Abstract
In the process of characterizing a custom-made affinity-purified antiserum for estrogen receptor beta (ERβ), ck5912, we used a number of common tests for specificity of ck5912 along with that of 8 commercially available ERβ antisera: Affinity Bioreagents PA1-310B, Invitrogen D7N, Upstate 06-629, Santa Cruz H150, Y19, L20, 1531, and Abcam 9.88. We tested their recognition of recombinant ERβ (rERβ) versus rERα, ERβ versus ERα transfected into cell lines, as well as labeling in wildtype (WT) versus estrogen receptor beta knockout (βERKO) and null (ERβSTL-/L-) mouse ovary, hypothalamus, and hippocampus. To our surprise, we found that while most of these antisera passed some tests, giving the initial impression of specificity, western blot analysis showed that all of them recognized apparently identical protein bands in WT, βERKO and ERβSTL-/L- tissues. We share these results with the goal of helping other researchers avoid pitfalls in interpretation that could come from use of these ERβ antisera.
Keywords: estrogen receptor beta, antibody, knockout mice, βERKO, ERβSTL-/L-
Introduction
The discovery of estrogen receptor β (ERβ; Kuiper et al., 1996) opened a new door to understanding physiological actions of estradiol. Neuroscientists who study hormone effects in the brain are particularly interested in ERβ because pharmacological and knockout studies point to ERβ as being important in learning and memory (Liu et al., 2008), anxiety (Imwalle et al., 2005, Tomihara et al., 2009), and aggression (Ogawa et al., 1999).
First cloned in rat and subsequently in human and mouse, the ERβ gene contains eight exons and shares a high degree of sequence homology with estrogen receptor alpha (ERα) in the DNA and ligand binding domains (Kuiper et al., 1996, Mosselman et al., 1996, Tremblay et al., 1997). Additionally, the ERβ gene undergoes alternative splicing leading to the expression of several isoforms. One splice variant, ERβ2, contains a 54 bp insert leading to an additional 18 amino acids; other splice variants, called delta variants, lack entire exons (Chu and Fuller, 1997, Lu et al., 1998). Like ERα, ERβ is traditionally thought of as a transcription regulator. However, estradiol also has many rapid effects on neurons that likely involve ER signaling outside of the nucleus and some pharmacological evidence implicates ERβ in these rapid, extranuclear effects (Zhao and Brinton, 2007, Kramár et al., 2009).
Understanding the function of ERβ requires knowing where it is located and what proteins it interacts with, which in turn, requires reliable and specific antibodies. Early studies with ERβ antisera showed some agreement but also some discrepancies between localization of ERβ immunoreactivity (Li et al., 1997, Shughrue and Mercenthaler, 2001) and mRNA (Shughrue et al. 1997), raising concerns about ERβ antisera (Warner et al., 2003, Shughrue and Mercenthaler, 2001). Then, in 2001, the Z8P ERβ antiserum showed consensus between ERβ mRNA and protein expression in many brain areas, including areas that previously were controversial (Shughrue and Mercenthaler, 2001). Unfortunately, however, Z8P is no longer available.
Our lab is particularly interested in ERβ function in the hippocampus, including its colocalization with other proteins. To facilitate studies of ERβ, we produced two ERβ antisera raised in chicken for use in conjunction with other commercially available antisera. The more promising of these was ck5912. In the process of characterizing ck5912, we performed a number of commonly used tests for its specificity, along with the specificity of 8 commercially available ERβ antisera. To our surprise, we found that while some of these antisera passed many tests, some did not. Most significantly, we found that all the ERβ antisera we tested detected immunoreactivity in tissues from two independently generated strains of ERβ knockout mice, the βERKO mouse (Krege et al., 1998) and Chambon's recently generated ERβ null mouse (ERβSTL-/L-; Antal et al., 2008). We share these findings with the goal of helping other researchers avoid pitfalls in interpretation that could arise from the use of these ERβ antisera.
Materials and Methods
Animals
All animal procedures were performed in accordance with the National Institute of Health Guide for the Care and Use of Laboratory Animals and were approved by the Northwestern University Institutional Animal Care and Use Committee. βERKO and C57/BL6J breeder mice were purchased from Jackson labs. Mice used for experiments were obtained by in-house breeding with genotype confirmation by Transnetyx. Tissues from ERβSTL-/L- and wildtype mice were a kind gift from Dr. Shaila Mani (Baylor College of Medicine, Houston, TX).
Antisera
For ERβ: antisera are summarized in Table 1. For GFP: Clontech JL8 mouse anti-GFP 632380 (1:1000).
Table 1.
Summary of ERβ antisera tested for specificity.
| Antisera | Company, catalog number | Antigen; dilution used | Host species |
|---|---|---|---|
| PA1-310B | Thermo Scientific (formerly Affinity Bioreagents), PA1-310B | Synthetic peptide corresponding to residues C(467) SSTEDSKNKESSQNLQSQ (485) of rat ERβ; 1:1000 |
Rabbit |
| ck5912 | Custom | Synthetic peptide corresponding to residues C(467) SSTEDSKNKESSQNLQSQ(485) of rat ERβ; 1:10,000 |
Chicken |
| D7N | Invitrogen (formerly Zymed), 51-7700 | 19 amino acid synthetic peptide derived from the C-terminus of human ERβ; 1:250 | Rabbit |
| H150 | Santa Cruz, sc-8974 | amino acids 1-150 of human ERβ; 1:1000 | Rabbit |
| 06-629 | Millipore (formerly Upstate), 06-629 | peptide (YAEPQKSPWCEARSLEHT) representing amino acids 54-71 of rat and mouse ERβ and amino acids 46-63 of human ERβ; 1:1000 | Rabbit |
| 1531 | Santa Cruz, sc-53494 | amino acids 256-505 of human ERβ; 1:250 | Mouse |
| Y19 | Santa Cruz, sc-6821 | N-terminus of mouse ERβ; 1:250 | Goat |
| L20 | Santa Cruz, sc-6822 | C-terminus of human ERβ; 1:250 | Goat |
| 9.88 | Abcam, ab16813 | Recombinant full-length human ERβ; 1:1000 | Mouse |
Western Blots
Mice were deeply anesthetized with sodium pentobarbital (80 mg/kg, i.p.) and perfused with ice-cold RIPA buffer lacking detergents (in mM: 50 Tris-HCl, 150 sodium chloride, 1 EDTA, 1 sodium orthovanadate, 0.1 phenylmethylsulfonyl fluoride, 50 sodium fluoride, 10 sodium pyrophosphate, 20 glycerophosphate, with 1 μg/ml leupeptin and 1 μg/ml aprotinin). Brains and ovaries were rapidly removed and placed on ice. Ovaries, hypothalamus, and hippocampi were dissected and homogenized in RIPA buffer containing 1% nonidet P40, 0.25% sodium deoxycholate, and 0.1% sodium dodecyl sulfate, incubated on ice for 25 minutes, and spun at 1000g for 10 minutes to remove large cell fragments and nuclear material. The supernatant was kept as the whole cell fraction. The protein sample was mixed with Laemmli sample buffer (62.5 mM Tris-HCl, pH 6.8, 25% glycerol, 2% SDS, 0.01% Bromophenol blue, 5% β-mercaptoethanol), boiled for 5 minutes, and separated on a 10% SDS-PAGE gel. After electrophoresis, proteins were transferred to polyvinylidene difluoride membranes (Millipore). The membrane was blocked in 5% nonfat milk and probed with primary antiserum (see Table 1). For preadsorption experiments, diluted antiserum was incubated with 1000 fold excess antigenic peptide and kept at 4°C overnight before probing membranes. Blots were then incubated with horseradish peroxidase coupled anti-rabbit, anti-goat, or anti-mouse IgG secondary antibody (Vector Laboratories) and proteins were visualized using enhanced chemiluminescence (ECL Plus, Amersham Biosciences). Recombinant estrogen receptor alpha (rERα) and recombinant estrogen receptor beta (rERβ) protein were purchased from Invitrogen.
