Abstract
Induction of autophagy in response to starvation is a highly conserved ability of eukaryotic cells, indicating a critical and ancient role of this process in adapting to nutrient conditions. The target of rapamycin (TOR) pathway is major conduit for such signals, and in most cell types TOR activity is necessary and sufficient to suppress autophagy under favorable growth conditions. Recent studies have begun to reveal how TOR activity is regulated in response to nutritional cues, and are shedding new light on the mechanisms by which TOR controls the autophagic machinery. In addition, a variety of signals, stressors and pharmacological agents that induce autophagy independent of nutrient conditions have been identified. In some cases these signals appear to have been spliced into the core TOR pathway, whereas others are able to bypass the control mechanisms regulated by TOR. Increasing evidence is pointing to an important role for both positive and negative feedback loops in controlling this pathway, leading to an emerging view that TOR signaling not only regulates autophagy but is also highly sensitive to cellular rates of autophagy and other TOR-dependent processes.
Introduction
Eukaryotic cells have evolved a number of defense mechanisms that allow survival in the face of environmental insults such as nutrient depletion, oxidative damage and conditions that disrupt protein folding. Chief amongst these responses is autophagy, a process whereby cytoplasmic constituents are engulfed within specialized double-membrane vesicles known as autophagosomes, for subsequent delivery to and degradation within the lysosome. Autophagy normally proceeds at a low, basal rate, playing a key role in the homeostatic clearance of old or damaged organelles and proteins. In response to starvation and other stresses, activation of autophagy can generate an internal source of nutrients that promotes survival in the absence of externally supplied nutrition. Both insufficient and excessive levels of autophagy have been shown to impact human health at multiple levels, and therefore understanding the cellular mechanisms that regulate this process is an important goal (reviewed in [1,2]).
TOR kinases are central regulators of multiple cellular responses to nutrient and growth factor signaling, including autophagy. The drug rapamycin induces autophagy in a wide variety of cell types and species by inhibiting the activity of TOR as part of the multi-component TOR Complex 1 (TORC1). This signaling complex includes TOR, the scaffolding protein Raptor, and additional proteins that also function with TOR in a second, rapamycin-insensitive complex not involved in autophagy (reviewed in [3]). Control of autophagy by TORC1 signaling is largely responsible for the potent effect of starvation as an autophagy inducer. The purpose of this review is to provide a summary of recent insights into the events upstream of TOR that regulate its function, and to discuss new developments in our understanding of how TOR interacts with the core set of proteins that directly control autophagosome formation and maturation. Signals that influence autophagy independently of TOR are also considered, with the goal of considering how specific steps of autophagy induction can be influenced by distinct molecular cues.
Regulation of TOR
Following the pioneering studies in the early 1990s that identified the TOR kinases as targets of rapamycin in yeast, experiments in mammalian cells and other model systems have defined several important regulatory steps and components that link TOR activity to upstream signals. Key amongst these regulatory factors are the small GTPase Rheb, which directly associates with TOR to stimulate its signaling activity, and the TSC1 and TSC2 proteins, which together inhibit Rheb through the GAP activity of TSC2. This TSC-Rheb-TOR trio has emerged as a core element of the pathway in metazoans, with the TSC1–TSC2 complex in particular acting as a convergence point for multiple signals that govern TOR. For example, factors that respond to cellular energy level (AMP-activated protein kinase, AMPK), growth factor signaling (Akt) or oxygen status (Redd1) have been shown to regulate TOR signaling through distinct, direct effects on TSC2 activity (reviewed in [4]).
Although this model is consistent with many experimental findings, it is now clear that some signals regulate TOR by other means. Perhaps most significantly, disruption of TSC genes does not prevent regulation of TOR activity by nutrient status [5], indicating that this conserved input must occur through an alternate mechanism. In addition, replacement of wild type Tsc1 and Tsc2 genes with mutant copies that lack Akt-dependent phosphorylation sites does not affect growth or viability in Drosophila [6,7]. These and other results have motivated efforts to identify additional regulatory mechanisms and factors acting upstream of TOR.
The hunt for nutrient-dependent signaling mechanisms has turned up a number of new potential regulators of this pathway. As TOR activity is highly sensitive to amino acid levels, particularly those of branched chain amino acids such as leucine, amino acid transporters have been considered as candidate factors involved in nutrient sensing. A number of proteins in the amino acid transporter family appear to act as amino acid sensors and signal transducers; often such factors have low transport activity, and are collectively referred to as transceptors (reviewed in [8]). In Drosophila, the proton-assisted transporter Path has the characteristics of a transceptor acting upstream of TOR [9]. path mutants are defective for cell growth and display genetic interactions with TOR. Expression of Path in a heterologous system leads to TOR activation, yet has only minimal transport capacity. In yeast, the general amino acid permease GAP1, which is regulated by TOR at the level of sorting to the plasma membrane, also has an amino acid sensor activity that is separable from its transport function, and is important for nutrient-dependent activation of cAMP-dependent protein kinase (PKA) [10].
The Rag family of GTPases has recently been identified as a new link between amino acids and TOR. These proteins function as novel heterodimeric GTPases, with the active dimer containing RagA or B (Gtr1 in yeast) and RagC or D (Gtr2) bound to GTP and GDP, respectively. In yeast, these proteins were previously shown to control exit from rapamycin-induced arrest as part of the EGO complex [11]. Three studies in mammalian cells, Drosophila and yeast now provide evidence that these GTPases are specific mediators of amino acid signaling to TOR [12*–14*]. Active Rag and Gtr heterodimers are able to bind and activate TORC1, through direct interactions with raptor, and this association is regulated by amino acid levels but not other inputs into TOR. Amino acids also specifically control the GTP loading of these proteins, and work in yeast identified Vam6 as the major guanine nucleotide exchange factor for Gtr1. In contrast to Rheb, Rag heterodimers do not appear to activate TOR directly; rather, Sabatini and coworkers demonstrated that amino acids and Rag proteins facilitate TOR-Rheb interaction, by mediating the intracellular trafficking of TOR to a Rheb-containing late endocytic compartment. This aspect of Rag/Gtr function may differ between yeast and mammalian cells, as GFP fusions to Gtr1, Vam6 and TOR1 co-localize to the vacuolar membrane and endocytic vesicles under both growth and starvation conditions [14*]. In addition, a metazoan-specific factor, the exocyst regulator RalA (yet another small GTPase), has been implicated in nutrient signaling to TOR downstream of Rheb in HeLa cells [15]. Nonetheless, together these findings indicate an unexpectedly high degree of conservation in nutrient signaling mechanisms, and underscore the importance of understanding how amino acid levels influence the activity of the Rag GTPases.
A number of other signaling molecules have also been shown to play a role in amino acid-dependent TOR activation. Gulati and colleagues found that amino acids induce an inward flux of Ca2+ in HeLa cells, leading to a calmodulin-dependent activation of the class III PI(3)K hVps34 [16]. This molecule has previously been reported to promote amino acid signaling to TOR, and has well established functions in the endocytic pathway, consistent with a potential involvement of Vps34 in the endocytic localization of Rheb or TOR. Interestingly, this function does not appear to be conserved in lower eukaryotes [17]. In other settings, increased cytosolic Ca2+ has been shown instead to suppress TOR signaling and induce autophagy, in a pathway involving Ca2+/calmodulin-dependent protein kinase-kinase and AMPK [18]. Finally, the Ste20 kinase ortholog MAP4K3 was recently shown to be required for activation of TOR in response to amino acids in Drosophila S2 cells [19]. Activity of this protein kinase is also sensitive to amino acid levels.
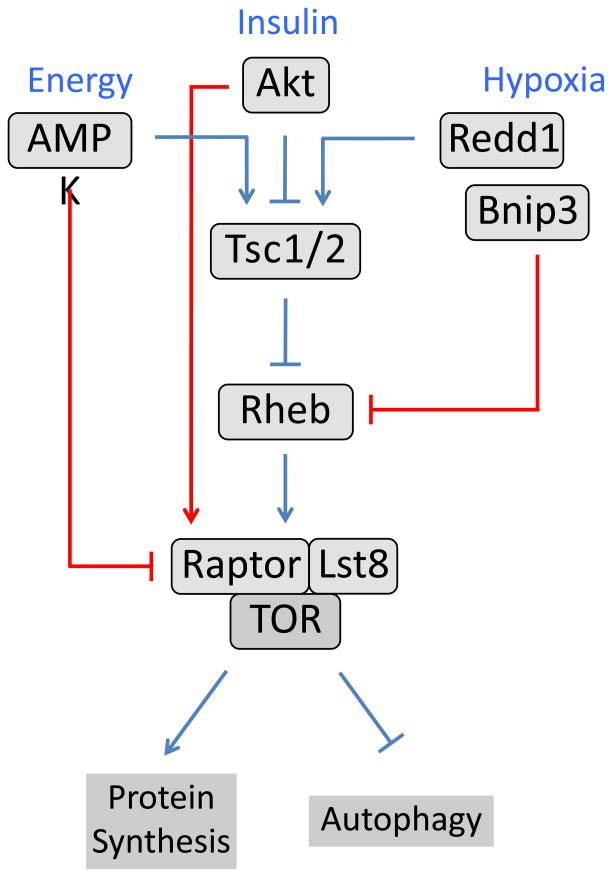
In addition to these new mechanisms of nutrient signaling, it has become clear that factors such AMPK and Akt can regulate TORC1 more directly than previously appreciated, through additional TSC-independent mechanisms (Figure 1). For example, AMPK can directly phosphorylate the TORC1 component Raptor, generating a docking site for the inhibitory 14-3-3 protein [20*]. In a parallel fashion, Akt can phosphorylate PRAS40, a Raptor binding protein that also acts as an inhibitor of TORC1. Akt-mediated phosphorylation of PRAS40 again promotes 14-3-3 binding, in this case leading to relief from PRAS40-mediated inhibition [21–24]. Thus Raptor is emerging as an important integration site for TORC1 regulation, in addition to its roles in substrate binding. Indeed, Raptor itself has been shown to act as a substrate for TOR in response to growth factors and amino acids, and phosphorylation of Raptor at S863 was found to be critical for full TORC1 activity [25,26]. Finally, inhibition of TOR signaling by hypoxia also signals through TSC-dependent and -independent routes, the latter through the hypoxia-inducible BH3 protein Bnip3, which can directly bind and inhibit GTP loading of Rheb [27].
