Summary of recent advances
Identifying the principles that describe the formation of protein oligomers and fibrils with distinct morphologies is a daunting problem. Here we summarize general principles of oligomer formation gleaned from molecular dynamics simulations of Aβ-peptides. The spectra of high free energy structures sampled by the monomer provide insights into the plausible fibril structures, providing a rationale for the “strain phenomenon.” Heterogeneous growth dynamics of small oligomers of Aβ16–22, whose lowest free energy structures are like nematic droplets, can be broadly described using a two-stage dock-lock mechanism. In the growth process, water is found to play various roles depending on the oligomer size, and peptide length and sequence. Water may be an explicit element of fibril structure linked to various fibril morphologies.
Introduction
According to the “amyloid hypothesis” [1], Amyloid Disease (AD) is caused by the accumulation of the Aβ-peptide which is a normal byproduct of the metabolism of amyloid precursor protein (APP). The cleavage of APP resulting in Aβ-peptide is achieved through the action of secretases [2]. The primary component of Alzheimer’s related amyloid plaques is the Aβ-peptide, a 39-to-43 amino acid polypeptide of known sequence [3, 4]. A variety of natural mutations that occur close to the secretase cleavage sites associated with variable AD pathology also produce large variations in the fibril growth rates [5–8]. The potential link between oligomer formation and amyloidogenic diseases has made it necessary to understand the energetics and dynamics of transitions from monomers to oligomers and beyond. Delineating the factors that contribute to the thermodynamics and kinetics of oligomer formation, which is an essential step in the cascade of events that turn the disordered “collapsed coil” form of Aβ-peptide monomers[9, 10] into fibrils with the characteristic cross β structure, is an important step in the discovery of methods that prevent their formation.
Significant advances have been made in the determination of high resolution crystal structures of a number of peptides that form amyloid-like fibrils [11, 12], and in the description of molecular events in the transition from disordered monomers to oligomers [13–16]. While the dynamics and phase diagram of full length aggregating proteins are expected to be considerably richer, the aggregation tendencies of smaller peptides are excellent model systems, which can be used to gain quantitative biophysical insights into oligomer formation. In recent years, a combination of experimental [11, 17–20] and computational studies [13, 21–25] have led to a microscopic picture of the oligomerization process of peptides including the role water of in their self-assembly. In this perspective, we focus on the role molecular dynamics simulations have played in elucidating some of the general principles that govern the process of protein aggregation with particular emphasis on the initial events in the assembly of Aβ-peptides and their implications for the growth into mature amyloid-fibrils.
General considerations in peptide and protein aggregation
The routes to protein aggregation are intimately related to the folding landscape of proteins [26] as a function of several external factors protecting and denaturing osmolytes, presence of crowding agents, and protein concentration. The hallmark of amyloid forming peptides and proteins is that they access one or more “assembly-competent” structures induced by denaturation, stress or thermal fluctuations with lifetimes sufficient to allow for inter-protein interactions to occur. The molecular details of the steps leading to the formation of amyloid fibrils remain unknown because the species along the aggregation pathways are highly dynamic and are likely to be metastable. The overall growth process exhibits the characteristics of a nucleation growth process[27]. Once the critical nucleus (whose characteristics depend on sequence as well as external conditions) forms, the fibril formation process is essentially downhill in free energy. From this perspective the qualitative scenarios for explaining aggregation kinetics are in place. However, the details of the process, including the dependence of oligomer formation on the specifics of the sequence and the structural features of the intermediates in the multiple stages leading to the nucleus, are not understood.
Despite the complexity of the aggregation process several theoretical studies [21, 22, 28] show that the spectra of the states sampled by the monomers can provide insight into the tendency of specific sequences to form amyloid structures. Two extreme scenarios, which follow from the energy landscape perspective of aggregation [8], can be envisioned in the description of the early events in protein aggregation [29]. According to Scenario I, which applies to Aβ peptides, fibril formation requires partial unfolding of the native state [30] or partial folding of the unfolded state. Both events, which are likely to involve crossing free energy barriers lead to the transient population of an ensemble of assembly-competent structures N*. According to Scenario II, which describes aggregation of PrPSCS [31], the ensemble of N* structures has a lower free energy than the structures in the native state ensemble thus making the folded (functional state) state metastable.
The scenarios based on the energy landscape perspective provide a plausible connection to the strain phenotypes that have been extensively studied especially in yeast prion biology [32, 33]. Originally found in the context of wasting diseases [34] and mammalian prions, strain phenotypes, which grow from the same protein but lead to different heritable states, are found even in peptide fibrils [35] and amyloids grown from Aβ-peptides [17, 36]. At what stage of the growth of fibrils is a particular strain “encoded” in the structure? The suggestion that the N* structures are aggregation prone implies that the strain phenotypes may be encoded in the monomer structures themselves. We speculate that the various N* structures can form oligomers with different structures, which can subsequently lead to fibrils with structurally distinct fibrils. Certainly, it is unlikely that that information for polymorphism in amyloid fibrils is found in post-nucleus structures, which makes monomers or low order oligomers the likely candidates.
Folding spectra of Aβ monomers
The analysis of protein aggregation in terms of N* leads to two key predictions. The first is that ordered aggregation starts in all likelihood from one of the structures that encompass the N* ensemble. The second is that the ease of aggregation is related to the probability of populating the N* species, which implies that the free energy barriers that separate the lowest free energy basin (unfolded U or folded N states) and N* conformations should dictate the growth kinetics. A number of studies have focused on the characteristic structures that are sampled by the monomer in the hope of gleaning insights into their amyloidogenic tendencies [21, 22, 28, 37, 38]. Before discussing the free energy spectra of monomers it is useful to describe the arrangements of Aβ in two well-known fibril models. The solid-state NMR-based structures [39, 40] for the fibrils of Aβ1–40 (Tycko model) include, as a key structural element, a bend involving residues V24VGSN27. The structural motif with the V24GSN27 turn and intrapeptide salt bridge between D23 and K28 ensures that isolated charges are not buried in the low-dielectric interior of the fibril. Such a structural motif when stacked in parallel leads to a fibril that satisfies the “amyloid self-organization principle” that the stability of amyloid fibril arises by maximizing the number of hydrophobic and favorable electrostatic interactions (formation of salt-bridges and hydrogen bonds)[21].