Ovary immunohistochemistry
Mice were deeply anesthetized with sodium pentobarbital (80 mg/kg, i.p.) and perfused with 4% paraformaldehyde/3.75% acrolein in phosphate buffer. Ovaries were removed, postfixed for 1 hour in paraformaldehyde, cryoprotected, and sectioned. Immunostaining was performed using a standard avidin-biotin peroxidase method as previously described with slight modifications (Rudick et al., 2003). Tissue was incubated in primary antiserum overnight and in secondary antiserum for 1 hour. Sections were counterstained with hematoxylin and coverslipped.
Cell culture, transfection and immunostaining
HT22 cells were a kind gift from Dr. Pamela Maher (The Scripps Research Institute, La Jolla, CA). Cells were grown on 100 mm tissue culture dishes or glass coverslips and maintained in DMEM media supplemented with 10% fetal calf serum and 1% Pen-Strep (Invitrogen) at 37°C in a 10% CO2 atmosphere. Cell density was maintained at ≤70% confluence and cells were split using 0.05% trypsin/0.53 mM EDTA (Invitrogen). HT22 cells were transiently transfected with plasmid expression vectors containing inserts for GFP, ERα-GFP, or ERβ2-GFP (kind gift of Dr. Toni Pak, Loyola University Chicago Stritch School of Medicine, Maywood, IL) using Lipofectamine 2000 according to the manufacturer's instructions. Twenty-fours after transfection, cells were fixed for immunocytochemistry or collected for western blot. For western blots, cells were scraped into RIPA buffer containing 1% nonidet P40, 0.25% sodium deoxycholate, and 0.1% sodium dodecyl sulfate, and spun to obtain whole cell fractions as above. For immunocytochemistry, cells were fixed for 15 minutes in methanol at −20°C. After rinsing with phosphate buffered saline (PBS), cells were incubated for 1 hour in 3% goat serum, 10% BSA, and 0.3% DMSO in PBS to block nonspecific staining. Cells were then incubated overnight with primary antiserum in 1% goat serum, 2% BSA, and 0.3% DMSO in PBS. Cells were rinsed and incubated with chicken or rabbit IgG coupled to Alexa Fluor 568 for ERβ or mouse IgG coupled to Alexa Fluor 488 for GFP. Cells were coverslipped and then imaged using a PerkinElmer Ultraview spinning disc laser confocal microscope. Experiments were also done using human embryonic kidney (HEK) 293 cells. Methods were as above except that cells were maintained in 5% CO2 and split using mechanical dissociation.
Reverse transcriptase PCR (rtPCR)
WT and βERKO mice were deeply anesthetized with sodium pentobarbital (80 mg/kg, i.p.). Ovary, hypothalamus, and hippocampus were removed immediately and placed on ice. RNA was extracted from tissue using a Trizol plus purification kit (Invitrogen) according to the manufacturer's instructions. First strand cDNA was generated from 1 μg RNA by reverse transcriptase using the SuperScript III First-Strand Synthesis System (Invitrogen). PCR amplification was performed on 3 μl of cDNA using primers 5′-GCCAATCATCGCTTCTCTAT-3′ and 5′-CCCTCTTTGCTCTTACTGTCCTCT-3′, as described (Krege et al., 1998). ERβ was amplified for 30 or 40 cycles as follows: 94°C for 1 minute, 55°C for 1 minute, and 72°C for 1 minute. Water was used as a negative control. Amplified DNA was run on a 1.75% agarose gel. Hippocampal DNA samples were cut from the gel and sent to ACGT, Inc. for sequencing. Following amplification, hypothalamicc DNA samples were purified using a QIAquick PCR purification kit (Qiagen) and sent to ACGT, Inc. for sequencing.
Results
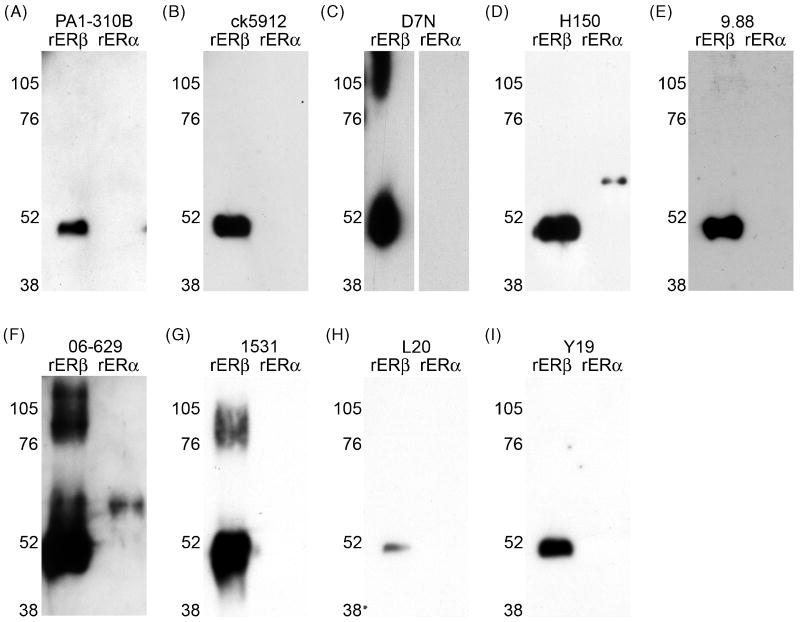
Our initial test of ERβ antisera was to confirm specificity for rERβ versus rERα using western blot. Gels were loaded with 0.1 μg rERβ and rERα, and then probed with one of 9 ERβ antisera: PA1-310B, ck5912, D7N, H150, 9.88, 06-629, 1531, L20, or Y19 (Fig. 1A-I). All antisera detected a band of ∼50 kDa for rERβ. D7N, 06-629, and 1531 also recognized a band >76 kDa (Fig. 1C, F, G), which could correspond to a cluster of rERβ; however this was not seen with the other antisera. Additionally, both H150 and 06-629 failed this initial test in that they both detected rERα (Fig. 1D, F).
Figure 1.
Western blots of recombinant ERβ (rERβ) and recombinant ERα (rERα) probed with anti-ERβ antisera. (A) PA1-310B anti-ERβ, (B) ck5912 anti-ERβ, (C) D7N anti-ERβ, (D) H150 anti-ERβ, (E) 9.88 anti-ERβ, (F) 06-629 anti-ERβ, (G) 1531 anti-ERβ, (H) L20 anti-ERβ, and (I) Y19 anti-ERβ. All antisera recognized rERβ with a band of ∼50 kDa, as appropriate. D7N, 06-629, and 1531 additionally recognized higher molecular weight bands, which could correspond to a cluster of rERβ. H150 and 06-629 also recognized rERα with a faint band of ∼68 kDa.
