Abstract
Background
Trypanosoma brucei gambiense is the causative agent of chronic Human African Trypanosomiasis or sleeping sickness, a disease endemic across often poor and rural areas of Western and Central Africa. We have previously published the genome sequence of a T. b. brucei isolate, and have now employed a comparative genomics approach to understand the scale of genomic variation between T. b. gambiense and the reference genome. We sought to identify features that were uniquely associated with T. b. gambiense and its ability to infect humans.
Methods and Findings
An improved high-quality draft genome sequence for the group 1 T. b. gambiense DAL 972 isolate was produced using a whole-genome shotgun strategy. Comparison with T. b. brucei showed that sequence identity averages 99.2% in coding regions, and gene order is largely collinear. However, variation associated with segmental duplications and tandem gene arrays suggests some reduction of functional repertoire in T. b. gambiense DAL 972. A comparison of the variant surface glycoproteins (VSG) in T. b. brucei with all T. b. gambiense sequence reads showed that the essential structural repertoire of VSG domains is conserved across T. brucei.
Conclusions
This study provides the first estimate of intraspecific genomic variation within T. brucei, and so has important consequences for future population genomics studies. We have shown that the T. b. gambiense genome corresponds closely with the reference, which should therefore be an effective scaffold for any T. brucei genome sequence data. As VSG repertoire is also well conserved, it may be feasible to describe the total diversity of variant antigens. While we describe several as yet uncharacterized gene families with predicted cell surface roles that were expanded in number in T. b. brucei, no T. b. gambiense-specific gene was identified outside of the subtelomeres that could explain the ability to infect humans.
Author Summary
Sleeping sickness, or Human African Trypanosomiasis, is a disease affecting the health and productivity of poor people in many rural areas of sub-Saharan Africa. The disease is caused by a single-celled flagellate, Trypanosoma brucei, which evades the immune system by periodically switching the proteins on its surface. We have produced a genome sequence for T. brucei gambiense, which is the particular subspecies causing most disease in humans. We compared this with an existing reference genome for a non-human infecting strain (T. b. brucei 927) to identify genes in T. b. gambiense that might explain its ability to infect humans and to assess how well the reference performs as a universal plan for all T. brucei. The genome sequences differ only due to rare insertions and duplications and homologous genes are over 95% identical on average. The archive of surface antigens that enable the parasite to switch its protein coat is remarkably consistent, even though it evolves very quickly. We identified genes with predicted cell surface functions that are only present in T. b. brucei and have evolved rapidly in recent time. These genes might help to explain variation in disease pathology between different T. brucei strains in different hosts.
Introduction
Trypanosoma brucei subsp. gambiense is the causative agent of Human African Trypanosomiasis (HAT), or sleeping sickness, which is a vector-borne disease restricted to rural areas of sub-Saharan Africa. Trypanosomiasis in humans and livestock imposes substantial morbidity, representing a major impediment of agricultural production in the affected areas [1], and is fatal where untreated. The World Health Organization estimated in 1998 that up to 60 million people are at risk in approximately 250 distinct foci [2], although under-reporting has been estimated as high as 40% in some foci [3]. T. b. gambiense is the most clinically relevant sub-species, causing over 90% of all human disease. The gambiense disease is typically chronic, often lasting several years with few severe signs and symptoms until the late stage of nervous system involvement. T. b. gambiense is sensitive to treatment with pentamidine (early stage) and eflornithine (late stage), drugs which are frequently ineffective against T. b. rhodesiense [4], although the underlying biochemical reasons for these differences are unknown. Combination therapies against the late stage disease have performed encouragingly [5] but few drugs are available. Furthermore, unpleasant and in some cases severe side effects often result in poor patient compliance. Hence, new molecular targets are required to supply current drug discovery programmes [6].
T. brucei is subdivided into three subspecies based on infectivity to humans, pathogenicity and geographical distribution. T. b. gambiense and T. b. rhodesiense are human pathogens, causing Human African Trypanosomiasis (HAT) in West/Central and East Africa respectively. T. b. brucei cannot by definition infect humans and is found in a wide range of wild and domestic mammals. The human pathogens have also been found in various animal species and HAT caused by T. b. rhodesiense in East Africa is recognized as a zoonosis. T. b. gambiense comprises two groups; a genetically homogeneous group to which the majority of isolates belong (group 1), and a second represented by a handful of isolates from West Africa (group 2). Group 1 T. b. gambiense strains have the smallest genomes in the T. brucei species complex, having 71–82% of the highest DNA content measured for T. b. brucei [7]–[8]. Pulse-field gel analysis of T. b. gambiense chromosomes shows that few if any mini-chromosomes are present compared to the estimated 100 in T. b. brucei and T. b. rhodesiense, and the mini-chromosomes are also of a smaller size–average 25 kb in T. b. gambiense compared to 100 kb in T. b. brucei and T. b. rhodesiense [8]–[9].
Perhaps as a consequence of this reduced genome, T. b. gambiense also has a restricted repertoire of Variant Surface Glycoprotein (VSG) genes [8], [10]–[12]. At any time, bloodstream form trypanosomes possess a surface glycoprotein coat formed through the expression of a single gene from a large archive of VSGs [13]. This coat obfuscates the host immune system by shielding the invariant surface epitopes from view and, when an immune response is inevitably raised against the VSG monolayer and the active VSG is replaced by another, it allows parasites expressing the novel variant to escape the immune response [13]. This periodic VSG ‘switching’, or in situ activation, is facilitated by transposition of inactive VSG into a dedicated expression site at the telomeres by gene conversion [13]–[15]. Although VSG repertoire is clearly very large [16]–[17], it is not known how VSG diversity accumulates over time and between strains. The SRA gene encodes a truncated VSG-like protein [18]; it is located within one specific VSG expression site and is expressed in Human serum-resistance clones of T. b. rhodesiense only [19]. Innate immunity to trypanosomes in Humans is conferred by a trypanolytic factor, apoL1 [20] and SRA has acquired a role in neutralizing the toxic effects of this protein [21]. Hence, when transcriptionally activated, SRA enables particular T. b. rhodesiense clones to infect Humans [22]. SRA is absent in T. b. gambiense [23] and the underlying basis for the trait of human infectivity here is as yet unknown. In T. b. gambiense, as yet the only example of a subspecies-specific gene is TgsGP, which encodes a 47 kDa VSG-like receptor protein, and is expressed in the flagellar pocket of bloodstream stage cells [24]. However, TgsGP is not associated with human infectivity in T. b. gambiense [25].
We produced an improved, high-quality draft genome sequence for T. b. gambiense DAL927 with the twin aims of identifying subspecies-specific genomic features that might contribute to our understanding of phenotypic variation and assessing the scale of genomic variation across T. brucei. This was achieved through comparison with the T. b. brucei 927 reference genome and we sought to evaluate the proficiency of this reference, ahead of the next generation of genome sequencing projects that will compare multiple isolates to scrutinize genetic divergence and genomic rearrangements in relation to disease. Our analyses show that the genome sequence of T. b. gambiense corresponds closely in gene order and content to the T. b. brucei 927 genome. Intraspecific genomic variation is largely associated with tandem or segmental duplications, among which we identify several subspecies-specific isoforms. Our final objective was to compare the VSG repertoires of T. b. brucei and T. b. gambiense, and so provide the first global perspective of how VSG diversity evolves on a genome scale. Details of the genome project describing the ‘Minimum Information for Genome Sequences’ are available online (http://genomesonline.org/GOLD_CARDS/Gi00917.html).
Methods
Accession numbers
The sequence of the Trypanosoma brucei gambiense genome has been submitted to the EMBL database under accession numbers FN554964- FN554974 inclusive.
Trypanosome stocks
The T. b. gambiense strain MHOM/CI/86/DAL972 was isolated from a patient in Côte d'Ivoire in 1986 and has been used routinely in laboratory studies since this time [26]. Bloodstream form trypanosomes were fed to tsetse in vitro and procyclics from infected midguts were established in culture and subsequently optically cloned. Procyclic form trypanosomes were grown in Cunningham's medium supplemented with 10% v/v heat-inactivated foetal calf serum, 5 µg/ml hemin and 10 µg/ml gentamycin at 27°C. High molecular weight DNA was purified by standard methods of phenol-chloroform extraction and alcohol precipitation.
T. b. gambiense genome sequencing and assembly
T. b. gambiense DNA was randomly sheared, size-selected DNA purified and subcloned into pUC19 plasmids (1.4 kb–4 kb inserts), as well as BAC vectors as previously described [27]. Inserts were sequenced by random sequencing using dye-terminator chemistry on ABI 3730 sequencing machines from both ends to generate paired end reads. There were 369,043 passed paired-end reads, producing roughly eight-fold coverage of the whole genome. Sequence reads were assembled using Phrap (www.phrap.org; P. Green, unpublished). Automated in-house software (Auto-Prefinish) was used to identify primers and clones for additional sequencing to close physical and sequence gaps by oligo-walking. Manual base checking and finishing was carried out using Gap4 (http://www.mrc-lmb.cam.ac.uk/pubseq/manual/gap4_unix_1.html). Regions containing repeat sequences or with an unexpected read depth were manually inspected. The assembled contigs were iteratively ordered and orientated against the T. brucei 927 genome sequence, with manual checking. Aided by information from orientated read-pairs, together with additional sequencing from selected large insert clones, we re-examined regions with apparent breaks in chromosomal colinearity for potential assembly errors.
