Abstract
Cell-cell fusion is a crucial and highly regulated event in the genesis of both form and function of many tissues. One particular type of cell fusion, myoblast fusion, is a key cellular process that shapes both the formation and repair of muscle. Despite its importance for human health, the mechanisms underlying this process are still not well understood. The purpose of this review is to highlight the recent literature pertaining to myoblast fusion and to focus on a comparison of these studies across several model systems, particularly the fly, zebrafish and mouse. Advances in technical analysis and imaging have allowed identification of new fusion genes and propelled further characterization of previously identified genes in each of these systems. Among the cellular steps identified as critical for myoblast fusion is migration, recognition, adhesion, membrane alignment and membrane pore formation and resolution. Importantly, striking new evidence indicates that orthologous genes govern several of these steps across these species. Taken together, comparisons across three model systems are illuminating a once elusive process, providing exciting new insights and a useful framework of genes and mechanisms.
Cell-cell fusion is an essential and highly coordinated event that occurs in numerous contexts during the development of diverse organisms. This powerful morphogenetic process forms and shapes developing tissues and organs as well as promotes tissue homeostasis. While a number of cell types undergo fusion during the lifetime of an organism, the majority of cells in the body remain mononucleate, suggesting that fusion is tightly regulated and must be properly restricted to a subset of cell types. Accordingly, aberrant cell fusion has recently been shown to have a role in carcinogenesis and tumor progression (Chen et al., 2007; Duelli and Lazebnik, 2007; Oren-Suissa and Podbilewicz, 2007; Podbilewicz, 2006).
Furthermore, cell-cell fusion serves as a mechanism to allow developing cells and tissues to adopt properties or perform functions not possible by their mononucleate predecessors. During fertilization, for example, the fusion of sperm and egg, two haploid cells, is required to create a single diploid cell, or zygote. During human placental development, trophoblast fusion is required for the implantation and maintenance of the developing embryo. Additionally, bone and muscle development and repair are dependent upon fusion; for the former, fusion of macrophages occurs to form osteoclasts, which have the ability to resorb calcified tissue, whereas for the latter, individual myoblasts fuse to form large syncytia capable of producing various muscle groups required for force generation. Recent data has also shown that stem cells can undergo fusion leading to genetic reprogramming of somatic cells (Chen et al., 2007; Chen and Olson, 2005; Oren-Suissa and Podbilewicz, 2007; Podbilewicz, 2006; Sapir et al., 2008). The diversity of tissues in which fusion occurs underscores the importance of this process to proper development, yet the underlying cellular mechanisms and subcellular behaviors underlying cell-cell fusion remain poorly understood.
Observations made in several cell-cell fusion systems, namely muscle, the hypodermis of C. elegans and sperm-egg, suggest a common set of cellular behaviors underlie the fusion process. These begin with the recognition and adhesion of the two cells that will fuse. Once the cells adhere, the membranes of the two cells become aligned, bringing the lipid bilayers in close proximity. A proposed fusogen, a membrane fusion effector protein, is trafficked to the site of fusion, leading to pore formation between the fusing cells. Whether one or several pores form and expand appears to depend on the system analyzed (Doberstein et al., 1997; Mohler et al., 1998). Nevertheless, the data suggests that membrane vesiculation is a possible intermediate in pore expansion, recycling these membranes to other areas of the cell. Once cytoplasmic continuity is achieved, the cell contents, including nuclei, are mingled in the resulting single cell (Chen, 2008). While we refer the reader to several recent reviews on fusion in general (Chen et al., 2007; Duelli and Lazebnik, 2007; Oren-Suissa and Podbilewicz, 2007; Podbilewicz, 2006; Sapir et al., 2008; Shemer and Podbilewicz, 2003), our goal in this review is to discuss cell-cell fusion within the context of skeletal muscle development.
From the head to the feet, a variety of skeletal muscles control movement in organisms of all sizes. Skeletal muscles generally consist of bundles of multinucleated myofibers and can be distinguished by morphological properties, such as size, shape, orientation, innervation and attachment sites. These unique properties allow for considerable muscle diversity generating muscles to produce a variety of movements and functions. Although data are emerging on how specific muscle shape and orientation are achieved, significant progress has been made in understanding the generation of muscle fiber size, which relies on the earlier iterative fusion of differentiated myoblasts common to all skeletal muscle. Knowledge of this process has particular relevance to the treatment of wasting of specific muscle groups due to diseases, such as muscular dystrophy and cancer cachexia, or to aging or atrophy.
Currently, three model organisms are employed to study myoblast fusion: the fruit fly Drosophila melanogaster, the zebrafish, Danio rerio and the mouse, Mus musculus (Figure 1). Because much of our knowledge to date has come from studies in Drosophila, we begin by briefly describing the salient features of the development of the larval body wall muscles to provide a context for myoblast fusion studies. Following this introduction, we discuss the data on recently identified muscle fusion genes in light of the conserved cellular behaviors required for fusion—migration, adhesion, actin regulation at the site of adhesion/fusion, vesicle trafficking at this site, membrane breakdown and reset of the fusion machinery for the next fusion (Figure 2). We then turn to the zebrafish and mouse, briefly describing muscle development in these systems and comparing what is known about the cell biology and molecular control of fusion in these models. Hence, in doing a cross-species comparison, we aim to gain a clearer understanding of the overall data pertaining to myoblast fusion and the molecular mechanisms and intracellular events that drive this process.
Figure 1. Myofibers in a variety of model systems form from the fusion of mononucleated muscle precursors.
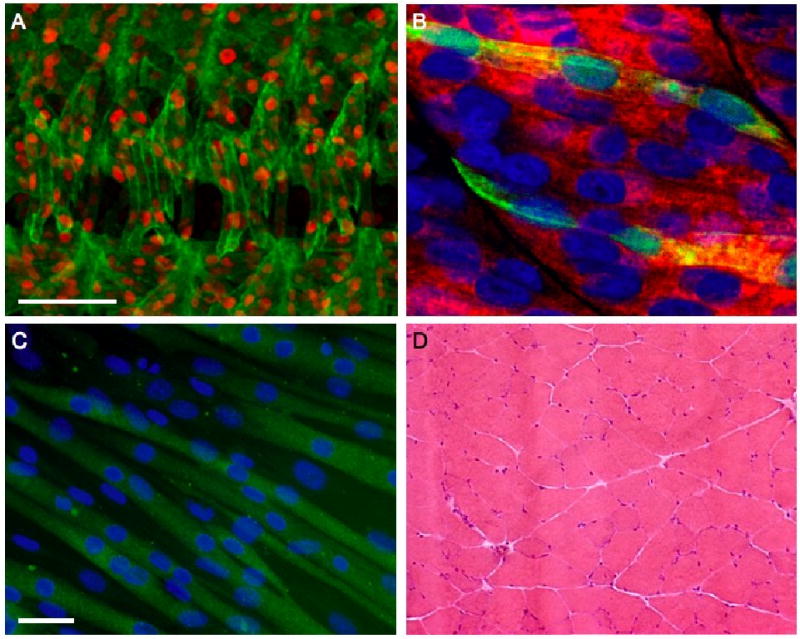
(A-D) Nuclei were visualized using a nuclear DsRed transgene (red) (A), DAPI (blue) (B, C) or hematoxylin (purple) (D). (A) Four hemisegments of a Drosophila embryo were analyzed by immunohistochemistry using antibodies against tropomyosin (green). (B) The syncytial fast-twitch muscle fibers of a wild-type zebrafish embryo were labeled with antibodies against fast myosin light chain (red). Two fast muscle fibers are highlighted by GFP expression (green) from a skeletal muscle actin∷gfp transgene. (C) C2C12 myoblasts, a satellite cell-derived mouse myoblast cultured cell line, were analyzed by immunohistochemistry using antibodies against myosin heavy chain (green). The scale bar represents 40 μm. (D) Cross sections of a major leg muscle, the gastrocnemius, of an adult mouse stained with hematoxylin and eosin. Note that the nuclei are peripherally located and, unlike in other panels, the multinucleate nature of the myofiber is not clearly evident from this section.
Figure 2. Cellular and subcellular behaviors that occur during myoblast fusion.
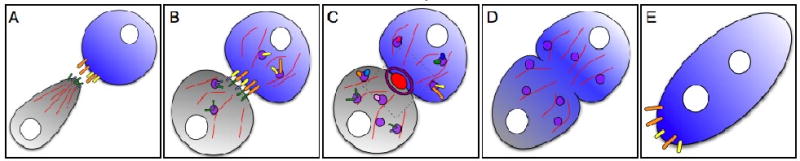
Several cellular steps occur during myoblast fusion; each step coincides with a defined series of subcellular events. (A) Cells first migrate toward their fusing partner, concurrently with actin polymerization (red lines) and expression of transmembrane attractants (small rectangles) to guide the migrating cell. (B) The cells then touch and adhere, leading to localization of the cell type-specific transmembrane proteins. (C) This leads to an accumulation of actin (red oval), and the formation of the FuRMAS (purple ring) at the site of fusion. Subsequently, a number of fusion proteins known collectively as the “fusion machinery” are localized to the site of fusion, presumably through vesicular trafficking. The area outlined by the grey box is examined in more detail in Figure 3. (D) This is followed by membrane breakdown and the removal of vesiculating membrane and the fusion machinery components. (E) Finally, the cell must reset for the next round of fusion by expressing appropriate levels of transmembrane attractant. This process will repeat iteratively until the final muscle or fiber size is achieved.
Drosophila myogenesis
In the Drosophila embryo, a repeated pattern of 30 distinct somatic muscle fibers is present in each abdominal hemisegment (Bate, 1990) (Figure 1A). Despite their similarities, such as shared expression of contractile proteins and neurotransmitter receptors, each muscle fiber can be distinguished by its size, shape, orientation, number of nuclei, innervation and tendon attachment sites (Baylies et al., 1998; Frasch, 1999). The acquisition of these muscle specific properties during myogenesis depends upon the prior specification of two classes of myoblasts: founder cells (FCs) and fusion competent myoblasts (FCMs) from each mesodermal hemisegment. Each muscle is prefigured by a single FC, which seeds muscle formation by fusing with surrounding FCMs.
FCs are a diverse population of myoblasts. Each FC has a unique identity that is characterized by the expression of transcription factors, such as Krüppel, Slouch, Even-skipped, Apterous and Nautilus (Baylies et al., 1998; Beckett, 2006; Carmena, 2006; Frasch, 1999). The specific combination of transcriptional regulators expressed in each FC determines the unique morphology of the final muscle. This is evidenced in embryos in which genes involved in fusion have been mutated: FCs express differentiation markers such as myosin heavy chain (MHC) and extend towards their attachment sites but fail to fuse, resulting in mononucleate FCs that have attempted to adopt their normal muscle morphology. In contrast to FCs, FCMs are considered a more uniform and naïve population of myoblasts, which express the transcription factor Lameduck (Lmd) (Duan et al., 2001; Furlong et al., 2001; Ruiz-Gomez et al., 2002). Upon fusion with a FC or growing myotube, the nuclei of the newly incorporated FCMs lose expression of Lmd and express the transcriptional regulators of the FC to which they have fused (Baylies et al., 1998; Beckett and Baylies, 2006; Knirr et al., 1999).
Recent evidence, however, suggests that FCMs have considerably more diversity than was first appreciated, which could impact the fusion process (Beckett and Baylies, 2007; Estrada et al., 2006; Richardson et al., 2008a). For example, three-dimensional analysis of the Drosophila somatic mesoderm reveals that the particular organization of FCs and FCMs during the developmental stages when fusion is occurring can impact the fusion process (Beckett and Baylies, 2007). FCs are located in the most external layers of the somatic mesoderm. FCMs comprise several cell layers; the most external FCMs contact FCs or epithelial tissue and the more internal FCMs contact primarily other FCMs (Beckett and Baylies, 2007). During the initial stages of fusion, bi-nucleate muscle precursors are initially detected externally without any appreciable alteration of the FCM arrangement (Beckett and Baylies, 2007). This suggests that FCs fuse with the nearest FCMs, those located in the most external cell layers of the somatic mesoderm; internal FCMs, which do not directly contact FCs, likely fuse later. As fusion proceeds, the more internally located FCMs adopt a migratory morphology, presumably in order to move through the somatic mesoderm to locate and fuse with FCs in the external layers (Beckett and Baylies, 2007). Thus, the position of an FCM within the hemisegment may impact its behavior. Interestingly, fusion also occurs without the expected concomitant reduction in FCMs by stage 14 (Beckett and Baylies, 2007), suggesting that FCMs, which were previously presumed to be post-mitotic, may be undergoing cell division. Indeed, a small subpopulation of FCMs were shown to undergo division during stages 12-13 (Beckett and Baylies, 2007). Molecular diversity among FCMs is supported by data, which demonstrated that Hbs, an FCM-specific gene, is expressed in a subset of FCMs during fusion (Artero et al., 2001). Validated genetic and genomic meta-analysis has also indicated molecular diversity among FCMs. In this case, the expression of FCM-specific genes, such as Sns and D-WIP, were shown to differ among FCM subpopulations at specific timepoints (Estrada et al., 2006). This new understanding of FCM diversity opens exciting possibilities for determining the differential genetic requirements in FCM subsets that maybe responsible for these cellular behaviors. Hence, muscle formation requires both FCs and FCMs and the identity and arrangement of these specific myoblast populations may impact the fusion process.
Myoblast fusion in the Drosophila embryo occurs over a 5.5 hour period during late embryogenesis (stages 12-15; 7.5-13 hrs after egg laying [AEL]). The size of each muscle is determined by the number of fusion events, ranging from as few as 2 to as many as 24 in an individual muscle (Bate, 1990). While there is some variation in the final size of individual muscles, a characteristic mean number of nuclei has been determined in several muscles (Beckett and Baylies, 2007; Menon et al., 2005). Additionally, FCs do not all begin to fuse at the same time; it is likely that the timing of their specification impacts the time at which they start to fuse. Likewise, while the exact time when each individual muscle stops fusing is not known, all fusion is completed by the end of stage 15 (Beckett and Baylies, 2007). Analysis of the fusion profile of individual muscles has shown that fusion occurs in two temporal phases. During the first 3 hours of fusion (stages 12-13), 9-27% of fusion events occur, while the remaining 73-91% of fusion events occur in the final 2.5 hours of the process (stages 14-15). Further work indicates that the frequency with which fusion events occur increases during these later stages of development (Beckett and Baylies, 2007; Richardson et al., 2008a).
There is also a second period of myogenesis during pupal development in which muscle fibers are assembled to facilitate the more complex behaviors of the adult fly (Roy and VijayRaghavan, 1999). During this time, adult muscle precursors, which have been set aside in the embryo, will divide to produce pools of myoblasts that will then fuse to existing larval muscles fibers as well as forming de novo fibers. Although this provides another system to assay myoblast fusion, this review will focus exclusively on the fusion events during embryonic myogenesis, which has been the best characterized Drosophila model.
In addition to its well-studied specification process, defined myoblast populations and distinct cellular organization, Drosophila affords a number of technical advantages for studying myoblast fusion. In the fly, advances in imaging, such as TEM and time-lapse microscopy (Doberstein et al., 1997; Richardson et al., 2007), have enhanced our ability to define the steps of fusion. Furthermore, the ease of genetic screens and mutational analysis has uncovered a large number of genes (Table 1, Figure 3) required for proper fusion to occur. We discuss genes that have been recently identified in light of the distinct cell behaviors required for fusion (Figure 2).
Table 1. A partial list of gene products shown to play a role in myoblast fusion.
| Drosophila Protein | Vertebrate Protein | Proposed Function in Drosophila and Cellular/Subcellular Behavior | Models testeda | Actin focus classb | References |
|---|---|---|---|---|---|
| Sns | Nephrin | Myoblast adhesion; Figure 2B | d, m, zc | 1 | Kestila et al.,, 1998; Bour et al.,, 2000; Sohn et al.,, 2009 |
| Hbs | Nephrin | Myoblast adhesion; Figure 2B | d, m, zc | Unknown | Kestila et al.,, 1998; Artero et al.,, 2001; Dworak et al.,, 2001 Sohn et al.,, 2009 |
| Duf (Kirre) | Kirrel family | Myoblast adhesion/attraction; Figure 2A, B | d, z | 1 | Ruiz-Gomez et al., 2000; Strunkelnberg et al., 2001; Srinivas et al., 2007 |
| Rst (Irre) | Kirrel family | Myoblast adhesion/attraction; Figure 2A, B | d | 1 | Strunkelnberg et al., 2001 |
| Rols (Ants) | Tanc1 | Adaptor, signaling; recycling | d | 1 | Menon and Chia 2001;Chen and Olson 2001; Rau et al., 2001; Menon et al., 2005 |
| Rac1, Rac2, Mtl | Rac1, Rac2, Rac3, RhoG | Cytoskeleton, SCAR/WAVE complex regulation; Figure 2C | dd ze, me | 3 | Luo et al., 1994;Hakeda-Suzuki et al., 2002; Chen et al., 2003; Srinivas et al., 2007; Vasyutina et al., 2009 |
| Kette (Hem) | Nap1 | Cytoskeleton, SCAR/WAVE complex regulation; Figure 2C | d, m | 3 | Richardson et al., 2007 |
| SCAR | WAVE | Cytoskeleton, Arp2/3 activation; Figure 2C | d | 3 | Richardson et al., 2007 |
| WASp | WASp | Cytoskeleton, Arp2/3 activation; Figure 2C | d | 2 | Schafer et al., 2007; Unpublished observations |
| Arp2/3 | Arp2/3 | Cytoskeleton, Actin polymerization; Figure 2C | d | Unknown | Richardson et al., 2007 |
| Mbc | Dock1/Dock5 | Cytoskeleton, Rac activation; Figure 2C | d, z, m | 3 | Rushton et al., 1995; Erickson et al., 1997; Moore et al., 2007; Pajcini et al., 2008; Laurin et al., 2008 Galletta et al., 1999; |
| Crk | Crk/Crk-like | Adaptor, signaling Figure 2C | df, z | Unknown | Balagopalan et al., 2006; Kim et al., 2007; Moore et al., 2007 |
| Blow | No known homolog | Unknown | d | 3 | Doberstein et al., 1997; Schroter et al., 2004 |
| Sltr/D-WIP (Vrp1) | WIP | Cytoskeleton, WASP regulation; Figure 2C | d | 2 | Massarwa et al., 2007; Kim et al., 2007 |
| Loner (Siz) | IQSec1/Brag2/GEP100 | Cytoskeleton, Arf6/Rac regulation; Figure 2C | d, m | 2 | Chen et al., 2003; Richardson et al., 2007 |
| Sing | (Caveolin-3)g | Vesicle fusion; Figure 2C | d, m | Unknown | Galbiati et al., 1999; Volonte et al., 2003; Estrada et al., 2007; |
d: Drosophila; z: zebrafish; m: mouse;
Actin foci classes from Richardson et al., (2007): 1, no/reduced number of foci; 2, increased number of normal-sized foci; 3, increased number of enlarged foci;
mutation results in a muscle defect, not fusion defect;
mutation of Rac1, Rac2 required for fusion defect;
mutation of Rac1 is required for fusion defect;
inferred;
Caveolin-3 is speculated to be the functional ortholog of singles bar (Estrada et al., 2007: Peckham 2008); however, this has not been functionally tested.
