SUMMARY
The inner ear epithelium, with its complex array of sensory, non-sensory, and neuronal cell types necessary for hearing and balance, is derived from a thickened patch of head ectoderm called the otic placode. Mouse embryos lacking both Fgf3 and Fgf10 fail to initiate inner ear development because appropriate patterns of gene expression fail to be specified within the pre-otic field. To understand the transcriptional “blueprint” initiating inner ear development, we used microarray analysis to identify prospective placode genes that were differentially expressed in control and Fgf3-/-;Fgf10-/- embryos. Several genes in the down-regulated class, including Hmx3, Hmx2, Foxg1, Sox9, Has2, and Slc26a9 were validated by in situ hybridization. We also assayed candidate target genes suggested by other studies of otic induction. Two placode markers, Fgf4 and Foxi3, were down-regulated in Fgf3-/-;Fgf10-/- embryos, whereas Foxi2, a cranial epidermis marker, was expanded in double mutants, similar to its behavior when WNT responses are blocked in the otic placode. Assays of hindbrain Wnt genes revealed that only Wnt8a was reduced or absent in FGF-deficient embryos, and that even some Fgf3-/-;Fgf10-/+ and Fgf3-/- embryos failed to express Wnt8a, suggesting a key role for Fgf3, and a secondary role for Fgf10, in Wnt8a expression. Chick explant assays showed that FGF3 or FGF4, but not FGF10, were sufficient to induce Wnt8a. Collectively, our results suggest that Wnt8a provides the link between FGF-induced formation of the pre-otic field and restriction of the otic placode to ectoderm adjacent to the hindbrain.
Keywords: FGF signaling, otic placode induction, WNT signaling, microarray analysis
INTRODUCTION
The inner ear is one of the most sophisticated vertebrate sensory organs, relaying both acoustical and motion/balance information to the brain. The entire structure, its intricate auditory and vestibular compartments, and innervating neurons, develops from the otic placode, a localized thickening of the caudalmost cranial ectoderm. All vertebrate cranial sensory organs derive from ectodermal placodes. Evidence suggests that these placodes emerge from a common ‘pan-placodal field’ that, shortly after gastrulation, surrounds the developing cranial neural plate in a narrow, horseshoe-shaped band. This field is defined as a distinct entity based on a characteristic set of expressed genes, including members of the Dlx, Eya, and Six gene families (Ohyama, 2009; Ohyama et al., 2007; Schlosser, 2006; Streit, 2007).
To form specific sensory placodes, the ectoderm must first acquire competence to respond to inductive signals (Groves and Bronner-Fraser, 2000). Many of the genes expressed in the pan-placodal field are transcription factors that may act individually or combinatorially as competence factors, preparing the ectoderm to receive subsequent inductive signals from the surrounding tissues (Martin and Groves, 2006; Schlosser and Ahrens, 2004). After acquiring competence, a second step in placode development is a differential response of competent preplacodal ectoderm to a unique code of local inducing signals that specify particular placode identity along the anteroposterior axis of the head. Thus, placode induction consists of a series of consecutive inductive events rather than binary fate assignments (Jacobson, 1966; McCabe and Bronner-Fraser, 2009; Ohyama, 2009; Ohyama et al., 2007; Schlosser, 2006). Much effort is focused on elucidating the nature of these signals and the transcriptional effectors they, in turn, activate within the target ectoderm to induce the subsequent development of a given placode.
Fibroblast growth factor (FGF) family members play pivotal roles in inner ear induction in several species, although the identity and sources of the FGFs often differ among species (Ohyama et al., 2007; Schimmang, 2007; Wright and Mansour, 2003b). Our studies and others showed that FGF3 and FGF10 are the most proximal signals that induce otic placodal fate in mice. Both factors are expressed near the pre-otic field (in the hindbrain and mesenchyme, respectively) at the time of otic placode induction, and mice lacking both Fgf3 and Fgf10 either fail to form otic vesicles altogether, or form microvesicles that do not differentiate into ear structures (Alvarez et al., 2003; Wright and Mansour, 2003a). Similarly, chick Fgf3 and Fgf19 are required for expression of otic placode marker genes (Freter et al., 2008). Fgf8 is also required in otic placode induction in the mouse and chick, functioning indirectly to induce mesodermal Fgf10 in mouse and Fgf19 in chick (Ladher et al., 2005; Zelarayan et al., 2007).
Ladher and colleagues (2000) first reported the involvement of hindbrain WNT signaling, in addition to FGF signaling, in chick otic placode induction. Exposing explants of presumptive chick otic ectoderm with WNT8A (previously called WNT8C)-expressing cells and FGF19-soaked beads resulted in the expression of several otic markers, whereas neither factor alone was sufficient. By studying transgenic TCF/Lef-lacZ reporter mice, Ohyama and colleagues (2006) found that WNT signaling is received by a subset of cells within the pre-otic field, those that reside most proximal to the neural tube, whereas the more lateral ectoderm displays little or no reporter activity. Importantly, they found that WNT signaling plays a role in refining the ventral boundary of the otic placode within the larger Pax2+ pre-otic field that is initially established by FGF induction and encompasses both the prospective otic placode and the ventrolateral ectoderm that has epibranchial placode and epidermal fates. Conditional inactivation of ß-catenin, a key effector of canonical WNT signaling, resulted in the formation of smaller otic vesicles and expanded the non-otic ectoderm. In contrast, constitutive activation of WNT signaling using activated ß-catenin, caused an expansion of the otic domain at the expense of non-otic ectodermal fates. Similarly, in chick, DKK1-mediated inhibition of ectodermal WNT signaling, or expression of activated ß-catenin, caused reduction or expansion, respectively, of the otic domain (Freter et al., 2008). These data suggest that WNT signaling modulates the otic placode-non-otic ectoderm fate decision and, as such, represents a third incremental step in the specification of the otic placode. It remains to be determined which WNT molecule(s) are required in vivo, and whether the FGF and WNT signals act in series or in parallel to instruct and refine placode induction.
To gain insight into these questions and to begin to understand the FGF-induced genetic program, or “blueprint”, for otic development, we isolated by microdissection the prospective otic ectoderm from control and Fgf3-/-;Fgf10-/- embryos at the time of otic induction (4-8 somite-stages). We subjected the RNA to microarray analysis to identify ectodermal genes that were differentially expressed, and found a small set of prospective targets. Several genes in the down-regulated class were validated by in situ hybridization. Among these were several transcription factor-encoding genes required at later stages of otic development, but not previously associated with placode induction in the mouse. Two additional validated targets, Has2 and Slc26a9, are novel candidates for genes that may play roles in inner ear development at inductive or later stages. We also assayed candidate genes suggested by other studies of otic induction, including those described above. Two placode markers, Fgf4 and Foxi3, were down-regulated in Fgf3-/-;Fgf10-/- embryos, whereas Foxi2, a cranial epidermis marker, was expanded in the double mutants, similar to its behavior when WNT signaling is blocked in the otic placode. Assays of hindbrain Wnt genes revealed that only Wnt8a was reduced or absent in FGF-deficient embryos, and that even some Fgf3-/-;Fgf10+/- and Fgf3-/- embryos failed to express Wnt8a, suggesting a key role for Fgf3, and a secondary role for Fgf10, in maintaining Wnt8a. Moreover, chick explant assays showed that FGF3 and FGF4, but not FGF10, were sufficient to induce Wnt8a. Taken together, our results show that FGF signaling is both necessary and sufficient to induce Wnt8a, and suggest that Wnt8a expression links FGF-induced formation of the pre-otic field with restriction of the otic placode to the ectoderm adjacent to the hindbrain.
MATERIALS AND METHODS
All mouse studies complied with protocols approved by the University of Utah Institutional Animal Care and Use Committee.
Generation of Fgf3 and Fgf10 conditional alleles
Fgf3- and Fgf10-genomic DNA-containing phages were isolated by recombination cloning from the λKO2 library (Zhang et al., 2002). One phage bore an approximate 9.0 kb fragment of Fgf3, with a loxP site located in the intron 3.8 kb downstream of exon 2, and the other carried an approximate 10.0 kb fragment Fgf10, with a loxP site located in the intron 0.85 kb downstream of exon 2. After recovery of the phage inserts, we generated targeting vectors, each of which had a tetracycline response element (TRE) with or without an antisense CMV promoter element, followed by loxP and FRT sites, and NeoR and KanR expression cassettes inserted in an intron 0.58 kb and 0.30 kb upstream of Fgf3 and Fgf10 exon 2, respectively. Each of the four targeting vectors was linearized and electroporated into R1-45 ES cells, which were selected in G-418 and ganciclovir as described (Li et al., 2007). Correct homologous recombination between the vectors and the genome was determined by Southern blot hybridization using 5’ and 3’ probes flanking the targeting sequences. Following introduction into C57Bl/6 embryos, correctly targeted cell lines generated highly chimeric males that transmitted the targeted alleles to offspring. Phenotypic observations of targeted homozygotes showed that the initially targeted alleles were hypomorphic (data not shown). To remove the intronic selection cassettes, heterozygotes were crossed to FLPe-expressing mice (Gt(ROSA)26Sortm1(FLP1)Dym, The Jackson Laboratory [JAX] strain #003946) (Farley et al., 2000). The resulting conditional strains were verified by Southern blot hybridization and PCR analyses (not shown) and have an intronic TRE with or without an antisense CMV promoter, and exon 2, comprised in both cases of 104 bp, is flanked by loxP sites. These alleles are designated c (no CMV promoter) or Pc (with the intronic CMV promoter). Each of the four homozygous conditional strains had normal viability, but Fgf3Pc/Pc mice invariably showed slight tail kinks—a mild version of the Fgf3 null short-tail phenotype (Alvarez et al., 2003; Hatch et al., 2007; Mansour et al., 1993). All experiments reported here involved the “c” alleles. For global removal of exon 2, hypomorphic mice were crossed to the Cre deleter strain, BALB/c-Tg(CMV-cre)1Cgn/J (JAX stock #003465) (Schwenk et al., 1995). Mice/embryos homozygous for the exon 2 deletions (designated Δ2 alleles) or compound heterozygotes with established null alleles (Fgf3tm1Mrc or Fgf10tm1Wss) (Mansour et al., 1993; Min et al., 1998) were indistinguishable from the established null homozygotes (data not shown).
