Abstract
Fungal natural products containing benzodiazepinone- and quinazolinone-fused ring systems can be assembled by nonribosomal peptide synthetases (NRPS) using the conformationally restricted β-amino acid anthranilate as one of the key building blocks. We validated that the first module of the acetylaszonalenin synthetase of Neosartorya fischeri NRRL 181 activates anthranilate to anthranilyl-AMP. With this as starting point, we then used bioinformatic predictions about fungal adenylation domain selectivities to identify and confirm an anthranilate-activating module in the fumiquinazoline A producer Aspergillus fumigatus Af293 as well as a second anthranilate-activating NRPS in N. fischeri. This establishes an anthranilate adenylation domain code for fungal NRPS and should facilitate detection and cloning of gene clusters for benzodiazepine- and quinazoline-containing polycyclic alkaloids with a wide range of biological activities.
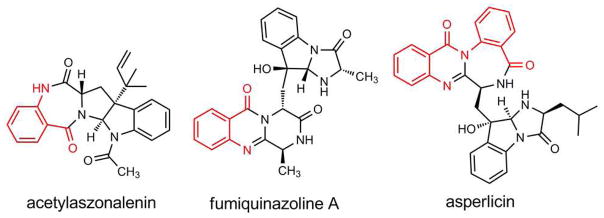
A variety of quinazoline- and benzodiazepine-containing natural product scaffolds of substantial structural complexity are produced by fungi (1), most likely via nonribosomal peptide synthetase (NRPS) assembly lines (2). These include the pentacyclic scaffold of acetylaszonalenin containing a fused 6-5-5-7-6 ring framework, fumiquinazoline A which contains a 6-6-6 core tethered to a 6-5-5 tricycle, and asperlicin which is defined by a 6-6-7-6 core tethered to 6-5-5 framework (Figure 1, additional examples provided in Figure S1). Inspection of these metabolites indicates that anthranilate (2-aminobenzoate, Ant) is a likely building block in all these scaffolds.
Figure 1.
Examples of fungal natural products that utilize anthranilic acid as a building block (Ant derived portion highlighted in red).
While anthranilate is a known primary metabolite in prokaryotes and single cell eukaryotes, e.g. in tryptophan biosynthesis, it is a non-proteinogenic aryl β-amino acid rather than a standard α-amino acid. One of the hallmarks of NRPS assembly lines is their ability to select and incorporate amino acid monomers other than the canonical twenty proteinogenic amino acids (3). The planarity of the 1,3-substituted carboxy and amino groups imposed by the aryl ring makes anthranilate a building block capable of subsequent cyclization by amide bond formation to create the benzodiazepinone and the quinazolinone frameworks of these classes of bioactive natural products. Radioisotope feeding studies have validated anthranilate incorporation by producing fungi (4–7), and much effort has been dedicated to the chemical synthesis of Ant-containing fungal alkaloids (8); however, little is known regarding the NRPS-based selection and incorporation of Ant by fungal NRPSs or the utilization of Ant by the NRPS machinery for subsequent intramolecular cyclization leading to polycyclic scaffold formation.
Identification of anthranilate-activating domains and modules in fungal NRPS assembly lines should be a path to identification of the genes and their encoded proteins responsible for the biosynthesis of this class of natural products. To that end, in this work we have utilized a combination of biochemical and bioinformatics approaches to identify fungal adenylation domains which selectively activate the aryl β-amino acid anthranilate. Starting with the known gene cluster for acetylaszonalenin from Neosartorya fischeri NRRL 181 (9), we have validated that module one of the two module AnaPS activates anthranilate as anthranilyl-AMP (Ant-AMP) and subsequently loads the anthranilyl group onto the phosphopantetheine (PPT) prosthetic group of its cognate thiolation domain (Figure 2). Then we have used the presumed ten amino acid code from the adenylation domain of AnaPS module one to predict and validate that a module encoded in the fumiquinazoline-producing Aspergillus fumigatus Af293 activates and loads anthranilate. As a third test we further examined the genome of Neosartorya fischeri NRRL 181 and found another predicted Ant-activating NRPS module which we have heterologously expressed and purified from E. coli and confirmed anthranilate-dependent activity.
Figure 2.
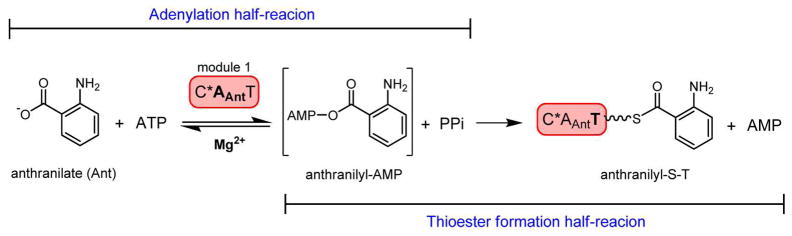
Scheme for the chemical steps of anthranilic acid activation (by adenylation) and anthranilyl transfer (by thiolation) catalyzed by the A- and T-domains of fungal NRPS module 1 constructs characterized in this work. The wavy line of the anthranilyl-S-T intermediate corresponds to the phosphopantetheine (PPT) prosthetic group.
EXPERIMENTAL PROCEDURES
Materials and general methods
Fungals strains: N. fischeri NRRL 181 was obtained from the Fungal Genetics Stock Center (FGSC #A1164), A. fumigatus Af293 was a gift from the R.A. Cramer lab (University of Montana). Radiolabeled chemicals: [Carboxy-14C]anthranilate (52 mCi/mmol), [7-14C]benzoate (55 mCi/mmol), [7-14C]salicylate (47 mCi/mmol), [(U)-14C]L-valine (250 mCi/mmol), and [1-14C]acetyl-CoA (53 mCi/mmol) were obtained from American Radiolabeled Chemicals. [32P]-PPi (105 mCi/mmol) was obtained from Perkin Elmer. 15N-Ant and 15N5-ATP were obtained from Cambridge Isotope Laboratories and Sigma-Aldrich, respectively. Other aryl or amino acids were obtained from Sigma-Aldrich. Stocks for all compounds other than radiolabeled chemicals were prepared at 50 mM concentration with 35% DMSO in water. PCR reactions were carried out using Phusion High-Fidelity PCR MasterMix (New England Biolabs). Oligonucleotide primers were purchased from Integrated DNA Technologies (Coralville, IA). Manipulation of plasmid DNA was performed in E. coli XL1 Blue (Stratagene), and plasmid DNA prepared using the QIAprep Spin Mini Kit (Qiagen). Automated DNA sequencing was performed by Genewiz (South Plainfield, NJ). MALDI-TOF peptide mass fingerprinting was performed by the Molecular Biology Core Facilities of the Dana-Farber Cancer Institute (Boston, MA).
Fungal growth and gene cloning
Spores of Neosartorya fischeri NRRL 181 were prepared following growth on malt extract agar for 10 days at 25°C and used to inoculate 50 mL of potato dextrose broth. Following liquid culture for 3 days at 28°C, cells were collected by filtration through Whatman 54 filter paper, washed with 0.6M MgCl2, flash frozen in liquid N2, and lyophilized overnight. Approximately 100 mg of the freeze-dried cells were used for gDNA isolation with the Qiagen DNeasy Plant Mini Kit according to the manufacturer’s protocol. PCR amplification of the region encoding AnaPS (NFIA_055290) module 1 C*AT (residues 1-847) was then performed with the following primers: 5′ GACGACGACAAGATacaatcactacaatgactccgcagaatgg 3′ (forward), 5′ GAGGAGAAGCCCGGTTAtccgttgctcaagttcgcagc 3′ (reverse); lowercase type signifies bases complementary to gene, bold type indicates a stop codon. The amplified DNA was gel-purified using the QIAquick Gel Extraction Kit (Qiagen), cloned using the pET32 Ek/LIC Vector Kit (EMDchemicals), and a 52bp intron removed by deletion using the QuikChange XL Site-Directed Mutagenesis Kit (Stratagene) and the following partial overlapping primers: 5′ tcgacgtcgagtatcacgatatcaacctactcctgcagtccaacc 3′ (forward), 5′ gatatcgtgatactcgacgtcgagaagacatctcccgttctgg 3′ (reverse); underline denotes primer-primer complementary (overlapping) sequence. The final construct encoding AnaPS C*A1T1 as an N-terminal thioredoxin (Trx)-His6-fusion was confirmed by DNA sequencing.
Cloning of NFIA_057960 module 1 C*AT from N. fischeri NRRL 181 was performed using the gDNA as described above for AnaPS. PCR amplification of module 1 (encoding residues 1-848) was accomplished with the following primers: 5′ GGTATTGAGGGTCGCatggcgaccgagcagtactccc 3′ (forward), 5′ AGAGGAGAGTTAGAGCCTTAtgcccttggctctgcagctc 3′ (reverse). Following ligation independent cloning with the pET30 Xa/LIC Vector Kit (EMDchemicals), a 54bp intron was removed by site-directed mutagenesis with the following primer pair: 5′ aactggaagagacttgcagcattattcttgttgcaacaaacatagac 3′ (forward), 5′ gctgcaagtctcttccagttcaagcgtctgttcaaaactatcc 3′ (reverse). The final NFIA_057960 C*A1T1 construct with an N-terminal His6-fusion tag was confirmed by DNA sequencing.
Growth of Aspergillus fumigatus Af293 was accomplished in a manner similar to that described above for N. fischeri, but liquid cultures were incubated at 37°C for 18–24 hours prior to harvesting cells by filtration and flash freezing. The Qiagen RNeasy Plant Mini Kit was used for RNA extraction and cDNA synthesis was accomplished using the ThermoScript RT-PCR system (Invitrogen) with on-column DNA digestion using the RNase-Free DNase Set (Qiagen). PCR amplification of AFUA_6g12080 module 1 C*AT (encoding residues 1-885) was then performed using the following primers: 5′ GGTATTGAGGGTCGCatggaacgtttggaagcaaagaatc 3′ (forward), 5′ AGAGGAGAGTTAGAGCCTTAatgttgtgcggctcctgagttg 3′ (reverse). Cloning of the amplified DNA was accomplished using the pET30 Xa/LIC Vector Kit (EMDchemicals). The final construct encoding AFUA_6g12080 C*A1T1 as an N-terminal His6-fusion was confirmed by DNA sequencing.
Protein expression and purification
All protein constructs were overproduced in E. coli BL21-Gold(DE3) cells (Stratagene) in a similar manner: 4–6 Liters of cells were grown at 37°C in LB (supplemented with the appropriate antibiotic) to an OD600 between 0.3–0.8, and the temperature lowered to 16°C prior to induction with 0.2 mM IPTG. Cells were harvested 18–24 hours post-induction, suspended in lysis buffer (25 mM Tris-HCl [pH 7.5], 300 mM NaCl, 10% glycerol, 1x protease inhibitor cocktail [SigmaFast, EDTA-free]), and lysed using a EmulsiFlex-C5 homogenizer (Avestin). Insoluble material was removed by centrifugation (35,000g) and soluble protein applied to 1–2 mL of Ni-NTA agarose (Qiagen) equilibrated in lysis buffer. Ni-affinity purification was performed by batch binding protein for 30 min at 4°C, washing Ni-resin with lysis buffer containing 10–20 mM imidazole, and then eluting protein using a step gradient from 100–500 mM imidazole. For AFUA_6g12080 and NFIA_057960 C*AT, the elutions containing target protein were pooled and concentrated using a microcentrifugal device (30K MWCO, Amicon), and concentrated protein either flash-frozen in liquid N2 (AFUA_6g12080) or used for further purification (AnaPS and NFIA_057960). Post-Ni-NTA chromatography, AnaPS Trx-C*AT protein was further purified by concentrating the pooled Ni-NTA elutions to a volume 1/10 of original and the resulting precipitate dissolved in Ni-bind buffer + 1 M imidazole. NFIA_057960 C*AT was further purified by a combination of anion-exchange (MonoQ HR 10/100, GE Healthcare) and gel-filtration (Superdex 200 10/300, GE Healthcare). For anion-exchange, concentrated protein was diluted 10-fold in Q-buffer A (25 mM Tris-HCl [pH 8.0], 1 mM EDTA, 1 mM TCEP, 10% glycerol) and a linear gradient to 1 M NaCl was ran over 20 column volumes. Fractions containing target protein were then combined and concentrated to 0.5 mL for injection onto a Superdex 200 column equilibrated in 25 mM Tris-HCl (pH 7.5), 100 mM NaCl, 1 mM TCEP, and 10% glycerol.
ATP-PPi exchange assays
Typical reactions (100 μL) contained 5 mM ATP, 5 mM MgCl2, 6 mM Na4[32P]PPi (0.28 μCi), 0.1–5 μM C*AT enzyme, and 1 mM substrate in buffer (50 mM Tris-HCl [pH 7.5], 40 mM NaCl, 5 mM TCEP, and 10% glycerol). Reactions were incubated at 25°C for 1.5 hours then quenched by addition of a charcoal solution (1.6% w/v activated charcoal, 100 mM sodium pyrophosphate, 3.5% perchloric acid in water). The charcoal was pelleted by centrifugation and washed with a solution containing 100 mM sodium pyrophosphate and 3.5% perchloric acid, and the charcoal-bound radioactivity detected by liquid scintillation counting.