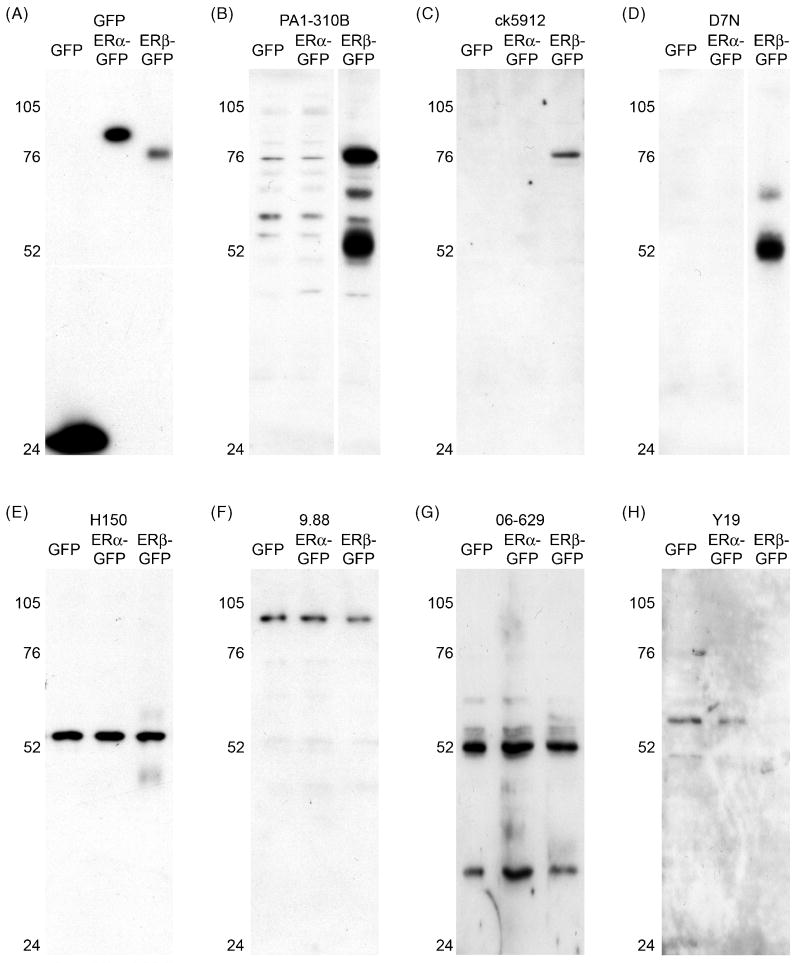
We next tested specificity in cultured cells using western blot and immunocytochemistry. We transfected GFP, ERα-GFP, or ERβ-GFP into the immortalized murine hippocampal cell line, HT22, which is devoid of functional estrogen receptors (Fitzpatrick et al., 2002). Probing western blots with anti-GFP confirmed successful transfection (Fig. 2A). Anti-GFP showed a band at ∼27 kDa in GFP-only transfected cells, at ∼83 kDa (the predicted molecular weight of GFP-tagged ERα) in ERα-GFP transfected cells, and at ∼78 kDa (the predicted molecular weight of GFP-tagged ERβ) in ERβ-GFP transfected cells.
Figure 2.
Detection of ERβ in transfected HT22 cells by western blot. (A-H) Western blots of HT22 cells transfected with GFP, ERα-GFP, or ERβ-GFP and probed with anti-GFP or various anti-ERβ antisera: (A) GFP is detected in all transfected cells, as appropriate. (B) PA1-310B anti-ERβ detected a prominent, appropriately sized band of ∼78 kDa in ERβ-GFP transfected cells, and also a prominent band at ∼52 kDa, which could reflect ERβ cleaved from GFP. PA1-310B also detected faint bands in ERα-GFP transfected cells. (C) ck5912 anti-ERβ detected a single appropriately sized band of ∼78 kDa only in ERβ-GFP transfected cells. (D) D7N anti-ERβ detected a single a band of ∼52 kDa in ERβ-GFP transfected cells, which could reflect ERβ cleaved from GFP. (E) H150 anti-ERβ detected a single band of inappropriate size (∼55 kDa) in all transfected cells. (F) 9.88 anti-ERβ detected a single band of inappropriate size (∼102 kDa) in all transfected cells. (G) Similar to H150, 06-629 anti-ERβ detected a band close to 52 kDa in all transfected cells, as well as band of ∼38 kDa in all transfected cells. (H) Y19 anti-ERβ detected faint bands in GFP- and ERα-GFP transfected cells.
Consistent with previous work (Sheldahl et al., 2008), PA1-310B detected an appropriately sized band of ∼78 kDa in ERβ-GFP transfected cells, and also a band at ∼52 kDa, which could reflect ERβ cleaved from GFP (Fig. 2B). The upper band was also seen faintly in GFP and ERα-GFP cell extracts. Ck5912 also recognized a band of ∼78 kDa and only in ERβ-GFP transfected cells (Fig. 2C), which is the expected result. D7N detected one main band in ERβ-GFP transfected cells, but its molecular weight suggested ERβ cleaved from GFP (Fig. 2D). H150 detected an incorrectly sized single band of ∼55 kDa in all transfected cell extracts (Fig. 2E), similar to a previous report (Sheldahl et al., 2008). Abcam 9.88 detected a band of ∼100 kDa in all cell extracts (Fig. 2F). 06-629 detected a band of ∼52 kDa, as well as a ∼38 kDa band in all cell extracts (Fig. 2G). Y19 detected very faint bands only in GFP and ERα-GFP transfected cells (Fig. 2H), and required long exposure times to visualize. Efforts to obtain more definitive results for Y19 by optimizing the protocol were unsuccessful. L20 and 1531 failed to detect any immunoreactivity in transfected cells (not shown). Results were identical when the same experiments were done in HEK 293 cells (not shown).
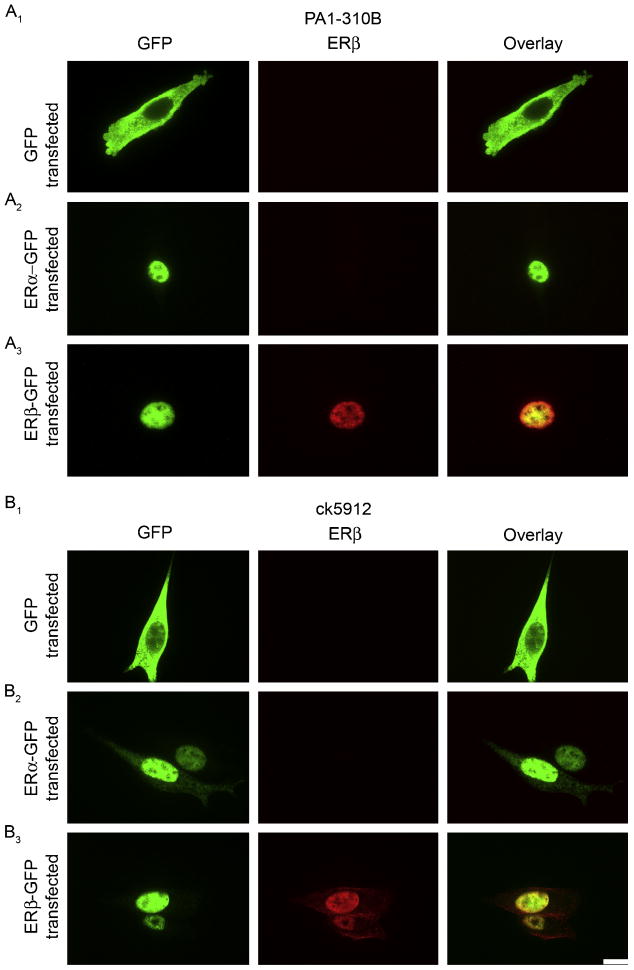
Because PA1-310B and ck5912 looked the best by western blot and because PA1-310B is a commonly used ERβ antiserum, we next tested both of these antisera with immunocytochemistry in HT22 cells. Confocal imaging of anti-GFP staining showed diffuse, cytoplasmic labeling in GFP-only transfected cells and nuclear labeling for both ERα-GFP and ERβ-GFP transfected cells (Fig. 3A1). PA1-310B showed no immunoreactivity in ERα-GFP transfected cells (Fig. 3A2) and nuclear labeling in ERβ-GFP transfected cells (Fig. 3A3). Results were identical for ck5912 (Fig. 3B). Thus, PA1-310B and ck5912 both appeared promising.