Generation and genotyping of Fgf3/Fgf10 global null embryos
Global deletion of both conditional alleles was accomplished using Hprt1tm1(cre)Mnn (Tang et al., 2002), kindly supplied by M. Capecchi. Standard genetic crosses were used to generate Fgf3Δ2/+;Fgf10Δ2/+;HprtCre/+ females, which were crossed to Fgf3c/c;Fgf10c/c males. Allele-specific PCR assays for genotyping included the following primers:
Hprt: SLM10 (5’-GCCTGCTTGCCGAATATCATGG-3’), SLM544 (5’-CCTGATTTTATTTCTATAGGACTGAAAGAC-3’), and SLM545 (5’-TAAGTTAATTATACTTACACAGTAGCTCTTC) produced 200 bp wild-type and 550 bp Cre insertion allele fragments.
Fgf3: SLM608 (5’-GGACGTATGAACGAGTGTATAGATGG-3’), SLM609 (5’-AGGGATGGTCCTACAGACTTGCAG-3’), and SLM485B (5’ GGTTCCTCGATCAAACTCTGG-3’) produced 615 bp deletion allele and 480 bp conditional allele, and 379 bp wild-type fragments.
Fgf10: SLM492B (5’-GTACCGAGCTCGACTTTCAC-3’), SLM479B (5’-GTCTTTTTGACTGAAACCTCAC-3’), and SLM411B (5’-ATCCTTGGGAGGCAGGATAACC-3’) produced 450 bp deletion allele, 277 bp conditional allele, and 175 bp wild-type fragments.
Crossing Fgf3Δ2/+;Fgf10Δ2/+;HprtCre/Y males to double conditional females was compromised by frequent CRE-mediated interchromosomal recombination between the similarly oriented loxP sites present in the Fgf3Δ2 and Fgf10Δ2 loci, leading to balanced and unbalanced translocations between chromosomes 7 and 11 (data not shown). Thus, it is inadvisable to attempt to generate Fgf3Δ2/+;Fgf102/+;HprtCre/+ females from such males.
Otic placode RNA isolation and microarray analysis
Timed matings between Fgf3Δ2/+;Fgf10Δ2/+;HprtCre females and Fgf3c/c;Fgf10c/c males were initiated, and embryos were harvested on embryonic day (E)8.5. The yolk sac was collected for genotyping. The otic region, including the prospective placodal ectoderm, neural ectoderm, and underlying mesendoderm was dissected from 4-8-somite embryos using tungsten knives. Isolation of placodal ectoderm was accomplished by modification (Y. Saijoh, personal communication) of the enzymatic procedure described by Hogan et al. (Hogan et al., 1994). Briefly, the bilateral “tri-layer” otic region fragments were rinsed in ice-cold PBS and moved to 50 μl of PT solution (25 mg/ml pancreatin [Sigma], 5 mg/ml trypsin [Sigma], and 5 mg/ml polyvinylpyrrolidone MW360 [Sigma] in Tyrode’s solution) for 5 minutes on ice to promote germ layer separation. The fragments were transferred to ice-cold HEPES-buffered DMEM with 10% FBS for 2-5 minutes, then the placodal ectoderm and associated neural ectoderm were teased away from the mesendoderm with tungsten needles, and the placodal ectoderm was freed by resection of the attached neural ectoderm with a tungsten knife. The two placodal ectoderm fragments from each embryo (~100 microns each) were aspirated into 30 μl RLT buffer (Qiagen MicroRNA Easy kit), vortexed for 1 minute, and stored at -70°C. All tissue isolates were stored separately prior to genotyping.
RNA was extracted (Qiagen MicroRNA Easy kit) from three biological replica pools; each comprising ten Fgf3Δ2/+;Fgf10Δ2/+ or ten Fgf3Δ2/Δ2;Fgf10Δ2/Δ2 placodes. After elution, RNA integrity was determined on a Bioanalyzer (Agilent). Recovery was ~100 ng/pool. RNA was subjected to a single linear amplification labeling reaction (Fgf3Δ2/+;Fgf10Δ2/+ control with Cy3, and Fgf3Δ2/Δ2;Fgf10Δ2/Δ2 mutant with Cy5) and hybridized to mouse whole genome microarray slides (Agilent GPL4134) using the standard Agilent 2-color gene expression hybridization protocol. Slides were scanned (Agilent G2505B) at 5 μm resolution using an extended dynamic range protocol, and images were processed with Agilent Feature Extraction software 9.5.1.1. The Lowess-normalized log2 ratio of Cy3/Cy5 was calculated. Genes significantly up- or down-regulated in all three mutant pools were identified by rank products analysis (Breitling et al., 2004). The full dataset was deposited with GEO (GSE18702).
Riboprobe preparation and in situ hybridization
Digoxigenin-labeled antisense cRNA probes were generated from plasmids carrying cDNA fragments according to standard procedures, or following direct PCR amplification of 3’ UTRs from genomic DNA (adapted from Ambion Technical Bulletin 154). For the latter, a 28-base T7 RNA polymerase promoter (5’-GGATCCTAATACGACTCACTATAGGGAG-3’) was incorporated at the 5’ end of the reverse primer.
Cloned mouse cDNAs used to prepare riboprobes included Erm (Li et al., 2007), Wnt8a (Bouillet et al., 1996), Wnt1, Wnt3a, and Hmx3 (Hatch et al., 2007), Spry1 (Minowada et al., 1999), Fgf4 (Wright and Mansour, 2003b), Sox9 (Wright et al., 1995), and Wnt6 (Gavin et al., 1990) as described in the cited publications. An 886 bp Has2 3’ UTR clone was purchased from OpenBiosystems (GenBank accession AI592649, clone #633823).
3’ coding region probes for Foxg1 and Cldn4 were generated by cloning PCR-amplified DNA fragments into pCRII-TOPO (Invitrogen). The amplicons included residues 2615-2976 of Foxg1 (GenBank accession NM_001160112) and residues 392-749 of Cldn4 (GenBank accession NM_009903).
Probes for the 3’ UTRs of Foxi3, Foxi2, and Hmx2 were PCR-amplified from genomic DNA using T7 promoter-tagged reverse primers. The primer sequences were: Foxi3 forward primer SLM656: 5’-ACTACAACCCTTTCTCTGGTGGC-3‘, reverse primer SLM655: 5’-T7-CGGACTTTGCTCACAGTAATCAAGC-3‘; Foxi2 forward primer SLM648: 5’-GCTTTGGGTTTGCCTTACTTGAC-3‘, reverse primer SLM647: 5’-T7-CAGCACACCAGGTAGGAACAACAC-3‘; Hmx2 forward primer SLM594: 5’-AGAATCGCCGCAACAAGTGG-3‘, reverse primer SLM595: 5’-T7-GAGAGCCTCCCCTTCCAAAATAG-3‘; Slc26a9 forward primer SLM691: 5’-TGACTTCCAGCCTTTAGAGTGAG-3‘, reverse primer SLM692: 5’-T7-CCAGTTTGCCCGAGTTTACATTAG-3‘. PCR products of 530 bp, 594 bp, 534 bp, and 554 bp respectively, were directly transcribed using T7 RNA polymerase.
Whole-mount in situ hybridization was performed on embryos isolated from timed pregnancies essentially as described (Wilkinson, 1992). Stained embryos were post-fixed in 4% paraformaldehyde, cryoprotected in 15% sucrose/7.5% gelatin, and cryosectioned at 14 μm (Sechrist et al., 1995).
Collagen gel cultures
The collagen gel matrix for tissue explant culture was made by mixing on ice, 25 μl of 10x DMEM, 40 μl of 7.5% sodium bicarbonate, 5 μl 1M HEPES buffer (Invitrogen), and 200 μl of 2.5 mg/ml rat collagen (Roche). The matrix was gently stirred until it reached a pale pink color and was kept on ice until used.
Hamburger and Hamilton (HH) stage 4-5 ectoderm, located rostral to Hensen’s node, and consisting of both neural and non-neural ectoderm, was used as the test tissue (region “Et”, Ladher et al., 2000, Fig. 2). Rostral ectodermal isolates were recombined with 0.1% BSA/PBS control or 0.25 μM recombinant human FGF (R&D Biosystems) coated Affi-blue gel beads (Bio-Rad Laboratories) in a 10 μl drop of collagen gel matrix, which was incubated at 37°C until the matrix gelled. Tissue/bead recombinants were cultured overnight at 37°C in a humidified chamber (5% CO2/95% air) in medium consisting of Neurobasal Media (Invitrogen) supplemented with B27 serum-free supplement (Invitrogen), 200 mM L-Glutamine, and 1:1000 penicillin and streptomycin and then fixed in 4% PFA at 4°C overnight. Explants were analyzed by in situ hybridization for expression of chick Wnt8a (Hume and Dodd, 1993).