HPLC and LC/MS-based assays
Reactions (100 μL) to investigate C*AT-catalyzed Ant-AMP formation contained 50 mM HEPES (pH 7.4), 5 mM ATP, 10 mM MgCl2, 1 mM DTT, 1 mM anthranilate, and varying concentrations of C*AT enzyme depending on the construct tested (10 μM AFUA_6g12080 C*AT, 0.1 to 2 μM NFIA_057960 and AnaPS C*AT). Control reactions were also prepared in which ATP, anthranilate, or enzyme was omitted. Reactions were incubated at 25°C for 2 hours, then quenched by filtration (3K MWCO filter device). HPLC analysis of the flow-through from filtered samples (20 μL injections) was performed on a Beckman Coulter System Gold equipped with a C18 reverse phase column (Alltech, 150 × 4.6 mm), and detection at 251 nm. Solvent systems A (water + 0.1% formic acid) and B (MeCN + 0.1 % formic acid) were used to apply a linear gradient from 0–20% B over 20 minutes, followed by 20–95% B over 1 min, and a hold at 95% B for 5 min with a flow rate of 1 mL/min. The identity of the ATP, ADP, AMP, and anthranilate peaks were confirmed by running authentic standards, and the identity of Ant-AMP was determined by collecting the peak and performing high-resolution ESI-TOF mass spectrometry. For high-resolution LC/MS analysis an Agilent Technologies 6520 Accurate-Mass Q-TOF instrument was used to analyze 10 μL sample injections onto a Gemini-NX C18 column (50 × 2.0 mm). A linear gradient from 2–100% B was ran over 20 minutes using the abovementioned solvent system. Unlabeled Ant and ATP were used to confirm Ant-AMP formation for AnaPS Trx-C*AT and NFIA_057960; whereas all possible combinations of 15N-labeled and unlabeled ATP and Ant were used with AFUA_6g12080 C*AT to corroborate the identity of Ant-AMP.
To study the stability of Ant-AMP in solution following microfiltration, the filtrate from an Ant-AMP experimental reaction (as described above) was incubated at 25°C for 0.25, 2.5, 5, 10, 24, and 48 hours prior to injection for HPLC analysis (251 nm detection). Quantification of Ant-AMP half-life was performed by integrating the Ant-AMP peaks obtained for each timepoint and fitting the integration area data points to a 1st order decay equation using GraphPad Prism.
Continuous PPi-release assay
Kinetic analysis of AFUA_6g12080 C*AT was performed using the EnzChek Pyrophosphate Assay Kit (Molecular Probes) essentially as instructed in the manufacturer’s protocol but with 0.1 mL reaction volumes in a 96-well microplate format. Reactions contained 50 mM HEPES (pH 7.4), 10 mM MgCl2, 1 mM DTT, 5% glycerol, 0.2 mM MESG, 1 U/mL purine nucleoside phosphorylase (PNP), 0.03 U/mL inorganic pyrophosphatase (IPP), 5 μM enzyme, and varying concentrations of substrates (ATP and aryl acids as listed in Table 3). Reactions were initiated by addition of substrate after a 10 min incubation period at 25°C, and activity monitored continuously with a SpectraMax Plus384 UV-Vis spectrometer with detection at 360 nm. A reaction lacking substrate was prepared for each trial and the initial rate obtained was subtracted from the (+) substrate rate data. Triplicate measurements were taken for each concentration of substrate. The net initial rate (obtained in OD/min) was converted to reaction velocity (as μmol/min) using an extinction coefficient of 11,000 (M·cm)−1 for 2-amino-6-mercapto-7-methylpurine (10), and path length of 0.3 cm as determined for the 100 μL reaction volumes and microplates used.
Table 3.
Kinetic parameters for AFUA_6g12080 m1 C*AT determined by coupled PPi-release assaya.
| Substrate | Km (μM) | Vmax (nmol min−1) | kcat (min−1) | kcat/Km (min−1 mM−1) |
|---|---|---|---|---|
| ATP | 230 (± 63) | 0.31 (± 0.024) | 0.63 (± 0.048) | 2.7 |
| Anthranilic acid | 18 (± 2.5) | 0.28 (± 0.0093) | 0.57 (± 0.019) | 32 |
| Salicylic acid | 17 (± 2.6) | 0.20 (± 0.0059) | 0.40 (± 0.012) | 24 |
| 2-chlorobenzoic acid | 29 (± 4.5) | 0.34 (± 0.012) | 0.68 (± 0.024) | 23 |
| 4-chlorobenzoic acid | 32 (± 2.7) | 0.69 (± 0.014) | 1.39 (± 0.028) | 48 |
| Benzoic acid | 74 (± 9.3) | 0.51 (± 0.018) | 1.0 (± 0.036) | 14 |
| 3-aminobenzoic acid | 416 (± 69) | 1.2 (± 0.086) | 2.32 (± 0.172) | 5.6 |
| 4-aminobenzoic acid | 1900 (± 130) | 1.2 (± 0.035) | 2.43 (± 0.07) | 1.3 |
It is of note that enzyme turnover and continuous release of PPi reflect the off-pathway release (or “leakage”) of the acyl-adenylate intermediates, and paradoxically the less tightly bound acyl-adenylates may give the fastest rates. Therefore, preferred substrates may yield relatively small Vmax and kcat values as their corresponding acyl-adenylates are expected to be tightly held for productive acyl-group transfer to the PPT of the cognate T-domain (35). By comparison, the acyl-adenylate of less preferred substrates have a higher tendency to leak from the A-domain active site resulting in faster turnover and larger Vmax and kcat values (35).
Phosphopantetheinylation of the AFUA_6g12080 module 1 T-domain with [1-14C]acetyl-CoA
Reactions (50 μL) contained 100 μM [1-14C]acetyl-CoA (0.25 μCi), 100 nM B. subtilis Sfp (11), and 5 μM enzyme in buffer (50 mM Tris-HCl [pH 7.5], 5 mM MgCl2, 40 mM NaCl, 5 mM TCEP, and 5% glycerol). Reaction were initiated by addition of Sfp and incubated at 25°C for 0.75, 1.5, 2.5, 5, 10, or 45 minutes prior to quenching by adding 0.5 mL 10% TCA (with 50 μg BSA for visualization of precipitated protein). Protein precipitate was pelleted by centrifugation, washed twice with 10% TCA, and dissolved in 80% formic acid for liquid scintillation counting. A “zero” timepoint was taken by processing a reaction prior to the addition of Sfp. A ratio of nmoles radioactivity counted to nmoles protein was used to calculate % conversion based on one equivalent of [1-14C]acetyl-CoA labeling one equivalent of C*AT protein (1:1 stoichiometry).
Module 1 C*AT T-domain loading assays with 14C-labeled substrates
First, In vitro phosphopantetheinylation of the module 1 C*AT constructs to generate holo-T-domain was carried out in 25 μL reactions by incubating varying concentration of C*AT protein (5–80 μM), 2 μM B. subtilis Sfp, and 1 mM CoA for 30 min at 25°C. Then, 6–25 μL of the “holo”-C*AT preparation was added to reaction buffer (50 mM Tris-HCl [pH 7.5], 5 mM MgCl2, 40 mM NaCl, and 5 mM TCEP) containing 5 mM ATP, and reactions (50 μL) initiated by addition of 50 μM 14C-labeled substrate (Ant [0.13 μCi], benzoate [0.2 μCi], Sal [0.13 μCi], or L-valine [0.3 μCi]). For autoradiography of SDS-PAGE, after 15 min incubation at 25°C, samples were quenched by adding 1x-SDS sample buffer. Following SDS-PAGE, radiolabeled protein was detected using a BAS-III imaging plate (Fuji Film, 48–96 hour exposure) and a Typhoon 9400 phosphorimager (GE Healthcare). For timepoint analysis with Orf12080 C*AT, following 25°C incubation samples were taken at 0.5, 1, 2.5, 5, 10, and 30 minutes by quenching with 10% TCA. The precipitated protein was collected by centrifugation, washed twice with 10% TCA and dissolved in 80% formic acid, and the amount of enzyme-bound radioactivity detected by liquid scintillation counting. Percentage loading was calculated as the mole fraction of 14C-substrate covalently transferred to holo-C*AT protein assuming 1:1 stoichiometry.
Bioinformatics
Putative Ant-activating fungal NRPS modules were identified by performing a BLASTP search of the Aspergillus Comparative Database (available from the Broad Institute) using AnaPS C*A1T1 (residues 1-847) as the query sequence. The results from BLASTP yielded over one-hundred putative NRPSs from the eight sequenced genomes searched. The adenylation domain 10AA code of the highest scoring hits was extracted as described in Results, and then compared to the predicted 10-residue specificity sequence from AnaPS A1 to identify possible positive matches. The ten A-domains identified were classified as positive matches based on the conservation of the 10AA code Pos1 and Pos8 glycines, the conservation of residues with small sidechains at Pos6 and Pos7, and the conservation of hydrophobicity at other residue positions.
Homology modeling of AnaPS A1 (residues 246-800) and AFUA_6g12080 A1 (residues 243-800) was performed using the online server HHpred (12). Due to the high-degree of structural similarity between adenylate-forming enzymes of this class (e.g. NRPS A-domains, firefly luciferase, and acyl-CoA synthetases/ligases) (13), there were several structural templates available for modeling. The best template identified by HHpred was PheA (PDB ID, 1AMU), with other high-scoring templates including A3 of the fungal siderophore synthetase SidN (3ITE), the A-domain of the surfactin synthetase termination module SrfAC (2VSQ), firefly luciferase (2D1S), D-alanine: D-alanyl carrier protein ligase DltA (3E7W), the bacterial aryl-acid activating A-domain DhbE (1MDB), and acyl-CoA ligases/synthetases that activate acetate (1PG4), benzoate (2V7B), and 4-chlorobenzoate (1T5H). Single-template homology models were generated for all templates listed, and 10AA code residues were identified by structural alignment.
RESULTS
Module 1 of AnaPS from N. fischeri NRRL 181 activates and loads anthranilic acid
Bioinformatics analysis identifies AnaPS module 1 as the putative Ant-activating module
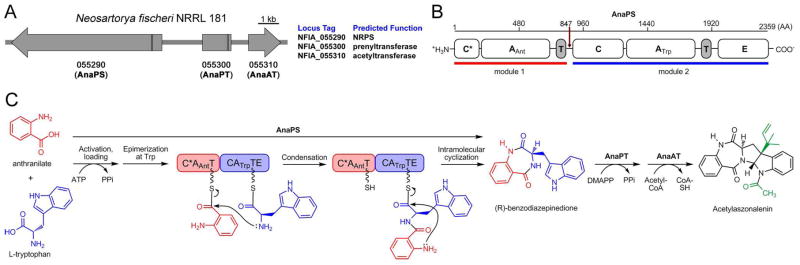
The gene cluster for acetylaszonalenin biosynthesis from Neosartorya fischeri NRRL 181 was reported in early 2009 by Shu-Ming Li and coworkers (Figure 3A) (9). AnaPS was postulated to be the nonribosomal peptide synthetase responsible for activation and condensation of anthranilic acid and tryptophan in the formation of an (R)-benzodiazepinedione intermediate which is converted on to acetylaszonalelin by enzymatic prenylation and N-acetylation of the Trp indole (Figure 3C) (9). Bioinformatics analysis of AnaPS, including sequence alignment and structure prediction, suggests the linear domain organization: C*-A1-T1-C2-A2-T2-E (Figure 3B; C, condensation; A, adenylation; T, thiolation; E, epimerization; C* indicates a truncated and presumably inactive C-domain). Module 2 of AnaPS (C2-A2-T2-E) is deduced to be Trp-selective based on: 1) similarity of extracted specificity-residues of the A2-domain to other characterized Trp-activating A-domains (e.g. 70% identity to the 10-residue specificity sequence from comA module2, complestatin NRPS), and 2) the presence of a terminal E-domain for epimerization at the α-atom of the tryptopanyl-S-T2 intermediate (L- to D-Trp) to give the observed (R)-stereogenic center of the benzodiazepinedione. Consequently, module 1 of AnaPS (C*-A1-T1) is predicted to be responsible for the selection, activation, and loading of anthranilic acid.
Figure 3.
Acetylaszonalenin biosynthesis from Neosartorya fischeri NRRL 181. (A) Annotated three-gene cluster. Vertical black lines in the Orfs demarcate introns. (B) Domain organization of the bimodular NRPS AnaPS, illustrating predicted domain boundaries as determined from bioinformatics analysis. Red arrow marks the “cutoff” point used for cloning module 1 C*AT. (C) Proposed route of acetylaszonalelin biosynthesis. Previous work has validated the conversion of (R)-benzodiazepinedione to acetylaszonalenin by AnaAT and AnaPT, and in this work we validate the activation and loading of anthranilate by AnaPS module 1. The loading and epimerization of Trp, and the condensation and cyclization reactions are hypothetical.