Figure 1. Mechanisms of signaling redundancy in the TOR pathway.
Multiple inputs regulate activity of the TOR/Raptor/Lst8 complex (TORC1) through redundant upstream signaling pathways. Interactions leading to activation of a downstream component are indicated by arrows; those that inhibit are indicated by a bar. The canonical Tsc1/Tsc2-dependent TOR pathway is indicated by the blue lines. Recently described Tsc1/Tsc2-independent signals are shown in red. Decreases in cellular energy level (ATP/AMP ratio) cause phosphorylation of Raptor by the AMP-dependent protein kinase, leading to 14-3-3 mediated inhibition of TORC1 [20]. In response to insulin and other growth factors, Akt phosphorylates and inactivates PRAS40 (not shown), an inhibitor of TORC1 [21–24]. Decreases in cellular oxygen levels lead to increased expression of hypoxia-induced genes, including Bnip3, which binds and inhibits the small GTPase Rheb [27].
Rheb has also recently been shown to have both direct and indirect effects on TORC1 signaling. Sato and colleagues found that the direct, binding-dependent effect of Rheb on TOR does not act on TOR’s intrinsic kinase activity; instead, Rheb stimulates TORC1 signaling by promoting interaction with its substrates [28]. An indirect effect of Rheb on TORC1 signaling involves Rheb-mediated, TOR-independent activation of phospholipase-D1 (PLD1). PLD1 and its product phosphatidic acid were shown to be necessary and sufficient for TOR activation downstream of Rheb [29]. Altogether, these findings indicate that TOR’s upstream regulators have evolved multiple, redundant and integrated mechanisms of control over TORC1 activity.
REGULATION OF AUTOPHAGY DOWNSTREAM OF TOR
Atg1 kinase complex
In yeast, autophagosomes generally arise from a single locus in the cell, known as the preautophagosomal structure or phagophore assembly site (PAS). This structure also serves as a nucleation point for the cytoplasm-to-vacuole (CVT) pathway, a selective form of autophagy that serves a biosynthetic role under vegetative conditions to deliver select hydrolases to the vacuole. The observation that most Atg proteins localize to this structure has facilitated order-of-action experiments, in which mutations in Atg genes are tested for effects on PAS localization of each Atg protein. Such studies have led to the elucidation of a hierarchical framework of Atg interactions [30].
One of the earliest and most important steps leading to autophagy induction downstream of TOR is mediated by a protein complex containing the Ser/Thr kinase Atg1 and two scaffolding proteins, Atg13 and Atg17 [31–33]. This complex is required for the PAS localization of nearly all other Atg proteins, placing it atop the Atg/PAS hierarchy. Moreover, assembly and activity of the yeast Atg1/13/17 complex is intimately connected to TOR signaling. Under growth conditions, TOR-dependent phosphorylation of Atg13 prevents complex assembly. Nutrient depletion or rapamycin treatment results in rapid dephosphorylation of Atg13, assembly of the complex, increase in Atg1 kinase activity and induction of autophagy [33]. As Atg1 associates both with proteins such as Atg17 that are specifically required for non-selective autophagy, as well as with CVT-specific proteins such as Atg11, it is thought that Atg1 may act as a switch between these different types of autophagy.
How does Atg1 regulate the recruitment of downstream Atg components to the PAS? Surprisingly, the kinase activity of Atg1 is not required for the PAS recruitment of most Atg proteins. Rather, the Atg1 protein itself appears to play a structural role in this process [34**–36]. In yeast cells carrying kinase-defective Atg1 mutations, other Atg proteins actually show a higher level of PAS association, implicating a role for Atg1 kinase activity in release or disassembly of these components from the PAS. In addition, although some autophagosomes are able to form in these mutants, they are significantly smaller than in wild type, suggesting that Atg1 kinase activity affects the rate at which the autophagosomal membrane expands [35**]. It is notable that recruitment of Atg proteins to the PAS by kinase-defective Atg1 is stimulated by starvation [34**], suggesting that TOR signaling regulates aspects of Atg1 function in addition to its kinase activity.
Further insight into Atg1 function may be found in the observation that PAS localization of the peripheral membrane protein Atg2 does require Atg1 kinase activity [34**]. Atg2 forms a complex with two other Atg proteins: the PI(3)P-binding protein Atg18, which has been identified as an in vitro substrate of Atg1 [37], and the integral membrane protein Atg9, which may track the source of autophagic membrane. PAS localization of Atg2 and Atg18 is required for Atg9 to dissociate efficiently from the PAS [36]. Atg18 is recruited to growing autophagic membranes through its interaction with PI(3)P, which accumulates on the inner surface and growing edges of autophagosomes [38] through the activity of Atg6/Vps34 PI(3)-kinase complexes. Thus, the kinase activities of Atg1 and Vps34 appear to converge on an Atg2/18/9 complex, potentially playing an important role in delivering membrane to autophagosomes as they grow from their sites of nucleation.
Our understanding of the role and regulation of Atg1 complexes in mammalian cells has not yet reached a level of precision comparable to that of the yeast system. Nonetheless, orthologs of Atg1 (Ulk1/2), Atg13 and Atg17 (FIP200) have recently been described in a variety of multicellular organisms including mouse, Drosophila, and C. elegans ([39**–44**]. These factors have been shown to form a multi-protein complex required for autophagy induction downstream of TOR, analogous to the yeast Atg1 complex. Despite this functional conservation across a wide evolutionary timescale, these proteins display some intriguing regulatory differences from the yeast system. For example, the metazoan Atg1/Ulk1 complex is generally stable under both fed and starved conditions, and hyperphosphorylation of Atg13 does not lead to complex disassembly. Also in contrast to yeast, metazoan Atg13 phosphorylation is not dramatically higher under non-autophagic conditions, and in some systems is actually induced by starvation, in an Atg1/Ulk1-dependent manner. TOR signaling and phosphorylation also has a significant effect on the stability of Atg1/Ulk1 and Atg13 protein in these systems [44**,45]. In addition, metazoan Atg1/Ulk1 complexes have been found to include factors with no obvious yeast counterparts, such as Atg101, which binds to and stabilizes Atg13 [46], [47]. Finally, direct physical interaction has been demonstrated between TORC1 and Atg1/Ulk1 complexes in mammalian and Drosophila cells. Proteomic-based studies have begun to identify sites of phosphorylation on these components [45], and further such analyses of TOR-dependent sites and identification of Atg1/Ulk1 substrates will be important to define mechanisms by which phosphorylation regulates signaling through this complex.
Overexpression of wild type Atg1 leads to high levels of autophagy in fed Drosophila cells, indicating that Atg1 activity promotes not only the initial steps of autophagy induction, but can be sufficient to drive a full autophagic response in this system [48*]. One or more components of a Vps34/Atg14L/Beclin1 complex may represent an additional target of Atg1-dependent regulation, as activation of Vps34 at sites of autophagosome formation fails to occur in Drosophila cells mutant for Atg1 [48*]. In Dictyostelium, loss of Atg1 does not cause a block in autophagy induction, but instead leads to an accumulation of defective autophagosomes [49], indicating that in this system the role of Atg1 in autophagosome maturation predominates over its role in induction. This function of Atg1 in later steps of autophagy may include effects on the endocytic pathway, which is required for autophagosome maturation in metazoan cells. Atg1/Ulk1 has been shown to associate with and regulate a number of factors involved in endocytic and vesicular trafficking including Syntenin, SynGAP, Unc-76 and Synaptotagmin [50,51]. Whether these or related factors have a role in autophagy remains to be explored. Finally, Atg1/Ulk1 was recently shown to be required for a novel, alternative form of autophagy that is independent of Atg5 and Atg7 (and, by inference, independent of the group of autophagy genes that act downstream of these regulators) [52], pointing to the likely relevance of additional Atg1 functions beyond recruitment of these factors to the autophagosome.
