Structure
Six3 (also known as six homeobox-3), a vertebrate homolog of the Drosophila 'sine oculis' (so) gene, is a member of the evolutionarily conserved SIX family. The members of SIX family are found in diverse organisms including flatworms, fruit fly, medaka fish, chickens, frogs, zebrafish, mice and humans (Oliver et al., 1995).
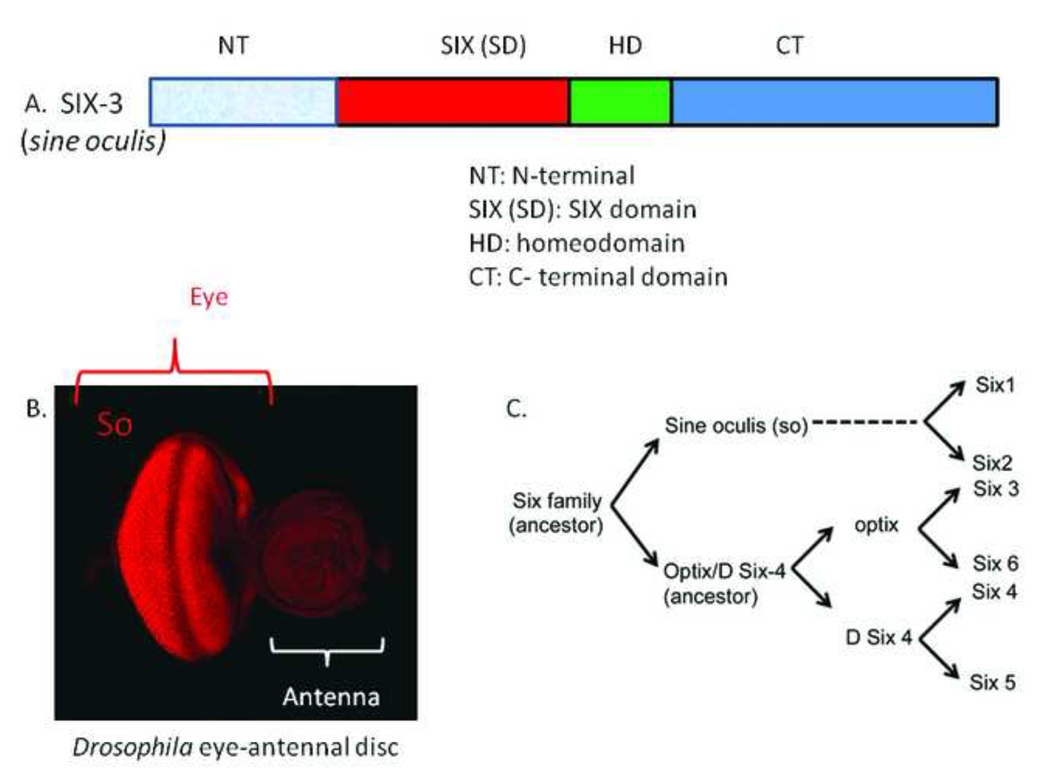
Members of the SIX gene family encode transcription factors characterized by the presence of evolutionarily conserved DNA-binding homeodomain and an upstream SIX domain, which may be involved both in determining DNA-binding specificity and in mediating protein-protein interactions (Fig. 1A). DNA binding activity of so is mediated through the Prd class of homeobox nucleic acid recognition domain (HD). SIX proteins contain lysine residue at position fifty (K50), which assign SIX3 to an Orthodenticle sub-group within the Prd class. The site bound by SIX3 contains the traditional ATTA homeodomain core recognition sequence. Even though there are individual differences, the consensus binding sites for SIX proteins in flies and worms are TGATAC and GGGTATCA. The SIX domain which is the second interaction domain in the six3 protein, is 146 amino acids long, and lies just 5’ and directly adjacent to the HD. This domain is involved in mediating protein-protein interactions.
Fig. 1.
A. Six3 encodes a transcription factor with two evolutionarily conserved domains, SD, the protein- protein interaction domain and HD (the homeobox recognition domain), responsible for DNA binding. The Six domain is directly adjacent to HD on 5' end. B. Immunohistochemical localization of So protein in developing eye imaginal disc of Drosophila Larva. So is localized both in differentiated retinal cells as well as retinal precursor cells. C. The comparison of gene structures and sequence has led to creation of three subclasses: each containing one of the fly genes so, optix and DSIX4. The neighbor joining tree analysis and blast searches of extant Six family members from 12 Drosophilids help understand the path of evolution in Six class of genes.
Function
In Drosophila, So is expressed in the eye imaginal disc (a monolayer epithelium that develops into the adult eye of fly) in nuclei of both differentiating retinal photoreceptors as well as retinal precursor cells (Fig. 1B). The SD domain of So physically interacts with transcriptional coactivator Eyes absent (Eya) to promote eye development in Drosophila. This interaction is conserved across the animal kingdom.
The evolution of six 1–6 class of genes in the vertebrate lineage involves a genome duplication event (Fig. 1C).
Genes in the six family are shown to be expressed in head, retina, lens, ear, nose, brain, kidney, muscle and gonads, and play major role in vertebrate and insect development or maintenance of the differentiated state of tissues. SIX3 and SIX6, are the major SIX proteins in the retina. SIX3 is a member of complex network of eye field transcription factors that regulate retina and lens development. SIX3 is expressed at multiple points during the development of the vertebrate eye. It is first localized to the optic vesicle and optic stalk but then expands to include the neural retina and developing lens placode (Oliver et al., 1995). In humans, northern blot analysis revealed that the SIX3 gene was expressed only in the eye. In human embryos, expression of SIX3 was detected as early as 5–7 weeks of gestation and found to be maintained in the eye throughout the entire period of fetal development. At 20 weeks of gestation, expression of SIX3 in the human retina was detected in the ganglion cells and in cells of the inner nuclear layer. In vertebrates the nuclear protein SIX3 recruits EYA4, a transcriptional coactivator, one of the four orthologs of Drosophila gene eyes absent.