Expression and purification of AnaPS module 1
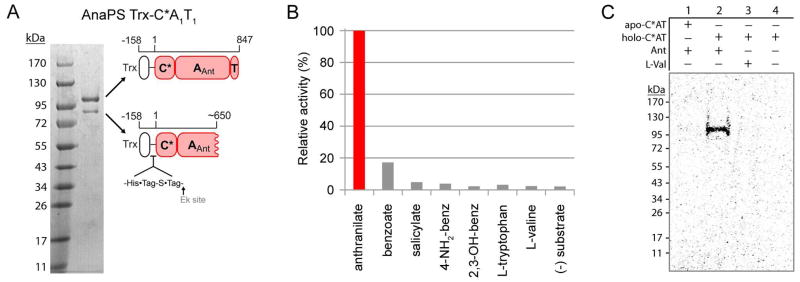
The region encoding AnaPS C*-A1-T1 (residues 1-847) was PCR amplified from N. fischeri NRRL 181 genomic DNA and, following removal of a single 52 bp intron, cloned into a pET32 Ek/LIC E. coli expression vector to provide an N-terminal thioredoxin (trx)-His6 fusion protein (AnaPS Trx-C*AT, 110 kDa). Thioredoxin (14) was chosen as a fusion partner following failed attempts to produce soluble C*-A1-T1 or A1-T1 protein as “His6-tag only” constructs in E. coli. Ni-affinity chromatography afforded protein that was only partially purified, with Trx-C*AT eluting with a host of contaminants (Figure S2). Peptide mass fingerprinting was used to identify the most prominent contaminating bands as degradation fragments and E. coli chaperones. The co-purification of chaperones with Trx-C*AT may indicate the difficulty of folding the target protein in E. coli. To further purify the target protein, we found that concentration-induced protein precipitation of the Ni-NTA elutions was effective to enrich for both the full-length and a C-terminal degradation fragment of Trx-C*AT (Figure 4A). The dissolved protein pellet was used directly for experimental validation of anthranilate-dependent activity.
Figure 4.
Purification and characterization of AnaPS module 1. (A) Purity of the thioredoxin fusion protein of AnaPS module 1 (Trx-C*A1T1) post Ni-NTA chromatography and concentration-induced protein precipitation. Cartoon representation is used to indicate the identity and domain-composition of the two prominent bands present in the gel as determined by peptide mass fingerprinting. (B) ATP-[32P]PPi exchange assay data obtained for Trx-C*AT. 100% relative activity for anthranilate-dependent exchange corresponds to 6200 CPM. (C) Autoradiograph of SDS-PAGE gel illustrating the covalent loading of [carboxy-14C]anthranilate onto the holo-T-domain of Trx-C*AT.
ATP-[32P]PPi exchange assay demonstrates that AnaPS module 1 activates anthranilate
The first test of AnaPS Trx-C*AT function was performed using the classical ATP-[32P]PPi exchange assay, by which the adenylation half of the two-step reaction catalyzed by an A-T NRPS pair may be monitored. The substrate-dependent and reversible formation of acyl-AMP, catalyzed by the A-domain of Trx-C*AT, was monitored by addition of [32P]PPi and detection of the exchanged radiolabel as [32P]ATP by absorption into activated charcoal and liquid scintillation counting. The level of exchange following 1.5 hour incubation was detected and demonstrates that anthranilate supports 6-fold higher exchange over benzoate, while salicylate (2-hydroxybenzoate), 4-aminobenzoate, 2,3-dihydroxybenzoate, and the α-amino acids L-tryptophan or L-valine yield only background-level activity (Figure 4B). AnaPS module 1 thus activates the aryl acid Ant as anthranilyl-AMP.
Detection of Ant-AMP formation and release by AnaPS module 1
As anticipated from the results of the ATP-PPi exchange assay, reconstitution of the adenylation half reaction by combining AnaPS Trx-C*AT, ATP, Mg2+, and anthranilic acid in buffer results in the formation of Ant-AMP detectable as a released product by LC/MS. A peak elutes at 5.6 min with the corresponding high-resolution MS showing molecular ion peaks in agreement with Ant-AMP formation: [M+H]+ (observed 467.1094, expected 467.1075) and [2M+H]+ (observed 933.2105, expected 933.2150) (Figure S3A). Though the acyl-adenylate formed by NRPS A-domains is considered to be a tightly bound intermediate to allow productive acyl-group transfer to the phosphopantetheinyl arm of its cognate T-domain (15–18), in the absence of the appropriate acyl-group acceptor it is not surprising that release of the acyl-AMP intermediate from enzyme slowly occurs. In agreement with this observation, release of aminoacyl-AMP was also observed with the phenylalanine activating subunit of gramicidin synthetase 1 (GrsA-PheA) (18).
In cis loading of anthranilate onto the T-domain of AnaPS module 1
In order to reconstitute and monitor the thiolation half-reaction catalyzed by the A-T pair of AnaPS Trx-C*AT; 14C-labeled anthranilate or L-valine (as a negative control) was combined with Trx-C*AT, B. subtilis Sfp (11), CoA, Mg2+, and ATP. Including Sfp and CoA allows enzymatic phosphopanthetheinylation of the T1-domain to generate holo-C*AT (19). In this assay the 14C-labeled acyl group of the acyl-adenylate formed by the A-domain is transferred in cis to the holo-T-domain to form a covalent thioester linkage, and detection of labeled protein achieved by SDS-PAGE autoradiography. As illustrated in Figure 4C, radiolabeled protein is detected when [14C]Ant is combined with holo-Trx-C*AT (lane 2), indicating that the tandem A1-T1 pair of Trx-C*AT is catalytically competent to select, activate, and load Ant as anthranilyl-S-T1.
Bioinformatics-based identification of fungal Ant-activating NRPS modules
Experimental validation that module 1 of AnaPS is Ant-selective provided the foundation for a bioinformatics-based approach to identify other Ant-activating NRPS modules from sequenced fungal genomes. The basis of this approach is the prediction and matching of the AnaPS A1 ten residue specificity sequence (also known as the 10AA code or “signature sequence”) to the extracted 10AA code of other fungal A-domains. The prediction of specificity-determining residues was originally based on mapping known A-domain sequences to the crystal structure of PheA (20–22), and has been developed into automated web-based resources such as NRPS predictor (23). Although signature sequence extraction has been utilized extensively and successfully for the prediction of bacterial A-domain substrate specificity (21), the application of this method to predict fungal A-domain specificity has been reported to be more difficult (24–26).
The amino acid sequence of AnaPS A1 was submitted to online servers including NRPS predictor (23) and the NRPS-PKS database (27) for automated extraction of specificity-defining residues, resulting in the preliminary assignment of GALFFAAGVK as the 10AA code (Table 1). The online servers did not match the extracted 10AA code to any A-domain of known specificity, suggesting uniqueness of the AnaPS A1 code for Ant-activation. The 10AA code assigned to AnaPS A1 is supported by multiple sequence alignment to seventeen bacterial amino acid activating A-domains (Figure S4).
Table 1.
Comparison of the 10-residue specificity sequences for select NRPS adenylation domains.
| Name/locus (module) | Pos1a (235) | Pos2 (236) | Pos3 (239) | Pos4 (278) | Pos5 (299) | Pos6 (301) | Pos7 (322) | Pos8 (330) | Pos9 (331) | Pos10 (517) | Substrateb |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gramicidin S GrsA-PheA | D | A | W | T | I | A | A | I | C | K | L-Phe |
| Surfactin SrfA-B (A5) | D | L | T | K | V | G | H | I | G | K | L-Asp |
| Bacillibactin DhbE | N | Y | S | A | Q | G | V | L | V | K | DHB |
| Mycobactin MbtA | N | F | C | A | Q | G | V | L | L | K | Sal |
| AnaPS/NFIA_055290 (A1) | G | A | L | F | F | A | A | G | V | K | Ant |
| AFUA_6g12080 (A1) | G | V | I | I | L | A | A | G | I | K | Ant |
| NFIA_057960 (A1) | G | I | I | L | G | A | A | G | I | K | Ant |
| NFIA_058030 (A1) | G | M | I | I | V | A | G | G | I | K | Ant (?) |
| NFIA_043670 (A1) | G | M | I | L | V | A | A | G | I | K | Ant (?) |
| ACLA_017890 (A1) | G | V | I | F | L | A | A | G | V | K | Ant (?) |
| ACLA_076770 (A1) | G | V | I | F | V | A | G | G | V | K | Ant (?) |
| ACLA_095980 (A1) | G | V | I | I | L | A | G | G | L | K | Ant (?) |
| AN9226.2 (A1) | G | M | I | I | V | A | A | G | I | K | Ant (?) |
| ATEG_07358 (A1) | G | I | I | L | F | G | V | G | V | K | Ant (?) |
| CONSENSUSc | G | Xh | I/L | Xh | Xh | A/G | A/G | G | Xh | K | Ant |
Residue positions (Pos) are numbered according to GrsA-PheA.
Abbreviations: DHB, 2,3-dihydroxybenzoic acid; Sal, Salicylic acid (2-hydroxybenzoic acid); Ant, anthranilic acid (2-aminobenzoic acid).
The abbreviation “Xh” stands for variable hydrophobic residues
Prediction of the 10AA code of AnaPS A1 proved useful in providing a sequence feature for the identification of putative Ant-activating modules by genome mining. A search of the Aspergillus Comparative Database (the Broad Institute) resulted in the identification of nine other fungal A-domains that matched the AnaPS A1 10AA code at positions 1 and 8 (as glycines) and at other positions in chemical property (hydrophobic or small sidechains), including by locus tag: AFUA_6g12080 A1, NFIA_057960 A1, NFIA_043670 A1, NFIA_058030 A1, ACLA_017890 A1, ACLA_079770 A1, ACLA_095980 A1, ANID_09226.1 A1, and ATEG_07358.1 A1 (Table 1, Figure S5).
Homology modeling of AnaPS A1 and AFUA_6g12080 A1
In order to further validate the predicted anthranilate 10AA code and to obtain preliminary structural insight into anthranilate binding, homology modeling of AnaPS A1 and AFUA_6g12080 A1 was performed using HHpred (Figure 5) (12).
Figure 5.
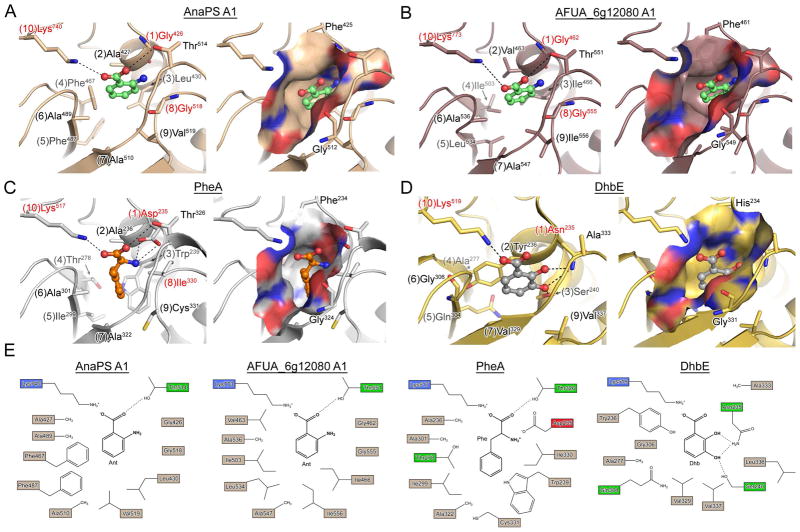
Homology models and crystal structures of NRPS adenylation domains. (A) and (B) are homology models of the proposed Ant-activating A-domains of AnaPS module 1 and AFUG_6g12080 module 1, respectively (PheA used as template). Anthranilic acid was manually docked into the binding pocket based on the PheA and DhbE structures and is rendered as green ball-and-stick. (C) Crystal structure of PheA (PDB ID: 1AMU) bound to AMP and L-Phe (orange ball-and-stick). (D) Crystal structure of DhbE (PDB ID: 1MDB) bound to 2,3-dihyhydroxybenzoyl-AMP (2,3-DHB rendered as grey ball-and-stick). For (A)-(D), left image shows specificity-determining residues as sticks, with potential hydrogen bond and/or ionic interactions drawn as dashed lines. Numbers in parenthesis preceding the residue label indicate residue position according to the 10AA specificity code. Right image presents a molecular surface representation of the substrate binding pocket colored according to electrostatic properties (blue = positive, red = negative), and includes two residues which are conserved among A-domains as Gly and Phe/His/Tyr. (E) 2D representation of the substrate selectivity residues for panels (A)–(D).
