We show that the putative N-peptide binding area in Sec1p domain 1 is important for Mso1p binding and that Mso1p can interact with Sso1p and Sso2p. Our results suggest that Mso1p mimics N-peptide binding to facilitate membrane fusion.
Abstract
Sec1p/Munc18 (SM) family proteins regulate SNARE complex function in membrane fusion through their interactions with syntaxins. In addition to syntaxins, only a few SM protein interacting proteins are known and typically, their binding modes with SM proteins are poorly characterized. We previously identified Mso1p as a Sec1p-binding protein and showed that it is involved in membrane fusion regulation. Here we demonstrate that Mso1p and Sec1p interact at sites of exocytosis and that the Mso1p–Sec1p interaction site depends on a functional Rab GTPase Sec4p and its GEF Sec2p. Random and targeted mutagenesis of Sec1p, followed by analysis of protein interactions, indicates that Mso1p interacts with Sec1p domain 1 and that this interaction is important for membrane fusion. In many SM family proteins, domain 1 binds to a N-terminal peptide of a syntaxin family protein. The Sec1p-interacting syntaxins Sso1p and Sso2p lack the N-terminal peptide. We show that the putative N-peptide binding area in Sec1p domain 1 is important for Mso1p binding, and that Mso1p can interact with Sso1p and Sso2p. Our results suggest that Mso1p mimics N-peptide binding to facilitate membrane fusion.
INTRODUCTION
During exocytosis, an evolutionarily conserved molecular machinery regulates transport vesicle targeting, tethering, and fusion at the plasma membrane. In the yeast Saccharomyces cerevisiae this machinery involves the action of the exocyst tethering complex, a Rab family small GTPase Sec4p, and the Sec1/Munc-18 (SM) family protein Sec1p (Novick and Guo, 2002; Toonen and Verhage, 2007; He and Guo, 2009). After tethering and docking, fusion of transport vesicles with the plasma membrane is mediated by the exocytic SNARE (soluble N-ethylmaleimide–sensitive factor attachment protein) complex composed of Snc1p/2p, Sec9p, and Sso1p/2p proteins (Jahn and Scheller, 2006). Despite numerous studies, the molecular interactions and how transport vesicle docking proceeds to SNARE complex–mediated membrane fusion are largely unknown.
The SM protein family members are central regulators of SNARE complex function (Gallwitz and Jahn, 2003; Kauppi et al., 2004; Toonen and Verhage, 2007). They appear to use three apparently different binding modes with the SNARE family proteins (Toonen and Verhage, 2007). First, several SM family members interact with their cognate SNARE complexes through binding to a N-terminal peptide of syntaxins (Bracher and Weissenhorn, 2002; Dulubova et al., 2002; Yamaguchi et al., 2002; Peng and Gallwitz, 2002; Latham et al., 2006; Carpp et al., 2006; Toonen and Verhage, 2007; Hu et al., 2007). Second, the S. cerevisiae Sec1p seems to interact predominantly with an assembled ternary SNARE complex (Carr et al., 1999; Scott et al., 2004; Togneri et al., 2006). Third, Munc18-1 and Vps45 bind closed conformations of syntaxin1 or Tlg2p, respectively (Misura et al., 2000; Furgason et al., 2009). However, recent results imply that Munc18-1 also can interact with a ternary SNARE complex and, at the same time, bind the N-terminal peptide of syntaxin1 (Dulubova et al., 2007; Shen et al., 2007; Deak et al., 2009). In contrast to syntaxins that are involved in exocytosis in higher eukaryotes, yeast Sso1p and Sso2p do not possess a N-terminal peptide. Thus, exocytic Sso protein–containing SNARE complexes may need the contribution of additional components for their efficient assembly and function in vivo. Candidates for such regulators could be SM protein interacting proteins.
In addition to syntaxins, only a few SM-binding proteins are known and, to large extent, detailed understanding on their interaction with SM proteins is lacking. Mint1, Mint2, Doc2, Granulophilin/Slp4, and phospholipase D proteins interact with Munc18-1 in mammalian cells (Okamoto and Sudhof, 1997; Verhage et al., 1997; Lee et al., 2004). In yeast, two direct SM family protein interactors, Mso1p and Vac1p, have been identified. Vac1p is a phosphatidyl-inositol-3-phosphate–binding protein that interacts with Vps45p and is required for proper vacuole maintenance (Weisman and Wickner, 1992). The Sec1p-interacting protein Mso1p was identified as a multicopy suppressor for the sec1-1 temperature-sensitive mutant (Aalto et al., 1997). Deletion of MSO1 in vegetatively growing haploid cells leads to vesicle accumulation at the site of cell growth and completely inhibits fusion of precursor vesicles in the de novo plasma membrane (prospore membrane) formation in sporulating diploid cells (Aalto et al., 1997; Jantti et al., 2002; Knop et al., 2005). These results suggest a positive role for Mso1p in membrane fusion. Mso1p binds Sec1p with a N-terminal peptide and it copurifies with Sec1p, Ssop/Sec9p/Sncp SNARE complexes, and Sec15p (Knop et al., 2005; Castillo-Flores et al., 2005). When combined with conditional mutations in SEC1, SEC2 and SEC4, MSO1 deletion is lethal (Aalto et al., 1997; Knop et al., 2005). Thus, genetic and biochemical interactions position Mso1p functionally in the interface of the exocyst, Sec4p, and SNARE complexes (Knop et al., 2005).
To understand how Mso1p and Sec1p interact to regulate membrane fusion, we generated a collection of sec1 mutants and analyzed their interactions with Mso1p and their functionality in vivo. Our studies show that the putative syntaxin N-peptide binding area in Sec1p domain 1 is important for Mso1p binding. Furthermore, our results indicate that Mso1p can interact with Sso1p and Sso2p. Collectively, the results suggest that Mso1p, through its binding to Sec1p domain 1, stabilizes Sec1p interaction with the SNARE complexes and facilitates membrane fusion.
MATERIALS AND METHODS
Strains
The yeast strains used are shown in Table 1. Standard growth media were used (Sherman, 1991). H3325, H3327, H3329, and H3331 were obtained by mating H2659 with H305, H306, H1127, and H1128 followed by tetrad dissection. Construction of H3476 and H3479 was achieved by transformation of H2905 with wt SEC1 containing plasmid B578, followed by tetrad dissection to obtain a haploid strain where the sole copy of SEC1 is expressed from a URA3 marker containing plasmid. To generate H3582 and H3584, MSO1 was hemagglutinin (HA)-tagged in H3476 and H3479 by transformation with a PCR cassette generated using pYM24 (B2966) as the template (Janke et al., 2004). The LEU2 plasmids bearing the sec1 insertion, or point mutations, were transformed into H3582, followed by 5-fluoroorotic acid (5-FOA) treatment to evict the wt SEC1 in the URA3 plasmid. MSO1 was subsequently deleted in these strains by transformation with a PCR cassette generated using pFA6-natNT2 (B3022) as the template (Janke et al., 2004).
Table 1.
Yeast strains
| Name | Genotype | Source |
|---|---|---|
| H304 | MATaleu2-3,112 ura3-52 | P. Novick |
| H305 | MATasec1-1 ura3-52 | P. Novick |
| H306 | MATasec1-11 ura3-52 | P. Novick |
| H758 | MATα sec6-4 leu2-3,112 ura3-52 | P. Novick |
| H759 | MATasec9-4 leu2-3,112 ura3-52 | P. Novick |
| H761 | MATasec15-1 leu2-3,112 ura3-52 | P. Novick |
| H891 | MATα sec18-1 leu2-3,112 trp1-289 ura3-52 | R. Schekmann |
| H1127 | MATα sec2-41 leu2-3,112 ura3-52 | P. Novick |
| H1128 | MATα sec4-8 leu2-3,112 ura3-52 | P. Novick |
| H1152 | MATasso2-1 leu2-3,112 trp1-1 ura3-1 sso1::HIS3 ade2-1 his3-11,15 can1-100 | H. Ronne |
| H1910 | MATaura3 trp1 his3 6lexAop-LEU2 | E. Golemis |
| H2530 | MATa/MATα leu2-3,112/leu2-3,112 ura3-52/ura3-52 | This study |
| H2598 | MATalys2 ura3 HO::hisG | M. Knop |
| H2599 | MATα lys2 ura3 LEU2::hisG HO::LYS2 | M. Knop |
| H2657 | MATaleu2-3,112 ura3-52 mso1::MSO1-3HA::kanMX6 | Knop et al. (2005) |
| H2658 | MATaleu2-3,112 ura3-52 mso1::hphMX4 GAL+ | Knop et al. (2005) |
| H2659 | MATα ura3-52 his4-619 mso1::MSO1-3HA::kanMX6 | Knop et al. (2005) |
| H2905 | MATa/MATα his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 met15Δ0/MET15 LYS2/lys2Δ0 ura3Δ0/ura3Δ0 sec1::kanMX4/SEC1 | EUROSCARF |
| H3325 | MATasec1-1 ura3-52 His4-619 mso1::MSO1-3HA::KanMX6 | This study |
| H3327 | MATasec1-11 ura3-52 His4-619 mso1::MSO1–3HA::KanMX6 | This study |
| H3329 | MATasec2-41leu2-3 ura3-52 mso1::MSO1-3HA::KanMX6 | This study |
| H3331 | MATasec4-8 leu2-3,112 ura3-52 mso1::MSO1-3HA::KanMX6 | This study |
| H3366 | MATaura3-52 lys2-801 trp1del63 his3del200 leu2del1 SEC1::SEC1-3HA-kanMX | M. Knop |
| H3367 | MATaura3-52 lys2-801 trp1del63 his3del200 leu2del1 mso1del::hphMX SEC1::SEC1-3HA-kanMX | M. Knop |
| H3419 | MATα his3-del200 leu2-3,112 ura3-52 ABP1-RFP::HIS3 | D. Drubin |
| H3463 | MATaLSP1::LSP1-RFP-KanMX leu2-3,112 ura3-52 | This study |
| H3466 | MATaLSP1::LSP1-RFP-KanMX pil1::natNT2 leu2-3,112 ura3-52 | This study |
| H3476 | MATahis3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 sec1::kanMX4 [YCpSEC1-URA3] | This study |
| H3479 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 sec1::kanMX4 [YCpSEC1-URA3] | This study |
| H3582 | MATahis3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 sec1::kanMX4 MSO1::MSO1-3HA-hphMX [YCpSEC1-URA3] | This study |
| H3584 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 sec1::kanMX4 MSO1::MSO1-3HA-hphMX [YCpSEC1-URA3] | This study |
Plasmids
Plasmids used are shown in the Supplementary Table 1. For bimolecular fluorescence complementation (BiFC), PCR-amplified SEC1 wt or the site-directed mutants were cloned as BamHI/SalI fragments into plasmid B2986. Similarly, MSO1 was cloned as a BglII/XhoI fragment into p416 METYC-CDC42 (B3031) (Cole et al., 2007) by replacing CDC42 with MSO1. The SEC1 wt and the L25D mutant were ligated to B3021 after releasing them from B2930 and B3060 with BamHI/SalI digestion. As the non-Sec1-binding controls, DNA fragments encoding N-terminal deletants of MSO1 (amino acids 59-210 or 136-210) were amplified with BglII and XhoI sites and cloned into B3031 to generate B3043 and B3353, respectively. Furthermore, the MSO1 fragments for amino acids 1-58, and 1-135 were amplified with BglII and XhoI sites and cloned into B3031. For stronger expression, full-length MSO1 or MSO1(136-210) were amplified with SpeI and SalI restriction sites and cloned into B2985 containing an ADH1 promoter (B2918 and B3012, respectively). To generate B3307, the SSO1 open reading frame (ORF) was amplified by PCR with BamHI and EcoRI sites and cloned into the BiFC vector B3018. For B3309, the SSO2 ORF was amplified by PCR with BamHI/BamHI sites and subcloned into B3018.