Genome annotation
The human-curated annotation of the T. b. brucei 927 reference genome was transferred to the assembled T. b. gambiense genome on the basis of BLASTp matches and positional information using custom perl scripts. Subsequently, gene structure and functional annotation were manually inspected and further edited, where appropriate, using the Artemis software [28], as previously detailed [27]. The annotation of the T. b. gambiense genome can be viewed and searched via GeneDB (http://www.genedb.org/) and comparative chromosome maps for T. b. brucei and T. b. gambiense are available at TritrypDB (http://tritrypdb.org; [29]). Chromosomal sequences have been submitted to EMBL with the following accession numbers: FN554964-FN554974 inclusive.
Variation detection from sequence data
The T. b. gambiense capillary shotgun reads were aligned against the T. b. brucei 927 reference genome using SSAHA2 (http://www.sanger.ac.uk/Software/analysis/SSAHA2/). We discarded reads that mapped to more than one location on the reference genome, as well as pairs of reads that did not map in the correct orientation or to within 20% of the expected insert size of the library. In-house perl scripts were used to identify single nucleotide polymorphisms (SNPs) from the SSAHA alignments that adhered to a modified version of the Neighbourhood Quality Standard (NQS, [30]); we term this AltNQS. According to NQS, an acceptable SNP (or fixed difference) has a phred quality score of ≥23 and the 5 bases on either side of the SNP position have a quality score of ≥15. However, these strict criteria do not allow for multiple mismatches within the 11 bp window. To accommodate the higher levels of polymorphism, our AltNQS adheres to the same rules as NQS but allows for multiple SNPs within the 11 bp alignment window as long as the base quality of each SNP has a phred score of at least 23. To identify regions with significantly high SNP density on each chromosome, non-overlapping windows of 10 kb with at least 50% of read coverage were selected for analysis. For these windows, SNP density was calculated as the number of SNPs divided by the number bases covered in that 10 kb window. Using random sampling we estimated the mean and 97.5% confidence limit of mean SNP density. Regions with a value above the 97.5% quantile were identified as having significantly high SNP density values.
Tandem repeat recombination analysis
Tandem gene arrays in the T. b. brucei 927 genome with >3 gene copies have previously been defined, and are known to contain polymorphism that is affected by recombination [31]. We assessed the variation among tandem gene duplicates to identify subspecies-specific genes. For each of these arrays, the coding and 3′ UTR sequences were gathered from the corresponding regions of the T. b. gambiense genome sequence. The downstream limit of the 3′ UTR was defined by the polypyrimidine termination motif [32]. All T. b. brucei and T. b. gambiense sequences were aligned in ClustalX [33] and manually adjusted. Those arrays showing no variation or only corresponding isoforms in both subspecies (i.e., simple orthology) were discarded, leaving just those cases where a disparity in sequence diversity was apparent. To detect any ambiguity in phylogenetic relationships among sequences, each of these alignments was analyzed using SplitsTree v4.3 [34], which applies a Neighbour-Net method [35]) to estimate a phylogenetic network. Genetic distances were corrected for variation in base composition after excluding phylogenetically-uninformative characters. Each alignment was also analyzed using the pair-wise homoplasy index (PHI) test [36] that can detect multiple phylogenetic signals within an alignment and is robust in the presence of rate heterogeneity. A third method, the genetic algorithm for recombination detection (GARD, [37]) was applied to estimate the number and placement of recombination breakpoints along each alignment.
Comparison of the variant surface glycoprotein (VSG) repertoire
1258 predicted VSG protein sequences encoded in the T. b. brucei genome were compared with the T. b. gambiense 972 read library using pair-wise BLASTp searches. These included 36 VSG-related (VR) sequences that are structurally distinct from the bulk of canonical VSG [17]. Initially, all VSG-like sequences were extracted from the T. b. gambiense read library using BLASTx against whole VSG protein sequences. Each T. b. brucei VSG protein sequence was then individually BLAST-searched against this subset of VSG-like reads to determine its closest match in T. b. gambiense. A reciprocal comparison was carried out to confirm the relationship. To determine if a given gene was most closely related to a paralog in T. b. brucei or to an ortholog in T. b. gambiense, each T. b. brucei VSG protein sequence was also compared a combined database of VSG gene models and VSG-like reads using BLASTp. BioLayout Express 3D [38] was used to visualize the relative genetic distances between the 1258 T. b. brucei VSG sequences, using the BLAST scores derived from comparisons of each gene with all others, and a 70% cutoff to simplify the resulting network graph. To determine if VSG diversity is sub-structured according to life stage, nine VSG sequences known to be associated with metacyclic expression sites were BLAST-searched against all other (bloodstream-expressed) VSG and added to the network.
Results/Discussion
The T. b. gambiense genome was whole-genome shotgun sequenced to eight-fold coverage by paired-end capillary sequencing of plasmid and bacterial artificial chromosome (BAC) clones, resulting in an improved high-quality draft sequence. In comparison with the T. b. brucei 927 reference sequence, the two genomes are very similar in composition and structure, such that no protein coding sequence unique to T. b. gambiense could be found. However, coding sequences unique to T. b. brucei were found and the two genomes displayed other subtle differences in the diversity of repetitive regions such as segmental duplications, tandem gene arrays and strand switch regions, which document the scale of genomic variation across T. brucei subspecies.
The T. b. brucei reference is an effective template for the T. b. gambiense genome sequence
The draft genome assembly consists of 1768 contigs larger than 2 kb, amounting to 32.6 Mb of data. Of these, 281 contigs, totaling 22.1 Mb, were ordered and orientated against the T. b. brucei 927 reference genome. The remaining contigs encode additional copies of tandemly arrayed gene families as well as genes typically associated with subtelomeres such as expression site associated genes (ESAGs), variant surface glycoprotein (VSG) genes and the ingi transposable element. The gene models and annotation of an initial set of 9898 coding sequences located on core chromosomes (i.e., not in subtelomeres) were transferred to the T. b. gambiense genome on the basis of BLASTp matches and positional information using custom perl scripts.
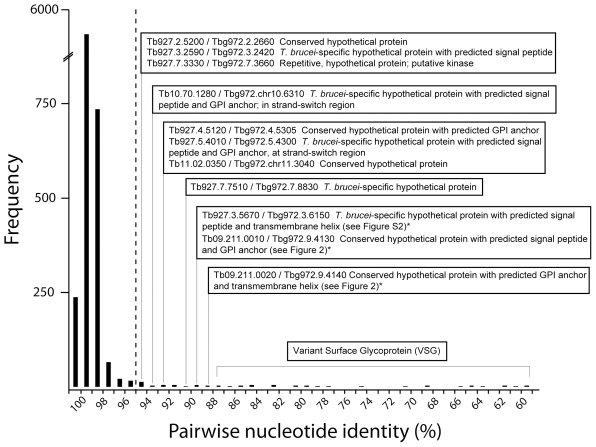
When compared, the T. b. brucei and T. b. gambiense genome sequences are very similar in terms of content, gene order and sequence identity. The absence of potentially gambiense-specific sequences was confirmed by examining a Phrap assembly of those capillary reads that did not map against the T. brucei 927 reference genome. Analysis of ∼40,000 unmapped sequence reads using BLASTx showed that among them were features homologous to VSG, ESAG and RHS genes, as well as ingi retrotransposons, but no additional coding sequences that were missing from T. b. brucei. We examined the divergence of coding sequences and a frequency histogram of percentage nucleotide identity (Fig. 1) shows that 86.4% of genes vary by less than 1% from their T. b. brucei ortholog (mean average nucleotide identity = 99.2%). Non-coding regions were more divergent, which is unsurprising given that they are probably under weaker purifying selection, but still remained 95.4% identical on average. However, against this general background of correspondence there are 69 pairs of orthologs that display significantly greater evolutionary change, (i.e., they are among the 5% most divergent orthologs with a nucleotide identity <95.2%). 35 of these gene pairs are VSG sequences; these surface glycoproteins are exposed to frequent gene conversion and evolve rapidly [16]–[17], so naturally, they display lower sequence identities of ∼60–85%. However, they still display reciprocal top BLAST hits with T. b. brucei sequences. Also among these divergent gene pairs are 17 uncharacterized genes, 10 of which are predicted to encode cell-surface targeted proteins. For example, Tb927.5.4010/Tbg972.5.4300 (92.7% identical) and Tb10.70.1280/Tbg972.10.6310 (93.7% identical) are both located at strand-switch regions and encode hypothetical proteins with predicted signal peptides and GPI anchor sites. These genes, which appear to be evolving very quickly, are not found in either Leishmania major or T. cruzi, indicating that they are specific to African trypanosomes.