A different structural model (Luhrs model) for Aβ1–42 fibril [41], which maintains the basic strand-bend-strand motif of the Tycko model, suggests that residues [17–42] form in-register parallel β sheets formed from a minimum of two peptides. In this model the side chains of strand 1 (β1 spanning residues (18–26)) from the nth peptide inter digitate with those of strand two (β2 that runs from residues (31–42)) of the (n − 1)th peptide [41]. The arrangement of strands in this model is some what reminiscent of a domain swap mechanism, which has been proposed as a generic way in which ordered structures can form [42]. A natural consequence of the Luhrs model is that the bend in the monomer involves residues S26NKGA30 with K28 being positioned in such a way that it can form a salt-bridge with D23 from β1 of the neighboring peptide. The possibility of inter-peptide salt-bridge was also proposed by Tycko and coworkers [18].
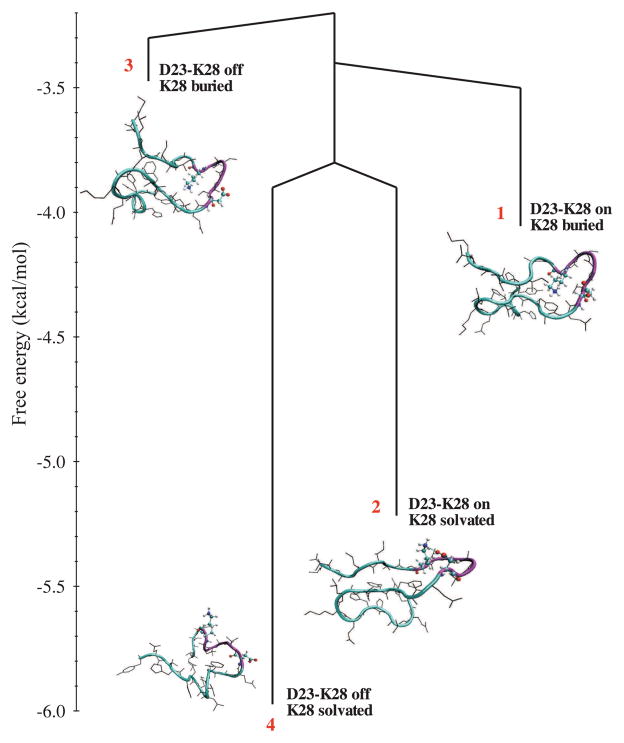
Several experimental and simulation studies have examined the interactions that stabilize the “folded” (lowest free energy states) of the Aβ21–30 fragment[37, 43–45]. Detailed MD studies of Aβ10–35 and Aβ9–40 monomers validate a key prediction of the “N* postulate,” namely, structural elements resembling those in the fibrils, manifest themselves in soluble monomers [21, 38]. Using extensive simulations and novel analysis of the data, it was shown that the formation of a stable structure with an intact D23-K28 salt bridge and the VGSN turn is highly improbable in the monomer. Our results suggest that overcoming the large barrier to desolvation of D23 and K28, which can only occur at finite peptide concentration, must be an early event in the oligomer formation. The spectrum of conformational states of Aβ10–35 (Fig. 1) suggests that a variety of high free energy states are accessible to the monomer. Among them the structures that belong to Basin 4, with the D23-K28 salt-bridge and V24GSN27 formed, resemble those found in the Tycko model for Aβ1–40. Similar conclusions were reached in MD simulations of Aβ9–40 monomer (see Fig. 5 in [38]) and Aβ21–30 monomer [43]. It is likely that in the interaction-driven aggregation process such a monomer structure will be accessed relatively early in the fibril formation process.
Figure 1.
Free energy spectrum of Aβ10–35 monomer obtained from MD simulations. States with the disrupted salt bridge are more favorable, and a large barrier makes the transition between the formed and disrupted substates improbable. Burying K28 in the peptide interior is an unfavorable process. The number of microstates associated with each of the four basins is indicated in parentheses. D23-K28 on stands for salt bridge present, while D23-K28 off stands for salt bridge broken. Reprinted from [21].
Interestingly, the salt-bridge is absent and K28 is in the interior in the ensemble of structures in Basin 3. The burial of charged residue is compensated by additional electrostatic interactions between the ammonium group of K28 and the backbone carbonyl oxygen of F20 and E22 and the hydrophobic interactions between the aliphatic side chain of K28 and the side chains of V24 and I31. It appears that the burial of K28 necessarily distorts the V24GSN27 turn, and perhaps displaces the turn region to S26NKGA30, as in the Luhrs model [41]. The ensemble of structures in Basin 3 has a significant overlap with the fibril model for Aβ1–42. We speculate that if the monomers in Basin 3 are packed to form fibrils, with intact S26SNKGA30, then the uncompensated charge on K28 can only be accommodated by an intermolecular D23-K28 salt bridge. The N* postulate explains the plausible differences in the different fibril morphologies in terms of the monomer seeds from which they are likely to grow.
We should stress that the N* ensemble alone cannot determine the diverse structures observed in the fibrils. The heterogeneous fibril morphologies with helical twists and striations, domain-swapped fibrils, and distinct symmetry arrangements can only be predicted using interactions between multiple chains. These intriguing variations even among fibrils grown from identical sequences cannot be anticipated by focusing on the conformational diversity of the monomers alone. After all, “More is Different” [46]. Nevertheless, the structures in the different basins of a monomer suggest potential candidates whose packing might provide insights into some of the morphologies observed in amyloid fibrils.
Free energy landscape for Aβ10–35 dimer formation
In order to characterize the early stages of Aβ peptide aggregation pathway, formation of the Aβ10–35-peptide dimer was studied in aqueous solution[47]. Dimer structures were evaluated for stability relative to the separated monomeric peptides, using computed estimates of the desolvation and electrostatic interaction energies, in an effort to identify putative stable dimer structures. The potential of mean force associated with the dimerization of the peptides in aqueous solution was computed using umbrella sampling and classical molecular dynamics simulation at constant temperature and pressure.
Two extreme models for monomer association – one which supposes that the principal mechanism stabilizing the dimer structure is the burial of hydrophobic surface and the other that supposes that the electrostatic interaction is the primary associative stabilizing interaction – were examined. It was found that the former leads to more energetically favorable dimerization[47]. It is more efficient to remove the entropically unfavorable structured water between the opposing hydrophobic regions of the two monomers than to stabilize the monomer solely through electrostatic interactions.