For the yeast two-hybrid analysis, the fragments encoding cytosolic parts of SSO1, SNC1, and SNC2 were amplified by PCR with EcoRI and XhoI restriction sites and cloned into B1226. The SSO2 was cut out from B1487 with EcoRI and XhoI and ligated into B1226. Sequences encoding Sec1p domain 1 and domain 3B were amplified by PCR with EcoRI and XhoI sites and ligated into B1226, generating B3272 and B3271, respectively. To generate B3400 and 3401, the MSO1(38-94) and MSO1(38-132) were amplified adding NcoI and XhoI restriction sites and cloned into B1231. For B3364, the MSO1(136-210) was amplified adding BglII and XhoI sites and cloned into B1232 that had been linearized with BamHI and XhoI.
For the SEC1 insertion library, wt SEC1 was first cloned into pBluescript SK− as a HindIII/XbaI fragments. The plasmid B2856 was generated by site-directed mutagenesis (QuickChange, Stratagene, La Jolla, CA) to introduce StuI and BglII restriction sites before the start codon and after the stop codon in SEC1. For the yeast two-hybrid assay, the B3073 plasmid was generated from pB42AD-B by removal of the NotI site with the Klenow enzyme (New England Biolabs, Beverly, MA). The plasmid SEC1pB42AD-BΔNotI (B3074) containing the wt SEC1 fragment was generated by cloning a StuI and BglII fragment from B2856. For in vivo studies, wt SEC1, or the insertion mutant versions, were subcloned into B707 where the NotI restriction site had been removed by filling-in with Klenow enzyme. Point mutations in SEC1 were introduced by site-directed mutagenesis using plasmid B2856 as the template. The SEC1 containing the BglII/StuI fragment was cloned into B3072 and B3074 to replace the wt SEC1. The sequenced sec1 insertion mutants were cloned into B3072 as BglII and StuI fragments. All DNA fragments generated by PCR were sequenced.
SEC1 Insertion Library
A SEC1 pentapeptide insertion mutant library was generated using the Mutation Generation System (Finnzymes, Espoo, Finland) that exploits a MuA transposase–catalyzed in vitro transposition reaction and generates five amino acid insertions into proteins (Haapa et al., 1999; Poussu et al., 2004). Four standard transposition reactions were performed, each with 500 ng of B2856 as a target and 100 ng of cat-Mu(NotI) as a transposon donor (Poussu et al., 2004). After incubation at 30°C for 4 h and inactivation at 75°C for 10 min, reactions were pooled, extracted with phenol/chloroform, ethanol-precipitated, and resuspended in water. Approximately 1 × 106 Escherichia coli DH10B transformants were pooled and grown in LB-Ap-Cm medium at 37°C for 2 h. Plasmid DNA from the pool (pJJHS1) was isolated and digested with StuI and BglII, followed by preparative electrophoresis on a 0.8% SeaPlaque GTG (Lonza, Basel, Switzerland) agarose gel in TAE buffer. The 3.2-kb DNA fragment pool, corresponding to transposon insertions into SEC1-encoding DNA segment, was isolated by electroelution and ligated into StuI- and BglII-cut B3073, followed by electroporation into DH10B cells. Plasmid DNA was prepared from ∼7 × 104 colonies (pJJHS2). Most of the transposon DNA was then eliminated from the plasmid pool by NotI cutting, followed by agarose gel purification of the plasmid backbone and recircularization by ligation. Ligated plasmids were electroporated into DH10B cells to generate the final SEC1 insertion mutant library for which DNA was isolated from ∼6 × 104 colonies (pJJHS4).
Yeast Two-Hybrid Assay
The yeast two-hybrid assay with the EGY48 (H1910) strain was performed as described previously (Golemis et al., 1998). The plasmids B1227 (pSH18-34), B1228 (pRFHM1), and B1229 (pSH17-4) were used as positive and negative controls. Four independent transformants were initially examined for growth on SC-Ura-Leu-His-Trp + 1% raffinose + 2% galactose. The blue/white screening was performed on X-gal plates (SD-Ura-His-Trp + 100 mM sodium phosphate (pH 7) + 0.6% glucose + 1.4% galactose + 0.2 mg/ml X-gal). For the sec1 insertion library screen EGY48 cells were transformed with plasmids B1227 and B1254 and the sec1 insertion library (pJJHS4). After transformation the cells were regrown in nonselective medium for 4 h and then directly plated on the X-gal plates. Three pools, each containing 150 white, light blue, and blue colonies, were created and the sites of insertions mapped as described previously (Pajunen et al., 2007).
In Vitro Binding Assays
His6-tagged Mso1p (amino acids 35-210) was produced in E. coli and purified using Ni-NTA agarose (Poussu et al., 2005). Glutathione S-transferase (GST)-fusion proteins Sso1p (amino acids 1-265) and Sso2p (amino acids 1-269) were purified with glutathione-Sepharose (Amersham, Uppsala, Sweden; Jantti et al., 2002), followed by thrombin cleavage. Mso1p was immobilized on CM5 biosensor chip (Amersham) using amino-coupling chemistry according to manufacturer's instructions. Sso1p and Sso2p were injected in 10 mM HEPES-KOH, 150 mM NaCl, 0.005% surfactant P20, pH 7.4, with flow rate of 40 μl/min for 5 min, followed by a 10-min dissociation time. The results were evaluated by 1:1 Langmuir binding model in Biacore Evaluation Software 3.1. The obtained equilibrium binding level responses are given in resonance units.
Antibodies
The anti-HA (12CA5) was purchased from Roche (Indianapolis, IN) and the myc-tag antibodies (9E10) were obtained from Kristiina Takkinen (VTT Technical Research Center, Otaniemi, Finland). The anti-Sec1p antibody used was the affinity-purified anti-Sec1p (no. 57; Scott et al., 2004), from James McNew (Rice University, Houston, TX). Anti-Sec9p-NT antibodies (Brennwald et al., 1994) were obtained from Patrick Brennwald (University of North Carolina at Chapel Hill). Anti-Sso1/2p (K8) and Mso1p (R285) antibodies have been previously described (Aalto et al., 1997; Jantti et al., 2002). Dilutions used for Western blotting were 1:2000 anti-HA, 1:5000 anti-Sec1p (no. 57), 1:1000 anti-Sec9p-NT, 1:20000 anti-Sso1p/2p (K8), and 1:2000 anti-Mso1p.
Immunoprecipitations
Immunoprecipitations were performed as described previously (Knop et al., 2005). For Western blot analysis, proteins were transferred onto nitrocellulose membranes, and the bound antibodies were visualized with the ECL detection system (Pierce, Rockford, IL). Each immunoprecipitation was performed three to five times, quantified using Bio-Rad Quantity One software (Sundbyberg, Sweden) and normalized for the amount of immunoprecipitated Mso1p-HA.
Fluorescence Microscopy
Cells were grown to OD600 0.8-1 at the permissive temperature. The temperature-sensitive cells were shifted to 37°C for 1 h before microscopy with the Olympus Provis microscope with a Plan Apo 60× 1.40 NA oil ph3 objective and bright-field and FITC filters (Olympus, Tokyo, Japan). The software used for recording images was AnalySIS. The exposure time for the bimolecular fluorescence complementation signal was 5–10 s. Images were prepared using Adobe Photoshop7 software (San Jose, CA). Fluorescence intensities of the Mso1p–Sec1p interaction signal in the cytosol of the mother cell were quantified by measuring the volume intensity of equal-sized squares of 50–120 cells with Bio-Rad Quantity One software. BiFC interaction intensities were quantified by measuring the mean gray value of random points on the plasma membrane using ImageJ 1.42 (http://rsb.info.nih.gov/ij/). For each condition the signals from at least 50 cells were quantified.
Homology Model of Sec1p
The structures of yeast Sly1p (1mqs) (Bracher and Weissenhorn, 2002), rat Sec1 (1dn1) (Burkhardt et al., 2008), and Munc18c (2pjx) (Hu et al., 2007) were retrieved from the RCSB Protein Data Bank, superimposed, and used as templates for the modeling of Sec1p. Prime 1.6 (Schrödinger, Portland, OR) was used to align the Sec1p sequence with the structure-based alignment of the three templates and squid Sec1 (1fvf) (Bracher and Weissenhorn, 2001). Corrections to this alignment were introduced manually, taking the positions of secondary structure elements in the templates into account. 1dn1 was used as template for residues 1-470 and 586-644, 1mqs for 471-508 and 544-585, and 2pjx for 509-543. The overall sequence identity between Sec1p and the structural templates ranges from 22 to 26%. Major differences between the sequences of Sec1p and the templates are in the C-terminal segments. These areas were omitted from the alignment and model building so that the final structural model of Sec1p consists of amino acids M1 to R643. Prime 1.6 was used to build loops for 27 insertion and deletion regions, and after this, new side chains were placed, and their conformations were optimized.