Single-template models based on the highest scoring template PheA (PDB ID: 1AMU) illustrate that the residues defining the substrate-binding pocket match those originally extracted for the 10AA code (compare to Table 1). The agreement between extracted and PheA-modeled 10AA code residues was substantiated by additional modeling of AnaPS A1 based on structures of other adenylate-forming enzymes. Models of AnaPS A1 based on PDB IDs: 3ITE, 2VSQ, 2D1S, 1PG4, and 2V7B all provided a substrate binding pocket defined by residues matching the predicted Ant 10AA code. Homology modeling of AnaPS A1 based on DhbE did not match the predicted 10AA code for position 3, 8, and 9 residues, likely due to the insertion/deletion of residues in the DhbE sequence that neighbor these positions (Figure S5). As these insertions/deletions are unique to aryl-acid activating A-domains that select for Sal or 2,3-DHB, and as AnaPS A1 possesses higher sequence similarity to PheA (43%) over DhbE (30%), we believe that the AnaPS A1 homology model based on PheA provides a more accurate representation of the putative Ant binding pocket.
The homology models of AnaPS A1 and AFUA_6g12080 A1 illustrate that the hydrophobic specificity-determining residues provide interactions and a molecular surface that is complementary in shape and chemical nature for binding the aromatic ring of anthranilate (Figure 5A and 5B). Additionally, the modeling provides a means to rationalize the strict conservation of the Pos1 and Pos8 glycines of the predicted anthranilate 10AA code (corresponding to G426, G518 of AnaPS A1 or G462, G555 of AFUA_6g12080 A1). The location of the 2-amino group of the docked anthranilate requires the presence of residues with small or no sidechains at Pos1 and Pos8 to avoid steric clash. Indeed, if the Pos1 and Pos8 glycines of AnaPS A1 or AFUA_6g12080 A1 are computationally mutated to the corresponding residues of PheA (Asp235 and Ile330), the side chains of these residues directly overlay onto the 2-amino group of the docked Ant molecule. Interestingly, with glycines present at Pos1 and 8, there are no residue sidechains identified by modeling that would provide specific interaction with the 2-amino group of Ant. In contrast, specific interaction with the 2-hydroxy group of 2,3-DHB is provided by a conserved Pos1 Asn (Figure 5D), whose amide functionality is flipped ~180° compared to the Pos1 Asp carboxylate of PheA in order to avoid clash with the substrate 2-OH substituent.
Module 1 of AFUA_6g12080 from A. fumigatus Af293 is selective for anthranilic acid
Bioinformatics analysis identifies module 1 of AFUA_6g12080 as a putative Ant-activating module
To test the predictive power of the AnaPS A1 10AA specificity code for identification of Ant-activating NRPS modules from fungal genomes, we focused our initial efforts on characterizing AFUA_6g12080 (Orf12080) from Aspergillus fumigatus Af293. Orf12080 is part of an eight gene cluster that we predict could be involved in the production of the Ant-containing alkaloid fumiquinazoline A (see SI Discussion and Figure S6 for details). Bioinformatics analysis suggests that Orf12080 is a trimodular NRPS consisting of the linear domain organization: C*-A1-T1-C2-A2-T2-E2-C3-A3-T3-C (an assignment in good agreement with previous work (28)), and identified Orf12080 C*-A1-T1 as the module predicted to select, activate, and load anthranilate. Orf12080 C*-A1-T1 (defined as amino acids 1-885) shares 34%/54% identity/similarity to module 1 of AnaPS, and the extracted 10AA specificity code of Orf12080 A1 is GVIILAAGIK (Table 1).
Expression and purification of Orf12080 m1 C*AT
The region encoding amino acids 1-885 of Orf12080 was PCR amplified from A. fumigatus Af293 cDNA and cloned into a pET30 Xa/LIC E. coli expression vector for production as an N-terminal His6-fusion protein (100 kDa). Ni-affinity purification yielded moderate levels of soluble protein (~5 mg/L culture) with minimal impurities. These results are in contrast to the observed low-level expression of soluble AnaPS Trx-C*AT protein and the poor degree of purification for Trx-C*AT post Ni-NTA chromatography (Figure S2). Of additional note is that three of the four most prominent contaminants observed for the Ni-NTA purified Orf12080 C*AT match the banding pattern and migration profile of the E. coli chaperones identified in the AnaPS Trx-C*AT preparation (Figure S2).
Ant-AMP formation and release by Orf12080 m1 C*AT
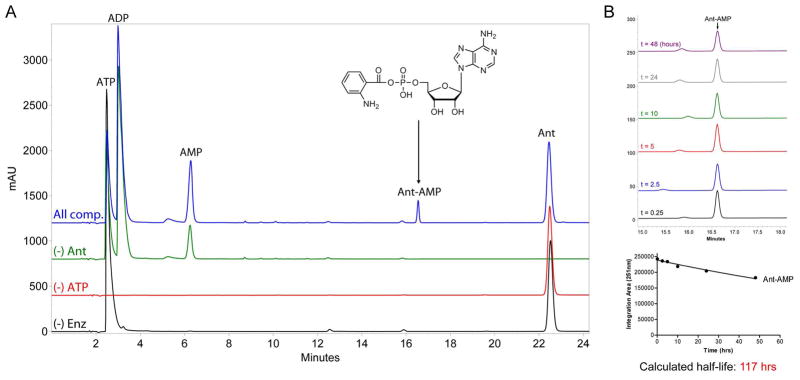
HPLC and LC/MS-based assays were used to detect Ant-AMP formation by Orf12080 C*AT as a released product, and to monitor Ant-AMP stability in the reaction medium. HPLC analysis of reactions containing purified protein, ATP, Mg2+, and anthranilic acid in buffer show a peak with a retention time of ~16.6 minutes that is not present in control reactions (Figure 7A). The identity of this peak as Ant-AMP was established by high-resolution mass spectrometry: [M+H]+ (observed 467.1079, expected 467.1075) and [2M+H]+ (observed 933.2066, expected 933.2150) (Figure S3B). The presence of ADP in the reaction lacking substrate may reflect the ATPase activity of contaminating E. coli chaperones (29). Additional confirmation of anthranilyl-AMP formation was obtained by LC/MS analysis of reactions containing combinations of 15N-Ant and/or 15N5-ATP, with 15N enrichment being detected from both the anthranilate and adenine components (Figure S3B). A listing of the high-resolution MS data obtained for all combinations of unlabeled- and 15N-labeled substrates tested is provided in Table 2. To investigate the stability of Ant-AMP in buffered solution following release from enzyme an HPLC-based time course study was performed (Figure 7B). Ant-AMP peak integration and curve fitting to a 1st order decay equation provided a calculated Ant-AMP half-life of 117 hours (nearly 5 days); surprisingly long as aminoacyl-adenylates have been characterized to be highly labile, and indicative that the released product resists water-mediated hydrolysis and does not spontaneously close to a beta lactam.
Figure 7.
HPLC-based determination of anthranilyl-AMP formation by AFUA_6g12080 module 1 C*AT and stability of Ant-AMP following release from enzyme. (A) Overlay of HPLC chromatograms (251 nm detection) obtained from 20 μL injections of control (black, red, and green traces) and experimental (blue trace) reactions monitoring the in vitro adenylation activity of C*AT with anthranilic acid as substrate. All reactions contained 50 mM HEPES (pH 7.4), 10 mM MgCl2, and 1 mM DTT; and varied combinations of 10 μM C*AT (“Enz”), 5 mM ATP, and 1 mM Ant as indicated. The chemical identity of peaks was determined by running authentic standards and high-resolution mass spectrometry. (B) The stability of Ant-AMP in reaction buffer following release from enzyme. Top image, overlay of HPLC chromatograms (251 nm detection) obtained at different timepoints post microcentrifugal filtration (10K molecular weight cutoff) of a sample prepared similar to the “All components” reaction presented in panel (A). Bottom image, quantification of Ant-AMP peak area for each timepoint used for determination of Ant-AMP stability in terms of calculated half-life.
Table 2.
High-resolution LC-MS validation of Ant-AMP formation.
| Protein assayed | Species Detected | Observed Mass | Target Mass | Difference (ppm) | |
|---|---|---|---|---|---|
| AFUA_6g12080C*AT | Ant-AMP | [M+H]+ | 467.1079 | 467.1075 | 0.85 |
| (15N)Ant-AMP | [M+H]+ | 468.1072 | 468.1045 | 5.76 | |
| Ant-(15 N5)AMP | [M+H]+ | 472.0936 | 472.0926 | 2.12 | |
| (15N)Ant-(15N5)AMP | [M+H]+ | 473.0919 | 473.0897 | 4.65 | |
Substrate selectivity of Orf12080 m1 C*AT monitored by ATP-[32P]PPi exchange assay
In order to investigate the substrate-dependent adenylation activity of Orf12080 C*AT, an end-point assay using ATP-[32P]PPi exchange was performed in a manner similar to that described for AnaPS Trx-C*AT. A variety of carboxy acids were tested, and the results provided in Figure 8A illustrate that Orf12080 C*AT exhibits fairly broad specificity for aryl acid substrates: though anthranilate promotes the highest level of exchange (~214,000 CPM), benzoate supports a level of activity nearly equivalent to Ant, while benzoic acid derivatives including 2- and 4-chloro, 3-hydroxy, and 3-aminobenzoate all promote significant levels of exchange (activities 30–70% relative to anthranilate). In contrast, the L-amino acids valine and alanine, as well as the aryl acids 2,3-DHB, 4-aminobenzoate, and salicylate promote only low-level exchange (2–8% relative to anthranilate, background level without substrate is 1%). The results obtained indicate that Ant is the preferred substrate for Orf12080 module 1.
Figure 8.
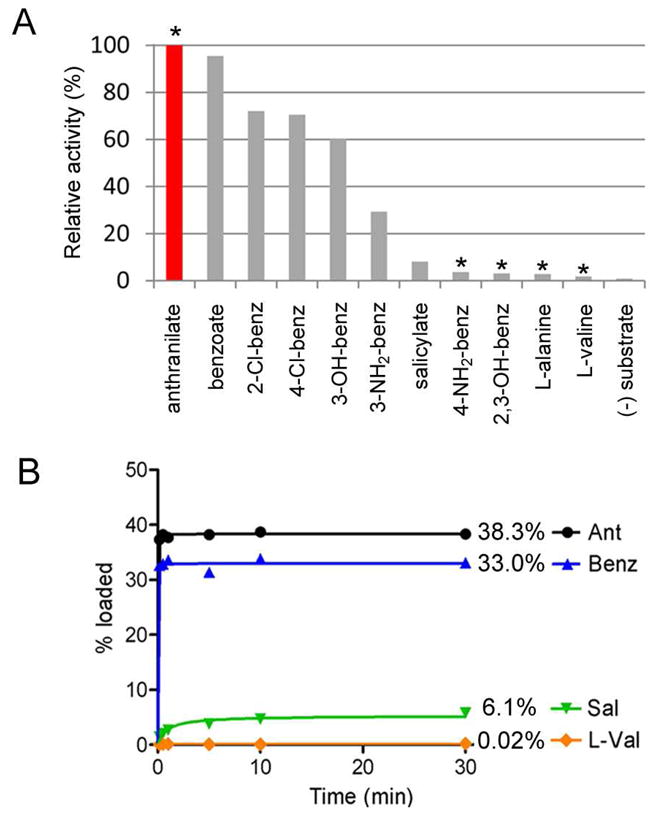
Biochemical characterization of AFUA_6g12080 module 1 C*AT. (A) ATP-[32P]PPi exchange data obtained for the adenylation activity of AFUA_6g12080 C*AT using various aryl and L-amino acids. Asterisks mark molecules which are known components of fungal primary metabolism (see Discussion). (B) Timecourse comparison for the in vitro loading of 14C-labeled anthranilate, benzoate, salicylate, and L-valine onto the holo-T-domain of C*AT.
In vitro phosphopantetheinylation of Orf12080 C*AT and loading of carboxy acid substrates
The phosphopanthetheinylation of the T-domain of Orf12080 C*AT to generate holo-protein was assessed using the enzyme Sfp from B. subtilis (11) and [1-14C]acetyl-CoA (used as a surrogate phosphopantetheine [PPT] donor). A time course experiment was performed to follow the covalent labeling process and shows that by ten minutes the phosphopantetheinylation of Orf12080 C*AT is essentially complete with a total of 28% conversion to [14C]acetyl-PPT-S-T.
The loading of radiolabeled anthranilate onto holo-Orf12080 C*AT was then performed to investigate in cis thiolation activity. Holo-C*AT protein was generated by incubating purified protein with Sfp and unlabeled CoA, then combined directly with [carboxy-14C]anthranilate, ATP, and Mg2+ in reaction buffer to reconstitute adenylate formation and acyl-group transfer to the PPT of holo-protein. A time course study was performed to follow the covalent loading process by liquid scintillation counting of TCA-precipitated protein. The results provided in Figure 8B show that 38% of Orf12080 C*AT is loaded with [14C]Ant under the conditions assayed. As expected for single-turnover reactions (e.g. thiolation occurring just once for each catalytically competent C*AT molecule in solution), loading of anthranilate occurs very quickly and is complete before the first timepoint (0.5 min). Additionally, there is no apparent loss of the loaded anthranilate from its thioester linkage on the T-domain during the timeframe investigated. A control reaction lacking ATP yields 2% loading. A control reaction lacking Sfp and CoA in the pre-incubation step, and so unable to generate holo-C*AT in vitro, yields 7% loading of Orf12080 C*AT with anthranilate. The loading of Ant onto C*AT protein was qualitatively assessed by SDS-PAGE autoradiography (Figure S8), corroborating the results of the timecourse experiments. These results indicate that some of the C*AT protein is phosphopantetheinylated by endogenous E. coli PPTases during expression, and are in good agreement with the [14C]acetyl-CoA labeling experiment: 7% endogenous PPT plus 28% in vitro PPT yielding 35% total PPT available for loading, close to the 38% detected for loading of [14C]Ant.
