnip3 has been shown also to inhibit mTOR, suggesting a path to cell death by protein synthesis inhibition (Li et al., 2007).
How Does p8 Regulate FoxO3 Activity?
FoxO localization is mostly determined by growth factors presence, and additional levels of regulation are represented by protein–protein interactions, acetylations, and phosphorylations (Arden, 2007; Calnan and Brunet, 2008). Some stressors, such as AMPK, can affect FoxO3 transcriptional activity without influencing its localization (Greer et al., 2007). p8 is primarily nuclear in subconfluent cells but can localizes throughout the cell in high density grown or in G0-arrested cells (Valacco et al., 2006). Our evidence show that p8 silencing leads to an increase in basal nuclear FoxO3 and that p8 can be found in complex with FoxO3. It is possible that p8 interferes either with FoxO3 shuttling or with its interactions with other partners.
We have shown previously that p8 associates with anf and mmp9 promoters in the cardiovascular system (Goruppi et al., 2007). Even though multiple FoxO consensus are present on bnip3 promoter (Mammucari et al., 2007), our evidence do not point to a direct p8 competition with FoxO3 for the same DNA target, because p8 lacks transactivation activity toward the minimal consensus reporter. Finally, FoxO3 induces autophagy by stimulating the expression of several atgs and other genes; it is unlikely that Bnip3 represents the only FoxO3's target whose expression is repressed by p8. The effects on these additional autophagic genes remain to be determined.
Our research evidences a possible conundrum: p8 is transiently induced by autophagic stimuli while its presence represses the autophagy. Thus, p8 might represent an additional level of control toward FoxO3 functions, possibly acting as a feedback loop, which integrates signals sensing both growth factors and low AMP levels. Accordingly, AICAR and 2DG, but not serum deprivation, stabilize posttranslationally p8 polypeptide, and the effects of p8 knockdown on cell viability are enhanced if cells are deprived of growth factors.
In the long-term, FoxO3-induced up-regulation of atgs has been proposed to be necessary to maintain active the autophagic machinery (Klionsky et al., 2008). Because prolonged autophagy by an energy stressor results in p8 down-regulation, p8 might participate in the integration of transient versus a more protracted cellular autophagy, acting as a transient feedback repressor for untimely FoxO3 transcription.
p8 Knockout Mice Develop a Decreased Left Ventricular Function
p8/Nupr1 is a stress-induced protein that is turned on by the cell's stress management program at times of emergencies (Chowdhury et al., 2009; Goruppi and Iovanna, 2009). Little is known regarding its basal functions in vivo. In the knockout mice, p8 absence does not cause evident phenotypes, and animals were reported to develop normally (Vasseur et al., 2002b). Because we hypothesized previously a function for p8 in heart failure, our investigation was focused on the role of p8 in the heart under basal conditions. We show here that p8 silencing causes autophagy and apoptosis in cardiomyocytes and cardiac fibroblasts, two major heart cellular components, thus extending the relevance of our findings to the cardiovascular system.
Activation of autophagy plays a causative role in cardiomyopathy. Autophagic cell death as cause of cellular degeneration has been implicated in patients with hypertrophied and failing myocardium secondary to ischemia or dilated cardiomyopathy (De Meyer and Martinet, 2008). Incomplete autophagy in lysosome-associated protein 2-deficient mice provokes autophagosome accumulation and cardiomyopathy, whereas atg5 deletion in the adult mice leads to hypertrophy and contractile disfunction (Nakai et al., 2007; De Meyer and Martinet, 2008). The general consensus indicates that at baseline autophagy represents a homeostatic mechanism for maintaining cardiomyocyte size and cellular structure (Rothermel and Hill, 2008b); further study will determine whether in p8 mice changes in the cellular architecture or increased apoptosis are responsible for the functional phenotype.