RESULTS
Mso1p and Sec1 Interact at Sites of Secretion
Mso1p localizes in the bud tip and in the septum of dividing yeast cells (Knop et al., 2005; Castillo-Flores et al., 2005). On the other hand, the Mso1p-interacting protein Sec1p and the SNARE proteins Sso1p, Sso2p, and Sec9p localize, in addition to the growing bud, along the mother cell plasma membrane (Brennwald et al., 1994; Scott et al., 2004). To evaluate the Mso1p–Sec1p interaction site in vivo, we made use of the BiFC technique (Hu et al., 2005; Kerppola, 2006; Skarp et al., 2008). Plasmids for expression of fusion proteins of Mso1p and the C-terminal fragment of yellow fluorescent protein (YFP; YFP(C)-Mso1) and Sec1p and the N-terminal fragment of Venus [Sec1-Venus(N)] were transformed to yeast cells followed by analysis of the YFP signal. The in vivo function of these fusion proteins was verified by their ability to rescue mutant phenotypes in the respective genes (data not shown).
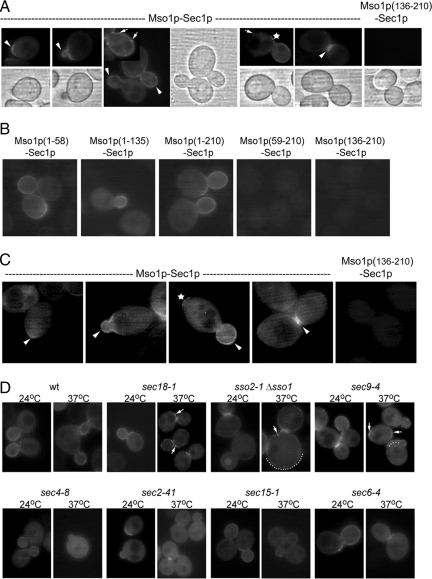
In haploid, vegetatively grown cells, the Mso1p–Sec1p interaction site localizes to the bud tip, the growing bud, and the bud neck (Figure 1A, arrowheads). In the growing bud, a dotty signal (Figure 1A, arrows) along the plasma membrane was frequently observed. A similar signal at the plasma membrane was also detected for the Mso1p(1-58) fragment and Sec1p (Figure 1B). The 1-58 fragment contains the minimal Sec1p-binding sequence (amino acids 38-59) of Mso1p (Knop et al., 2005). In cells expressing Sec1p-Venus(N) and a mutant version of Mso1p lacking the Sec1p-binding domain [YFP(C)-Mso1p(59-210) and YFP(C)-Mso1p(136-210)], negligible YFP signal was observed (Figure 1, A and B). In 32% of the budding cells, Mso1p–Sec1p interaction was also detected in the mother cell plasma membrane on one side of the bud (Figure 1A, star). This signal is likely to correspond to a former or a newly selected bud site that typically localizes axially next to the current bud site in haploid cells. The Mso1p–Sec1p interaction was also analyzed in vegetatively grown diploid cells that follow a bipolar budding mode. Similarly to haploid cells, Mso1p–Sec1p interaction localized to the bud tip, growing bud, and the septum (Figure 1C, arrowheads). In 44% of budding cells, Mso1p–Sec1p interaction was observed on the pole opposite to the growing bud (Figure 1C, star). No signal was detected at the cell poles in diploid cells undergoing cytokinesis (Figure 1C). This indicates that the Mso1p–Sec1p complexes detected by BiFC do not represent terminally plasma membrane locked complexes, but are dynamic by nature.
Figure 1.
Localization of the Mso1p–Sec1p interaction site in vivo. (A) Live cell imaging of vegetatively grown haploid (H304) cells expressing YFP(C)-Mso1p (B3044) or YFP(C)-Mso1p(136-210) (B3064) with Sec1p-Venus(N) (B2930). (B) Live cell imaging of vegetatively grown haploid cells (H304) expressing YFP(C)-Mso1p(1-58) (B3355), YFP(C)-Mso1p(1-135) (B3354), YFP(C)-Mso1p(1-210) (B3044), YFP(C)-Mso1p(59-210) (B3353), or YFP(C)-Mso1p(136-210) (B3064) with Sec1p-Venus(N) (B2930), and (C) diploid (H2530) cells expressing Mso1p-Venus(C) (B2918) or Mso1p(136-210)-Venus(C) (B3012) with Sec1p-Venus(N) (B2930). Mso1p–Sec1p interaction is detected preferentially at the bud tip, plasma membrane of the growing bud, and the bud neck (arrowheads). Regularly, the interaction signal appeared dott-like (arrows). The interaction site was adjacent to the bud or at the opposite pole of the cells (star). (D) Mso1p–Sec1p interaction is altered in different secretion mutants. Indicated mutants cells expressing YFP(C)-Mso1p (B3044) with Sec1p-Venus(N) (B2930) were grown to OD600 0.8–1 at 24°C, split, and either left at 24°C or shifted to 37°C for 1 h before investigation. Mislocalization and clusters of the interaction signal are pointed out by arrows or a dotted line.
To identify factors that affect Mso1p–Sec1p interaction, we analyzed the Mso1p–Sec1p BiFC signal in different temperature-sensitive mutants functionally linked to Sec1p. At the permissive temperature 24°C, Mso1p–Sec1p complex localization was similar both in wild-type and in sec18-1, sec9-4, sec4-8, sec2-41, sec15-1, and sec6-4 cells (Figure 1D; Table 2). In sec18-1 cells, defective in cis-SNARE complex disassembly at the restrictive temperature, Mso1p–Sec1p complexes accumulated in dots (Figure 1D, arrows). The dots could represent accumulations of SNARE complexes. The sso2-1 Δsso1 mutant has an abnormal morphology, with large cells attached to each other because of abnormal cytokinesis (Jantti et al., 2002). When incubated at the restrictive temperature, the signal for Mso1p–Sec1p interaction throughout the whole plasma membrane became more pronounced, and at the same time, the signal in the septum was reduced (Figure 1D, dotted line, Table 2). This suggests a defect in the polarization of Mso1p–Sec1p complexes in these cells. In sec9-4 cells, the Mso1p–Sec1p interaction signal remained at the septum after a shift to the restrictive temperature. However, a lower signal in the bud and a partial mislocalization throughout the plasma membrane was also observed (Figure 1D, arrows and dotted line; Table 2).
Table 2.
Quantification of Mso1p–Sec1p interaction site in different secretion mutants
| Strains; temperature | Distribution of the Mso1p–Sec1p interaction signal (%) |
||||
|---|---|---|---|---|---|
 |
 |
 |
 |
 |
|
| Growing bud | Septum | Plasma membrane | Cytosol | No signal | |
| wt; 24°C | 51.8 | 11.0 | 0.0 | 2.5 | 34.7 |
| wt; 37°C | 44.5 | 8.4 | 0.0 | 12.5 | 34.5 |
| sec18-1; 24°C | 48.0 | 14.0 | 4.0 | 0.0 | 34.0 |
| sec18-1; 37°C | 16.2 | 35.3 | 11.1 | 0.0 | 37.4 |
| sso2-1 Δsso1; 24°C | 12.5 | 16.7 | 47.2 | 0.0 | 23.6 |
| sso2-1 Δsso1; 37°C | 5.8 | 6.7 | 58.4 | 0.8 | 28.3 |
| sec9-4; 24°C | 42.0 | 21.4 | 0.0 | 0.0 | 36.6 |
| sec9-4; 37°C | 8.2 | 33.7 | 12.2 | 2.0 | 43.9 |
| sec4-8; 24°C | 42.9 | 10.7 | 0.0 | 7.1 | 39.3 |
| sec4-8; 37°C | 1.7 | 8.5 | 0.0 | 45.8 | 44.0 |
| sec2-41; 24°C | 37.3 | 17.6 | 0.0 | 7.8 | 37.3 |
| sec2-41; 37°C | 0.0 | 15.0 | 0.0 | 45.0 | 40.0 |
| sec15-1; 24°C | 39.1 | 15.2 | 0.0 | 3.3 | 42.5 |
| sec15-1; 37°C | 16.9 | 16.9 | 0.0 | 6.2 | 60.0 |
| sec6-4; 24°C | 48.3 | 13.8 | 0.0 | 0.0 | 37.9 |
| sec6-4; 37°C | 3.3 | 13.1 | 0.0 | 1.6 | 82.0 |
A prominent change in the Mso1p–Sec1p interaction site was observed in sec4-8 and sec2-41 cells after 1-h incubation at the restrictive temperature. Mutations in the small GTPase Sec4p abolish coimmunoprecipitation of Mso1p with SNARE complexes, but not with Sec1p (Knop et al., 2005). In line with these findings, in sec4-8 and sec2-41 cells incubated at the restrictive temperature, the Mso1p–Sec1p complexes no longer accumulated at the sites of secretion in vivo. Instead, compared with cells grown at 24°C, an greater than fivefold increase in fluorescence signal was detected in the cytosol (Figure 1D; Table 2). This suggests that Mso1p and Sec1p still can interact, but are not associated with the plasma membrane. This change in distribution may be due to disassembly of SNARE complexes in sec2-41 and sec4-8 cells (Grote and Novick, 1999; Grote et al., 2000). In the exocyst subunit mutants (sec15-1, sec6-4), the Mso1p–Sec1p interaction signal at the plasma membrane was reduced at the restrictive temperature, but was not completely abolished (Figure 1D; Table 2). Typically, Mso1p–Sec1p interaction signal persisted in the septum during cytokinesis of most of the temperature-sensitive mutants. This suggests slower dynamics for Mso1p–Sec1p complex turnover at that location.
Mso1p Stability Is Dependent on Sec1p
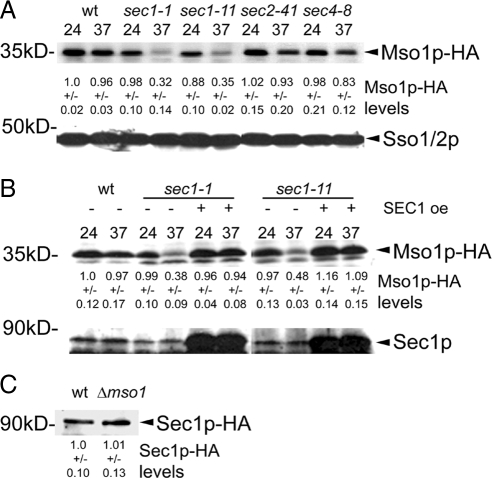
Previously, SM family protein Sly1p binding with its interaction partner Ufe1p was shown to protect Ufe1p from degradation (Braun and Jentsch, 2007). Mso1p and Sec1p are normally found complexed at the plasma membrane (Figure 1A). When plasma membrane association is disrupted, e.g., by the inactivation of Sec4p, the Mso1p–Sec1p complex appears to be stable in the cytosol (Figure 1D). These results, together with the previous biochemical data (Knop et al., 2005) indicate that the life cycles of these two proteins are closely coupled. To test whether Mso1p stability is dependent on Sec1p, Mso1p was carboxy-terminally HA-tagged at its own genomic locus in sec1-1 and sec1-11 cells, where Sec1p can be functionally inactivated by a temperature shift to 37°C. When the amount of Mso1p-HA and Sec1-1p and Sec1-11p was analyzed by Western blotting in these cells, a clear reduction in Mso1p levels was observed (68 and 65%, respectively; Figure 2A). This suggests that in cells expressing these Sec1p mutants, Mso1p becomes unstable and is degraded. At the same time, the expression levels for Sec1p, Sso1p/2p (Figure 2, A and B), and Sec9p (data not shown) did not change significantly. In wild-type and sec2-41 and sec4-8 strains, only a slight change in Mso1p levels was observed (4, 7, and 17% reduction, respectively) at 37°C. On overexpression of wt SEC1 in sec1-1 and sec1-11 cells, Mso1p levels were restored to wild-type levels (Figure 2B). These results underscore the importance of an active Sec1p for Mso1p stability. We next studied a possible effect of Mso1p deletion on Sec1p levels. As shown in Figure 2C, Sec1p levels are not significantly affected by MSO1 deletion. This result is in line with the nonessential role of Mso1p for vegetative haploid cell growth.