This finding agrees with the experimental observation that the mutation E22Q – where a charged glutamic acid residue is replaced by a polar glutamine residue – increases the propensity for amyloid formation [7, 48] and our previous computational studies of solvation of the E22Q mutant and WT peptides [49]. In more recent simulations of the Aβ16–35 peptide monomers and dimers, no significant secondary structure formation was observed, while the key E22DVGSNK28 region is observed to form a “loop” structure similar to that observed in shorter fragments and peptide fibrils. It would be profitable to carry out a more detailed energy landscape analysis as has been done for the aggregation of human transthyretin protein fragments[50] and KFFE tetrapeptide[51] to better evaluate the energetics of the aggregation ensemble.
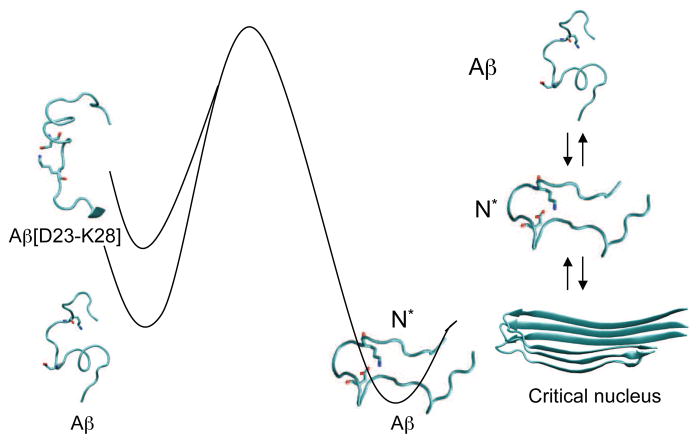
Recent experiments have shown that the congener, Aβ1–40 [D23-K28], in which the side chains of residues Asp23 and Lys28 are linked by a lactam bridge, forms amyloid fibrils that are structurally similar to the wild type (WT) Aβ peptide at a rate that is nearly one thousand times faster than the WT [52]. All-atom molecular dynamics simulations of the WT dimer, as well as a monomer and dimers of Aβ10–35 [D23-K28] with constrained D23-K28 salt bridge in explicit solvent, have been used to explore the origin of the observed enhanced rate of fibril formation [38].
Those simulations show that the assembly-competent monomers (N*), with strand conformations in the residues spanning the N and C termini and a bend involving residues D23VGSNKG29, are populated to a greater extent in Aβ10–35 [D23-K28] than in the WT, which has negligible probability of forming N*. The salt bridge in N*, whose topology is similar to that found in the fibril, is hydrated. The reduction in the free energy barrier to fibril formation in Aβ10–35 [D23-K28], compared to the WT is attributed to entropic restrictions that arise from the salt bridge constraint (see Fig. 2). A decrease in the entropy of the unfolded state and the lesser penalty for conformational rearrangement, including the formation of the salt bridge in Aβ peptides with D23-K28 constrained, results in a reduction in the kinetic barrier in the constrained Aβ1–40 [D23-K28] compared to the WT peptide.
Figure 2.
Free energy landscape of Aβ1–40 and the Aβ1–40 [D23-K28] peptide congener. By constraining the D23 and K28 residues to be proximate, through a theoretical constraint or formation of a covalent β-lactam bond, the free energy barrier between the monomer and the aggregation competent N* is reduced. Reprinted from [38].
Although a number of factors determine the growth of fibrils, the decrease in the free energy barrier of formation of N* in the Aβ1–40 [D23-K28] congener, relative to the WT peptide, is a major factor in the rate enhancement for fibril formation [52]. Qualitatively similar results were obtained using simulations of Aβ9–40 peptides. These results support the N* conjecture that mutations or other constraints that preferentially enhance the population of N* species would enhance aggregation rates.
Probing the structural characteristics of oligomers of Aβ-peptides
The unstable nature of oligomers makes it difficult to determine their dynamics and structures. Simulations using coarse-grained (CG) models [25, 53–55] have revealed that the formation of oligomers and subsequently fibrils involves a number of distinct stages during which the monomers, oligomers, and protofilaments undergo substantial conformational changes. Interestingly, it has been proposed the mutations that diminish β-strand propensity in the monomeric peptides may diminish fibril formation (by reducing population of N*) while enhancing the formation of potentially toxic oligomeric structures[55]. Typically, it is found that disordered oligomers form readily and the peptides then adopt ordered conformations even when the size of the oligomer is less than the critical nucleus size [13, 15, 47]. While the results from the CG models establish the generic features of protein aggregation, detailed all-atom MD simulations are often necessary to identify the driving force for protein aggregation and the formation of ordered structures.
Oligomer growth mechanism of Aβ16–22 fragments
The fragment Aβ16–22 (K16LVFFAE22) that encompasses the Central Hydrophobic Cluster (CHC) L17VFFA21 is predominantly a random coil in isolation [13] with some tendency for the hydrophobic residues (especially V18) to adopt β conformation[56]. In the fibril state the peptides are arranged in an anti-parallel manner [57], which results in salt-bridge formation and maximization of hydrophobic interaction between the residues in the CHC. Trimers of Aβ16–22 coalesce to rapidly form disordered aggregates driven primarily by hydrophobic interactions between the CHC residues. In the process, the peptide transiently adopts α-helical conformations even though there is no evidence for the isolated monomer to be found in the α-helical basin [13]. At longer times the peptides are arranged in an anti-parallel fashion as in the fibril with substantial excursions to other basins of attraction in which the peptides adopt alternate structures [15, 58–60]. The mutants G16LVFFAG22 and K16SVSSAE22 are unstable [13], thus establishing the importance of both the hydrophobic and electrostatic interactions in stabilizing the oligomer and presumably the fibrils [61].
The growth mechanism of (Aβ16–22)n for n > 3 was probed by monitoring the reaction (Aβ16–22)n−1 + Aβ → (Aβ16–22)n for n ranging from 4 to 6 has provided a detailed picture of the growth dynamics of oligomers[15]. In this reaction the unstructured monomer is added to a preformed “fluid-like” template composed of (n − 1) monomers. The lowest free energy structures of the oligomers (Aβ16–22)n resemble nematic droplets with the β strands aligned along a director resulting in a value of ~ 0.9 for the liquid crystal order parameter. The process of adding a monomer to a preformed nematic droplet can be globally described by a two-stage Dock-Lock (DL) mechanism, which was first suggested to describe the addition of a monomer to a growing fibril [62].
