Actin arginylation regulates lamella formation in motile fibroblasts, but the underlying molecular mechanisms are unknown. Here, we found that actin regulation by arginylation affects its biochemical properties and binding of actin-associated proteins, modulating the overall structural organization of actin filaments in the cell.
Abstract
Actin arginylation regulates lamella formation in motile fibroblasts, but the underlying molecular mechanisms are unknown. To understand how arginylation affects the actin cytoskeleton, we investigated the biochemical properties and the structural organization of actin filaments in wild-type and arginyltransferase (Ate1) knockout cells. We found that Ate1 knockout results in a dramatic reduction of the actin polymer levels in vivo accompanied by a corresponding increase in the monomer level. Purified nonarginylated actin has altered polymerization properties, and actin filaments from Ate1 knockout cells show altered interactions with several associated proteins. Ate1 knockout cells have severe impairment of cytoskeletal organization throughout the cell. Thus, arginylation regulates the ability of actin to form filaments in the whole cell rather than preventing the collapse of preformed actin networks at the cell leading edge as proposed in our previous model. This regulation is achieved through interconnected mechanisms that involve actin polymerization per se and through binding of actin-associated proteins.
INTRODUCTION
Posttranslational protein arginylation consists of the addition of arginine (Arg) onto proteins and is mediated by arginyltransferase (Ate1; Kaji et al., 1963; Kaji, 1968; Balzi et al., 1990), an evolutionarily conserved enzyme that is required for normal development in mammals (Kwon et al., 2002). Ate1 knockout (KO) mice die of severe defects in cardiovascular development and angiogenesis (Kwon et al., 2002). Many proteins are arginylated in vivo during normal metabolism and stress responses (Soffer, 1971, 1975; Kopitz et al., 1990; Lee et al., 2005; Decca et al., 2006, 2007; Wong et al., 2007). Remarkably, over a third of identified arginylated proteins are cytoskeletal, suggesting a special role for arginylation in the regulation of cytoskeleton-related events. One of these proteins, actin, is among the most essential and abundant intracellular proteins (Karakozova et al., 2006).
β- and γ-Actins are the major structural components of the actin cytoskeleton in nonmuscle cells (Rubenstein and Spudich, 1977; Vandekerckhove and Weber, 1978; Otey et al., 1987). Despite high levels of similarity, the β-, but not the γ- isoform of actin is N-terminally arginylated in vivo (Karakozova et al., 2006). Both isoforms share two additional arginylation sites located on the same surface of the folded protein as the actin N-terminus (Wong et al., 2007); however, it is not clear whether both actin isoforms in vivo are arginylated on these sites. In the absence of arginylation, actin forms aggregates during polymerization in extracts. Cultured fibroblasts derived from Ate1 KO mice have collapsed lamella, which can be rescued by reintroduction of N-terminally arginylated β-actin into the Ate1 KO cells (Karakozova et al., 2006). Thus, N-terminal arginylation regulates the functions of actin in vivo and is important for normal cell morphology and migration; however, the effects of arginylation on the actin cytoskeleton in a living cell and the underlying molecular mechanisms for actin regulation by arginylation are unknown.
Here we addressed the role of arginylation in the regulation of the actin cytoskeleton in vivo and found that the absence of arginylation results in a dramatic decrease in intracellular actin polymer levels, suggesting a severe impairment of actin polymerization in arginylation-deficient cells. We further found that this impairment is caused by changes in actin polymerization properties and the structure of the resulting filaments, as well as the ability of actin to interact with several distinct actin-associated proteins that modulate its in vivo functions. These changes suggest a global role of arginylation in cell motility–dependent cytoskeleton polymerization and turnover.
MATERIALS AND METHODS
Purification and Baculovirus Expression of Actin
Wild-type (WT) and Ate1 KO mouse embryonic fibroblast (MEF) actin was purified by a DNase-I affinity chromatography method as described before (Schafer et al., 1998) with some modifications. For each preparation, 1 g of wet cell pellet was lysed in 10 ml of extraction buffer (1 M Tris, pH 7.0, 0.6 M KCl, 0.5 mM ATP, 1 mM DTT, 0.5 mM MgCl2, and 0.1 mM PMSF) using a Potter-Elvehjem homogenizer, followed by addition of 4% Triton X-100 and 0.1% Tween-20 and further homogenization. The resulting lysates were sonicated by five 15-s pulses to shear DNA, incubated at 4°C for 30 min with occasional swirling, clarified by sequential centrifugation at 25,000 × g for 20 min and 100,000 × g for 1 h at 4°C, and dialyzed two times against 1l of buffer A (2 mM Tris, pH 8.0, 0.2 mM CaCl2, 50 mM KCl, 0.2 mM ATP, and 0.5 mM DTT). The dialyzed lysate was clarified by centrifugation at 17,500 × g for 20 min at 4°C, loaded onto a DNase-I agarose column (prepared by coupling DNase-I [Roche, Indianapolis, IN]) to aminolink matrix [Pierce, Rockford, IL]) according to the manufacturer's protocol), preequilibrated with buffer G (2 mM Tris, pH 8.0, 0.2 mM ATP, 0.5 mM DTT, and 0.2 mM CaCl2), washed with 10 column volumes of buffer G, and eluted with chilled 50% formamide in buffer G into 4 volumes of chilled buffer G to adjust the formamide concentration to 10%. The eluted fractions were immediately loaded onto a DE-52 column equilibrated with buffer D (2 mM Tris, pH 8.0, 0.2 mM CaCl2, 100 mM KCl, 0.2 mM ATP, and 0.5 mM DTT), washed with 10 column volumes of buffer D, and batch-eluted with buffer D containing 400 mM NaCl. Peak fractions were then dialyzed against buffer G.
Baculovirus expression and purification of arginylated and nonarginylated actin was performed as described in Yates et al. (2007) using mouse M-β and R-β ubiquitin-actin fusion constructs made as described in Karakozova et al. (2006).
Biochemical Fractionation of Actin and Estimation of G- and F-Actin Ratios
MEFs were harvested and lysed in F-actin stabilization buffer (50 mM PIPES, pH 6.9, 50 mM NaCl, 5 mM MgCl2, 5 mM EGTA, 5% glycerol, 0.1% NP40, 0.1% Triton X-100, 0.1% Tween 20, 0.1% 2-mercaptoethanol, 1 mM ATP, and protease inhibitor cocktail; Sigma, St. Louis, MO; P8340). The resulting lysates were sedimented at 37°C at 200 × g for 5 min, 1,500 × g for 15 min, 16,000 × g for 15 min, and 66,000 × g for 60 min. Protein concentrations in all the fractions were normalized by the estimated protein concentration in the 200 × g supernatant as determined by the Bio-Rad protein assay reagent (Richmond, CA), and aliquots of supernatant and pellet fractions from each centrifugation step were analyzed by SDS-PAGE and Western blot. Actin proteins present in all the fractions were quantified by Western blots using antibodies to total actin (Cytoskeleton, Denver, CO; AAN01), β-actin (Sigma; A1978), and γ-actin (a gift from Dr. J. C. Bulinski, Columbia University).
Actin Polymerization and Activity Assays
A pyrene actin assay (Cooper and Pollard, 1982) was performed as previously described.
Actin assembly rates were determined from the slope of the pyrene fluorescence curves within the period of fast fluorescence increase, and actin critical concentration was estimated by plotting actin concentrations versus actin assembly rates and determination of the x-axis intersection point by extrapolation of the curves.
For lag phase measurements and estimation of actin nucleation rates, a total of 1 μM WT and KO actin containing 20% pyrene actin was polymerized in triplicates. To estimate the lag phase and actin nucleation rate, a theoretical model (Nishida and Sakai, 1983), and a 4th order polynomial fit were used to fit each curve and the point (time: T0) at the maximum slope (Sm) and averaged over three trials. The results of these two tests agreed with each other within error; numbers obtained based on the Nishida and Sakai, (1983) model are stated in the legend to Figure S3. This result was also in agreement with an estimation performed by plotting −n(1 − F(t)/F(∞)) against time (t) to obtain the intercept with the x-axis corresponding to the lag phase time as described in Lal et al. (1984).