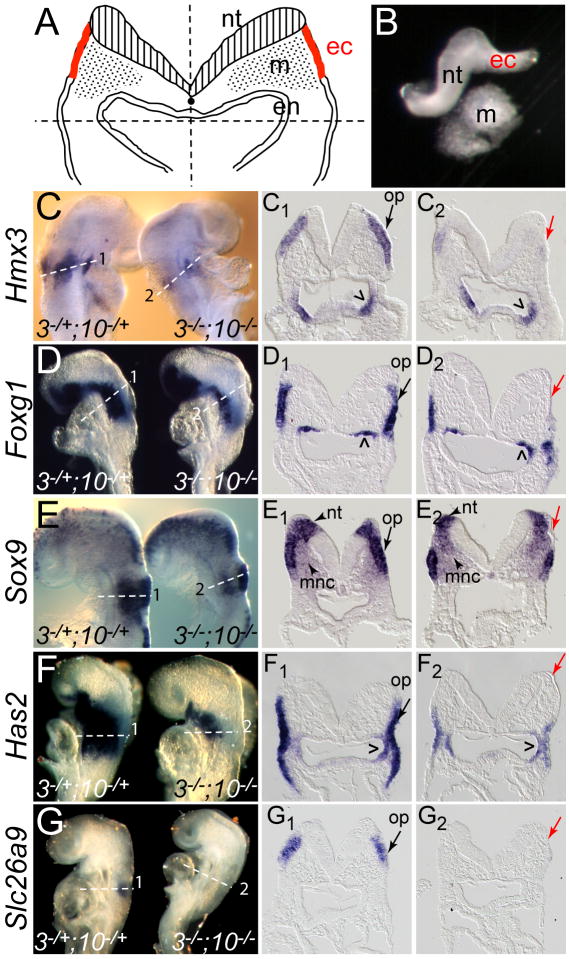
Fig. 2. Isolation of tissue and validation by in situ hybridization of microarray candidate genes down-regulated in placodal ectoderm of Fgf3/Fgf10-deficient embryos.
(A) Schematic depiction of microdissection of the placodal region of an E8.5 5-8-somite embryo. Embryos were first bisected along the midline (vertical dashed line). The dorsal, presumptive otic region, was isolated from the ventral aspect with a second cut (horizontal dashed line), generating two fragments containing neural tube (nt), placodal ectoderm (ec), and mesendoderm (m, en). (B) Protease treatment of each hemisected otic region released the mesendoderm from the neural tube and attached placodal ectoderm, which was then severed from the nt and recovered for RNA extraction. (C-G) 7-8-somite Fgf3Δ2/+;Fgf10Δ2/+ (3-/+;10-/+) and Fgf3Δ2/Δ2;Fgf10Δ2/Δ2 (3-/-;10-/-) embryos hybridized with riboprobes for Hmx3 (C), Foxg1 (D), Sox9 (E), Has2 (F), and Slc26a9 (G). Anterior is to the top. Transverse sections taken through the placodal region (planes numbered and indicated with dashed lines in panels C-G) show otic placode (op) expression of each gene in double heterozygotes (C1-G1, black arrows) and loss of gene expression in the corresponding (thin) ectoderm in double mutants (C2-G2, red arrows). Carets and arrowheads indicate Fgf-independent expression in pharyngeal endoderm, neural tube (nt), and migrating neural crest (mnc), respectively.
RESULTS
Conditional alleles enable efficient generation of Fgf3/Fgf10 double null embryos for isolation and microarray analysis of prospective otic ectodermal RNA
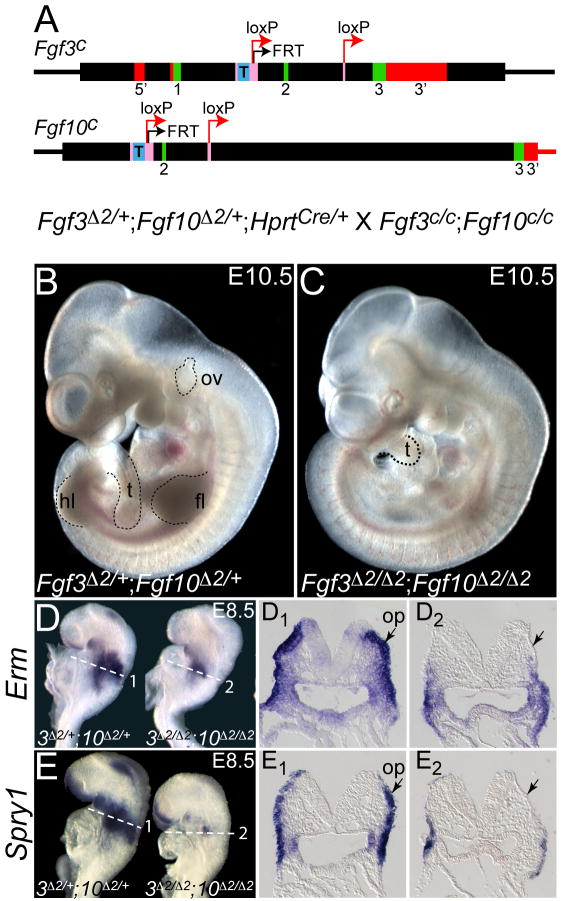
To enable conditional or global deletion of Fgf3 and/or Fgf10, we generated targeted alleles in which the 104 bp exon 2 of each gene was flanked by loxP sites (Fig. 1A; “c” alleles). Single or double homozygous conditional animals were normal (data not shown). In each case, germline deletion of exon 2 yielded a null allele (designated Δ2 or –, for brevity), and the gross phenotypes of homozygous null animals were indistinguishable from those generated by other means (Abler et al., 2009; data not shown; Hatch et al., 2007). For efficient generation of double mutants we crossed Fgf3Δ2/+;Fgf10Δ2/+;HprtCre/+ females with Fgf3c/c;Fgf10c/c males. Since CRE was deposited into oocytes, the “c” alleles were deleted to “Δ2” alleles in all offspring, regardless of whether they inherited HprtCre. Thus, one quarter of the E10.5 embryos were double heterozygotes (Fgf3Δ2/+;Fgf10Δ2/+) that had a normal phenotype (Fig. 1B), and one quarter were double homozygotes (Fgf3Δ2/Δ2;Fgf10Δ2/Δ2) that, as expected, had shortened tails, lacked limbs, and had little or no otic vesicle development (Fig. 1C). To evaluate the efficacy of FGF signaling disruption at the otic placode stage, we stained E8.5 (7-8 somites) control and double mutant embryos with FGF signaling target genes, Erm and Spry1. Both genes were normally expressed in a domain that includes, but is larger than, the prospective otic placode (Fig. 1D,D1,E,E1), and were strongly reduced in the dorsal (presumptive otic) portion of double mutant ectoderm (Fig. 1D,D2,E,E2), demonstrating successful disruption of FGF signaling in the prospective otic domain. Retention of Erm and Spry1 in the ventral ectoderm of double mutants suggests that this region continued to receive FGF signals, which could originate from the ventral ectoderm itself (e.g. Fgf4, see Fig. 3B,B1, below) or from the pharyngeal endoderm (e.g. Fgf8, (Ladher et al., 2005); Fgf15, (Wright et al., 2004); or Fgf4, see Fig. 3B,B1, below).
Fig. 1. Use of Fgf3 and Fgf10 conditional alleles to produce double null mutants that lack otic vesicles at E10.5 and FGF/MAPK signaling markers at E8.5.
(A) Each targeted conditional allele has loxP sites (red arrows) flanking exon 2. An FRT site (black arrow) remains from FLP-mediated removal of a Neo expression cassette. A tetracycline response element (T) is located upstream of exon 2. CRE-mediated deletion of the 104 bp exon 2 from the conditional (c) alleles generates the null alleles (designated Δ2). Thick boxes indicate the sequences included within the targeting vectors; exons are indicated in green; 5’ and 3’ UTRs in red; introns in black; recombinase recognition sequences in pink; and the T in blue. The cross generating double mutants at a frequency of 25% is indicated below the allele diagrams. (B) E10.5 Fgf3Δ2/+;Fgf10Δ2/+ embryo shows a normal phenotype. (C) E10.5 Fgf3Δ2/Δ2;Fgf10Δ2/Δ2 embryo lacks limbs and otic vesicles, and has a shortened tail. Dashed lines demark limbs (fl, forelimb; hl, hindlimb), tail (t), and otic vesicle (ov) where present. Whole-mount E8.5 (7-somite) embryos were probed with FGF signaling indicators, Erm (D) and Spry1 (E). Genotype is indicated below each embryo. (D1, E1) Transverse sections taken through the placodal region (planes numbered and indicated with dashed lines in D and E) show otic placode (op) expression (arrows) of each gene in double heterozygotes and loss of gene expression in the corresponding (thin) ectoderm in double mutants (D2, E2, arrows).