The role of autophagy in the heart is still controversial. In an ischemia/reperfusion (I/R) model, the up-regulation of autophagy can protect from cell death, as either pharmacological inhibition or by interfering with atg5 functions will worsen the consequences. Interestingly, much of the I/R damages seem mediated by Bnip3 up-regulation, as its overexpression causes myocytes autophagy and cell death, whereas its silencing rescues both (Hamacher-Brady et al., 2006; Hamacher-Brady et al., 2007). The inability to up-regulate autophagy during sustained pressure overload in mice with genetic disruption of atg5 in the heart causes left ventricular dilation at a time when the wt animals develop compensatory hypertrophy (Nakai et al., 2007). Determining the effects of p8 absence following heart pressure overload or in an animal swimming model will contribute to the understanding of the functions carried out by basal versus induced autophagy in the heart and the role of FoxO3 transcription in this process. In addition, multiple pathophysiological conditions are associated with induction of autophagy. Fully understanding p8's multifaceted functions might open new avenues to the understanding of pathophysiological conditions associated with induction of autophagy, such as cardiovascular pathologies and cancer.
Supplementary Material
ACKNOWLEDGMENTS
We thank Marco Sandri (Padova University) and Alessia Di Nardo (Harvard University) for reagents and Joseph Alroy (Tufts University) for discussion and suggestions. C. C. was supported by “la ligue contre le cancer.” This work was supported by funds from American Heart Association grant SDG 0830359N (to S. G.) and National Institutes of Health grant R01-CA112399 (to J.M.K.).
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-09-0818) on February 24, 2010.
REFERENCES
- Arden K. C. FoxOs in tumor suppression and stem cell maintenance. Cell. 2007;128:235–237. doi: 10.1016/j.cell.2007.01.009. [DOI] [PubMed] [Google Scholar]
- Attaix D., Bechet D. FoxO3 controls dangerous proteolytic liaisons. Cell Metab. 2007;6:425–427. doi: 10.1016/j.cmet.2007.11.005. [DOI] [PubMed] [Google Scholar]
- Baehrecke E. H. Autophagy: dual roles in life and death? Nat. Rev. Mol. Cell Biol. 2005;6:505–510. doi: 10.1038/nrm1666. [DOI] [PubMed] [Google Scholar]
- Brunet A., Bonni A., Zigmond M. J., Lin M. Z., Juo P., Hu L. S., Anderson M. J., Arden K. C., Blenis J., Greenberg M. E. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- Calnan D. R., Brunet A. The FoxO code. Oncogene. 2008;27:2276–2288. doi: 10.1038/onc.2008.21. [DOI] [PubMed] [Google Scholar]
- Carracedo A., et al. The stress-regulated protein p8 mediates cannabinoid-induced apoptosis of tumor cells. Cancer Cell. 2006;9:301–312. doi: 10.1016/j.ccr.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Chowdhury U. R., Samant R. S., Fodstad O., Shevde L. A. Emerging role of nuclear protein 1 (NUPR1) in cancer biology. Cancer Metastasis Rev. 2009;28:225–232. doi: 10.1007/s10555-009-9183-x. [DOI] [PubMed] [Google Scholar]
- Clark D. W., Mitra A., Fillmore R. A., Jiang W. G., Samant R. S., Fodstad O., Shevde L. A. NUPR1 interacts with p53, transcriptionally regulates p21 and rescues breast epithelial cells from doxorubicin-induced genotoxic stress. Curr. Cancer Drug Targets. 2008;8:421–430. doi: 10.2174/156800908785133196. [DOI] [PubMed] [Google Scholar]
- Codogno P., Meijer A. J. Autophagy and signaling: their role in cell survival and cell death. Cell Death Differ. 2005;12(suppl 2):1509–1518. doi: 10.1038/sj.cdd.4401751. [DOI] [PubMed] [Google Scholar]
- De Meyer G. R., Martinet W. Autophagy in the cardiovascular system. Biochim. Biophys. Acta. 2008;1793:1485–1495. doi: 10.1016/j.bbamcr.2008.12.011. [DOI] [PubMed] [Google Scholar]