To measure actin elongation, actin seeds were prepared by polymerizing 20 μM purified muscle actin for 30 min at room temperature and shearing by pipetting. 0.5 μM seeds and 0.4 μM pyrene actin were added to the reaction.
To measure actin polymerization by sedimentation, different concentrations of purified G-actin were supplemented with 10 mM HEPES, pH 7.1, 135 mM KCl, 10 mM NaCl, 2 mM MgCl2, 10 mM EGTA, and 0.2 mM ATP to induce polymerization and incubated at room temperature for 4 h, followed by centrifugation at 100,000 × g for 1 h at 17°C and quantification of actin in the pellets by SDS-PAGE and Coomassie blue staining. Pure nonmuscle actin (Cytoskeleton) was used as a concentration standard.
For sucrose gradient fractionation, 7 μM of WT and KO actin was polymerized overnight at room temperature, layered onto 5 ml of 10–60% step gradient of sucrose (prepared as steps of 10, 20, 30, 40, 50, and 60%), and centrifuged for 3 h at 34,000 rpm (Beckman Instruments, Fullerton, CA; SW 50.1 rotor) at 25°C. After the completion of the run, 200-μl fractions were collected starting at the top of the gradient and analyzed by SDS-PAGE and Coomassie blue staining. To determine the seeding capacity of the bottom gradient fraction, 0.8 μM KO actin and 0.2 μM pyrene actin copolymerized in the presence of 17 nM KO actin from fraction 25 of the sucrose gradient containing actin aggregates.
For electron microscopy (EM), in vitro polymerized actin filaments were loaded onto glow discharge-cleaned carbon-coated grids (Ted Pella, Irvine, CA), negatively stained with 1% uranyl acetate and imaged with transmission electron microscopy.
Gelsolin-binding and -severing Assays
To measure gelsolin-severing activity, 30 μg of WT or KO actin was polymerized for 5 h in the presence of 5 μM phalloidin. Stabilized actin polymers were then incubated with or without 6 μg of gelsolin for 5 min in the presence of 1.5 μM free Ca2+ at room temperature. The reaction mix was layered onto a 20% sucrose cushion containing 5 μM phalloidin and centrifuged at 100,000 × g for 45 min. The pellet was washed with polymerization buffer (10 mM HEPES, pH 7.1, 135 mM KCl, 10 mM NaCl, 2 mM MgCl2, and 2 mM EGTA), resuspended in SDS gel loading buffer, and analyzed by SDS-PAGE.
To measure gelsolin binding, 2.0 μM WT or KO actin was polymerized for 24 h (to convert ATP-F-actin completely to ADP-F-actin) in the presence of 2 μM phalloidin. Stabilized ADP-F actin polymers were diluted to 1.6 μM with actin polymerization buffer containing 16 μM phalloidin and 200 nM free Ca2+. F-actin was incubated at room temperature with different concentrations of gelsolin for 5 min and centrifuged at 100,000 × g for 30 min at 20°C. The pellet was washed once with actin polymerization buffer supplemented with 16 μM phalloidin. Actin and gelsolin present in the pellet was analyzed by Western blotting.
Capping Protein Pulldown Assay
To examine capping protein binding activity, 2.0 μM WT or KO actin was polymerized for 7 h at room temperature in actin polymerization buffer. The F-actin was then diluted to 1.6 μM in actin polymerization buffer supplemented with 0.5 mM fresh ATP and incubated with 330 nM capping protein (recombinant mouse α1 β2 isoform; Yang et al., 2005) at room temperature for 5 min. The F-actin was centrifuged at 100,000 × g for 30 min at 20°C. The pellet was washed once with actin polymerization buffer supplemented with 0.5 mM ATP. Actin and capping protein in the pellet were analyzed by Western blotting.
Electron Microscopy of Cytoskeleton Platinum Replicas
Platinum replicas of the cytoskeleton were prepared as described before (Svitkina and Borisy, 1998; Svitkina, 2007) using 1% Triton X-100, 4% PEG (MW 35,000 Da) and 2 μM phalloidin in PEM buffer (100 mM PIPES, pH 6.9, 1 mM MgCl2, and 1 mM EGTA), as the extraction solution.
For filament end counting, images of the same fields were taken at two different angles by tilting the grid by 10° (+) and 10° (−) while keeping the center of the field static, to create an angle differential of 20°. The images were pseudocolored as red (+10°) and cyan (−10°), superimposed using the Color Combine tool of the MetaMorph imaging software (Universal Imaging, West Chester, PA) and viewed through red-cyan stereo glasses, enabling clear visualization and manual counting of the filament ends. The numbers of filament ends were normalized to the local cytoskeleton density in each image, defined as the gray value of the cytoskeleton above the background threshold.
Isolation and Analysis of Actin-binding Proteins
Cells were collected by scraping, lysed in HEK buffer (20 mM HEPES-NaOH, pH 7.5, 1 mM EDTA, 50 mM KCl, and protease inhibitor cocktail; Sigma; P8340) and centrifuged at 16,000 × g to remove cellular debris. Supernatant fractions were supplemented with glycerol to a final concentration of 10% and clarified by centrifugation at 100,000 × g for 45 min at 4°C. High-speed supernatant fractions were supplemented with 5 mM MgCl2, 0.2 mM ATP, and 2.5 μM phalloidin and incubated at 4°C overnight to allow actin polymerization. The mix was layered over a cushion containing 15% sucrose and 10% glycerol in HEK buffer and centrifuged at 66,000 × g for 90 min at 4°C. The pellets were washed with HEK buffer containing 10% glycerol and 5 mM MgCl2, resuspended in two-dimensional (2D) gel loading buffer, and analyzed by 2D electrophoresis.
2D Gel Electrophoresis and Comparison of the Levels of Actin-binding Proteins in WT and KO Cells
2D electrophoresis was performed by Kendrick Laboratories (Madison, WI) as described previously (O'Farrell, 1975). For computerized comparison, duplicate silver-stained gels were analyzed as described in Wong et al. (2007). For spot excision and identification, similarly run Coomassie blue–stained gels were manually compared with the silver-stained gels and spots showing prominent differences that were clearly visible by Coomassie staining were excised and identified by mass spectrometry. Student's t test values were generated by the software for fold changes averaged from two gels.
The levels of actin-binding proteins (ABPs) in total lysates were estimated by Western blot analysis using rabbit polyclonal antibodies specific to capping protein (described in Mejillano et al., 2004), gelsolin (a gift from Dr. H. L. Yin, University of Texas), drebrin (StressGen, San Diego, CA; NBA-110E), twinfilin (a gift from Dr. P. Lappalainen, University of Helsinki), or protein disulphide isomerase (PDI; Stressgen).
Capping Protein Staining
WT and Ate1 KO MEF cytosol was extracted using extraction buffer (1% Triton X-100, 4% PEG, and 2 μM phalloidin in PEM buffer) followed by fixation in 0.2% glutaraldehyde in 0.1 M Na-cacodylate buffer (pH 7.3). Cells were blocked using NaBH4 and stained with polyclonal capping protein antibody (a gift from Dr. D. Schafer, University of Virginia). Cells were counterstained with rhodamine-phalloidin to visualize the actin cytoskeleton.
Statistical Analysis
In all the experiments where quantitative measurements were made, the variability in data point values were measured and represented as SD or SEM, as indicated in the text and figure legends. Student's t test was used to calculate p values.