Other pathways downstream of TOR
In yeast, two major signaling branches downstream of TOR have been identified, one regulated by the AGC kinase Sch9, which functions analogously to the mammalian TORC1 substrate S6K, and a second branch mediated by Tap42, a regulator of protein phosphatases of the PP2A family. Recent studies have shown that both of these factors play important roles in autophagy regulation, but that whereas Sch9-mediated autophagy is independent of TOR, the Tap42 branch mediates TOR signaling to Atg1/13.
A clear understanding of the role of Tap42 in autophagy, and in TOR signaling generally, has been hindered by disparate results amongst the different studies characterizing this protein, due perhaps to the nature of the tap42 alleles analyzed. Originally identified through a temperature-sensitive mutation that confers rapamycin resistance to yeast cells, Tap42 was shown to bind PP2A and the PP2A-related phosphatase Sit4 in a TOR-dependent manner, and genetic data indicated that Tap42 promotes the growth-related function of these phosphatases [53]. In contrast, later studies found that Sit4 antagonizes TOR-and Tap42-dependent functions, such as phosphorylation of the GATA transcription factor Gln3 and the kinase Npr1 [54,55]. Although mutation of Tap42 was initially reported to have no effect on autophagy induction [33], a subsequent study showed that disruption of Sit4 blunts the induction of autophagy in response to rapamycin [56]. Recently, the role of these factors in autophagy has been comprehensively reexamined using multiple Tap42 alleles and more quantitative autophagy measurements. Yorimitsu and co-workers found that Tap42 and PP2A play an essential role in suppressing autophagy under growth conditions [57]. Autophagy was shown to be induced by loss of PP2A or Tap42 and suppressed by PP2A overexpression; these effects were Atg1-dependent and were not observed with Sit4. These results are consistent with a recent large-scale proteomic analysis of TOR-dependent protein phosphorylation, which identified rapamycin-sensitive phosphorylation sites on Atg13 as being dependent on Tap42 [58].
At present, evidence of a role for Tap42 in autophagy regulation is lacking in higher eukaryotes, and the potential role of this factor in mTOR signaling in general is poorly characterized. Disruption of Tap42 in Drosophila does not result in defects in cell growth or S6K phosphorylation characteristic of TOR inactivation, and does not cause induction of autophagy [59]. The mammalian Tap42 ortholog, known as alpha4, interacts with PP2A and related phosphatases, but this interaction is not sensitive to rapamycin or nutrient status [60,61]. However, a complex containing mTOR, alph4, PP2A and the STAT1 transcription factor was recently described in human cells, and it was demonstrated that depletion of mTOR, alpha4 or PP2A led to nuclear accumulation of STAT1 and induction of STAT1-responsive genes [62]. This is the first evidence indicating that these proteins can function together to regulate at least a subset of TOR’s effects in mammalian cells.
Other known outputs of TOR signaling are also likely to contribute to autophagy regulation. Induction of autophagy in response to starvation is associated with increased expression of number of autophagy-related genes, most strikingly Atg8, whose product may otherwise become limiting due to its degradation in the autolysosome. TOR signaling contributes to these transcriptional effects, as a similar upregulation of Atg genes is observed in cells treated with rapamycin ([63–65]. The transcription factors that mediate these effects have been identified in only a few cases; for example, the TOR-responsive transcriptional activator Gln3 has been shown to directly induce Atg14 expression in response to rapamycin [66]. In mammalian cells, the P53 family member P73 appears to play a role in autophagy related gene activation downstream of TOR [67]. It is unclear whether these TOR-dependent effects on Atg gene expression are a limiting step in autophagy induction, or whether these transcriptional effects of TOR play more of a supporting role.
Finally, TOR signaling has a variety of well-established effects on endocytic uptake and trafficking [68,69], and through these functions is likely to influence the efficiency of autophagosome maturation and fusion with the endo-lysosomal compartment. Recent studies in mammalian and yeast cells suggest these effects of TOR may be more direct than previously appreciated. Biochemical fractionation and in vivo localization approaches have shown that TOR is closely associated with several distinct membrane compartments, including a novel vesicular fraction consisting of a distinct form of detergent-resistant membrane [12*,70*,71*]. Proteomic analyses of these membrane fractions identified a number of endocytic regulators, several of which were shown by genetic analysis to function with TOR. As described above, Rheb is also closely associated with endocytic membranes and may influence endocytic trafficking through TOR-dependent and -independent mechanisms [72]. Thus, in addition to providing a platform for TOR regulation, localization of TOR and associated factors to vesicular compartments likely reflect an intimate involvement of this pathway in endocytic functions.
FEEDBACK IN TOR-MEDIATED AUTOPHAGY REGULATION
Levels of autophagy either above or below a normal range have been found to be detrimental at both a cell autonomous and organismal level [73,74]. Moreover, the rate of autophagy most beneficial to the cell or organism can vary with environmental conditions in a dynamic way. Thus, nutrient withdrawal will often induce a burst of autophagy initially, followed by a lower sustained rate during longer periods of starvation. Several mechanisms contribute to these kinetics of autophagy induction and maintenance, and are critical to keeping the levels of autophagy within a viable range.
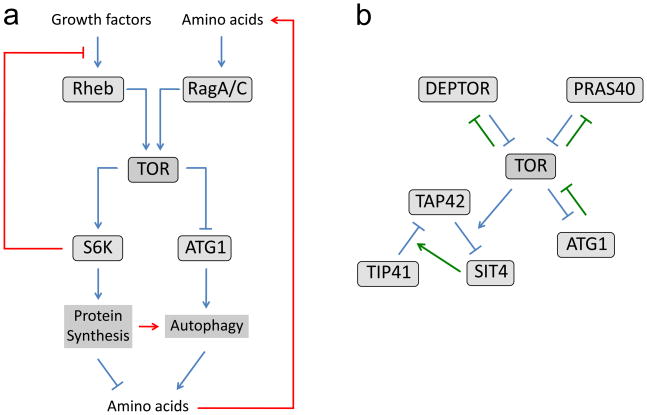
Like many biochemical reactions, autophagy is suppressed by its end products: the rise in intracellular amino acids due to autophagic proteolysis and efflux of amino acids from the lysosome results in up-regulation of TOR signaling, and thus down-regulation of autophagy (Figure 2a). For example, cell types such as hepatocytes with high autophagic capacity are able to partially maintain TOR activity under starvation conditions, in an autophagy-dependent manner [75]. In other cell types, TOR activity can be sustained in the absence of external amino acids by protein synthesis inhibitors, which cause an increase in intracellular amino acid levels; this effect, too, is dependent on autophagy [76]. Importantly, perturbation of this negative feedback loop can result in an overcompensation response that ultimately harms the cell. Ramachandran and coworkers recently showed that defective assembly of the V-ATPase proton pump is the underlying cause of XMEA, an autophagy-associated myopathic disease [77*]. Mutations in the V-ATPase assembly chaperone VMA21 caused increased lysosomal pH and defective autophagic degradation. The ensuing reduction in free intracellular amino acids caused diminished TOR activity, leading to increased but ultimately ineffective induction of autophagy; autophagosomes were able to form, but were unable to properly mature and instead caused extensive cell vacuolation.
Figure 2. Mechanisms of feedback signaling in the TOR pathway.
(a) Negative feedback mechanisms that limit activation of autophagy are shown in red. Protein degradation within the autolysosome raises the intracellular levels of amino acids, leading to activation of TOR through the RagA/RagC small GTPases and other nutrient signaling pathways. TOR activity is self-limited through an S6K-mediated inhibition of IRS1 (not shown), an essential component in the insulin/PI3K/Akt pathway upstream of Rheb. S6K is also involved in a self-limiting pathway in autophagy induction downstream of TOR, probably through its function in protein synthesis. Inhibition of TOR leads to both activation of Atg1 and inactivation of S6K; both of these kinases play positive roles in starvation-induced autophagy. (b) Feed-forward mechanisms in TOR signaling, indicated by green lines, may lead to amplification and stabilization of initially modest changes in TOR activity. DEPTOR, PRAS40 and Atg1 are each inhibited by TOR mediated phosphorylation and in turn inhibit TOR activity. In yeast, TOR mediates activation of Tap42, which leads to downregulation of the phosphatase Sit4 and decreased activity of the Tap42 inhibitor Tip41, resulting in further activation of Tap42.
Additional homeostatic effects on TOR signaling result from a number of negative feedback signaling loops intrinsic to the wiring of the TOR pathway. In yeast, TOR promotes expression of ribosomal biogenesis genes through phosphorylation and activation of the transcription factor Sfp1; disruption of this factor leads to increased TOR activity as measured by phosphorylation of Sch9 [78*]. The best-characterized negative loop in the mammalian TOR pathway involves downregulation of PI3K/AKT signaling in response to activation of TOR. This is mediated in part by S6K-dependent reduction in the levels and activity of IRS-1, an essential bridge between the insulin receptor and PI3K [79,80]. S6K is also involved in negative feedback in FGF-and PDGF-mediated signaling, as it is both activated by these factors and can inhibit their signaling effects [81,82]. Similarly, activation of targets downstream of S6K such as PP1gamma has recently been shown to decrease TOR-dependent phosphorylation and activity of S6K through a negative feedback loop [83]. Thus, numerous independent mechanisms collaborate to ensure that activity of this pathway remains within a prescribed range.