Figure 2.
Mso1p stability is depended on Sec1p. (A) The wild-type, sec1-1, sec1-11, sec2-41 and sec4-8 strains where the sole copy of MSO1 is HA tagged were grown to an OD600 of 1 at 24°C, split, and either maintained at 24°C or shifted to 37°C for 1 h before lysate preparation. Lysates were subjected to Western blotting and detection with anti-HA and anti-Sso1p/2p antibodies. (B) Overexpression of SEC1 restores Mso1p levels. A SEC1 overexpression vector or an empty vector was transformed to wild-type and sec1-1 and sec1-11 cells expressing Mso1p-HA. The temperature shift experiment was performed as is in (A). Detection was done with anti-HA and anti-Sec1p antibodies. (C) Sec1p level is not affected by MSO1 deletion. HA-tagged Sec1p was detected in wild-type and Δmso1 cells. (A–C) The quantification of the ECL signals, normalized against the wild-type 24°C lysate, are shown underneath each lane.
Mapping of Mso1p-binding Site in Sec1p
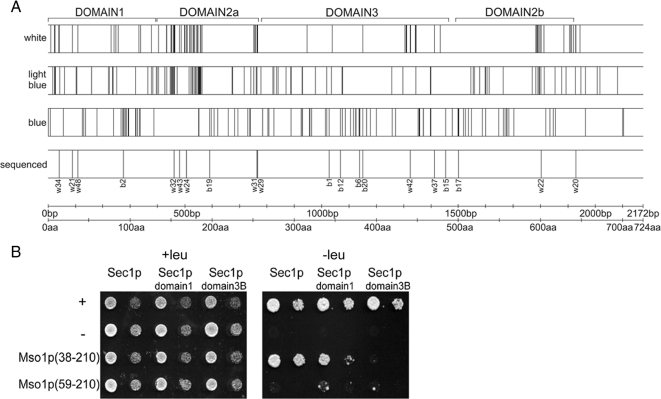
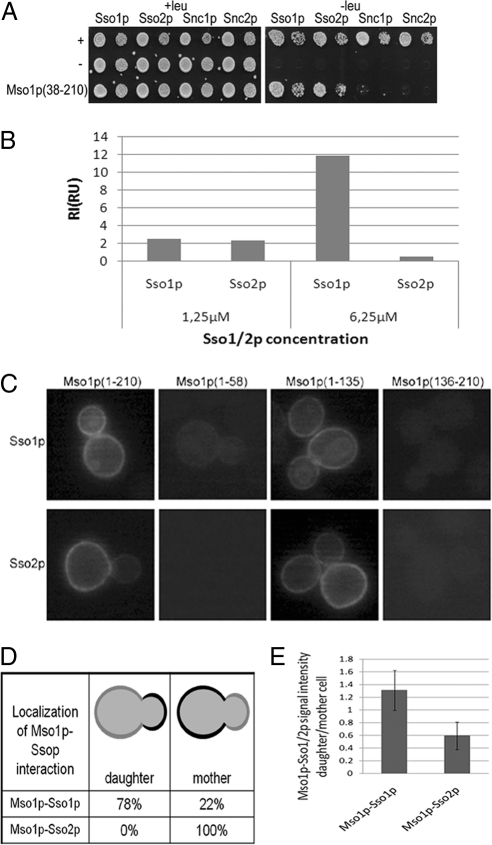
The amino acids 38-59 at the amino terminus of Mso1p are necessary and sufficient for Sec1p binding (Knop et al., 2005). However, the Mso1p-binding site in Sec1p is unknown. To study this, we made use of a Mu-transposition–assisted mutagenesis method that results in five amino acid insertions randomly in the target protein (Taira et al., 1999; Poussu et al., 2004; Pajunen et al., 2007). A plasmid library encoding such mutant Sec1 proteins was created and used as a prey in a yeast two-hybrid screen with Mso1p as the bait. Sec1p insertion mutants (n = 1189) were obtained, 6% of which were noninteracting (white colonies), 53% weakly interacting (light blue), and 41% strongly interacting (dark blue) with Mso1p.
The distribution of the insertion sites in SEC1 was mapped in 150 clones per interaction mode category. Mutations resulting in no interaction with Mso1p were mainly clustered, whereas mutations that allowed interaction with Mso1p (blue and light-blue) were distributed rather evenly throughout the gene (Figure 3A and Supplementary Figure S1). Because of clear differences in percentages of the obtained mutants, it can be assumed that only the noninteracting pool was saturated. The two most pronounced clusters of mutations that weakened the interaction with Mso1p were located at the beginning of domain 2a and at the end of domain 2b (Figure 3A). It is possible that these mutations cause Sec1p misfolding, because crystal structures of Sec1p homologues show that domain 2 is formed by cooperative folding of amino- and carboxy terminal parts of the polypeptide (Misura et al., 2000; Bracher and Weissenhorn, 2001; Bracher and Weissenhorn, 2002). It is thus possible that these regions do not represent the domain interacting with Mso1p, but that rather the overall folding of Sec1p was impaired. Another cluster of mutations that resulted in white colonies was located at the end of domain 2a, bridging to domain 3. Furthermore, a cluster of noninteracting mutations was localized at the end of domain 3 close to the location of the known sec1-1 (G443E) and sec1-11 (R432P) mutations. At the same time, domain 3 contained several insertion mutations that did not affect Mso1p interaction (Supplementary Figure S2, A and B). Several noninteracting mutations also localized at the beginning of the Sec1p C-terminal tail, which bears no homology to other SM proteins. In addition to these mutations, domain 1 contained several noninteracting insertion mutations (Figure 3).
Figure 3.
Mapping of the Mso1p-binding site in Sec1p. (A) Map of the Mso1p interacting (blue), weakly interacting (light blue), and noninteracting (white) sec1 mutations. The insertion sites are indicated by black vertical lines within the SEC1 gene. Twenty mutants were sequenced to confirm the mapping. The domain borders of Sec1p are indicated above of the gene. (B) Yeast two-hybrid assay between different Mso1p fragments and Sec1p domains 1 and 3. Mso1p (59-210) is lacking the Sec1p-binding area (Knop et al., 2005). As an additional negative control, Bicoid (B1228) and as a positive control Gal4 (B1229) were used. Two 10-fold dilutions of cells were grown with or without leucine.
Sec1p domain 1 and domain 3B have a predicted globular structure. Possible interactions of domain 1 and 3 with Mso1p were analyzed directly by the yeast two-hybrid assay (Figure 3B). As a positive control we used Mso1p(38-210), which binds to the full-length Sec1p (Knop et al., 2005). The Sec1p domain 1 showed binding to Mso1p, whereas no interaction with Sec1p domain 3B was observed. Negligible interaction between Mso1p(59-210) lacking the previously reported minimal Sec1p-binding area (Knop et al., 2005) and Sec1p domain 1 was observed. These results suggest that Mso1p can interact with Sec1p domain 1.
Mso1p Interaction with Sec1p Is Affected by Mutations in the Putative Syntaxin N-Peptide-binding Area
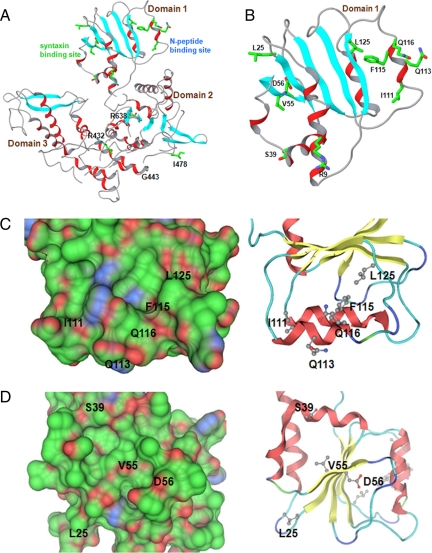
To better define the Mso1p interaction site, a more precise understanding of the S. cerevisiae Sec1p structure was needed. Therefore, a model of Sec1p structure was created based on the structures of homologues SM family proteins (Figure 4). The model pointed out two potential protein interaction interfaces in domain 1 that correspond to the previously identified N-peptide and syntaxin-binding sites in SM proteins (Figure 4, B–D). When the insertion mutation sites were placed in this model, several mutations that abolished Mso1p–Sec1p interaction, localized in two α-helices adjacent to the hydrophobic pocket, known to be important for syntaxin N-peptide binding in SM proteins (Supplementary Figure S2, A and B).
Figure 4.
A model for Sec1p structure. (A) The overall structure of Sec1p. Two potential protein-binding interfaces can be identified: the putative syntaxin binding and the N-peptide–binding site. (B) A detailed view of domain 1 where the potential N-peptide and syntaxin-like binding sites are located. Helices are depicted in red and strands in light blue. Only side chains of residues that were subjected to site targeted mutagenesis are shown. (C and D) Surface and ribbon presentations of the putative N-peptide– (C) and syntaxin- (D) binding sites. The Connolly surface is colored green for hydrophobic, red for hydrogen-bonding, and blue for mildly polar atoms. Only positions of residues mutated in the present study are indicated.