Figure 3. Current working model of the genes required for myoblast fusion in Drosophila.
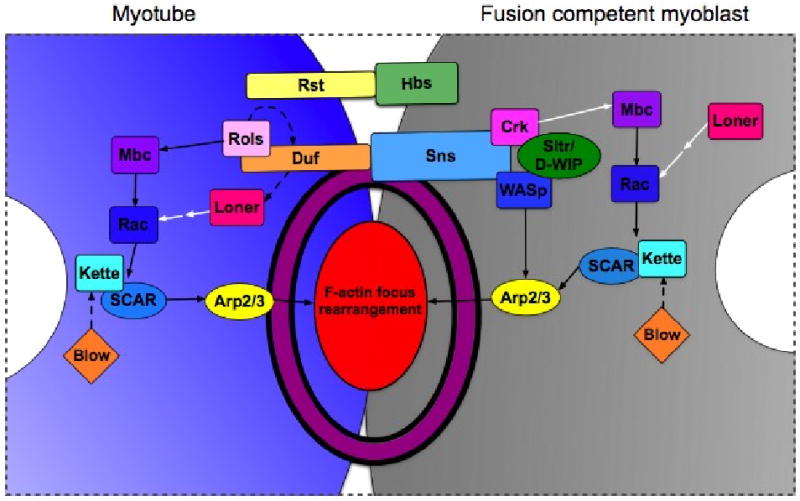
A simplified model that has been updated to reflect recently identified fusion genes and to illustrate the conservation of proteins to the zebrafish and mouse myoblast fusion paradigms. Note that nuclei are in white, myotube in is grey and the FCM is in blue. The actin focus and FuRMAS are depicted as a red oval and a purple ring, respectively. Rectangles represent proteins that have a conserved role in myoblast fusion in multiple systems. Ovals represent proteins with known homologs in vertebrates that have no role in fusion described to date. Diamonds represent proteins that have no known homologs in vertebrates. Solid arrows denote well-characterized biochemical interactions, dashed arrows indicate genetic and/or suggested biochemical interactions and white arrows designate interactions that are suggested from work on orthologous proteins in other contexts. While this depiction suggests specific interacting partners for each transmembrane protein, there is evidence that this may not be the case as interactions between Duf and Hbs have been shown to mediate cell adhesion in vitro. Additionally, the reader is cautioned that, although strong biochemical exists for Mbc-mediated Rac activation and for the Rac→ SCAR complex→ Arp2/3 pathway, there is not yet evidence for a complete pathway linking Mbc to Arp2/3-dependent actin polymerization.
Migration
Several lines of evidence support the importance of migration to myoblast fusion. For example, the migration of myoblasts prior to fusion has been observed and documented in both primary and immortalized mammalian culture systems (Horsley and Pavlath, 2004). Given the conservation of the basic cellular processes underlying fusion from flies to mice, migration was also thought to be an important step in Drosophila myogenesis.
Data documenting the arrangement of FCs and FCMs (Beckett and Baylies, 2007) and cell shape changes of FCMs in fixed materials (Bour et al., 2000) provided initial support for migratory behavior of FCMs. However, time-lapse analysis (Richardson et al., 2007) clearly documented movement of FCMs towards FCs/myotubes in vivo. Labeled using an actin∷GFP reporter, FCMs exhibited dynamic cell shape changes, transforming from a rounded to a teardrop-like morphology. Moreover, FCMs extended their filopodia directionally toward developing FC/myotubes (Richardson et al., 2007). These observations are in agreement with previous data which show that both Dumbfounded/Kirre (Duf) and Roughest/IrreC (Rst), FC-specific Immunoglobulin (Ig) domain-containing single pass transmembrane proteins, serve as attractants for FCMs (Ruiz-Gomez et al., 2000; Strunkelnberg et al., 2001) (Table 1) (Figure 3). Ectopic expression of these proteins was sufficient to alter the migratory path of FCMs and direct them to the source of the attractant although the cells failed to fuse. It is believed that the ability of FCMs to recognize these attractants, migrate and subsequently adhere to FCs/myotubes is mediated by the FCM-specific Ig domain-containing transmembrane proteins, Sticks and Stones (Sns) and Hibris (Hbs) (Artero et al., 2001; Bour et al., 2000; Dworak et al., 2001) (Table 1) (Figure 3).
The dynamic cell behaviors revealed by time-lapse microscopy suggest that cytoskeletal rearrangements are crucial for proper migration in Drosophila. Consistent with this, recent work indicates that Kette, a conserved member of the SCAR/WAVE regulatory complex, has a role in myoblast migration (Gildor et al., 2009). SCAR/WAVE is known to regulate Apr2/3 (Ibarra et al., 2005; Smith and Li, 2004; Vartiainen and Machesky, 2004), a potent activator of actin nucleation, thus linking migration and the cytoskeleton (Figure 3). Initially, SCAR is located diffusely throughout the cytoplasm; however, when cells adopt the teardrop-like migratory shape, SCAR becomes asymmetrically localized and is enriched in lamellipodia (Gildor et al., 2009). This localization is lost in kette mutants and many cells remain rounded and unfused, implying that SCAR/WAVE localization is involved in regulating changes in morphology and migratory behavior (Gildor et al., 2009). The small GTPase Rac has also been shown to regulate SCAR/WAVE. Rac triple mutants (rac1, rac2, mtl) display a severe fusion defect and a more rounded cellular morphology, which suggests that these FCMs are unable to migrate (Gildor et al., 2009; Hakeda-Suzuki et al., 2002; Luo et al., 1994; Richardson et al., 2007).
However, the requirement for Kette and SCAR in migration might be more complicated than originally thought. The role of Kette in myoblast migration was also investigated using the ectopic migration assay described above. When they were exposed to ectopically expressed Duf, kette mutant FCMs were able to migrate to the source of attractant in numbers comparable to wild type (B. Richardson, unpublished) (Figure 4). These data indicate that kette mutant FCMs are able to migrate. However, aspects of migration, like migrational velocity, may be affected in this mutant background, such that differences in staging may show different results. Resolution of this conflicting data will require live imaging to determine how Kette is required for migration.
Figure 4. kette mutant FCMs can migrate towards ectopically expressed Dumbfounded (Duf), which acts as an FCM attractant in vivo.
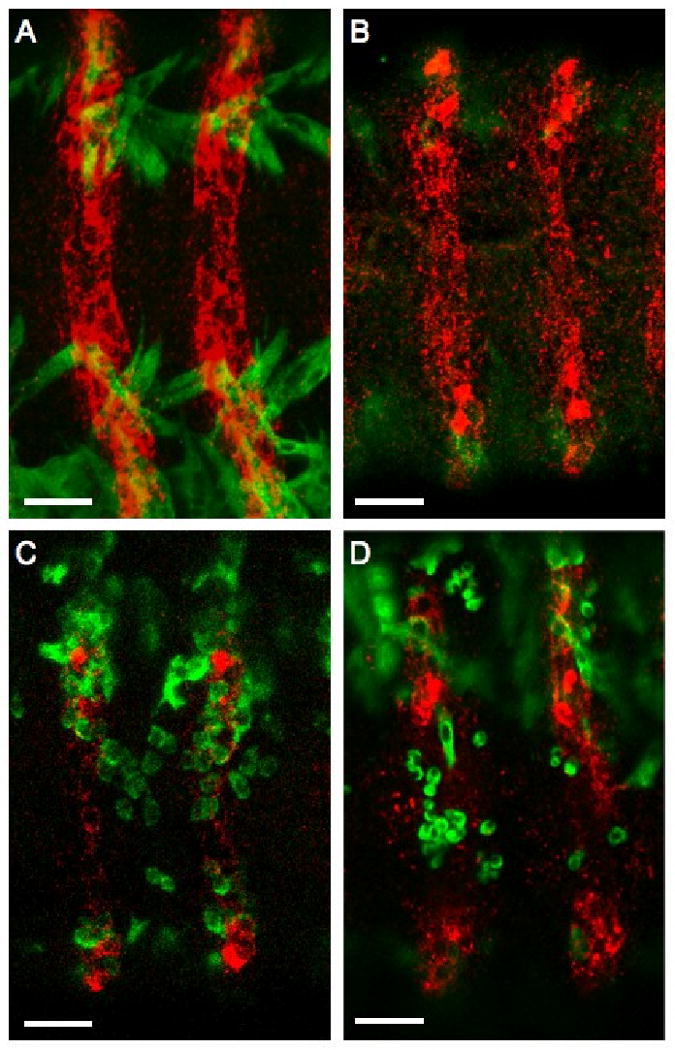
(A) wild type, (B) kette mutant, (C) Wingless (Wg)-GAL4 > UAS-Duf and (D) Wg-GAL4 > UAS-Duf; kette embryos. Myofibers and FCMs are in green; Wg epidermal cells, the source of the attractant Duf in the experimental embryos are in red. In wild type (A) and kette mutant (B) embryos, no myoblasts are present in the ventral ectoderm. However, when the expression of Duf is driven by Wg GAL4 in wild-type embryos (C), FCMs migrate ectopically into the ventral ectoderm. When Duf is expressed in kette mutant embryos (D), FCMs are capable of migrating towards the source of the attractant. The scale bar represents 20 μm.
Recognition and Adhesion
In addition to being required for migration, Duf, Rst, Sns and Hbs function in the recognition and adhesion of myoblasts (Artero et al., 2001; Bour et al., 2000; Dworak et al., 2001; Ruiz-Gomez et al., 2000; Strunkelnberg et al., 2001), suggesting that although, migration, recognition and adhesion are often thought of as functionally distinct steps during myoblast fusion, they are actually mechanistically and genetically linked at this level (Table 1). Duf and Rst function redundantly in FCs and only through deletion of both proteins is fusion severely disrupted (Strunkelnberg et al., 2001). In this case, myoblasts fail to properly recognize and adhere to one other, leading to a block in fusion. Duf and Rst both adhere to Sns, which, in conjunction with Hbs, has been shown to mediate recognition and adhesion in FCMs (Bour et al., 2000; Dworak et al., 2001; Galletta et al., 2004). These proteins are hypothesized to interact via their multiple extracellular Ig domains, but the exact mechanism of this interaction remains unclear.
Recent data have shown that Hbs functions as a positive regulator of myoblast fusion and has partially redundant function with Sns (Shelton et al., 2009). In sns mutants, some residual myoblast fusion occurs (1-2 per muscle) and only through the loss of both Sns and Hbs is myoblast fusion completely blocked. Additional genetic studies support this positive role for Hbs (Menon et al., 2005; Shelton et al., 2009). Thus, it appears that both genes mediate recognition and adhesion, much like Duf and Rst in FCs. Although this cell type-specific regulation appears straightforward, it is somewhat more complicated. Rst is initially expressed in FCs and some FCMs; however, Rst has not been shown to have a functional role in FCMs and is not capable of directing fusion in the absence of Sns and Hbs (Shelton et al., 2009; Strunkelnberg et al., 2001). Moreover, Hbs is only expressed transiently in FCMs with the majority of expression in the somatic mesoderm declining by stage 14, prior to the completion of most fusion events (Artero et al., 2001).
Myoblast recognition and subsequent adhesion results in the organization of Duf and Sns into a ring-like structure, the FuRMAS (fusion-restricted myogenic-adhesive structure) (Kesper et al., 2007; Onel and Renkawitz-Pohl, 2009) (Figure 3). This structure is believed to serve as a signaling center, triggering signaling cascades from the membrane to intracellular proteins, which leads to the recruitment of the fusion machinery to this site (Kesper et al., 2007; Kim et al., 2007; Onel and Renkawitz-Pohl, 2009; Richardson et al., 2007). Extensive analysis by serial deletion and site-directed mutagenesis of the Sns cytodomain has revealed redundant functional domains that direct myoblast fusion. This study suggests that Sns is rendered nonfunctional only when multiple domains are deleted simultaneously. Additionally, removal of all tyrosine residues, which are known to be phosphorylated, does not completely impair Sns function (Kocherlakota et al., 2008), indicating complex interactions with adapter proteins that are still not well understood. Interactions with different adaptor proteins may explain how these transmembrane proteins play multiple roles during myoblast fusion.
Actin regulation at the site of adhesion/fusion
A specific actin structure, termed the “focus,” has been identified at the site of fusion through fixed and time-lapse imaging approaches (Kesper et al., 2007; Kim et al., 2007; Richardson et al., 2007) (Figures 2, 3). The formation of this F-actin structure depends upon the adhesion proteins discussed above; without adhesion, the actin focus does not form. Current data has also demonstrated that the dissolution of the actin focus directly precedes the fusion event. In fact, when analyzed using live imaging, fusion has never been observed in the absence of foci formation and dissolution (Richardson et al., 2007). Precise measurement of the size and duration of the actin focus in wild-type embryos has shown that the average focus size is 1.9 μm (ranging from 0.7-4.5 μm) and that the average duration of the focus is 11.9 minutes (ranging from 5.7 – 29.5 minutes) (Richardson et al., 2007). A second structure at the site of fusion, the FuRMAS, surrounds the actin foci and is speculated to limit the size of the fusion site; consistent with this, the FuRMAS has been reported to range in size from 1-5 μm in fixed samples (Kesper et al., 2007; Onel and Renkawitz-Pohl, 2009). Whether reported ranges in sizes reflect the growth of the FuRMAS and/or actin focus or the inherent natural variation of the size of this structure between myoblasts has not been determined. Live imaging of the dynamics of the FuRMAS, rather than fixed samples, will be required to address this issue.
In addition to providing a physical marker for the site of fusion (Figures 2, 3), the actin focus has also proven to be a valuable tool for classifying fusion mutants (Richardson et al., 2007). Previous studies of essential fusion genes were hindered due to the similarities between mutant phenotypes, which consist of unfused myoblasts and small muscles with one or few nuclei. These phenotypes, as a result of their similarity to one another, often gave little mechanistic insight into the role of a protein during fusion. However, analysis of the actin foci in fusion mutants uncovered interesting differences between mutants and distributed them into three classes based on their size and number (Richardson et al., 2007; Richardson et al., 2008b) (Table 1). Interestingly, these three classes also grouped mutants based on the process they affect during fusion. In the first class of fusion mutants, which includes the transmembrane proteins required for myoblast recognition and adhesion and a known regulator of Duf (Sns, Rst, Duf, Rols), the foci number is greatly reduced, suggesting that adhesion is critical for actin focus formation. The second class of mutants, which currently consists of only three known genes (D-WIP, Loner, WASp) (Gildor et al., 2009; Richardson et al., 2007; Richardson et al., 2008b), contains normal-sized foci, implying that these genes may play focus-independent roles during fusion. Finally, the third class, which contains the majority of known fusion genes that impinge on actin cytoskeletal regulation, results in enlarged foci (Rac, Kette, Scar, Mbc, Blow, Sing), indicating that these genes are required for focus regulation (Richardson et al., 2007). The importance of actin at the fusion site is underscored by the requirement for foci formation and dissolution prior to fusion as well as these differences in focus behavior among fusion mutants. Accordingly, a great deal of recent research has been directed to understanding the molecular mechanisms of actin foci regulation at the site of fusion (Table 1, Figure 3).
Currently two major families of nucleation-promoting factors (NPFs): SCAR/WAVE (Berger et al., 2008; Richardson et al., 2007) and WASp (Gildor et al., 2009; Kim et al., 2007; Massarwa et al., 2007; Schafer et al., 2007) have been shown to regulate Arp2/3 during fusion in Drosophila (Figure 3). Kette, a conserved member of the SCAR/WAVE regulatory complex, regulates myoblast fusion through stabilization of SCAR, which positively regulates Arp2/3 leading to actin branching. In wildtype embryos, SCAR is typically localized to the lamellepodial extensions of FCMs that appear migratory and also to the site of fusion in adherent FCMs. As discussed previously, kette mutant myoblasts that do not adhere to a FC fail to properly localize SCAR to lamellipodial extensions. However, kette mutant myoblasts that do adhere fail to localize SCAR to the site of fusion and exhibit enlarged foci (Gildor et al., 2009; Richardson et al., 2007). These distinct localization patterns during different steps of fusion suggest functional roles for Kette in both migration and fusion, providing yet another molecular link between the two processes. scar maternal zygotic mutant embryos also have enlarged foci indicating that this pathway is required for actin reorganization involved in the dissolution of the actin focus (Richardson et al., 2007).