Figure 1. Frequency distribution of pairwise sequence divergence between 6929 single-copy gene orthologs in T. b. brucei and T. b. gambiense.
Divergence values to the right of the dashed line are statistically significant; the identities of selected divergent gene pairs are noted. An asterisk * denotes genes belonging to a T. b. brucei subspecies-specific tandem gene array (see Figure 2 and Supplementary Figure S2).
A source of variation with potentially important functional consequences is allelic polymorphism. We detected high-confidence SNPs and fixed differences by mapping the T. b. gambiense reads to the T. brucei 927 reference sequence. Our analysis focused on the non-repetitive component of the genome as firstly, non-identical repeats can appear indistinguishable from SNPs and secondly, repeated regions may be subject to unusual selective pressures (see below). After excluding these sequences, we identified a total of 224,568 putative fixed differences from 19.4 Mb of non-repetitive sequence, i.e. a diversity (π) of 0.0116 nucleotides per site. 92,794 of these differences were in coding regions, 49% of which were non-synonymous. To confirm that the variation identified when mapping the T. b. gambiense reads against the T. b. brucei 927 were not in fact false-positives due to heterozygosity within the T. b. brucei 927 reference sequence itself we also used the available capillary read data from the T. b. brucei 927 genome project to identify polymorphism within the published “haploid” consensus. Unfortunately, this was only possible for the four chromosomes (1, 9–11) that were originally produced by shotgun sequencing, (rather than a clone walking strategy), since these contain data from two homologous chromosomes at a given locus. From the SSAHA alignments, we identified 23,804 SNPs in 10.8 Mb of map-able sequence (π = 0.0022), of which 1,187 had the same heterozygous alleles in both the T. b. brucei 927 and the T. b. gambiense genome, indicating a false-positive rate of 5%. We identified 298 regions exhibiting higher than average diversity along the megabase chromosome. It is noteworthy, that this analysis excluded all telomere proximal regions owing to their highly repetitive nature. Whereas telomeres are well established in many species as sites of sequence variation and rearrangement [39]–[40], the presence of interstitial regions of high diversity in addition to the sub-telomeres is striking.
Disruptions to chromosomal colinearity are rare and reveal few subspecies-specific features
On rare occasions the otherwise consistent chromosomal colinearity is disrupted by sequence inversions and insertion-deletion events (indels). In many cases indels coincided with sequence gaps, making it difficult to confirm genuine rearrangements. Nevertheless, chromosome 10 provides two examples, between 275–330 kb and 3250–3350 kb, of 55 and 110 kb segmental inversions respectively. Gene order within these inverted regions remains conserved. Typically, indels have two principal causes: transposable elements and internal VSG ‘islands’. Transposable elements such as ingi and RIME sequences recombine in trypanosome genomes and are responsible for several rearrangements [27]. On chromosome 9, a 7 kb insertion occurs in T. b. brucei due to an ingi element (at 1.24 Mb) not present in T. b. gambiense. Similarly, a 29 kb indel follows Tb11.02.5830 where an expression site-associated gene (ESAG) and a trans-sialidase gene have been inserted into T. b. gambiense at the corresponding position to a RIME sequence in T. b. brucei. By their nature, such rearrangements frequently occur in repetitive regions of the genome and, consequently, are difficult to resolve in genome assemblies. This therefore does not preclude that further events will be identified in the future.
Another source of genomic variation concerns core chromosomal VSG and ESAG genes. VSG genes are predominantly found in subtelomeric arrays, on intermediate or mini-chromosomes [27], [13]. In addition, VSG/ESAG genes are less commonly found non-telomerically as ‘islands’, often on the opposing strand to neighbouring loci. These genes (or pseudogenes) may be: (i) atypical VSGs that do not encode all elements for accurate folding or post-translational modification; (ii) VR genes; or (iii) canonical VSG genes, imported from the subtelomere or mini-chromosome through segmental duplication. An example of the latter is a segmental insertion including 8 VSG genes that affects chromosome 9 in T. b. gambiense (Tbg972.2.570–640), since the VSG sequences are unrelated to each other and therefore, have not resulted from recent tandem duplications. In total, 17 such VSG/ESAG islands were noted in both genomes, only 6 of which were unique to one subspecies or other, including a segmental duplication in T. b. brucei of an atypical VSG combined with an insertion or deletion of ESAGs (Supplementary Fig. S1). Clearly, VSG/ESAG islands are among the more dynamic features of core chromosomes, yet where they are conserved between T. b. brucei and T. b. gambiense they contain orthologous gene sequences, indicating that they not exposed to frequent gene conversion processes like VSGs elsewhere.
Beyond transposable elements and VSG ‘islands’, other differences in gene order are caused by a class of small, putative coding sequences of unknown function (103 cases). These genes encode hypothetical proteins with a predicted length of 151–274 amino acids and which have no database matches to any experimentally characterized protein. Transcriptomic data (George Cross, Rockefeller University, unpublished data; Veitch et al., University of Glasgow, submitted) suggest that some of these putative genes are at least transcribed, although no product has yet been identified in proteomic assays to date (Aswini Panigrahi, SBRI, pers. comm.). Regardless of which genome encodes the putative gene, homologous sequences of high identity are found in the other genome at the corresponding positions, but without the open reading frame. Hence, they may be non-coding RNA genes or other non-coding conserved elements of undiscovered function. These features are annotated to ensure completeness, and they may yet reveal functional importance, but our view is that they are unlikely to produce proteins and will not be considered further.
An isolate-specific locus: a putative iron-ascorbate oxidoreductase in T. b. brucei 927
Our comparative analysis identified only a single coding sequence, a putative iron-ascorbate oxidoreductase (Tb09.211.4990), which is absent from the genomic repertoire of T. b. gambiense. We did not identify the TgsGP locus, which is known to be unique to T. b. gambiense [24] because it is located in the subtelomere and these regions were not fully assembled. However, sequence identical to TgsGP was identified among the unassembled reads. Thus, it is possible that other subspecies-specific genes exist within the subtelomeres that are not recorded here. Tb09.211.4990 is preceded upstream on chromosome 9 by a strand-switch region and downstream by both retrotransposon-like proteins and the splice-leader RNA tandem array. This region is conserved in T. b. gambiense, but the oxidoreductase is absent. The gene is absent from the more distantly related kinetoplastids Leishmania major and T. cruzi, as well as 9 out of 11 other T. b. brucei strains and a representative group 2 T. b. gambiense (STIB 386) that we examined with PCR primers specific to this oxidoreductase (data not shown). When compared phylogenetically with other iron-ascorbate oxidoreductases in T. brucei, (principally the tandem gene array at the right-hand terminus of chromosome 2, e.g. Tb927.2.6180), this protein is clearly structurally distinct (only 80% amino acid identity) and constitutes an evolutionarily old lineage. This suggests that Tb09.211.4990 is gained and lost at the population level, and that it provides additional functionality to T. b. brucei 927 and two other T. b. brucei strains in which it has been found.
Segmental duplications of putative membrane proteins contain subspecies-specific gene copies
The comparison of gene content did not identify widespread subspecies-specific loci, and found no obvious differences that could explain the distinct phenotypes of T. brucei subspecies. For example, ornithine decarboxylase, the target of eflornithine to which T. b. gambiense is uniquely sensitive, is present in single, diploid copy in both genomes and displays only a single non-synonymous substitution (N137I). We did, however, detect substantial variation within families of certain uncharacterized genes that could have important functional consequences. Such differences in co-linearity involve either the expansion of a single-copy gene in one subspecies to a tandem pair in the other, or a difference in the number of duplicates where there is a tandem array in both subspecies. Current methods of genome assembly tend to detect the first scenario (i.e., single copy vs. many) but have limitations in accurately quantifying copy number and in distinguishing between copy number and allelic variation. In fact, while the number of repeat units assembled can be arbitrary, the variation among tandem gene duplicates can be accurately assessed from genome sequence data for the two subspecies. In 20 cases, a single-copy feature (be it a single gene or chromosomal segment) in T. b. gambiense exists in multiple, tandem copies in T. b. brucei, while 8 cases of the converse were observed (Table 1). For the majority of these cases, the tandem duplicates were identical and the duplication did not result in any novel, unique sequence. But in 8 cases in T. b. brucei, the extra duplicates contained sequence variation that might represent subspecies-specific isoforms. In four additional cases, the would-be unique sequences were found among sequence reads of the apparently single-copy subspecies, indicating that it had been omitted from the assembly (marked by an asterisk in Table 1).