For comparison, loading experiments were also performed with the 14C-labeled salicylate, benzoate, and L-valine (Figure 8B). Benzoate is loaded at a rate and to a total level (33%) comparable to anthranilate. Salicylate is loaded much more slowly than anthranilate, taking greater than ten minutes to go to completion, and is only loaded to ~6% by the last timepoint. As expected from previous results obtained for AnaPS Trx-C*AT (Figure 4C), no loading of L-valine is observed. These results are diagnostic of substrate preference exhibited by Orf12080 C*AT for anthranilate for the overall process of adenylation-thiolation.
Kinetic parameters for Orf12080 m1 C*AT determined by coupled PPi-release assay
A continuous, coupled spectrophotometric assay was used to determine the kinetics of apo Orf12080 C*AT catalyzed aryl-AMP formation by monitoring PPi release from the adenylation domain. This assay employs a coupled enzyme system where conversion of PPi to Pi provides for phosphorolysis of the guanosine analog MESG and an increase in absorbance at 360 nm. Initial velocity data for PPi release was used to produce saturation curves fit to the Michaelis-Menten equation (SI Figure 7), providing a kcat of 0.57 min−1 and Km of 18 μM for Ant, a kcat of 0.63 min−1 and Km of 230 μM for ATP, and parameters for other aryl acids as listed in Table 4. The low kcat values for Orf12080 C*AT as determined by the PPi-release assay indicate slow turnover likely due to the rate-limiting slow release of the acyl-adenylate from the A-domain active site (15, 17, 30). Due to the off-pathway kcat values, a measure of substrate preference was estimated by Km comparison. It is notable that the Km values obtained by the PPi-release assay for Orf12080 C*AT with substrates Ant and ATP are comparable to values reported for other adenylate-forming enzymes (31–34). Ranking the compounds tested according to Km values indicates that Orf12080 C*AT has equal preference for Ant and Sal as substrates (Table 4), this result is in contrast to the preference of C*AT for Ant over Sal as determined by the ATP-PPi exchange and substrate loading assays. Importantly, among the aryl acid substrates tested of presumed physiological relevance to fungal metabolism (anthranilate and 4-aminobenzoate, see Discussion), the kinetic parameters obtained for Orf12080 C*AT demonstrate a clear preference for anthranilate as substrate, yielding a Km that is 100-fold lower and a kcat/Km value that is 20-fold higher than that obtained for 4-aminobenzoate.
Table 4.
Examples of fungal natural products assembled in part from anthranilate (Ant).
| Compound | AA building blocks | Producer | References |
|---|---|---|---|
| Acetylaszonalenin | Ant, Trp | N. fischeri | (9) |
| 5-N-acetylardeemin | Ant, Ala, Trp | A. terreus | (69) |
| Glyantrypine | Ant, Trp, Gly | A. clavatus | (70) |
| Alantrypinone | Ant, Trp, Ala | A. terreus | (46) |
| Fumiquinazoline A | Ant, Trp, Ala + Alaa | A. fumigatus | (44) |
| Fiscalin A | Ant, Trp, Val + Alaa | N. fischeri | (71) |
| Tryptoquivaline | Ant, Trp, Val + 2-Me-Alaa | A. clavatus | (47) |
| Asperlicin | Ant, Ant, Trp + Leua | A. alliaceus | (42) |
| Benzomalvin C | Ant, Ant, Phe | Penicillium sp. | (41) |
| Circumdatin F | Ant, Ant, Ala | A. ochraceus | (72) |
| Sclerotigenin | Ant, Ant, Gly | P. sclerotigenum | (73) |
| Verrucine A | Ant, Gln, Phe | P. verrucosum | (74) |
| Auranthine | Ant, Gln, Ant | P. aurantiogriseum | (4) |
| Cyclopenin | Ant, Phe | P. cyclopium | (75) |
The amino acids listed before the “+” sign are postulated to form a cyclized tripeptide core structure that is further modified by incorporation of the amino acid following the “+” sign (see Figure S6B for an example).
Module 1 of NFIA_057960 from N. fischeri NRRL 181 selectively activates anthranilic acid
Cloning, expression, and, purification of NFIA_057960 module 1 C*AT
As a second test of the predictive power of the AnaPS 10AA code to identify Ant-activating NRPS modules from fungi, we chose to study module 1 of NFIA_057960 from N. fischeri NRRL 181. NFIA_057960 is predicted to be a trimodular NRPS with the domain organization C*-A1-T1-C2-A2-T2-C3-A3-T3-C; the domain organization is similar to AFUA_6g12080, but in NFIA_057960 there is no E-domain present in module 2. Module 1 of NFIA_057960 (residues 1-848) was identified as the putative Ant-activating module, possessing a 10AA specificity code of GIILGAAGIK (Table 1), and sharing 35%/51% identity/similarity in overall amino acid sequence to module 1 of N. fischeri AnaPS.
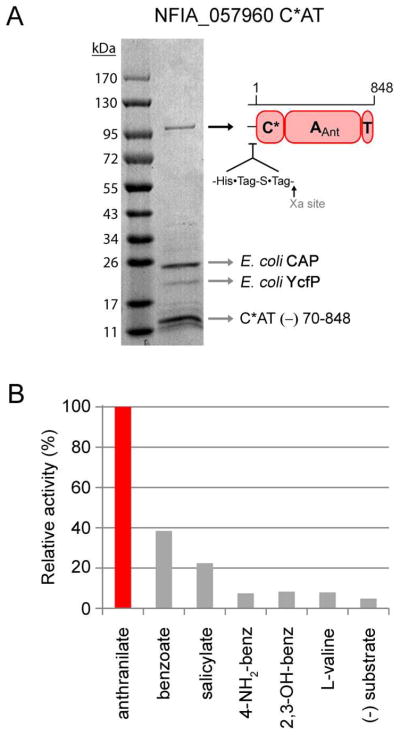
The region encoding residues 1-848 of NFIA_057960 was cloned from genomic DNA into the E. coli expression vector pET30 Xa/LIC for overproduction of C*A1-T1 as an N-terminal His6-tagged fusion protein (100 kDa). The yield of soluble protein was low, and as the purity level following Ni-affinity chromatography was poor (see Figure S2), anion-exchange and gel-filtration chromatography were performed to futher purify the protein (Figure 9A). Although the final purity achieved is less than ideal, the major contaminating bands post-three-step purification are not expected to possess activity that would interfere with subsequent C*AT enzyme characterization.
Figure 9.
Purification and characterization of NFIA_057960 module 1 C*AT. (A) Purity of the C*AT protein following Ni-NTA, anion-exchange, and gel-filtration chromatography. The identity of the ~100 kDa band as full-length C*AT protein, and the identities of the low-molecular-weight impurities indicated by arrows were determined by mass fingerprinting (CAP, catabolite gene activator protein; YcfP, conserved hypothetical protein; C*AT (−) 70-848, fragment of full-length protein missing residues ≈ 70-848). (B) Adenylation activity of C*AT with a panel of aryl-acids assessed by ATP-[32P]PPi exchange assay. L-valine is provided as a negative control.
Characterization of NFIA_057960 module 1 C*AT
Enzyme activity and specificity toward anthranilate were verified using ATP-[32P]PPi exchange assay. An end-point assay was performed (reactions quenched after 1.5 hrs) and the results suggest that NFIA_057960 exhibits a 2–14 fold preference for anthranilate over other aryl-acids tested (Figure 9B). Although the total exchange activity promoted by anthranilate (~6500 CPM) is low compared to the activity measured for AFUA_6g12080 C*AT, this level of activity is significantly above background (316 CPM for a no-substrate control) and as such is highly diagnostic for the anthranilate-specific adenylation activity of NFIA_057960 C*AT. Anthranilate-dependent enzyme activity was further supported by reconstitution of Ant-AMP formation by NFIA_057960 C*AT and detection as a released product by LC/MS (Figure S3C). These results provide experimental support that module 1 of NFIA_057960 is anthranilate-activating, and help validate the 10AA code described in this work for the identification of fungal Ant-specific A-domains.
DISCUSSION
Fungal metabolites with benzodazepinone and quinazolinone polycyclic scaffolds
Fungi biosynthesize a variety of polycyclic natural products that may arise from short nonribosomal peptide synthetase assembly lines (28, 36). The morphing of linear peptide chains into architecturally constrained molecules with high functional group densities underlie the high affinities and biological selectivities reported with several classes of fungal natural products, both as therapeutics and toxins (such as penicillin (37), gliotoxin (38), and cyclopiazonic acid (39)). Many bioactive polycylic compounds produced by fungi contain quinazolinone and/or benzodiazepinone scaffold elements including the hexacylic chemosensitizing agent N-acetylardeemin (40), the pentacyclic mycotoxin acetylaszonalenin (9), the tetracyclic substance P inhibitor benzomalvin (41), the tetracyclic scaffold of the cholecystokinen (CCK) receptor antagonist asperlicin (42, 43), the tricyclic framework of the antitumor fumiquinazolines and fiscalins (44, 45), the bridged-tricyclic insecticidal agent alantrypinone (46), and the bicylic core of the mycotoxin tryptoquivaline (47) (Figure 1, Figure S1). We hypothesize that a central monomer building block used to biosynthesize these polycyclic fungal peptide/alkaloid frameworks is the nonproteinogenic amino acid anthranilate (2-aminobenzoate). Anthranilate is a known component of bacterial, fungal, and plant primary metabolism, arising from enzymatic amination of chorismate by anthranilate synthases and serving as the framework for the indole ring of the amino acid tryptophan (48–50). While no anthranilate-specific adenylation domains of NRPS modules have been previously characterized, inspection of the molecules presented in Figure 1 and Figure S1 clearly identifies the anthranilate-derived moieties.
A substantial effort has been dedicated to the chemical synthesis of fungal natural products containing quinazolinone and benzodiazepinone scaffolds (8, 51), though an understanding of their natural assembly has been generally limited to precursor feeding studies (4–7). In at least one case, cyclopeptine production by Penicillium cylopium, protein crude extracts were used to demonstrate ATP-dependent benzodiazepine alkaloid assembly through a thiol-template mechanism characteristic of NRPSs (52). More recently, the biosynthetic gene cluster responsible for acetylaszonalenin biosynthesis from Neosartorya fischeri was identified by genome mining, and the enzymes responsible for the post-NRPS acetylation and prenylation tailoring reactions were characterized (9). However, the basis of fungal NRPS selection and incorporation of anthranilate, the mechanism of morphing the NRPS-templated intermediate into a multicylic scaffold, and many of the additional tailoring reactions necessary to yield the final natural product remain to be investigated. In comparison, genetic analysis has elucidated several biosynthetic routes to bacterial benzodiazepine natural products which incorporate substituted anthranilates including anthramycin (incorporating 4-methyl-3-hydroxyanthranilate) (53, 54), sibiromycin (4-methyl-3,5-dihydroxyanthranilate) (55), tomaymycin (4,5-hydroxyanthranilate) (56), and diazepinomicin (3-hydroxyanthranilate) (57). With the exception of diazepinomicin, the assembly of the core tricyclic scaffold is proposed to be NRPS-mediated.
In this work we took an initial step to understand the nonribosomal assembly of Ant-containing fungal alkaloids through identification, purification, and characterization of three NRPS modules predicted to specifically select, activate, and load anthranilate.
Identification and characterization of Ant-activating NRPS modules
By association with the known biosynthetic pathway for the Ant-containing metabolite acetylaszonalenin from N. fischeri NRRL 181 (9), AnaPS was used as our initial target for characterization of Ant-activating NRPS modules from fungi. From bioinformatic analysis we predicted that module 1 of AnaPS would be Ant-specific and therefore that this bimodular NRPS should make an Ant-Trp-S-T2-tethered intermediate en route to acetylaszonalenin (Figure 3). Expression of AnaPS module 1 C*A1T1 as a thioredoxin-fusion protein allowed experimental validation of anthranilate-specific adenylation activity and installment of the activated anthranilate onto the module 1 T-domain. AnaPS A1 is the first validated fungal adenylation domain for activation of the rigid 2-aminobenzoate monomer, and allowed us to examine the ten residue specificity-code of AnaPS A1 as a tool for the identification of additional fungal Ant-activating NRPS modules.