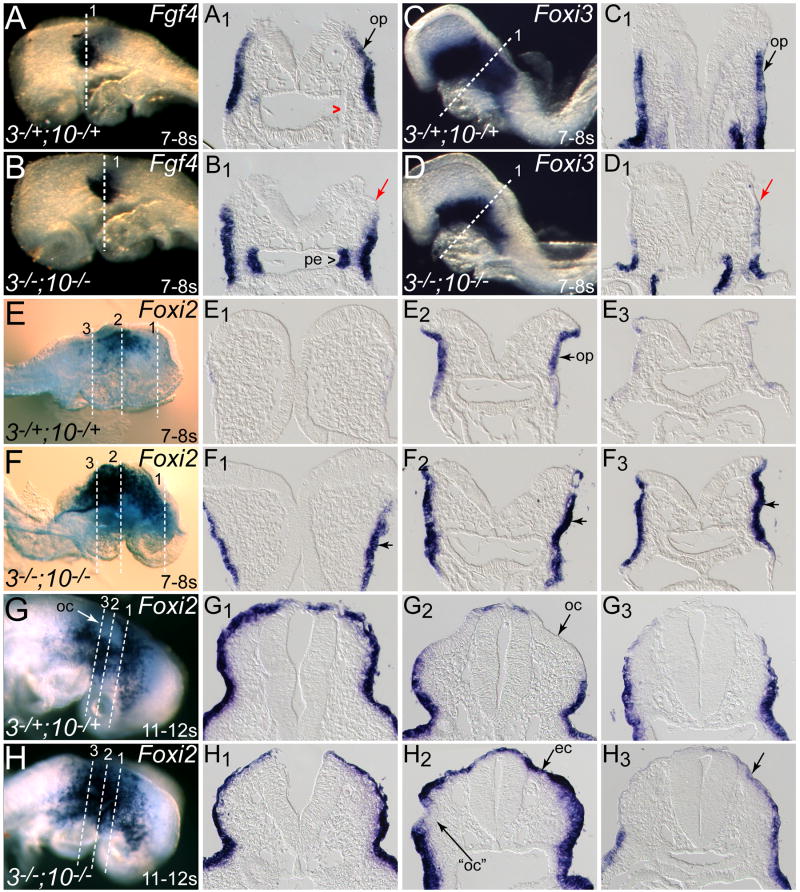
Fig. 3. Other placode-expressed genes are differentially affected in Fgf3/Fgf10-deficient ectoderm.
Somite-matched E8.5 Fgf3Δ2/+;Fgf10Δ2/+ (3-/+;10-/+) and Fgf3Δ2/Δ2;Fgf10Δ2/Δ2 (3-/-;10-/-) embryos were hybridized with probes for Fgf4 (A,B), Foxi3 (C,D), and Foxi2 (E-H). Anterior is to the left in A-D and to the right in E-H. Transverse sections taken through the otic region (planes numbered and indicated with dashed lines in panels A-H) show otic placode (op) expression of each gene in double heterozygotes (A1,C1,E2,G2, black arrows; red caret indicates lack of expression in pharyngeal endoderm). Fgf4 was absent from dorsal double mutant ectoderm (B1, red arrow), but up-regulated in pharyngeal endoderm (B1, caret) and Foxi3 was absent from dorsal ectoderm in double mutants (D1, red arrow). Foxi2 expression was expanded both anteriorly (F1) and posteriorly (F3) in 7-8 somite double mutants, and overall expression in the placodal region was more intense (F2). In 11-12 somite embryos, Foxi2 expression was restricted from the otic cup (oc) in control embryos (G, G1,G2,G3), whereas Foxi2 was present throughout the dorsal ectoderm (ec) in double mutants (H,H1,H2,H3). The ventrally localized cup-like structure (“oc”), which may represent the precursor to one of the microvesicles occasionally seen in double mutants, showed incomplete clearing of Foxi2 expression (H2).
Microarray analysis reveals candidate targets of FGF signaling
Previous studies showed that expression of several otic placode marker genes, including Pax2, Gbx2, Dlx5, and Pax8, was not detected in dorsal ectoderm of Fgf3/Fgf10 double null embryos generated from standard double heterozygous intercrosses (Alvarez et al., 2003; Wright and Mansour, 2003a). To enable a more comprehensive assessment of FGF-regulated otic ectodermal genes we compared gene expression profiles of three pools each of prospective otic ectoderm microdissected from 4-8-somite Fgf3Δ2/+;Fgf10Δ2/+ and Fgf3Δ2/Δ2;Fgf10Δ2/Δ2 embryos (Fig. 2A,B). Rank products analysis yielded a list of 64 unique transcripts significantly up-regulated and 28 down-regulated by at least 1.5-fold in the FGF-deficient ectodermal RNA pools (Table S1). Technical difficulties in perfecting embryo staging and tissue isolation are likely to account for the high degree of variability between transcript levels detected in pools of the same genotype, which in turn may account for the small number of genes ultimately identified as significantly regulated in all three comparisons.
Validation of FGF-dependent ectodermal target genes
To validate candidate down-regulated transcripts we compared gene expression in Fgf3Δ2/+;Fgf10Δ2/+ and Fgf3Δ2/Δ2;Fgf10Δ2/Δ2 7-8-somite embryos by whole mount in situ hybridization. Hmx3, Foxg1, Sox9, Has2, and Slc26a9 were all clearly expressed in control otic ectoderm (Fig. 2C,C1,D,D1,E,E1,F,F1G,G1), but were absent from Fgf3/Fgf10-deficient dorsal ectoderm (Fig. 2C,C2,D,D2,E,E2,F,F2G,G2) which, unlike control dorsal ectoderm, failed to thicken. Retention of Foxg1, Sox9, and Has2 in double mutant ventral ectoderm (Fig. 2D2,E2,F2) was consistent with retention of FGF signaling indicators in this region. Other sites of gene expression (e.g. pharyngeal endodermal Hmx3, Foxg1, and Has2) were unaffected in double mutants (Fig. 2C1,C2,D1,D2,F1,F2). Indeed, expression of Sox9 in the neural tube and migrating neural crest was unaffected in double mutants (Fig. 2E1,E2), indicative of the high degree of ectodermal target tissue purification achieved by the dissections. We also tested Hmx2 expression and found that it, too, was absent from dorsal double mutant ectoderm (data not shown). In contrast, two placode-expressed genes (Cldn4 and Zic4) and one ubiquitously expressed gene (Dusp16) were unchanged in double mutants (data not shown). Several other ubiquitously or relatively non-specifically expressed genes (Runx2, Sbsn, Yawae, and Gpr83) were not examined in double mutants. Finally, Kifc1 and Dcc may represent false positives, as neither gene was expressed in control placodes, but could be detected elsewhere. Taken together, these results indicate that our strategy of gene expression profiling successfully enriched for otic genes that are directly or indirectly dependent on FGF signaling, revealing at least six genes not previously associated with FGF responses in the mouse otic placode.
Other ectodermal genes regulated by FGF signaling include Fgf4 and Foxi genes
We next examined the expression of several other potential targets of FGF signaling in otic placode induction, including Fgf4, which previous studies showed is expressed in a restricted portion of the prospective placode in 4-11-somite embryos (Fig. 3A,A1 and Wright et al., 2003) and Foxi3 and Foxi2, which mark the prospective otic and non-otic ectoderm, respectively (Fig. 3C,C1,E,E2) (Ohyama and Groves, 2004). Like the down-regulated candidates identified in the microarray screen, Fgf4 expression was absent from the dorsal ectoderm of 7-8-somite double mutants (Fig. 3B,B1). Interestingly, although Fgf4 expression was unchanged in ventral double mutant ectoderm, it appeared up-regulated in pharyngeal endoderm (Fig. 3B,B1). As endodermal Fgf4 is normally strongly up-regulated at the 9-somite stage, it is unclear whether this finding is significant, or merely a result of somite counting ambiguities. Foxi3 expression was also eliminated from dorsal ectoderm in all five 4-7-somite stage double mutant embryos (Fig. 3D,D1), but was only slightly reduced in the two 8-9-somite stage double mutants examined (not shown). In contrast, Foxi2 expression was expanded in double mutant ectoderm in 7-8-somite embryos (Fig. 3F,F1,F2,F3). Furthermore, by the 11-12-somite stage, when Foxi2 expression was excluded from the otic cup of control embryos (Fig. 3G,G2), it was expressed throughout an inappropriately enlarged ectodermal domain in double mutant embryos (Fig. 3H,H2). Even when a small ventrally localized otic cup-like structure formed in double mutants (Fig. 3H2), some Foxi2 expression encroached into the thickened region, suggesting that it did not have a true otic character, much like the microvesicles found previously in older double mutants (Wright and Mansour, 2003a).
FGF signaling is required for hindbrain Wnt8a expression
The expansion of Foxi2 in Fgf3Δ2/Δ2;Fgf10Δ2/Δ2 ectoderm is similar to the response of head ectoderm when WNT signaling to the Pax2 lineage is blocked by Tg(Pax2-cre) otic conditional ablation of ß-catenin (Ohyama et al., 2006). Hindbrain WNT signals are implicated in otic induction (Freter et al., 2008; Ladher et al., 2000; Ohyama et al., 2006; Park and Saint-Jeannet, 2008). To determine whether FGF signals function upstream of hindbrain WNT signals during otic induction, we assayed hindbrain Wnt genes in control and FGF-deficient mutants. Wnt6 (Fig. 4A) and Wnt3a (Fig. 4C) were expressed as early as the 7-somite stage, and Wnt1 (data not shown) was expressed from the 12-somite stage in the dorsal neural ectoderm, from which the neural crest emigrates; however, expression of these three Wnt genes was unaffected in Fgf3Δ2/Δ2;Fgf10Δ2/Δ2 embryos (Fig. 4B,D; Wnt1 data not shown). In contrast, Wnt8a, which is expressed in hindbrain rhombomere (r)4 at least as early as the 4-somite stage and diminished dramatically by the 9-somite stage (Fig. 4E; Bouillet et al., 1996 and L.D.U. and S.L.M, unpublished), was significantly reduced (Fig. 4F, n = 2), or in some cases absent (n = 2), in the double mutants (Table 1). Hindbrain Wnt8a was also absent from some Fgf3-/-;Fgf10-/+ (Fig. 4H, n = 3) and Fgf3-/- embryos (Fig. 4J, n = 2). However, Wnt8a expression was normal in all but one Fgf3-/+;Fgf10-/- embryo (Fig. 4G, n = 4) and in all Fgf10-/- embryos (Fig. 4I, n = 2), suggesting that FGF signaling is directly or indirectly required for hindbrain Wnt8a expression, and that Fgf3 plays a more significant role in this process than does Fgf10.