According to the DL mechanism, in the first stage the monomer rapidly and non-specifically docks onto the preformed nematic droplet. In the locking stage the monomer adopts the β-strand conformation of the template nematic droplet. The qualitative aspects of the DL mechanism capture essential features of the time-dependent changes in the β -strand content of the peptides in the nematic droplet and the monomer for the reaction (Aβ16–22)5 + Aβ → (Aβ16–22)6 [15]. In the absence of interaction with the nematic droplet the probability of the monomer adopting β-strand is negligible. Interaction with the nematic droplet first results in an increase in the end-to-end distance with a concomitant increase in the β-strand content. The high initial β-strand content of the nematic droplet is maintained during the course of the simulation.
The β-strand content of the added monomer grows in two stages. In the first phase, the β-strand content increases substantially from its initial low value, which shows that most of the growth occurs immediately upon docking. The extent of strand formation continues to increase over a period of tens of ns during which there are large changes in the structure of the nascent monomer. In the second stage the monomer adopts a β-strand conformation on a very long time scale.
A few comments about the DL mechanism are in order. (1) Although discussed in the context of addition of Aβ16–22 to a preformed nematic droplet the global description of the growth process in terms of a broad two stage dynamics is applicable to other systems as well [63]. The locking time scale, which increases as the number of peptides increases, can be approximately described using the Lifschitz-Slyazov growth mechanism, i.e., τlock ≈ τ0(N)M3 [28] where M is the number of peptides, and the prefactor τ0(N) depends on the length of the peptide. (2) The description of growth dynamics in terms of a two stage DL mechanism is simplistic. When examined carefully, the assembly of the oligomers consists of multiple stages characterized by a range of time scales. In addition, there is considerable structural heterogeneity in the growth of oligomers (and indeed fibrils [63]) that cannot be captured by the DL mechanism. The nuances discovered in computer simulations can only be captured using single molecule experiments [64] and theoretical models that capture the structural fluctuations in the monomers and the oligomers (or fibrils) as they grow.
The role of water in Aβ fibril formation
Simulations of (Aβ16–22)3 formation [13] showed the ordered state can form in multiple ways. The rapid formation of disordered oligomers is typically driven by interaction between hydrophobic residues in the CHC. The observation that the interior of the small orientationally disordered structures is dry implies that expulsion of water molecules occurs on time scales that are far shorter than the timescale on which ordering of the peptides occur. Mutation of F19 renders the oligomers unstable [13], which further supports the conclusion that the lack of water in the interior of Aβ16–22 is largely due to side chain contacts (see Figs. 3a and 3c) between the residues in the CHC (L17VFFA21). The antiparallel orientation requires the formation of the salt bridge between K16 from one peptide and E22 from another, which underscores the importance of both the hydrophobic and electrostatic interactions in stabilizing the ordered state. In contrast, there are very few stable hydrogen bonds that persist between the peptides [15].
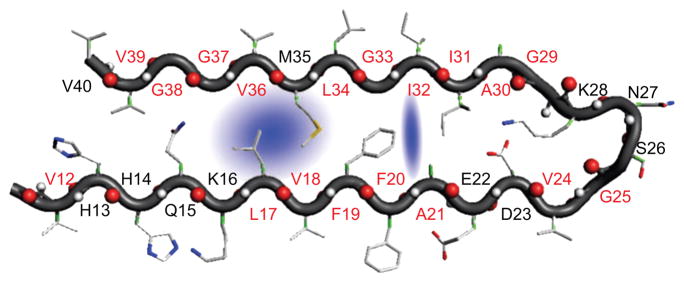
Figure 3.
Structure of the hydrated monomer in Aβ1–40 fibril. The blue shade represents trapped water molecules that are localized in a hydrophobic pocket. Reprinted from [68].
The expectation that water might play a subtle role as n increases was clearly demonstrated in the assembly of protofilaments of Aβ16–22 [65]. In some of the trajectories water is expelled early prior to assembly. In other trajectories, the two processes are observed to be coincident. The predominant interactions that mediate protofilament formation are hydrophobic in nature with interactions involving Phe playing a major role, as was previously shown in the context of oligomer formation [13, 66, 67]
It is remarkable that atomically detailed structures of fibrils grown from a large number of small peptides [11] show that the interior of two sheets is “bone dry.” This finding has lead to the suggestion that a key structural motif of fibrils could be pair of peptides held together by a “steric zipper” in which the sides chains are fully inter-digitated. As the size of the peptide increases the complexity of the assembly dynamics must increase, including the way water molecules mediate oligomer and fibril formation. In addition, given the presence of multiple fibril morphologies and the observed heterogeneities in the oligomer formation, we expect many variations in the way water mediates amyloid formation. Subtle roles played by water are starting to be elucidated. Using 2D IR spectroscopy it was recently shown that water molecules (roughly 1.2 per monomer) are trapped in Aβ1–40 (Fig. 3) fibrils [68]. The formation of water channels near the salt bridge (D23-K28) has been observed in simulations of a solid state NMR-derived structural model [69, 70]. However, the experimental finding that there are water molecules in the hydrophobic pocket (L17, V18, L34, and V36) that interact with the amide backbone of L17 and L34 is a surprise [68]. There are two possible explanations for this finding. If mobile water molecules are not part of the fully mature fibrils, it is likely that the fibril structures with trapped waters are metastable. Only by performing careful kinetic experiments can one assess if the water-trapped fibrils undergo further rearrangements. Alternatively, it is possible that these structures represent another fibril morphology. In light of these results [68], it is likely that in Luhrs model incorporating inter-peptide D23-K28 salt-bridge is soaked with mobile water molecules. The disparate experimental reports show that many of the questions pertaining to the roles discrete water molecules play in the formation of oligomers and fibrils formed from full length Aβ-peptides stand unanswered. In this regard, oligomer formation in reverse micelles with varying hydration levels [71] should provide a quantitative basis for describing how water mediates amyloid assembly.
Conclusions
Studies of amyloid-forming small peptides have provided valuable lessons, which may be useful in shedding light on the intriguing questions surrounding the considerably more complex process of fibril formation in proteins. Revealing the additional complexities that invariably arise when considering longer proteins will require a combination of new computational tools and experiments that can provide detailed structural and kinetic data. It is also necessary to bridge the gap between the biology and biophysics of fibril formation in order to decipher the structural basis of functional amyloids as well as those implicated in diseases. Finally, we suspect that the idiosyncratic role water plays in leading to distinct strains, which is surely one of the most perplexing aspect of fibril formation, will challenge researchers from all disciplines.