Upstream regulators of Kette/SCAR complex, including Myoblast city (Mbc) and Rac, have also been shown to have a role in myoblast fusion. Additionally, biochemical evidence has demonstrated that Mbc together with ELMO, function as an atypical, bipartite GEF to directly regulate Rac1 in vivo; overexpression of Mbc and ELMO in vivo demonstrated a genetic interaction, which induced downstream events leading to disrupted myoblast fusion. Additional experiments in the Drosophila eye suggest interactions between Mbc and ELMO lead to increased activity of Rac (Geisbrecht et al., 2008). Upon activation, Rac, in turn, activates the Kette/SCAR complex (Miki and Takenawa, 2003; Nolan et al., 1998; Smith and Li, 2004). Although strong biochemical exists for Mbc-mediated Rac activation and for the activation of Arp2/3 through the sequential activation of Rac and the SCAR complex, there is not yet biochemical evidence for a complete pathway linking Mbc to Arp2/3-dependent actin polymerization during myoblast fusion. Additional experiments are needed to determine if the activity of Mbc, mediated by Rac and SCAR, results in Arp2/3-dependent actin branching in myoblast fusion in vivo.
The second actin regulatory pathway in Drosophila is mediated by the WASp family of NPFs (Gildor et al., 2009; Kim et al., 2007; Massarwa et al., 2007; Schafer et al., 2007). In this case, Drosophila WASp Interacting Protein (D-WIP)/Solitary (Sltr) is recruited to the site of fusion and interacts with WASp through a WBD (WASp binding domain). WASp then stimulates Arp2/3 resulting in actin branching. Loss of either D-WIP/Sltr or WASp causes a severe fusion defect (Kim et al., 2007; Massarwa et al., 2007). Biochemical evidence has also demonstrated that D-WIP/Sltr is capable of interacting with the Sns adapter protein Crk (Kim et al., 2007), providing a potential direct link from the transmembrane receptors to actin polymerization. Interestingly, both D-WIP/sltr and wasp mutants both have normal-sized actin foci, while analysis of wasp, kette double mutant embryos revealed enlarged foci (Gildor et al., 2009), suggesting that the two families of NPFs act sequentially during myoblast fusion, with the SCAR pathway acting earlier than the D-WIP/WASp pathway (Gildor et al., 2009; Onel and Renkawitz-Pohl, 2009).
Vesicle trafficking at the site of fusion
The importance of vesicles in myoblast fusion was first noted using transmission electron microscopy (TEM) to examine myoblast fusion at the ultrastructural level (Doberstein et al., 1997). Using this approach, several structures were identified and ordered into a timeline of fusion based on their prevalence during different developmental stages. Following FC/FCM contact, paired electron-dense vesicles, or “pre-fusion complexes,” were identified at the putative site of fusion. These vesicles, containing an unknown material, align along apposed membranes of fusing myoblasts. Subsequently, electron-dense plaques, which are presumed to be formed from the contents of the pre-fusion complexes, appear (Figure 2). The prominence of these vesicles during fusion suggested an important role for trafficking in this process and, as with analysis of the actin foci, TEM analysis of available fusion mutants have revealed distinct blocks in these steps, providing important insight into the mechanism of several fusion genes.
Support for the role of actin in regulating vesicular transport has come from recent work on D-WIP/Sltr (Table 1, Figure 3). EM analysis of D-WIP/sltr mutant embryos reveals potential defects in vesicular targeting as electron dense vesicles were observed not only at the site of fusion but also between adjacent fusion competent cells, which is not observed in wild-type embryos (Kim et al., 2007). This is concurrent with actin misregulation at the site of fusion, suggesting that actin may provide a positional cue during vesicular transport. However, exactly how these vesicles are targeted to the site of fusion and the role of actin in this process remains controversial. Additionally, it has been shown that vesicles in D-WIP/sltr mutants persist and that plaques are not observed, suggesting a defect in the ability of vesicles to fuse with the plasma membrane (Kim et al., 2007).
A second example of the link between actin cytoskeletal regulation and trafficking is illustrated by Rolling pebbles/Antisocial (Rols/Ants), an intracellular adapter protein (Chen and Olson, 2001; Menon and Chia, 2001; Rau et al., 2001) (Table 1, Figure 3). Rols is initially present in FCs and is required for myoblast fusion in vivo. rols mutant embryos have severe fusion defects with only one to two fusions occurring per FC. In rols mutant embryos, normal-sized, but decreased numbers of foci form (Richardson et al., 2007), suggesting a role for Rols in regulating cell-cell contacts. Consistent with this, Rols is crucial for the recycling of Duf to the cell membrane in order to promote subsequent rounds of fusion (Menon et al., 2005). Upon adhesion, Rols is translocated from the cytoplasm to the site of fusion in a Duf-dependent manner, where it has been shown to physically link Duf to components and regulators of the cytoskeleton (Chen and Olson, 2001; Menon and Chia, 2001), including D-Titin and Mbc, a guanine nucleotide exchange factor (GEF) with the ability to regulate the small GTPase, Rac (Erickson et al., 1997; Kiyokawa et al., 1998; Nolan et al., 1998). Thus, Rols functions as a link between the membrane and the actin cytoskeleton via its regulation of the localization of a specific transmembrane protein, Duf.
Singles bar (sing), which was initially identified in a genetic screen, also appears to play a role in vesicular trafficking during myoblast fusion in Drosophila (Estrada et al., 2007) (Table 1). Sing encodes a multipass transmembrane MARVEL domain protein and is present in both FCs and FCMs during fusion. MARVEL domain proteins in vertebrates have been shown to be involved in tight junction formation and vesicular trafficking (Sanchez-Pulido et al., 2002). Consistently, TEM analysis of sing mutants reveals an increased number of pre-fusion complexes, which contain an overall greater number of vesicles, implying an involvement of Sing in the progression of fusion beyond the pre-fusion complex (Estrada et al., 2007). The inability to properly target vesicles to or fuse vesicles with the membrane suggests that Sing may be a functional component of vesicles required for myoblast fusion. Further study is required to determine the exact molecular mechanism of Sing during fusion.
Loner/Schizo, a guanine nucleotide exchange factor (GEF), which is localized near, but not at, the site of fusion has also been suggested to meditate Rac localization (Richardson et al., 2007) (Table 1, Figure 3). Loner is present in both FC/myotubes and FCMs (Richardson et al., 2007) and has been shown to interact with the intracellular domain of Duf (Chen et al., 2003). Although Loner can regulate another small GTPase, Arf6, in vitro (Chen et al., 2003), its targets in vivo remain unclear, as arf6 null flies are viable and myogenesis is not perturbed (Dyer et al., 2007). loner mutants also have normal-sized foci; however, the foci fail to resolve, leading to a strong fusion defect and suggest that Loner is involved in other essential fusion processes (Richardson et al., 2007), the nature of which is currently unclear. Given the ability of Loner to act upon Arf6, it is tempting to speculate about its potential involvement in Arf-dependent processes, such as trafficking and actin cytoskeletal reorganization (D'Souza-Schorey and Chavrier, 2006).
Membrane breakdown at the site of fusion and reset
Using TEM analysis in Drosophila, small fusion pores can be observed at the site of fusion (Berger et al., 2008; Doberstein et al., 1997; Kim et al., 2007). These membrane pores expand, with the vesiculating membrane from the site being removed (Figure 2), leading to membrane breakdown, cytoplasmic continuity and ultimately, the addition of another nucleus to the growing myotube.
Work in other fusion systems, such as the epidermis and vulva of C. elegans has identified “fusogens”, the EFF-1 and AFF-1 proteins, that are responsible for the formation and/or expansion of membrane pores, respectively (Sapir et al., 2007). To date, transmission electron microscopy (TEM) data in Drosophila embryonic muscle suggest that multiple pores form, enlarge and coalesce (Doberstein et al., 1997), while similar experiments in C. elegans hypodermis suggest a single pore forms and is subsequently expanded (Mohler et al., 1998).
Research to date has failed to identify a myoblast fusogen in any system. However, recent data in Drosophila suggests that actin may provide a force-generating mechanism at the site of fusion (Berger et al., 2008; Gildor et al., 2009). To investigate this, several groups have performed ultrastructural analysis of actin regulatory fusion genes at the site of fusion to determine if the membrane remains intact. In several mutants, including blow, sing, and kette, the membrane remains continuous, indicating that these proteins are required for earlier events at the site of fusion (Doberstein et al., 1997; Estrada et al., 2007; Schroter et al., 2004).
In contrast, analysis of a mutant allele of Arp3, one of the integral components of the Arp2/3 complex, has revealed fusion pore formation at the aligned membranes at the site of fusion (Berger et al., 2008). This suggests that Arp2/3-mediated actin polymerization may be required for expansion of the fusion pores in order to complete fusion. Several studies analyzing the role of D-WIP/Sltr using EM suggest a potential role for actin in fusion pore expansion, although the results remain somewhat controversial. One study reported that D-WIP/sltr mutants progress to the final stage of fusion and have fusion pores between aligned cells at the site of fusion (Massarwa et al., 2007). This was supported by cytoplasmic leak experiments, which tested whether GFP translated in the FC/myotube could be detected in adhering FCMs. In D-WIP/sltr mutants, leak was detected, supporting the conclusion that there are fusion pores that form but are unable to expand to complete fusion. This result further suggests that there may be a requirement for D-WIP/Sltr WASp-mediated Arp2/3 activation for actin polymerization in fusion pore formation. However, a conflicting study reported finding an intact membrane at the site of fusion in D-WIP/sltr mutants (Kim et al., 2007), which was supported by their own cytoplasmic leak assay. These results were obtained with a different mutant allele of D-WIP/sltr and by using a different EM fixation method. Whether the different genetic background, different fixation, or different alleles contributed to these discrepancies remains to be resolved.
Subsequently, the nucleus of the FCM is incorporated into the cell body of the FC/myotube and will undergo a nuclear identity shift, turning off expression of Lmd, an FCM-specific identity gene, and adopting the transcriptional profile of its fusing partner (Baylies et al., 1998; Beckett and Baylies, 2006). An underappreciated, but extremely important aspect of myoblast fusion, which is absent in several other systems of fusion is the observation that muscle cells repeat these steps multiple times. The requirement to undergo successive fusion events indicates that there must be a cellular mechanism by which the developing myotube is able to conclude one fusion and reset itself to undergo the next fusion event. Though little is currently known about this interesting aspect of fusion it remains an exciting area to be addressed in future research.
Models of Drosophila myoblast fusion
Currently, there are two prevailing models of Drosophila myoblast fusion. The first model proposes that fusion occurs in two distinct steps and each step requires not only a particular subset of gene products but also distinct subcellular events. This two-step model proposes that the first step of fusion produces a bi- or tri-nucleate precursor cell and that the second step of fusion includes the remaining fusions required to generate the final muscle size. Support for this model comes predominantly from mutant analysis and TEM. For a more comprehensive discussion of this model, we refer the readers to several reviews (Onel and Renkawitz-Pohl, 2009; Richardson et al., 2008a).
The second model, known as the two-phase model, proposes that all genes and subcellular events identified to date are required for all fusions. This model is based on the observation that all fusion mutants are capable of occasional fusions. Furthermore, in fusion mutants that commonly have two to three fusions per FC, there are also FCs that undergo no fusion, suggesting that the ability of some fusion mutants to undergo several rounds of fusion could be due to maternally contributed gene products and not a specific stepwise requirement. This model also suggests that the primary difference between early and late fusion is the frequency with which they occur. This is based on the observation that fusion occurs in two temporal phases: rare and limited fusion in the first phase (stage 12-13) and more frequent fusion in the second phase (stage 14-15). Increased fusion is coincident with the mobilization and migration of more internally located FCMs, which may provide a mechanism to transition between the two phases. Recently, there have been attempts to merge these two models; however, more genetic and functional data are required (Onel and Renkawitz-Pohl, 2009).
A conserved paradigm from fly to fish to mouse?
Experimentation in Drosophila has yielded a basic structure of events and molecules necessary for myoblast fusion. Recent work in vertebrate systems has begun to test the validity of the Drosophila paradigm in vertebrates, which is discussed below. This work has revealed a dramatic conservation of genes involved in the process; yet, whether the exact function of these genes during the fusion process is conserved remains to be investigated. Additionally, new genes that have not been implicated in Drosophila myoblast fusion have emerged as a result of the investigation of myoblast fusion in vertebrates. As with the discussion of the recent results from Drosophila, we organize the recent findings from the vertebrate models, zebrafish and mouse, in the same process-based format used above.
Vertebrate myogenesis
Mouse
In the mammalian embryo, skeletal muscle arises from mesodermal precursor cells from within the somites. Somites are transient, segmented blocks of mesoderm that develop in pairs on the sides of the neural tube and the notochord during embryogenesis. The dorsal region of the somite, the dermomyotome, is further regionalized into the dermatome and myotome (Christ, 1995; Stockdale et al., 2000). Cells of the myotome receive instructional signals from the surrounding notochord, neural tube, and dorsal ectoderm to become myogenic precursors, eventually giving rise to the skeletal muscle (Chen et al., 2005; Christ, 1995; Dietrich et al., 1998; Geetha-Loganathan P. et al., 2008; Ikeya and Takada, 1998; McDermott et al., 2005; Pourquié et al., 1996; Stockdale et al., 2000; Tajbakhsh et al., 1998; Tajbakhsh S. and M., 2000). This specification requires the upregulation of MyoD and Myf5, basic helix-loop-helix (bHLH) transcriptional activators of the myogenic regulatory factor family (MRF), which operate in an intricate transcriptional regulatory network with the paired domain and homeobox-containing transcription factors Pax3 and Pax7 (Buckingham and Relaix, 2007; Pownall et al., 2002). As this review focuses on the latter aspect of myogenesis, fusion, we will not deal with specification in detail here.
Once they have exited the cell cycle and begin to express the cadre of muscle specific transcription factors, myoblasts can then fuse with one another to generate syncytial myofibers. The development of mature myofibers in the mammalian embryo occurs in two temporally distinct periods: primary myogenesis, in which the initial myofibers are generated and secondary myogenesis, in which additional fibers are formed. Muscle growth also occurs postnatally. This process is also accomplished by fusion of myoblasts into myofibers and is mediated by a subset of myoblasts, termed satellite cells, which are derived from the same progenitor pool as those that form adult muscle. In contrast to myoblasts that participate in primary and secondary myogenesis, satellite cells fail to differentiate and remain associated with the surface of the developing myofiber as quiescent cells and line the basal lamina of mature muscle fibers. (Gros et al., 2005; Relaix et al., 2005).
In vitro data suggests that each period of muscle development can be further partitioned into two phases of fusion. In the first phase, individual myoblasts undergo fusion with one another to generate nascent myotubes, which contain few nuclei. The second phase of fusion is characterized by further maturation of the nascent myofiber during which the myofiber increases in size and begins to express contractile proteins. This increase in size is a direct result of the incorporation of additional differentiated myoblasts into the nascent myotube (Figure 1C, D). Although these phases appear similar, different molecules have been shown to regulate the two phases of fusion in vitro (Horsley and Pavlath, 2004) (Table 2). For example, the membrane proteins β1-integrin (Schwander et al., 2003), VLA-4, VCAM (Rosen et al., 1992) and caveolin-3 (Galbiati et al., 1999) have been shown to play a role in the fusion of myoblasts with one another; whereas the NFATC2 pathway (Horsley et al., 2001), activated by calcium and calmodulin, and IL-4, a secreted cytokine (Horsley and Pavlath, 2004), are critical for the fusion of myoblasts with nascent myotubes.
Table 2. A partial list of recently identified mammalian gene products involved in fusion.
| Mammalian gene product | Proposed function | Localization | References |
|---|---|---|---|
| Caveolin-3 | Myoblast-myoblast fusion | Membrane | Galbiati et al., 1999; Volonte et al., 2003 |
| Netrin-3 | Myogenic differentiation; myotube formation | Membrane | Kang et al., 2004 |
| Neogenin (Netrin receptor) | Myogenic differentiation; myotube formation | Membrane | Kang et al., 2004 |
| Myoferlin | Myoblast-myotube fusion; endocytic recycling; membrane fusion at sites of contact | Membrane | Doherty et al., 2005; Doherty et al., 2008 |
| Mannose receptor | Myoblast migration/attraction | Membrane | Jansen and Pavlath, 2006 |
| Trio | Cytoskeleton, Rac1, RhoA/G activation | Cytoplasm | Charrasse et al., 2007 |
| DGK-ζ | Cytoskeleton, Actin reorganization | Membrane | Abramovici and Gee, 2007 |
| Prostacyclin | Regulation of myoblast migration/promotion of fusion | Secreted | Bondesen et al., 2007 |
| CD164 | Myoblast migration | Membrane | Bae et al., 2008 |
| EHD2 | Endocytic recycling, interaction with myoferlin | Cytoplasm, membrane | Doherty et al., 2008 |
| Cdc42 | Cytoskeleton, WASp activation | Ubitquitous | Vasyutina et al., 2009 |
| MOR23 | Myoblast migration and adhesion; regeneration | Membrane | Griffin et al., 2009 |
In addition to their role in postnatal muscle growth, satellite cells have a well-known function in the regeneration and repair of damaged skeletal muscle. In response to muscle damage as a result of natural causes (i.e. physical exertion, direct trauma) or inherent genetic predisposition to defects in the skeletal muscle (i.e. dystrophies), satellite cells become activated (Campion, 1984; Grounds et al., 2002; Hawke and Garry, 2001). Subsequently, these cells proliferate, with some exiting the cell cycle, differentiating and fusing with the injured myofiber to repair the damage. Satellite cells can also fuse with one another to generate nascent myofibers; in fact, the regeneration process is largely based on this type of fusion (Horsley and Pavlath, 2004; Morgan and Partridge, 2003; Wagers and Conboy, 2005).