Table 1. Species-specific segmental duplications in (a) T. b. gambiense and (b) T. b. brucei.
| (a) T. b. gambiense locus | Chr | Repeat unit in T. b. brucei: | Description | Distance | Unique isoforms | ||
| Length | Genes | Copies | |||||
| Tbg972.3.6170∧ | 3 | 1803 | 1 | 5 | Hypothetical protein | 0.0363 | 2 |
| Tbg972.4.4370 | 4 | 3758 | 2 | 2 | Glycosyltransferase | 0.0592 | 1 |
| Hypothetical protein | 0.00025 | 0 | |||||
| Tbg972.5.710 | 5 | 3379 | 1 | 3 | Adenylate cyclase | 0.1004 | 1* |
| Tbg972.5.4870 | 5 | 6483 | 1 | 2 | Adenylate cyclase | 0.00135 | 0 |
| Tbg972.6.30 | 6 | 1462 | 1 | 2 | Hypothetical protein | 0.0005 | 0 |
| Tbg972.6.140–170 | 6 | 7402 | 4 | 3 | GPEET2 procyclin precursor | 0.0227 | 1 |
| EP3-2 procyclin | 0.0022 | 0 | |||||
| EP3-3 procyclin | 0.131 | 0 | |||||
| Tbg972.6.1030# | 6 | 3865 | 2 | 5 | PPIase cyclophilin-type isomerase | 0.0068 | 0 |
| Hypothetical protein | 0.1564 | 4 | |||||
| Tbg972.7.1230–1240 | 7 | 4162 | 2 | 2 | Hypothetical protein | 0.001 | 0 |
| Hypothetical protein | 0.0584 | 1 | |||||
| Tbg972.7.6460 | 7 | 2147 | 1 | 2 | Cell-cycle associated MOB1 | 0.008 | 1* |
| Tbg972.8.540 | 8 | 10346 | 1 | 2 | Hypothetical protein | 0.0003 | 0 |
| Tbg972.8.740 | 8 | 6215 | 1 | 3 | Vacuolar-type Ca2+-ATPase | 0.0839 | 1* |
| Tbg972.8.1230 | 8 | 2137 | 1 | 5 | gp63 | 0.0077 | 1* |
| Tbg972.8.1705 | 8 | 1623 | 1 | 2 | Hypothetical protein | 0.5671 | 1 |
| Tbg972.8.7610 | 8 | 2650 | 1 | 2 | Trans-sialidase | 0.0222 | 0 |
| Tbg972.9.1930–1950 | 9 | 6350 | 3 | 2 | Ribosomal protein S7 | 0.0016 | 0 |
| Hypothetical protein | 0.0037 | 0 | |||||
| Nicotinamidase | 0 | 0 | |||||
| Tbg972.9.4160–4130† | 9 | 5729 | 3 | 5 | Hypothetical protein | 0.015 | 0 |
| Hypothetical protein | 0.488 | 4 | |||||
| Hypothetical protein | 0.409 | 4 | |||||
| Tbg972.chr10.5040 | 10 | 3163 | 1 | 2 | Elongation factor 2 | 0.0004 | 0 |
| Tbg972.chr11.1620 | 11 | 1290 | 1 | 2 | ESAG | 0.481 | 1 |
| Tbg972.chr11.8655 | 11 | 1325 | 1 | 2 | Hypothetical protein | 0.0183 | 0 |
| Tbg972.chr11.12640 | 11 | 1310 | 1 | 2 | Activated protein kinase c receptor | 0.001 | 0 |
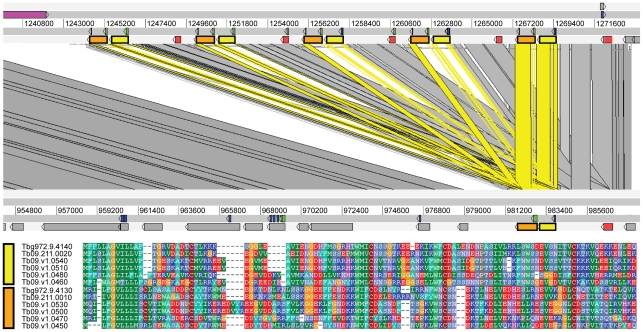
The genes involved in these T. b. brucei-specific segmental duplications are as yet uncharacterized, but their features suggest that they are potential sources of subspecies-specific factors and interesting opportunities for further research. They are evolutionarily novel since they are not conserved in either T. cruzi or L. major; several encode proteins with predicted cell surface roles; and some are among the fastest evolving of all T. brucei genes. For example, a tandem gene array of hypothetical genes encoding cysteine-rich secretory proteins is shown in Supplementary Fig. S2; these are homologous to a single gene (Tbg972.3.6170) at the corresponding position on chromosome 3 in T. b. gambiense. From the relative strength of BLAST hits between homologs, it is clear that the first gene in the array and the singleton in T. b. gambiense are orthologs, while the additional copies in T. b. brucei (absent from the T. b. gambiense read library) are unique paralogs. Indeed, they have evolved considerably, sharing only 55.1% amino acid identity with the upstream orthologs. Similarly, Figure 2 shows a single segment on chromosome 9 in T. b. gambiense (Tbg972.9.4160, 4140 and 4130) that corresponds to five tandem repeats in T. b. brucei. Among gene duplicates of the second and third coding sequences, which encode hypothetical transmembrane and GPI-anchored proteins respectively, there is considerable sequence variation (average nucleotide identities of 51.2% and 59.1% respectively). As in Supplementary Fig. S2, the 5′-most segment in T. b. brucei is orthologous to the T. b. gambiense genes, but the downstream copies are structurally divergent. A third example of segmental duplication with subsequent divergence of tandem copies occurs on chromosome 6 and concerns a hypothetical protein with a predicted signal peptide and GPI anchor (Supplementary Fig. S3).
Figure 2. Segmental duplication on chromosome 9 in T. b. brucei.
A single segment in T. b. gambiense comprising three coding sequences (Tbg972.9.4160, 4140 and 4130) corresponds to a three-gene segmental duplication (5 repeats) on chromosome 9 in T. b. brucei. The first coding sequence (shaded red) is a conserved, hypothetical gene encoding a putative secretory protein and all copies are identical. The second (shaded yellow) and third (shaded orange) coding sequences are tandem-duplicate, conserved hypothetical genes encoding putative membrane-bound proteins. Both second and third genes contain substantial sequence variation in T. b. brucei; the upstream-most copies are orthologous to the T. b. gambiense genes, but none of the remaining variants were identified among T. b. gambiense sequence reads. The segmental duplication is preceded immediately upstream by an INGI-mediated insertion (shaded purple).
Such segmental duplications provide rare examples of subspecies-specific gene paralogs or isoforms. It remains to be seen how common, and how ephemeral, such copy number variation is among T. brucei strains generally. But these cases are especially interesting because they do not simply concern gene dosage. In fact, with divergence in protein sequence often between 30–40% among paralogs, the effects on protein function could be considerable. Not only have these genes multiplied in number in very recent evolutionary time, this has been accompanied by rapid structural divergence in their predicted cell surface gene products, suggesting a role for adaptive change. Such protein isoforms could contribute to the observed differences between group 1 T. b. gambiense and other T. brucei isolates in the host-parasite relationship, both in the mammalian and insect hosts.
Tandem gene arrays frequently contain subspecies-specific sequence mosaics
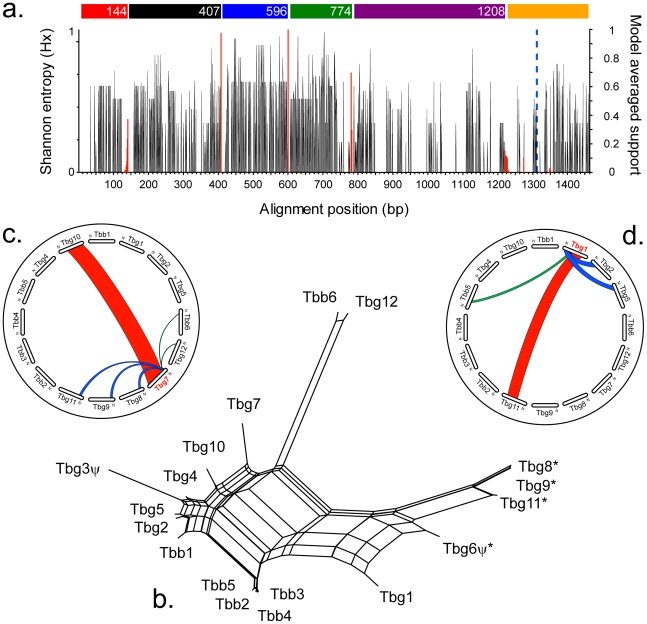
Tandem gene arrays in the T. b. brucei genome usually contain sequence variants and analysis of tandem duplicate variation using T. b. brucei sequences alone showed that divergence frequently results in sequence mosaics and concerted evolution within genomes [31]. After discounting the minority of invariant tandem arrays in T. b. gambiense, 35 tandem gene arrays that contained sequence variation were compared with their T. b. brucei homologues, demonstrating that 27 arrays contained subspecies-specific gene copies (Table 2). In 5/49 instances subspecies-specific copies displayed unique sequence motifs, suggesting differential assortment of the ancestral gene repertoire between the daughter subspecies. Elsewhere, subspecies-specific copies were recombinants of other duplicates. Tests for recombination carried out on multiple alignments of gene copies from both subspecies demonstrated that sequence mosaics occurred in 31/35 data sets as exemplified by the array of invariant surface glycoproteins on chromosome 2 (ISG; Tb927.2.3270–3320) (Fig. 3). The ISG array comprises 6 and 12 gene copies in T. b. brucei and T. b. gambiense, respectively. GARD analysis detected at least five recombination breakpoints (Fig. 3a) and the recombinant nature of ISG is reflected in a highly reticulated phylogenetic network (Fig. 3b). This also identifies potential subspecies-specific recombinants, for instance, the proximity of ‘Tbg7’ to ‘Tbg10’ reflects the overall similarity of these copies, but closer inspection shows that small sections of homology exist with other copies, i.e., ‘Tbg8/9’(Fig. 3c). Similarly, the intermediate position of Tbg1 reflects its affinities with multiple, unrelated sequences (Fig. 3d).