Injection of six3 RNA into medaka fish embryos caused ectopic pax6 and Rx2 expression in midbrain and cerebellum, resulting in the formation of ectopic retinal primordia. Misexpresion of six3 in chick eye using retrovirus disrupts the integrity of corneal endothelium and affects corneal transparency. Gain-of-function studies suggested that six3 plays a role in eye anterior segment morphogenesis. Injected mouse six3 mRNA initiated ectopic expression of endogenous medaka six3, uncovering a feedback control of six3 expression. Initiation of ectopic retina formation demonstrated a pivotal role for six3 in vertebrate retina development and hinted at a conserved regulatory network underlying vertebrate and invertebrate eye development. Six3 plays a role in regulation of anterior eye segment development in vertebrates.
Six3 is involved in the proliferation of retinal precursor cells. Many cell cycle regulators are regulated in cell cycle dependent manner. SIX3 is a partner of DNA replication-inhibitor Geminin (Gem). Gem inhibits cell-cycle progression by sequestering Cdt1, the key component for the assembly of the pre-replication complex. SIX3 competes with Cdt1 directly to bind to Gem. SIX3 and Gem act antagonistically in controlling proliferation and differentiation. During early stages of development, high levels of SIX3 inhibit Gem activity, which allows proliferation and prevents premature neuronal differentiation. Later, Gem promote cell-cycle exit, a necessary prerequisite for the initiation of neuronal differentiation. At this stage Gem does not overlap with SIX3 and is maintained in the differentiating central neural retina. Recently, it has been seen that Drosophila cdc25 homolog, string, and mammalian cyc A are direct transcriptional targets of So and SIX1.
Six3 is a major player in lens development. Six3 normally exerts its effect by directly activating pax6, a gene considered to be the "master regulator of eye development." In the absence of six3, pax6 and Sox2 are down-regulated and fail to coordinate the activity of a series of additional genes that cooperate to form the lens (Liu et al., 2006). Six3 has also been implicated in newt lens regeneration (Grogg et al., 2005). Lens regeneration is mediated by transdifferentiation of the dorsal iris pigment epithelial cells, but never from the same cells in the ventral iris. Overexpression of six3 (with concomitant addition of retinoic acid) in the ventral iris can elicit lens transdifferentiation from that site (Grogg et. al., 2005). Six3 is involved in a feedback loop regulation with pax6. However, the gene network differs somewhat in Drosophila and vertebrates. For example, six3, which is important for lens development and activates pax6, is an ortholog of Drosophila optix, which in turn is not part of the ey subcircuit (so is). In other words in parts of the eye of some vertebrates six-3 expression does depend on pax6 and in others it does not. Also, feedback mechanisms involved in regulation of ey and so by dac are present in Drosophila but absent in the mouse.
Disease involvement
Six3 has not been directly involved with eye diseases. This could be explained by its role in the pax6 circuit as discussed above. However, it has been associated with holoprosencephaly (HPE) (Wallis, et al., 1999). Six3 is one of the earliest genes to be expressed in the anterior forebrain. SIX3 regulates Sonic hedgehog, Wnt, BMP and Nodal signaling in forebrain and is thought to enable these cells to adopt anterior cell fate. Interestingly, the map position of human six3 overlaps the locations of dominant disorder holoprosencephaly type 2 with ocular phenotypes. HPE exhibit signs like single central incisor, hypotelorism, microcephaly, or other craniofacial findings that can be present with or without associated brain malformations. Several evidences from studies indicated that misregulation of SIX transcription and/or posttranslational modification is an underlying cause for a wide range of primary cancers and metastatic lesions. The role of SIX proteins in cell cycle is significant and may be important in tumorigenesis and cancer biology.
Future Studies
The elucidation of the 3-D structure of six3 will be of paramount interest in delineating how it interacts with other members of the pax6 circuit. Also more elaborate studies of the physical interaction with other important members of the circuit, such as eya and dach will shed light of how the circuit is structured and how regulation is achieved in order to control differentiation and morphogenesis of the eye.
Acknowledgments
A.S. is supported by Ohio Cancer Research Associate grant. P.A.T is supported by NIH grant EY10540. We apologize to authors whose work cannot be cited due to limits for reference citation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Grogg MW, Call MK, Okomato M, Vergara MN, Del Rio-Tsonis K, Tsonis PA. BMP inhibition-driven regulation of SIX-3 underlies induction of newt lens regeneration. Nature. 2005;438(7069):858–862. doi: 10.1038/nature04175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Lagutin OV, Mende M, Streit A, Oliver G. Six3 activation of Pax6 expression is essential for mammalian lens induction and specification. EMBO J. 2006;25:5383–5395. doi: 10.1038/sj.emboj.7601398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver G, Mailhos A, Wehr R, Copeland NG, Jenkins NA, Gruss P. SIX3, a murine homologue of the sine oculis gene, demarcates the most anterior border of the developing neural plate and is expressed during eye development. Development. 1995;121(12):4045–4055. doi: 10.1242/dev.121.12.4045. [DOI] [PubMed] [Google Scholar]
- Wallis DE, Roessler E, Hehr U, Nanni L, Wiltshire T, Richieri-Costa A, Gillessen-Kaesbach G, Rommens J, Muenke M. Mutations in homeodomain of the human SIX3 gene cause holoprosencephaly. Nat. Genet. 1999;22(2):196–198. doi: 10.1038/9718. [DOI] [PubMed] [Google Scholar]