Interestingly, the genes underlying the fusion of activated satellite cells to the damaged myotube appear to overlap with those that are involved in the fusion of myoblasts during development (Table 2). For example, the expression of M- and N-cadherin, which play roles in muscle development in vitro and in vivo (Donalies et al., 1991; George-Weinstein et al., 1997; Hollnagel et al., 2002; Radice et al., 1997), are also upregulated in activated satellite cells (Irintchev et al., 1994; Moore and Walsh, 1993), suggestive of a role for the cadherins in muscle repair. However, there are different mechanisms at play during repair that are absent or unnecessary for myoblast fusion during development. Evidence for this has come from studies analysis of both processes in desmin (Smythe et al., 2001) and Il-4 (Horsley et al., 2003) mutant mice. In both situations, mutant mice are able to form muscle properly; however, the repair process is impaired (Horsley et al., 2003; Li et al., 1997; Smythe et al., 2001). Taken together, this suggests that understanding the mechanisms that underlie fusion during muscle development may inform aspects of satellite cell biology and repair, but differences between these processes exist.
Zebrafish
In comparison to those of mouse, the somite and the mature myotome of the zebrafish are relatively simple in their cellular composition. The dermomyotome is absent; however, the existence of an analogous structure to the dermomyotome, a layer of flattened cells termed external cells, has been suggested (Hammond et al., 2007; Hollway et al., 2007; Stellabotte and Devoto, 2007). These cells express a similar set of genes as in mouse (Devoto et al., 2006) and participate in the growth of muscles (Hollway et al., 2007; Stellabotte and Devoto, 2007), implying that the external cells of the zebrafish are analogous to the amniotic dermomyotome. Additionally, several pieces of evidence have suggested the existence of a satellite cell population in zebrafish, including the identification of a population of external cells that express Pax7, a marker commonly used to identify satellite cells, which can power the growth of the myotome (Devoto et al., 2006; Hammond et al., 2007; Hollway et al., 2007; Stellabotte and Devoto, 2007).
By 24 hours post-fertilization, a stereotypical pattern of differentiated muscle fibers can be observed in the zebrafish myotome (Figure 1B), which is comprised of muscle fibers of two distinct types, slow- and fast-twitch. These are analogous to the slow- and fast-twitch muscle fibers found in higher vertebrates but absent in some invertebrates, such as the Drosophila embryo. These distinct fibers originate from two discrete lineages of the presomitic mesoderm. Whether muscle progenitors contribute to the slow- or to the fast-twitch muscle fiber population is determined by their mediolateral position with respect to the midline. For example, muscle progenitors residing medially, termed adaxial cells, require Hedgehog (Hh) signaling from the neural tube and notochord for their specification (Baxendale et al., 2004; Blagden et al., 1997; Devoto et al., 1996; Du et al., 1997; Lewis et al., 1999; Roy et al., 2001). With the onset of somitogenesis, the slow muscle precursors, which express a combination of slow and fast isoforms of myosin heavy chain (MyHC), are incorporated into the somites. Subsequently, they migrate radially past the more laterally located somitic cells to eventually appear on the surface of the myotome, where they complete their differentiation into slow-twitch muscle fibers. (Baxendale et al., 2004; Blagden et al., 1997; Bryson-Richardson et al., 2005; Devoto et al., 1996; Du et al., 1997; Roy et al., 2001; Xu et al., 2000). In contrast, the more laterally positioned somatic muscle precursors differentiate into densely packed fast-twitch muscles that make up the bulk of the myotome (Blagden et al., 1997; Devoto et al., 1996; Du et al., 1997; Roy et al., 2001) (Figure 1).
It was originally thought that all zebrafish somitic myoblasts matured into single-celled muscle fibers. This notion has since been revised through more sophisticated lineage tracing studies, as well as detailed characterization of the expression patterns of slow and fast muscle-specific marker genes (Roy et al., 2001). It is now recognized that while the muscle fibers of the slow-twitch type differentiate into mononucleate muscles, the fast-twitch muscles comprise syncytial fibers – a feature that is typical of skeletal muscles. Moreover, the observation that chimeric fast-twitch muscles readily form in zebrafish embryos constructed with differentially labeled donor and host cells, has clearly established the syncytial nature of the fast-twitch muscles (Roy et al., 2001). More recently, analysis of fast-twitch muscle fiber morphogenesis using live imaging techniques and precise quantification of cell shape changes have uncovered a dynamic increase in fast myoblast size prior to their fusion to form syncytia (Snow et al., 2008). These findings have now opened up the possibility for a systematic analysis of vertebrate myoblast fusion in vivo, at a cellular and genetic resolution that has thus far only been possible with the Drosophila embryo.
Despite differences in the developmental program that specifies fusion substrates in Drosophila, zebrafish and mouse, the cellular steps leading to fusion are conserved (Figure 2). In light of these parallels, here, as in Drosophila, we organize our discussion of recently identified genes in a process-based format.
Migration
Myoblasts are specified in the somites (Christ, 1995; Stockdale et al., 2000); however, muscle is found throughout the embryonic and adult body, often in sites quite distant from the somites. Thus, the migration of myoblasts must be an important mechanism to disperse muscle precursors. In mouse, there are two migratory periods that are essential for myoblast fusion. First, precursor cells migrate from sites of myogenesis (i.e. somites) to sites of future musculature, such as the limbs and diaphragm reviewed in (Vasyutina and Birchmeier, 2006)). This initial migration has been the focus of the majority of studies. In the second phase of migration, myoblasts migrate in search of a fusion partner. The importance of this second migration to fusion has been highlighted only recently as an important mechanism in bringing myoblasts in close enough proximity so that can they can fuse (Bae et al., 2008; Bondesen et al., 2007; Jansen and Pavlath, 2006; Mylona et al., 2006) (Table 2).
The actin cytoskeleton plays an important role in the migration of mouse myoblasts. Treatment of differentiating mammalian myoblasts with latrunculin A or cytochalasin D, which interfere with F-actin remodeling by binding to actin monomers or actin branches, respectively (Coué et al., 1987; Dhawan and Helfman, 2004; Nowak et al., 2009), adversely affect myoblast migration in vitro (Constantin et al., 1995; Sanger and Holtzer, 1972) consistent with a requirement of the actin cytoskeleton in the migration of myoblasts.
In Drosophila, Rac triple mutant FCMs remain rounded, which suggests that these myoblasts fail to migrate (Gildor et al., 2009; Hakeda-Suzuki et al., 2002; Richardson et al., 2007). Interestingly, the activity of Rac1 in the mouse does not appear to be required for the migration of one population of muscle precursors to the limb bud. Similarly, Cdc42 does not appear to be required for the first phase of migration in vivo (Vasyutina et al., 2009). Their requirement has not been tested in the second phase of fusion, leaving open the possibility that other proteins are required for the first migratory period. Additionally, the Rho GTPases may be able to compensate for the loss of the other, explaining why precursors are able to migrate in single mutants.
Recently, a role for the odorant receptor, MOR23, in regulating the migration of myoblasts has been demonstrated in vitro and in vivo (Griffin et al., 2009) (Table 2), (Figure 6). Odorant receptors (ORs), members of the G-protein-coupled receptor (GPCR) family, are chemosensors, which detect small odorant molecules (Young and Trask, 2002). While most are highly expressed during proliferation, a number of ORs are expressed during myoblast fusion and may control overlapping or discrete steps in this process (Griffin et al., 2009). Consistent with a role in migration, MOR23 expression is upregulated during the period in which migration occurs and knockdown of MOR23 expression using siRNA decreased migration in primary muscle cell cultures. Furthermore, MOR23 siRNA in primary cultures exhibited a defect in fusion, whereas overexpression of MOR23 increased the number of myotubes in culture, reinforcing the importance of migration to the overall fusion process. Interestingly, MOR23 also regulates the chemotaxis and motility of mouse sperm in vitro (Fukuda et al., 2004). Thus, MOR23 plays a conserved role in the migration of multiple cell types, and ORs, in general, may play a wider variety of roles in multiple tissues that previously thought, opening up an exciting new mechanism of migration not described in any muscle system to date.
Figure 6. Venn diagram depicting a subset of genes that have been analyzed in each model system.
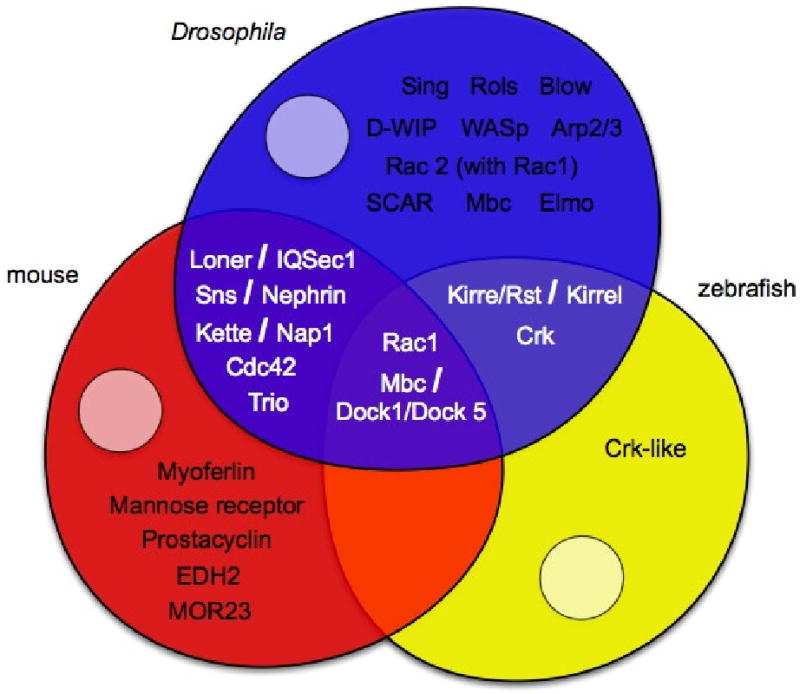
The model systems Drosophila, mouse and zebrafish are illustrated as blue, red and yellow partially overlapping myoblasts. A subset of genes analyzed to date are listed in each myoblast. Genes unique to a model organism are located in non-overlapping, monochromatic regions of the suitable myoblast. Genes that have been studied in multiple models are localized to the appropriate overlapping regions. Genes that have been analyzed in more than one system are phenotypically compared in Table 3.
Recognition and Adhesion
In Drosophila, two distinct types of myoblasts, FCs and FCMs, must recognize one another and do so by expressing different transmembrane proteins on their surfaces (Artero et al., 2001; Bour et al., 2000; Dworak et al., 2001; Ruiz-Gomez et al., 2000; Strunkelnberg et al., 2001). While the gene products that are required for recognition and adhesion in Drosophila, namely Duf, Rst, Sns and Hbs, are known, the analogous process and proteins in vertebrate myoblast fusion are just beginning to emerge with the recent publication of two compelling findings in both zebrafish and mouse systems. These studies add additional genes to the fusion paradigms of both organisms as well as illuminate the existence of conserved orthologs of the Drosophila transmembrane Ig domain-containing proteins (Table 1).
Zebrafish
The zebrafish genome encodes orthologs of the Drosophila Duf/Kirre/Rst/Irre and Sns/Hbs families of Ig domain-containing proteins (Kramer-Zucker et al., 2005; Sohn et al., 2009; Srinivas et al., 2007) (Table 1). Although a number of Kirrel (Kirre-like) genes are present in the zebrafish genome, only Kirrel is specifically expressed within the somites and restricted to the muscle precursors, which eventually give rise to the fast-twitch muscle fibers (Srinivas et al., 2007).
In embryonic fast-twitch myoblasts, Kirrel localizes to the membranes and is rapidly down-regulated as fusion proceeds. Analysis of zebrafish embryos in which Kirrel is knocked down (Kirrel morphants) has established that its activity is critical for the fusion of fast-twitch myoblasts (Srinivas et al., 2007). In striking resemblance to Drosophila embryos lacking Duf/Kirre and Rst/Irre function (Strunkelnberg et al., 2001), Kirrel morphants showed a significant block in fast-twitch precursor fusion; instead of the arrays of syncytial fast-twitch fibers that are characteristic of wild-type embryos (Figure 5A), Kirrel morphants showed a preponderance of unfused myoblasts within the myotome (Figure 5C). These subsequently differentiated into mononucleate fast-twitch muscles (Srinivas et al., 2007), reminiscent of unfused FCs in Drosophila (Ruiz-Gomez et al., 2000; Strunkelnberg et al., 2001) (Table3).
Figure 5. The activities of Rac1 and Kirrel/Nephrin are required for vertebrate myoblast fusion.
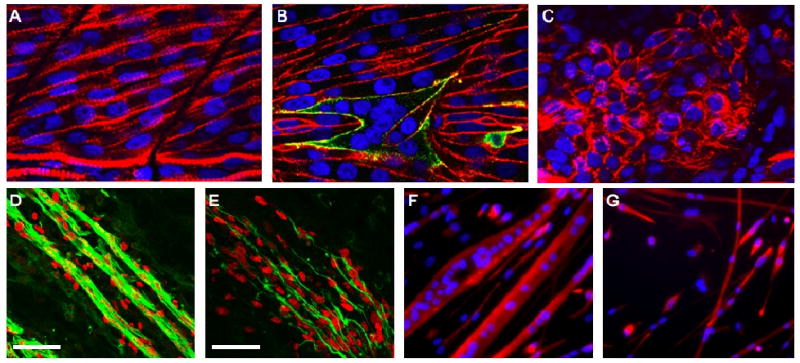
(A-C) The zebrafish fast-twitch syncytium in a control embryo (A), an embryo expressing constitutively active Rac1 (caRac1) (B) and a kirrel morphant zebrafish embryo (C) were analyzed by immunohistochemistry using antibodies against myosin heavy chain (red) and hemagglutinin to label Rac1 (green) in (B). (A) The fast-twitch muscle fibers of a wild-type zebrafish embryo contain multiple nuclei. (B) The fast-twitch syncytium in zebrafish embryos expressing caRac1 are hyperfused, containing more nuclei than in wild-type syncytium. (C) In kirrel morphants, there are large numbers of unfused fast-twitch precursors. (D-E) Longitudinal sections of myofibers from the proximal forelimb of control (D) and conditional Rac1 mutant (E) mice at E13.5 were analyzed by immunohistochemistry using antibodies against desmin (green) and MyoD (red). The scale bar represents 50 μm. (E) Myofibers in conditional Rac1 mutant mice (E) are short and thin, indicative of a fusion impairment in vivo, in comparison to control myofibers, which form long, multinucleated fibers (D). (F-G) Myoblasts isolated from control (F) and nephrin null (G) neonatal mice were analyzed by immunohistochemistry using antibodies against desmin (red). (F) Control myocytes form large, multinucleated cells after 4 days of differentiation in vitro. (G) Myocytes from nephrin null mice differentiate, but fail to fuse during the same time period. Nuclei were visualized with DAPI (blue) (A-C, F, G) or MyoD (D, E).
Table 3. Comparison of fusion phenotypes in genes that have been analyzed in at least two model organisms.
| Gene Name (Drosophila/ Vertebrate) | Drosophila | mouse | zebrafish |
|---|---|---|---|
| wild type | |||
| rac1,2, mtl/rac11 | |||
| mbc/dock1, 5 | |||
| trio/trio | |||
| loner/Iqsec (Brag2)* | |||
| kette/Nap1* | |||
| sns, hbs/nephrin | |||
| ca cdc422/cdc421 | |||
| crk/crk* | |||
| kirre, rst/kirrel** | |||
| ca rac12/ca Rac |
Key:  no/reduced fusion
no/reduced fusion  normal fusion
normal fusion  hyperfusion
hyperfusion
Conditional Rac1 and Cdc42 mutant mice (Rac1fl/fl; Lbx1cre and Cdc42fl/fl; Lbc1cre);
Constitutively active (ca) Rac1 (Drac1V12) and Cdc42 (cdc42V12) alleles; Single and double asterisks indicate phenotype resulting from protein knockdown using shRNA or morpholinos, respectively. See text for references.
Presently, it is unclear whether zebrafish Kirrel functions by heterotypic interaction with other Kirrel or Nephrin (Sns orthologs) family members, or whether homotypic binding is sufficient for fusion. In support of the latter scenario, Nephrin is not expressed in zebrafish somites, but localized to the pronephros, the zebrafish equivalent of kidneys (Kramer-Zucker et al., 2005). In spite of this, however, a recent publication has suggested a role for Nephrin in zebrafish muscle development. Myosepta in nephrin morphants were less organized and shorter, a phenotype consistent with muscle defects (Sohn et al., 2009). Whether this difference in size is attributable to a fusion defect is not clear, but the results leave open the possibility for an interaction between these proteins for other aspects of muscle development in the zebrafish. Furthermore, analysis of additional proteins, which are discussed below, suggest that the adhesion of myoblasts in the mouse is not governed exclusively by one protein family as it is in Drosophila. Thus, it remains to be seen whether other proteins mediate this process in zebrafish.
Mouse
Early in vitro work has indicated that the cadherins mediate the recognition and adhesion process between myoblasts in the mouse (Donalies et al., 1991; George-Weinstein et al., 1997); however, additional genes must be involved as M- and N-cadherin are not essential in vivo (Hollnagel et al., 2002; Radice et al., 1997). Other data has pointed to a contributing role for integrin family members and other transmembrane proteins in myoblast adhesion and alignment (Galbiati et al., 1999; Horsley and Pavlath, 2004; Rosen et al., 1992; Schwander et al., 2003) (Table 2). Taken together, the existing data suggests that one protein family is not solely responsible for the recognition and adhesion of mouse myoblasts to one another, perhaps reflecting the additional complexities that underlie myoblast fusion in the mouse when compared to Drosophila.