Table 2. Evidence for recombination within variable tandem gene arrays conserved in T. b. gambiense and T. b. brucei.
| Recombination tests: | |||||||||||||||||
| Annotated copies: | Shared isoforms: | Unique isoforms: | PHI | LRT: | GARD: | ||||||||||||
| Array | Description | Sample identifier | Chr | Tbb | Tbg | Tbb | Tbg | Mean | Observed | p | Sequences | Sites | Breakpoints | Sequences | Sites | Breakpoints | |
| 5 | Pteridine transporter | Tb927.1.2880 | 1 | 3 | 3 | 2 | 1 | 0 | 0.367 | 0.078 | <0.001 | 6 | 3619 | 2 | 5 | 3619 | 9 |
| 6 | Hypothetical protein | Tb927.1.4540 | 1 | 6 | 4 | 2 | 0 | 1 | 0.053 | 0.005 | <0.001 | 9 | 1462 | 0 | 18 | 1462 | 8* |
| 9 | 65 kDa invariant surface glycoprotein | Tb927.2.3270 | 2 | 6 | 10 | 2 | 0 | 1 | 0.384 | 0.161 | <0.001 | 13 | 3226 | 8 | 10 | 3226 | 6* |
| 12 | Hypothetical protein | Tb927.2.5290 | 2 | 8 | 24 | 5 | 2 | 0 | 0.348 | 0.190 | <0.001 | 29 | 1790 | 10 | 35 | 1790 | 3* |
| 12bi | Adenosine transporter | Tb927.2.6150 | 2 | 6 | 3 | 3 | 3 | 0 | 0.415 | 0.289 | <0.001 | 9 | 1407 | 10 | 9 | 1407 | 14* |
| 12bii | Iron-ascorbate oxidoreductase | Tb927.2.6180 | 2 | 5 | 2 | 1 | 1 | 0 | 0.038 | 0.011 | <0.001 | 4 | 960 | 0 | 7 | 960 | 3 |
| 13 | Hypothetical protein | Tb927.3.2550 | 3 | 4 | 4 | 3 | 1 | 1 | 0.328 | 0.038 | <0.001 | 6 | 1536 | 0 | 8 | 1536 | 4 |
| 17 | Hypothetical protein | Tb927.3.4070 | 3 | 5 | 5 | 5 | 0 | 0 | 0.157 | 0.096 | <0.001 | 8 | 2343 | 6 | 10 | 2343 | 9 |
| 22 | Hypothetical protein | Tb927.4.3220 | 4 | 3 | 2 | 0 | 3 | 2 | 0.457 | 0.285 | <0.001 | 6 | 2515 | 10 | 6 | 2515 | 9* |
| 25 | Adenylate cyclase, GRESAG4 | Tb927.4.3880 | 4 | 3 | 4 | 3 | 0 | 1 | 0.179 | 0.062 | <0.001 | 7 | 5171 | 2 | 8 | 5171 | 9* |
| 28 | Adenylate cyclase, GRESAG4 | Tb927.4.4470 | 4 | 7 | 7 | 5 | 0 | 0 | 0.314 | 0.085 | <0.001 | 13 | 4716 | 11 | 13 | 4716 | 4* |
| 36 | Oligosaccharyl transferase subunit | Tb927.5.890 | 5 | 3 | 1 | 1 | 2 | 0 | 0.391 | 0.274 | 0.001 | 4 | 2475 | 0 | 4 | 2475 | 9 |
| 41i | Procyclin | Tb927.6.450 | 6 | 3 | 2 | 2 | 1 | 0 | 0.004 | 0.008 | 0.998 | 6 | 390 | 0 | 6 | 390 | 3 |
| 42 | Adenylate cyclase, GRESAG4 | Tb927.6.760 | 6 | 5 | 4 | 1 | 1 | 1 | 0.154 | 0.044 | <0.001 | 4 | 4701 | 2 | 9 | 4701 | 11 |
| 50 | Trypanothione peroxidase 1 | Tb927.7.1120 | 7 | 3 | 2 | 2 | 1 | 0 | 0.232 | 0.230 | 0.467 | 5 | 510 | 1 | 5 | 510 | 5 |
| 50bi | Hypothetical protein | Tb927.7.360 | 7 | 5 | 2 | 1 | 1 | 1 | 0.210 | 0.126 | <0.001 | 7 | 1170 | 1 | 7 | 1170 | 4 |
| 55 | Retrotransposon-hotspot protein | Tb927.7.2030 | 7 | 10 | 20 | 5 | 2 | 1 | 0.570 | 0.185 | <0.001 | 25 | 2769 | 12 | 31 | 2769 | 3* |
| 61 | Hypothetical protein | Tb927.7.5930 | 7 | 8 | 6 | 6 | 0 | 0 | 0.116 | 0.092 | <0.001 | 14 | 1755 | 15 | 14 | 2134 | 5* |
| 62 | Adenylate cyclase, GRESAG4 | Tb927.7.6080 | 7 | 5 | 4 | 3 | 0 | 0 | 0.501 | 0.062 | <0.001 | 8 | 5355 | 2 | 8 | 5355 | 12 |
| 62ai | Hypothetical protein | Tb927.7.6110 | 7 | 4 | 3 | 3 | 1 | 0 | 0.030 | 0.020 | <0.001 | 7 | 663 | 1 | 7 | 663 | 6 |
| 62aii | Hypothetical protein | Tb927.7.6120 | 7 | 3 | 2 | 1 | 0 | 1 | 0.142 | 0.166 | 0.588 | 5 | 534 | 0 | 5 | 534 | 1 |
| 70 | 1,2-alpha-mannosidase IB | Tb927.8.2900 | 8 | 5 | 5 | 4 | 0 | 0 | 0.049 | 0.005 | <0.001 | 6 | 1941 | 0 | 10 | 2020 | 7 |
| 75 | Hypothetical protein | Tb927.8.6700 | 8 | 4 | 3 | 3 | 1 | 0 | 0.038 | 0.019 | <0.001 | 7 | 2276 | 5 | 7 | 2276 | 6 |
| 80 | Amino acid transporter | Tb927.8.7600 | 8 | 10 | 5 | 4 | 2 | 1 | 0.081 | 0.033 | <0.001 | 14 | 1686 | 14 | 15 | 1686 | 5* |
| 80a | Adenylate cyclase, GRESAG4 | Tb927.8.7860 | 8 | 8 | 4 | 2 | 3 | 0 | 0.215 | 0.142 | <0.001 | 10 | 4323 | 10 | 11 | 4323 | 11* |
| x2 | Amino acid transporter | Tb927.8.4740 | 8 | 5 | 6 | 4 | 1 | 0 | 0.256 | 0.058 | <0.001 | 11 | 1704 | 1 | 11 | 1704 | 7 |
| 84 | Hypothetical protein | Tb09.160.4630 | 9 | 3 | 2 | 2 | 1 | 0 | 0.199 | 0.009 | <0.001 | 5 | 1576 | 0 | 5 | 1576 | 6 |
| 93 | Brucei alanine-rich protein, BARP | Tb09.244.2530 | 9 | 14 | 13 | 8 | 2 | 0 | 0.314 | 0.286 | <0.001 | 25 | 1534 | 12 | 27 | 1534 | 3* |
| 103 | Hypothetical protein | Tb10.70.0040 | 10 | 4 | 4 | 3 | 1 | 1 | 0.078 | 0.061 | <0.001 | 8 | 1557 | 4 | 8 | 1557 | 6 |
| 105 | Hexose transporter | Tb10.6k15.2040 | 10 | 3 | 4 | 2 | 1 | 1 | 0.026 | 0.016 | <0.001 | 7 | 1862 | 4 | 7 | 2243 | 7 |
| 106 | Procyclin-associated gene | Tb11.01.6210 | 10 | 4 | 2 | 2 | 2 | 0 | 0.092 | 0.047 | <0.001 | 6 | 1185 | 0 | 6 | 1434 | 4 |
| 111 | Hypothetical protein | Tb10.61.1420 | 10 | 3 | 3 | 3 | 0 | 0 | 0.143 | 0.100 | <0.001 | 6 | 1102 | 0 | 6 | 1102 | 5 |
| 112 | DNA polymerase kappa | Tb11.01.0080 | 11 | 10 | 7 | 4 | 0 | 0 | 0.332 | 0.167 | <0.001 | 16 | 2205 | 9 | 16 | 2205 | 10* |
| 113 | Cation transporter | Tb11.01.0725 | 11 | 4 | 4 | 4 | 0 | 0 | 0.005 | 0.003 | <0.001 | 10 | 1212 | 0 | 10 | 2458 | 2 |
| 126 | Major surface glycoprotein, gp63 | Tb11.02.5640 | 11 | 4 | 2 | 2 | 2 | 0 | 0.305 | 0.396 | 1 | 4 | 1864 | 6 | 6 | 1864 | 6 |
NB: Alignments of tandem gene duplicates without evidence of recombination under the PHI test are shown in italics.