RESULTS
Arginylation Regulates Actin Polymer In Vivo
Our previous studies suggested that nonarginylated actin forms bundles that interfere with normal actin function (Karakozova et al., 2006), but the effect of arginylation on intracellular actin polymers was never characterized in vivo. To address the question of whether arginylation regulates the levels or distribution of actin polymer in cells, we examined immortalized embryonic fibroblasts derived from WT and Ate1 KO mouse embryos (Karakozova et al., 2006) by rhodamine-phalloidin staining and fluorescence microscopy.
We previously found (Karakozova et al., 2006) that Ate1 KO fibroblasts appear smaller than WT cells, but have similar volume, suggesting that these cells are poorly spread during normal culture conditions. Despite this spreading defect, the overall distribution of F-actin seen at the light microscope level appeared largely unperturbed in Ate1 KO cells (Figure 1A). However, measurement of the total rhodamine-phalloidin fluorescence per cell revealed a dramatic reduction in the levels of F-actin staining in Ate1 KO cells, resulting in a ∼2.5-fold decrease of average fluorescence intensity compared with WT (Figure 1B). Such a decrease could be due to a corresponding decrease in the actin polymer level in Ate1 KO cells or to a decreased ability of nonarginylated F-actin to interact with phalloidin. The latter possibility is unlikely, because the sites of actin interaction with phalloidin have been shown to be located around the nucleotide binding cleft, away from the N-terminus and the outer surface predicted to be affected by arginylation (Vandekerckhove et al., 1985; Lorenz et al., 1993). Therefore it appears likely that reduced rhodamine-phalloidin fluorescence in Ate1 KO cells is due to decreased actin polymer level in these cells.
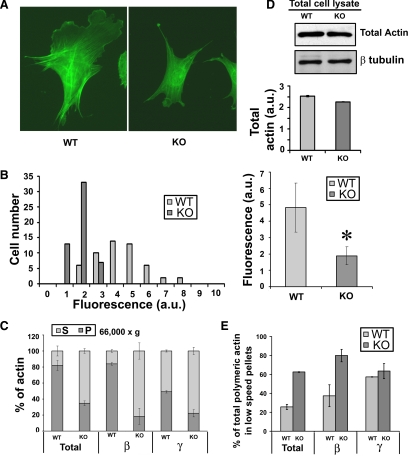
Figure 1.
Arginylation regulates actin polymer level in vivo. (A) Staining of wild-type (WT) and Ate1 knockout (KO) cells with rhodamine-phalloidin. (B) Frequency histogram showing the distribution of the total fluorescence levels in individual (left) and averaged (right) WT and Ate1 KO cells (n = 54; error bars, SD between the readings shown on the left; p < 0.0001). (C) Fractionation of actin from the lysates of WT and KO cells by high-speed centrifugation (see Figure S1 for the other centrifugation steps and Supplemental Table 1 for the percentages plotted in the diagrams). Bars show percentages of actin present in the supernatant (G-actin, light gray) and pellet (F-actin, dark gray) for total actin and individual actin isoforms, estimated by Western blot quantification using antibodies specific to total, β- and γ-actin, as labeled. Error bars, SD for two independent experiments. (D) Western blot analysis of WT and Ate1 KO cell lysates using antibody to total actin and β-tubulin as a loading control. Bars represent a quantification of total actin adjusted to the loading control, averaged from three independent experiments. (E) Percentage of total, β- and γ-actin in the lower speed centrifugation steps (representing bundles and aggregates) calculated from the centrifugation steps shown in C and in Figure S1. Error bars, SD for two independent experiments. See also Supplemental Table 1 for the percentages used to obtain the bar diagrams shown in C and E.
To confirm this hypothesis, we performed a biochemical fractionation of extracts from WT and KO cells by differential centrifugation (Figure 1C). Cell lysates were centrifuged sequentially at 200 × g (to remove nuclei, debris, and large protein aggregates), 1500 × g, 16,000 × g, and 66,000 × g (to pellet actin filaments). Supernatants and pellets from each step were normalized by protein concentration, and relative amounts of actin present in each fraction were quantified by Western blot and calculated as percentages of the total actin at each centrifugation step (see Figure S1 and Supplementary Table 1).
Western blots with antibodies against total actin showed that the major difference between WT and KO cell extracts during such fractionation was observed at the highest sedimentation speed, which separates F- and G-actin (Figure 1C). Although in WT cells the majority of actin (∼80%) was found in the 66,000 × g pellets, in Ate1 KO cells the fraction of actin found in the high-speed pellets was significantly lower (∼30%). These results are consistent with the results of phalloidin staining and suggest that Ate1 KO cells indeed contain a significantly lower amount of actin polymer.
We have previously found that although cultured mouse fibroblasts used in these assays contain an ∼1:1 ratio of β- and γ-actin, only β-, but not γ-actin undergoes N-terminal arginylation (Karakozova et al., 2006; Wong et al., 2007). To test whether the reduction in the intracellular actin polymer was biased toward any of these two actin isoforms, we probed the fractions from differential centrifugation steps with isoform-specific antibodies against β- or γ-actin. Although there was no significant difference between WT and KO actin sedimented at lower speeds (Figure S1), in high-speed fractions the differences in polymer levels were much more prominent for β- compared with γ-actin (Figure 1C), suggesting that the arginylation-dependent effect on the actin polymer level is biased toward β-actin containing polymers. Because β-actin is the only isoform found to be arginylated on the N-terminus, it is possible that this isoform-specific effect is regulated by N-terminal arginylation. At the same time it is possible that some of these effects are regulated by the other two arginylation sites on one or both isoforms.
To determine whether the decrease in the amount of actin polymer in Ate1 KO cells is caused by defects in actin polymerization or a reduction in the overall amount of actin protein in Ate1 KO cells, we measured the levels of the total actin protein in the lysates of WT and KO cells, normalized to total protein concentration by Western blot (Figure 1D). We found that in both cell types, actin protein levels were approximately the same.
We have previously shown that KO actin filaments polymerized in cell extracts undergo bundling and aggregation (Karakozova et al., 2006), suggesting that actin in Ate1 KO cells may form similar bundles and aggregates in vivo that sediment at lower centrifugation speeds. Given that WT and Ate1 KO cells had similar amounts of actin sedimenting at lower centrifugation speeds (Figure S1), although actin polymer sedimenting at high centrifugation speed in WT cells was significantly higher compared with KO (Figure 1D), the fraction of the overall actin polymer found in higher order structures, such as bundles and aggregates (precipitated at 200 ×, 1500 ×, and 16,000 × g), should be significantly higher in Ate1 KO cells. To estimate this, we quantified the percentage of actin polymer in the lower speed sedimentation fractions (200 ×, 1500 ×, and 16,000 × g) compared with the total amount of actin polymer sedimented at all speeds. We found that this percentage is ∼2.5 times higher in KO compared with WT (Figure 1E): a difference that could be explained by the overall reduction in the actin polymer level combined with the increased bundling or aggregation among the remaining polymer. Moreover, similar to the effect on the total actin polymer level, this reduction was strongly biased toward the β-actin isoform (Figure 1E).
Arginylation Regulates Polymerization of Pure Actin
To determine whether the decreased amount of F-actin in Ate1 KO cells is accompanied by changes in the polymerization capacity of nonarginylated actin, we characterized the in vitro properties of actin purified from WT and KO cell extracts (referred to as WT and KO actin, Figure 2A) by three different assays (Figure 2, B and C). The critical concentration of polymerization determined by high-speed sedimentation was similar for WT and KO, ∼0.4 μM (Figure 2B, left). At the same time, the rate of spontaneous actin polymerization measured by pyrene fluorescence (Kouyama and Mihashi, 1981; Cooper and Pollard, 1982) showed that KO actin had a steeper slope compared with the WT actin, when actin concentration versus polymerization rate was plotted (Figure 2B, right). To address whether this increased rate of polymerization was due to increased nucleation or elongation, we analyzed the length of the lag phase in the spontaneous polymerization curves and found that the lag phase for KO actin was ∼1.4 times shorter than WT, suggesting an increased nucleation rate (Figure S3). We also performed a pyrene fluorescence assay in the presence of prepolymerized actin seeds to measure elongation independent of nucleation. Remarkably, in this assay, KO actin polymerized slower than WT (Figure 2C and Figure S2), indicating that KO actin has a lower elongation rate.