The mouse genome also encodes orthologous proteins for Duf/Kirre/Rst and Sns/Hbs (Sellin et al., 2003; Sun et al., 2003; Ueno et al., 2003). To date, murine Kirrel and Nephrin proteins have been well-characterized as components of the slit-diaphragm, a specialized cell-cell adhesion complex that makes up the filtration barrier of the kidney (Pätäri-Sampo et al., 2006; Tryggvason, 2001) or expressed in the β cells of the pancreatic islets (Sun et al., 2003). A recent study, however, has indicated that Nephrin, an Sns ortholog, may play a conserved role in mammalian myoblast fusion. Nephrin is expressed in developing mouse skeletal muscle and human fetal muscle cells undergoing fusion and is upregulated in two murine models of human muscular dystrophies. Analysis of primary myoblasts isolated from Nephrin null mice established a fusion impairment in vitro (Figure 5F, G) (Table3), indicating that Nephrin plays role in vertebrate skeletal muscle fusion. Interestingly, cell-mixing experiments demonstrated an asymmetrical requirement for Nephrin during fusion; Nephrin is required in myoblasts but not in myotubes (Sohn et al., 2009). Thus, Nephrin may play a role that is analogous to that of Drosophila Sns, which is expressed on the surface of FCMs exclusively (Bour et al., 2000).
As in zebrafish, the protein/s with which mammalian Nephrin interacts has not been identified. In the slit-diaphragm, Nephrin has been shown to interact with itself (Gerke et al., 2003; Khoshnoodi et al., 2003; Pätäri-Sampo et al., 2006) as well as with Neph1 and Neph2 (orthologs of Drosophila Duf/Kirre) (Barletta et al., 2003; Gerke et al., 2003; Pätäri-Sampo et al., 2006). Whether the Duf orthologs play any role in myoblast fusion has not been examined. Given that zebrafish Kirrel is conserved in fast-twitch myoblast fusion (Srinivas et al., 2007), it is likely that they do. However, Neph1 deficient mice do not display a muscle phenotype (Donoviel et al., 2001), suggesting the possibility that Neph1 and Neph2 play redundant functions in myoblast recognition. Additionally, various transmembrane proteins, such as the mannose receptor, CDO and BOC have been shown to regulate fusion in the mouse in vitro (Cole et al., 2004; Jansen and Pavlath, 2006; Kang et al., 2002; Wegorzewska et al., 2003) (Table 2); thus, the possibility that Nephrin associates with a transmembrane protein that is not orthologous to Sns/Rst or Duf/Kirre to promote myoblast fusion in mice remains a possibility.
Great strides have been recently made in identifying the genes responsible for myoblast recognition in zebrafish and mouse. It will be important to explore the role of and relationship between Kirrel and Nephrin family proteins in myoblast fusion in both vertebrate systems. This study is required if we are to gain a complete understanding of the mechanisms that underlie individual cellular behaviors that contribute to the overall fusion of myoblasts and continue to draw parallels, where they exist, between vertebrate and Drosophila myoblast fusion. Taken together, this research indicates that the once elusive Sns/Rst and Duf/Kirre equivalents have been identified in vertebrate models, opening up the possibility of exploring many interesting questions and demonstrating for the first time the conservation of the Drosophila recognition proteins in vertebrates.
Actin regulation at the site of fusion
As discussed in previous reviews and above, genetic and cell biological evidence in Drosophila points to the careful orchestration of the actin cytoskeleton and its many regulators in the cellular activities that underlie successful myoblast fusion (Berger et al., 2008; Kesper et al., 2007; Massarwa et al., 2007; Richardson et al., 2007; Schafer et al., 2007). For example, analysis of the actin focus, which marks the site of fusion in Drosophila, has indicated that many of the regulators of the actin cytoskeleton, including Rac, Mbc and Kette are localized to this structure and likely play a role in its regulation (Richardson et al., 2007). Thus, it is no surprise that recent research has been focused on testing whether the actin regulators previously identified in Drosophila are conserved during vertebrate myoblast fusion. An equivalent actin focus structure has not been identified in zebrafish or mouse; however, orthologs of the Drosophila proteins exist in both systems (Figure 6), (Table 1) and, in many cases, their role in fusion has recently been addressed. Thus, for the sake of clarity and continuity, we have discussed them accordingly.
Zebrafish
The requirement of Rac1 and Rac2 for myoblast fusion has been clearly established in Drosophila (Hakeda-Suzuki et al., 2002; Luo et al., 1994). Recent work has demonstrated that Rac1 also plays a conserved role in the fusion of zebrafish fast-twitch precursors. In Rac1 morphant embryos, fusion among fast-twitch myoblasts is severely compromised (Srinivas et al., 2007). However, in contrast to constitutively active Rac mutants in Drosophila in which myoblast fusion is impaired (Luo et al., 1994), constitutive Rac1 (caRac1) activity in the zebrafish myotome triggers unrestrained fusion of fast-twitch myoblasts, resulting in abnormally large syncytia (Figure 5B), (Table 3). In addition to this hyper-fusion defect, fast-twitch myofibers in caRac1 embryos also appeared to deviate from the normal pattern of fast-twitch myofibers (Figure 5A). Whereas fast-twitch myofibers are arranged in multiple layers in wild-type embryos with a stereotypical pattern across the width of the myotome, the large muscle syncytia of embryos expressing caRac1 are oriented randomly and extend unsystematically across the myotome (Figure 5A, B). This orderly arrangement of fast fibers within the wild-type myotome indicates that fusion among the fast myoblasts is not random, but must be a spatially-regulated process. Thus, regulated Rac1 activity is essential for ensuring the proper amount and directionality of fast-twitch myoblast fusion in the zebrafish (Srinivas et al., 2007).
The cellular basis of this hyper-fusion phenotype, however, is unclear. For example, it is not known whether myoblasts expressing caRac1 can fuse with neighboring wild-type myoblasts to undergo uncontrolled fusion or whether the myoblasts expressing caRac1 undergo uncontrolled fusion exclusively with one another to generate the large syncytia. Time-lapse analysis of the fusion dynamics of myoblasts expressing constitutively active Rac1 may help to clarify these issues.
In Drosophila, the importance Mbc, a noncanonical GEF for Rac1, in myoblast fusion has been demonstrated (Erickson et al., 1997; Nolan et al., 1998; Rushton et al., 1995). The zebrafish genome encodes two closely related orthologs of Mbc, Dock1 and Dock5 (Moore et al., 2007). As in Drosophila mbc mutant embryos (Erickson et al., 1997; Nolan et al., 1998; Rushton et al., 1995), fusion of the fast-twitch myoblasts is significantly reduced in Dock1 and Dock5 morphant embryos, indicating that Dock1 and Dock5 are functionally conserved in myoblast fusion in zebrafish (Table 3). Furthermore, Dock1 and Dock5 equally contribute to myoblast fusion in the zebrafish myotome (Moore et al., 2007).
Mammalian Dock1 was originally identified in a screen by virtue of its physical interaction with the adaptor protein Crk (Hasegawa et al., 1996). Dock family proteins associate with Crk in a variety of signaling contexts (Galletta et al., 2004; Hasegawa et al., 1996; Kiyokawa et al., 1998). This physical interaction, together with the role of Dock1 and Dock5 in the zebrafish fusion paradigm, suggested a role for Crk in myoblast fusion. To explore whether the zebrafish orthologs play a role in myoblast fusion, morpholinos directed against Crk and Crk-like (Crkl) were injected into zebrafish embryos, resulting in a substantial block in the formation of syncytial fast-twitch myotubes (Moore et al., 2007), indicating that both Crk and Crkl play a role in zebrafish fast-twitch myoblast fusion.
Whether this affect requires their direct interaction with Dock1 and Dock5 is currently unclear. There is some indication in Drosophila that Crk could have a role in regulating myoblast fusion through mechanisms that are independent of its physical association with Mbc (Balagopalan et al., 2006); thus, the possibility of a Dock-independent role for Crk and Crk-like in the fusion of zebrafish fast-twitch myoblasts cannot be ruled out.
Mouse
Evidence in mammalian in vitro models has also supported a role for the actin cytoskeleton in mammalian myoblast fusion. For example, time-lapse imaging and a variety of other experimental techniques have illuminated the actin-based behaviors of differentiating myoblasts in vitro (Kawamura et al., 2004; Nowak et al., 2009; Ohtake et al., 2006; Steffen et al., 2006; Steffen et al., 2004). Furthermore, assays that measure distinct roles for the actin cytoskeleton during myoblast fusion have been developed, thus, providing an important framework for dissecting the cell biology of myoblast fusion in mammals (Nowak et al., 2009), which is especially important when considering the many actin regulators and actin-based processes involved in myoblast fusion.
A requirement for Rac in the fusion paradigm of mammals had not, until recently, been experimentally demonstrated. One reason for this is the complication of in vivo analysis in the mouse as a result of the importance of the small GTPase for early developmental events. rac1 deficient mouse embryos die before E9.5 (Sugihara et al., 1998), precluding analysis of myoblast fusion. As a result, an early study of Rac function in mammalian myoblast fusion, which concluded that Rac was required for fusion, was in an in vitro culture model and utilized a Rac-specific inhibitor rather than genetic perturbations (Charrasse et al., 2007). However, a conditional Rac1 knockout mouse model was recently employed to analyze the effects of Rac1 knockout in migrating hypaxial muscle precursors. Analysis of conditional Rac1 mutants revealed short, thin myofibers in the limbs, indicative of a fusion impairment in vivo (Figure 5D, E) and established the conservation of Rac to the mammalian fusion process (Vasyutina et al., 2009) (Table 1). Interestingly, as in zebrafish, genetic ablation of Rac1 alone was sufficient to reveal a fusion defect in mice (Srinivas et al., 2007; Vasyutina et al., 2009), whereas mutation of both Rac1 and Rac2 are required to obtain a fusion defect in Drosophila (Hakeda-Suzuki et al., 2002) (Table 3). Primary myoblasts isolated from conditional knockout mice were able to appropriately recruit early markers of adherence, α- and β-catenin, while the accumulation of polymerized actin, vinculin and Arp2/3 at the site of myoblast-myoblast adhesion was reduced (Vasyutina et al., 2009). This is unsurprising given the role of Rac in activating Arp2/3 to drive actin polymerization (Miki and Takenawa, 2003; Smith and Li, 2004).
In Drosophila, actin foci in Rac triple mutants are enlarged. This has been taken to mean that Rac participates in the dissolution of foci in Drosophila (Richardson et al., 2007). In contrast, actin at the site of contact between mouse myoblasts is reduced (Vasyutina et al., 2009). Whether this difference indicates different roles for the Rac proteins in Drosophila versus mouse or is reflective of the fact that a site of fusion/actin focus has not yet been identified in mouse needs to be assessed further.
A second member of the Rho family of GTPases, Cdc42, plays a well-characterized role in actin-based filopodia (Heasman and Ridley, 2008) and has also been recently implicated in mammalian myoblast fusion (Vasyutina et al., 2009). Like Rac1, Cdc42 is required for early embryogenesis in mouse (Chen et al., 2000). In Drosophila, constitutively active Cdc42 does not affect myoblast fusion, but does exert effects on myotube morphology: myotubes are spindle-like rather than tubular (Luo et al., 1994). However, conditional mutation of murine Cdc42, which was achieved in the same manner as in the Rac1 conditional mouse discussed above, interferes with myoblast fusion in vivo, demonstrating that the requirements of the fusion process in mammals are not likely to be the same as in Drosophila in all cases (Table 3). As in Rac1 mutant myoblasts, Cdc42 mutant myoblasts are able to properly recruit α- and β-catenin, but not vinculin and polymerized actin. In contrast, however, Arp2/3 accumulation at sites of myoblast-myoblast contact is unchanged (Vasyutina et al., 2009), suggesting that Cdc42 and Rac1 play nonredundant roles in myoblast fusion.
Although its role has not been described in zebrafish, it is interesting to speculate what role, if any, Cdc42 plays in the fusion of fast-twitch myoblasts in zebrafish. Cdc42 may play a vertebrate- or a mammalian-specific role in myoblast fusion, which may hint at its function in myoblast fusion. Zebrafish, as a non-amniotic vertebrate, is uniquely positioned to address this question, providing a tangible situation in which vertebrate models should be used in combination.
Recent data has also implicated the murine homologs of Mbc-Dock1 and Dock5-in myoblast fusion in vivo and in vitro (Laurin et al., 2008; Pajcini et al., 2008). Analysis of the role of mammalian Dock1 in myoblast fusion in vivo reveals an essential role for Dock1 in development. Dock1 null embryos present with hallmarks of defective primary myogenesis, showing a dramatic and general reduction in muscle content as a result of impaired myoblast fusion (Laurin et al., 2008). Taken together, these data demonstrate that Dock1, a component of the fusion machinery discovered in Drosophila, also has a conserved role in higher vertebrates (Laurin et al., 2008; Pajcini et al., 2008). In contrast, analysis of Dock5 mutant mice reveals a nonessential role for the protein during embryogenesis; however, there appears to be a functionally redundant role for Dock5 in myoblast fusion in vivo (Laurin et al., 2008).
In addition to a role for the mammalian homologs of Mbc in murine myoblast fusion, an additional GEF, Trio, also plays a role in mammalian myoblast fusion (Charrasse et al., 2007; O'Brien et al., 2000). Interestingly, Drosophila Trio does not play a role in myoblast fusion in vivo (Hakeda-Suzuki et al., 2002) (Table 3). The knockdown of Trio in mouse, which can act as a dual GEF for Rac1 and RhoA/G (Bellanger et al., 1998; Blangy et al., 2000; Debant et al., 1996; Seipel et al., 1999), results in a block in fusion in vitro (Charrasse et al., 2007) and defects in secondary skeletal muscle formation in vivo (O'Brien et al., 2000). Thus, Dock1/5 and Trio, although both GEFs for Rho GTPases, play different roles in mouse myoblast fusion in vivo (Laurin et al., 2008; O'Brien et al., 2000), highlighting the importance of spatial and temporal regulation of a number of GTPases during myoblast fusion. What particular aspect each GTPase and its cognate GEF governs during fusion is not fully understood. This is further complicated by the possibility that one GEF could functionally, at least to a degree, substitute for one another. For example, in the mouse, Dock1 and Dock 5 appear to play functionally redundant roles in vivo (Laurin et al., 2008). This differs when compared to the requirement for Dock1 and Dock5 in myoblast fusion in zebrafish where both proteins are required equally for the fusion process (Moore et al., 2007) (Table 3). Better understanding of the intracellular behaviors that each pair regulates, as well as determining additional regulators, effectors and the cellular output of each GTPase will be important steps in understanding the mechanisms that underlie fusion.
In Drosophila, the identification of the site of fusion, marked by the actin focus (Richardson et al., 2007) precipitated the discovery of a large amount of data, resulting in new methodology for studying fusion mutants and changing the landscape of the field dramatically. Thus, identifying the site of fusion, which has been lacking in vertebrates, is an important step in moving forward in mouse and zebrafish alike. The analysis of one mutant in particular, Nap1, the mammalian homolog of Kette and member of the WAVE actin-remodeling complex (Rakeman and Anderson, 2006; Weiner et al., 2006) (Table 1), provides some indication that an equivalent structure exists (Nowak et al., 2009). Nap1 is required for myoblast fusion in vitro and appears to play a role in actin cytoskeletal remodeling at sites of contact between myoblasts (Table 3). Actin cytoskeletal remodeling was observed with the aid of an indirect marker of actin cytoskeletal dynamics: the pleckstrin homology (PH) domain of PLC-δ1 fused to GFP (PH∷GFP) (Tall et al., 2000). The PH domain of PLC-δ1 binds specifically to phosphatidylinositol 4,5-bisphosphate (PIP2), which is highly associated with sites of actin cytoskeletal remodeling (Coppolino et al., 2002; Insall and Weiner, 2001; Nowak et al., 2009; Rozelle et al., 2000; Tall et al., 2000). In contrast to the behavior of the PH∷GFP reporter in wild-type myoblasts, live imaging of fusing Nap1 knockdown myoblasts revealed that the PH∷GFP reporter perdured at sites of contact between myoblasts (Nowak et al., 2009). This is analogous to the persistence of the actin focus in kette mutants in Drosophila (Richardson et al., 2007), suggesting the conservation of Nap1 during myoblast fusion between mammals and Drosophila (Table 1) as well as placing Nap1 function at the step of membrane remodeling (Nowak et al., 2009).
The identification of an actin focus has not yet been unequivocally demonstrated in any vertebrate model; although actin has been found at proposed sites of fusion (Duan and Gallagher, 2009). Recent work has suggested that a transient accumulation of PIP2 may act as an indirect marker for the actin focus in mouse. Additionally, the localization of proteins known to be at the actin focus in Drosophila to this site, for example Dock1 (Nowak et al., 2009), provides further evidence that it is. However, whether PH∷GFP accumulations represent the same structure as the actin focus is still a matter of speculation. Nevertheless, this reporter provides new insight to membrane dynamics during myoblast fusion and offers a new avenue for research in the other model systems.
Vesicle trafficking at the site of fusion
TEM data has revealed vesicles at sites of fusion in Drosophila and mammalian model systems (Doberstein et al., 1997); hence, trafficking may be involved in membrane coalescence. However, the nature of these vesicles is unknown and their role in fusion is not understood. A recent publication suggests that endocytic recycling could be an important mechanism for the localization of proteins required for myoblast fusion (Doherty et al., 2008), suggesting a novel mechanism for the intracellular control of myoblast fusion.
Myoferlin, a member of the ferlin family and homolog of human dysferlin, is found at the interface between fusing myoblasts (Davis et al., 2002; Doherty et al., 2005). Myoferlin is highly expressed in developing skeletal muscle and plays a role in myoblast-myofiber fusion during development and regeneration (Doherty et al., 2005) (Figure 6), (Table 2). Recently, Myoferlin was shown to interact directly with the eps15 homology domain protein, EHD2 (Doherty et al., 2008). Members of the EHD family have been implicated in the endocytic recycling of membrane proteins (Daumke et al., 2007; Naslavsky and Caplan, 2005; Rapaport et al., 2006), thus, this interaction suggests a role for Myoferlin in recycling during myoblast fusion. Consistent with this, in myoferlin null myoblasts, transferrin was internalized normally, but displayed delayed endocytic recycling; transferrin was found in internal aggregations. Similarly, expression of a dominant negative EDH2 impaired myoblast fusion and resulted in the sequestration of Myoferlin in internal compartments, indicating a requirement in vitro for EHD2 in myoblast fusion and suggesting that improper trafficking of Myoferlin could be responsible for this defect (Doherty et al., 2008) (Figure 6), (Table 2).