*Due to data complexity, GARD analyses timed out before convergence had been achieved, returning a minimum number of breakpoint.
Figure 3. Analysis of sequence variation among T. brucei 65 kDa invariant surface glycoprotein genes.
Gene copies are numbered consecutively in positional order from left to right on the chromosome, beginning with Tbb1 (Tb927.2.3270) and Tbg1 (Tbg972.2.1130) respectively. a. Sequence variation (expressed as Shannon entropy score, left scale) along a multiple sequence alignment, combined with recombination breakpoints (red lines, right scale) inferred by GARD analysis. A dotted blue line marks the boundary between coding and 3′ UTR regions. Coloured bars above the chart indicate recombination tracts as identified by GARD. b. A phylogenetic network including sequences unique to one subspecies (marked with an asterisk *). Annotated pseudogenes are indicated by ψ. c. A chart showing the affinities of sequence Tbg7 only; all sequences are represented in a circle, coloured bars connect Tbg7 (or regions thereof) to its closest relative among other sequences. Different colours are used to denote different affinities. d. Affinity chart for Tbg1.
Subtelomeres are compositionally similar
Some of the hardest genome regions to reliably assemble are subtelomeres, since they usually contain numerous high-copy gene families, as well as simple and complex sequence repeats. The fluidity of subtelomeric assemblies perhaps reflects some reality about the true mutability of subtelomeric regions, since they are known to vary widely in length between trypanosome strains [41]. In comparing ∼1.3 Mbp of subtelomeric sequence immediately contiguous to the chromosomal cores between the two subspecies, it is clear that they are highly similar in composition and gene order. In both T. b. brucei and T. b. gambiense the largest component of subtelomeric genes comprises VSGs (67.8% and 44.4%, respectively), followed by ESAGs (13.4%, 15.8%), and transposable element-related genes (7.6%, 13.5%). Adenylate cyclases (2.2%, 3.0%) and glycosyltransferases (1.1%, 1.5%) are also prominent features in both genomes. Beyond these subtelomeric regions, previous comparisons of telomeric VSG expression sites in various T. brucei strains and subspecies have established that the essential components are ubiquitous [42]–[43]. Hence, although T. brucei telomeres are known to evolve rapidly and display widespread karyotypic variation, the composition of regions beyond core chromosomes remains consistent across the species.
VSG sequence types are conserved between T. brucei subspecies
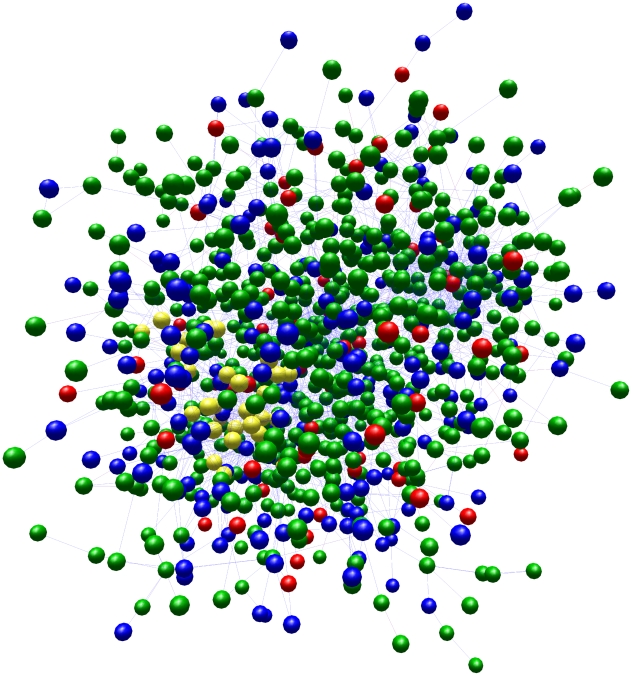
As the relative divergence (Fig. 1) and antigenic variability of different T. brucei strains is of diagnostic and clinical importance, we investigated the diversity between the VSG repertoires in the two subspecies by comparing all of the 1258 VSG sequences annotated to date in the T. b. brucei 927 genome with the unassembled sequence reads from the T. b. gambiense genome. Hence, it should be noted that we are comparing whole genes from T. b. brucei with fragments from T. b. gambiense. Among VSG genes with reciprocal top hits in the T. b. gambiense read library, the average amino acid identity is 43.3%, but with substantial variation (SD = 21.35, n = 692). Clearly, the substitution rate affecting VSG nucleotide sequences is relatively high, due either to positive selection, or a relaxation of purifying selection. Yet VSGs do not evolve so quickly as to abolish detectable orthology between subspecies; 692 VSG genes had a reciprocal top BLAST hit with a T. b. gambiense sequence read, indicating that 55% of the T. b. brucei repertoire (or parts thereof) were conserved in T. b. gambiense. Furthermore, 1061 VSG genes (84%) had reciprocal BLAST hits or very close matches, (i.e., within the top three BLAST hits for the matching T. b. gambiense read). The network representation emphasizes the global perspective of the VSG repertoire in T. b. brucei 927 relative to T. b. gambiense (Fig. 4). 197 VSG without close matches to T. b. gambiense reads are distributed throughout the network, indicating that they do not share a common origin and represent losses in T. b. gambiense.
Figure 4. Variant surface glycoprotein (VSG) repertoire of T. b. brucei (927) represented as a three-dimensional network graph.
1258 T. b. brucei VSG protein sequences were compared using pairwise BLAST searches. BLAST scores were used to arrange VSG into a graph using BioLayout Express 3D 3.0. 968 individual VSG are represented as coloured spheres and are joined by edges to all other nodes with which they share >70% amino acid identity. The program minimizes the distance required to arrange all nodes such that related nodes are arranged closest to one another. Nodes are shaded by type: orthologous sequence in T. b. gambiense (blue), orthologous sequence in T. b. gambiense, but closest relative in T. b. brucei (green), no corresponding sequence in T. b. gambiense (red), metacyclic-stage VSG (purple) and VSG-related (VR) proteins (yellow).
As the T. b. brucei 927 subtelomeric sequences are incomplete, its VSG set is partial, and therefore further T. b. brucei-specific VSG sequences may be identified in future. That said, our analysis consistently demonstrated that VSG genes have corresponding sequences in both subspecies, though 787 (63%) were better related to other T. b. brucei genes than any T. b. gambiense read, suggesting that a gene duplication or gene conversion event had occurred since separation of the subspecies. We sought to identify phylogenetic structure, or discernable subsets, among VSG to establish limitations on gene conversion. The structural distinction between canonical VSG and VR proteins is already established [17] and is consistent with the location of VRs outside of the subtelomeres and their lack of pseudogenes. Accordingly, the VRs cluster together (yellow shading) in the network. They contrast with the otherwise diffuse arrangement of canonical VSG; sequences do not cluster by chromosomal location or by developmental stage expression since metacyclic-VSG are distributed throughout. VSG specific to T. b. brucei (red shading) do not belong to a single lineage, and equally, there is no evidence for evolutionary expansion of a particular subset. Instead, these data indicate that gene conversion has occurred frequently among subtelomeric VSG in the recent past, unlimited by genomic location and resulting in occasional gene loss.
The complement of variant surface glycoproteins in T. b. gambiense has previously been reported as being smaller, or more restricted, than that of T. b. brucei, based on a smaller overall genome size and reduced subtelomeric components [8]. This could indicate fewer discrete VSG sequence types or just fewer copies of an equal number of types. The data presented here suggest that if T. b. gambiense had a smaller VSG repertoire, this is likely to reflect quantity rather than sequence types. Hutchinson et al. [44] reported that, while protein sequences had diverged consistent with subspecies-specific adaptation, 14 expressed VSG genes in the T. b. brucei Tororo strain all had close homologs in both T. b. brucei 927 and T. b. gambiense. Our data support the idea that the VSG repertoire is relatively stable across T. brucei subspecies, but that VSG genes have an inherently high substitution rate resulting in rapid sequence divergence relative to other genes. Corresponding VSG sequence types are thus likely to be found in any T. brucei strain, making it realistic to catalogue all VSG types and to monitor their expression in the field. Thus, the apparent lack of genetic hypervaribility concerning VSG in T. brucei seems simpler than other systems of antigenic variation, such as the var surface glycoproteins in Plasmodium falciparum, where frequent switching of expressed antigens is combined with genetic hypervariability and there is minimal overlap between repertoires between isolates [45]–[47]. Like P. falciparum, T. brucei ‘shuffles its deck’ with every infection, but, unlike Plasmodium, it always uses the same pack of cards.