Figure 3.
Detection of ERβ in transfected HT22 cells by immunocytochemistry. (A) Representative images of PA1-310B immunostaining in HT22 cells transfected with (A1) GFP, (A2) ERα-GFP, or (A3) ERβ-GFP. PA1-310B anti-ERβ detected nuclear labeling only in ERβ-GFP transfected cells, as appropriate. (B) Representative images of ck5912 immunostaining in HT22 cells transfected with (B1) GFP, (B2) ERα-GFP, or (B3) ERβ-GFP. Ck5912 anti-ERβ detected nuclear labeling only in ERβ-GFP transfected cells, as appropriate. Scale bar is 10 μm and applies to all frames.
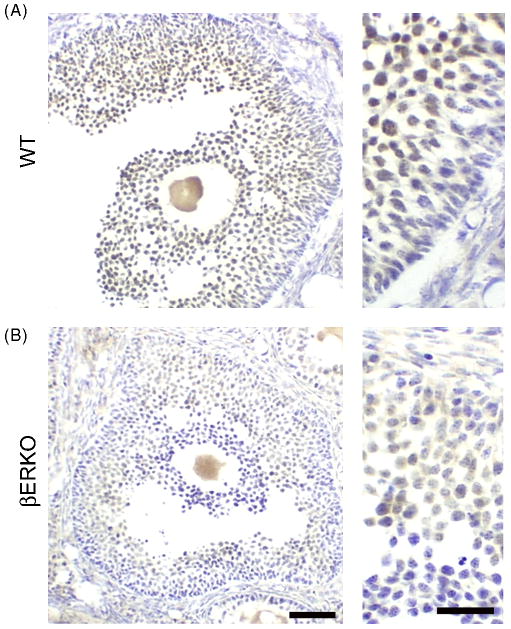
We next investigated immunolabeling in C57/B6 wildtype (WT) and βERKO mouse ovary. In contrast to Krege et al. 1998, we found that PA1-310B showed labeling in granulosa cells from both WT (Fig. 4A) and βERKO (Fig. 4B) ovaries, although staining was less intense in βERKOs. This surprising result prompted us to further investigate labeling in βERKO tissue.
Figure 4.
ERβ immunoreactivity in wildtype (WT) and ERβ knockout mouse (βERKO) ovaries. Representative photomicrographs from (A) WT and (B) βERKO ovaries labeled with PA1-310B anti-ERβ. ERβ immunoreactivity is detected in granulosa cells of both WT and βERKO mice, although less intense in βERKO. Scale bars are 50 μm and 25 μm, left and right panels, respectively.
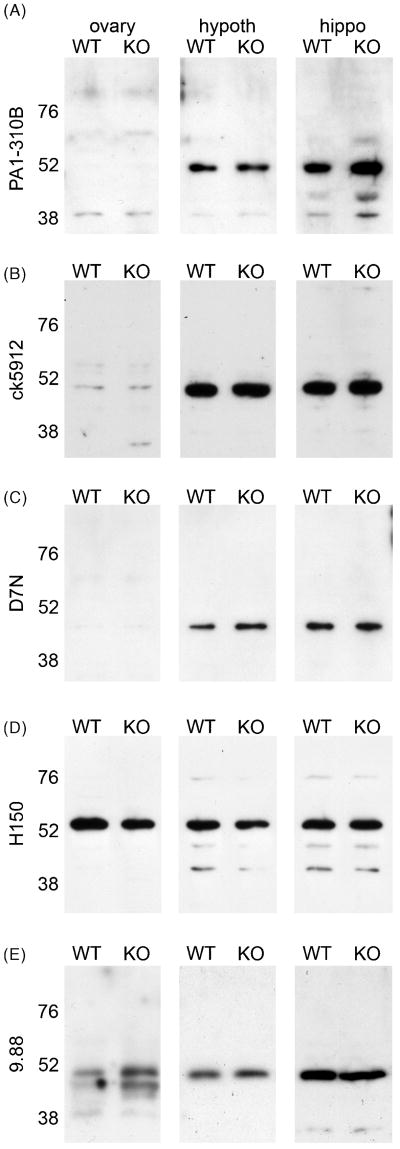
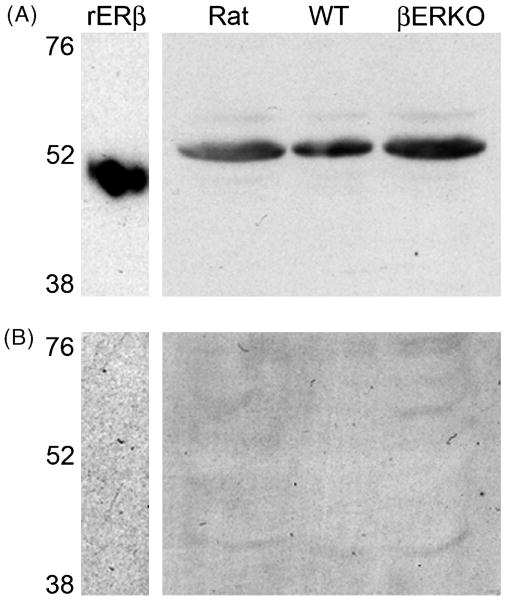
We made whole cell extracts from WT and βERKO (KO) ovary, hypothalamus, and hippocampus and probed western blots with a similar panel of antisera as we used in previous specificity tests. While labeling was often faint in ovary samples, importantly, there was no apparent difference in either the molecular weight or intensity of any bands between WT and KO tissues with any antisera. PA1-310B, ck5912 and 9.88 detected a single band close to 52 kDa, while D7N detected a band slightly below and H150 detected a band slightly above 52 kDa in WT and KO tissue (Fig. 5A-E). 06-629 labeled multiple bands between 52-76 kDa in WT and KO tissues (not shown). Additionally, 1531 detected strong bands of ∼55 kDa in βERKO and WT ovary and, along with Y19 and L20, detected multiple faint bands in both WT and βERKO hypothalamus and hippocampus (not shown). We further tested ck5912 by preadsorption with the immunogenic peptide. Ck5912 detected a single band at ∼50 kDa for rERβ and a band at ∼52 kDa in extracts from rat, WT, and βERKO mouse hypothalamus (Fig. 6A). In each case, labeling was completely eliminated by preadsorption of the antiserum with the immunogenic peptide (Fig. 6B). A similar experiment was performed for PA1-310B on rat tissue and preadsorption also eliminated staining (not shown).