Given the colocalization of Myoferlin and EHD2 to the site of fusion, it can be argued that the vesicles seen in ultrastructural analysis are from the endocytic compartment. That the recycling but not the internalization of transferrin is perturbed (Doherty et al., 2008) suggests the possibility that other receptors for myoblast fusion are similarly affected by the mutation of EHD2. Given this, it is easy to speculate that one function of EHD2 is in the recycling of the transmembrane proteins responsible for recognition and adhesion of myoblasts to one another. However, this has yet to be experimentally tested. Furthermore, ferlin proteins contain multiple C2 domains, which are capable of membrane association via their ability to bind phospholipids (Davis et al., 2002; Doherty et al., 2005); the C2 domains of myoferlin are most similar to those of synaptotagmin, which is thought to participate in the union of two independent membranes at nerve terminals (Hui et al., 2006; Tang et al., 2006). Taken together, this data suggests a role for myoferlin in the fusion of endocytic vesicles at the site of myoblast-myoblast fusion.
In Drosophila, Loner is localized at the site of fusion adjacent to the actin focus and does not appear to play a role in actin regulation at the site of fusion (Chen et al., 2003; Richardson et al., 2007). What role Loner is playing in myoblast fusion has been unclear, though it has been identified as a GEF for Arf6 in vitro. However, recent data in the mouse is beginning to shed light on the role of IQSec1, the mammalian ortholog of Loner, in myoblast fusion. IQSec1 (Brag2, GEP100) plays a conserved role in mammalian myoblast fusion: IQSec1 knockdown myoblasts exhibit a severe fusion defect in vitro (Tables 1 and 3). In vitro data in the mouse suggests that IQSec1 can act as a GEF for Arf6 (Dunphy et al., 2007; Pajcini et al., 2008). Consistent with the known roles of Arf6 in intracellular trafficking, paxillin, a component of focal adhesions, is improperly localized in IQSec1-knockdown myoblasts. Interestingly, the morphology of IQSec1-depleted myoblasts was distinct from that of the Dock180 knockdown myoblasts (Pajcini et al., 2008), supporting data from Drosophila that suggests Loner and Mbc have independent roles in myoblast fusion (Richardson et al., 2007).
The union of the membrane of two separate myoblasts to generate one multinucleate cell is typically considered the final step of fusion. For most fusing cell types, it is. However, the generation of a final, mature myofiber requires that the steps of fusion repeat until the final muscle size is achieved. Thus, an essential aspect of fusion is the ability of a post-fusion cell to relocate appropriate cell-surface proteins to the membrane and reset intracellular pathways within the myoblasts to prepare for additional rounds of fusion. Little is known about this process of resetting because, until recently, little was known about the proteins and mechanisms that underlie fusion. However, recent findings, which have been reviewed here, have demonstrated that this no longer is the case. The discovery, in recent years, that many vertebrate orthologs of known fusion genes in Drosophila play a role in vertebrate myoblast fusion (Tables 1 and 3) and that novel genes and mechanisms (Tables 2 and 3) are involved suggests that we know a good deal about the fusion process. Further insight, thus, will help us to begin to understand the mechanisms that need to be reset. This area of myoblast fusion biology remains unexplored, but only for now.
Perspectives
The studies discussed above have highlighted the cellular steps to myoblast fusion: migration, recognition and adhesion, membrane breakdown and ultimately incorporation of new nuclei (Figure 2). Research in Drosophila, zebrafish and mammals has demonstrated some degree of evolutionary conservation in the molecular regulation of these steps (Figure 6), (Tables 1 and 3). However, like many other developmental processes, there also exist many points of divergence between the different systems, which illustrates the need for multiple models to understand the complex series of events that occur during myoblast fusion (Figure 6), (Tables 2 and 3). In addition, the increasing complexity and muscle diversity as one moves from fly to zebrafish to mouse underscores the challenges and exciting potential for future studies. Comparison of data gathered across multiple models will identify gaps in knowledge and can be used to guide future studies. For example, a large number of fusion genes have been identified in Drosophila that remain to be tested in zebrafish and mouse (Figure 6). Similarly, technologies successfully used in one system, TEM and time-lapse microscopy, can be modified for use in other models to further our knowledge of the events that underlie fusion. Some of the areas where we can expect to witness significant advances in the coming years include the identification of additional components of the fusion pathway, better evaluation of the interaction between the components that have already been identified, and use of sophisticated microscopy to delve further into the cellular and sub-cellular details that underlie myoblast fusion. The Drosophila, zebrafish and mammalian systems each have unique capabilities, ranging from specialized imaging approaches to genetic screens and genetic manipulation; only through the incorporation of data from all these systems can we expect to understand the complex process of myoblast fusion in its entirety.
Acknowledgments
We thank Grace Pavlath, Emanuela Gussoni and Carmen Birchmeier for providing panels for our figures (panel D for Figure 1; panels F, G and D, E Figure 5, respectively). We also would like to thank Brian Richardson and the members of the Baylies lab for helpful discussions and comments on the manuscript. This work was supported by the Sloan Kettering Institute and NIH grant GM 78318 to M.B. and the Institute of Molecular and Cell Biology and the Agency for Science Technology and Research (A*STAR), Singapore, to S.R.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Artero RD, Castanon I, Baylies MK. The immunoglobulin-like protein Hibris functions as a dose-dependent regulator of myoblast fusion and is differentially controlled by Ras and Notch signaling. Development. 2001;128:4251–64. doi: 10.1242/dev.128.21.4251. [DOI] [PubMed] [Google Scholar]
- Bae GU, Gaio U, Yang YJ, Lee HJ, Kang JS, Krauss RS. Regulation of myoblast motility and fusion by the CXCR4-associated sialomucin, CD164. J Biol Chem. 2008;283:8301–9. doi: 10.1074/jbc.M706730200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balagopalan L, Chen MH, Geisbrecht ER, Abmayr SM. The CDM superfamily protein MBC directs myoblast fusion through a mechanism that requires phosphatidylinositol 3,4,5-triphosphate binding but is independent of direct interaction with DCrk. Mol Cell Biol. 2006;26:9442–55. doi: 10.1128/MCB.00016-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barletta GM, Kovari IA, Verma RK, Kerjaschki D, Holzman LB. Nephrin and Neph1 co-localize at the podocyte foot process intercellular junction and form cis hetero-oligomers. J Biol Chem. 2003;278:19266–71. doi: 10.1074/jbc.M301279200. [DOI] [PubMed] [Google Scholar]
- Bate M. The embryonic development of larval muscles in Drosophila. Development. 1990;110:791–804. doi: 10.1242/dev.110.3.791. [DOI] [PubMed] [Google Scholar]
- Baxendale S, Davison C, Muxworthy C, Wolff C, Ingham PW, Roy S. The B-cell maturation factor Blimp-1 specifies vertebrate slow-twitch muscle fiber identity in response to Hedgehog signaling. Nat Genet. 2004;36:88–93. doi: 10.1038/ng1280. [DOI] [PubMed] [Google Scholar]
- Baylies MK, Bate M, Ruiz Gomez M. Myogenesis: a view from Drosophila. Cell. 1998;93:921–7. doi: 10.1016/s0092-8674(00)81198-8. [DOI] [PubMed] [Google Scholar]
- Beckett K, Baylies MK. The development of the Drosophila larval body wall muscles. Int Rev Neurobiol. 2006;75:55–70. doi: 10.1016/S0074-7742(06)75003-6. [DOI] [PubMed] [Google Scholar]
- Beckett K, Baylies MK. 3D analysis of founder cell and fusion competent myoblast arrangements outlines a new model of myoblast fusion. Dev Biol. 2007;309:113–25. doi: 10.1016/j.ydbio.2007.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckett K, B MK. International Review of Neurobiology. Vol. 75. Elsevier; Academic Press; San Diego: 2006. The Development of Drosophila larval body wall muscle The Fly Neuromuscular Junction: Structure and Function; pp. 55–70. [DOI] [PubMed] [Google Scholar]
- Bellanger JM, Lazaro JB, Diriong S, Fernandez A, Lamb N, Debant A. The two guanine nucleotide exchange factor domains of Trio link the Rac1 and the RhoA pathways in vivo. Oncogene. 1998;16:147–52. doi: 10.1038/sj.onc.1201532. [DOI] [PubMed] [Google Scholar]
- Berger S, Schafer G, Kesper DA, Holz A, Eriksson T, Palmer RH, Beck L, Klambt C, Renkawitz-Pohl R, Onel SF. WASP and SCAR have distinct roles in activating the Arp2/3 complex during myoblast fusion. J Cell Sci. 2008;121:1303–13. doi: 10.1242/jcs.022269. [DOI] [PubMed] [Google Scholar]
- Blagden CS, Currie PD, Ingham PW, Hughes SM. Notochord induction of zebrafish slow muscle mediated by Sonic hedgehog. Genes & Development. 1997;11:2163–75. doi: 10.1101/gad.11.17.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blangy A, Vignal E, Schmidt S, Debant A, Gauthier-Rouvière C, Fort P. TrioGEF1 controls Rac- and Cdc42-dependent cell structures through the direct activation of rhoG. J Cell Sci. 2000;113(Pt 4):729–39. doi: 10.1242/jcs.113.4.729. [DOI] [PubMed] [Google Scholar]
- Bondesen BA, Jones KA, Glasgow WC, Pavlath GK. Inhibition of myoblast migration by prostacyclin is associated with enhanced cell fusion. The FASEB Journal. 2007;21:3338–3345. doi: 10.1096/fj.06-7070com. [DOI] [PubMed] [Google Scholar]
- Bour BA, Chakravarti M, West JM, Abmayr SM. Drosophila SNS, a member of the immunoglobulin superfamily that is essential for myoblast fusion. Genes Dev. 2000;14:1498–511. [PMC free article] [PubMed] [Google Scholar]
- Bryson-Richardson RJ, Daggett DF, Cortes F, Neyt C, Keenan DG, Currie PD. Myosin heavy chain expression in zebrafish and slow muscle composition. Dev Dyn. 2005;233:1018–22. doi: 10.1002/dvdy.20380. [DOI] [PubMed] [Google Scholar]
- Buckingham M, Relaix F. The role of Pax genes in the development of tissues and organs: Pax3 and Pax7 regulate muscle progenitor cell functions. Annu Rev Cell Dev Biol. 2007;23:645–73. doi: 10.1146/annurev.cellbio.23.090506.123438. [DOI] [PubMed] [Google Scholar]
- Campion D. The muscle satellite cell: a review. Int Rev Cytol. 1984:225–51. doi: 10.1016/s0074-7696(08)62444-4. [DOI] [PubMed] [Google Scholar]
- Carmena A, Baylies MK. The Development of the Drosophila Larval Somatic Musculature. In: Sink H, editor. Drosophila Muscle Development. Sink Landes Press; 2006. pp. 79–92. [Google Scholar]
- Charrasse S, Comunale F, Fortier M, Portales-Casamar E, Debant A, Gauthier-Rouvière C. M-cadherin activates Rac1 GTPase through the Rho-GEF trio during myoblast fusion. Mol Biol Cell. 2007;18:1734–43. doi: 10.1091/mbc.E06-08-0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AE, Ginty DD, Fan CM. Protein kinase A signalling via CREB controls myogenesis induced by Wnt proteins. Nature. 2005;433:317–22. doi: 10.1038/nature03126. [DOI] [PubMed] [Google Scholar]
- Chen EH, editor. Cell Fusion: Overviews and Methods. Humana Press; 2008. [Google Scholar]
- Chen EH, Grote E, Mohler W, Vignery A. Cell-cell fusion. FEBS Lett. 2007;581:2181–93. doi: 10.1016/j.febslet.2007.03.033. [DOI] [PubMed] [Google Scholar]
- Chen EH, Olson EN. Antisocial, an intracellular adaptor protein, is required for myoblast fusion in Drosophila. Dev Cell. 2001;1:705–15. doi: 10.1016/s1534-5807(01)00084-3. [DOI] [PubMed] [Google Scholar]
- Chen EH, Olson EN. Unveiling the mechanisms of cell-cell fusion. Science. 2005;308:369–73. doi: 10.1126/science.1104799. [DOI] [PubMed] [Google Scholar]
- Chen EH, Pryce BA, Tzeng JA, Gonzalez GA, Olson EN. Control of myoblast fusion by a guanine nucleotide exchange factor, loner, and its effector ARF6. Cell. 2003;114:751–62. doi: 10.1016/s0092-8674(03)00720-7. [DOI] [PubMed] [Google Scholar]
- Chen F, Ma L, Parrini MC, Mao X, Lopez M, Wu C, Marks PW, Davidson L, Kwiatkowski DJ, Kirchhausen T, Orkin SH, Rosen FS, Mayer BJ, Kirschner MW, Alt FW. Cdc42 is required for PIP(2)-induced actin polymerization and early development but not for cell viability. Curr Biol. 2000;10:758–65. doi: 10.1016/s0960-9822(00)00571-6. [DOI] [PubMed] [Google Scholar]
- Christ B, O CP. Early stages of chick somite development. Anat Embryol (Berl) 1995:381–396. doi: 10.1007/BF00304424. [DOI] [PubMed] [Google Scholar]
- Cole F, Zhang W, Geyra A, Kang JS, Krauss RS. Positive regulation of myogenic bHLH factors and skeletal muscle development by the cell surface receptor CDO. Developmental Cell. 2004;7:843–54. doi: 10.1016/j.devcel.2004.10.009. [DOI] [PubMed] [Google Scholar]
- Constantin B, Imbert N, Besse C, Cognard C, Raymond G. Cultured rat skeletal muscle cells treated with cytochalasin exhibit normal dystrophin expression and intracellular free calcium control. Biology of the Cell. 1995;85:125–135. doi: 10.1016/0248-4900(96)85273-7. [DOI] [PubMed] [Google Scholar]
- Coppolino MG, Dierckman R, Loijens J, Collins RF, Pouladi M, Jongstra-Bilen J, Schreiber AD, Trimble WS, Anderson R, Grinstein S. Inhibition of phosphatidylinositol-4-phosphate 5-kinase Ialpha impairs localized actin remodeling and suppresses phagocytosis. J Biol Chem. 2002;277:43849–57. doi: 10.1074/jbc.M209046200. [DOI] [PubMed] [Google Scholar]
- Coué M, Brenner SL, Spector I, Korn ED. Inhibition of actin polymerization by latrunculin A. FEBS Lett. 1987;213:316–8. doi: 10.1016/0014-5793(87)81513-2. [DOI] [PubMed] [Google Scholar]
- D'Souza-Schorey C, Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nat Rev Mol Cell Biol. 2006;7:347–58. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- Daumke O, Lundmark R, Vallis Y, Martens S, Butler P, McMahon H. Architectural and mechanistic insights into an EHD ATPase involved in membrane remodelling. Nature. 2007;449:923–927. doi: 10.1038/nature06173. [DOI] [PubMed] [Google Scholar]
- Davis DB, Doherty KR, Delmonte AJ, McNally EM. Calcium-sensitive phospholipid binding properties of normal and mutant ferlin C2 domains. J Biol Chem. 2002;277:22883–8. doi: 10.1074/jbc.M201858200. [DOI] [PubMed] [Google Scholar]
- Debant A, Serra-Pagès C, Seipel K, O'Brien S, Tang M, Park SH, Streuli M. The multidomain protein Trio binds the LAR transmembrane tyrosine phosphatase, contains a protein kinase domain, and has separate rac-specific and rho-specific guanine nucleotide exchange factor domains. Proc Natl Acad Sci USA. 1996;93:5466–71. doi: 10.1073/pnas.93.11.5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devoto SH, Melançon E, Eisen JS, Westerfield M. Identification of separate slow and fast muscle precursor cells in vivo, prior to somite formation. Development. 1996;122:3371–80. doi: 10.1242/dev.122.11.3371. [DOI] [PubMed] [Google Scholar]
- Devoto SH, Stoiber W, Hammond CL, Steinbacher P, Haslett JR, Barresi MJF, Patterson SE, Adiarte EG, Hughes SM. Generality of vertebrate developmental patterns: evidence for a dermomyotome in fish. Evolution & Development. 2006;8:101–110. doi: 10.1111/j.1525-142X.2006.05079.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhawan J, Helfman DM. Modulation of acto-myosin contractility in skeletal muscle myoblasts uncouples growth arrest from differentiation. J Cell Sci. 2004;117:3735–48. doi: 10.1242/jcs.01197. [DOI] [PubMed] [Google Scholar]
- Dietrich S, Schubert FR, Healy C, Sharpe PT, Lumsden A. Specification of the hypaxial musculature. Development. 1998;125:2235–49. doi: 10.1242/dev.125.12.2235. [DOI] [PubMed] [Google Scholar]