A search of the eight genomes present in the Aspergillus Comparative Database (the Broad Institute) identified nine additional putative Ant-activating A-domains based on similarity of the extracted 10AA specificity sequence to that determined for AnaPS A1 (Table 1). Intriguingly, all nine A-domains identified are part of the first module of their associated NRPS assembly lines, and all are predicted to possess the domain organization C*A1T1. Of the nine identified, we chose two NRPSs for experimental characterization of anthranilate selectivity: AFUA_6g12080 from A. fumigatus Af293 and a second from N. fischeri NRRL 181, NFIA_057960. Experimental validation of anthranilate activation (as anthranilyl-AMP) was demonstrated for both C*A1T1 constructs, while anthranilate loading (as Ant-S-T1 thioester) was confirmed for Orf12080 C*A1T1. The characterization of these two constructs provides experimental support (as a “statistical duplicate”) to validate that the AnaPS A1 10AA code may be successfully applied for the prediction of fungal Ant-activating NRPS modules.
The predicted 10AA specificity-determining code of fungal Ant-activating A-domains
A consensus sequence for the predicted fungal anthranilate 10AA code may be derived from the ten representative members identified in this work as: GXh(I/L)XhXh(A/G)(A/G)GXhK, where Xh represents positions defined by variable hydrophobic amino acids (Table 1). Two conserved and unique features of the predicted anthranilate 10AA code are glycines at Pos1 and Pos8. Homology modeling of the module 1 A-domains of AnaPS and AFUA_6g12080 indicate the Pos1 and Pos2 glycines may be necessary to create space for accommodation of the 2-amino substituent of anthranilate for substrate binding. A strictly conserved Lys is present at Pos10 (a universal feature of carboxy acid activating adenylation enzymes) (22, 25, 58), while functional conservation of residues with sidechains that are hydrophobic or small is observed at other 10AA code positions. Homology modeling of AnaPS A1 and Orf12080 A1 demonstrates that distinct compositions of hydrophobic/small residues at the non-conserved positions of the 10AA code can effectively provide a molecular surface that is complementary to binding the compact benzene ring of anthranilate (Figure 5A, 4B).
The Pos1 residue of the NRPS 10AA code has demonstrated importance in substrate binding through interaction with the α-amino (Pos1 Asp) (20) or aryl acid 2-OH (Pos1 Asn) (59) substituent that neighbors the substrate carboxylate. For the Ant-activating A-domains identified in this work the Pos1 residue is strictly conserved as a glycine. Due to the predicted orientation of the backbone carbonyl (as part of an α-helix) and the lack of sidechain functionally, this Pos1 Gly is not predicted to provide interaction with the 2-NH2 group beyond steric contact (Figure 5). It may be that other binding-pocket residues not identified by our bioinformatics approaches are involved in 2-NH2 group interaction, or that other modes of substrate selectivity are in place. Of note is that Ala is the predicted Pos1 residue among bacterial A-domains that activate substituted anthranilates in pyrrolobenzodiazepine biosynthesis (54) (Figure S5). Neither Gly nor Ala would provide specific interaction with the aryl acid 2-NH2 substituent, in contrast to residues that might be expected at this position such as Ser/Thr. This comparison indicates that a Pos1 Gly may be a hallmark feature of fungal NRPS A-domains selective for Ant.
Among nearly 400 NRPS A-domains of known specificity (23), the only others predicted to possess a glycine at Pos1 are known to activate D-2-hydroxyisovaleric acid (D-Hiv) in enniatin (60, 61), beauvericin (62), and bassionolide (63) biosynthesis. The consensus sequence for these D-Hiv activating A-domains has been assigned as GALx(I/V)VG(S/T)IK, where the “x” is variable (M, L, or H) (62). Comparison of the Ant and D-Hiv 10AA codes reveals that there are only two positions of distinction: 1) Pos6, Ant code Ala/Gly vs. D-Hiv code Val, and 2) Pos8, Ant code Gly vs. D-Hiv code Ser/Thr. Of these two differences, the Pos8 change may be the most useful for differentiating between Ant and D-Hiv A-domain substrate selectivities based on 10AA code matching. Importantly, the ability of the three module 1 C*AT constructs to selectively activate Ant over D-Hiv was tested by ATP-PPi exchange, with results consistent with their identification as anthranilate synthetases (Figure S9).
Substrate selectivity of Ant-activating fungal NRPS modules
The results of ATP-PPi exchange assay for the module 1 C*AT constructs show good agreement for substrate selectivity among common molecules tested: Ant > benzoate > Sal > 4-NH2-benzoate ≈ 2,3-DHB ≈ L-Val ≈ background (Figures 4B, 8A, and 9B). Anthranilate promotes the highest level of exchange for all three C*AT constructs tested, confirming the expected Ant-specific activity of AnaPS module 1 and supporting that Ant is the physiological substrate for the module 1 constructs of Orf12080 and NFIA_057960. After anthranilate, benzoate promotes the next highest level of exchange with relative activities of 17% for AnaPS Trx-C*AT, 95% for AFUA_6g12080 C*AT, and 38% for NFIA_057960 C*AT. The ability of the C*AT enzymes to utilize benzoate as a substrate is expected as the absence of additional aryl-ring substituent functionality would allow productive binding in the enzyme active-site. The high degree of specificity for anthranilate over salicylate is somewhat surprising as the chemical difference between these two molecules is seemingly small (2-NH2 vs. 2-OH). Both substituents are characterized as benzene ring-activating groups and both are polar functional groups with hydrogen bonding potential. Although a structure-based rationale for the preference for Ant over Sal is presently unclear, it is of note that a similar degree of selectivity for Ant over Sal has been observed for the Ant-CoA ligase PqsA from Pseudomonas aeruginosa (32). L-valine, used as a negative control, suggests that the C*AT enzymes can effectively distinguish between standard α-amino acids and the planar β-amino acid Ant.
An expanded panel of potential substrates was tested with AFUA_6g12080 C*AT, showing that in addition to anthranilate and benzoate, significant exchange activity is also promoted by 2-Cl-benzoate, 4-Cl-benzoate, 3-OH-benzoate, and 3-NH2-benzoate (Figure 8A). Tolerance by adenylate-forming enzymes to activate noncognate aryl acids has been reported previously (15, 32, 64–66). Importantly, among the aryl acids found to be efficiently activated by Orf12080 C*AT only anthranilate is reported to be a common constituent of fungal metabolism. The additional aryl acids tested which are known components of fungal metabolism include 4-aminobenzoate (a building block used in folate biosynthesis) (67), and 2,3-dihydroxybenzoate (arising from anthranilate catabolism) (68); however, relative to anthranilate these molecules are only activated to 4% and 3%, respectively (Figure 8A). Similar low-level activity for 4-aminobenzoate and 2,3-DHB is also observed for AnaPS Trx-C*AT and NFIA_057960 C*AT. Therefore, the selective pressure introduced by the metabolic pool may generally only require distinguishing between 2-amino- (Ant), 4-amino-, and 2,3-dihydroxy-substituted benzoic acids, which all three module 1 C*AT constructs are able to accomplish very effectively.
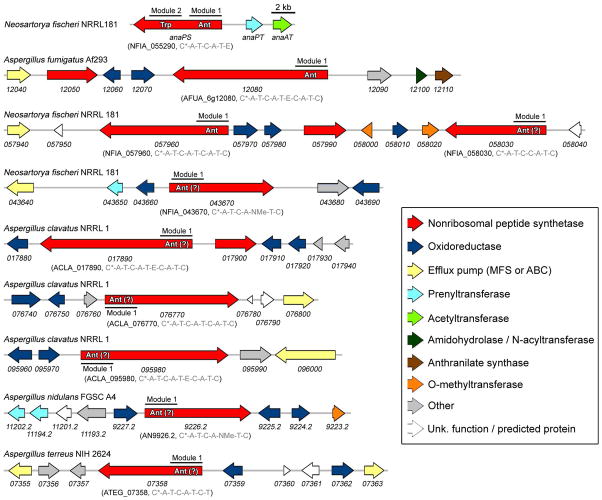
Gene clusters from Aspergillus fungi predicted to contain Ant-activating modules
Ant-containing alkaloids have been isolated from a variety of fungi of the phylum Ascomycota (1, 8), most notably by fungi of the genus Aspergillus (teleomorph Neosartorya) and Penicillium (Table 4).
The first seven compounds listed in Table 4 (shaded grey) have been isolated from fungi that share genus and species classification with a strain whose genome has been sequenced. However, the sequenced strain is a verified producer of the compound listed in just two cases: acetylaszonalenin from N. fischeri NRRL 181 (9), and fumiquinazoline A from A. fumigatus Af293 (76). By genome mining a biosynthetic gene cluster for acetylaszonalenin biosynthesis has previously been reported (9), and in this work we identify a possible cluster for fumiquinazoline biosynthesis (see Supporting Discussion). Figure 10 shows eight other gene clusters from Neosartorya fischeri or Aspergillus clavatus, nidulans, and terreus, where anthranilate activation is predicted to be the chain-initiating step for the NRPS-based assembly of polycyclic alkaloids. Interestingly, Ant-containing metabolites or putative Ant-activating modules were not identified from A. flavus, A. niger, or A. oryzae.
Figure 10.
Organization of fungal biosynthetic gene clusters containing, or proposed to contain, anthranilic acid-activating nonribosomal peptide synthetases. The locus tag and NRPS domain organization (from N- to C-terminus) is provided in parenthesis. The minimum domains constituting a single NRPS module include C*/C-A-T. Additional domains sometimes present as part of a single module are an E-domain (epimerization), or an NMe-domain (N-methylation). The N-terminal C* domains of NFIA_058030, NFIA_043670, and AN9226.2 are about one-third the size of the C* domains of the other NRPSs (which are predicted to be defined by ~200 residues).
Sequence analysis of the putative Ant-activating NRPS genes indicates bimodular or trimodular organization, a finding consistent with their involvement in assembling the di- or tripeptide scaffolds of Ant-containing fungal metabolites. The neighboring monomodular NRPSs found in the AFUA_6g12080, NFIA_057960, and ACLA_017890 clusters may select for a single amino acid for subsequent coupling to the polyclic Ant-containing scaffold produced by their multimodular NRPS counterparts (see Figure S6B for an example). Several Ant-containing natural products may be assembled by such partnered NRPS systems, including the asperlicins, fumiquinazolines, fiscalins, and tryptoquivalines (8). With the exception of the three-gene acetylaszonelin cluster, all clusters identified contain one or more genes encoding putative oxidoreductases. These enzymes could function in tailoring reactions such as oxidative coupling via epoxidation of the 2,3-double bond of Trp indole (aperlicin, fumiquinazoline A, fiscalin A, trytoquivaline), epoxidation of an α-β unsaturated phenylalanyl moiety (benzomalvin C, cyclopenin), oxidation of position 2 of the Trp indole (alantrypinone), or aromatic hydroxylation of anthranilate-derived moieties (circumdatins C-E, G and H) (8). A putative anthranilate synthase is found in only one of the ten clusters, and so it may be that the metabolic pool of anthranilate derived from Trp biosynthesis suffices for the production of several Ant-containing alkaloids.
A conserved feature among the putative Ant-activating NRPS genes is an amino-terminal C*-domain and a carboxy-terminal C-domain. The C* is predicted to be non-functional as sequence alignment and structural modeling suggests that the N-terminal sub-domain (which possesses a conserved HHxxxDG motif for enzyme activity (77–79)) is completely missing. The crystal structure of the multidomain SrfA-C shows a significant interaction interface between the tandem C- and A-domains (80), suggesting that the C* of the putative Ant-activating NRPSs may play a structural role for protein stability. The C-terminal condensation domain, in place of a thioesterase or “reductive” domain normally present for release of the peptidyl-S-T intermediate, is fairly common for fungal NRPSs (81). One proposed role of terminal C-domains is to mediate chain release by catalyzing inter- or intramolecular cyclization via amide bond formation (82), which is what we predict occurs in the acetylaszonalenin (Figure 3C) and fumiquinazoline (Figure S6B) pathways. In the case of acetylaszonalenin biosynthesis the terminal E-domain of AnaPS, which is related in both sequence and structure to the C-domain (79), could serve as a bifunctional epimerase/cyclase.
Two of the predicted Ant-activating NRPSs, AN9926.2 and NFIA_043670, are sometimes annotated as (or predicted to be) enniatin synthetases. This may be due to the similar size of the encoded NRPSs (bimodular) and the fact that the two putative Ant-activating NRPSs contain an embedded N-methylation domain first identified for the enniatin and beauvericin synthetases. However, a closer comparison of domain organization and 10AA code matching suggests that these two NRPSs produce N-methylated Ant-containing alkaloids (e.g. benzomalvin or cyclopenin, Figure S1) and not N-methylated cyclodepsipeptides such as the enniatins. The domain organization of the enniatin and beauvericin synthetases is: C-A-T-C-A-NMe-T-T-C, where the N- and C-terminal C-domains and tandem T-T domains are hallmark features for the proposed route of iterative cyclodepsipeptide biosynthesis (61, 62). In comparison, the domain organization of NFIA_043670 and AN9926.2 is C*-AT-C-A-NMe-T-C, which lacks the tandem T-domains and contains a C* domain at the N-terminus may produce a cyclic dipeptide in collinear fashion. Additionally, the predicted specificity sequences for AN9926.2 and NFIA_043760 are 80% identical (100% similar) and 70% identical (100% similar) to the validated Ant-activating A-domain of AFUA_6g12080, respectively.