Figure 5.
Anti-ERβ antisera recognize nearly identical bands on western blots from wildtype (WT) and ERβ knockout (βERKO, KO) mouse tissues. Representative western blots of ovary, hypothalamus (hypoth), and hippocampus (hippo) from WT and βERKO (KO) probed with (A) PA1-310B anti-ERβ, (B) ck5912 anti-ERβ, (C) D7N anti-ERβ, (D) H150 anti-ERβ, or (E) 9.88 anti-ERβ.
Figure 6.
ERβ immunoreactivity is eliminated by preadsorption of the antiserum with the antigenic peptide. (A) Representative western blot of rERβ, rat, wildtype and estrogen receptor beta knockout (βERKO) hypothalamic tissue probed with ck5912 anti-ERβ. One prominent band was detected for rERβ, rat, WT and βERKO tissues. (B) Preadsorption with the antigenic peptide eliminated all labeling.
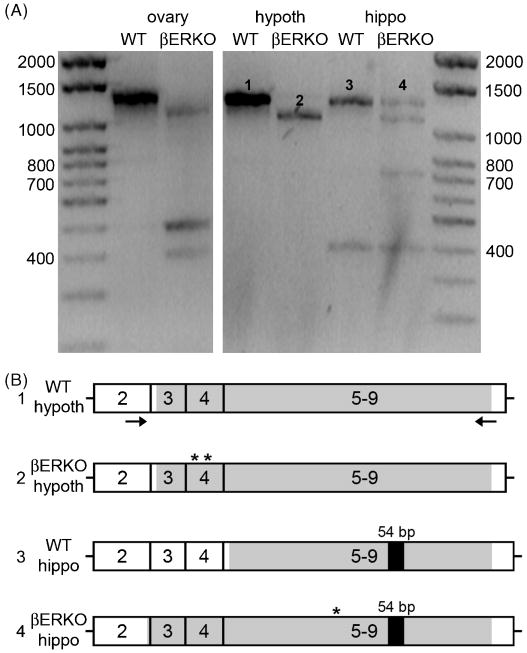
Labeling in βERKO tissues was a serious concern. To exclude the possibility that residual ERβ was expressed in βERKO tissues, we performed rtPCR for ERβ mRNA on WT and βERKO ovary, hypothalamus, and hippocampus (Fig. 7A). DNA sequencing was then performed on WT and βERKO hypothalamus and hippocampus. The expected full-length product of 1,291 bp was found in WT ovary, hypothalamus, and hippocampus and surprisingly, also in βERKO hippocampus (Fig. 7A). βERKO ovary, hypothalamus, and hippocampus contained lower base pair products. Sequencing showed that while the WT sequences contained no stop codons and therefore were likely to be translatable, βERKO sequences contained stop codons. For hypothalamus, partial sequences of ∼940 bp were obtained for both WT and βERKO samples. While the WT hypothalamic sequence was translatable, the βERKO hypothalamic sequence contained 2 stop codons in exon 4 (Fig. 7B). For hippocampus, WT and the full-length βERKO product, which corresponded to the product from WT hippocampus (Fig. 7A), were sent for sequencing. A sequence of 708 bp was obtained for WT and 1,077 bp for βERKO hippocampus. Similar to hypothalamus, the WT hippocampal transcript contained no stop codons whereas the βERKO hippocampal transcript is not likely to be translatable due to a stop codon in exon 5 (Fig. 7B). Thus, rtPCR and sequencing analysis indicated that ERβ protein is not expressed in βERKO tissues. Interestingly, both hippocampal transcripts contained the 54 bp insert corresponding to ERβ2 (Fig. 7B).
Figure 7.
rtPCR for ERβ mRNA in wildtype (WT) and ERβ knockout mouse (βERKO) ovary, hypothalamus (hypoth), and hippocampus (hippo). (A) Representative gel electrophoresis of the rtPCR products. WT ovary, hypothalamus, and hippocampus and βERKO hippocampus contained the expected full-length product of 1,291 bp. Additionally, βERKO ovary, hypoth, and hippo contained lower base pair products. Numbers indicate the transcripts sent for sequence analysis. (B) Representation of the rtPCR products from WT and βERKO mRNA that were sequenced showing exons 2-9. The black arrows indicate the primers used, the shaded regions indicate the portion for which sequence data were obtained, and asterisks indicate stop codons. Sequences obtained from WT hypoth and hippo PCR products did not contain stop codons and are therefore likely to be translated. However, both βERKO hypoth and hippo products contained stop codons. Interestingly both hippocampal samples contained the 54 bp insert corresponding to ERβ2.
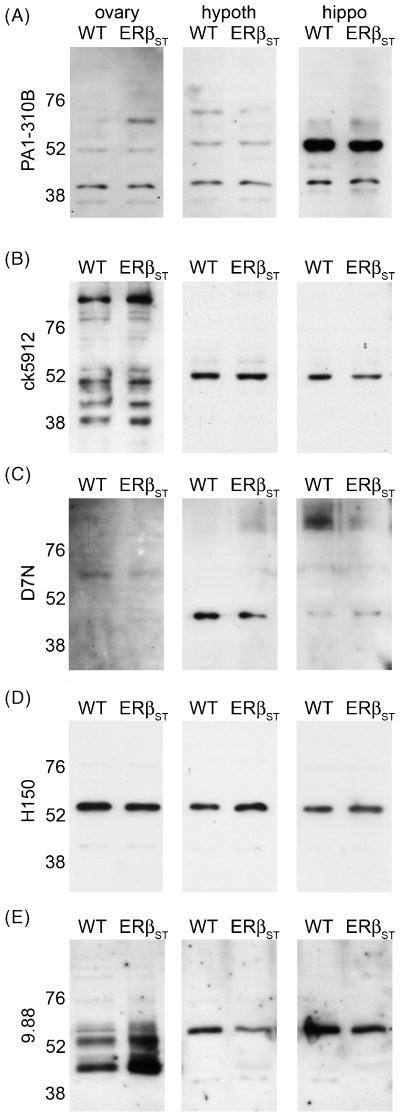
To corroborate results with βERKO mice, we also made whole cell extracts from wildtype (WT) and ERβ null (ERβSTL-/L-, Antal et al., 2008) mouse tissues and probed western blots with a panel of ERβ antisera. Similar to results with βERKO tissue, all of the ERβ antisera we tested recognized the same bands in tissue from WT and ERβSTL-/L- mice. PA1-310B detected prominent bands at ∼52 and ∼40 kDa in ovary, hypothalamic, and hippocampal extracts from both WT and ERβSTL-/L- (Fig. 8A). Ck5912 detected several bands in ovary, and a single band at ∼52 kDa in WT and ERβSTL-/L- hypothalamic and hippocampal extracts (Fig. 8B). D7N detected a faint band of ∼60 kDa in WT and ERβSTL-/L- ovary and a ∼45 kDa band in hypothalamus and hippocampus (Fig. 8C). Similar to results from cell culture, H150 recognized a band of ∼55 kDa in all extracts from WT and ERβSTL-/L- tissue (Fig. 8D). 9.88 detected multiple bands in ovary and a single band of ∼55 kDa in WT and ERβSTL-/L- hypothalamic and hippocampal extracts (Fig. 8E). 06-629, 1531, Y19, and L20 detected multiple bands in WT and ERβSTL-/L- tissues (not shown). Thus, while all ERβ antisera detected a band or bands at molecular weights appropriate for ERβ, the observations of apparently identical labeling in WT and both βERKO and ERβ null tissues are impossible to overlook and raise serious concerns about the specificity of all 9 antisera we tested.