S6K also plays a more direct role in limiting autophagy responses. Disruption of S6K through null mutation of dS6K in Drosophila or RNA-mediated interference of S6K1 and S6K2 in human cells has been shown to reduce starvation-induced autophagy, indicating that the function of S6K in autophagy runs counter to the rest of the TOR pathway [84]. Inhibition of TOR signaling thus leads to both activation of autophagy and inactivation of S6K, a positive regulator of autophagy. Differences in the dynamics of autophagy induction vs. S6K suppression in response to nutrient withdrawal may play a role in limiting high levels of autophagy to an initial induction phase, followed by a lower, sustained response under chronic starvation conditions. Indeed, dS6K activity becomes limiting for autophagy under conditions of long term TOR inactivation but not acute starvation [84].
In contrast to these examples of self-limiting feedback in the TOR pathway, a number of self-amplifying feed-forward mechanisms have recently been identified that may be important to promote rapid and discrete changes in TOR activation state (Figure 2b). In yeast, the activity of Tap42 is inhibited by a Tap42 associated factor, Tip41; suppression of TOR activity by rapamycin results in increased Tip41–Tap42 binding and a further decrease in signaling [54]. Similarly, the ability of PRAS40 to bind and inhibit mTOR is antagonized by TOR-mediated phosphorylation of PRAS40 [85*], again leading to an amplification of small changes in TOR activity. TOR signaling is also inhibited by another recently identified mTOR-associated factor known as DEPTOR, and this effect is amplified by TOR-dependent negative regulation of DEPTOR expression [86]. Interestingly, loss of this DEPTOR loop in multiple myeloma cells contributes to their survival by relieving S6K-dependent negative feedback inhibition of PI3KAkt, indicating that these amplifying and restraining effects can interact in complex ways.
Finally, components of the autophagic machinery itself have been shown to have a strong impact on TOR activity. Genetic disruption of Atg1/Ulk1 or Atg13 causes increased phosphorylation of TOR substrates in Drosophila and human cells, suggesting that this target of TOR activity can in turn inhibit TOR [41**,87]. Consistent with this idea, overexpression of Atg1 in Drosophila leads to decreased TOR activity and reduced cell growth, effects that are stimulated by co-expression of Atg13 [44**,48*]. Atg1 was shown to cause a redistribution of TOR to a punctate perinuclear localization, consistent a trafficking-mediated mechanism. Alternatively, the Ulk1 interacting protein FIP200 has previously been shown to bind and inhibit the TSC1/TSC2 complex [88]; changes in Atg1/Ulk1 levels or activity may affect levels of FIP200 available for this interaction. The interaction between Ulk1 and raptor noted above is also consistent with a direct effect of Atg1/Ulk1 on TORC1 activity. Regardless of mechanism, initially small increases in the activity or abundance of Atg1 complexes would be expected to promote their further activation by antagonizing TOR-mediated inhibition. Altogether, these self-amplifying effects may allow cells to switch between relatively stable states of autophagic activity, and may provide a counter force to the inertial effects of negative feedback.
AUTOPHAGY REGULATION: TOR AND BEYOND
Numerous studies over the past few years have found that autophagy can be influenced by a remarkable number of genes, pathways, cellular perturbations and chemical compounds. Given the central role of TOR signaling in autophagy control, many of these studies have examined whether changes in TOR activity can account for the effects of these factors. Two general themes are emerging from such investigations. First, studies of autophagy from a broad array of perspectives is leading to the discovery a number of new and sometimes unexpected inputs to TOR. Second, studies of factors that regulate autophagy independent of TOR are helping to distinguish between molecular events that are critical for this process, versus steps that can be substituted for or bypassed altogether.
A number of new TOR regulators have been identified through cell-based screens for small molecules that induce autophagy. Balgi and colleagues [89] identified four such compounds, each of which cause specific, reversible inhibition of TORC1. These drugs act with distinct kinetics and appear to intersect with the TOR pathway at different points upstream. Several natural products also appear to induce autophagy through inhibition of TOR signaling, including caffeine [90], resveratrol [91], cannabinoids [92], and curcumin [93]. The sphingolipids ceramide and sphingosine 1-phosphate (S1P) have also been shown to induce autophagy, and both cause a reduction in TOR activity, apparently through distinct mechanisms [94,95]. The effects of TOR on autophagy may also impact the survival of oncogene-transformed tumor cells, as the SV40 small T antigen leads to autophagy induction through PP2A-dependent activation of AMPK upstream of TOR [96]. Autophagy promotes survival of these cells under conditions of glucose deprivation, mimicking the starvation state of pre-angiogenic tumors; thus TOR may have tumor-suppressive effects at this stage of tumorigenesis. The tumor suppressor p53 has also been found to control cell survival in part through effects on autophagy. Interestingly, p53 can act both as an inducer of autophagy through transcriptional effects in the nucleus [97], and as an inhibitor of autophagy through a novel cytosolic function of p53 [98**]. Thus, both activation and loss of p53 lead to autophagy induction, and both cases are associated with activation of AMPK and inhibition of TOR. Altogether, these studies are showing that TOR activity is responsive to a wider range of signals than previously appreciated. Several of these factors seem to play an important role in the normal physiological regulation of autophagy by TOR. For example, starvation leads to activation of sphingosine kinase and proteosomemediated degradation of p53, and both of these events are required for starvation-induced autophagy [94,98**]. Finally, a number of these factors have been shown to have a strong influence on lifespan, and recent evidence suggests that this is mediated in part through effects on TOR signaling and autophagy [99,100].
TOR activity is both necessary and sufficient to suppress autophagy in a wide range of cell types and contexts. Signals that induce autophagy without inactivating TOR therefore pose a challenge and opportunity to understanding the key steps required to initiate autophagosome formation, and different mechanisms by which those steps can be regulated. In yeast, PKA has been identified as an important regulator of autophagy in response to glucose levels: activation of PKA blocks induction of autophagy in starved cells, and loss of PKA activity can cause autophagy induction under growth conditions. The effects of TOR and PKA on autophagy appear to be independent, as inhibition of one pathway does not affect activity of the other [101*–103]. Interestingly, however, these pathways converge on similar targets: as in the TOR pathway, PKA signaling has significant effects on the phosphorylation and activity of the Atg1/Atg13 complex. PKA and TOR appear to regulate distinct sites of phosphorylation on Atg1 and Atg13, and to affect these proteins in different ways. Whereas TOR controls interaction between Atg1 and Atg13, PKA was found to affect localization of these proteins to the PAS [102**,104]. These results suggest that autophagy can be induced by promoting either interaction or localization of Atg1 and Atg13, but that both may not be necessary. Consistent with this interpretation, inactivation of both TOR and PKA signaling leads to a more efficient induction of autophagy than inhibition of either pathway alone [101,102**].
In human cells, it is unclear whether additional pathways similarly impinge on Atg1/Ulk1, although the roles of this kinase in non-autophagic processes such as axon outgrowth and cytoskeletal regulation are consistent with regulation by alternative signals. However, several examples of TOR-independent autophagy have been described. In separate screens carried out by the Rubinsztein and Yuan groups for small molecule regulators of autophagy, a large number of compounds that activate autophagy independent of TOR were identified [105*,106*]. Williams and coworkers showed that several of these molecules affect a cAMP-, IP3-and Ca2+-mediated cycle [106*], analogous to the feed-forward mechanisms affecting TOR signaling. Although how these factors regulate the core autophagy machinery has not yet been determined, the IP3 receptor was recently shown to directly bind and regulate Beclin 1 [107]. As in yeast, the effects of these TOR-independent and TOR-dependent pathways are additive (reviewed in [108]).
Another TOR-independent pathway of autophagy control is regulated by the FOXO3 transcription factor, which acts in parallel to TOR downstream of Akt [109,110]. In skeletal muscle, activation of FOXO leads to induction of autophagy through increased expression of a number of autophagy related genes. A key autophagic target of FOXO3 is Bnip3, a member of the BH3-only family of proteins which have been shown to promote autophagy by relieving the inhibition of Beclin 1 by Bcl-2 [111]. The transcriptional pathway mediated by FOXO3 appears to play an especially important role in skeletal muscle, as inactivation of TOR signaling by rapamycin or mTOR shRNA does not induce autophagy in these cells [110]. Interestingly, it was previously reported that TOR-independent autophagy is induced in cultured myotubes starved for leucine, normally an essential activator of TOR [112]. These results suggest that non-canonical functions and regulation of TOR may have evolved in skeletal muscle to meet the unique metabolic demands of this tissue, although it should be noted that similar TOR-independent effects of amino acids on autophagy have also been described in other cell types [113].
Together these findings point to Beclin 1/Vps34 complexes as potential common targets of multiple TOR-independent autophagy signals. As discussed above, TOR-and Atg1-mediated signals also appear to act upstream of this complex. Co-overexpression of components of this complex can be sufficient to induce autophagy [17], and both nutrient and ER-stress signals appear to regulate the activity of Beclin 1/Vps34 in part through a Jun-kinase dependent pathway [114]. Activation of Vps34 at discrete regions of the endoplasmic reticulum has recently been suggested to dictate the location of assembly sites for autophagosome formation [115].