Acknowledgments
We are grateful to M. S. Li, Edward O’Brien, Eva Rivera, G. Reddy, and B. Tarus for useful discussions. This work was supported by a generous grant from the National Institutes of Health (GM076688–08).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and annotations
- 1**.Shankar GM, Li S, Mehta TH, Garcia-Munoz A, Shepardson NE, Smith I, Brett FM, Farrell MA, Rowan MJ, Lemere CA, Regan CM, Walsh DM, Sabatini BL, Selkoe DJ. Amyloid-β protein dimers isolated directly from Alzheimer’s brains impair synaptic plasticity and memory. Nat Med. 2008;14:837–842. doi: 10.1038/nm1782. Aβ dimers were shown to be the cause of long term potentiation and long term depression in rats. Learning behavior in rats was greatly disrupted by injection of soluble Aβ-dimers. Taken together these studies show that soluble oligomers by themselves can lead to many of the symptoms associated with AD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolfe M, Guénette SY. APP at a glance. J Cell Sci. 2007;120:3157–3161. doi: 10.1242/jcs.03481. [DOI] [PubMed] [Google Scholar]
- 3.Roher AE, Ball MJ, Bhave SV, Wakade AR. β-Amyloid from Alzheimer disease brains inhibits sprouting and survival of sympathetic neurons. Biochem Biophys Res Commun. 1991;174:572–579. doi: 10.1016/0006-291x(91)91455-l. [DOI] [PubMed] [Google Scholar]
- 4.Glenner GG, Wong CW. Alzheimer’s disease: Initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem Biophys Res Commun. 1984;120:885–890. doi: 10.1016/s0006-291x(84)80190-4. [DOI] [PubMed] [Google Scholar]
- 5.Lee JP, Stimson ER, Ghilardi JR, Mantyh PW, Lu YA, Felix AM, Llanos W, Behbin A, Cummings M, Criekinge MV, Timms W, Maggio JE. 1H NMR of Aβ amyloid peptide congeners in water solution. Conformational changes correlate with plaque competence. Biochemistry. 1995;34:5191–5200. doi: 10.1021/bi00015a033. [DOI] [PubMed] [Google Scholar]
- 6.Esler WP, Stimson EV, Ghilardi JR, Lu Y, Felix A, Mantyh PW, Lee JP, Maggio JE. Point substitution in the central hydrophobic cluster of human β-amyloid congener disrupts peptide folding and abolishes plaque competence. Biochem. 1996;35:13914–13921. doi: 10.1021/bi961302+. [DOI] [PubMed] [Google Scholar]
- 7.Esler WP, Stimson ER, Lachenmann MJ, Ghilardi JR, Lu Y, Vinters HV, Mantyh PW, Lee JP, Maggio JE. Activation barriers to structural transition determine deposition rates of Alzheimer’s disease. J Struct Biol. 2000;130:174–183. doi: 10.1006/jsbi.2000.4276. [DOI] [PubMed] [Google Scholar]
- 8.Massi F, Straub JE. Energy landscape theory for Alzheimer’s amyloid β-peptide fibril elongation, Proteins: Structure. Function and Genetics. 2001;42:217–229. doi: 10.1002/1097-0134(20010201)42:2<217::aid-prot90>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 9.Massi F, Peng JW, Lee JP, Straub JE. Simulation study of the structure and dynamics of the Alzheimer’s amyloid peptide congener in solution. Biophys J. 2001;80:31–44. doi: 10.1016/S0006-3495(01)75993-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sgourakis NG, Yan Y, McCallum SA, Wang CY, Garcia AE. The Alzheimer’s peptides Aβ 40 and 42 adopt distinct conformations in water: A combined MD/NMR study. J Mol Bio. 2007;368:1448–1457. doi: 10.1016/j.jmb.2007.02.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nelson R, Sawaya MR, Balbirnie M, Madsen AO, Riekel C, Grothe R, Eisenberg D. Structure of the cross-β spine of amyloid-like fibrils. NATURE. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12**.Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, MAI, Thompson MJ, Balbirnie M, Wiltzius JW, McFarlane HT, Madsen AO, Riekel C, Eisenberg D. Atomic structures of amyloid cross-β spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. Identified steric zippers, in which the side chains of two peptides from distinct sheets are fully inter digitated, as possible building blocks of amyloid fibrils. The interface between the steric zippers are necessarily devoid of water. [DOI] [PubMed] [Google Scholar]
- 13.Klimov DK, Thirumalai D. Dissecting the assembly of A β (16–22) amyloid peptides into antiparallel β sheets. Structure. 2003;11:295–307. doi: 10.1016/s0969-2126(03)00031-5. [DOI] [PubMed] [Google Scholar]
- 14.Gsponer J, Haberthur U, Caflisch A. The role of side-chain interactions in the early steps of aggregation: Molecular dynamics simulations of an amyloid-forming peptide from the yeast prion Sup35. Proc Natl Acad Sci. 2003;100:5154–5159. doi: 10.1073/pnas.0835307100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15**.Nguyen PH, Li MS, Stock G, Straub JE, Thirumalai D. Monomer adds to preformed structured oligomers of Aβ-peptides by a two-stage dock-lock mechanism. Proc Natl Acad Sci. 2007;104:111–116. doi: 10.1073/pnas.0607440104. The first convincing demonstration that growth of oligomers occurs by a dock-dock mechanism. The authors also suggested that, in addition to growth of oligomers by a global dock-lock mechanism, there are hidden complexities associated with the conformational changes from a monomer to an oligomer. In particular, water plays an important role in the Aβ assembly. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16*.Miller Y, Ma B, Nussinov R. Polymorphism of Alzheimer’s Aβ (17–42) (p3) Oligomers: The Importance of the Turn Location and Its Conformation. Biophys J. 2009;97:1168–1177. doi: 10.1016/j.bpj.2009.05.042. A nice study of the structural aspects of polymorphism in oligomers in an important fragment of Aβ peptide. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17**.Petkova AT, Leapman RD, Guo ZH, Yau WM, Mattson MP, Tycko R. Self-propagating, molecular-level polymorphism in Alzheimer’s β-amyloid fibrils. Science. 2005;307:262–265. doi: 10.1126/science.1105850. It is shown that changes in growth conditions can lead to different morphologies in the fibrils of Aβ [1–40] peptides, and that the corresponding molecular structures are distinct. More importantly, they showed that the toxicity also varies greatly and depends on the precise morphology. In seeded experiments the molecular structure is passed on from generation to generation. These findings have clear implications for the strain phenomenon. [DOI] [PubMed] [Google Scholar]