Figure 8.
Anti-ERβ antisera recognize nearly identical bands on Western blots from wildtype (WT) and ERβ null (ERβSTL-/L-, ERβST) mouse tissues. Representative western blots of ovary, hypothalamus (hypoth), and hippocampus (hippo) from WT and ERβST mice probed with (A) PA1-310B anti-Rβ, (B) ck5912 anti-ERβ, (C) D7N anti-ERβ, (D) H150 anti-ERβ, or (E) 9.88 anti-ERβ.
Discussion
We initially set out to characterize a new affinity purified anti-ERβ antiserum produced in chicken, ck5912. We included 8 commercially available ERβ antisera as controls: Affinity Bioreagents PA1-310B, Invitrogen D7N, Upstate 06-629, Santa Cruz H150, L20, Y19, and 1531, and Abcam 9.88. We first tested recognition of recombinant ERα and ERβ, then detection of ERα and ERβ in transfected cell lines, followed by the most important test, labeling in tissues from WT versus βERKO and ERβSTL-/L- mice. During this process, we found that ck5912, along with the 8 control antisera, recognized apparently identical protein bands by western blot in WT, βERKO, and ERβSTL-/L- ovary, hypothalamus, and hippocampus.
To date, 5 lines of ERβ-deficient mice have been produced. The first βERKO mouse was originally generated by Krege et al. in 1998; this same line was duplicated at the Karolinska Institute and used in studies of bone (Windahl et al., 1999), brain (Wang et al., 2001; 2003), and prostate (Imamov et al., 2004). Three other independent lines were also made, by Chambon and colleagues (Dupont et al., 2000), by Shughrue et al. (2002) at Wyeth, and the ERβSTL-/L- line also by Chambon's group (Antal et al, 2008). Given an initial focus on the role of ERβ in reproductive function, the original characterizations of ERβ-deficient mice rarely investigated ERβ expression in brain. One exception was the mouse produced at Wyeth. Using the Z8P antiserum that is no longer available, Shughrue et al. (2002) showed a lack of ERβ immunoreactivity in the hypothalamic paraventricular nucleus, a region that expresses ERβ intensely in WT mice. Also, using this same mouse, a lack of ERβ in brain capillary endothelial cells was reported based on western blots probed with the D7N antiserum (Razandi et al, 2004). Unfortunately, the Wyeth mouse is currently unavailable for further experiments. Still, subsequent studies utilizing the Krege et al. (1998) βERKO mouse have shown several neural differences from WT that suggest ERβ expression in brain is disrupted, including alterations in neuronal migration, hypocellularity in regions known to express ERβ mRNA, as well as deficits in social recognition and spatial learning (Wang et al., 2001; Rissman et al., 2002; Choleris et al., 2003; Wang et al., 2003). These findings, along with its commercial availability, made the βERKO mouse a good choice for characterization of the ck5912 antiserum. We also used tissues from the ERβ null mouse, ERβSTL-/L-, to confirm results obtained with βERKO mice. As with βERKOs, the original analysis of ERβ expression in ERβSTL-/L- mice focused on reproductive tissues. rtPCR on ERβSTL-/L- uterine and ovarian tissue showed no ERβ transcripts containing sequences beyond exon 3, and a frame shift-induced stop codon at nucleotide 599 predicts a severely truncated protein (Antal et al., 2008). Western blots of testis and prostate tissue probed using an antiserum raised to aa 465-485 of ERβ showed multiple bands in both WT and ERβSTL-/L-, but notably, ERβSTL-/L- tissues lacked a band at 61 kDa that was present in WT (Antal et al., 2008).
The antisera we tested recognized protein bands ranging from 45-60 kDa, within the range of known ERβ isoforms (LaVoie et al., 2002; Lewandowski et al., 2002). These antisera were generated against a diverse set of epitopes and were produced in various host species including, rabbit, mouse, chicken, and goat (Table 1). It is therefore highly unlikely that the labeling seen in βERKO and ERβSTL-/L- tissues is an artifact of secondary reagent labeling of IgG bands. Most of the antisera we tested recognized rERβ and not rERα, giving an initial impression of specificity. However two of them, H150 and 06-629, also recognized rERα. Both H-150 and 06-629 were raised against an N-terminal sequence in the A/B domain of ERβ, which shares low sequence homology with ERα (Kuiper et al., 1996, Tremblay et al., 1997). Therefore, the source of their cross-reactivity with ERα is unclear. In the case of H150, it is unlikely that ERα accounts for the labeling seen in tissue, since the immunolabeled band was consistently ∼55 kDa, too low for ERα (68 kDa).
In contrast to other antisera, PA1-310B and ck5912 initially looked promising based on specificity for rERβ and ERβ transfected into HT22 or HEK 293 cells. However, in subsequent tests, these and other antisera showed identical immunoreactivity in WT compared with βERKO and ERβSTL-/L- mice. The original characterization of the βERKO mouse reported a lack of nuclear labeling in ovarian granulosa cells using PA1-310B anti-ERβ (Krege et al., 1998). Yet, in our experiments we saw faint immunolabeling using this same antiserum. It is possible that variation between lots of antiserum and/or in the experimental protocols used accounts for this discrepancy (Lorincz and Nusser, 2008).
The βERKO mouse was generated by disrupting exon 3 of ERβ and the ERβSTL-/L- null mouse was generated by excising exon 3. Because a splice variant of ERβ that lacks exon 3 (ERβΔ3) has been characterized in rat and human (Petersen et al., 1998; Poola et al., 2002), it was important to exclude the possibility that an ERβΔ3 variant was expressed in βERKO tissues, which might account for the immunolabeling we observed. In previous work, rtPCR was preformed on βERKO ovarian and prostate tissue and this revealed transcript variants encoding truncated proteins (Krege et al., 1998). We performed a similar experiment as Krege et al. (1998) on WT and βERKO ovary, and because neither Krege et al., (1998) nor Antal et al. (2008) investigated ERβ expression in brain, we also included hypothalamus and hippocampus. Sequence analysis showed that neither the βERKO hypothalamic nor hippocampal transcripts were likely to account for protein bands of apparently identical molecular weight and intensity in WT and βERKO extracts. Both hypothalamic and hippocampal βERKO transcripts contained stop codons that would lead to severely truncated proteins. Thus, while it is conceivable that a truncated ERβ variant could be expressed in βERKOs, one would expect to see a clear difference in molecular weight(s) and we observed no differences. It also seems very unlikely that all of the antisera we tested recognize specifically an ERβΔ3 isoform. The more likely explanation for our results is that an as yet uncharacterized protein similar to ERβ cross reacts with many ERβ antisera.