- Doberstein SK, Fetter RD, Mehta AY, Goodman CS. Genetic analysis of myoblast fusion: blown fuse is required for progression beyond the prefusion complex. J Cell Biol. 1997;136:1249–61. doi: 10.1083/jcb.136.6.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty KR, Cave A, Davis DB, Delmonte AJ, Posey A, Earley JU, Hadhazy M, McNally EM. Normal myoblast fusion requires myoferlin. Development. 2005;132:5565–75. doi: 10.1242/dev.02155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty KR, Demonbreun AR, Wallace GQ, Cave A, Posey AD, Heretis K, Pytel P, McNally EM. The endocytic recycling protein EHD2 interacts with myoferlin to regulate myoblast fusion. J Biol Chem. 2008;283:20252–60. doi: 10.1074/jbc.M802306200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donalies M, Cramer M, Ringwald M, Starzinski-Powitz A. Expression of M-cadherin, a member of the cadherin multigene family, correlates with differentiation of skeletal muscle cells. Proc Natl Acad Sci USA. 1991;88:8024–8. doi: 10.1073/pnas.88.18.8024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoviel DB, Freed DD, Vogel H, Potter DG, Hawkins E, Barrish JP, Mathur BN, Turner CA, Geske R, Montgomery CA, Starbuck M, Brandt M, Gupta A, Ramirez-Solis R, Zambrowicz BP, Powell DR. Proteinuria and perinatal lethality in mice lacking NEPH1, a novel protein with homology to NEPHRIN. Mol Cell Biol. 2001;21:4829–36. doi: 10.1128/MCB.21.14.4829-4836.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du SJ, Devoto SH, Westerfield M, Moon RT. Positive and negative regulation of muscle cell identity by members of the hedgehog and TGF-beta gene families. J Cell Biol. 1997;139:145–56. doi: 10.1083/jcb.139.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan H, Skeath JB, Nguyen HT. Drosophila Lame duck, a novel member of the Gli superfamily, acts as a key regulator of myogenesis by controlling fusion-competent myoblast development. Development. 2001;128:4489–500. doi: 10.1242/dev.128.22.4489. [DOI] [PubMed] [Google Scholar]
- Duan R, Gallagher PJ. Dependence of myoblast fusion on a cortical actin wall and nonmuscle myosin IIA. Dev Biol. 2009;325:374–85. doi: 10.1016/j.ydbio.2008.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duelli D, Lazebnik Y. Cell-to-cell fusion as a link between viruses and cancer. Nat Rev Cancer. 2007;7:968–76. doi: 10.1038/nrc2272. [DOI] [PubMed] [Google Scholar]
- Dunphy JL, Ye K, Casanova JE. Nuclear functions of the Arf guanine nucleotide exchange factor BRAG2. Traffic. 2007;8:661–72. doi: 10.1111/j.1600-0854.2007.00561.x. [DOI] [PubMed] [Google Scholar]
- Dworak HA, Charles MA, Pellerano LB, Sink H. Characterization of Drosophila hibris, a gene related to human nephrin. Development. 2001;128:4265–76. doi: 10.1242/dev.128.21.4265. [DOI] [PubMed] [Google Scholar]
- Dyer N, Rebollo E, Dominguez P, Elkhatib N, Chavrier P, Daviet L, Gonzalez C, Gonzalez-Gaitan M. Spermatocyte cytokinesis requires rapid membrane addition mediated by ARF6 on central spindle recycling endosomes. Development. 2007;134:4437–47. doi: 10.1242/dev.010983. [DOI] [PubMed] [Google Scholar]
- Erickson MR, Galletta BJ, Abmayr SM. Drosophila myoblast city encodes a conserved protein that is essential for myoblast fusion, dorsal closure, and cytoskeletal organization. J Cell Biol. 1997;138:589–603. doi: 10.1083/jcb.138.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada B, Choe SE, Gisselbrecht SS, Michaud S, Raj L, Busser BW, Halfon MS, Church GM, Michelson AM. An integrated strategy for analyzing the unique developmental programs of different myoblast subtypes. PLoS Genet. 2006;2:e16. doi: 10.1371/journal.pgen.0020016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada B, Maeland AD, Gisselbrecht SS, Bloor JW, Brown NH, Michelson AM. The MARVEL domain protein, Singles Bar, is required for progression past the pre-fusion complex stage of myoblast fusion. Dev Biol. 2007;307:328–39. doi: 10.1016/j.ydbio.2007.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasch M. Controls in patterning and diversification of somatic muscles during Drosophila embryogenesis. Curr Opin Genet Dev. 1999;9:522–9. doi: 10.1016/s0959-437x(99)00014-3. [DOI] [PubMed] [Google Scholar]
- Fukuda N, Yomogida K, Okabe M, Touhara K. Functional characterization of a mouse testicular olfactory receptor and its role in chemosensing and in regulation of sperm motility. J Cell Sci. 2004;117:5835–45. doi: 10.1242/jcs.01507. [DOI] [PubMed] [Google Scholar]
- Furlong EE, Andersen EC, Null B, White KP, Scott MP. Patterns of gene expression during Drosophila mesoderm development. Science. 2001;293:1629–33. doi: 10.1126/science.1062660. [DOI] [PubMed] [Google Scholar]
- Galbiati F, Volonte D, Engelman JA, Scherer PE, Lisanti MP. Targeted down-regulation of caveolin-3 is sufficient to inhibit myotube formation in differentiating C2C12 myoblasts. Transient activation of p38 mitogen-activated protein kinase is required for induction of caveolin-3 expression and subsequent myotube formation. J Biol Chem. 1999;274:30315–21. doi: 10.1074/jbc.274.42.30315. [DOI] [PubMed] [Google Scholar]
- Galletta BJ, Chakravarti M, Banerjee R, Abmayr SM. SNS: Adhesive properties, localization requirements and ectodomain dependence in S2 cells and embryonic myoblasts. Mech Dev. 2004;121:1455–68. doi: 10.1016/j.mod.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Geetha-Loganathan P, Nimmagadda S, Scaal M, Huang R, B C. Wnt signaling in somite development. Ann Anat. 2008;190:208–22. doi: 10.1016/j.aanat.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Geisbrecht ER, Haralalka S, Swanson SK, Florens L, Washburn MP, Abmayr SM. Drosophila ELMO/CED-12 interacts with Myoblast city to direct myoblast fusion and ommatidial organization. Dev Biol. 2008;314:137–49. doi: 10.1016/j.ydbio.2007.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George-Weinstein M, Gerhart J, Blitz J, Simak E, Knudsen KA. N-cadherin promotes the commitment and differentiation of skeletal muscle precursor cells. Developmental Biology. 1997;185:14–24. doi: 10.1006/dbio.1997.8542. [DOI] [PubMed] [Google Scholar]
- Gerke P, Huber TB, Sellin L, Benzing T, Walz G. Homodimerization and heterodimerization of the glomerular podocyte proteins nephrin and NEPH1. J Am Soc Nephrol. 2003;14:918–26. doi: 10.1097/01.asn.0000057853.05686.89. [DOI] [PubMed] [Google Scholar]
- Gildor B, Massarwa R, Shilo BZ, Schejter ED. The SCAR and WASp nucleation-promoting factors act sequentially to mediate Drosophila myoblast fusion. EMBO Rep. 2009;10:1043–50. doi: 10.1038/embor.2009.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin CA, Kafadar KA, Pavlath GK. MOR23 promotes muscle regeneration and regulates cell adhesion and migration. Dev Cel. 2009 doi: 10.1016/j.devcel.2009.09.004. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros J, Manceau M, Thomé V, Marcelle C. A common somitic origin for embryonic muscle progenitors and satellite cells. Nature. 2005;435:954–958. doi: 10.1038/nature03572. [DOI] [PubMed] [Google Scholar]
- Grounds MD, White JD, Rosenthal N, Bogoyevitch MA. The role of stem cells in skeletal and cardiac muscle repair. J Histochem Cytochem. 2002;50:589–610. doi: 10.1177/002215540205000501. [DOI] [PubMed] [Google Scholar]
- Hakeda-Suzuki S, Ng J, Tzu J, Dietzl G, Sun Y, Harms M, Nardine T, Luo L, Dickson BJ. Rac function and regulation during Drosophila development. Nature. 2002;416:438–42. doi: 10.1038/416438a. [DOI] [PubMed] [Google Scholar]
- Hammond CL, Hinits Y, Osborn DPS, Minchin JEN, Tettamanti G, Hughes SM. Signals and myogenic regulatory factors restrict pax3 and pax7 expression to dermomyotome-like tissue in zebrafish. Dev Biol. 2007;302:504–21. doi: 10.1016/j.ydbio.2006.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa H, Kiyokawa E, Tanaka S, Nagashima K, Gotoh N, Shibuya M, Kurata T, Matsuda M. DOCK180, a major CRK-binding protein, alters cell morphology upon translocation to the cell membrane. Mol Cell Biol. 1996;16:1770–6. doi: 10.1128/mcb.16.4.1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawke TJ, Garry DJ. Myogenic satellite cells: physiology to molecular biology. J Appl Physiol. 2001;91:534–51. doi: 10.1152/jappl.2001.91.2.534. [DOI] [PubMed] [Google Scholar]
- Heasman SJ, Ridley AJ. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat Rev Mol Cell Biol. 2008;9:690–701. doi: 10.1038/nrm2476. [DOI] [PubMed] [Google Scholar]
- Hollnagel A, Grund C, Franke WW, Arnold HH. The cell adhesion molecule M-cadherin is not essential for muscle development and regeneration. Mol Cell Biol. 2002;22:4760–70. doi: 10.1128/MCB.22.13.4760-4770.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollway GE, Bryson-Richardson RJ, Berger S, Cole NJ, Hall TE, Currie PD. Whole-somite rotation generates muscle progenitor cell compartments in the developing zebrafish embryo. Developmental Cell. 2007;12:207–19. doi: 10.1016/j.devcel.2007.01.001. [DOI] [PubMed] [Google Scholar]
- Horsley V, Friday BB, Matteson S, Kegley KM, Gephart J, Pavlath GK. Regulation of the growth of multinucleated muscle cells by an NFATC2-dependent pathway. J Cell Biol. 2001;153:329–38. doi: 10.1083/jcb.153.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsley V, Jansen KM, Mills ST, Pavlath GK. IL-4 acts as a myoblast recruitment factor during mammalian muscle growth. Cell. 2003;113:483–94. doi: 10.1016/s0092-8674(03)00319-2. [DOI] [PubMed] [Google Scholar]
- Horsley V, Pavlath GK. Forming a Multinucleated Cell: Molecules That Regulate Myoblast Fusion. Cells Tissues Organs. 2004;176:67–78. doi: 10.1159/000075028. [DOI] [PubMed] [Google Scholar]
- Hui E, Bai J, Chapman ER. Ca2+-triggered simultaneous membrane penetration of the tandem C2-domains of synaptotagmin I. Biophys J. 2006;91:1767–77. doi: 10.1529/biophysj.105.080325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra N, Pollitt A, Insall RH. Regulation of actin assembly by SCAR/WAVE proteins. Biochem Soc Trans. 2005;33:1243–6. doi: 10.1042/BST0331243. [DOI] [PubMed] [Google Scholar]
- Ikeya M, Takada S. Wnt signaling from the dorsal neural tube is required for the formation of the medial dermomyotome. Development. 1998;125:4969–76. doi: 10.1242/dev.125.24.4969. [DOI] [PubMed] [Google Scholar]
- Insall RH, Weiner OD. PIP3, PIP2, and cell movement--similar messages, different meanings? Developmental Cell. 2001;1:743–7. doi: 10.1016/s1534-5807(01)00086-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irintchev A, Zeschnigk M, Starzinski-Powitz A, Wernig A. Expression pattern of M-cadherin in normal, denervated, and regenerating mouse muscles. Dev Dyn. 1994;199:326–37. doi: 10.1002/aja.1001990407. [DOI] [PubMed] [Google Scholar]
- Jansen KM, Pavlath GK. Mannose receptor regulates myoblast motility and muscle growth. J Cell Biol. 2006;174:403–13. doi: 10.1083/jcb.200601102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JS, Mulieri PJ, Hu Y, Taliana L, Krauss RS. BOC, an Ig superfamily member, associates with CDO to positively regulate myogenic differentiation. EMBO J. 2002;21:114–24. doi: 10.1093/emboj/21.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura K, Takano K, Suetsugu S, Kurisu S, Yamazaki D, Miki H, Takenawa T, Endo T. N-WASP and WAVE2 acting downstream of phosphatidylinositol 3-kinase are required for myogenic cell migration induced by hepatocyte growth factor. J Biol Chem. 2004;279:54862–71. doi: 10.1074/jbc.M408057200. [DOI] [PubMed] [Google Scholar]
- Kesper DA, Stute C, Buttgereit D, Kreiskother N, Vishnu S, Fischbach KF, Renkawitz-Pohl R. Myoblast fusion in Drosophila melanogaster is mediated through a fusion-restricted myogenic-adhesive structure (FuRMAS) Dev Dyn. 2007;236:404–15. doi: 10.1002/dvdy.21035. [DOI] [PubMed] [Google Scholar]
- Khoshnoodi J, Sigmundsson K, Ofverstedt LG, Skoglund U, Obrink B, Wartiovaara J, Tryggvason K. Nephrin promotes cell-cell adhesion through homophilic interactions. Am J Pathol. 2003;163:2337–46. doi: 10.1016/S0002-9440(10)63590-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Shilagardi K, Zhang S, Hong SN, Sens KL, Bo J, Gonzalez GA, Chen EH. A critical function for the actin cytoskeleton in targeted exocytosis of prefusion vesicles during myoblast fusion. Dev Cell. 2007;12:571–86. doi: 10.1016/j.devcel.2007.02.019. [DOI] [PubMed] [Google Scholar]
- Kiyokawa E, Hashimoto Y, Kobayashi S, Sugimura H, Kurata T, Matsuda M. Activation of Rac1 by a Crk SH3-binding protein, DOCK180. Genes Dev. 1998;12:3331–6. doi: 10.1101/gad.12.21.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knirr S, Azpiazu N, Frasch M. The role of the NK-homeobox gene slouch (S59) in somatic muscle patterning. Development. 1999;126:4525–35. doi: 10.1242/dev.126.20.4525. [DOI] [PubMed] [Google Scholar]
- Kocherlakota KS, Wu JM, McDermott J, Abmayr SM. Analysis of the cell adhesion molecule sticks-and-stones reveals multiple redundant functional domains, protein-interaction motifs and phosphorylated tyrosines that direct myoblast fusion in Drosophila melanogaster. Genetics. 2008;178:1371–83. doi: 10.1534/genetics.107.083808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer-Zucker AG, Wiessner S, Jensen AM, Drummond IA. Organization of the pronephric filtration apparatus in zebrafish requires Nephrin, Podocin and the FERM domain protein Mosaic eyes. Developmental Biology. 2005;285:316–29. doi: 10.1016/j.ydbio.2005.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurin M, Fradet N, Blangy A, Hall A, Vuori K, Côté J. The atypical Rac activator Dock180 (Dock1) regulates myoblast fusion in vivo. Proc Natl Acad Sci USA. 2008 doi: 10.1073/pnas.0805546105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis KE, Currie PD, Roy S, Schauerte H, Haffter P, Ingham PW. Control of muscle cell-type specification in the zebrafish embryo by Hedgehog signalling. Dev Biol. 1999;216:469–80. doi: 10.1006/dbio.1999.9519. [DOI] [PubMed] [Google Scholar]
- Li Z, Mericskay M, Agbulut O, Butler-Browne G, Carlsson L, Thornell LE, Babinet C, Paulin D. Desmin is essential for the tensile strength and integrity of myofibrils but not for myogenic commitment, differentiation, and fusion of skeletal muscle. J Cell Biol. 1997;139:129–44. doi: 10.1083/jcb.139.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Liao YJ, Jan LY, Jan YN. Distinct morphogenetic functions of similar small GTPases: Drosophila Drac1 is involved in axonal outgrowth and myoblast fusion. Genes & Development. 1994;8:1787–802. doi: 10.1101/gad.8.15.1787. [DOI] [PubMed] [Google Scholar]
- Massarwa R, Carmon S, Shilo BZ, Schejter ED. WIP/WASp-based actin-polymerization machinery is essential for myoblast fusion in Drosophila. Dev Cell. 2007;12:557–69. doi: 10.1016/j.devcel.2007.01.016. [DOI] [PubMed] [Google Scholar]
- McDermott A, Gustafsson M, Elsam T, Hui CC, Emerson CP, Borycki AG. Gli2 and Gli3 have redundant and context-dependent function in skeletal muscle formation. Development. 2005;132:345–57. doi: 10.1242/dev.01537. [DOI] [PubMed] [Google Scholar]
- Menon SD, Chia W. Drosophila rolling pebbles: a multidomain protein required for myoblast fusion that recruits D-Titin in response to the myoblast attractant. Dumbfounded Dev Cell. 2001;1:691–703. doi: 10.1016/s1534-5807(01)00075-2. [DOI] [PubMed] [Google Scholar]
- Menon SD, Osman Z, Chenchill K, Chia W. A positive feedback loop between Dumbfounded and Rolling pebbles leads to myotube enlargement in Drosophila. J Cell Biol. 2005;169:909–20. doi: 10.1083/jcb.200501126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki H, Takenawa T. Regulation of actin dynamics by WASP family proteins. J Biochem. 2003;134:309–13. doi: 10.1093/jb/mvg146. [DOI] [PubMed] [Google Scholar]
- Mohler WA, Simske JS, Williams-Masson EM, Hardin JD, White JG. Dynamics and ultrastructure of developmental cell fusions in the Caenorhabditis elegans hypodermis. Curr Biol. 1998;8:1087–90. doi: 10.1016/s0960-9822(98)70447-6. [DOI] [PubMed] [Google Scholar]
- Moore CA, Parkin CA, Bidet Y, Ingham PW. A role for the Myoblast city homologues Dock1 and Dock5 and the adaptor proteins Crk and Crk-like in zebrafish myoblast fusion. Development. 2007;134:3145–53. doi: 10.1242/dev.001214. [DOI] [PubMed] [Google Scholar]