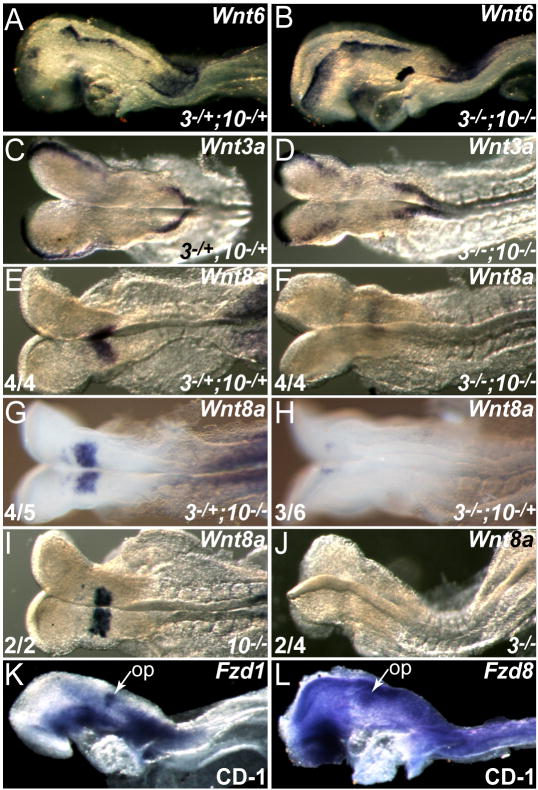
Fig. 4. Wnt8a is the only hindbrain Wnt that is dependent on Fgf3 and Fgf10 expression, and the otic field expresses at least two WNT receptor (Fzd) genes.
Somite-matched E8.5 control Fgf3Δ2/+;Fgf10Δ2/+ (3-/+;10-/+; A,C,E) and Fgf3Δ2/Δ2;Fgf10Δ2/Δ2 (3-/-;10-/-; B,D,F) embryos were hybridized with probes for Wnt6 (A,B), Wnt3a (C,D), and Wnt8a (E,F). Wnt6 and Wnt3a were expressed in control neural plates (A,C), and expression was unchanged in double mutant embryos (B,D). r4 expression of Wnt8a (E) was reduced or absent (F) in all 4 double mutants. Fgf3+/Δ2;Fgf10Δ2/Δ2 (3+/-;10-/-; G) and Fgf10Δ2/Δ2 (10-/-; I) embryos showed normal levels of Wnt8a in 4/5 and 4/4 embryos respectively. In contrast, Fgf3Δ2/Δ2;Fgf10+/Δ2 (3-/-;10+/-; H) and Fgf3Δ2/Δ2 (3-/-; J) showed reduced Wnt8a expression in 3/6 and 2/4 embryos respectively. WNT receptor genes Fzd1 (K) and Fzd8 (L) were expressed in the otic placode (op).
Table 1. Wnt8a expression in r4 at 5-8 somites depends on Fgf expression.
5-8-somite mouse embryos of the indicated genotypes were hybridized with the Wnt8a probe and subjectively scored for the intensity of r4 labeling as normal, reduced, or absent. The number of embryos in each category is indicated.
| Genotype | ||||||||
|---|---|---|---|---|---|---|---|---|
| Wnt8a expression (r4) | 3-/+;10-/+ control | 3-/-;10-/- | 3-/-;10-/+ | 3-/+;10-/- | 3-/+ control | 3-/- | 10-/+ control | 10-/- |
| Normal | 4 | 0 | 3 | 4 | 3 | 2 | 4 | 2 |
| Reduced | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 |
| Absent | 0 | 2 | 3 | 0 | 0 | 2 | 0 | 0 |
| Total | 4 | 4 | 6 | 5 | 3 | 4 | 4 | 2 |
To determine whether WNT receptors are expressed in the prospective otic placode, we surveyed expression of several Fzd genes and found that Fzd1 is most specific to the placode (Fig. 4K), but expression of Fzd8 could also be detected (Fig. 4L).
FGF signaling is sufficient to induce Wnt8a in a chick ectodermal explant assay
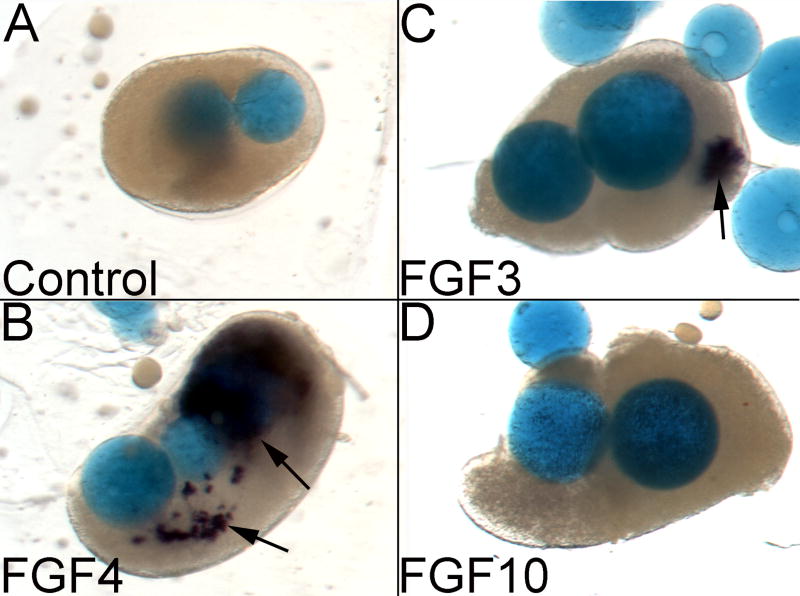
Human FGF19 induces chick Wnt8a in ectodermal explants that include neural tissue (Ladher et al., 2000). To determine whether additional otic region FGFs are sufficient to induce Wnt8a expression, we combined rostral, ectodermal isolates from HH stage 4-5 chick embryos with FGF protein-coated beads in collagen gel culture. The explants were cultured overnight and then assayed for Wnt8a expression via in situ hybridization (Table 2, Fig. 5). Weak Wnt8a expression was detected in only one of 10 explants exposed to control beads. However, both FGF4- and, to a lesser extent, FGF3-coated beads, induced expression of Wnt8a (Fig. 5A-C). FGF4 strongly induced Wnt8a nearly all of the time (11 of 12 explants), whereas FGF3 induced a weak signal less frequently (4 of 12 explants). In contrast, FGF10 almost never induced Wnt8a (1 out 12 explants, Fig. 5D).
Table 2. FGFs induce Wnt8a expression in chick explant culture.
Explants of chick HH stage 4-5 rostral ectoderm were cultured in the absence (control) or presence of FGF protein-soaked beads and assayed for chick Wnt8a expression. The fraction of all explants showing any Wnt8a expression is indicated in the first column, and the number of explants exhibiting strong (++), weak (+), or no (-) expression of Wnt8a is indicated is indicated in subsequent columns.
| Beads | Wnt8a pos./total | Wnt8a ++ | Wnt8a + | Wnt8a - |
|---|---|---|---|---|
| Control | 1/10 | 0 | 1 | 9 |
| FGF4 | 11/12 | 10 | 1 | 0 |
| FGF3 | 4/12 | 0 | 4 | 8 |
| FGF10 | 1/12 | 0 | 1 | 11 |
++ = strong expression
+ = weak expression
- = no expression
Fig. 5. FGF4 or FGF3, but not FGF10, induces Wnt8a in chick ectodermal explants.
HH stage 4-5 ectodermal explants were cultured with control or FGF protein-coated beads, and Wnt8a expression was assessed by in situ hybridization. Control beads (A) were unable to induce Wnt8a, whereas FGF4 induced robust expression of Wnt8a (B, arrows) and FGF3 induced weak Wnt8a expression in approximately 1/3 of the explant cultures (C, arrow). In contrast, FGF10 did not induce Wnt8a (D). See Table 2 for quantification of the results.
DISCUSSION
Induction of the mouse otic placode from pre-otic ectoderm located adjacent to the hindbrain requires FGF3 and FGF10, which are expressed in the hindbrain and head mesenchyme, respectively. In the absence of these signals, the otic vesicle either does not form, or only a small, rudimentary vesicle is induced (Alvarez et al., 2003; Wright and Mansour, 2003a). Although in some circumstances FGFs modulate target tissue proliferation and/or survival, in the case of Fgf3/Fgf10-deficient embryos, these processes are unperturbed in the prospective otic territory (Wright and Mansour, 2003a). Instead, FGF signaling controls gene expression within the pre-otic ectoderm, and the small set of known FGF target genes, identified serendipitously, are presumably contained within a larger set of genes that serve as a molecular blueprint directing the subsequent differentiation of the otic anlage. Here we identified and validated additional FGF targets that are new candidates for genes involved in otic placode induction and/or subsequent FGF-regulated processes in ear development. In addition, we uncovered a mechanism linking FGF and WNT signaling in series in the refinement of the otic territory (Fig. 6).