Our results are in stark contrast to those obtained with the Z8P antiserum, which as noted above, did not label in tissues from ERβ deficient mice (Shughrue et al., 2002). Thus, based on this test and the good correspondence between Z8P immunoreactivity and ERβ mRNA, it is likely that brain areas labeled with Z8P do express ERβ protein. Shughrue and Merchenthaler (2001) used Z8P to generate a comprehensive atlas of nuclear ERβ immunoreactivity in the brain and reported labeling in many brain areas including (but not limited to) strong labeling in the preoptic area and hypothalamus, bed nucleus of the stria terminalis, and amygdala, moderate labeling in the neocortex and hippocampus, as well as in areas of the mid- and hindbrain. In addition to nuclear labeling, Z8P also has been used to show extranuclear ERβ immunoreactivity both in vitro and in vivo (Kalita et al., 2005; Milner et al., 2005; Jelks et al., 2007). Thus, it remains a strong possibility that ERβ is responsible for at least some of the rapid effects of estradiol in the brain.
We are not the first to report a lack of specificity in commercially available antisera. For example, Grimsey et al. (2008) tested several commercially available anti-cannabinoid CB1 receptor antisera and found that they failed to specifically immunolabel CB1 receptor transfected into HEK 293 cells. Similar to our findings, several commercially available antisera for muscarinic receptors have been shown to label identically in tissue from wildtype and muscarinic receptor knockout mice by western blot and immunohistochemistry (Pradidarcheep et al., 2008). Additionally, multiple galanin receptor antisera were reported to label in galanin receptor knockout mice (Lu and Bartfai, 2009). These reports, along with our own study on ERβ antisera, highlight the need for careful characterization of antisera used for protein localization or functional studies.
In conclusion, our results indicate that, while many ERβ antisera do recognize rERβ and ERβ expressed in cultured cells, they also recognize some protein(s) other than known ERβ variants in vivo that is/are present in βERKO and ERβSTL-/L- mouse brain. Thus, the results of experiments utilizing these antisera should be interpreted with appropriate caution.
Acknowledgments
This work was supported by the Northwestern University Institute for Women's Health Research, National Institute of Neurological Disorders and Stroke Grant R01 NS037324, National Institute of Mental Health Grant T32 MH067564.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Melissa A. Snyder, Email: m-ramous@northwestern.edu.
Tereza Smejkalova, Email: t-smejkalova@northwestern.edu.
Paul M. Forlano, Email: pforlano@brooklyn.cuny.edu.
Catherine S. Woolley, Email: cwoolley@northwestern.edu.
References
- Antal MC, Krust A, Chambon P, Mark M. Sterility and absence of histopathological defects in nonreproductive organs of a mouse ERbeta-null mutant. Proc Natl Acad Sci USA. 2008;105:2433–2438. doi: 10.1073/pnas.0712029105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choleris E, Gustafsson JA, Korach KS, Muglia LJ, Pfaff DW, Ogawa S. An estrogen-dependent four-gene micronet regulating social recognition: a study with oxytocin and estrogen receptor-alpha and -beta knockout mice. Proc Natl Acad Sci U S A. 2003;100:6192–6197. doi: 10.1073/pnas.0631699100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S, Fuller PJ. Identification of a splice variant of the rat estrogen receptor beta gene. Mol Cell Endocrinol. 1997;132:195–199. doi: 10.1016/s0303-7207(97)00133-0. [DOI] [PubMed] [Google Scholar]
- Dupont S, Krust A, Gansmuller A, Dierich A, Chambon P, Mark M. Effect of single and compound knockouts of estrogen receptors alpha (ERalpha) and beta (ERbeta) on mouse reproductive phenotypes. Development. 2000;127:4277–4291. doi: 10.1242/dev.127.19.4277. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick JL, Mize AL, Wade CB, Harris JA, Shapiro RA, Dorsa DM. Estrogen-mediated neuroprotection against beta-amyloid toxicity requires expression of estrogen receptor alpha or beta and activation of the MAPK pathway. J Neurochem. 2002;82:674–682. doi: 10.1046/j.1471-4159.2002.01000.x. [DOI] [PubMed] [Google Scholar]
- Grimsey NL, Goodfellow CE, Scotter EL, Dowie MJ, Glass M, Graham ES. Specific detection of CB1 receptors; cannabinoid CB1 receptor antibodies are not all created equal! J Neurosci Methods. 2008;171:78–86. doi: 10.1016/j.jneumeth.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Imamov O, Morani A, Shim GJ, Omoto Y, Thulin-Andersson C, Warner M, Gustafsson JA. Estrogen receptor beta regulates epithelial cellular differentiation in the mouse ventral prostate. Proc Natl Acad Sci USA. 2004;101:9375–9380. doi: 10.1073/pnas.0403041101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imwalle DB, Gustafsson JA, Rissman EF. Lack of functional estrogen receptor beta influences anxiety behavior and serotonin content in female mice. Physiol Behav. 2005;84:157–163. doi: 10.1016/j.physbeh.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Jelks KB, Wylie R, Floyd CL, McAllisster AK, Wise P. Estradiol targets synaptic proteins to induce glutamatergic synapse formation in cultured hippocampal neurons: critical role of estrogen receptor α. J Neurosci. 2007;27:6903–6913. doi: 10.1523/JNEUROSCI.0909-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalita K, Szymczak S, Kaczmarek L. Non-nuclear estrogen receptor b and a in the hippocampus of male and female rats. Hippocampus. 2005;15:404–412. doi: 10.1002/hipo.20066. [DOI] [PubMed] [Google Scholar]
- Kramár EA, Chen LY, Brandon NJ, Rex CS, Liu F, Gall CM, Lynch G. Cytoskeletal changes underlie estrogen's acute effects on synaptic transmission and plasticity. J Neurosci. 2009;29:12982–12993. doi: 10.1523/JNEUROSCI.3059-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krege JH, Hodgin JB, Couse JF, Enmark E, Warner M, Mahler JF, Sar M, Korach KS, Gustafsson JA, Smithies O. Generation and reproductive phenotypes of mice lacking estrogen receptor beta. Proc Natl Acad Sci USA. 1998;95:15677–15682. doi: 10.1073/pnas.95.26.15677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiper GG, Enmark E, Pelto-Huikko M, Nilsson S, Gustafsson JA. Cloning of a novel receptor expressed in rat prostate and ovary. Proc Natl Acad Sci USA. 1996;93:5925–5930. doi: 10.1073/pnas.93.12.5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaVoie HA, DeSimone DC, Gillio-Meina C, Hui YY. Cloning and characterization of porcine ovarian estrogen receptor beta isoforms. Biol Reprod. 2002;66:616–623. doi: 10.1095/biolreprod66.3.616. [DOI] [PubMed] [Google Scholar]