- Furuyama T., Nakazawa T., Nakano I., Mori N. Identification of the differential distribution patterns of mRNAs and consensus binding sequences for mouse DAF-16 homologues. Biochem. J. 2000;349:629–634. doi: 10.1042/0264-6021:3490629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goruppi S., Bonventre J. V., Kyriakis J. M. Signaling pathways and late-onset gene induction associated with renal mesangial cell hypertrophy. EMBO J. 2002;21:5427–5436. doi: 10.1093/emboj/cdf535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goruppi S., Chiaruttini C., Ruaro M. E., Varnum B., Schneider C. Gas6 induces growth, beta-catenin stabilization, and T-cell factor transcriptional activation in contact-inhibited C57 mammary cells. Mol. Cell Biol. 2001;21:902–915. doi: 10.1128/MCB.21.3.902-915.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goruppi S., Iovanna J. L. The stress-inducible protein p8 is involved in several physiological and pathological processes. J. Biol. Chem. 2009;285:1577–1581. doi: 10.1074/jbc.R109.080887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goruppi S., Kyriakis J. M. The pro-hypertrophic basic helix-loop-helix protein p8 is degraded by the ubiquitin/proteasome system in a protein kinase B/Akt- and glycogen synthase kinase-3-dependent manner, whereas endothelin induction of p8 mRNA and renal mesangial cell hypertrophy require NFAT4. J. Biol. Chem. 2004;279:20950–20958. doi: 10.1074/jbc.M312401200. [DOI] [PubMed] [Google Scholar]
- Goruppi S., Patten R. D., Force T., Kyriakis J. M. Helix-loop-helix protein p8, a transcriptional regulator required for cardiomyocyte hypertrophy and cardiac fibroblast matrix metalloprotease induction. Mol. Cell Biol. 2007;27:993–1006. doi: 10.1128/MCB.00996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer E. L., Oskoui P. R., Banko M. R., Maniar J. M., Gygi M. P., Gygi S. P., Brunet A. The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. J. Biol. Chem. 2007;282:30107–30119. doi: 10.1074/jbc.M705325200. [DOI] [PubMed] [Google Scholar]
- Hamacher-Brady A., Brady N. R., Gottlieb R. A., Gustafsson A. B. Autophagy as a protective response to Bnip3-mediated apoptotic signaling in the heart. Autophagy. 2006;2:307–309. doi: 10.4161/auto.2947. [DOI] [PubMed] [Google Scholar]
- Hamacher-Brady A., Brady N. R., Logue S. E., Sayen M. R., Jinno M., Kirshenbaum L. A., Gottlieb R. A., Gustafsson A. B. Response to myocardial ischemia/reperfusion injury involves Bnip3 and autophagy. Cell Death Differ. 2007;14:146–157. doi: 10.1038/sj.cdd.4401936. [DOI] [PubMed] [Google Scholar]
- Hardie D. G. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat. Rev. Mol. Cell Biol. 2007;8:774–785. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- Herrero-Martin G., Hoyer-Hansen M., Garcia-Garcia C., Fumarola C., Farkas T., Lopez-Rivas A., Jaattela M. TAK1 activates AMPK-dependent cytoprotective autophagy in TRAIL-treated epithelial cells. EMBO J. 2009;28:677–685. doi: 10.1038/emboj.2009.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmeister A., et al. The HMG-I/Y-related protein p8 binds to p300 and Pax2 trans-activation domain-interacting protein to regulate the trans-activation activity of the Pax2A and Pax2B transcription factors on the glucagon gene promoter. J. Biol. Chem. 2002;277:22314–22319. doi: 10.1074/jbc.M201657200. [DOI] [PubMed] [Google Scholar]
- Klionsky D. J., et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu M., Thompson C. B. Autophagy: basic principles and relevance to disease. Annu. Rev. Pathol. 2008;3:427–455. doi: 10.1146/annurev.pathmechdis.2.010506.091842. [DOI] [PubMed] [Google Scholar]
- Levine B., Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wang Y., Kim E., Beemiller P., Wang C. Y., Swanson J., You M., Guan K. L. Bnip3 mediates the hypoxia-induced inhibition on mammalian target of rapamycin by interacting with Rheb. J. Biol. Chem. 2007;282:35803–35813. doi: 10.1074/jbc.M705231200. [DOI] [PubMed] [Google Scholar]