- 18.Petkova AT, Yau WM, Tycko R. Experimental constraints on quaternary structure in Alzheimer’s β-amyloid fibrils. Biochemistry. 2006;45:498–512. doi: 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cannon MJ, Williams AD, Wetzel R, Myszka DG. Kinetic analysis of β-amyloid fibril elongation. Anal Biochem. 2004;328:67–75. doi: 10.1016/j.ab.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 20.Platt GW, Xue WF, Homans SW, Radford SESE. Probing Dynamics within Amyloid Fibrils Using a Novel Capping Method. Ang Chemie-Iint Ed. 2009;48:5705–5707. doi: 10.1002/anie.200901343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21**.Tarus B, Straub JE, Thirumalai D. Dynamics of Asp23-Lys28 salt-bridge formation in A β (10–35) monomers. J Am Chem Soc. 2006;128:16159–16168. doi: 10.1021/ja064872y. By exhaustively exploring the free energy spectra of Aβ peptides the authors showed that the high free energy N* structures have a great degree of overlap with the monomer conformation in the fibril. This study provides a theoretical basis for probing the energy landscape of monomers of amyloidogenic peptides. [DOI] [PubMed] [Google Scholar]
- 22.Baumketner A, Shea JE. The structure of the Alzheimer amyloid β 10–35 peptide probed through replica-exchange molecular dynamics simulations in explicit solvent. J Mol Biol. 2007;366:275–285. doi: 10.1016/j.jmb.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 23.Takeda T, Klimov DK. Interpeptide interactions induce helix to strand structural transition in A β peptides. Proteins-Struct Funct Bioinformatics. 2009;77:1–13. doi: 10.1002/prot.22406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24**.Takeda T, Klimov DK. Replica Exchange Simulations of the Thermodynamics of A β Fibril Growth. Biophys J. 2009;96:442–452. doi: 10.1016/j.bpj.2008.10.008. By studying the temperature dependence of Aβ fibril growth using implicit solvent simulations, the authors observed thermodynamics consistent with the dock-lock mechanism. Interestingly, they show that the promiscuous docking process occurs over a wide temperature range, whereas the routes in the locking stage, which requires adaptation of β-strand structure commensurate with the underlying fibril morphology, is restricted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25*.Fawzi NL, Yap EH, Okabe Y, Kohlstedt KL, Brown SP, Head-Gordon T. Contrasting disease and nondisease protein aggregation by molecular simulation. Acc Chem Res. 2008;41:1037–1047. doi: 10.1021/ar800062k. An interesting summary of simulations of aggregation kinetics in protein L and G (non-disease related) and Aβ [1–40] peptides. The authors argue that the size of the critical nucleus for Aβ [1–40] is between 6–10 monomers. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bartlett AI, Radford SE. An expanding arsenal of experimental methods yields an explosion of insights into protein folding mechanisms. Nat Strucy Mol Biol. 2009;16:582–588. doi: 10.1038/nsmb.1592. [DOI] [PubMed] [Google Scholar]
- 27.Harper JD, Lansbury PT. Models of amyloid seeding in Alzheimer’s disease and scrapie: Mechanistic truths and physiological consequences of time-dependent stability of amyloid proteins. Annu Rev Biochem. 1997;66:385–407. doi: 10.1146/annurev.biochem.66.1.385. [DOI] [PubMed] [Google Scholar]
- 28*.Li MS, Klimov DK, Straub JE, Thirumalai D. Probing the mechanisms of fibril formation using lattice models. J Chem Phys. 2008;129:175101–175101. doi: 10.1063/1.2989981. A simple cubic lattice model, for which exact enumeration of all conformations can be made, shows that an aggregation prone conformation is the first excited state in the spectrum of the chosen sequence, thus validating the N* hypothesis. It is shown that fibril growth occurs by the Lifshitz-Slazov mechanism. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thirumalai D, Klimov DK, Dima RI. Emerging ideas on the molecular basis of protein and peptide aggregation. Curr Opin Struct Biol. 2003;13:146–159. doi: 10.1016/s0959-440x(03)00032-0. [DOI] [PubMed] [Google Scholar]
- 30.Fink AL. Protein aggregation: folding aggregates, inclusion bodies and amyloid. Folding & Design. 1998;3:R9–R23. doi: 10.1016/S1359-0278(98)00002-9. [DOI] [PubMed] [Google Scholar]
- 31.Dima RI, Thirumalai D. Probing the instabilities in the dynamics of helical fragments from mouse PrPc. Proc Natl Acad Sci. 2004;101:15335–15340. doi: 10.1073/pnas.0404235101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanaka M, Collins S, Toyama BH, Weissman JS. The physical basis of how prion conformations determine strain phenotypes. Nature. 2006;442:585–589. doi: 10.1038/nature04922. [DOI] [PubMed] [Google Scholar]
- 33**.Tessier PM, Lindquist S. Unraveling infectious structures, strain variants and species barriers for the yeast prion [PSI+] Nat Struct & Mol Biol. 2009;16:598–605. doi: 10.1038/nsmb.1617. An excellent summary of the current knowledge of the relationship between conformational misfolding and prion formation using the yeast system as an example. The molecular origins of strain variants are succinctly explained. The generality of the ideas to other systems is emphasized. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bessen RA, Kocisko DA, Raymond GJ, Nandan S, Lansbury PT, Caughey B. Non-genetic propagation of strain-specific properties of Scrapie Prion Protein. Nature. 1995;375:698–700. doi: 10.1038/375698a0. [DOI] [PubMed] [Google Scholar]