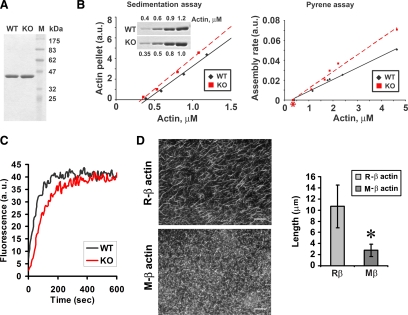
Figure 2.
Arginylation regulates polymerization properties of pure actin. (A) Coomassie blue–stained SDS-PAGE of actin purified from WT and Ate1 KO fibroblasts. (B) Left, sedimentation-based actin polymerization assay using lower actin concentrations to determine the critical concentration for polymerization. To perform the assay, actin at each concentration was polymerized at room temperature for 4 h, followed by centrifugation at 100,000 × g for 1 h at 17°C and quantification of actin in the pellets by SDS-PAGE and Coomassie blue staining. Inset, the Coomassie blue–stained SDS-PAGE of the sedimented actin, which was quantified and plotted against the corresponding actin concentrations on the graph. Right, actin polymerization rates measured using a pyrene actin assay. The assembly rates derived as the slopes of the individual pyrene actin curves in the rapid growth phase were plotted against the corresponding concentrations of actin. The critical concentration values (denoted with an asterisk) were plotted on the graph as determined in sedimentation assays (left). Data were combined from two independent experiments. (C) Pyrene actin polymerization assay in the presence of seeds to determine the elongation rate. G-Actin concentration used to obtain the curve was 5 μm. See Figure S2 for the curves at other actin concentrations. (D) Left, rhodamine-phalloidin staining of filaments polymerized from arginylated (R) and nonarginylated (M) actin. Scale bar, 20 μm. Right, average lengths of filaments formed from M- and R-actin determined by measuring images shown on right. Error bars, SD for 100 measurements (p < 0.0001).
Taken together, these results suggest that KO actin has faster nucleation but slower elongation compared with WT. This effect should lead to significant changes in the overall length of the actin filaments. Indeed, faster nucleation/slower elongation would result in a formation of a large number of seeds and delayed filament growth on these seeds, resulting in a larger amount of shorter filaments with KO actin compared with WT. In addition, it is possible that KO actin forms unstable, “brittle” filaments whose breakage could further alter the polymerization kinetics and may contribute to the formation of shorter filaments in KO compared with WT.
To test these results by an independent assay, we expressed unmodified and N-terminally arginylated β actin (termed M- and R-actin, respectively) in a baculovirus expression system (Figure S4) and performed polymerization assays with these actin proteins. Consistent with endogenous actin, M-actin (equivalent to KO actin) showed a greater increase in pyrene fluorescence per unit of polymerized actin compared with R-actin (equivalent to WT actin; Figure S5A). Control experiments showed that these two actins had similar ability to inhibit DNase-I activity, confirming that these two actins had proper activity and folding (Figure S5B). Examination of the overall morphology of M- and R-actin filaments stained by rhodamine-phalloidin showed that, consistent with the results of polymerization assays, M-actin indeed formed larger numbers of shorter filaments (overall length about four times shorter) compared with R-actin (Figure 2D).
To test whether polymerized KO actin has other ultrastructural defects, we next analyzed the structure of spontaneously polymerized actin purified from WT and KO cells by negative staining EM (Figure 3A). Actin in both preparations was capable of forming filaments with apparently normal structure (Figure 3A two top rows). In denser areas of the grids, both actins formed bundles and clusters (Figure 3A, third row from the top); however, at higher magnification, KO actin in these areas appeared structurally different from WT (Figure 3A, bottom three panels) and apparently consisted of abnormally clustered protein rather than bundled filaments. To quantify this effect and estimate which fraction of the KO and WT actin is found in this bundled and aggregated state, we fractionated prepolymerized actin filaments by sucrose density gradient centrifugation under the conditions, in which single actin filaments sediment in the middle of the gradient and actin bundles pellet as a distinct fraction at the bottom (see Figure 3B for gradient profiles and EM images of the peak fractions containing single filaments or aggregates). A significantly higher fraction of KO actin sedimented at the bottom of the gradient compared with WT. In addition, the KO actin filament peak appeared wider than WT, suggesting a tendency of KO actin filaments to aggregate, trailing toward the bottom of the gradient.
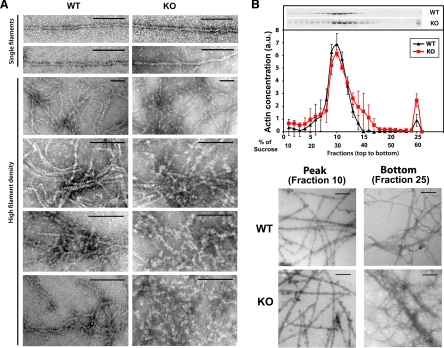
Figure 3.
Purified arginylation-free actin forms bundles and aggregates that are structurally distinct from normal filaments. (A) Negative staining EM images of actin filaments polymerized from pure WT (left column) and KO (right column) actin. Scale bars, 100 nm. (B) Sucrose density gradient fractionation of actin filaments polymerized from actins purified from WT and KO cells. Top, gel of the gradient fractions and protein level profile across the gradient. Bottom, negative staining EM images of the gradient peak (fraction 10) and bottom (fraction 25). Error bars, SD for two experiments.
The formation of abnormal actin clusters may have additional effects on actin polymerization: if these clusters have the capacity to nucleate actin filaments, such nucleation would contribute to an additional means by which KO actin yields filaments that, in addition to being shorter, are also less organized and less localized to the leading edge. To test this idea and to find out whether aggregated actin can serve as a nucleator, we compared KO actin polymerization dynamics in the absence and presence of the aggregates found in the bottom fraction of the sucrose gradient (fraction 25 in Figure 3B). We found that addition of fraction 25 indeed increases the actin polymerization rate and alters actin polymerization dynamics, suggesting that this fraction can serve as seeds for nucleation of new actin filaments (Figure S6).
Arginylation Regulates Actin-associated Protein Levels and Their Binding to F-Actin
The actin cytoskeleton is highly dynamic and requires the action of more than a hundred ABPs that take part in its formation and remodeling in vivo (Sheterline et al., 1995; Pollard et al., 2000). To determine whether actin arginylation state regulates its interaction with ABPs, we performed a 2Dgel comparison of actin preparations from WT and KO cells, obtained by phalloidin-induced polymerization in clarified cell extracts, followed by sedimentation of F-actin–ABP complexes. Several protein spots exhibited prominent changes between the two preparations (Figure 4, A and B). Gelsolin, drebrin, and PDI were significantly decreased in the KO sample, whereas twinfilin and capping protein subunits α and β were increased (Figure 4B).
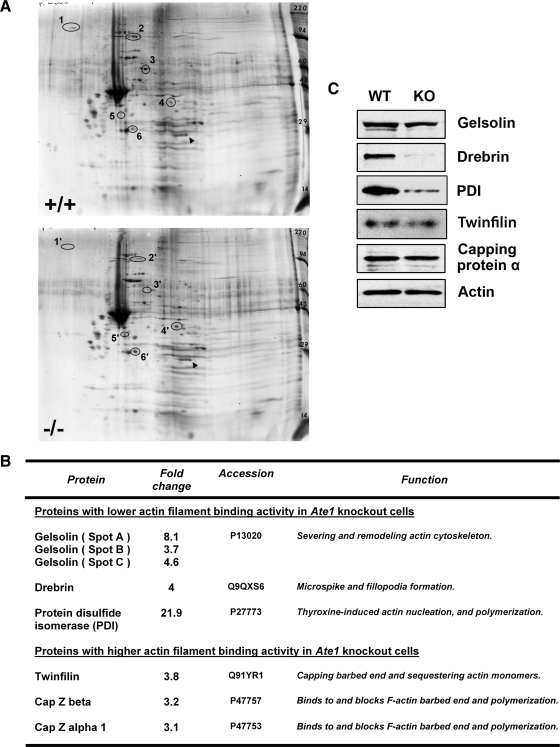
Figure 4.