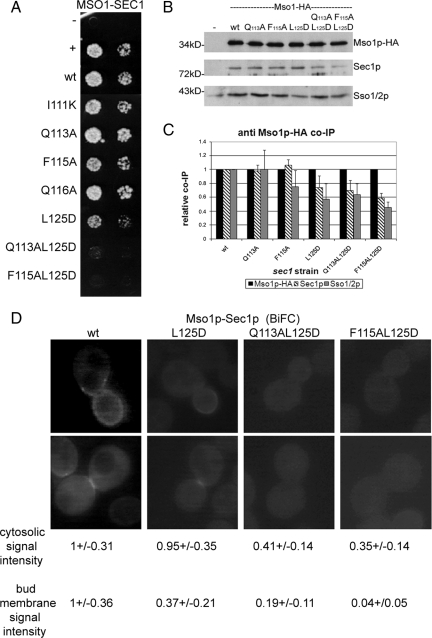
To test the functionality of the putative N-peptide binding area for Mso1p interaction, mutations Q113A in a residue at the mouth of the potential pocket, F115A in a central conserved pocket residue, and L125D in a conserved pocket-forming residue were generated (Figure 4, B and C). At the same time, double mutants Q113A L125D and F115A L125D were made. Two additional mutations (I111K and Q116A), not predicted to be directly involved in the pocket formation, were also introduced.
When Sec1p proteins with mutations affecting the putative N-peptide–binding area, or residues close by, were assayed in the yeast two-hybrid assay for Mso1p binding, Q113A and Q116A mutants displayed binding with Mso1p that was comparable to that of the wt Sec1p (Figure 5A). At the same time, I111K and F115A mutants showed slightly reduced binding, whereas the L125D mutant clearly affected the association of Sec1p with Mso1p. When Q113A or F115 were combined with the L125D mutation, Sec1p interaction with Mso1p was abolished (Figure 5A).
Figure 5.
Mso1p binding is affected by mutations in the putative N-peptide binding area in Sec1p. (A) Yeast two-hybrid assay between Mso1p and different Sec1p mutants. As a negative control Bicoid (B1228) and as a positive control Gal4 (B1229) were used. Two 10-fold dilutions of cells grown on medium without leucine are shown. (B and C) Mso1p coimmunoprecipitates less efficiently with Sec1p N-peptide domain mutants. Cells with endogenously HA-tagged Mso1p expressing different sec1 mutants were grown to OD600 = 1, lysed, and subjected to anti-HA immunoprecipitations. Immunoprecipitates were subjected to Western blotting and detection with anti-HA, Sec1p, and Sso1p/2p antibodies. (B) A representative Western of the immunoprecipitations. (C) Quantification of independent immunoprecipitation normalized to the amount of immunoprecipitated Mso1p-HA. SDs are shown for each immunoprecipitation. (D) Localization of the Mso1p interaction site with Sec1p mutants L125D, Q113A L125D, and F115 L125D. Haploid vegetatively grown cells (H304) expressing YFP(C)-Mso1p (B3044) and different mutant versions of Sec1p-Venus(N) [wt (B2930), L125D (B3263), Q113A L125D (B3264), and F115A L125D (B3265)] were investigated by fluorescence microscopy. The BiFC signal was quantified as described in Materials and Methods.
We next analyzed coimmunoprecipitation of these mutants with Mso1p-HA (expressed from its own promoter) in strains where the mutant version of Sec1p were the sole copy of Sec1p (expressed from its own promoter). The results show that although Q113A and F115A mutations have little effect (98 ± 9 and 106 ± 9%, respectively) on Mso1p coimmunoprecipitation, the L125D (74 ± 17, 26% less), Q113A L125D (70 ± 15%, 30% less), and F115A L125D (59 ± 7%, 41% less) mutations caused reduced Sec1p binding (Figure 5, B and C). For Sso protein coimmunoprecipitation with Mso1p, the Q113A mutation had no effect (100 ± 28%). However, when compared with wt Sec1p expressing cell lysates, F115A (75 ± 24), L125D (57 ± 23%), Q113A L125D (64 ± 16%), and F115A L125D (45 ± 9%) all caused reduction in Sso protein coimmunoprecipitation with Mso1p (Figure 5, B and C). Western blot analysis showed that the mutant Sec1 proteins were expressed at the wt Sec1p level in these cells (Supplementary Figure S3). This indicates that the reduced coimmunoprecipitation was not due to lower abundance of these proteins in the lysates.
To evaluate the in vivo effect of mutations in the putative N-peptide binding area, the growth of cells expressing these mutants (I111K, Q113A, F115A, Q116A, L125D, Q113A L125D, and F115A L125D) as their sole copy of Sec1p was monitored. In these strains, temperature sensitivity was observed at 38°C for sec1 (L125D), at 37°C for sec1 (Q113A L125D), and at 34°C for sec1 (F115A L125D) (Supplementary Figure S4A). Given that these mutations reduce or abolish Mso1p binding, deletion of Mso1p was not expected to have a significant additive effect in these strains. Indeed, deletion of MSO1 did not markedly change temperature sensitivity of these strains (Supplementary Figure S4B).
We next subjected Sec1p wt, L125D, and the double mutants Q113L L125D and F115A L125D to BiFC analysis for Mso1p interaction. Compared with the wt Sec1p, L125D mutation caused a 63% weaker signal with Mso1p at the plasma membrane (Figure 5D). This signal was, nevertheless, properly localized in the bud and in the septum of dividing cells. In case of the Q113L L125D and F115A L125D mutants, the BiFC signal for interaction with Mso1p was significantly reduced at the plasma membrane (81 and 96%, respectively) and in the cytosol (59 and 65%, respectively; Figure 5D). These signal intensities match the level of the Sec1p nonbinding control Mso1p(136-210) (data not shown, see Figure 1A). This indicates that these mutations severely affect Mso1p–Sec1p interaction at the plasma membrane in vivo.
Mso1p interaction with Sec1p is required for membrane fusion during sporulation of diploid S. cerevisiae cells (Knop et al., 2005). When diploid cells expressing only the mutant version of Sec1p were assayed for their ability to sporulate, it was evident that cells expressing mutations I111K, Q113A, Q116A, and F115A did not differ from wild-type cells (Table 3). However, L125D and Q113L L125D, and F115A L125D expressing cells did not sporulate. These results, together with the yeast two-hybrid and immunoprecipitation experiments, suggest that the putative syntaxin N-peptide–binding area in Sec1p domain 1 is involved in Mso1p binding.
Table 3.
Sporulation of sec1 mutants
| Mutant | Sporulation |
|---|---|
| WT | + |
| R9A | + |
| S39K | + |
| V55D | − |
| D56A | + |
| I111K | + |
| Q113A | + |
| F115A | + |
| Q116A | + |
| L125D | − |
| Q113A L125D | − |
| F115L L125D | − |
Mutations L25D and V55D in Sec1p Affect SNARE Complex Binding
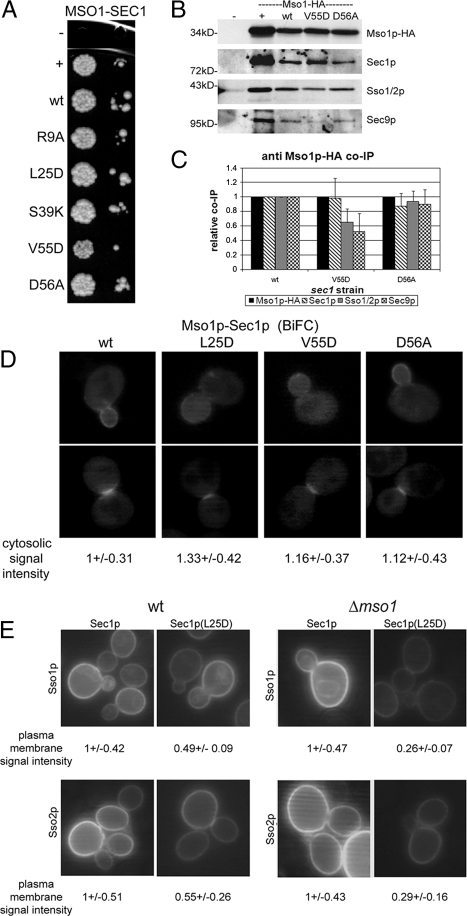
We next studied the role of the second potential protein interaction interface in domain 1 for Mso1p binding. A set of mutations were generated in this area. Mutations S39K and D56A target amino acids that, based on the structural data, are expected to face the SNARE complex (Figure 4, A, B, and D). In addition, a mutation in the neighboring residue V55D was introduced. Furthermore, we generated the L25D mutation, which is in a less conserved region further away from the syntaxin interaction pocket (Figure 4D). Previously, a mutation affecting the same position (W28) in Munc18-1 was shown to participate in syntaxin 1 binding (Misura et al., 2000). In addition, a mutation R9A, located at the “saddle-point” between the syntaxin and the putative N-peptide binding areas, was generated (Figure 4B).
In the yeast two-hybrid assay a slight decrease in Mso1p interaction was observed for Sec1p(V55D), whereas no effect for other mutants tested was seen (Figure 6A). To address the functionality of the different sec1 mutants in vivo, we created a strain where MSO1 is HA tagged and the sec1 mutants were the only copy of Sec1p expressed. The sec1 (L25D) strain was not viable. This suggests that, based on homology with the Munc18 W28 (Misura et al., 2000; Burkhardt et al., 2008), Sec1p L25D is likely to face Sso proteins and may therefore significantly affect Sec1p–SNARE complex interaction. The mutations V55D and D56A affect the same potential binding surface (Figure 4D). However, only Sec1p (V55D) showed reduced binding to Mso1p in the yeast two-hybrid assay and rendered the strain temperature sensitive (Figure 6A; Supplementary Figure S5A). When MSO1 was deleted from this strain, an increase in the temperature sensitivity in the sec1(V55D) strain was observed. At the same time, the wild-type and other sec1 mutant strains remained the same (Supplementary Figure S5B). When MSO1 was overexpressed in sec1(V55D) cells, a full rescue of the temperature-sensitive phenotype was observed (Supplementary Figure S5C). This suggests that Mso1p interaction with the N-peptide binding area is important for the functional stabilization of Sec1p (V55D).
Figure 6.