- 35**.Wiltzius JJ, Landau M, Nelson R, Sawaya MR, Apostol MI, Goldschmidt L, Soriaga AB, Cascio D, Rajashankar K, Eisenberg D. Molecular mechanisms for protein-encoded inheritance. Nat Struct Mol Biol. 2009;16:973–978. doi: 10.1038/nsmb.1643. Based on the crystal structures of fibrils from several peptides, the authors suggest two mechanisms for generating prion-like strains. One, termed packing polymorphism, arises from different packing arrangement from the same segment of protein, which is the usual way of thinking about strains. The other, segmental polymorphism, refers to distinct β sheets formed from different parts of the protein. These studies expand the routes to prion strains. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36**.Paravastu AK, Leapman RD, Yau WM, Tycko R. Molecular structural basis for poly-morphism in Alzheimer’s β-amyloid fibrils. Proc Natl Acad Sci. 2008;105:18349–18354. doi: 10.1073/pnas.0806270105. The authors discovered that Aβ [1–40] can adopt a twisted morphology with a three fold symmetry around the fibril axis. While the arrangement of monomers in this model is the same as in the striated structure, the two structures differ in the overall symmetry and quaternary interactions. These variations provide a structural basis for describing polymorphism. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37*.Murray MM, Krone MG, Bernstein SL, Baumketner A, Condron MM, Lazo ND, Teplow DB, Wyttenbach T, Shea JE, Bowers MT. Amyloid β-Protein: Experiment and Theory on the 21–30 Fragment. J Phys Chem B. 2009;113:6041–6046. doi: 10.1021/jp808384x. Investigation of the structures of Aβ [21–30] using a combination of ion mobility mass spectrometry and molecular simulations showed that the peptide has a bend and a perpendicular turn in the backbone that is stabilized by a network of interactions involving D23. These studies further establish that structuring around this charged residue occurs as the fibril forms in the full length peptide. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reddy G, Straub JE, Thirumalai D. Influence of Preformed Asp23-Lys28 Salt Bridge on the Conformational Fluctuations of Monomers and Dimers of A β Peptides with Implications for Rates of Fibril Formation. J Phys Chem B. 2009;113:1162–1172. doi: 10.1021/jp808914c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tycko R. Insights into the amyloid folding problem from solid-state NMR. Biochemistry. 2003;42:3151–3159. doi: 10.1021/bi027378p. [DOI] [PubMed] [Google Scholar]
- 40.Petkova AT, Ishii Y, Balbach JJ, Antzutkin ON, Leapman RD, Delaglio F, Tycko R. A structural model for Alzheimer’s β-amyloid fibrils based on experimental constraints from solid state NMR. Proc Natl Acad Sci. 2002;99:16742–16747. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luhrs T, Ritter C, Adrian M, Riek-Loher D, Bohrmann B, Doeli H, Schubert D, Riek R. 3D structure of Alzheimer’s amyloid-β (1–42) fibrils. Proc Natl Acad Sci. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo ZF, Eisenberg D. Runaway domain swapping in amyloid-like fibrils of T7 endonuclease I. Proc Natl acad Sci. 2006;103:8042–8047. doi: 10.1073/pnas.0602607103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tarus B, Straub JE, Thirumalai D. Structures and free-energy landscapes of the wild type and mutants of the Aβ (21–30) peptide are determined by an interplay between intrapeptide electrostatic and hydrophobic interactions. J Mol Bio. 2008;379:815–829. doi: 10.1016/j.jmb.2008.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen W, Mousseau N, Derreumaux P. The conformations of the amyloid–β (21–30) fragment can be described by three families in solution. J Chem Phys. 2006;125:084911. doi: 10.1063/1.2337628. [DOI] [PubMed] [Google Scholar]
- 45.Krone M, Baumketner A, Bernstein S, Wyttenbach T, Lazo N, Teplow D, Bowers M, Shea JE. Effects of familial Alzheimer’s disease mutations on the folding nucleation of the amyloid β-protein. J Mol Bio. 2008;381:221–228. doi: 10.1016/j.jmb.2008.05.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anderson PW. More is different - Broken Symmetry and Nature of Hierarchical Structure of Science. Science. 1972;177:393–396. doi: 10.1126/science.177.4047.393. [DOI] [PubMed] [Google Scholar]
- 47**.Tarus B, Straub JE, Thirumalai D. Probing the initial stage of aggregation of the A β (10–35)-protein: Assessing the propensity for peptide dimerization. J Mol Biol. 2005;345:1141–1156. doi: 10.1016/j.jmb.2004.11.022. Using a protocol based on shape complementarity and molecular dynamics simulations, it was shown that the assembly of dimers from unstructured Aβ [10–35] monomers occurs largely by hydrophobic interactions. Expulsion of water from the interface, which involves crossing a free energy barrier, is likely to be a key early step. [DOI] [PubMed] [Google Scholar]
- 48.Miravalle L, Tokuda T, Chiarle R, Giaccone G, Bugiani O, Tagliavini F, Frangione B, Ghiso J. Substitutions at codon 22 of Alzheimer’s Aβ peptide induce diverse conformational changes and apoptotic effects in human cerebral endothelial cells. J Biol Chem. 2000;275:27110–27116. doi: 10.1074/jbc.M003154200. [DOI] [PubMed] [Google Scholar]
- 49.Massi F, Klimov D, Thirumalai D, Straub JE. Charge states rather than propensity for β-structure determine enhanced fibrillogenesis in wild-type Alzheimer’s β-amyloid peptide compared to E22Q Dutch mutant. Protein Sci. 2002;11:1639–1647. doi: 10.1110/ps.3150102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li D, Han L, Huo SH. Structural and pathway complexity of β-strand reorganization within aggregates of human transthyretin(105–115) peptide. J Phys Chem B. 2007;111:5425–5433. doi: 10.1021/jp0703051. [DOI] [PubMed] [Google Scholar]
- 51.Strodel B, Wales DJ. Implicit solvent models and the energy landscape for aggregation of the amyloidogenic KFFE peptide. J Chem Theor Comp. 2008;4:657–672. doi: 10.1021/ct700305w. [DOI] [PubMed] [Google Scholar]
- 52*.Sciarretta KL, Gordon DJ, Petkova AT, Tycko R, Meredith SC. Aβ40-lactam(D23/K28) Models a Conformation Highly Favorable for Nucleation of Amyloid. Biochemistry. 2005;44:60036014. doi: 10.1021/bi0474867. The aggregation rate is found to increase by a factor of nearly 1000 in Aβ [1–40] monomers in which there is a lactam bond that links D23 and K28. It was shown in [28] that entropic restrictions only account for a factor of about 200 increase in rates. It is likely that there is reduction in the free energy barrier to nucleation in the lactam construct compared to the wild type. [DOI] [PubMed] [Google Scholar]