Arginylation regulates ABPs and their interaction with F-actin. (A) Silver-stained 2D gels of actin filaments in complex with ABP obtained from WT and Ate1 KO fibroblasts by phalloidin-induced polymerization in clarified cell lysates followed by sedimentation. Gel pH range is 4.5–10, increasing left to right. Spots corresponding to those circled in the figure were excised from similarly run Coomassie-stained 2D gels and identified by mass spectrometry as (1) drebrin, (2) gelsolin, (3) PDI, (4) twinfilin, (5) capping protein α, and (6) capping protein β. (B) Table with identified proteins that are altered between the two gels. Fold-change p-values determined by Student's t test generated by gel comparison software based on the analysis of duplicate silver-stained gels were as follows: gelsolin spot A, 0.001; gelsolin spot B, 0.015; gelsolin spot C, 0.002; drebrin, 0.015; PDI, 0.028; twinfilin, 0.240; capping protein β, 0.041; and capping protein α, 1: 0.061. (C) Western blot analysis of WT and Ate1 KO total cell lysates.
To test the possibility that the differential abundance of these proteins in the two preparations reflects their overall abundance in the WT and KO cells, we analyzed the intracellular levels of these proteins by Western blot (Figure 4C). The levels of gelsolin, twinfilin, and capping protein were similar in both cell types (with gelsoin levels lower by a very small margin in the Ate1 KO cells; Figure S7C); however, drebrin and PDI were down-regulated to a considerable extent in Ate1 KO cells. Control experiments showed that this down-regulation was not seen in whole extracts of E12.5 Ate1 KO embryos and embryonic tissues (Figure S7, A and B), suggesting that drebrin and PDI down-regulation was cell type–specific or related to the differentiation events that follow fibroblast isolation from the embryo. Thus, the decreased amounts of drebrin and PDI in the KO F-actin preparation likely reflect their decreased intracellular level in response to Ate1 KO. At the same time, the altered levels of gelsolin, twinfilin, and capping protein in our F-actin preparation were due to their differential binding capacity for the WT and KO actin filaments.
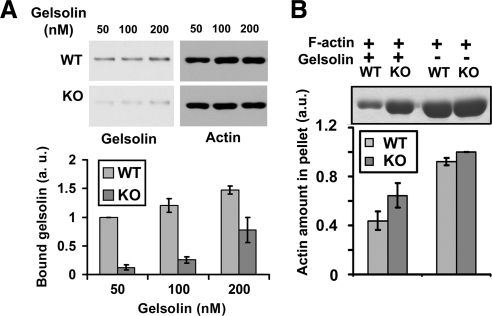
To confirm these results, we performed the following assays. First, to measure gelsolin interaction with WT and KO actin, we performed sedimentation of WT and KO actin filaments, prepolymerized under conditions similar to those used in the binding experiments, in the presence of gelsolin at low or high Ca2+ concentrations (Figure 5). At low Ca2+, gelsolin binds to the actin filaments but its severing activity is inhibited, allowing direct measurement of its interaction with the actin filaments (Figure 5A). At high Ca2+, the addition of gelsolin induces actin filament severing, and, after pelleting through a sucrose cushion, severed fragments of the actin filaments remain in the supernatant along with gelsolin. As a result, the amount of actin in the pellet fraction is reduced in proportion to the strength of gelsolin-induced severing (Figure 5B). Consistent with the results of pulldowns in the extracts, both assays confirmed that gelsolin indeed interacts more strongly with WT actin filaments.
Figure 5.
Arginylation regulates gelsolin–actin binding and gelsolin-induced severing of the actin filaments. (A) Cosedimentation assays in the presence of low Ca2+ at increasing gelsolin–actin ratios confirm that gelsolin binding to WT actin is stronger than to KO actin. Error bars, SEM of three independent measurements. Two-tailed unpaired t test; p < 0.0001 for 50 nM gelsolin, 0.0019 for 100 nM gelsolin, and 0.0412 for 200 nM gelsolin. (B) Cosedimentation assays in the presence of high Ca2+ (evidenced by the severing-dependent reduction of pelleting of WT actin in the presence of gelsolin) show higher severing activity of gelsolin toward WT actin. Error bars, SEM of four independent measurements; p = 0.0458.
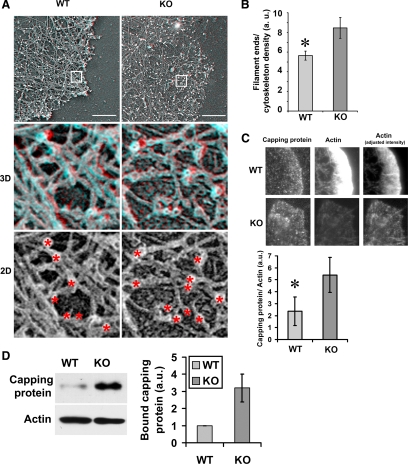
Second, to test whether the binding of the capping protein to the actin cytoskeleton is indeed higher in Ate1 KO cells compared with WT, we double-stained detergent-extracted WT and KO cells with capping protein and rhodamine-phalloidin and used the resulting images to measure the ratio of capping protein to the actin polymer level (see Figure 7C). We also performed a cosedimentation assay to directly test the binding of capping protein to the prepolymerized actin filaments in vitro (Figure 7D). These measurements showed that, consistent with the results of cosedimentation assay in cell extracts, the amount of capping protein bound to the actin cytoskeleton in vitro and in the lamellar region of the Ate1 KO cells was on average ∼2.3 times higher than in WT. Although it is possible that this difference is due at least in part to the increased number of exposed actin filament ends in vitro and in vivo, rather than to the increased affinity of the capping protein for the KO actin, it is clear that the binding of the capping protein to actin is higher in the absence of arginylation.
Figure 7.
Absence of arginylation increases capping protein binding and the number of filament ends in the leading edge actin network. (A) Top, electron micrographs of platinum replicas of the lamellipodial actin network in a WT and a KO cell with filament ends marked as white dots. Middle, stereo electron micrographs of the magnified regions boxed in the top panels, showing individual actin filament ends. (Red-cyan glasses should be used to view these images, with red on the left eye). Bottom, electron micrographs of the same regions as in the middle panels without the stereo effect, with actin filament ends marked by red asterisks. Scale bars, 500 nm. (B) Bar diagram showing the average number of visible filament ends normalized to total cytoskeleton density and averaged from three different images (error bars, SD; p = 0.0136). (C) Top, areas of the leading edge in WT and KO cells stained with an antibody to the capping protein (left panels) and actin, presented as identically scaled images to show the difference in the actin intensity between WT and KO (middle panels), and the images of the same area adjusted to the optimal brightness for each condition (right panels). Bottom, quantification of the ratios of capping protein to actin in WT and KO cells (error bars, SD of measurements from 32 WT and 31 KO cells; p < 0.0001). (D) Western blot and quantification of the levels of purified capping protein bound to WT and KO F-actin in an in vitro sedimentation assay. Error bars, the average of three independent measurements; two-tailed unpaired t test; p = 0.0534.
Arginylation Regulates the Ultrastructure of the Actin Network
Because we found that arginylation regulates several important properties of actin, the absence of arginylation is expected to result in ultrastructural changes in the intracellular actin networks. To test this hypothesis, we analyzed WT and KO cells by EM of platinum replicas of the cytoskeleton.