SNARE complex, but not Mso1p binding is affected in sec1 (L25D) and sec1 (V55D) mutants. (A) Yeast two-hybrid assay between Mso1p and different Sec1p syntaxin-binding mutants. As a negative control Bicoid (B1228) and as a positive control Gal4 (B1229) was used. The interactions were scored of two 10-fold dilutions on medium lacking leucine. In B and C less coimmunoprecipitation of Sso1p/2p and Sec9p is observed in Sec1p- (V55D) and Sec1p- (D56A) expressing cells. MSO1-HA cells expressing different sec1 mutants were grown until a OD600 = 1, lysed, and subjected to anti-HA immunoprecipitations. Immunoprecipitates were analyzed by Western blotting, and the blots detected with anti-HA, Sec1p, Sso1p/2p, and Sec9 antibodies. (B) A representative Western blot of the immunoprecipitations. (C) Quantification of independent immunoprecipitations, normalized to the amount of immunoprecipitated Mso1p-HA. SDs are shown for each immunoprecipitation. (D) Localization of the Mso1p interaction site with Sec1p mutants L25D, V55D, and D56A in vivo. Haploid, vegetatively grown cells (H304) expressing YFP(C)-Mso1p (B3044) and different mutant versions of Sec1p-Venus(N) [wt (B2930), L25D (B3260), V55D (B3261), and D56A (B3262)] were investigated by fluorescence microscopy. (E) Sec1p(L25D) BiFC interaction with Sso1p and Sso2p is sensitive to deletion of MSO1. Haploid vegetatively grown cells [wt (H304) or Δmso1 (H2658)] expressing Sec1p-YFP(C) wt (B3308) or the L25D (B3312) mutant together with either YFP(N)-Sso1p (B3307) or YFP(N)-Sso2p (B3309) were investigated by fluorescence microscopy for YFP signal. The BiFC signal was quantified as described in Materials and Methods..
When V55D and D56A mutant cell lysates were subjected to immunoprecipitations, the Mso1p-HA levels immunoprecipitated were similar (Figure 6, B and C). Compared with wild-type lysates, Mso1p-HA immunoprecipitations from sec1(D56A) strain lysates showed marginal changes in the amount of coprecipitating Sec1p (87 ± 18%), Sso1p/2p (94 ± 14%), and Sec9p (90 ± 20%; Figure 6, B and C). In V55D lysates, no significant change for coimmunoprecipitation of Sec1p (98 ± 27%) with Mso1p was observed. However, a drop in coimmunoprecipitation for Sso1p/2p (65 ± 19%, 35% less) and Sec9p (52 ± 25%, 48% less) was observed (Figure 6, B and C). Considering that the Sec1p interaction with the SNAREs is essential, it is plausible that the significantly reduced binding of Sec1p (V55D) with Sso1p/2p and Sec9p accounts for the temperature-sensitivity of the sec1 (V55D) strain.
To analyze Mso1p interactions with the syntaxin binding surface mutants, cells were transformed with plasmids expressing YFP(C)-Mso1p, together with wild-type or the mutant versions L25D, V55D, or D56A of Sec1-Venus(N). In line with the immunoprecipitation results, Mso1p–Sec1p interaction was detected by BiFC at the bud tip growing bud and the septum of dividing cells (Figure 6D). The characteristic daughter cell plasma membrane–associated BiFC signal for Mso1p–Sec1p interaction was reduced in V55D expressing cells and was almost undetectable in L25D mutant cells. At the same time, a 12–33% increase in the cytoplasm signal was detected in these mutants (Figure 6D), suggesting that while Mso1p–Sec1p complexes are not disrupted, they are less membrane associated.
Based on the structural data, the L25D mutation is likely to be important for Sec1p interaction with syntaxins (Misura et al., 2000). To address the observed positive effect of Mso1p on Sec1p mutants, we analyzed the BiFC signal between Sec1p (L25D) and Sso1p and Sso2p in wild-type and Δmso1 cells. When sec1(L25D) was expressed in wild-type cells together with Sso1p or Sso2p, compared with wt Sec1p–Ssop signal, a 50% lower BiFC signal was detected at the plasma membrane on average (Figure 6E). When the same interaction was measured in Δmso1 cells, an additional 25% drop in the plasma membrane signal intensity was observed (Figure 6E). These data suggests that Mso1p can provide additional affinity and stability for Sec1p–Sso1p/Sso2p binding.
Mso1p Can Interact with Sso Proteins
The role of the putative domain 1 N-peptide–binding area in Mso1p interaction and the stabilizing effect of Mso1p on Sec1p-Ssop interaction prompted us to analyze whether Mso1p can associate with Sso1p and Sso2p. To test this, interaction of Mso1p with the cytosolic domains of Sso1p and Sso2p was first analyzed by the yeast two-hybrid assay. Interaction between Mso1p(38-210) and Sso1p, and repeatedly slightly less with Sso2p, was observed (Figure 7A). No interaction was detected between Mso1p and the cytosolic domains of v-SNAREs Snc1p or Snc2p (Figure 7A). The interaction between the Sec1p-binding deficient Mso1p(59-210) mutant and Sso proteins persisted, whereas reduced interaction was observed for Mso1p(136-210) (Supplementary Figure S6A). Deletion of the Mso1p C-terminus up to amino acid 94 did not reduce binding with Sso proteins. This suggests that amino acids 59-94 contain a motif that can interact with Sso1p and Sso2p in the yeast two-hybrid assay.
Figure 7.
Mso1p interacts with Sso1p and Sso2p. (A) Yeast two-hybrid assay between Mso1p and Sso1p, Sso2p, Snc1p, and Snc2p. As negative control Bicoid (B1228) and as positive control Gal4 (B1229) was used. Two 10-fold dilutions on medium with or without leucine are shown. (B) In vitro analysis of Sso protein interaction with Mso1p. The interactions were analyzed by the surface plasmon resonance technique as described in Materials and Methods. (C) Mso1p interacts with Sso1p/2p at the plasma membrane. Haploid vegetatively grown cells expressing YFP(C)-Mso1p (B3044), YFP(C)-Mso1p(1-58) (B3355), YFP(C)-Mso1p(1-135) (B3354), YFP(C)-Mso1p(136-210) (B3043), and YFP(N)-Sso1p/2p (B3307, B3309) were investigated by fluorescence microscopy. (D) Quantification of the Mso1p–Ssop localization. (E) Quantification of the Mso1p-Ssop plasma membrane signal intensity in daughter versus mother cell.
We next used surface plasmon resonance technique to test for possible direct interactions between Mso1p and Sso proteins in vitro. For this, His6-tagged Mso1p was expressed in E. coli, purified, and coupled to a sensor chip. The cytosolic fragments of untagged Sso1p and Sso2p were then flown on top of the immobilized Mso1p, and binding of Sso proteins with Mso1p was recorded. For 6.5 μM Sso1p weak binding with Mso1p was observed (Figure 7B; Supplementary Figure 6B). Under the same conditions, no binding for Sso2p with Mso1p was observed in this assay.
To analyze the interactions of Mso1p with Sso1p and Sso2p in vivo, we made use of the BiFC assay. When full-length Mso1p or Mso1p(1-135) was expressed, a BiFC signal was observed at the plasma membrane for Sso1p and Sso2p binding (Figure 7C). No signal was observed for Mso1p(1-58) and Mso1p(136-210) interaction with Ssos (Figure 7C). This result is in line with the yeast two-hybrid data, suggesting that Mso1p amino acids 59 to 94 contribute to Sso interaction (Supplementary Figure S6). Interestingly, the Mso1p(1-210)–Sso1p BiFC signal was consistently stronger at the daughter cell plasma membrane (Figure 7, C and D). At the same time, the signal for Mso1p–Sso2p interaction was stronger at the mother cell plasma membrane and weaker at the daughter cell plasma membrane (Figure 7, C and D). Quantification of the BiFC signal in these cells revealed that compared with Mso1p–Sso2p, Mso1p–Sso1p BiFC complexes displayed more than twofold enrichment in the daughter cells (Figure 7E).
DISCUSSION
Members of the SM protein family are central regulators of membrane fusion (Jahn and Scheller, 2006; Toonen and Verhage, 2007). Several SM family members bind a N-terminal peptide of syntaxins (Hu et al., 2007; Munson and Bryant, 2009). However, the role of the N-peptide binding in SM or SNARE protein complex regulation is currently unclear. Recent results indicate that the N-peptide is needed for neurotransmission in C. elegans (Johnson et al., 2009). However, abolishment of the N-terminal peptide binding between Sly1p-Sed5p and Vps45p-Tlg2p does not result in any obvious in vivo phenotypes in yeast (Peng and Gallwitz, 2004; Carpp et al., 2006). In contrast to syntaxin1, Sed5p or Tlg2p, yeast syntaxins Sso1p and Sso2p do not have a N-terminal peptide for Sec1p binding and are thus likely to be regulated in a different way.
We previously identified Mso1p as a Sec1p-binding protein and showed that it is required for membrane fusion (Aalto et al., 1997; Knop et al., 2005). In the present study, we show that Mso1p and Sec1p interact at the plasma membrane of the growing daughter cell and at the septum of dividing cells. This interaction site fits with the previously published localization of GFP-tagged forms of Mso1p and Sec1p (Carr et al., 1999; Knop et al., 2005). The localization of Mso1p–Sec1p complexes at the plasma membrane was sensitive to inactivation of Sec4p (Figure 1D). This finding is in line with the results showing that in sec4 mutant cell lysates, Mso1p–Sec1p complexes are stable, but they do not interact with plasma membrane–associated SNARE complex subunits (Knop et al., 2005).
Our results show that Sec1p domain 1 is important for Mso1p binding in vivo. The combined use of yeast two-hybrid, BiFC, immunoprecipitation, and genetic techniques show that mutations in the putative N-peptide binding area in Sec1p domain 1 affect Mso1p–Sec1p interaction. In addition, our results show that domain 1 mutations affecting Mso1p interaction lead to inhibition of prospore membrane formation (Table 2). Mso1p binding to Sec1p is essential for the homotypic fusion of prospore membrane precursor vesicles (Knop et al., 2005). The sporulation inhibition by the N-peptide–binding area mutations suggests an important role for this interaction surface in Ssop-Sncp-Sec9p complex–mediated membrane fusion.
The minimal Mso1p peptide that can mediate Sec1p interaction, both in vivo and in vitro (Knop et al., 2005) does not display obvious sequence similarity with the syntaxin N-peptides. An interesting target for future studies will be to reveal how closely Mso1p binding with the domain 1 resembles the binding modes of the syntaxin N-terminal peptides. To resolve this matter, the three-dimensional structure of Mso1p–Sec1p complexes needs to be determined.