In sum, herein we describe the biochemical validation of anthranilate-dependent acyl-adenylate formation and acyl-group loading onto thiolation domains by module 1 constructs of AnaPS (NFIA_055290), AFUA_6g12080, and NFIA_057960. This work supports the predictive method based on the Ant 10AA code used to identify the gene clusters provided in Figure 10, and sets the stage for the identification/characterization of gene clusters from both sequenced and unsequenced genomes involved in the biosynthesis of anthranilate-containing natural products.
Supplementary Material
Figure 6.
Purified recombinant AFUA_6g12080 C*A1T1 following heterologous expression in E. coli and Ni-affinity chromatography. The identity of the indicated band as the full-length protein was confirmed by peptide mass fingerprinting.
Acknowledgments
We thank Professor Robert A. Cramer for providing A. fumigatus Af293. We thank Professor Michael A. Fischbach for helpful discussions. We thank Thomas Gerken for providing A. fumigatus Af293 cDNA and Elizebeth Sattely for purified Sfp. We are grateful to Wenjun Zhang and Michael Acker for their careful reading of the manuscript.
Abbreviations and Textual Footnotes
- 10AA
ten amino acid
- 2,3-DHB
2,3-dihydroxybenzoic acid
- ADP
adenosine-5′-diphosphate
- AMP
adenosine-5′-monophosphate
- Ant
anthranilate (2-aminobenzoate)
- ATP
adenosine-5′-triphospate
- BSA
bovine serum albumin
- CoA
coenzyme A
- CPM
counts per minute
- D-Hiv
D-2-hydroxyisovaleric acid
- DMSO
dimethyl sulfoxide
- DTT
dithiothreitol
- EDTA
ethylenediaminetetraacetic acid
- ESI
electrospray ionization
- FPLC
fast protein liquid chromatography
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- HPLC
high-performance liquid chromatography
- IPTG
isopropyl-β-D-galactopyranoside
- LC/MS
liquid chromatography/mass spectrometry
- LB
Luria-Bertani medium
- MALDI-TOF
matrix-assisted laser desorption ionization time-of-flight
- MeCN
acetonitrile
- MESG
2-amino-6-mercapto-7-methylpurine riboside
- Ni-NTA
nickel nitrilotriacetic acid-agarose
- NRPS
nonribosomal peptide synthetase
- Orf
open reading frame
- PCR
polymerase chain reaction
- PDB ID
Protein Data Bank identifier
- Pi
inorganic phosphate
- PPi
inorganic pyrophosphate
- PPT
4′-phosphopantetheine
- PPtase
phosphopantetheinyl transferase
- Q-TOF
quadrupole time-of-flight
- Sal
salicylic acid (2-hydroxybenzoic acid)
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- TCA
trichloroacetic acid
- TCEP
tris(2-carboxyethyl)phosphine
- Tris
Tris(hydroxymethyl)aminomethane
- Trx
thioredoxin
Footnotes
This work was supported in part by NIH Grant GM49338 (to C.T.W.) and by Award Number F32GM090475 from the National Institute of General Medical Sciences (to B.D.A.).
SUPPORTING INFORMATION AVAILABLE
Discussion of a putative fumiquinazoline A gene cluster and hypothetical biosynthetic route. Figures providing additional examples of Ant-containing fungal alkaloids (Figure S1), gel of Ni-NTA purified module 1 C*AT proteins (Figure S2), high-resolution LC/MS data for Ant-AMP (Figure S3), sequence alignments of NRPS A-domains (Figures S4 and S5), depiction of putative fumiquinazoline A gene cluster and biosynthetic route (Figure S6), saturation curves for Orf12080 C*AT kinetics (Figure S7), autoradiograph of SDS-PAGE gel showing loading of 14C-Ant onto Orf12080 C*AT (Figure S8), and ATP-PPi exchange data comparing the activity promoted by Ant vs. D-Hiv (Figure S9). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.D’Yakonov A, Telezhenetskaya M. Quinazoline alkaloids in nature. Chem Nat Compd. 1997;33:221–267. [Google Scholar]
- 2.Finking R, Marahiel MA. Biosynthesis of nonribosomal peptides. Annu Rev Microbiol. 2004;58:453–488. doi: 10.1146/annurev.micro.58.030603.123615. [DOI] [PubMed] [Google Scholar]
- 3.Sattely ES, Fischbach MA, Walsh CT. Total biosynthesis: in vitro reconstitution of polyketide and nonribosomal peptide pathways. Nat Prod Rep. 2008;25:757–793. doi: 10.1039/b801747f. [DOI] [PubMed] [Google Scholar]
- 4.Yeulet SE, Mantle PG. Biosynthesis of auranthine by Penicillium aurantiogriseum. FEMS Microbiol Lett. 1987;41:207–210. [Google Scholar]
- 5.Houck DR, et al. On the biosynthesis of asperlicin and the directed biosynthesis of analogs in Aspergillus alliaceus. J Antibiot (Tokyo) 1988;41:882–891. doi: 10.7164/antibiotics.41.882. [DOI] [PubMed] [Google Scholar]
- 6.Penn J, Purcell M, Mantle PG. Biosynthesis of glyantrypine by Aspergillus clavatus. FEMS Micro Lett. 1992;92:229–233. doi: 10.1016/0378-1097(92)90714-y. [DOI] [PubMed] [Google Scholar]
- 7.Nover L, Luckner M. On the biosynthesis of cyclopenin and cyclopenol, benzodiazepine alkaloids from Penicillium cyclopium Westling. Eur J Biochem. 1969;10:268–273. doi: 10.1111/j.1432-1033.1969.tb00684.x. [DOI] [PubMed] [Google Scholar]
- 8.Mhaske SB, Argade NP. The chemistry of recently isolated naturally occurring quinazolinone alkaloids. Tetrahedron. 2006;62:9787–9826. [Google Scholar]
- 9.Yin WB, Grundmann A, Cheng J, Li SM. Acetylaszonalenin biosynthesis in Neosartorya fischeri: identification of the biosynthetic gene cluster by genomic mining and functional proof of the genes by biochemical investigation. J Biol Chem. 2009;284:100–109. doi: 10.1074/jbc.M807606200. [DOI] [PubMed] [Google Scholar]
- 10.Webb MR. A continuous spectrophotometric assay for inorganic phosphate and for measuring phosphate release kinetics in biological systems. Proc Natl Acad Sci USA. 1992;89:4884–4887. doi: 10.1073/pnas.89.11.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quadri LEN, et al. Characterization of Sfp, a Bacillus subtilis Phosphopantetheinyl Transferase for Peptidyl Carrier Protein Domains in Peptide Synthetaseses. Biochemistry. 1998;37:1585–1595. doi: 10.1021/bi9719861. [DOI] [PubMed] [Google Scholar]
- 12.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucl Acids Res. 2005;33:W244–248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmelz S, Naismith JH. Adenylate-forming enzymes. Curr Opin Struct Biol. 2009;19:666–671. doi: 10.1016/j.sbi.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.LaVallie ER, et al. A thioredoxin gene fusion expression system that circumvents inclusion body formation in the E. coli cytoplasm. Nat Biotech. 1993;11:187–193. doi: 10.1038/nbt0293-187. [DOI] [PubMed] [Google Scholar]
- 15.Rusnak F, Faraci WS, Walsh CT. Subcloning, expression, and purification of the enterobactin biosynthetic enzyme 2,3-dihydroxybenzoate-AMP ligase: demonstration of enzyme-bound (2,3-dihydroxybenzoyl)adenylate product. Biochemistry. 1989;28:6827–6835. doi: 10.1021/bi00443a008. [DOI] [PubMed] [Google Scholar]
- 16.Ehmann DE, Shaw-Reid CA, Losey HC, Walsh CT. The EntF and EntE adenylation domains of Escherichia coli enterobactin synthetase: Sequestration and selectivity in acyl-AMP transfers to thiolation domain cosubstrates. Proc Natl Acad Sci USA. 2000;97:2509–2514. doi: 10.1073/pnas.040572897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keating TA, Suo Z, Ehmann DE, Walsh CT. Selectivity of the yersiniabactin synthetase adenylation domain in the two-step process of amino acid activation and transfer to a holo-carrier protein domain. Biochemistry. 2000;39:2297–2306. doi: 10.1021/bi992341z. [DOI] [PubMed] [Google Scholar]
- 18.Luo L, Walsh CT. Kinetic Analysis of Three Activated Phenylalanyl Intermediates Generated by the Initiation Module PheATE of Gramicidin S Synthetase. Biochemistry. 2001;40:5329–5337. doi: 10.1021/bi015518+. [DOI] [PubMed] [Google Scholar]
- 19.Lambalot RH, et al. A new enzyme superfamily -- the phosphopantetheinyl transferases. Chem Biol. 1996;3:923–936. doi: 10.1016/s1074-5521(96)90181-7. [DOI] [PubMed] [Google Scholar]
- 20.Conti E, Stachelhaus T, Marahiel MA, Brick P. Structural basis for the activation of phenylalanine in the non-ribosomal biosynthesis of gramicidin S. EMBO J. 1997;16:4174–4183. doi: 10.1093/emboj/16.14.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Challis GL, Ravel J, Townsend CA. Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem Biol. 2000;7:211–224. doi: 10.1016/s1074-5521(00)00091-0. [DOI] [PubMed] [Google Scholar]
- 22.Stachelhaus T, Mootz HD, Marahiel M. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem Biol. 1999;6:493–505. doi: 10.1016/S1074-5521(99)80082-9. [DOI] [PubMed] [Google Scholar]
- 23.Rausch C, Weber T, Kohlbacher O, Wohlleben W, Huson DH. Specificity prediction of adenylation domains in nonribosomal peptide synthetases (NRPS) using transductive support vector machines (TSVMs) Nucl Acids Res. 2005;33:5799–5808. doi: 10.1093/nar/gki885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.von Döhren H. Biochemistry and General Genetics of Nonribosomal Peptide Synthetases in Fungi. Molecular Biotechnolgy of Fungal beta-Lactam Antibiotics and Related Peptide Synthetases. 2004:217–264. doi: 10.1007/b99262. [DOI] [PubMed] [Google Scholar]
- 25.Bushley K, Ripoll D, Turgeon BG. Module evolution and substrate specificity of fungal nonribosomal peptide synthetases involved in siderophore biosynthesis. BMC Evol Biol. 2008;8:328. doi: 10.1186/1471-2148-8-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Torsten S, et al. Nonribosomal peptide synthesis in Schizosaccharomyces pombe and the architectures of ferrichrome-type siderophore synthetases in Fungi. Chembiochem. 2006;7:612–622. doi: 10.1002/cbic.200500301. [DOI] [PubMed] [Google Scholar]
- 27.Ansari MZ, Yadav G, Gokhale RS, Mohanty D. NRPS-PKS: a knowledge-based resource for analysis of NRPS/PKS megasynthases. Nucl Acids Res. 2004;32:W405–413. doi: 10.1093/nar/gkh359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stack D, Neville C, Doyle S. Nonribosomal peptide synthesis in Aspergillus fumigatus and other fungi. Microbiology. 2007;153:1297–1306. doi: 10.1099/mic.0.2006/006908-0. [DOI] [PubMed] [Google Scholar]
- 29.Evstigneeva ZG, Solov’eva NA, Sidel’nikova LI. Structures and functions of chaperones and chaperonins. Appl Biochem Biotechnol. 2001;37:1–13. [PubMed] [Google Scholar]
- 30.Luo L, Burkart MD, Stachelhaus T, Walsh CT. Substrate Recognition and Selection by the Initiation Module PheATE of Gramicidin S Synthetase. Journal of the American Chemical Society. 2001;123:11208–11218. doi: 10.1021/ja0166646. [DOI] [PubMed] [Google Scholar]
- 31.Altenschmidt U, Oswald B, Fuchs G. Purification and characterization of benzoate-coenzyme A ligase and 2-aminobenzoate-coenzyme A ligases from a denitrifying Pseudomonas sp. J Bacteriol. 1991;173:5494–5501. doi: 10.1128/jb.173.17.5494-5501.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coleman JP, et al. Pseudomonas aeruginosa PqsA is an anthranilate-coenzyme A ligase. J Bacteriol. 2008;190:1247–1255. doi: 10.1128/JB.01140-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bains J, Boulanger MJ. Biochemical and Structural Characterization of the Paralogous Benzoate CoA Ligases from Burkholderia xenovorans LB400: Defining the Entry Point into the Novel Benzoate Oxidation (box) Pathway. Journal of Molecular Biology. 2007;373:965–977. doi: 10.1016/j.jmb.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 34.Mootz HD, Marahiel MA. The tyrocidine biosynthesis operon of Bacillus brevis: complete nucleotide sequence and biochemical characterization of functional internal adenylation domains. J Bacteriol. 1997;179:6843–6850. doi: 10.1128/jb.179.21.6843-6850.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Couch R, O’Connor SE, Seidle H, Walsh CT, Parry R. Characterization of CmaA, an Adenylation-Thiolation Didomain Enzyme Involved in the Biosynthesis of Coronatine. J Bacteriol. 2004;186:35–42. doi: 10.1128/JB.186.1.35-42.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoffmeister D, Keller NP. Natural products of filamentous fungi: enzymes, genes, and their regulation. Nat Prod Rep. 2007;24:393–416. doi: 10.1039/b603084j. [DOI] [PubMed] [Google Scholar]