- Lewandowski S, Kalita K, Kaczmarek L. Estrogen receptor beta. Potential functional significance of a variety of mRNA isoforms. FEBS Lett. 2002;524:1–5. doi: 10.1016/s0014-5793(02)03015-6. [DOI] [PubMed] [Google Scholar]
- Li X, Schwartz PE, Rissman EF. Distribution of estrogen receptor-beta-like immunoreactivity in rat forebrain. Neuroendocrinology. 1997;66:63–67. doi: 10.1159/000127221. [DOI] [PubMed] [Google Scholar]
- Liu F, Day M, Muñiz LC, Bitran D, Arias R, Revilla-Sanchez R, Grauer S, Zhang G, Kelley C, Pulito V, Sung A, Mervis RF, Navarra R, Hirst WD, Reinhart PH, Marquis KL, Moss SJ, Pangalos MN, Brandon NJ. Activation of estrogen receptor-beta regulates hippocampal synaptic plasticity and improves memory. Nat Neurosci. 2008;11:334–343. doi: 10.1038/nn2057. [DOI] [PubMed] [Google Scholar]
- Lorincz A, Nusser Z. Specificity of immunoreactions: the importance of testing specificity in each method. J Neurosci. 2008;28:9083–9086. doi: 10.1523/JNEUROSCI.2494-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B, Leygue E, Dotzlaw H, Murphy LJ, Murphy LC, Watson PH. Estrogen receptor-beta mRNA variants in human and murine tissues. Mol Cell Endocrinol. 1998;138:199–203. doi: 10.1016/s0303-7207(98)00050-1. [DOI] [PubMed] [Google Scholar]
- Lu X, Bartfai T. Analyzing the validity of GalR1 and GalR2 antibodies using knockout mice. Naunyn Schmiedebergs Arch Pharmacol. 2009;379:417–420. doi: 10.1007/s00210-009-0394-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner TA, Ayoola K, Drake CT, Herrick SP, Tabori NE, McEwen BS, Warrier S, Alves SE. Ultrastructural localization of estrogen receptor immunoreactivity in the rat hippocampal formation. J Comp Neurol. 2005;491:81–95. doi: 10.1002/cne.20724. [DOI] [PubMed] [Google Scholar]
- Mosselman S, Polman J, Dijkema R. ER beta: identification and characterization of a novel human estrogen receptor. FEBS Lett. 1996;392:49–53. doi: 10.1016/0014-5793(96)00782-x. [DOI] [PubMed] [Google Scholar]
- Ogawa S, Chan J, Chester AE, Gustafsson JA, Korach KS, Pfaff DW. Survival of reproductive behaviors in estrogen receptor beta gene-deficient (betaERKO) male and female mice. Proc Natl Acad Sci USA. 1999;96:12887–12892. doi: 10.1073/pnas.96.22.12887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradidarcheep W, Labruyère WT, Dabhoiwala NF, Lamers WH. Lack of specificity of commercially available antisera: better specifications needed. J Histochem Cytochem. 2008;56:1099–1111. doi: 10.1369/jhc.2008.952101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen DN, Tkalcevic GT, Koza-Taylor PH, Turi TG, Brown TA. Identification of estrogen receptor beta2, a functional variant of estrogen receptor beta expressed in normal rat tissues. Endocrinology. 1998;139:1082–1092. doi: 10.1210/endo.139.3.5840. [DOI] [PubMed] [Google Scholar]
- Poola I, Abraham J, Baldwin K. Identification of ten exon deleted ERbeta mRNAs in human ovary, breast, uterus and bone tissues: alternate splicing pattern of estrogen receptor beta mRNA is distinct from that of estrogen receptor alpha. FEBS Lett. 2002;516:133–138. doi: 10.1016/s0014-5793(02)02521-8. [DOI] [PubMed] [Google Scholar]
- Razandi M, Pedram A, Merchenthaler I, Greene GL, Levin ER. Plasma membrane estrogen receptors exist and functions as dimers. Mol Endocrinol. 2004;18:2854–2865. doi: 10.1210/me.2004-0115. [DOI] [PubMed] [Google Scholar]
- Rissman EF, Heck AL, Leonard JE, Shupnik MA, Gustafsson JA. Disruption of estrogen receptor beta gene impairs spatial learning in female mice. Proc Natl Acad Sci U S A. 2002;99:3996–4001. doi: 10.1073/pnas.012032699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudick CN, Woolley CS. Selective estrogen receptor modulators regulate phasic activation of hippocampal CA1 pyramidal cells by estrogen. Endocrinology. 2003;144:179–187. doi: 10.1210/en.2002-220581. [DOI] [PubMed] [Google Scholar]
- Sheldahl LC, Shapiro RA, Bryant DN, Koerner IP, Dorsa DM. Estrogen induces rapid translocation of estrogen receptor beta, but not estrogen receptor alpha, to the neuronal plasma membrane. Neuroscience. 2008;153:751–761. doi: 10.1016/j.neuroscience.2008.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shughrue PJ, Lane MV, Merchenthaler I. Comparative distribution of estrogen receptor-alpha and -beta mRNA in the rat central nervous system. J Comp Neurol. 1997;388:507–525. doi: 10.1002/(sici)1096-9861(19971201)388:4<507::aid-cne1>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Shughrue PJ, Merchenthaler I. Distribution of estrogen receptor beta immunoreactivity in the rat central nervous system. J Comp Neurol. 2001;436:64–81. [PubMed] [Google Scholar]
- Shughrue PJ, Askew GR, Dellovade TL, Merchenthaler I. Estrogen-binding sites and their functional capacity in estrogen receptor double knockout mouse brain. Endocrinology. 2002;143:1643–1650. doi: 10.1210/endo.143.5.8772. [DOI] [PubMed] [Google Scholar]
- Tomihara K, Soga T, Nomura M, Korach KS, Gustafsson JA, Pfaff DW, Ogawa S. Effect of ER-beta gene disruption on estrogenic regulation of anxiety in female mice. Physiol Behav. 2009;96:300–306. doi: 10.1016/j.physbeh.2008.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay GB, Tremblay A, Copeland NG, Gilbert DJ, Jenkins NA, Labrie F, Giguère V. Cloning, chromosomal localization, and functional analysis of the murine estrogen receptor beta. Mol Endocrinol. 1997;11:353–365. doi: 10.1210/mend.11.3.9902. [DOI] [PubMed] [Google Scholar]
- Wang L, Andersson S, Warner M, Gustafsson JA. Morphological abnormalities in the brains of estrogen receptor beta knockout mice. Proc Natl Acad Sci U S A. 2001;98:2792–2796. doi: 10.1073/pnas.041617498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Andersson S, Warner M, Gustafsson JA. Estrogen receptor (ER)beta knockout mice reveal a role for ERbeta in migration of cortical neurons in the developing brain. Proc Natl Acad Sci U S A. 2003;100:703–708. doi: 10.1073/pnas.242735799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner M, Wang L, Weihua Z, Cheng G, Sakaguchi H, Saji S, Nilsson S, Kiesselbach T, Gustafsson JA. Analysis of estrogen receptor expression in tissues. Methods Enzymol. 2003;364:448–463. doi: 10.1016/s0076-6879(03)64025-5. [DOI] [PubMed] [Google Scholar]
- Windahl SH, Vidal O, Andersson G, Gustafsson JA, Ohlsson C. Increased cortical bone mineral content but unchanged trabecular bone mineral density in female ERbeta(-/-) mice. J Clin Invest. 1999;104:895–901. doi: 10.1172/JCI6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Brinton RD. Estrogen receptor alpha and beta differentially regulate intracellular Ca(2+) dynamics leading to ERK phosphorylation and estrogen neuroprotection in hippocampal neurons. Brain Res. 2007;1172:48–59. doi: 10.1016/j.brainres.2007.06.092. [DOI] [PubMed] [Google Scholar]