CONCLUSIONS
One set of questions that emerges from these recent studies concerns why cells have evolved multiple pathways to control autophagy. Part of the answer may lie in the pleiotropic nature of TOR’s many functions. Under some conditions, it may be critical to maintain biosynthesis or other TOR-dependent activities even as a cell activates autophagy. A range of different signals may also allow induction of qualitatively different types of autophagy, with different classes of substrates or cellular regions becoming targeted depending on the inducing signal. Identification of the pathways targeted by different inducing compounds, and analysis of the physiological conditions under which these pathways are utilized, will help to clarify this issue.
For autophagy to become a more useful therapeutic tool, it will be critical to better understand the specific steps affected by different components of both TOR-dependent and -independent autophagy pathways, as conditions that may benefit from increased autophagy may actually worsen if autophagy is induced unsuccessfully [77*]. Looking ahead, there are still significant gaps in our understanding of how the recently identified nutrient-dependent inputs to TOR – Vps34, Rags, MAP4K3, RalA, and likely others – are integrated and coordinated with one another, and how these factors themselves are regulated by nutrients. Downstream of TOR, further studies are required to determine how the Atg1/Atg13 complex is regulated by TOR, the mechanistic role this complex plays in autophagosome formation, and how TOR and Atg1/Atg13 interact with Beclin 1/Vps34 complexes. Together, this information will provide a comprehensive picture of the steps leading from changes in nutrient levels to the activation of autophagosome formation, and should lead toward new therapies based on precise control and targeting of autophagy.
Acknowledgments
Supported by National Institutes of Health grant RO1 GM62509.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.He C, Klionsky DJ. Regulation Mechanisms and Signaling Pathways of Autophagy. Annu Rev Genet. 2009 doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhaskar PT, Hay N. The two TORCs and Akt. Dev Cell. 2007;12:487–502. doi: 10.1016/j.devcel.2007.03.020. [DOI] [PubMed] [Google Scholar]
- 4.Avruch J, Long X, Ortiz-Vega S, Rapley J, Papageorgiou A, Dai N. Amino acid regulation of TOR complex 1. Am J Physiol Endocrinol Metab. 2009;296:E592–602. doi: 10.1152/ajpendo.90645.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith EM, Finn SG, Tee AR, Browne GJ, Proud CG. The tuberous sclerosis protein TSC2 is not required for the regulation of the mammalian target of rapamycin by amino acids and certain cellular stresses. J Biol Chem. 2005;280:18717–18727. doi: 10.1074/jbc.M414499200. [DOI] [PubMed] [Google Scholar]
- 6.Dong J, Pan D. Tsc2 is not a critical target of Akt during normal Drosophila development. Genes Dev. 2004;18:2479–2484. doi: 10.1101/gad.1240504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schleich S, Teleman AA. Akt phosphorylates both Tsc1 and Tsc2 in Drosophila, but neither phosphorylation is required for normal animal growth. PLoS One. 2009;4:e6305. doi: 10.1371/journal.pone.0006305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hundal HS, Taylor PM. Amino acid transceptors: gate keepers of nutrient exchange and regulators of nutrient signaling. Am J Physiol Endocrinol Metab. 2009;296:E603–613. doi: 10.1152/ajpendo.91002.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goberdhan DC, Meredith D, Boyd CA, Wilson C. PAT-related amino acid transporters regulate growth via a novel mechanism that does not require bulk transport of amino acids. Development. 2005;132:2365–2375. doi: 10.1242/dev.01821. [DOI] [PubMed] [Google Scholar]
- 10.Donaton MC, Holsbeeks I, Lagatie O, Van Zeebroeck G, Crauwels M, Winderickx J, Thevelein JM. The Gap1 general amino acid permease acts as an amino acid sensor for activation of protein kinase A targets in the yeast Saccharomyces cerevisiae. Mol Microbiol. 2003;50:911–929. doi: 10.1046/j.1365-2958.2003.03732.x. [DOI] [PubMed] [Google Scholar]
- 11.Dubouloz F, Deloche O, Wanke V, Cameroni E, De Virgilio C. The TOR and EGO protein complexes orchestrate microautophagy in yeast. Mol Cell. 2005;19:15–26. doi: 10.1016/j.molcel.2005.05.020. [DOI] [PubMed] [Google Scholar]
- 12*.Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. identify the GTR/Rag family of small GTPases as key mediators of amino acid signaling to TOR in yeast, Drosophila and mammalian cells. The yeast guanine nucleotide exchange factor Vam6 acts as a positive regulator of this complex. In mammalian cells, Rag GTPases promote redistribution of TOR to Rheb-positive endosomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13*.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10:935–945. doi: 10.1038/ncb1753. identify the GTR/Rag family of small GTPases as key mediators of amino acid signaling to TOR in yeast, Drosophila and mammalian cells. The yeast guanine nucleotide exchange factor Vam6 acts as a positive regulator of this complex. In mammalian cells, Rag GTPases promote redistribution of TOR to Rheb-positive endosomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14*.Binda M, Peli-Gulli MP, Bonfils G, Panchaud N, Urban J, Sturgill TW, Loewith R, De Virgilio C. The Vam6 GEF controls TORC1 by activating the EGO complex. Mol Cell. 2009;35:563–573. doi: 10.1016/j.molcel.2009.06.033. identify the GTR/Rag family of small GTPases as key mediators of amino acid signaling to TOR in yeast, Drosophila and mammalian cells. The yeast guanine nucleotide exchange factor Vam6 acts as a positive regulator of this complex. In mammalian cells, Rag GTPases promote redistribution of TOR to Rheb-positive endosomes. [DOI] [PubMed] [Google Scholar]
- 15.Maehama T, Tanaka M, Nishina H, Murakami M, Kanaho Y, Hanada K. RalA functions as an indispensable signal mediator for the nutrient-sensing system. J Biol Chem. 2008;283:35053–35059. doi: 10.1074/jbc.M805822200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gulati P, Gaspers LD, Dann SG, Joaquin M, Nobukuni T, Natt F, Kozma SC, Thomas AP, Thomas G. Amino acids activate mTOR complex 1 via Ca2+/CaM signaling to hVps34. Cell Metab. 2008;7:456–465. doi: 10.1016/j.cmet.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Juhász G, Hill JH, Yang Y, Sass M, Baehrecke EH, Backer JM, Neufeld TP. The class III PI(3)K Vps34 promotes autophagy and endocytosis but not TOR signaling in Drosophila. J Cell Biol. 2008;181:655–666. doi: 10.1083/jcb.200712051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoyer-Hansen M, Bastholm L, Szyniarowski P, Campanella M, Szabadkai G, Farkas T, Bianchi K, Fehrenbacher N, Elling F, Rizzuto R, et al. Control of macroautophagy by calcium, calmodulin-dependent kinase kinase-beta, and Bcl-2. Mol Cell. 2007;25:193–205. doi: 10.1016/j.molcel.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 19.Findlay GM, Yan L, Procter J, Mieulet V, Lamb RF. A MAP4 kinase related to Ste20 is a nutrient-sensitive regulator of mTOR signalling. Biochem J. 2007;403:13–20. doi: 10.1042/BJ20061881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20*.Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. Using a proteomic approach, the authors identify the TORC1 component Raptor as a direct target of AMPK. Phosphorylation of Raptor by AMPK mediates a TSC2-independent signal essential for inhibiton of TORC1 by energy stress. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oshiro N, Takahashi R, Yoshino K, Tanimura K, Nakashima A, Eguchi S, Miyamoto T, Hara K, Takehana K, Avruch J, et al. The proline-rich Akt substrate of 40 kDa (PRAS40) is a physiological substrate of mammalian target of rapamycin complex 1. J Biol Chem. 2007;282:20329–20339. doi: 10.1074/jbc.M702636200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vander Haar E, Lee SI, Bandhakavi S, Griffin TJ, Kim DH. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat Cell Biol. 2007;9:316–323. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- 23.Thedieck K, Polak P, Kim ML, Molle KD, Cohen A, Jeno P, Arrieumerlou C, Hall MN. PRAS40 and PRR5-like protein are new mTOR interactors that regulate apoptosis. PLoS One. 2007;2:e1217. doi: 10.1371/journal.pone.0001217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L, Harris TE, Roth RA, Lawrence JC., Jr PRAS40 regulates mTORC1 kinase activity by functioning as a direct inhibitor of substrate binding. J Biol Chem. 2007;282:20036–20044. doi: 10.1074/jbc.M702376200. [DOI] [PubMed] [Google Scholar]
- 25.Foster KG, Acosta-Jaquez HA, Romeo Y, Ekim B, Soliman GA, Carriere A, Roux PP, Ballif BA, Fingar DC. Regulation of mTOR complex 1 (mTORC1) by raptor S863 and multi-site phosphorylation. J Biol Chem. 2009 doi: 10.1074/jbc.M109.029637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang L, Lawrence JC, Jr, Sturgill TW, Harris TE. Mammalian target of rapamycin complex 1 (mTORC1) activity is associated with phosphorylation of raptor by mTOR. J Biol Chem. 2009;284:14693–14697. doi: 10.1074/jbc.C109.002907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y, Wang Y, Kim E, Beemiller P, Wang CY, Swanson J, You M, Guan KL. Bnip3 mediates the hypoxia-induced inhibition on mammalian target of rapamycin by interacting with Rheb. J Biol Chem. 2007;282:35803–35813. doi: 10.1074/jbc.M705231200. [DOI] [PubMed] [Google Scholar]