Conclusion
T. b. gambiense is the most important human-infective form of T. brucei and currently endemic throughout central Africa. In producing a draft genome sequence for T. b. gambiense this study attempted to identify genetic causes for human infectivity in T. b. gambiense, as well as assess the scale of intraspecific genomic variation. Genomic conservation between T. b. gambiense and the T. b. brucei validates the use of T. brucei brucei as a model for studying the unculturable T. b. gambiense. Specifically, intraspecific genomic divergence is typically <1% in coding regions; gene gain and loss is associated with rare segmental duplications; indels are few and generally caused by transposable elements or VSG/ESAG ‘islands’; and 84% of surface antigens are represented whole or in part in both subspecies. The VSG repertoire is essentially conserved at the level of modular protein domains, which are reorganized by gene conversion into novel mosaics in each strain. Therefore our data are likely to anticipate the archive present in the genomes of other strains, and a definition of total VSG diversity should be achievable through the addition of further sequences in the near future.
Comparative genomics has identified species-specific genes in other eukaryotic pathogens that display interspecific pathological variations, including Leishmania spp. [48]; Candida albicans and C. dubliniensis [49]; and Plasmodium knowlesi and P. vivax [50]. In applying a similar rationale here, we found no obvious candidate for a gene analogous to SRA that could account for human-infectivity in T. b. gambiense. However, since both SRA and TgsGP are homologous to VSG genes and subtelomeric, such a gene might not be apparent from comparison of the core chromosomes and could still exist within the subtelomeres of T. b. gambiense. Alternatively, rather than differences in gene content per se, phenotypic variability could be due to individual SNPs or indels, or to differences in gene expression. Given that innate immunity to trypanosomes in Humans is based on the uptake of high-density lipoprotein particles, which contain apoL1 and stimulate trypanolysis [20]–[21], perhaps the likeliest cause of phenotypic variation relates to this process. Indeed, it is possible to select for resistance to trypanolysis by down-regulating TbHpHbR [51], which encodes an haptoglobin-haemoglobin surface receptor that normally scavenges haem from the host, but also facilitates the uptake of trypanolytic particles [52]. However, this gene is present in T. b. gambiense (Tbg972.6.120) and differs from its T. b. brucei counterpart (Tb927.6.440) by only 5 amino acid replacements (L210S, A293V, E369G, G370A and M398I).
Hence, the basis for human infectivity in T. b. gambiense remains debatable, and we must now consider that features shared by both subspecies have been modified in structure or expression in T. b. gambiense to provide the genotypic basis for resistance to trypanolysis. This issue apart, several putative cell-surface protein families that include subspecies-specific members have been identified in T. b. brucei. These proteins are previously unrecognized elements of the trypanosome surface that display both recent gene duplications and accelerated evolutionary rates and we speculate that they may have acquired novel functions. We also suggest the presence or absence of such hypothetical genes varies on a population scale, and might yet contribute to phenotypic variability in host range within T. brucei.
Supporting Information
Disruption to co-linearity on chromosome 9 concerning an internal VSG ‘island’.
(0.40 MB TIF)
Tandem duplication on chromosome 3 in T. b. brucei.
(0.46 MB TIF)
Tandem duplication on chromosome 6 in T. b. brucei.
(1.22 MB TIF)
Acknowledgments
We would like to thank the core sequencing and informatics teams at the Wellcome Trust Sanger Institute for their assistance.
Footnotes
The authors have declared that no competing interests exist.
The genome sequencing and annotation work was funded by the Wellcome Trust (grant number WT085775/Z/08/Z). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Simarro PP, Jannin J, Cattand P. Eliminating human African trypanosomiasis: where do we stand and what comes next? PLoS Med. 2008;5:e55. doi: 10.1371/journal.pmed.0050055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. Control and surveillance of African trypanosomiasis. Report of a WHO Expert Committee. World Health Organ Tech Rep Ser. 1998 [PubMed] [Google Scholar]
- 3.Fèvre EM, Wissmann BV, Welburn SC, Lutumba P. The burden of human african trypanosomiasis. PLoS Negl Trop Dis. 2008;2:e333. doi: 10.1371/journal.pntd.0000333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Nieuwenhove S. Gambiense sleeping sickness: re-emerging and soon untreatable? Bull World Health Organ. 2000;78:1283. [PMC free article] [PubMed] [Google Scholar]
- 5.Priotto G, Kasparian S, Mutombo W, Ngouama D, Ghorashian S, et al. Nifurtimox-eflornithine combination therapy for second-stage African Trypanosoma brucei gambiense trypanosomiasis: a multicentre, randomised, phase III, non-inferiority trial. Lancet. 2009;374:56–64. doi: 10.1016/S0140-6736(09)61117-X. [DOI] [PubMed] [Google Scholar]
- 6.Lowell JE, Earl CD. Leveraging biotech's drug discovery expertise for neglected diseases. Nat Biotechnol. 2009;27:323–329. doi: 10.1038/nbt0409-323. [DOI] [PubMed] [Google Scholar]
- 7.Kanmogne GD, Bailey M, Gibson WC. Wide variation in DNA content among isolates of Trypanosoma brucei ssp. Acta Trop. 1997;63:75–87. doi: 10.1016/s0001-706x(96)00600-6. [DOI] [PubMed] [Google Scholar]
- 8.Dero B, Zampetti-Bosseler F, Pays E, Steinert M. The genome and the antigen gene repertoire of Trypanosoma brucei gambiense are smaller than those of T. b. brucei. Mol Biochem Parasitol. 1987;26:247–256. doi: 10.1016/0166-6851(87)90077-6. [DOI] [PubMed] [Google Scholar]
- 9.Gibson WC. Will the real Trypanosoma b. gambiense please stand up. Parasitol Today. 1986;2:255–257. doi: 10.1016/0169-4758(86)90011-6. [DOI] [PubMed] [Google Scholar]
- 10.Pays E, Dekerck P, Van Assel S, Babiker EA, Le Ray D, et al. Comparative analysis of a Trypanosoma brucei gambiense antigen gene family and its potential use in epidemiology of sleeping sickness. Mol Biochem Parasitol. 1983;7:63–74. doi: 10.1016/0166-6851(83)90117-2. [DOI] [PubMed] [Google Scholar]
- 11.Paindavoine P, Zampetti-Bosseler F, Pays E, Schweizer J, Guyaux M, et al. Trypanosome hybrids generated in tsetse flies by nuclear fusion. EMBO J. 1986;5:3631–3636. doi: 10.1002/j.1460-2075.1986.tb04692.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gray AR. Variable agglutinogenic antigens of Trypanosoma gambiense and their distribution among isolates of the trypanosome collected in different places in Nigeria. Trans R Soc Trop Med Hyg. 1972;66:263–284. doi: 10.1016/0035-9203(72)90158-7. [DOI] [PubMed] [Google Scholar]
- 13.Pays E, Salmon D, Morrison LJ, Marcello L, Barry JD. Antigenic variation in Trypanosoma brucei, In: Barry JD, McCulloch R, Mottram J, Acosta-Serrano A, editors. Trypanosomes after the genome. Horizon Bioscience; 2007. pp. 339–372. [Google Scholar]
- 14.Borst P, Ulbert S. Control of VSG gene expression sites. Mol Biochem Parasitol. 2001;114:17–27. doi: 10.1016/s0166-6851(01)00243-2. [DOI] [PubMed] [Google Scholar]
- 15.Horn D. The molecular control of antigenic variation in Trypanosoma brucei. Curr Mol Med. 2004;4:563–576. doi: 10.2174/1566524043360078. [DOI] [PubMed] [Google Scholar]
- 16.Taylor JE, Rudenko G. Switching trypanosome coats: what's in the wardrobe? Trends Genet. 2006;22:614–620. doi: 10.1016/j.tig.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 17.Marcello L, Barry JD. Analysis of the VSG gene silent archive in Trypanosoma brucei reveals that mosaic gene expression is prominent in antigenic variation and is favored by archive substructure. Genome Res. 2007;17:1344–1352. doi: 10.1101/gr.6421207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Greef C, Hamers R. The serum resistance-associated (SRA) gene of Trypanosoma brucei rhodesiense encodes a variant surface glycoprotein-like protein. Mol Biochem Parasitol. 1994;68:277–284. doi: 10.1016/0166-6851(94)90172-4. [DOI] [PubMed] [Google Scholar]