Genetic results suggest a stabilizing role for Mso1p in Sec1p–SNARE complex binding. This idea is supported by the capability of MSO1 overexpression to rescue the temperature-sensitive phenotype of the Sso binding–deficient sec1 (V55D) mutant. A similar stabilizing role was revealed by the significantly reduced BiFC signal between the syntaxin-binding–deficient Sec1p(L25D) and Sso proteins at the plasma membrane when Mso1p was deleted (Figure 6E). The capability of Mso1p to facilitate the function of the syntaxin binding–deficient Sec1p mutants could be explained by its association with Sso proteins. Such an interaction is supported by several lines of evidence. Mso1p was shown by the yeast two-hybrid technique to interact with Sso proteins. In vitro analysis of Mso1p–Ssop interaction revealed a weak interaction between Sso1p and Mso1p. No interaction with Sso2p was detected in vitro. Yeast exocytosis involves Sec1p binding with an assembled SNARE complex composed of Sso1p/2p-Sec9p-Snc1p/2p (Carr et al., 1999; Scott et al., 2004; Togneri et al., 2006; Hashizume et al., 2009). Our data indicates that the Mso1p–Ssop interaction takes place within a larger protein complex in vivo (Knop et al., 2005; Figure 5). Because of insufficient folding or affinity, weak interactions between the subunits of a larger protein complex may be challenging to reproduce in vitro without the stabilizing contribution of other subunits. Similarly to Mso1p–Ssop binding, the in vitro interaction between isolated Sec1p and Sso proteins is weak (Scott et al., 2004; Togneri et al., 2006). Point mutations in Sec1p domain 1 did not result in complete loss of coimmunoprecipitation of Sec1p and Sso proteins with Mso1p (Figure 5, B and C). It is possible that the reason for this coimmunoprecipitation is at least partly explained by the affinity of Mso1p to Sso proteins.
A difference in Mso1p interaction with Sso1p and Sso2p is in line with the yeast two-hybrid data, where repeatedly, a weaker interaction between Mso1p and Sso2p was observed (Figure 7B). In addition, previous data indicate a difference in the functional association between Mso1p and Sso1p or Sso2p. Although MSO1 deletion in sso2-1 cells causes a synthetic phenotype even in the presence of the wt SSO1, no effect is observed for the viability of sso1-1 SSO2 cells deleted for MSO1 (Jantti et al., 2002). This suggests that Mso1p is important for full Sso1p functionality when the paralogous Sso2p is functionally compromised. At the same time, the opposite seems not to be the case (Jantti et al., 2002). A difference in the functional association between Mso1p and Sso proteins is further supported by the cooperation of Mso1p with Sso1p, but not with Sso2p in membrane fusion of prospore membrane precursor vesicles in meiotic diploid cells (Jantti et al., 2002; Knop et al., 2005).
The BiFC analysis indicated that amino acids 59-94 are important for Mso1p interaction with Sso proteins at the plasma membrane in vivo. Negligible interaction signal was detected for Mso1p(1-58) and Mso1p(136-210) with Sso1p or Sso2p (Figure 7C). However, the Mso1p(1-58) was clearly targeted to the plasma membrane, as an interaction between that fragment and Sec1p was readily visualized at the plasma membrane by BiFC (Figure 1A). Taking into account that Sso proteins are plasma membrane bound, our results show that a mere plasma membrane colocalization is not sufficient for the Mso1–Ssop BiFC signal. Interestingly, BiFC analysis revealed a qualitatively different distribution for Mso1p–Sso1p and Mso1p–Sso2p complexes. Mso1p–Sso1p complexes occupied predominantly the daughter cell plasma membrane, whereas Mso1p–Sso2p complexes were enriched in the mother cell. These findings support the selectivity of Mso1p interactions with Sso1p and Sso2p containing SNARE complexes.
In the present study, for the first time, protein interaction sites for the exocytic machinery are reported. Our results suggest that in vegetatively growing haploid cells there is a preference for association of Mso1p with Sso1p containing SNARE complexes at the fast growing daughter cell plasma membrane, rather than in the more quiescent mother cell. This type of differences would be impossible to detect by biochemical techniques analyzing protein-protein interactions in cell lysates. This underscores the usefulness of techniques like BiFC that enable spatial resolution of protein interactions sites.
SM protein interactions with SNARE complexes and syntaxins have received significant attention during the least 10 years. At the same time the binding modes of known SM protein interactors outside the SNARE protein family are largely uncharacterized. The present work is the first detailed analysis of interactions between a SM protein with its non-SNARE–binding partner. Our results suggest that Mso1p can act as a bridging factor between Sec1p domain 1 and Sso proteins and possibly, through this interaction, can stabilize Sec1p–SNARE complexes. The binding of the N-peptide with SM proteins is not essential in vegetatively grown haploid cells (Peng and Gallwitz, 2004; Carpp et al., 2006). Likewise, Mso1p–Sec1p interaction is nonessential for haploid cell viability. However, Mso1p binding with the domain 1 appears to be essential for homotypic membrane fusion of prospore membrane precursor vesicles in diploid cells. Membrane fusion initiation of these precursor vesicles, docked to the spindle pole body, is triggered during meiosis by a poorly characterized, temporally regulated signal (Moreno-Borchart and Knop, 2003). It is interesting to note, that this signal triggered membrane fusion may bear similarities with fusion of neurotransmitter loaded, docked vesicles at the presynaptic membrane. Syntaxin N-peptide binding with UNC-18 appears to be important for neurotransmission in C. elegans (Johnson et al., 2009). In case of Sso proteins, which do not possess such a peptide, Mso1p could mimic this role and therefore be essential for membrane fusion in meiotic cells. It appears, that constitutive exocytosis during haploid cell growth does not require such tight regulation by Mso1p.
Mso1p is homologous with the PTB-binding domains of the Munc18-binding Mint proteins (Knop et al., 2005). The functional role of Mint proteins in association with Munc18 is poorly understood. However, Mint proteins have been proposed to act as adaptors interlinking several proteins involved in neuronal exocytosis (Biederer and Sudhof, 2000). Our results suggest a similar role for Mso1p in the regulation of Sec1p and SNARE complexes. We propose that Mso1p acts as a facilitator of Sec1p function through its interactions with Sec1p and Sso proteins to ensure efficient membrane fusion.
Supplementary Material
ACKNOWLEDGMENTS
Pat Brennwald, David Drubin (University of California, Berkeley, Berkeley, CA), Jeffrey Gerst (Weizmann Institute of Science, Rehovot, Israel), Wei Guo (University of Pennsylvania, Philadelphia, PA), Doug Johnson (University of Vermont, Burlington, VT), Michael Knop (EMBL, Heidelberg, Germany), James McNew, and Peter Novick (University of California, San Diego, La Jolla, CA) are acknowledged for generously providing strains, plasmids, and/or antibodies. Members of the Jäntti lab are thanked for discussions, and Anna-Liisa Nyfors and Pirjo Rahkola are thanked for excellent technical assistance. Maria Aatonen is acknowledged for excellent cooperation for the Biacore analysis. This work was financially supported by the Academy of Finland (Grants 211171 and 124249 to J.J.), Viikki Graduate School in Biosciences, Magnus Ehrnrooth Foundation, Alfred Kordelin Foundation, and the Institute of Biotechnology.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-07-0546) on February 24, 2010.
REFERENCES
- Aalto M. K., Jantti J., Ostling J., Keranen S., Ronne H. Mso1p: a yeast protein that functions in secretion and interacts physically and genetically with Sec1p. Proc. Natl. Acad. Sci. USA. 1997;94:7331–7336. doi: 10.1073/pnas.94.14.7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biederer T., Sudhof T. C. Mints as adaptors: direct binding to neurexins and recruitment of munc18. J. Biol. Chem. 2000;275:39803–39806. doi: 10.1074/jbc.C000656200. [DOI] [PubMed] [Google Scholar]
- Bracher A., Weissenhorn W. Crystal structures of neuronal squid Sec1 implicate inter-domain hinge movement in the release of t-SNAREs. J. Mol. Biol. 2001;306:7–13. doi: 10.1006/jmbi.2000.4347. [DOI] [PubMed] [Google Scholar]
- Bracher A., Weissenhorn W. Structural basis for the Golgi membrane recruitment of Sly1p by Sed5p. EMBO J. 2002;21:6114–6124. doi: 10.1093/emboj/cdf608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun S., Jentsch S. SM-protein-controlled ER-associated degradation discriminates between different SNAREs. EMBO Rep. 2007;8:1176–1182. doi: 10.1038/sj.embor.7401105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennwald P., Kearns B., Champion K., Keranen S., Bankaitis V., Novick P. Sec9 is a SNAP-25-like component of a yeast SNARE complex that may be the effector of Sec4 function in exocytosis. Cell. 1994;79:245–258. doi: 10.1016/0092-8674(94)90194-5. [DOI] [PubMed] [Google Scholar]
- Burkhardt P., Hattendorf D. A., Weis W. I., Fasshauer D. Munc18a controls SNARE assembly through its interaction with the syntaxin N-peptide. EMBO J. 2008;27:923–933. doi: 10.1038/emboj.2008.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpp L. N., Ciufo L. F., Shanks S. G., Boyd A., Bryant N. J. The Sec1p/Munc18 protein Vps45p binds its cognate SNARE proteins via two distinct modes. J. Cell Biol. 2006;173:927–936. doi: 10.1083/jcb.200512024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr C. M., Grote E., Munson M., Hughson F. M., Novick P. J. Sec1p binds to SNARE complexes and concentrates at sites of secretion. J. Cell Biol. 1999;146:333–344. doi: 10.1083/jcb.146.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Flores A., Weinberger A., Robinson M., Gerst J. E. Mso1 is a novel component of the yeast exocytic SNARE complex. J. Biol. Chem. 2005;280:34033–34041. doi: 10.1074/jbc.M507142200. [DOI] [PubMed] [Google Scholar]