- 28.Sato T, Nakashima A, Guo L, Tamanoi F. Specific activation of mTORC1 by Rheb G-protein in vitro involves enhanced recruitment of its substrate protein. J Biol Chem. 2009;284:12783–12791. doi: 10.1074/jbc.M809207200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun Y, Fang Y, Yoon MS, Zhang C, Roccio M, Zwartkruis FJ, Armstrong M, Brown HA, Chen J. Phospholipase D1 is an effector of Rheb in the mTOR pathway. Proc Natl Acad Sci U S A. 2008;105:8286–8291. doi: 10.1073/pnas.0712268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- 31.Cheong H, Yorimitsu T, Reggiori F, Legakis JE, Wang CW, Klionsky DJ. Atg17 regulates the magnitude of the autophagic response. Mol Biol Cell. 2005;16:3438–3453. doi: 10.1091/mbc.E04-10-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kabeya Y, Kamada Y, Baba M, Takikawa H, Sasaki M, Ohsumi Y. Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol Biol Cell. 2005;16:2544–2553. doi: 10.1091/mbc.E04-08-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34**.Kawamata T, Kamada Y, Kabeya Y, Sekito T, Ohsumi Y. Organization of the Pre-autophagosomal Structure Responsible for Autophagosome Formation. Mol Biol Cell. 2008;19:2039–2050. doi: 10.1091/mbc.E07-10-1048. the authors examine starvation-mediated recruitment of Atg proteins to the phagophore assembly site (PAS) in atg11 mutant cells, in which the constitutive autophagy-like CVT pathway is disrupted. Both kinase and non-kinase functions of the Atg1/Atg13 complex are essential for normal flux of proteins through the PAS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35**.Cheong H, Nair U, Geng J, Klionsky DJ. The Atg1 Kinase Complex Is Involved in the Regulation of Protein Recruitment to Initiate Sequestering Vesicle Formation for Nonspecific Autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2008;19:668–681. doi: 10.1091/mbc.E07-08-0826. the authors examine starvation-mediated recruitment of Atg proteins to the phagophore assembly site (PAS) in atg11 mutant cells, in which the constitutive autophagy-like CVT pathway is disrupted. Both kinase and non-kinase functions of the Atg1/Atg13 complex are essential for normal flux of proteins through the PAS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sekito T, Kawamata T, Ichikawa R, Suzuki K, Ohsumi Y. Atg17 recruits Atg9 to organize the pre-autophagosomal structure. Genes Cells. 2009;14:525–538. doi: 10.1111/j.1365-2443.2009.01299.x. [DOI] [PubMed] [Google Scholar]
- 37.Ptacek J, Devgan G, Michaud G, Zhu H, Zhu X, Fasolo J, Guo H, Jona G, Breitkreutz A, Sopko R, et al. Global analysis of protein phosphorylation in yeast. Nature. 2005;438:679–684. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- 38.Obara K, Noda T, Niimi K, Ohsumi Y. Transport of phosphatidylinositol 3-phosphate into the vacuole via autophagic membranes in Saccharomyces cerevisiae. Genes Cells. 2008;13:537–547. doi: 10.1111/j.1365-2443.2008.01188.x. [DOI] [PubMed] [Google Scholar]
- 39**.Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, Mizushima N. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. identify components of the Atg1/Atg13 complex in metazoans, and characterize its interaction with TORC1. In contrast to yeast, this complex is relatively stable under both fed and starved conditions, and TORC1-mediated phosphorylation of Atg13 does not cause complex disassembly. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40**.Ganley IG, Lam du H, Wang J, Ding X, Chen S, Jiang X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284:12297–12305. doi: 10.1074/jbc.M900573200. identify components of the Atg1/Atg13 complex in metazoans, and characterize its interaction with TORC1. In contrast to yeast, this complex is relatively stable under both fed and starved conditions, and TORC1-mediated phosphorylation of Atg13 does not cause complex disassembly. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41**.Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. identify components of the Atg1/Atg13 complex in metazoans, and characterize its interaction with TORC1. In contrast to yeast, this complex is relatively stable under both fed and starved conditions, and TORC1-mediated phosphorylation of Atg13 does not cause complex disassembly. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42**.Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. identify components of the Atg1/Atg13 complex in metazoans, and characterize its interaction with TORC1. In contrast to yeast, this complex is relatively stable under both fed and starved conditions, and TORC1-mediated phosphorylation of Atg13 does not cause complex disassembly. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43*.Tian E, Wang F, Han J, Zhang H. epg-1 functions in autophagy-regulated processes and may encode a highly divergent Atg13 homolog in C. elegans. Autophagy. 2009;5:608–615. doi: 10.4161/auto.5.5.8624. identify components of the Atg1/Atg13 complex in metazoans, and characterize its interaction with TORC1. In contrast to yeast, this complex is relatively stable under both fed and starved conditions, and TORC1-mediated phosphorylation of Atg13 does not cause complex disassembly. [DOI] [PubMed] [Google Scholar]
- 44**.Chang YY, Neufeld TP. An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol Biol Cell. 2009;20:2004–2014. doi: 10.1091/mbc.E08-12-1250. identify components of the Atg1/Atg13 complex in metazoans, and characterize its interaction with TORC1. In contrast to yeast, this complex is relatively stable under both fed and starved conditions, and TORC1-mediated phosphorylation of Atg13 does not cause complex disassembly. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dorsey FC, Rose KL, Coenen S, Prater SM, Cavett V, Cleveland JL, Caldwell-Busby J. Mapping the phosphorylation sites of Ulk1. J Proteome Res. 2009;8:5253–5263. doi: 10.1021/pr900583m. [DOI] [PubMed] [Google Scholar]
- 46.Mercer CA, Kaliappan A, Dennis PB. A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy. Autophagy. 2009:5. doi: 10.4161/auto.5.5.8249. [DOI] [PubMed] [Google Scholar]
- 47.Hosokawa N, Sasaki T, Iemura S, Natsume T, Hara T, Mizushima N. Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy. 2009;5:973–979. doi: 10.4161/auto.5.7.9296. [DOI] [PubMed] [Google Scholar]
- 48*.Scott RC, Juhasz G, Neufeld TP. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr Biol. 2007;17:1–11. doi: 10.1016/j.cub.2006.10.053. This study demonstrates that overexpression of Drosophila Atg1 is sufficient to induce autophagy under fed conditions. Atg1 is shown to cause a downregulation of TORC1 activity, leading to a self-amplifying signaling loop. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tekinay T, Wu MY, Otto GP, Anderson OR, Kessin RH. Function of the Dictyostelium discoideum Atg1 kinase during autophagy and development. Eukaryot Cell. 2006;5:1797–1806. doi: 10.1128/EC.00342-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tomoda T, Kim JH, Zhan C, Hatten ME. Role of Unc51.1 and its binding partners in CNS axon outgrowth. Genes Dev. 2004;18:541–558. doi: 10.1101/gad.1151204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Toda H, Mochizuki H, Flores R, 3rd, Josowitz R, Krasieva TB, Lamorte VJ, Suzuki E, Gindhart JG, Furukubo-Tokunaga K, Tomoda T. UNC-51/ATG1 kinase regulates axonal transport by mediating motor-cargo assembly. Genes Dev. 2008;22:3292–3307. doi: 10.1101/gad.1734608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T, Komatsu M, Otsu K, Tsujimoto Y, Shimizu S. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461:654–658. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- 53.Di Como CJ, Arndt KT. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 1996;10:1904–1916. doi: 10.1101/gad.10.15.1904. [DOI] [PubMed] [Google Scholar]
- 54.Jacinto E, Guo B, Arndt KT, Schmelzle T, Hall MN. TIP41 interacts with TAP42 and negatively regulates the TOR signaling pathway. Mol Cell. 2001;8:1017–1026. doi: 10.1016/s1097-2765(01)00386-0. [DOI] [PubMed] [Google Scholar]