- 19.De Greef C, Chimfwembe E, Kihang'a Wabacha J, Bajyana Songa E, Hamers R. Only the serum-resistant bloodstream forms of Trypanosoma brucei rhodesiense express the serum resistance associated (SRA) protein. Ann Soc Belg Med Trop. 1992;72(Suppl 1):13–21. [PubMed] [Google Scholar]
- 20.Pays E, Vanhollebeke B. Mutual self-defence: the trypanolytic factor story. Microbes Infect. 2008;10:985–989. doi: 10.1016/j.micinf.2008.07.020. [DOI] [PubMed] [Google Scholar]
- 21.Pérez-Morga D, Vanhollebeke B, Paturiaux-Hanocq F, Nolan DP, Lins L, et al. Apolipoprotein L-I promotes trypanosome lysis by forming pores in lysosomal membranes. Science. 2005;309:469–472. doi: 10.1126/science.1114566. [DOI] [PubMed] [Google Scholar]
- 22.Xong HV, Vanhamme L, Chamekh M, Chimfwembe CE, Van Den Abbeele J, et al. A VSG expression site-associated gene confers resistance to human serum in Trypanosoma rhodesiense. Cell. 1998;95:839–846. doi: 10.1016/s0092-8674(00)81706-7. [DOI] [PubMed] [Google Scholar]
- 23.Radwanska M, Chamekh M, Vanhamme L, Claes F, Magez S, et al. The serum resistance-associated gene as a diagnostic tool for the detection of Trypanosoma brucei rhodesiense. Am J Trop Med Hyg. 2002;67:684–690. doi: 10.4269/ajtmh.2002.67.684. [DOI] [PubMed] [Google Scholar]
- 24.Berberof M, Pérez-Morga D, Pays E. A receptor-like flagellar pocket glycoprotein specific to Trypanosoma brucei gambiense. Mol Biochem Parasitol. 2001;113:127–138. doi: 10.1016/s0166-6851(01)00208-0. [DOI] [PubMed] [Google Scholar]
- 25.Felu C, Pasture J, Pays E, Pérez-Morga D. Diagnostic potential of a conserved genomic rearrangement in the Trypanosoma brucei gambiense-specific TGSGP locus. Am J Trop Med Hyg. 2007;76:922–929. [PubMed] [Google Scholar]
- 26.Dukes P, Kaukus A, Hudson KM, Asonganyi T, Gashumba JK. A new method for isolating Trypanosoma brucei gambiense from sleeping sickness patients. Trans R Soc Trop Med Hyg. 1989;83:636–639. doi: 10.1016/0035-9203(89)90379-9. [DOI] [PubMed] [Google Scholar]
- 27.Berriman M, Ghedin E, Hertz-Fowler C, Blandin G, Renauld H, et al. The genome of the African trypanosome Trypanosoma brucei. Science. 2006;309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- 28.Carver T, Berriman M, Tivey A, Patel C, Böhme U, et al. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics. 2008;24:2672–2676. doi: 10.1093/bioinformatics/btn529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aslett M, Aurrecoechea C, Berriman M, Brestelli J, Brunk BP, et al. TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic Acids Res. 2010;2010 38 (Database issue):D457–462. doi: 10.1093/nar/gkp851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Altshuler D, Pollara VJ, Cowles CR, Van Etten WJ, Baldwin J, et al. An SNP map of the human genome generated by reduced representation shotgun sequencing. Nature. 2000;407:513–516. doi: 10.1038/35035083. [DOI] [PubMed] [Google Scholar]
- 31.Jackson AP. Tandem gene arrays in Trypanosoma brucei: comparative phylogenomic analysis of duplicate sequence variation. BMC Evol Biol. 2007;7:54. doi: 10.1186/1471-2148-7-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benz C, Nilsson D, Andersson B, Clayton C, Guilbride DL. Messenger RNA processing sites in Trypanosoma brucei. Mol Biochem Parasitol. 2005;143:125–134. doi: 10.1016/j.molbiopara.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 33.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 34.Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 35.Bryant D, Moulton V. Neighbor-net: an agglomerative method for the construction of phylogenetic networks. Mol Biol Evol. 2004;21:255–265. doi: 10.1093/molbev/msh018. [DOI] [PubMed] [Google Scholar]
- 36.Bruen TC, Philippe H, Bryant D. A simple and robust statistical test for detecting the presence of recombination. Genetics. 2006;172:2665–2681. doi: 10.1534/genetics.105.048975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SD. GARD: a genetic algorithm for recombination detection. Bioinformatics. 2006;22:3096–3098. doi: 10.1093/bioinformatics/btl474. [DOI] [PubMed] [Google Scholar]
- 38.Goldovsky L, Cases I, Enright AJ, Ouzounis CA. BioLayout(Java): versatile network visualisation of structural and functional relationships. Appl Bioinformatics. 2005;4:71–74. doi: 10.2165/00822942-200504010-00009. [DOI] [PubMed] [Google Scholar]
- 39.Barry JD, Ginger ML, Burton P, McCulloch R. Why are parasite contingency genes often associated with telomeres? Int J Parasitol. 2003;33:29–45. doi: 10.1016/s0020-7519(02)00247-3. [DOI] [PubMed] [Google Scholar]
- 40.Scherf A, Lopez-Rubio JJ, Riviere L. Antigenic variation in Plasmodium falciparum. Annu Rev Microbiol. 2008;62:445–470. doi: 10.1146/annurev.micro.61.080706.093134. [DOI] [PubMed] [Google Scholar]
- 41.Callejas S, Leech V, Reitter C, Melville S. Hemizygous subtelomeres of an African trypanosome chromosome may account for over 75% of chromosome length. Genome Res. 2006;16:1109–1118. doi: 10.1101/gr.5147406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hertz-Fowler C, Figueiredo LM, Quail MA, Becker M, Jackson A, et al. Telomeric expression sites are highly conserved in Trypanosoma brucei. PLoS One. 2008;3:e3527. doi: 10.1371/journal.pone.0003527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Young R, Taylor JE, Kurioka A, Becker M, Louis EJ, et al. Isolation and analysis of the genetic diversity of repertoires of VSG expression site containing telomeres from Trypanosoma brucei gambiense, T. b. brucei and T. equiperdum. BMC Genomics. 2008;9:385. doi: 10.1186/1471-2164-9-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hutchinson OC, Picozzi K, Jones NG, Mott H, Sharma R, et al. Variant Surface Glycoprotein gene repertoires in Trypanosoma brucei have diverged to become strain-specific. BMC Genomics. 2007;8:234. doi: 10.1186/1471-2164-8-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Freitas-Junior LH, Bottius E, Pirrit LA, Deitsch KW, Scheidig C, et al. Frequent ectopic recombination of virulence factor genes in telomeric chromosome clusters of P. falciparum. Nature. 2000;407:1018–1022. doi: 10.1038/35039531. [DOI] [PubMed] [Google Scholar]
- 46.Tami A, Ord R, Targett GA, Sutherland CJ. Sympatric Plasmodium falciparum isolates from Venezuela have structured var gene repertoires. Malar J. 2003;2:7. doi: 10.1186/1475-2875-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bull PC, Buckee CO, Kyes S, Kortok MM, Thathy V, et al. Plasmodium falciparum antigenic variation. Mapping mosaic var gene sequences onto a network of shared, highly polymorphic sequence blocks. Mol Microbiol. 2008;68:1519–1534. doi: 10.1111/j.1365-2958.2008.06248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peacock CS, Seeger K, Harris D, Murphy L, Ruiz JC, et al. Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat Genet. 2007;39:839–847. doi: 10.1038/ng2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jackson AP, Gamble JA, Yeomans T, Moran GP, Saunders D, et al. Comparative genomics of the fungal pathogens Candida dubliniensis and C. albicans. Genome Res Published in Advance. 2009 doi: 10.1101/gr.097501.109. Sep 10, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pain A, Böhme U, Berry AE, Mungall K, Finn RD, et al. The genome of the simian and human malaria parasite Plasmodium knowlesi. Nature. 2008;455:799–803. doi: 10.1038/nature07306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Faulkner SD, Oli MW, Kieft R, Cotlin L, Widener J, et al. In vitro generation of human high-density-lipoprotein-resistant Trypanosoma brucei brucei. Eukaryot Cell. 2006;5:1276–1286. doi: 10.1128/EC.00116-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vanhollebeke B, De Muylder G, Nielsen MJ, Pays A, Tebabi P, et al. A haptoglobin-hemoglobin receptor conveys innate immunity to Trypanosoma brucei in humans. Science. 2008;320:677–681. doi: 10.1126/science.1156296. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Disruption to co-linearity on chromosome 9 concerning an internal VSG ‘island’.
(0.40 MB TIF)
Tandem duplication on chromosome 3 in T. b. brucei.
(0.46 MB TIF)
Tandem duplication on chromosome 6 in T. b. brucei.
(1.22 MB TIF)