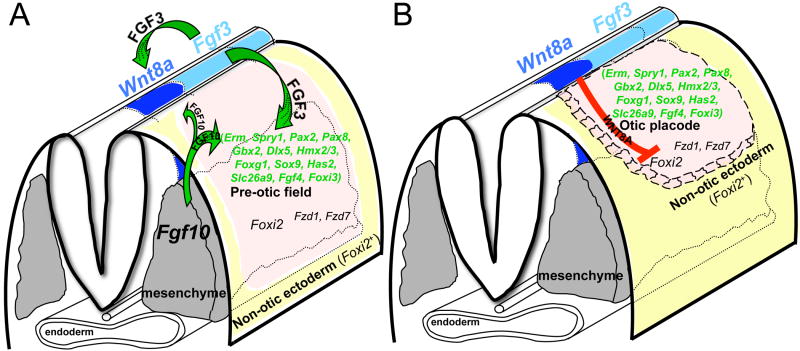
Fig. 6. Model for induction and resolution of the otic placode from pre-otic ectoderm.
(A) Oblique view of a pre-placodal-stage embryo illustrating the initiation of placode specification by hindbrain-expressed FGF3 (light blue) and mesenchyme-expressed FGF10 (grey) acting on the pre-otic field (pink) to induce gene expression (data from this and previous studies). Both FGFs are also required for induction and/or maintenance of hindbrain Wnt8a (dark blue); FGF3 being the more potent activator. (B) Oblique view of a placodal-stage embryo showing the proposed role of WNT8A interacting with FZD receptors, repressing Foxi2 and limiting otic placode fate to hindbrain-proximal ectoderm, with remaining ectoderm (yellow) assuming a non-otic fate (epibranchial placode or epidermis).
Microarray analysis reveals FGF target genes important for inner ear development
Microarray-based comparison of control and FGF3/10-deficient ectoderms revealed a relatively small set of up- and down-regulated FGF target gene candidates, with theoretical changes in expression of between 1.5- and 4-fold. We attribute the small set size to several factors, including small differences in staging between embryos that were subsequently pooled, the variation inherent in the use of microdissection and enzymatic digestions for isolating small tissue samples, and stochastic differences amplified in extremely small RNA sets. Consequently, it is not surprising that previously identified FGF-responsive placodal transcription factor genes (including Pax2, Pax8, Gbx2, and Dlx5) did not appear on the list, which likely represents a mere “tip of the iceberg”. Given these considerations, it is striking that six of the nine down-regulated candidate genes evaluated by in situ hybridization in both genotypes were strongly reduced or completely absent specifically from double mutant ectoderm. It remains to be determined, of course, whether these genes are directly or indirectly regulated by the FGFs, but their differential expression cannot be a trivial consequence of the absence of an otic placode in double mutants, given that they are expressed (and were evaluated) prior to placode-thickening.
Three transcription factor genes (Hmx3, Hmx2, and Foxg1) shown previously to be required for otic morphogenesis are FGF dependent and fall within this class. Targeted single and double mutagenesis of the tandemly-duplicated Hmx3 and Hmx2 genes revealed that they are required for normal vestibular development in mice (Hadrys et al., 1998; Mennerich et al., 1999; Wang et al., 2001; Wang et al., 2004; Wang and Lufkin, 2005; Wang et al., 1998). In addition, some patients with hemizygous deletions encompassing HMX2 and HMX3 present with vestibular dysfunction, congenital sensorineural hearing loss, and inner ear malformations, implicating these genes in human, as well as mouse, inner ear development (Miller et al., 2009). However, the abnormalities described are more consistent with roles in otic vesicle morphogenesis than in otic placode induction. A third member of this gene family, Hmx1, is expressed in the developing otic vesicle (Munroe et al., 2009; Yoshiura et al., 1998), but apparently has no unique role in inner ear development (Munroe et al., 2009). If Hmx1 is expressed in the placode, then triple mutant analysis with Hmx2 and Hmx3 could reveal a role for these genes in otic placode induction. Interestingly, consistent with our findings of FGF responsiveness of Hmx2 and Hmx3 during otic placode induction, ectodermal Nkx5.1 (Hmx3) is also down-regulated in zebrafish Fgf8 (ace) mutants (Adamska et al., 2000) and otocyst-expressed Hmx2 and Hmx3 are down-regulated in SU-5402-treated zebrafish embryos (Feng and Xu, 2009).
Foxg1, another otic placode-expressed transcription factor gene identified in our microarray study, is also expressed in and required for normal morphogenesis, as well as for innervation of the vestibular system (Hwang et al., 2009; Pauley et al., 2006). The Foxg1-/- abnormalities are related to, but much milder than the Fgf10-/- vestibular defects, suggesting that Foxg1 may be directly or indirectly regulated by FGF signaling during otic vesicle morphogenesis, similar to its behavior during placode induction.
Sox9, which encodes an SRY-box transcription factor, was also strongly down-regulated in Fgf3-/-;Fgf10-/- mutants. Sox9 is required for otic placode specification in Xenopus and zebrafish (Liu et al., 2003a; Saint-Germain et al., 2004) and its expression is clearly dependent upon FGF signaling; it is down-regulated both in Fgf3 and Fgf8-deficient zebrafish otic ectoderm (Liu et al., 2003a) and Fgf3 and Fgf10-deficient mouse otic ectoderm (this study). Importantly, Sox9 otic conditional mutants show normal otic placode induction, but the next step of otic development, namely, placode invagination to form a cup, fails (Barrionuevo et al., 2008). Thus, our differential expression screen is clearly capable of identifying FGF-regulated genes that are critical for both otic placode and vesicle morphogenesis.
Only two validated target genes, Has2 and Slc26a9, do not encode transcription factors. Has2 encodes hyaluronan synthase-2, which catalyzes the synthesis of hyaluronan, an extracellular glycosaminoglycan. In Xenopus inner ears, hyaluronan is found in the space between the mesenchyme and the axial protrusions of otic epithelium that eventually fuse to form the semicircular canals, where it serves as a propellant in the fusion process (Haddon and Lewis, 1991). Although the mechanism of semicircular canal formation differs somewhat between frogs and mice, hyaluronan and Has2 expression are also found in the developing mouse inner ear (McPhee et al., 1987; Tien and Spicer, 2005). Unfortunately, global Has2-/- mice die too early to address its roles in semicircular canal formation (Camenisch et al., 2000). It will be interesting to study semicircular canal morphogenesis using the recently reported Has2 conditional strain (Matsumoto et al., 2009). Has2-/- embryos do form an otic vesicle; therefore, Has2 does not seem to be required for placode induction. A role for Has2 in otic induction could be masked, however, by redundancy with other gene family members (Tien and Spicer, 2005).
Slc26a9, which encodes a chloride channel required for gastric acid secretion (Dorwart et al., 2008; Xu et al., 2008), has not been associated previously with otic expression or development. However, its relatives in the SLC26 family of solute carriers--Slc26a4, encoding Pendrin, and Slc26a5, encoding Prestin--play critical roles in mouse ear development and function (Everett et al., 2001; Liberman et al., 2002) and are mutated in human hearing loss subjects (Everett et al., 1997; Li et al., 1998; Liu et al., 2003b). Studies of auditory function in Slc26a9-/- mice are underway.
Expression of MAPK pathway targets, Fgf4, and Foxi genes is altered in Fgf3-/-;Fgf10-/- mutants
Concomitant with the microarray analysis, we also determined the FGF responsiveness of pre-otic field genes identified in other studies. Not surprisingly, common transcriptional targets of the MAPK pathway mediating FGF signals, Erm and Spry1, were absent from dorsal double mutant ectoderm. Similarly Fgf4, which is the only Fgf, other than Fgf3 itself, expressed in the mouse otic placode (McKay et al., 1996; Wright et al., 2003; Wright and Mansour, 2003a) was also lost from dorsal ectoderm. This suggests that FGF4 might be required downstream of FGF3 and FGF10 to maintain or augment the placodal response to FGF3 and FGF10. Mice lacking Fgf4 die during pre-implantation (Feldman et al., 1995), but Tg(Pax2-cre)-mediated otic conditional ablation of Fgf4 did not reveal a unique role for this FGF in otic induction or subsequent function of the inner ear (S. L. Mansour and E. P. Hatch, unpublished). Therefore, to determine the role of Fgf4 in the otic placode it will be necessary first to generate a conditional Fgf3-Fgf4 deletion allele, as these Fgf genes are located within 20 kb of one another.