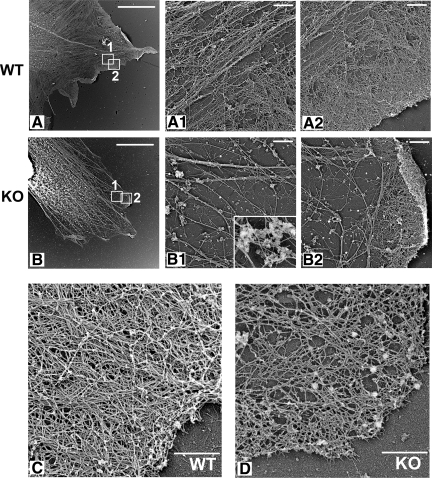
In agreement with light microscopy observations, actin filaments in Ate1 KO cells were scarce compared with WT, an effect which became especially prominent in the region of the leading lamella (Figure 6 and Figure S8). The zone of the dense lamellipodial network at the leading edge was reduced in Ate1 KO cells to roughly half in width compared with WT. At higher magnification, lamellipodial actin networks in Ate1 KO cells appeared scarce, disorganized, and contained prominent protein aggregates not usually found in WT cells (Figure 6B, 1 and 2, and inset in B1). At higher magnification, these protein aggregates appeared similar in morphology to those seen in vitro by negative staining EM (cf. bottom right images in Figure 3A and inset in Figure 6B1).
Figure 6.
Arginylation regulates actin network density and organization at the leading edge of the motile cells. Electron micrographs of the platinum replicas of the cytoskeleton in WT and Ate1 KO fibroblasts. (A and B) WT and Ate1 KO cell replicas at low magnification. (A1, A2, B1, and B2) Higher magnification images of the areas boxed in A and B, respectively, and represent the cell interior (A1 and B1) and leading edge (A2 and B2) from the WT and KO cells shown on the left. (C and D) Higher resolution images of lamellipodia from WT and Ate1 KO cells depicting the structural organization of actin network in these cells at the leading edge. Inset in B1, an enlarged image of a protein aggregate. Scale bars, (A and B) 10 μm and (A1, A2, B1, B2, C and D) 500 nm.
Shorter filaments and increased binding to capping proteins in KO actin seen in biochemical assays should result in the formation of shorter actin filaments in vivo, resulting in an increased number of visible filament ends, especially in the areas of high actin density such as the leading lamella. To test this possibility, we analyzed stereo images of platinum replicas of WT and KO cells in the area of the leading lamella with similar density of the actin filaments (Figure 7 and Figure S9) and counted the number of visible filament ends per local density of the cytoskeleton (Figure 7A). This analysis showed that Ate1 KO cells had an ∼50% higher number of filament ends in the lamellipodial actin network compared with WT cells (Figure 7B). Moreover, unlike WT, the filament ends in the KO cells were scattered throughout the lamellipodia rather than concentrated near the cell edge, suggesting that these ends may be capped and therefore unable to contribute to the pushing force that drives lamellipodia protrusion.
Additional defects observed in Ate1 KO cells included disorganization of the filopodia (Figure S10). Fewer filopodial actin bundles could be seen in the KO cells compared with the control cells, and the existing bundles showed defects that suggested abnormal formation and/or premature termination (Svitkina et al., 2003; Korobova and Svitkina, 2008).
DISCUSSION
The results of this study demonstrate that arginylation regulates actin polymer level in vivo, actin properties, and the ultrastructure of the intracellular actin networks. We have previously shown that β-actin arginylation regulates lamella formation (Karakozova et al., 2006). Here we show that arginylation also regulates overall actin polymerization, its interaction with ABPs, and actin network ultrastructure, expected to cause severe impairments in cell spreading and motility. This is the first demonstration of an arginylation-regulated mechanism that affects multiple functions of a protein in vivo, resulting in global intracellular effects.
We had previously hypothesized that a lack of arginylation causes a collapse of preformed actin networks at the leading edge (Karakozova et al., 2006). Here we tested this hypothesis and found that a lack of arginylation results in global, cell-wide defects in actin network formation and maintenance and leads to a dramatic reduction in actin polymer level, shortened filaments, and the formation of intracellular aggregates, defects that are by no means confined to the cell leading edge. These defects are rooted in different polymerization properties of WT and KO actin and their differential interaction with a prominent subset of ABPs that act in concert to regulate actin network in vivo. We have previously found that only β- but not γ-actin undergoes N-terminal arginylation and that two additional arginylation sites on the actin molecule may be modified on one or both nonmuscle actin isoforms. The present study suggests that the combined effects of actin regulation by arginylation are important not only for the leading lamella, but for the proper cytoarchitecture of the entire cytoplasm.
Because arginylation is predicted to affect the surface properties of actin monomers and filaments by placing one or several positive charges on their normally negatively charged surface, it is expected to affect the ability of actin to interact with other proteins that modulate its properties in vivo. Here we found that gelsolin, capping protein, and twinfilin show significant changes in binding to actin filaments with different arginylation states, expected to produce marked changes in the actin cytoskeleton. Gelsolin, primarily known as an F-actin–severing protein, is the major modulator of rapid actin remodeling; reduced gelsolin binding to nonarginylated F-actin could result in a decrease of actin subunit turnover and impaired lamellar activity of the cell. Indeed, gelsolin KO fibroblasts have decreased migration speeds (Witke et al., 1995), consistent with the fact that Ate1 KO fibroblasts have impaired motility in culture (Karakozova et al., 2006). It has been previously found that residues 1–10 at the actin N-terminus are essential for gelsolin binding (Feinberg et al., 1995; Burtnick et al., 1997), further suggesting that arginylation may be important in the regulation of this binding and the subsequent actin turnover, in vivo. Capping protein and twinfilin both act as barbed end capping proteins that inhibit actin dynamics and the formation of new actin polymer (Schleicher et al., 1984; Carlier and Pantaloni, 1997; Goode et al., 1998; Cooper and Schafer, 2000; Vartiainen et al., 2000; Palmgren et al., 2001; Ojala et al., 2002; Helfer et al., 2006), which could contribute to the reduced amount of actin polymer in Ate1 KO cells. Although capping protein has not been shown to bind to the actin interface predicted to be affected by N-terminal arginylation (Narita et al., 2006), the tendency of KO actin to form shorter filaments, by altered polymerization and/or breakage, likely increases the binding of these capping proteins to the filament barbed ends.
The increased binding of actin-capping proteins could also inhibit filopodia formation in KO cells by competing with the filopodia tip complexes and blocking the growth of the filopodial bundles, causing their premature termination (Svitkina et al., 2003). Filopodia formation could also be inhibited by the reduced amount of drebrin, a protein that has been shown to play a role in filopodia formation in neurons and other cell types (Shirao et al., 1994; Hayashi and Shirao, 1999; Peitsch et al., 2006). The role of PDI in the actin cytoskeleton functioning is not clear, but it has been shown that PDI associates with actin filaments upon thyroxine treatment, which also induces actin polymerization, suggesting that PDI is somehow involved in actin polymerization and that its down-regulation in Ate1 KO cells would affect actin polymer levels (Safran et al., 1992). Although it is not presently known how Ate1 KO down-regulates the expression of drebrin and PDI, it could be envisioned that some proteins involved in the regulation of their expression (at the transcription or translation level) are substrates of Ate1. It is possible that such regulation uses a feedback mechanism involving drebrin and PDI themselves, or their association with actin that could, for example, increase or decrease their metabolic stability and turnover.