- Moore R, Walsh FS. The cell adhesion molecule M-cadherin is specifically expressed in developing and regenerating, but not denervated skeletal muscle. Development. 1993;117:1409–20. doi: 10.1242/dev.117.4.1409. [DOI] [PubMed] [Google Scholar]
- Morgan JE, Partridge TA. Muscle satellite cells. Int J Biochem Cell Biol. 2003;35:1151–6. doi: 10.1016/s1357-2725(03)00042-6. [DOI] [PubMed] [Google Scholar]
- Mylona E, Jones KA, Mills ST, Pavlath GK. CD44 regulates myoblast migration and differentiation. Journal of Cellular Physiology. 2006;209:314–321. doi: 10.1002/jcp.20724. [DOI] [PubMed] [Google Scholar]
- Naslavsky N, Caplan S. C-terminal EH-domain-containing proteins: consensus for a role in endocytic trafficking, EH? J Cell Sci. 2005;118:4093–101. doi: 10.1242/jcs.02595. [DOI] [PubMed] [Google Scholar]
- Nolan KM, Barrett K, Lu Y, Hu KQ, Vincent S, Settleman J. Myoblast city, the Drosophila homolog of DOCK180/CED-5, is required in a Rac signaling pathway utilized for multiple developmental processes. Genes & Development. 1998;12:3337–42. doi: 10.1101/gad.12.21.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak SJ, Nahirney PC, Hadjantonakis AK, Baylies MK. Nap1-mediated actin remodeling is essential for mammalian myoblast fusion. J Cell Sci. 2009;122:3282–93. doi: 10.1242/jcs.047597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien SP, Seipel K, Medley QG, Bronson R, Segal R, Streuli M. Skeletal muscle deformity and neuronal disorder in Trio exchange factor-deficient mouse embryos. Proc Natl Acad Sci USA. 2000;97:12074–8. doi: 10.1073/pnas.97.22.12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtake Y, Tojo H, Seiki M. Multifunctional roles of MT1-MMP in myofiber formation and morphostatic maintenance of skeletal muscle. J Cell Sci. 2006;119:3822–32. doi: 10.1242/jcs.03158. [DOI] [PubMed] [Google Scholar]
- Onel SF, Renkawitz-Pohl R. FuRMAS: triggering myoblast fusion in Drosophila. Dev Dyn. 2009;238:1513–25. doi: 10.1002/dvdy.21961. [DOI] [PubMed] [Google Scholar]
- Oren-Suissa M, Podbilewicz B. Cell fusion during development. Trends Cell Biol. 2007;17:537–46. doi: 10.1016/j.tcb.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Pajcini KV, Pomerantz JH, Alkan O, Doyonnas R, Blau HM. Myoblasts and macrophages share molecular components that contribute to cell-cell fusion. The Journal of Cell Biology. 2008;180:1005–19. doi: 10.1083/jcb.200707191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pätäri-Sampo A, Ihalmo P, Holthöfer H. Molecular basis of the glomerular filtration: nephrin and the emerging protein complex at the podocyte slit diaphragm. Ann Med. 2006;38:483–92. doi: 10.1080/07853890600978149. [DOI] [PubMed] [Google Scholar]
- Podbilewicz B. Cell fusion. WormBook; 2006. pp. 1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourquié O, Fan CM, Coltey M, Hirsinger E, Watanabe Y, Bréant C, Francis-West P, Brickell P, Tessier-Lavigne M, Le Douarin NM. Lateral and axial signals involved in avian somite patterning: a role for BMP4. Cell. 1996;84:461–71. doi: 10.1016/s0092-8674(00)81291-x. [DOI] [PubMed] [Google Scholar]
- Pownall ME, Gustafsson MK, Emerson CP. Myogenic regulatory factors and the specification of muscle progenitors in vertebrate embryos. Annu Rev Cell Dev Biol. 2002;18:747–83. doi: 10.1146/annurev.cellbio.18.012502.105758. [DOI] [PubMed] [Google Scholar]
- Radice GL, Rayburn H, Matsunami H, Knudsen KA, Takeichi M, Hynes RO. Developmental defects in mouse embryos lacking N-cadherin. Developmental Biology. 1997;181:64–78. doi: 10.1006/dbio.1996.8443. [DOI] [PubMed] [Google Scholar]
- Rakeman AS, Anderson KV. Axis specification and morphogenesis in the mouse embryo require Nap1, a regulator of WAVE-mediated actin branching. Development. 2006;133:3075–83. doi: 10.1242/dev.02473. [DOI] [PubMed] [Google Scholar]
- Rapaport D, Auerbach W, Naslavsky N, Pasmanik-Chor M, Galperin E, Fein A, Caplan S, Joyner AL, Horowitz M. Recycling to the plasma membrane is delayed in EHD1 knockout mice. Traffic. 2006;7:52–60. doi: 10.1111/j.1600-0854.2005.00359.x. [DOI] [PubMed] [Google Scholar]
- Rau A, Buttgereit D, Holz A, Fetter R, Doberstein SK, Paululat A, Staudt N, Skeath J, Michelson AM, Renkawitz-Pohl R. rolling pebbles (rols) is required in Drosophila muscle precursors for recruitment of myoblasts for fusion. Development. 2001;128:5061–73. doi: 10.1242/dev.128.24.5061. [DOI] [PubMed] [Google Scholar]
- Relaix F, Rocancourt D, Mansouri A, Buckingham M. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature. 2005;435:948–53. doi: 10.1038/nature03594. [DOI] [PubMed] [Google Scholar]
- Richardson B, Beckett K, Baylies M. Visualizing new dimensions in Drosophila myoblast fusion. Bioessays. 2008a;30:423–31. doi: 10.1002/bies.20756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson BE, Beckett K, Nowak SJ, Baylies MK. SCAR/WAVE and Arp2/3 are crucial for cytoskeletal remodeling at the site of myoblast fusion. Development. 2007;134:4357–67. doi: 10.1242/dev.010678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson BE, Nowak SJ, Baylies MK. Myoblast fusion in fly and vertebrates: new genes, new processes and new perspectives. Traffic. 2008b;9:1050–9. doi: 10.1111/j.1600-0854.2008.00756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen GD, Sanes JR, LaChance R, Cunningham JM, Roman J, Dean DC. Roles for the integrin VLA-4 and its counter receptor VCAM-1 in myogenesis. Cell. 1992;69:1107–19. doi: 10.1016/0092-8674(92)90633-n. [DOI] [PubMed] [Google Scholar]
- Roy S, VijayRaghavan K. Muscle pattern diversification in Drosophila: the story of imaginal myogenesis. Bioessays. 1999;21:486–98. doi: 10.1002/(SICI)1521-1878(199906)21:6<486::AID-BIES5>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Roy S, Wolff C, Ingham PW. The u-boot mutation identifies a Hedgehog-regulated myogenic switch for fiber-type diversification in the zebrafish embryo. Genes & Development. 2001;15:1563–76. doi: 10.1101/gad.195801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozelle AL, Machesky LM, Yamamoto M, Driessens MH, Insall RH, Roth MG, Luby-Phelps K, Marriott G, Hall A, Yin HL. Phosphatidylinositol 4,5-bisphosphate induces actin-based movement of raft-enriched vesicles through WASP-Arp2/3. Curr Biol. 2000;10:311–20. doi: 10.1016/s0960-9822(00)00384-5. [DOI] [PubMed] [Google Scholar]
- Ruiz-Gomez M, Coutts N, Price A, Taylor MV, Bate M. Drosophila dumbfounded: a myoblast attractant essential for fusion. Cell. 2000;102:189–98. doi: 10.1016/s0092-8674(00)00024-6. [DOI] [PubMed] [Google Scholar]
- Ruiz-Gomez M, Coutts N, Suster ML, Landgraf M, Bate M. myoblasts incompetent encodes a zinc finger transcription factor required to specify fusion-competent myoblasts in Drosophila. Development. 2002;129:133–41. doi: 10.1242/dev.129.1.133. [DOI] [PubMed] [Google Scholar]
- Rushton E, Drysdale R, Abmayr SM, Michelson AM, Bate M. Mutations in a novel gene, myoblast city, provide evidence in support of the founder cell hypothesis for Drosophila muscle development. Development. 1995;121:1979–88. doi: 10.1242/dev.121.7.1979. [DOI] [PubMed] [Google Scholar]
- Sanchez-Pulido L, Martin-Belmonte F, Valencia A, Alonso MA. MARVEL: a conserved domain involved in membrane apposition events. Trends Biochem Sci. 2002;27:599–601. doi: 10.1016/s0968-0004(02)02229-6. [DOI] [PubMed] [Google Scholar]
- Sanger JW, Holtzer H. Cytochalasin B: effects on cell morphology, cell adhesion, and mucopolysaccharide synthesis (cultured cells-contractile microfilaments-glycoproteins-embryonic cells-sorting-out) Proc Natl Acad Sci USA. 1972;69:253–7. doi: 10.1073/pnas.69.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapir A, Avinoam O, Podbilewicz B, Chernomordik LV. Viral and developmental cell fusion mechanisms: conservation and divergence. Dev Cell. 2008;14:11–21. doi: 10.1016/j.devcel.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapir A, Choi J, Leikina E, Avinoam O, Valansi C, Chernomordik LV, Newman AP, Podbilewicz B. Dev Cell. Vol. 12. 2007. AFF-1, a FOS-1-regulated fusogen, mediates fusion of the anchor cell in C. elegans; pp. 683–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer G, Weber S, Holz A, Bogdan S, Schumacher S, Muller A, Renkawitz-Pohl R, Onel SF. The Wiskott-Aldrich syndrome protein (WASP) is essential for myoblast fusion in Drosophila. Dev Biol. 2007;304:664–74. doi: 10.1016/j.ydbio.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Schroter RH, Lier S, Holz A, Bogdan S, Klambt C, Beck L, Renkawitz-Pohl R. kette and blown fuse interact genetically during the second fusion step of myogenesis in Drosophila. Development. 2004;131:4501–9. doi: 10.1242/dev.01309. [DOI] [PubMed] [Google Scholar]
- Schwander M, Leu M, Stumm M, Dorchies OM, Ruegg UT, Schittny J, Müller U. Beta1 integrins regulate myoblast fusion and sarcomere assembly. Developmental Cell. 2003;4:673–85. doi: 10.1016/s1534-5807(03)00118-7. [DOI] [PubMed] [Google Scholar]
- Seipel K, Medley QG, Kedersha NL, Zhang XA, O'Brien SP, Serra-Pages C, Hemler ME, Streuli M. Trio amino-terminal guanine nucleotide exchange factor domain expression promotes actin cytoskeleton reorganization, cell migration and anchorage-independent cell growth. J Cell Sci. 1999;112(Pt 12):1825–34. doi: 10.1242/jcs.112.12.1825. [DOI] [PubMed] [Google Scholar]
- Sellin L, Huber TB, Gerke P, Quack I, Pavenstädt H, Walz G. NEPH1 defines a novel family of podocin interacting proteins. The FASEB Journal. 2003;17:115–7. doi: 10.1096/fj.02-0242fje. [DOI] [PubMed] [Google Scholar]
- Shelton C, Kocherlakota KS, Zhuang S, Abmayr SM. The immunoglobulin superfamily member Hbs functions redundantly with Sns in interactions between founder and fusion-competent myoblasts. Development. 2009;136:1159–68. doi: 10.1242/dev.026302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemer G, Podbilewicz B. The story of cell fusion: big lessons from little worms. Bioessays. 2003;25:672–82. doi: 10.1002/bies.10301. [DOI] [PubMed] [Google Scholar]
- Smith LG, Li R. Actin polymerization: riding the wave. Curr Biol. 2004;14:R109–11. [PubMed] [Google Scholar]
- Smythe G, Davies M, Paulin D, Grounds M. Absence of desmin slightly prolongs myoblast proliferation and delays funsion in vivo in regenerating grafts of skeletal muscle. Cell Tissue Res. 2001:287–294. doi: 10.1007/s004410100366. [DOI] [PubMed] [Google Scholar]
- Snow CJ, Goody M, Kelly MW, Oster EC, Jones R, Khalil A, Henry CA. Time-lapse analysis and mathematical characterization elucidate novel mechanisms underlying muscle morphogenesis. PLoS Genet. 2008;4:e1000219. doi: 10.1371/journal.pgen.1000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn RL, Huang P, Kawahara G, Mitchell M, Guyon J, Kalluri R, Kunkel LM, Gussoni E. A role for nephrin, a renal protein, in vertebrate skeletal muscle cell fusion. Proc Natl Acad Sci USA. 2009;106:9274–9. doi: 10.1073/pnas.0904398106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas BP, Woo J, Leong WY, Roy S. A conserved molecular pathway mediates myoblast fusion in insects and vertebrates. Nat Genet. 2007;39:781–6. doi: 10.1038/ng2055. [DOI] [PubMed] [Google Scholar]
- Steffen A, Faix J, Resch GP, Linkner J, Wehland J, Small JV, Rottner K, Stradal TEB. Filopodia formation in the absence of functional WAVE- and Arp2/3-complexes. Mol Biol Cell. 2006;17:2581–91. doi: 10.1091/mbc.E05-11-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen A, Rottner K, Ehinger J, Innocenti M, Scita G, Wehland J, Stradal TEB. Sra-1 and Nap1 link Rac to actin assembly driving lamellipodia formation. EMBO J. 2004;23:749–59. doi: 10.1038/sj.emboj.7600084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellabotte F, Devoto SH. The teleost dermomyotome. Dev Dyn. 2007;236:2432–43. doi: 10.1002/dvdy.21253. [DOI] [PubMed] [Google Scholar]
- Stockdale FE, Nikovits William, J, Christ B. Molecular and cellular biology of avian somite development. Developmental Dynamics. 2000;219:304–321. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1057>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Strunkelnberg M, Bonengel B, Moda LM, Hertenstein A, de Couet HG, Ramos RG, Fischbach KF. rst and its paralogue kirre act redundantly during embryonic muscle development in Drosophila. Development. 2001;128:4229–39. doi: 10.1242/dev.128.21.4229. [DOI] [PubMed] [Google Scholar]
- Sugihara K, Nakatsuji N, Nakamura K, Nakao K, Hashimoto R, Otani H, Sakagami H, Kondo H, Nozawa S, Aiba A, Katsuki M. Rac1 is required for the formation of three germ layers during gastrulation. Oncogene. 1998;17:3427–33. doi: 10.1038/sj.onc.1202595. [DOI] [PubMed] [Google Scholar]
- Sun C, Kilburn D, Lukashin A, Crowell T, Gardner H, Brundiers R, Diefenbach B, Carulli JP. Kirrel2, a novel immunoglobulin superfamily gene expressed primarily in beta cells of the pancreatic islets. Genomics. 2003;82:130–42. doi: 10.1016/s0888-7543(03)00110-1. [DOI] [PubMed] [Google Scholar]
- Tajbakhsh S, Borello U, Vivarelli E, Kelly R, Papkoff J, Duprez D, Buckingham M, Cossu G. Differential activation of Myf5 and MyoD by different Wnts in explants of mouse paraxial mesoderm and the later activation of myogenesis in the absence of Myf5. Development. 1998;125:4155–62. doi: 10.1242/dev.125.21.4155. [DOI] [PubMed] [Google Scholar]
- Tajbakhsh S, B M. The birth of muscle progenitor cells in the mouse: spatiotemporal considerations. Curr Top Dev Biol. 2000;48:225–68. doi: 10.1016/s0070-2153(08)60758-9. [DOI] [PubMed] [Google Scholar]
- Tall EG, Spector I, Pentyala SN, Bitter I, Rebecchi MJ. Dynamics of phosphatidylinositol 4,5-bisphosphate in actin-rich structures. Current Biology. 2000;10:743–46. doi: 10.1016/s0960-9822(00)00541-8. [DOI] [PubMed] [Google Scholar]
- Tang J, Maximov A, Shin OH, Dai H, Rizo J, Südhof TC. A complexin/synaptotagmin 1 switch controls fast synaptic vesicle exocytosis. Cell. 2006;126:1175–87. doi: 10.1016/j.cell.2006.08.030. [DOI] [PubMed] [Google Scholar]
- Tryggvason K. Neprhin: role in normal kidney and in disease. Adv Nephrol Necker Hosp. 2001;31:221–34. [PubMed] [Google Scholar]
- Ueno H, Sakita-Ishikawa M, Morikawa Y, Nakano T, Kitamura T, Saito M. A stromal cell-derived membrane protein that supports hematopoietic stem cells. Nat Immunol. 2003;4:457–63. doi: 10.1038/ni916. [DOI] [PubMed] [Google Scholar]
- Vartiainen MK, Machesky LM. The WASP-Arp2/3 pathway: genetic insights. Curr Opin Cell Biol. 2004;16:174–81. doi: 10.1016/j.ceb.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Vasyutina E, Birchmeier C. The development of migrating muscle precursor cells. Brain Struct Funct. 2006;211:37–41. doi: 10.1007/s00429-006-0118-9. [DOI] [PubMed] [Google Scholar]
- Vasyutina E, Martarelli B, Brakebusch C, Wende H, Birchmeier C. The small G-proteins Rac1 and Cdc42 are essential for myoblast fusion in the mouse. Proc Natl Acad Sci USA. 2009;106:8935–40. doi: 10.1073/pnas.0902501106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagers AJ, Conboy IM. Cellular and molecular signatures of muscle regeneration: current concepts and controversies in adult myogenesis. Cell. 2005;122:659–67. doi: 10.1016/j.cell.2005.08.021. [DOI] [PubMed] [Google Scholar]
- Wegorzewska M, Krauss RS, Kang JS. Overexpression of the immunoglobulin superfamily members CDO and BOC enhances differentiation of the human rhabdomyosarcoma cell line RD. Mol Carcinog. 2003;37:1–4. doi: 10.1002/mc.10121. [DOI] [PubMed] [Google Scholar]
- Weiner OD, Rentel MC, Ott A, Brown GE, Jedrychowski M, Yaffe MB, Gygi SP, Cantley LC, Bourne HR, Kirschner MW. Hem-1 complexes are essential for Rac activation, actin polymerization, and myosin regulation during neutrophil chemotaxis. PLoS Biol. 2006;4:e38. doi: 10.1371/journal.pbio.0040038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, He J, Wang X, Lim TM, Gong Z. Asynchronous activation of 10 muscle-specific protein (MSP) genes during zebrafish somitogenesis. Developmental Dynamics. 2000;219:201–215. doi: 10.1002/1097-0177(2000)9999:9999<::aid-dvdy1043>3.3.co;2-9. [DOI] [PubMed] [Google Scholar]
- Young JM, Trask BJ. The sense of smell: genomics of vertebrate odorant receptors. Hum Mol Genet. 2002;11:1153–60. doi: 10.1093/hmg/11.10.1153. [DOI] [PubMed] [Google Scholar]