- Maiuri M. C., Zalckvar E., Kimchi A., Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 2007;8:741–752. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- Malicet C., Dagorn J. C., Neira J. L., Iovanna J. L. p8 and prothymosin alpha: unity is strength. Cell Cycle. 2006;5:829–830. doi: 10.4161/cc.5.8.2686. [DOI] [PubMed] [Google Scholar]
- Mammucari C., et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Meijer A. J., Codogno P. Nutrient sensing: TOR's Ragtime. Nat. Cell Biol. 2008;10:881–883. doi: 10.1038/ncb0808-881. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Levine B., Cuervo A. M., Klionsky D. J. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai A., et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat. Med. 2007;13:619–624. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- Oltersdorf T., et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- Pattingre S., Tassa A., Qu X., Garuti R., Liang X. H., Mizushima N., Packer M., Schneider M. D., Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Polager S., Ofir M., Ginsberg D. E2F1 regulates autophagy and the transcription of autophagy genes. Oncogene. 2008;27:4860–4864. doi: 10.1038/onc.2008.117. [DOI] [PubMed] [Google Scholar]
- Quirk C. C., Seachrist D. D., Nilson J. H. Embryonic expression of the luteinizing hormone beta gene appears to be coupled to the transient appearance of p8, a high mobility group-related transcription factor. J. Biol. Chem. 2003;278:1680–1685. doi: 10.1074/jbc.M209906200. [DOI] [PubMed] [Google Scholar]
- Rothermel B. A., Hill J. A. Autophagy in load-induced heart disease. Circ. Res. 2008a;103:1363–1369. doi: 10.1161/CIRCRESAHA.108.186551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothermel B. A., Hill J. A. The heart of autophagy: deconstructing cardiac proteotoxicity. Autophagy. 2008b;4:932–935. doi: 10.4161/auto.6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar M., et al. Cannabinoid action induces autophagy-mediated cell death through stimulation of ER stress in human glioma cells. J. Clin. Invest. 2009;119:1359–1372. doi: 10.1172/JCI37948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambasivan R., Cheedipudi S., Pasupuleti N., Saleh A., Pavlath G. K., Dhawan J. The small chromatin-binding protein p8 coordinates the association of anti-proliferative and pro-myogenic proteins at the myogenin promoter. J. Cell Sci. 2009;122:3481–3491. doi: 10.1242/jcs.048678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valacco M. P., Varone C., Malicet C., Canepa E., Iovanna J. L., Moreno S. Cell growth-dependent subcellular localization of p8. J. Cell Biochem. 2006;97:1066–1079. doi: 10.1002/jcb.20682. [DOI] [PubMed] [Google Scholar]
- Vasseur S., Hoffmeister A., Garcia S., Bagnis C., Dagorn J. C., Iovanna J. L. p8 is critical for tumour development induced by rasV12 mutated protein and E1A oncogene. EMBO Rep. 2002a;3:165–170. doi: 10.1093/embo-reports/kvf023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasseur S., Hoffmeister A., Garcia-Montero A., Mallo G. V., Feil R., Kuhbandner S., Dagorn J. C., Iovanna J. L. p8-deficient fibroblasts grow more rapidly and are more resistant to Adriamycin-induced apoptosis. Oncogene. 2002b;21:1685–1694. doi: 10.1038/sj.onc.1205222. [DOI] [PubMed] [Google Scholar]
- Xie Z., Klionsky D. J. Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- Zalckvar E., Berissi H., Mizrachy L., Idelchuk Y., Koren I., Eisenstein M., Sabanay H., Pinkas-Kramarski R., Kimchi A. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep. 2009;10:285–292. doi: 10.1038/embor.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Ney P. A. Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ. 2009;16:939–946. doi: 10.1038/cdd.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Brault J. J., Schild A., Cao P., Sandri M., Schiaffino S., Lecker S. H., Goldberg A. L. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.