Foxi1 is a master regulator of otic development in zebrafish, and appears to function in a pathway parallel to that of FGF signaling (Hans et al., 2007; Solomon et al., 2004). Mouse Foxi1 is expressed and required at a later stage of inner ear development (Hulander et al., 2003), however, the expression patterns of mouse Foxi family members, Foxi3 and Foxi2, coincide spatiotemporally with the period of otic induction. Foxi3 is expressed in a broad ectodermal region beginning at E6.5, well before otic induction, and is maintained until approximately the 8-somite stage, when it starts to be down-regulated in the placode (Ohyama and Groves, 2004). Foxi3 was virtually absent from the dorsal ectoderm of 4-7-somite Fgf3-/-;Fgf10-/- embryos, but could be detected in 8-9-somite double mutants, suggesting that in the mouse, there are both FGF-dependent and independent pathways of Foxi3 activation. It will be interesting to learn whether mouse Foxi3 is the functional ortholog of zebrafish Foxi1 in otic placode induction, but even if so, there are clearly differences between the two species with respect to the relationship between Foxi1/3 genes and FGF signaling.
FGF signaling is required not only to specify the pre-otic domain, but also to regulate the WNT signaling that restricts the otic placode to dorsal ectoderm
Foxi2 is initially expressed throughout the cranial ectoderm, including in the presumptive placode and more ventral ectoderm that is fated to become epibranchial placode or epidermis, but is subsequently excluded from the dorsal ectoderm as otic placode fate is specified. Exclusion is particularly evident by 11-somites as the placode thickens in preparation for invagination (Ohyama and Groves, 2004). The expanded domain of ectodermal Foxi2 expression we found in 7-8-somite Fgf3-/-;Fgf10-/- mutants, and the failure of the hindbrain proximal ectoderm to extinguish Foxi2 expression at 11-12 somites is similar to the behavior of Foxi2 when WNT signal reception in the ectoderm is blocked by Tg(Pax2-cre)-mediated otic conditional ablation of ß-catenin (Ohyama et al., 2006). Since WNT signals, presumably emanating from the hindbrain, have long been implicated in otic induction (Freter et al., 2008; Ladher et al., 2000; Ohyama et al., 2006; Park and Saint-Jeannet, 2008), this result suggested that FGF signaling might control the WNT signal that restricts the otic domain. Indeed, we found that FGF signaling is required for r4 expression of Wnt8a, the only hindbrain-expressed gene affected among those tested here and previously (Wright and Mansour, 2003a), suggesting that WNT8A is the signal, or a component of the WNT signal, that restricts the otic placode to the hindbrain proximal portion of the Pax2-positive pre-otic field. We also found that Wnt8a expression was reduced or absent in some Fgf3-/-;Fgf10-/+ and Fgf3-/- embryos, but not in Fgf3-/+;Fgf10-/- or Fgf10-/- embryos, suggesting that Fgf3 may be the main regulator of Wnt8a (Fig. 6). Furthermore, our chick explant studies showed that FGF3 and, more potently, FGF4, were able to induce Wnt8a expression, whereas FGF10 was not. Taken together with the mouse loss of function data, these results show that FGF3 is both necessary and sufficient for Wnt8a expression, whereas FGF4 is sufficient but not uniquely necessary, and FGF10 is necessary but not sufficient. Preliminary data suggest that this FGF/WNT regulatory pathway is unidirectional, as r5-r6 expression of Fgf3 was unperturbed in embryos with Tg(Wnt1-cre)-mediated conditional deletion of ß-catenin (S. L. Mansour and E. P. Hatch, unpublished).
Canonical WNT/ß-catenin signaling relies on activation of Frizzled (Fzd) receptors (Angers and Moon, 2009; MacDonald et al., 2009; van Amerongen and Nusse, 2009) and consistent with this expectation we found two Fzd genes that are expressed in the prospective otic territory. Within the caudal cranial region, Fzd1 was exclusive to the otic placode, whereas Fzd8 was more widely expressed but was strongest within the otic domain. In chick, Fzd1, Fzd2, and Fzd7 are expressed at otic placode and otic vesicle stages (Sienknecht and Fekete, 2009; Stark et al., 2000), suggesting roles, at least for Fzd1, in otic development. Genetic loss-of-function analyses will be necessary to test the roles of Wnt8a and the Fzd genes in the restriction of the otic placode.
FGFs and WNTs have interactive roles at multiple stages of inner ear development. Otic induction is initiated by FGF signaling, with WNT limiting the final otic domain (Freter et al., 2008; Ladher et al., 2000; Ohyama et al., 2006; Park and Saint-Jeannet, 2008; this study; Whitfield and Hammond, 2007). In contrast, the first stage of otic vesicle morphogenesis (outgrowth of the endolymphatic duct) is initiated by WNT signaling and refined and maintained by FGF signaling (Hatch et al., 2007; Riccomagno et al., 2005). At later stages of inner ear development, non-canonical WNT signaling regulates the patterning of the cochlear sensory epithelium (Dabdoub and Kelley, 2005; Qian et al., 2007; Yamamoto et al., 2008), and FGF signaling directs outgrowth and morphogenesis of the semicircular canals (Chang et al., 2004; Ohuchi et al., 2005; Pauley et al., 2003). It will be interesting to learn whether there is WNT-FGF cross-regulation during either of these two processes.
Supplementary Material
Acknowledgments
We thank Jeff Barrow, Anne Boulet, Andy Groves, Andy McMahon, Charlie Murtaugh, Kathy Shim, and Terri Yamaguchi, who supplied plasmids used to generate riboprobes; Yukio Saijoh, Susan Chapman, and Ellen Huang, who provided technical advice and assistance; Brian Dalley and Brett Milash (University of Utah Microarray Core), who assisted with design and analysis of the microarray study; and Mario Capecchi, who supplied HprtCre mice and assisted with generation of Fgf10Pc mice. This work was supported by grants from the University of Utah (Catalyst, G.C.S, S.L.M), American Hearing Research Foundation (S.L.M), Deafness Research Foundation (L.D.U., C.N.P.) and the NIH (DC004185 to G.C.S., S.L.M. and DC005608 to S.L.M.).
Footnotes
Supplementary Material Table S1. Excel file containing genes significantly up- or down- regulated in FGF-deficient ectoderm.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abler LL, Mansour SL, Sun X. Conditional gene inactivation reveals roles for Fgf10 and Fgfr2 in establishing a normal pattern of epithelial branching in the mouse lung. Dev Dyn. 2009;238:1999–2013. doi: 10.1002/dvdy.22032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamska M, Leger S, Brand M, Hadrys T, Braun T, Bober E. Inner ear and lateral line expression of a zebrafish Nkx5-1 gene and its downregulation in the ears of FGF8 mutant, ace. Mech Dev. 2000;97:161–5. doi: 10.1016/s0925-4773(00)00414-7. [DOI] [PubMed] [Google Scholar]
- Alvarez Y, Alonso MT, Vendrell V, Zelarayan LC, Chamero P, Theil T, Bosl MR, Kato S, Maconochie M, Riethmacher D, Schimmang T. Requirements for FGF3 and FGF10 during inner ear formation. Development. 2003;130:6329–38. doi: 10.1242/dev.00881. [DOI] [PubMed] [Google Scholar]
- Angers S, Moon RT. Proximal events in Wnt signal transduction. Nat Rev Mol Cell Biol. 2009;10:468–77. doi: 10.1038/nrm2717. [DOI] [PubMed] [Google Scholar]
- Barrionuevo F, Naumann A, Bagheri-Fam S, Speth V, Taketo MM, Scherer G, Neubuser A. Sox9 is required for invagination of the otic placode in mice. Dev Biol. 2008;317:213–24. doi: 10.1016/j.ydbio.2008.02.011. [DOI] [PubMed] [Google Scholar]
- Bouillet P, Oulad-Abdelghani M, Ward SJ, Bronner S, Chambon P, Dolle P. A new mouse member of the Wnt gene family, mWnt-8, is expressed during early embryogenesis and is ectopically induced by retinoic acid. Mech Dev. 1996;58:141–52. doi: 10.1016/s0925-4773(96)00569-2. [DOI] [PubMed] [Google Scholar]
- Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 2004;573:83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- Camenisch TD, Spicer AP, Brehm-Gibson T, Biesterfeldt J, Augustine ML, Calabro A, Jr, Kubalak S, Klewer SE, McDonald JA. Disruption of hyaluronan synthase-2 abrogates normal cardiac morphogenesis and hyaluronan-mediated transformation of epithelium to mesenchyme. J Clin Invest. 2000;106:349–60. doi: 10.1172/JCI10272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W, Brigande JV, Fekete DM, Wu DK. The development of semicircular canals in the inner ear: role of FGFs in sensory cristae. Development. 2004;131:4201–11. doi: 10.1242/dev.01292. [DOI] [PubMed] [Google Scholar]
- Dabdoub A, Kelley MW. Planar cell polarity and a potential role for a Wnt morphogen gradient in stereociliary bundle orientation in the mammalian inner ear. J Neurobiol. 2005;64:446–57. doi: 10.1002/neu.20171. [DOI] [PubMed] [Google Scholar]