- 55.Beck T, Hall MN. The TOR signalling pathway controls nuclear localization of nutrient- regulated transcription factors. Nature. 1999;402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- 56.Schmelzle T, Beck T, Martin DE, Hall MN. Activation of the RAS/cyclic AMP pathway suppresses a TOR deficiency in yeast. Mol Cell Biol. 2004;24:338–351. doi: 10.1128/MCB.24.1.338-351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yorimitsu T, He C, Wang K, Klionsky DJ. Tap42-associated protein phosphatase type 2A negatively regulates induction of autophagy. Autophagy. 2009;5:616–624. doi: 10.4161/auto.5.5.8091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huber A, Bodenmiller B, Uotila A, Stahl M, Wanka S, Gerrits B, Aebersold R, Loewith R. Characterization of the rapamycin-sensitive phosphoproteome reveals that Sch9 is a central coordinator of protein synthesis. Genes Dev. 2009;23:1929–1943. doi: 10.1101/gad.532109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cygnar KD, Gao X, Pan D, Neufeld TP. The phosphatase subunit tap42 functions independently of target of rapamycin to regulate cell division and survival in Drosophila. Genetics. 2005;170:733–740. doi: 10.1534/genetics.104.039909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nanahoshi M, Nishiuma T, Tsujishita Y, Hara K, Inui S, Sakaguchi N, Yonezawa K. Regulation of protein phosphatase 2A catalytic activity by alpha4 protein and its yeast homolog Tap42. Biochem Biophys Res Commun. 1998;251:520–526. doi: 10.1006/bbrc.1998.9493. [DOI] [PubMed] [Google Scholar]
- 61.Yoo SJ, Jimenez RH, Sanders JA, Boylan JM, Brautigan DL, Gruppuso PA. The alpha4-containing form of protein phosphatase 2A in liver and hepatic cells. J Cell Biochem. 2008;105:290–300. doi: 10.1002/jcb.21830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fielhaber JA, Han YS, Tan J, Xing S, Biggs CM, Joung KB, Kristof AS. Inactivation of mammalian target of rapamycin increases STAT1 nuclear content and transcriptional activity in alpha4- and protein phosphatase 2A-dependent fashion. J Biol Chem. 2009;284:24341–24353. doi: 10.1074/jbc.M109.033530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reinke A, Chen JC, Aronova S, Powers T. Caffeine targets TOR complex I and provides evidence for a regulatory link between the FRB and kinase domains of Tor1p. J Biol Chem. 2006;281:31616–31626. doi: 10.1074/jbc.M603107200. [DOI] [PubMed] [Google Scholar]
- 64.Hardwick JS, Kuruvilla FG, Tong JK, Shamji AF, Schreiber SL. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc Natl Acad Sci U S A. 1999;96:14866–14870. doi: 10.1073/pnas.96.26.14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, Noda T, Ohsumi Y. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol. 1999;147:435–446. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chan TF, Bertram PG, Ai W, Zheng XF. Regulation of APG14 expression by the GATA-type transcription factor Gln3p. J Biol Chem. 2001;276:6463–6467. doi: 10.1074/jbc.M008162200. [DOI] [PubMed] [Google Scholar]
- 67.Rosenbluth JM, Pietenpol JA. mTOR regulates autophagy-associated genes downstream of p73. Autophagy. 2009;5:114–116. doi: 10.4161/auto.5.1.7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hennig KM, Colombani J, Neufeld TP. TOR coordinates bulk and targeted endocytosis in the Drosophila melanogaster fat body to regulate cell growth. J Cell Biol. 2006;173:963–974. doi: 10.1083/jcb.200511140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hackstein H, Taner T, Logar AJ, Thomson AW. Rapamycin inhibits macropinocytosis and mannose receptor-mediated endocytosis by bone marrow-derived dendritic cells. Blood. 2002;100:1084–1087. doi: 10.1182/blood.v100.3.1084. [DOI] [PubMed] [Google Scholar]
- 70*.Aronova S, Wedaman K, Anderson S, Yates J, 3rd, Powers T. Probing the membrane environment of the TOR kinases reveals functional interactions between TORC1, actin, and membrane trafficking in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:2779–2794. doi: 10.1091/mbc.E07-03-0274. use different approaches to examine the localization of TOR proteins in budding yeast. Together these papers support the conclusion that TORC1 is associated with detergent-resistant membranes near the vacuole, and that this complex is involved in membrane trafficking. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71*.Sturgill TW, Cohen A, Diefenbacher M, Trautwein M, Martin DE, Hall MN. TOR1 and TOR2 have distinct locations in live cells. Eukaryot Cell. 2008;7:1819–1830. doi: 10.1128/EC.00088-08. use different approaches to examine the localization of TOR proteins in budding yeast. Together these papers support the conclusion that TORC1 is associated with detergent-resistant membranes near the vacuole, and that this complex is involved in membrane trafficking. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Saito K, Araki Y, Kontani K, Nishina H, Katada T. Novel role of the small GTPase Rheb: its implication in endocytic pathway independent of the activation of mammalian target of rapamycin. J Biochem (Tokyo) 2005;137:423–430. doi: 10.1093/jb/mvi046. [DOI] [PubMed] [Google Scholar]
- 73.Kang C, You YJ, Avery L. Dual roles of autophagy in the survival of Caenorhabditis elegans during starvation. Genes Dev. 2007;21:2161–2171. doi: 10.1101/gad.1573107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Levine B. Cell biology: autophagy and cancer. Nature. 2007;446:745–747. doi: 10.1038/446745a. [DOI] [PubMed] [Google Scholar]
- 75.Shigemitsu K, Tsujishita Y, Hara K, Nanahoshi M, Avruch J, Yonezawa K. Regulation of translational effectors by amino acid and mammalian target of rapamycin signaling pathways. Possible involvement of autophagy in cultured hepatoma cells. J Biol Chem. 1999;274:1058–1065. doi: 10.1074/jbc.274.2.1058. [DOI] [PubMed] [Google Scholar]
- 76.Beugnet A, Tee AR, Taylor PM, Proud CG. Regulation of targets of mTOR signalling by intracellular amino acid availability. Biochem J. 2003 doi: 10.1042/BJ20021266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77*.Ramachandran N, Munteanu I, Wang P, Aubourg P, Rilstone JJ, Israelian N, Naranian T, Paroutis P, Guo R, Ren ZP, et al. VMA21 deficiency causes an autophagic myopathy by compromising V-ATPase activity and lysosomal acidification. Cell. 2009;137:235–246. doi: 10.1016/j.cell.2009.01.054. The authors demonstrate defects in lysosomal acidification are a cause of X-linked myopathy. Defective lysosomal hydrolysis causes a reduction in cellular amino acid concentrations, leading to decreased TOR signaling. This stimulates production of autophagosomes which are unable to undergo lysosomal fusion, leading to cellular vacuolation. [DOI] [PubMed] [Google Scholar]
- 78*.Lempiainen H, Uotila A, Urban J, Dohnal I, Ammerer G, Loewith R, Shore D. Sfp1 interaction with TORC1 and Mrs6 reveals feedback regulation on TOR signaling. Mol Cell. 2009;33:704–716. doi: 10.1016/j.molcel.2009.01.034. This paper identifies a novel feedback loop in the TOR signaling pathway involving Sfp1, a transcriptional activator of ribosomal protein and ribosome biogenesis genes in yeast. Disruption of Sfp1 leads to increased TOR-dependent phosphorylation of Sch9. Although Sfp1 binds directly to TORC1, this feedback mechanism appears to involve an indirect, transcription-dependent signal to TOR. [DOI] [PubMed] [Google Scholar]
- 79.Tremblay F, Marette A. Amino acids and insulin signaling via the mTOR/p70 S6 kinase pathway: A negative feedback mechanism leading to insulin resistance in skeletal muscle cells. J Biol Chem. 2001 doi: 10.1074/jbc.M106703200. [DOI] [PubMed] [Google Scholar]
- 80.Shah OJ, Wang Z, Hunter T. Inappropriate activation of the TSC/Rheb/mTOR/S6K cassette induces IRS1/2 depletion, insulin resistance, and cell survival deficiencies. Curr Biol. 2004;14:1650–1656. doi: 10.1016/j.cub.2004.08.026. [DOI] [PubMed] [Google Scholar]
- 81.Takai S, Hanai Y, Matsushima-Nishiwaki R, Minamitani C, Otsuka T, Tokuda H, Kozawa O. P70 S6 kinase negatively regulates fibroblast growth factor 2-stimulated interleukin-6 synthesis in osteoblasts: function at a point downstream from protein kinase C. J Endocrinol. 2008;197:131–137. doi: 10.1677/JOE-07-0560. [DOI] [PubMed] [Google Scholar]
- 82.Takai S, Tokuda H, Hanai Y, Kozawa O. Limitation by p70 S6 kinase of platelet-derived growth factor-BB-induced interleukin 6 synthesis in osteoblast-like MC3T3-E1 cells. Metabolism. 2007;56:476–483. doi: 10.1016/j.metabol.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 83.Djouder N, Metzler SC, Schmidt A, Wirbelauer C, Gstaiger M, Aebersold R, Hess D, Krek W. S6K1-mediated disassembly of mitochondrial URI/PP1gamma complexes activates a negative feedback program that counters S6K1 survival signaling. Mol Cell. 2007;28:28–40. doi: 10.1016/j.molcel.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 84.Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell. 2004;7:167–178. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 85*.Wang L, Harris TE, Lawrence JC., Jr Regulation of proline-rich Akt substrate of 40 kDa (PRAS40) function by mammalian target of rapamycin complex 1 (mTORC1)-mediated phosphorylation. J Biol Chem. 2008;283:15619–15627. doi: 10.1074/jbc.M800723200. This study identifies TOR-dependent phosphorylation sites in the TORC1 inhibitor PRAS40. As TOR is activated, phosphorylation of these sites leads to 14-3-3-mediated inhibition of PRAS40, resulting in further amplification of TORC1 activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Peterson TR, Laplante M, Thoreen CC, Sancak Y, Kang SA, Kuehl WM, Gray NS, Sabatini DM. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell. 2009;137:873–886. doi: 10.1016/j.cell.2009.