- Cole K. C., McLaughlin H. W., Johnson D. I. Use of bimolecular fluorescence complementation to study in vivo interactions between Cdc42p and Rdi1p of Saccharomyces cerevisiae. Eukaryot. Cell. 2007;6:378–387. doi: 10.1128/EC.00368-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deak F., Xu Y., Chang W. P., Dulubova I., Khvotchev M., Liu X. R., Sudhof T. C., Rizo J. Munc18-1 binding to the neuronal SNARE complex controls synaptic vesicle priming. J. Cell Biol. 2009;184:751–764. doi: 10.1083/jcb.200812026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulubova I., Khvotchev M., Liu S. Q., Huryeva I., Sudhof T. C., Rizo J. Munc18-1 binds directly to the neuronal SNARE complex. Proc. Natl. Acad. Sci. USA. 2007;104:2697–2702. doi: 10.1073/pnas.0611318104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulubova I., Yamaguchi T., Gao Y., Min S. W., Huryeva I., Sudhof T. C., Rizo J. How Tlg2p/syntaxin 16 ‘snares’ Vps45. EMBO J. 2002;21:3620–3631. doi: 10.1093/emboj/cdf381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furgason M.L.M., Macdonald C., Shanks S. G., Ryder S. P., Bryant N. J., Munson M. The N-terminal peptide of the syntaxin Tlg2p modulates binding of its closed conformation to Vps45p. Proc. Natl. Acad. Sci. USA. 2009;106:14303–14308. doi: 10.1073/pnas.0902976106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallwitz D., Jahn R. The riddle of the Sec1/Munc-18 proteins—new twists added to their interactions with SNAREs. Trends Biochem. Sci. 2003;28:113–116. doi: 10.1016/S0968-0004(03)00028-8. [DOI] [PubMed] [Google Scholar]
- Golemis E. A., Serebriiskii I., Finlay R. L., Kolonin M. G., Gyuris J., Brent R. Interaction trap/two-hybrid system to identify interacting proteins. In: Ausubel F. M., et al., editors. Current Protocols in Protein Science. New York: Wiley Interscience; 1998. [Google Scholar]
- Grote E., Carr C. M., Novick P. J. Ordering the final events in yeast exocytosis. J. Cell Biol. 2000;151:439–452. doi: 10.1083/jcb.151.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote E., Novick P. J. Promiscuity in Rab-SNARE interactions. Mol. Biol. Cell. 1999;10:4149–4161. doi: 10.1091/mbc.10.12.4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haapa S., Taira S., Heikkinen E., Savilahti H. An efficient and accurate integration of mini-Mu transposons in vitro: a general methodology for functional genetic analysis and molecular biology applications. Nucleic Acids Res. 1999;27:2777–2784. doi: 10.1093/nar/27.13.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashizume K., Cheng Y. S., Hutton J. L., Chiu C. H., Carr C. M. Yeast Sec1p functions before and after vesicle docking. Mol. Biol. Cell. 2009;20:4673–4685. doi: 10.1091/mbc.E09-02-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B., Guo W. The exocyst complex in polarized exocytosis. Curr. Opin. Cell Biol. 2009;21:537–542. doi: 10.1016/j.ceb.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C. D., Grinberg A. V., Kerppola T. K. Visualization of protein interactions in living cells using bimolecular fluorescence complementation (BiFC) analysis. Curr. Protoc. Protein Sci. 2005 doi: 10.1002/0471140864.ps1910s41. Unit 1910. DOI: 10.1002/0471140864.ps1910s41. [DOI] [PubMed] [Google Scholar]
- Hu S. H., Latham C. F., Gee C. L., James D. E., Martin J. L. Structure of the Munc18c/Syntaxin4 N-peptide complex defines universal features of the N-peptide binding mode of Sec1/Munc18 proteins. Proc. Natl. Acad. Sci. USA. 2007;104:8773–8778. doi: 10.1073/pnas.0701124104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn R., Scheller R. H. SNAREs—engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- Janke C., et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- Jantti J., Aalto M. K., Oyen M., Sundqvist L., Keranen S., Ronne H. Characterization of temperature-sensitive mutations in the yeast syntaxin 1 homologues Sso1p and Sso2p, and evidence of a distinct function for Sso1p in sporulation. J. Cell Sci. 2002;115:409–420. doi: 10.1242/jcs.115.2.409. [DOI] [PubMed] [Google Scholar]
- Johnson J. R., Ferdek P., Lian L. Y., Barclay J. W., Burgoyne R. D., Morgan A. Binding of UNC-18 to the N-terminus of syntaxin is essential for neurotransmission in Caenorhabditis elegans. Biochem. J. 2009;418:73–80. doi: 10.1042/BJ20081956. [DOI] [PubMed] [Google Scholar]
- Kauppi M., Jäntti J., Olkkonen V. M. The function of Sec1/Munc18 proteins—solution of the mystery in sight? In: Keränen S., Jäntti J., editors. Regulatory Mechanisms of Intracellular Membrane Transport. vol. 10. Heidelberg: Springer; 2004. pp. 115–143. [Google Scholar]
- Kerppola T. K. Visualization of molecular interactions by fluorescence complementation. Nat. Rev. Mol. Cell Biol. 2006;7:449–456. doi: 10.1038/nrm1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M., Miller K. J., Mazza M., Feng D. J., Weber M., Keranen S., Jantti J. Molecular interactions position Mso1p, a novel PTB domain homologue, in the interface of the exocyst complex and the exocytic SNARE machinery in yeast. Mol. Biol. Cell. 2005;16:4543–4556. doi: 10.1091/mbc.E05-03-0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham C. F., et al. Molecular dissection of the Munc18c/syntaxin4 interaction: implications for regulation of membrane trafficking. Traffic. 2006;7:1408–1419. doi: 10.1111/j.1600-0854.2006.00474.x. [DOI] [PubMed] [Google Scholar]
- Lee H. Y., Park J. B., Jang H. I., Chae Y .C., Kim J. H., Kim S. I., Suh P. G., Ryu S. H. Munc-18-1 inhibits phospholipase D activity by direct interaction in an epidermal growth factor-reversible manner. FASEB J. 2004;18:C21. doi: 10.1074/jbc.M310976200. [DOI] [PubMed] [Google Scholar]
- Misura K. M., Scheller R. H., Weis W. I. Three-dimensional structure of the neuronal-Sec1-syntaxin 1a complex. Nature. 2000;404:355–362. doi: 10.1038/35006120. [DOI] [PubMed] [Google Scholar]
- Moreno-Borchart A. C., Knop M. Prospore membrane formation: how budding yeast gets shaped in meiosis. Microbiol. Res. 2003;158:83–90. doi: 10.1078/0944-5013-00194. [DOI] [PubMed] [Google Scholar]
- Munson M., Bryant N. J. A role for the syntaxin N-terminus. Biochem. J. 2009;418:e1–e3. doi: 10.1042/BJ20082389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P., Guo W. Ras family therapy: Rab, Rho and Ral talk to the exocyst. Trends Cell Biol. 2002;12:247–249. doi: 10.1016/s0962-8924(02)02293-6. [DOI] [PubMed] [Google Scholar]
- Okamoto M., Sudhof T. C. Mints, Munc18-interacting proteins in synaptic vesicle exocytosis. J. Biol. Chem. 1997;272:31459–31464. doi: 10.1074/jbc.272.50.31459. [DOI] [PubMed] [Google Scholar]
- Pajunen M., Turakainen H., Poussu E., Peranen J., Vihinen M., Savilahti H. High-precision mapping of protein protein interfaces: an integrated genetic strategy combining en masse mutagenesis and DNA-level parallel analysis on a yeast two-hybrid platform. Nucleic Acids Res. 2007;35:e103. doi: 10.1093/nar/gkm563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng R. W., Gallwitz D. Sly1 protein bound to Golgi syntaxin Sed5p allows assembly and contributes to specificity of SNARE fusion complexes. J. Cell Biol. 2002;157:645–655. doi: 10.1083/jcb.200202006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng R. W., Gallwitz D. Multiple SNARE interactions of an SM protein: Sed5p/Sly1p binding is dispensable for transport. EMBO J. 2004;23:3939–3949. doi: 10.1038/sj.emboj.7600410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poussu E., Jantti J., Savilahti H. A gene truncation strategy generating N- and C-terminal deletion variants of proteins for functional studies: mapping of the Sec1p binding domain in yeast Mso1p by a Mu in vitro transposition-based approach. Nucleic Acids Res. 2005;33:e104. doi: 10.1093/nar/gni102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poussu E., Vihinen M., Paulin L., Savilahti H. Probing the alpha-complementing domain of E. coli beta-galactosidase with use of an insertional pentapeptide mutagenesis strategy based on Mu in vitro DNA transposition. Prot. Struct. Funct. Bioinform. 2004;54:681–692. doi: 10.1002/prot.10467. [DOI] [PubMed] [Google Scholar]
- Scott B. L., Van Komen J. S., Irshad H., Liu S., Wilson K. A., McNew J. A. Sec1p directly stimulates SNARE-mediated membrane fusion in vitro. J. Cell Biol. 2004;167:75–85. doi: 10.1083/jcb.200405018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J. S., Tareste D. C., Paumet F., Rothman J. E., Melia T. J. Selective activation of cognate SNAREpins by Sec1/Munc18 proteins. Cell. 2007;128:183–195. doi: 10.1016/j.cell.2006.12.016. [DOI] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. In: Guthrie C., Fink G. R., editors. Guide to Yeast Genetics and Molecular Biology. San Diego, CA: Academic Press; 1991. pp. 3–21. [Google Scholar]
- Skarp K. P., Zhao X., Weber M., Jantti J. Use of bimolecular fluorescence complementation in yeast Saccharomyces cerevisiae. Methods Mol. Biol. 2008;457:165–175. doi: 10.1007/978-1-59745-261-8_12. [DOI] [PubMed] [Google Scholar]
- Taira S., Tuimala J., Roine E., Nurmiaho-Lassila E. L., Savilahti H., Romantschuk M. Mutational analysis of the Pseudomonas syringae pv. tomato hrpA gene encoding Hrp pilus subunit. Mol. Microbiol. 1999;34:736–744. doi: 10.1046/j.1365-2958.1999.01635.x. [DOI] [PubMed] [Google Scholar]
- Togneri J., Cheng Y. S., Munson M., Hughson F. M., Carr C. M. Specific SNARE complex binding mode of the Sec1/Munc-18 protein, Sec1p. Proc. Natl. Acad. Sci. USA. 2006;103:17730–17735. doi: 10.1073/pnas.0605448103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toonen R.F.G., Verhage M. Munc18-1 in secretion: lonely Munc joins SNARE team and takes control. Trends Neurosci. 2007;30:564–572. doi: 10.1016/j.tins.2007.08.008. [DOI] [PubMed] [Google Scholar]
- Verhage M., de Vries K. J., Roshol H., Burbach J. P., Gispen W. H., Sudhof T. C. DOC2 proteins in rat brain: complementary distribution and proposed function as vesicular adapter proteins in early stages of secretion. Neuron. 1997;18:453–461. doi: 10.1016/s0896-6273(00)81245-3. [DOI] [PubMed] [Google Scholar]
- Weisman L. S., Wickner W. Molecular characterization of VAC1, a gene required for vacuole inheritance and vacuole protein sorting. J. Biol. Chem. 1992;267:618–623. [PubMed] [Google Scholar]
- Yamaguchi T., Dulubova I., Min S. W., Chen X. H., Rizo J., Sudhof T. C. Sly1 binds to Golgi and ER syntaxins via a conserved N-terminal peptide motif. Dev. Cell. 2002;2:295–305. doi: 10.1016/s1534-5807(02)00125-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.