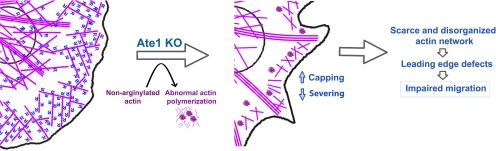
Based on our data, we propose a model of how arginylation regulates actin cytoskeleton in vivo (Figure 8). Nonarginylated actin at physiological concentrations forms abnormal aggregates, which are sequestered and disassembled back into the monomer pool, resulting in the lower levels of the intracellular actin polymer and the corresponding increase in the amount of monomers in the cytoplasm. Increased binding of capping proteins further lowers the polymer levels and decreased gelsolin-dependent actin turnover contributes to cytoskeletal abnormalities, resulting in a scarce and disorganized actin network in the cell body and at the leading edge. These effects, compounded by the general actin disorganization due to abnormal clustering of actin subunits and nucleation of new filaments off these clusters, result in leading edge collapse, causing severe impairments in cell migration.
Figure 8.
Arginylation is required for lamella formation, cell spreading, and locomotion. In a normal cell, arginylated actin facilitates the formation of a proper leading edge network and normal actin structures throughout the cell. Absence of arginylation results in actin forming abnormal aggregates, which are sequestered and disassembled back into the monomer pool, causing the overall reduction of the amount of the actin polymer. Increased binding of capping proteins further lowers the polymer levels, and decreased gelsolin-dependent actin turnover contributes to the cytoskeletal abnormalities, resulting in a scarce and disorganized actin network in the cell body and at the leading edge. These effects result in reduced cell spreading and lamella collapse, causing severe impairments in cell migration.
Previously we had reported that, in addition to actin, several ABPs are arginylated in vivo, including Arp3, filamin, spectrin, myosin 9, and talin (Wong et al., 2007). The role of arginylation of these, and other actin-associated proteins in actin cytoskeleton regulation remains to be characterized. It appears likely that arginylation of Arp3, a major actin nucleator at the cell leading edge, contributes to the formation of the proper leading edge network and that arginylation of other proteins, like filamin and spectrin, may facilitate normal actin arrangement in the cytoplasm. The current study takes the first steps toward uncovering the molecular mechanisms behind arginylation-dependent regulation of cell locomotion and embryonic development.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. D. Dong for actin lag phase calculations and help with the analysis of pyrene actin curves; Dr. Bing He for help with actin pyrene assays; C. Yuan at the Proteomics Core Facility of the University of Pennsylvania for mass spectrometry; J. Johansen and M. Hoelter (Kendrick Laboratories) for 2D gel electrophoresis and computerized gel comparisons; Drs. J. C. Bulinski, H. S. Yin, D. Schafer, and P. Lappalainen for the gift of antibodies; N. Shah and R. Meade for help with electron microscopy; Drs. Y. Goldman, E. Yarmola, and V. Sirotkin for helpful discussions; and Drs. A. Mogilner, V. Rodionov, E. Korn, and S. Kurosaka for critical reading of the manuscript. This work was supported by National Institutes of Health Grant 5R01HL084419, W. W. Smith Charitable Trust, and Philip Morris Research Management Group awards to A.K. J.F.D. is a HSFC New Investigator.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-09-0829) on February 24, 2010.
REFERENCES
- Balzi E., Choder M., Chen W. N., Varshavsky A., Goffeau A. Cloning and functional analysis of the arginyl-tRNA-protein transferase gene ATE1 of Saccharomyces cerevisiae. J. Biol. Chem. 1990;265:7464–7471. [PubMed] [Google Scholar]
- Burtnick L. D., Koepf E. K., Grimes J., Jones E. Y., Stuart D. I., McLaughlin P. J., Robinson R. C. The crystal structure of plasma gelsolin: implications for actin severing, capping, and nucleation. Cell. 1997;90:661–670. doi: 10.1016/s0092-8674(00)80527-9. [DOI] [PubMed] [Google Scholar]
- Carlier M. F., Pantaloni D. Control of actin dynamics in cell motility. J. Mol. Biol. 1997;269:459–467. doi: 10.1006/jmbi.1997.1062. [DOI] [PubMed] [Google Scholar]
- Cooper J. A., Pollard T. D. Methods to measure actin polymerization. Methods Enzymol. 1982;85(Pt B):182–210. doi: 10.1016/0076-6879(82)85021-0. [DOI] [PubMed] [Google Scholar]
- Cooper J. A., Schafer D. A. Control of actin assembly and disassembly at filament ends. Curr. Opin. Cell Biol. 2000;12:97–103. doi: 10.1016/s0955-0674(99)00062-9. [DOI] [PubMed] [Google Scholar]
- Decca M. B., Bosc C., Luche S., Brugiere S., Job D., Rabilloud T., Garin J., Hallak M. E. Protein arginylation in rat brain cytosol: a proteomic analysis. Neurochem. Res. 2006;31:401–409. doi: 10.1007/s11064-005-9037-z. [DOI] [PubMed] [Google Scholar]
- Decca M. B., Carpio M. A., Bosc C., Galiano M. R., Job D., Andrieux A., Hallak M. E. Post-translational arginylation of calreticulin: a new isospecies of calreticulin component of stress granules. J. Biol. Chem. 2007;282:8237–8245. doi: 10.1074/jbc.M608559200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg J., Benyamin Y., Roustan C. Definition of an interface implicated in gelsolin binding to the sides of actin filaments. Biochem. Biophys. Res. Commun. 1995;209:426–432. doi: 10.1006/bbrc.1995.1520. [DOI] [PubMed] [Google Scholar]
- Goode B. L., Drubin D. G., Lappalainen P. Regulation of the cortical actin cytoskeleton in budding yeast by twinfilin, a ubiquitous actin monomer-sequestering protein. J. Cell Biol. 1998;142:723–733. doi: 10.1083/jcb.142.3.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K., Shirao T. Change in the shape of dendritic spines caused by overexpression of drebrin in cultured cortical neurons. J. Neurosci. 1999;19:3918–3925. doi: 10.1523/JNEUROSCI.19-10-03918.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfer E., Nevalainen E. M., Naumanen P., Romero S., Didry D., Pantaloni D., Lappalainen P., Carlier M. F. Mammalian twinfilin sequesters ADP-G-actin and caps filament barbed ends: implications in motility. EMBO J. 2006;25:1184–1195. doi: 10.1038/sj.emboj.7601019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji H. Further studies on the soluble amino acid incorporating system from rat liver. Biochemistry. 1968;7:3844–3850. doi: 10.1021/bi00851a009. [DOI] [PubMed] [Google Scholar]
- Kaji H., Novelli G. D., Kaji A. A soluble amino acid-incorporating system from rat liver. Biochim. Biophys. Acta. 1963;76:474–477. [PubMed] [Google Scholar]
- Karakozova M., Kozak M., Wong C. C., Bailey A. O., Yates J. R., 3rd, Mogilner A., Zebroski H., Kashina A. Arginylation of beta-actin regulates actin cytoskeleton and cell motility. Science. 2006;313:192–196. doi: 10.1126/science.1129344. [DOI] [PubMed] [Google Scholar]
- Kopitz J., Rist B., Bohley P. Post-translational arginylation of ornithine decarboxylase from rat hepatocytes. Biochem. J. 1990;267:343–348. doi: 10.1042/bj2670343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korobova F., Svitkina T. Arp2/3 complex is important for filopodia formation, growth cone motility, and neuritogenesis in neuronal cells. Mol. Biol. Cell. 2008;19:1561–1574. doi: 10.1091/mbc.E07-09-0964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouyama T., Mihashi K. Fluorimetry study of N-(1-pyrenyl)iodoacetamide-labelled F-actin. Local structural change of actin protomer both on polymerization and on binding of heavy meromyosin. Eur. J. Biochem. 1981;114:33–38. [PubMed] [Google Scholar]