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lee SB, Kim S, Lee J, Park J, Lee G, Kim Y, Kim JM, Chung J. ATG1, an autophagy regulator, inhibits cell growth by negatively regulating S6 kinase. EMBO Rep. 2007;8:360–365. doi: 10.1038/sj.embor.7400917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gan B, Melkoumian ZK, Wu X, Guan KL, Guan JL. Identification of FIP200 interaction with the TSC1-TSC2 complex and its role in regulation of cell size control. J Cell Biol. 2005;170:379–389. doi: 10.1083/jcb.200411106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89*.Balgi AD, Fonseca BD, Donohue E, Tsang TC, Lajoie P, Proud CG, Nabi IR, Roberge M. Screen for chemical modulators of autophagy reveals novel therapeutic inhibitors of mTORC1 signaling. PLoS One. 2009;4 doi: 10.1371/journal.pone.0007124. e7124 use small molecule screening to identify molecules and pathways involved in autophagy regulation. These studies identified a number of compounds that activate autophagy through both TOR-dependent and -independent pathways. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Winter G, Hazan R, Bakalinsky AT, Abeliovich H. Caffeine induces macroautophagy and confers a cytocidal effect on food spoilage yeast in combination with benzoic acid. Autophagy. 2008;4:28–36. doi: 10.4161/auto.5127. [DOI] [PubMed] [Google Scholar]
- 91.Scarlatti F, Maffei R, Beau I, Codogno P, Ghidoni R. Role of non-canonical Beclin 1-independent autophagy in cell death induced by resveratrol in human breast cancer cells. Cell Death Differ. 2008;15:1318–1329. doi: 10.1038/cdd.2008.51. [DOI] [PubMed] [Google Scholar]
- 92.Salazar M, Carracedo A, Salanueva IJ, Hernandez-Tiedra S, Lorente M, Egia A, Vazquez P, Blazquez C, Torres S, Garcia S, et al. Cannabinoid action induces autophagy-mediated cell death through stimulation of ER stress in human glioma cells. J Clin Invest. 2009;119:1359–1372. doi: 10.1172/JCI37948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Aoki H, Takada Y, Kondo S, Sawaya R, Aggarwal BB, Kondo Y. Evidence that curcumin suppresses the growth of malignant gliomas in vitro and in vivo through induction of autophagy: role of Akt and extracellular signal-regulated kinase signaling pathways. Mol Pharmacol. 2007;72:29–39. doi: 10.1124/mol.106.033167. [DOI] [PubMed] [Google Scholar]
- 94.Lavieu G, Scarlatti F, Sala G, Carpentier S, Levade T, Ghidoni R, Botti J, Codogno P. Regulation of autophagy by sphingosine kinase 1 and its role in cell survival during nutrient starvation. J Biol Chem. 2006;281:8518–8527. doi: 10.1074/jbc.M506182200. [DOI] [PubMed] [Google Scholar]
- 95.Pattingre S, Bauvy C, Carpentier S, Levade T, Levine B, Codogno P. Role of JNK1-dependent Bcl-2 phosphorylation in ceramide-induced macroautophagy. J Biol Chem. 2009;284:2719–2728. doi: 10.1074/jbc.M805920200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kumar SH, Rangarajan A. Simian virus 40 small T antigen activates AMPK and triggers autophagy to protect cancer cells from nutrient deprivation. J Virol. 2009;83:8565–8574. doi: 10.1128/JVI.00603-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Feng Z, Zhang H, Levine AJ, Jin S. The coordinate regulation of the p53 and mTOR pathways in cells. Proc Natl Acad Sci U S A. 2005;102:8204–8209. doi: 10.1073/pnas.0502857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98**.Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D'Amelio M, Criollo A, Morselli E, Zhu C, Harper F, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008;10:676–687. doi: 10.1038/ncb1730. The authors identify a novel non-transcriptional function of p53 in suppressing basal levels of autophagy. Loss or inhibition of P53 led to induction of autophagy through a TOR-dependent mechanism, and this promoted survival of p53(−/−) cells under nutrient stress. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wanke V, Cameroni E, Uotila A, Piccolis M, Urban J, Loewith R, De Virgilio C. Caffeine extends yeast lifespan by targeting TORC1. Mol Microbiol. 2008;69:277–285. doi: 10.1111/j.1365-2958.2008.06292.x. [DOI] [PubMed] [Google Scholar]
- 100.Tavernarakis N, Pasparaki A, Tasdemir E, Maiuri MC, Kroemer G. The effects of p53 on whole organism longevity are mediated by autophagy. Autophagy. 2008;4:870–873. doi: 10.4161/auto.6730. [DOI] [PubMed] [Google Scholar]
- 101*.Yorimitsu T, Zaman S, Broach JR, Klionsky DJ. Protein kinase A and Sch9 cooperatively regulate induction of autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:4180–4189. doi: 10.1091/mbc.E07-05-0485. this paper characterizes mechanisms of autophagy regulation by PKA and TOR signaling in yeast. Both pathways are shown to converge on the Atg1/Atg13 complex, and regulate the phosphorylation of these proteins and distinct sites, with distinct effects on their function. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102**.Stephan JS, Yeh YY, Ramachandran V, Deminoff SJ, Herman PK. The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proc Natl Acad Sci U S A. 2009;106:17049–17054. doi: 10.1073/pnas.0903316106. this paper characterizes mechanisms of autophagy regulation by PKA and TOR signaling in yeast. Both pathways are shown to converge on the Atg1/Atg13 complex, and regulate the phosphorylation of these proteins and distinct sites, with distinct effects on their function. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Budovskaya YV, Stephan JS, Reggiori F, Klionsky DJ, Herman PK. The Ras/cAMP-dependent protein kinase signaling pathway regulates an early step of the autophagy process in Saccharomyces cerevisiae. J Biol Chem. 2004;279:20663–20671. doi: 10.1074/jbc.M400272200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Budovskaya YV, Stephan JS, Deminoff SJ, Herman PK. An evolutionary proteomics approach identifies substrates of the cAMP-dependent protein kinase. Proc Natl Acad Sci U S A. 2005;102:13933–13938. doi: 10.1073/pnas.0501046102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105*.Zhang L, Yu J, Pan H, Hu P, Hao Y, Cai W, Zhu H, Yu AD, Xie X, Ma D, et al. Small molecule regulators of autophagy identified by an image-based high-throughput screen. Proc Natl Acad Sci U S A. 2007;104:19023–19028. doi: 10.1073/pnas.0709695104. use small molecule screening to identify molecules and pathways involved in autophagy regulation. These studies identified a number of compounds that activate autophagy through both TOR-dependent and -independent pathways. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106*.Williams A, Sarkar S, Cuddon P, Ttofi EK, Saiki S, Siddiqi FH, Jahreiss L, Fleming A, Pask D, Goldsmith P, et al. Novel targets for Huntington's disease in an mTOR-independent autophagy pathway. Nat Chem Biol. 2008;4:295–305. doi: 10.1038/nchembio.79. use small molecule screening to identify molecules and pathways involved in autophagy regulation. These studies identified a number of compounds that activate autophagy through both TOR-dependent and -independent pathways. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Vicencio JM, Ortiz C, Criollo A, Jones AW, Kepp O, Galluzzi L, Joza N, Vitale I, Morselli E, Tailler M, et al. The inositol 1,4,5-trisphosphate receptor regulates autophagy through its interaction with Beclin 1. Cell Death Differ. 2009;16:1006–1017. doi: 10.1038/cdd.2009.34. [DOI] [PubMed] [Google Scholar]
- 108.Sarkar S, Ravikumar B, Floto RA, Rubinsztein DC. Rapamycin and mTOR-independent autophagy inducers ameliorate toxicity of polyglutamine-expanded huntingtin and related proteinopathies. Cell Death Differ. 2009;16:46–56. doi: 10.1038/cdd.2008.110. [DOI] [PubMed] [Google Scholar]
- 109.Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, Lecker SH, Goldberg AL. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 110.Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, Burden SJ, Di Lisi R, Sandri C, Zhao J, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 111.Maiuri MC, Criollo A, Tasdemir E, Vicencio JM, Tajeddine N, Hickman JA, Geneste O, Kroemer G. BH3-only proteins and BH3 mimetics induce autophagy by competitively disrupting the interaction between Beclin 1 and Bcl-2/Bcl-X(L) Autophagy. 2007;3:374–376. doi: 10.4161/auto.4237. [DOI] [PubMed] [Google Scholar]
- 112.Mordier S, Deval C, Bechet D, Tassa A, Ferrara M. Leucine limitation induces autophagy and activation of lysosome-dependent proteolysis in C2C12 myotubes through a mammalian target of rapamycin-independent signaling pathway. J Biol Chem. 2000;275:29900–29906. doi: 10.1074/jbc.M003633200. [DOI] [PubMed] [Google Scholar]
- 113.Kanazawa T, Taneike I, Akaishi R, Yoshizawa F, Furuya N, Fujimura S, Kadowaki M. Amino acids and insulin control autophagic proteolysis through different signaling pathways in relation to mTOR in isolated rat hepatocytes. J Biol Chem. 2004;279:8452–8459. doi: 10.1074/jbc.M306337200. [DOI] [PubMed] [Google Scholar]
- 114.Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008;30:678–688. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]