- 53.Dima RI, Thirumalai D. Exploring protein aggregation and self-propagation using lattice models: Phase diagram and kinetics. Prot Sci. 2002;11:1036–1049. doi: 10.1110/ps.4220102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nguyen HD, Hall CK. Molecular dynamics simulations of spontaneous fibril formation by random-coil peptides. Proc Natl Acad Sci. 2004;101:1618016185. doi: 10.1073/pnas.0407273101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55**.Bellesia G, Shea JE. Diversity of kinetic pathways in amyloid fibril formation. J Chem Phys. 2009;131:111102. doi: 10.1063/1.3216103. Using a coarse-grained off-lattice model the authors show that there are multiple routes to fibril formation. Interestingly, the model also suggests that non-fibrillar aggregates can also form in addition to the ordered cross β structures. [DOI] [PubMed] [Google Scholar]
- 56.Massi F, Straub JE. Probing the origins of increased activity of the E22Q “Dutch” mutant Alzheimer’s β-amyloid peptide. Biophys J. 2001;81:697–709. doi: 10.1016/S0006-3495(01)75734-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ma B, Nussinov R. Stabilities and conformations of alzheimer’s β-amyloid peptide oligomers (Aβ16–22, Aβ16–35, and Aβ10–35): Sequence effects. Proc Natl Acad Sci USA. 2002;99:14126–14131. doi: 10.1073/pnas.212206899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Santini S, Wei G, Mousseau N, Derreumaux P. Pathway complexity of Alzheimer’s β–Amyloid Aβ16–22 peptide assembly. Structure. 2004;12:1245–1255. doi: 10.1016/j.str.2004.04.018. [DOI] [PubMed] [Google Scholar]
- 59**.Gnanakaran S, Nussinov R, Garcia AE. Atomic-level description of amyloid β-dimer formation. J Am Chem Soc. 2006;128:2158–2159. doi: 10.1021/ja0548337. The complex energy landscape of dimer formation in Aβ [16–22] is illustrated using all atom simulations with replica exchange calculations. They find that many minima, with different structural arrangements of the dimer, are seen in the free energy landscape. [DOI] [PubMed] [Google Scholar]
- 60.Mousseau N, Derreumaux P. Exploring energy landscapes of protein folding and aggregation. Frontiers in Bioscience. 2008;13:4495–4516. doi: 10.2741/3019. [DOI] [PubMed] [Google Scholar]
- 61.Yun S, Urbanc B, Bitan G, Teplow DB, Stanley HE. Role of electrostatic interactions in amyloid β-protein (aβ) oligomer formation: A discrete molecular dynamics study. Biophys J. 2007;92:4064–4077. doi: 10.1529/biophysj.106.097766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Esler WP, Stimson ER, Jennings JM, Vinters VH, Ghilardi JR, Lee JP, Mantyh PW, Maggio JE. Alzheimer’s disease amyloid propagation by a template-dependent dock-lock mechanism. Biochemistry. 2000;39:6288–6295. doi: 10.1021/bi992933h. [DOI] [PubMed] [Google Scholar]
- 63**.Reddy G, Straub JE, Thirumalai D. Dynamics of locking of peptides onto growing amyloid fibrils. Proc Natl Acad Sci. 2009;106:11948–11953. doi: 10.1073/pnas.0902473106. The most detailed study to date on how a monomer from Sup35 adds on to a growing fibril to form a dry interface. Surprisingly, water molecules between the strands are expelled in a “quantized” manner in two distinct stages. This was contrasted with the growth of Aβ peptides in which water is expelled in a continuous manner. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64**.Ferreon ACM, Gambin Y, Lemke EAEA, Deniz AA. Interplay of alpha-synuclein binding and conformational switching probed by single-molecule fluorescence. Proc Natl acad Sci. 2009;106:5645–5650. doi: 10.1073/pnas.0809232106. The authors probed the conformational changes that occur as the unstructured α-synuclein interacts with membranes. They showed that in that process the monomer undergoes a series of conformational transitions, with the folding landscape consisting of two distinct α-helical structures. The single molecule FRET experiments used by these authors will be most useful in shedding light on the growth of oligomers in other systems as well. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65*.Krone MG, Hua L, Soto P, Zhou R, Berne BJ, Shea JE. Role of water in mediating the assembly of Alzheimer amyloid-β A β 16–22 protofilaments. J Am Chem Soc. 2008;130:11066–11072. doi: 10.1021/ja8017303. The authors used MD simulations to explore the various routes by which protofilaments assemble. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baumketner A, Shea JE. Free energy landscape for amyloidogenic tetrapeptide dimerization. Biophys J. 2005;89:1493–1503. doi: 10.1529/biophysj.105.059196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Soreq H, Gazit E. The structural basis of amyloid formation. Curr Alzheimer Res. 2008;5:232. doi: 10.2174/156720508784533349. [DOI] [PubMed] [Google Scholar]
- 68**.Kim YS, Liu L, Axelsen PH, Hochstrasser RM. 2D IR provides evidence for mobile water molecules in β-amyloid fibrils. Proc Natl Acad Sci. 2009;106:17751–17756. doi: 10.1073/pnas.0909888106. Using sophisticated 2D-IR spectroscopy the authors show that Aβ fibrils are soaked with water with each monomer carrying on average roughly 1.2 molecules. Surprisingly, the water molecules are predominantly in the hydrophobic pocket. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Buchete NV, Tycko R, Hummer G. Molecular dynamics simulations of Alzheimer’s β-amyloid protofilaments. J Mol Biol. 2005;353:804–821. doi: 10.1016/j.jmb.2005.08.066. [DOI] [PubMed] [Google Scholar]
- 70.Buchete NV, Hummer G. Structure and dynamics of parallel β-sheets, hydrophobic core, and loops in Alzheimer’s A β fibrils. Biophys J. 2007;92:3032–3039. doi: 10.1529/biophysj.106.100404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71**.Mukherjee S, Chowdhury P, Gai F. Effect of Dehydration on the Aggregation Kinetics of Two Amyloid Peptides. J Phys Chem B. 2009;113:531–535. doi: 10.1021/jp809817s. Using reverse micelles, which are used to control the extent of hydration, the authors beautifully illustrate that dimerization of Aβ is enhanced as the extent of hydration decreases. The implication of the studies under crowded in vivo conditions is explored. [DOI] [PMC free article] [PubMed] [Google Scholar]