- Kwon Y. T., Kashina A. S., Davydov I. V., Hu R. G., An J. Y., Seo J. W., Du F., Varshavsky A. An essential role of N-terminal arginylation in cardiovascular development. Science. 2002;297:96–99. doi: 10.1126/science.1069531. [DOI] [PubMed] [Google Scholar]
- Lal A. A., Korn E. D., Brenner S. L. Rate constants for actin polymerization in ATP determined using cross-linked actin trimers as nuclei. J. Biol. Chem. 1984;259:8794–8800. [PubMed] [Google Scholar]
- Lee M. J., Tasaki T., Moroi K., An J. Y., Kimura S., Davydov I. V., Kwon Y. T. RGS4 and RGS5 are in vivo substrates of the N-end rule pathway. Proc. Natl. Acad. Sci. USA. 2005;102:15030–15035. doi: 10.1073/pnas.0507533102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz M., Popp D., Holmes K. C. Refinement of the F-actin model against X-ray fiber diffraction data by the use of a directed mutation algorithm. J. Mol. Biol. 1993;234:826–836. doi: 10.1006/jmbi.1993.1628. [DOI] [PubMed] [Google Scholar]
- Mejillano M. R., Kojima S., Applewhite D. A., Gertler F. B., Svitkina T. M., Borisy G. G. Lamellipodial versus filopodial mode of the actin nanomachinery: pivotal role of the filament barbed end. Cell. 2004;118:363–373. doi: 10.1016/j.cell.2004.07.019. [DOI] [PubMed] [Google Scholar]
- Narita A., Takeda S., Yamashita A., Maeda Y. Structural basis of actin filament capping at the barbed-end: a cryo-electron microscopy study. EMBO J. 2006;25:5626–5633. doi: 10.1038/sj.emboj.7601395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida E., Sakai H. Kinetic analysis of actin polymerization. J. Biochem. 1983;93:1011–1020. doi: 10.1093/oxfordjournals.jbchem.a134224. [DOI] [PubMed] [Google Scholar]
- O'Farrell P. H. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Ojala P. J., Paavilainen V. O., Vartiainen M. K., Tuma R., Weeds A. G., Lappalainen P. The two ADF-H domains of twinfilin play functionally distinct roles in interactions with actin monomers. Mol. Biol. Cell. 2002;13:3811–3821. doi: 10.1091/mbc.E02-03-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otey C. A., Kalnoski M. H., Bulinski J. C. Identification and quantification of actin isoforms in vertebrate cells and tissues. J. Cell. Biochem. 1987;34:113–124. doi: 10.1002/jcb.240340205. [DOI] [PubMed] [Google Scholar]
- Palmgren S., Ojala P. J., Wear M. A., Cooper J. A., Lappalainen P. Interactions with PIP2, ADP-actin monomers, and capping protein regulate the activity and localization of yeast twinfilin. J. Cell Biol. 2001;155:251–260. doi: 10.1083/jcb.200106157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peitsch W. K., Bulkescher J., Spring H., Hofmann I., Goerdt S., Franke W. W. Dynamics of the actin-binding protein drebrin in motile cells and definition of a juxtanuclear drebrin-enriched zone. Exp. Cell Res. 2006;312:2605–2618. doi: 10.1016/j.yexcr.2006.04.017. [DOI] [PubMed] [Google Scholar]
- Pollard T. D., Blanchoin L., Mullins R. D. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu. Rev. Biophys. Biomol. Struct. 2000;29:545–576. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- Rubenstein P. A., Spudich J. A. Actin microheterogeneity in chick embryo fibroblasts. Proc. Natl. Acad. Sci. USA. 1977;74:120–123. doi: 10.1073/pnas.74.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safran M., Farwell A. P., Leonard J. L. Thyroid hormone-dependent redistribution of the 55-kilodalton monomer of protein disulfide isomerase in cultured glial cells. Endocrinology. 1992;131:2413–2418. doi: 10.1210/endo.131.5.1425439. [DOI] [PubMed] [Google Scholar]
- Schafer D. A., Jennings P. B., Cooper J. A. Rapid and efficient purification of actin from nonmuscle sources. Cell Motil. Cytoskelet. 1998;39:166–171. doi: 10.1002/(SICI)1097-0169(1998)39:2<166::AID-CM7>3.0.CO;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleicher M., Gerisch G., Isenberg G. New actin-binding proteins from Dictyostelium discoideum. EMBO J. 1984;3:2095–2100. doi: 10.1002/j.1460-2075.1984.tb02096.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheterline P., Clayton J., Sparrow J. Actin. Protein Profile. 1995;2:1–103. [PubMed] [Google Scholar]
- Shirao T., Hayashi K., Ishikawa R., Isa K., Asada H., Ikeda K., Uyemura K. Formation of thick, curving bundles of actin by drebrin A expressed in fibroblasts. Exp. Cell Res. 1994;215:145–153. doi: 10.1006/excr.1994.1326. [DOI] [PubMed] [Google Scholar]
- Soffer R. L. Enzymatic modification of proteins. 4. Arginylation of bovine thyroglobulin. J. Biol. Chem. 1971;246:1481–1484. [PubMed] [Google Scholar]
- Soffer R. L. Enzymatic arginylation of beta-melanocyte-stimulating hormone and of angiotensin II. J. Biol. Chem. 1975;250:2626–2629. [PubMed] [Google Scholar]
- Svitkina T. Electron microscopic analysis of the leading edge in migrating cells. Methods Cell Biol. 2007;79:295–319. doi: 10.1016/S0091-679X(06)79012-4. [DOI] [PubMed] [Google Scholar]
- Svitkina T. M., Borisy G. G. Correlative light and electron microscopy of the cytoskeleton of cultured cells. Methods Enzymol. 1998;298:570–592. doi: 10.1016/s0076-6879(98)98045-4. [DOI] [PubMed] [Google Scholar]
- Svitkina T. M., Bulanova E. A., Chaga O. Y., Vignjevic D. M., Kojima S., Vasiliev J. M., Borisy G. G. Mechanism of filopodia initiation by reorganization of a dendritic network. J. Cell Biol. 2003;160:409–421. doi: 10.1083/jcb.200210174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandekerckhove J., Deboben A., Nassal M., Wieland T. The phalloidin binding site of F-actin. EMBO J. 1985;4:2815–2818. doi: 10.1002/j.1460-2075.1985.tb04008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandekerckhove J., Weber K. Mammalian cytoplasmic actins are the products of at least two genes and differ in primary structure in at least 25 identified positions from skeletal muscle actins. Proc. Natl. Acad. Sci. USA. 1978;75:1106–1110. doi: 10.1073/pnas.75.3.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartiainen M., Ojala P. J., Auvinen P., Peranen J., Lappalainen P. Mouse A6/twinfilin is an actin monomer-binding protein that localizes to the regions of rapid actin dynamics. Mol. Cell. Biol. 2000;20:1772–1783. doi: 10.1128/mcb.20.5.1772-1783.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witke W., Sharpe A. H., Hartwig J. H., Azuma T., Stossel T. P., Kwiatkowski D. J. Hemostatic, inflammatory, and fibroblast responses are blunted in mice lacking gelsolin. Cell. 1995;81:41–51. doi: 10.1016/0092-8674(95)90369-0. [DOI] [PubMed] [Google Scholar]
- Wong C. C., Xu T., Rai R., Bailey A. O., Yates J. R., 3rd, Wolf Y. I., Zebroski H., Kashina A. Global analysis of posttranslational protein arginylation. PLoS Biol. 2007;5:e258. doi: 10.1371/journal.pbio.0050258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Pring M., Wear M. A., Huang M., Cooper J. A., Svitkina T. M., Zigmond S. H. Mammalian CARMIL inhibits actin filament capping by capping protein. Dev. Cell. 2005;9:209–221. doi: 10.1016/j.devcel.2005.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates S. P., Otley M. D., Dawson J. F. Overexpression of cardiac actin with baculovirus is promoter dependent. Arch. Biochem. Biophys. 2007;466:58–65. doi: 10.1016/j.abb.2007.07.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.