- 37.Schofield CJ, et al. Proteins of the penicillin biosynthesis pathway. Curr Opin Struct Biol. 1997;7:857–864. doi: 10.1016/s0959-440x(97)80158-3. [DOI] [PubMed] [Google Scholar]
- 38.Balibar CJ, Walsh CT. GliP, a multimodular nonribosomal peptide synthetase in Aspergillus fumigatus, makes the diketopiperazine scaffold of gliotoxin. Biochemistry. 2006;45:15029–15038. doi: 10.1021/bi061845b. [DOI] [PubMed] [Google Scholar]
- 39.Chang P-K, Ehrlich K, Fujii I. Cyclopiazonic acid biosynthesis of Aspergillus flavus and Aspergillus oryzae. Toxins. 2009;1:74–99. doi: 10.3390/toxins1020074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karwowski JP, et al. 5-N-Acetylardeemin, a novel heterocyclic compound which reverses multiple drug resistance in tumor cells. I Taxonomy and fermentation of the producing organism and biological activity. J Antibiot (Tokyo) 1993;46:374–379. doi: 10.7164/antibiotics.46.374. [DOI] [PubMed] [Google Scholar]
- 41.Sun HH, Barrow CJ, Sedlock DM, Gillum AM, Cooper R. Benzomalvins, new substance P inhibitors from a Penicillium sp. J Antibiot (Tokyo) 1994;47:515–522. doi: 10.7164/antibiotics.47.515. [DOI] [PubMed] [Google Scholar]
- 42.Chang RS, et al. A potent nonpeptide cholecystokinin antagonist selective for peripheral tissues isolated from Aspergillus alliaceus. Science. 1985;230:177–179. doi: 10.1126/science.2994227. [DOI] [PubMed] [Google Scholar]
- 43.Herranz R. Cholecystokinin antagonists: Pharmacological and therapeutic potential. Med Res Rev. 2003;23:559–605. doi: 10.1002/med.10042. [DOI] [PubMed] [Google Scholar]
- 44.Takahashi C, et al. Fumiquinazolines A–G, novel metabolites of a fungus separated from a Pseudolabrus marine fish. J Chem Soc, Perkin Trans. 1995;1:2345–2353. [Google Scholar]
- 45.Wang H, Ganesan A. Total synthesis of the fumiquinazoline alkaloids: solution-phase studies. J Org Chem. 2000;65:1022–1030. doi: 10.1021/jo9914364. [DOI] [PubMed] [Google Scholar]
- 46.Watanabe T, et al. Alantrypinone and its derivatives: synthesis and antagonist activity toward insect GABA receptors. Bioorg Med Chem. 2009;17:94–110. doi: 10.1016/j.bmc.2008.11.017. [DOI] [PubMed] [Google Scholar]
- 47.Clardy J, Springer JP, Buechi G, Matsuo K, Wightman R. Tryptoquivaline and tryptoquivalone, two tremorgenic metabolites of Aspergillus clavatus. J Am Chem Soc. 1975;97:663–665. doi: 10.1021/ja00836a045. [DOI] [PubMed] [Google Scholar]
- 48.Crawford IP. Evolution of a biosynthetic pathway: The tryptophan paradigm. Annu Rev Microbiol. 1989;43:567–600. doi: 10.1146/annurev.mi.43.100189.003031. [DOI] [PubMed] [Google Scholar]
- 49.Hutter R, Niederberger P, DeMoss JA. Tryptophan biosynthetic genes in eukaryotic microorganisms. Annu Rev Microbiol. 1986;40:55–77. doi: 10.1146/annurev.mi.40.100186.000415. [DOI] [PubMed] [Google Scholar]
- 50.Radwanski ER, Last RL. Tryptophan biosynthesis and metabolism: biochemical and molecular genetics. Plant Cell. 1995;7:921–934. doi: 10.1105/tpc.7.7.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sand P, et al. Naturally occurring benzodiazepines: current status of research and clinical implications. Eur Arch Psy Clin N. 2000;250:194–202. doi: 10.1007/s004060070024. [DOI] [PubMed] [Google Scholar]
- 52.Lerbs W, Luckner M. Cyclopeptine synthetase activity in surface cultures of Penicillium cyclopium. J Basic Microb. 1985;25:387–391. doi: 10.1002/jobm.3620250611. [DOI] [PubMed] [Google Scholar]
- 53.Hu Y, et al. Benzodiazepine biosynthesis in Streptomyces refuineus. Chem Biol. 2007;14:691–701. doi: 10.1016/j.chembiol.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 54.Phelan VV, Du Y, McLean JA, Bachmann BO. Adenylation enzyme characterization using γ-18O4-ATP pyrophosphate exchange. Chem Biol. 2009;16:473–478. doi: 10.1016/j.chembiol.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li W, Khullar A, Chou S, Sacramo A, Gerratana B. Biosynthesis of sibiromycin, a potent antitumor antibiotic. Appl Environ Microbiol. 2009;75:2869–2878. doi: 10.1128/AEM.02326-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li W, Chou S, Khullar A, Gerratana B. Cloning and characterization of the biosynthetic gene cluster for tomaymycin, an SJG-136 monomeric analog. Appl Environ Microbiol. 2009;75:2958–2963. doi: 10.1128/AEM.02325-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McAlpine JB, et al. Biosynthesis of diazepinomicin/ECO-4601, a Micromonospora secondary metabolite with a novel ring system. J Nat Prod. 2008;71:1585–1590. doi: 10.1021/np800376n. [DOI] [PubMed] [Google Scholar]
- 58.Branchini BR, Murtiashaw MH, Magyar RA, Anderson SM. The role of lysine 529, a conserved residue of the acyl-adenylate-forming enzyme superfamily, in firefly luciferase. Biochemistry. 2000;39:5433–5440. doi: 10.1021/bi9928804. [DOI] [PubMed] [Google Scholar]
- 59.May JJ, Kessler N, Marahiel MA, Stubbs MT. Crystal structure of DhbE, an archetype for aryl acid activating domains of modular nonribosomal peptide synthetases. Proc Natl Acad Sci USA. 2002;99:12120–12125. doi: 10.1073/pnas.182156699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Haese A, Pieper R, von Ostrowski T, Zocher R. Bacterial expression of catalytically active fragments of the multifunctional enzyme enniatin synthetase. J Mol Biol. 1994;243:116–122. doi: 10.1006/jmbi.1994.1634. [DOI] [PubMed] [Google Scholar]
- 61.Glinski M, Urbanke C, Hornbogen T, Zocher R. Enniatin synthetase is a monomer with extended structure: evidence for an intramolecular reaction mechanism. Arch Microbiol. 2002;178:267–273. doi: 10.1007/s00203-002-0451-1. [DOI] [PubMed] [Google Scholar]
- 62.Xu Y, et al. Biosynthesis of the cyclooligomer depsipeptide beauvericin, a virulence factor of the entomopathogenic fungus Beauveria bassiana. Chem Biol. 2008;15:898–907. doi: 10.1016/j.chembiol.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 63.Jirakkakul J, et al. Identification of the nonribosomal peptide synthetase gene responsible for bassianolide synthesis in wood-decaying fungus Xylaria sp. BCC1067. Microbiology. 2008;154:995–1006. doi: 10.1099/mic.0.2007/013995-0. [DOI] [PubMed] [Google Scholar]
- 64.Admiraal SJ, Walsh CT, Khosla C. The loading module of rifamycin synthetase is an adenylation-thiolation didomain with substrate tolerance for substituted benzoates. Biochemistry. 2001;40:6116–6123. doi: 10.1021/bi010080z. [DOI] [PubMed] [Google Scholar]
- 65.Geissler JF, Harwood CS, Gibson J. Purification and properties of benzoate-coenzyme A ligase, a Rhodopseudomonas palustris enzyme involved in the anaerobic degradation of benzoate. J Bacteriol. 1988;170:1709–1714. doi: 10.1128/jb.170.4.1709-1714.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gibson J, Dispensa M, Fogg GC, Evans DT, Harwood CS. 4-Hydroxybenzoate-coenzyme A ligase from Rhodopseudomonas palustris: purification, gene sequence, and role in anaerobic degradation. J Bacteriol. 1994;176:634–641. doi: 10.1128/jb.176.3.634-641.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cossins EA, Chen L. Folates and one-carbon metabolism in plants and fungi. Phytochemistry. 1997;45:437–452. doi: 10.1016/s0031-9422(96)00833-3. [DOI] [PubMed] [Google Scholar]
- 68.Subramanian V, Vaidyanathan CS. Anthranilate hydroxylase from Aspergillus niger: new type of NADPH-linked nonheme iron monooxygenase. J Bacteriol. 1984;160:651–655. doi: 10.1128/jb.160.2.651-655.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hochlowski JE, et al. 5-N-Acetylardeemin, a novel heterocyclic compound which reverses multiple drug resistance in tumor cells. II Isolation and elucidation of the structure of 5-N-acetylardeemin and two congeners. J Antibiot (Tokyo) 1993;46:380–386. doi: 10.7164/antibiotics.46.380. [DOI] [PubMed] [Google Scholar]
- 70.Penn J, Mantle PG, Bilton JN, Sheppard RN. Glyantrypine, a novel anthranilic acid-containing metabolite of Aspergillus clavatus. J Chem Soc, Perkin Trans. 1992;1:1495–1496. [Google Scholar]
- 71.Wong SM, et al. Fiscalins: new substance P inhibitors produced by the fungus Neosartorya fischeri. Taxonomy, fermentation, structures, and biological properties. J Antibiot (Tokyo) 1993;46:545–553. doi: 10.7164/antibiotics.46.545. [DOI] [PubMed] [Google Scholar]
- 72.Rahbk L, Breinholt J. Circumdatins D, E, and F: further fungal benzodiazepine analogues from Aspergillus ochraceus. J Nat Prod. 1999;62:904–905. doi: 10.1021/np980495u. [DOI] [PubMed] [Google Scholar]
- 73.Joshi BK, Gloer JB, Wicklow DT, Dowd PF. Sclerotigenin: a new antiinsectan benzodiazepine from the sclerotia of Penicillium sclerotigenum. J Nat Prod. 1999;62:650–652. doi: 10.1021/np980511n. [DOI] [PubMed] [Google Scholar]
- 74.Larsen TO, Franzyk H, Jensen SR. UV-guided isolation of verrucines A and B, novel quinazolines from Penicillium verrucosum structurally related to anacine from Penicillium aurantiogriseum. J Nat Prod. 1999;62:1578–1580. doi: 10.1021/np990251p. [DOI] [PubMed] [Google Scholar]
- 75.Bringmann G. A first biosynthetic proposal for the in vivo formation of naturally occurring diazepam-like 1,4-benzodiazepines. J Neur Transm. 1992;88:77–82. doi: 10.1007/BF01245038. [DOI] [PubMed] [Google Scholar]
- 76.Frisvad JC, Rank C, Nielsen KF, Larsen TO. Metabolomics of Aspergillus fumigatus. Med Mycol. 2009;47:53–71. doi: 10.1080/13693780802307720. [DOI] [PubMed] [Google Scholar]
- 77.Stachelhaus T, Mootz H, Bergendahl V, Marahiel M. Peptide bond formation in nonribosomal peptide biosynthesis. Catalytic role of the condensation domain. J Biol Chem. 1998;273:22773–22781. doi: 10.1074/jbc.273.35.22773. [DOI] [PubMed] [Google Scholar]
- 78.Samel SA, Schoenafinger G, Knappe TA, Marahiel MA, Essen L-O. Structural and functional insights into a peptide bond-forming bidomain from a nonribosomal peptide synthetase. Structure. 2007;15:781–792. doi: 10.1016/j.str.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 79.Keating TA, Marshall CG, Walsh CT, Keating AE. The structure of VibH represents nonribosomal peptide synthetase condensation, cyclization and epimerization domains. Nat Struct Mol Biol. 2002;9:522–526. doi: 10.1038/nsb810. [DOI] [PubMed] [Google Scholar]
- 80.Tanovic A, Samel SA, Essen L-O, Marahiel MA. Crystal structure of the termination module of a nonribosomal peptide synthetase. Science. 2008;321:659–663. doi: 10.1126/science.1159850. [DOI] [PubMed] [Google Scholar]
- 81.Eisfeld K. Non-ribosomal peptide synthetases of fungi. Physiology and Genetics. 2009:305–330. [Google Scholar]
- 82.Keating TA, et al. Chain termination steps in nonribosomal peptide synthetase assembly lines: directed acyl-enzyme breakdown in antibiotic and siderophore biosynthesis. Chembiochem. 2001;2:99–107. doi: 10.1002/1439-7633(20010202)2:2<99::AID-CBIC99>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.