- Dorwart MR, Shcheynikov N, Yang D, Muallem S. The solute carrier 26 family of proteins in epithelial ion transport. Physiology (Bethesda) 2008;23:104–14. doi: 10.1152/physiol.00037.2007. [DOI] [PubMed] [Google Scholar]
- Everett LA, Belyantseva IA, Noben-Trauth K, Cantos R, Chen A, Thakkar SI, Hoogstraten-Miller SL, Kachar B, Wu DK, Green ED. Targeted disruption of mouse Pds provides insight about the inner-ear defects encountered in Pendred syndrome. Hum Mol Genet. 2001;10:153–61. doi: 10.1093/hmg/10.2.153. [DOI] [PubMed] [Google Scholar]
- Everett LA, Glaser B, Beck JC, Idol JR, Buchs A, Heyman M, Adawi F, Hazani E, Nassir E, Baxevanis AD, Sheffield VC, Green ED. Pendred syndrome is caused by mutations in a putative sulphate transporter gene (PDS) Nat Genet. 1997;17:411–22. doi: 10.1038/ng1297-411. [DOI] [PubMed] [Google Scholar]
- Farley FW, Soriano P, Steffen LS, Dymecki SM. Widespread recombinase expression using FLPeR (flipper) mice. Genesis. 2000;28:106–10. [PubMed] [Google Scholar]
- Feldman B, Poueymirou W, Papaioannou VE, DeChiara TM, Goldfarb M. Requirement of FGF-4 for postimplantation mouse development. Science. 1995;267:246–9. doi: 10.1126/science.7809630. [DOI] [PubMed] [Google Scholar]
- Feng Y, Xu Q. Pivotal role of hmx2 and hmx3 in zebrafish inner ear and lateral line development. Dev Biol. 2009 doi: 10.1016/j.ydbio.2009.12.028. [DOI] [PubMed] [Google Scholar]
- Freter S, Muta Y, Mak SS, Rinkwitz S, Ladher RK. Progressive restriction of otic fate: the role of FGF and Wnt in resolving inner ear potential. Development. 2008;135:3415–24. doi: 10.1242/dev.026674. [DOI] [PubMed] [Google Scholar]
- Gavin BJ, McMahon JA, McMahon AP. Expression of multiple novel Wnt-1/int-1-related genes during fetal and adult mouse development. Genes Dev. 1990;4:2319–32. doi: 10.1101/gad.4.12b.2319. [DOI] [PubMed] [Google Scholar]
- Groves AK, Bronner-Fraser M. Competence, specification and commitment in otic placode induction. Development. 2000;127:3489–99. doi: 10.1242/dev.127.16.3489. [DOI] [PubMed] [Google Scholar]
- Haddon CM, Lewis JH. Hyaluronan as a propellant for epithelial movement: the development of semicircular canals in the inner ear of Xenopus. Development. 1991;112:541–50. doi: 10.1242/dev.112.2.541. [DOI] [PubMed] [Google Scholar]
- Hadrys T, Braun T, Rinkwitz-Brandt S, Arnold HH, Bober E. Nkx5-1 controls semicircular canal formation in the mouse inner ear. Development. 1998;125:33–9. doi: 10.1242/dev.125.1.33. [DOI] [PubMed] [Google Scholar]
- Hans S, Christison J, Liu D, Westerfield M. Fgf-dependent otic induction requires competence provided by Foxi1 and Dlx3b. BMC Dev Biol. 2007;7:5. doi: 10.1186/1471-213X-7-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch EP, Noyes CA, Wang X, Wright TJ, Mansour SL. Fgf3 is required for dorsal patterning and morphogenesis of the inner ear epithelium. Development. 2007;134:3615–25. doi: 10.1242/dev.006627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan B, Beddington R, Costantini FEL. A Laboratory Manual. ColdSpring Harbor Laboratory Press; Cold Spring Harbor: 1994. Manipulating the Mouse Embryo. [Google Scholar]
- Hulander M, Kiernan AE, Blomqvist SR, Carlsson P, Samuelsson EJ, Johansson BR, Steel KP, Enerback S. Lack of pendrin expression leads to deafness and expansion of the endolymphatic compartment in inner ears of Foxi1 null mutant mice. Development. 2003;130:2013–25. doi: 10.1242/dev.00376. [DOI] [PubMed] [Google Scholar]
- Hume CR, Dodd J. Cwnt-8C: a novel Wnt gene with a potential role in primitive streak formation and hindbrain organization. Development. 1993;119:1147–60. doi: 10.1242/dev.119.4.1147. [DOI] [PubMed] [Google Scholar]
- Hwang CH, Simeone A, Lai E, Wu DK. Foxg1 is required for proper separation and formation of sensory cristae during inner ear development. Dev Dyn. 2009;238:2725–34. doi: 10.1002/dvdy.22111. [DOI] [PubMed] [Google Scholar]
- Jacobson AG. Inductive processes in embryonic development. Science. 1966;152:25–34. doi: 10.1126/science.152.3718.25. [DOI] [PubMed] [Google Scholar]
- Ladher RK, Anakwe KU, Gurney AL, Schoenwolf GC, Francis-West PH. Identification of synergistic signals initiating inner ear development. Science. 2000;290:1965–7. doi: 10.1126/science.290.5498.1965. [DOI] [PubMed] [Google Scholar]
- Ladher RK, Wright TJ, Moon AM, Mansour SL, Schoenwolf GC. FGF8 initiates inner ear induction in chick and mouse. Genes Dev. 2005;19:603–13. doi: 10.1101/gad.1273605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Scott DA, Hatch E, Tian X, Mansour SL. Dusp6 (Mkp3) is a negative feedback regulator of FGF-stimulated ERK signaling during mouse development. Development. 2007;134:167–76. doi: 10.1242/dev.02701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XC, Everett LA, Lalwani AK, Desmukh D, Friedman TB, Green ED, Wilcox ER. A mutation in PDS causes non-syndromic recessive deafness. Nat Genet. 1998;18:215–7. doi: 10.1038/ng0398-215. [DOI] [PubMed] [Google Scholar]
- Liberman MC, Gao J, He DZ, Wu X, Jia S, Zuo J. Prestin is required for electromotility of the outer hair cell and for the cochlear amplifier. Nature. 2002;419:300–4. doi: 10.1038/nature01059. [DOI] [PubMed] [Google Scholar]
- Liu D, Chu H, Maves L, Yan YL, Morcos PA, Postlethwait JH, Westerfield M. Fgf3 and Fgf8 dependent and independent transcription factors are required for otic placode specification. Development. 2003a;130:2213–24. doi: 10.1242/dev.00445. [DOI] [PubMed] [Google Scholar]
- Liu XZ, Ouyang XM, Xia XJ, Zheng J, Pandya A, Li F, Du LL, Welch KO, Petit C, Smith RJ, Webb BT, Yan D, Arnos KS, Corey D, Dallos P, Nance WE, Chen ZY. Prestin, a cochlear motor protein, is defective in non-syndromic hearing loss. Hum Mol Genet. 2003b;12:1155–62. doi: 10.1093/hmg/ddg127. [DOI] [PubMed] [Google Scholar]
- MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour SL, Goddard JM, Capecchi MR. Mice homozygous for a targeted disruption of the proto-oncogene int-2 have developmental defects in the tail and inner ear. Development. 1993;117:13–28. doi: 10.1242/dev.117.1.13. [DOI] [PubMed] [Google Scholar]
- Martin K, Groves AK. Competence of cranial ectoderm to respond to Fgf signaling suggests a two-step model of otic placode induction. Development. 2006;133:877–87. doi: 10.1242/dev.02267. [DOI] [PubMed] [Google Scholar]
- Matsumoto K, Li Y, Jakuba C, Sugiyama Y, Sayo T, Okuno M, Dealy CN, Toole BP, Takeda J, Yamaguchi Y, Kosher RA. Conditional inactivation of Has2 reveals a crucial role for hyaluronan in skeletal growth, patterning, chondrocyte maturation and joint formation in the developing limb. Development. 2009;136:2825–35. doi: 10.1242/dev.038505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe KL, Bronner-Fraser M. Molecular and tissue interactions governing induction of cranial ectodermal placodes. Dev Biol. 2009;332:189–95. doi: 10.1016/j.ydbio.2009.05.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay IJ, Lewis J, Lumsden A. The role of FGF-3 in early inner ear development: an analysis in normal and kreisler mutant mice. Dev Biol. 1996;174:370–8. doi: 10.1006/dbio.1996.0081. [DOI] [PubMed] [Google Scholar]
- McPhee JR, Van de Water TR, Su HX. Hyaluronate production by the inner ear during otic capsule and perilymphatic space formation. Am J Otolaryngol. 1987;8:265–72. doi: 10.1016/s0196-0709(87)80045-5. [DOI] [PubMed] [Google Scholar]
- Mennerich D, Hoffmann S, Hadrys T, Arnold HH, Bober E. Two highly related homeodomain proteins, Nkx5-1 and Nkx5-2, display different DNA binding specificities. Biol Chem. 1999;380:1041–8. doi: 10.1515/BC.1999.130. [DOI] [PubMed] [Google Scholar]
- Miller ND, Nance MA, Wohler ES, Hoover-Fong JE, Lisi E, Thomas GH, Pevsner J. Molecular (SNP) analyses of overlapping hemizygous deletions of 10q25.3 to 10qter in four patients: evidence for HMX2 and HMX3 as candidate genes in hearing and vestibular function. Am J Med Genet A. 2009;149A:669–80. doi: 10.1002/ajmg.a.32705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min H, Danilenko DM, Scully SA, Bolon B, Ring BD, Tarpley JE, DeRose M, Simonet WS. Fgf-10 is required for both limb and lung development and exhibits striking functional similarity to Drosophila branchless. Genes Dev. 1998;12:3156–61. doi: 10.1101/gad.12.20.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minowada G, Jarvis LA, Chi CL, Neubuser A, Sun X, Hacohen N, Krasnow MA, Martin GR. Vertebrate Sprouty genes are induced by FGF signaling and can cause chondrodysplasia when overexpressed. Development. 1999;126:4465–75. doi: 10.1242/dev.126.20.4465. [DOI] [PubMed] [Google Scholar]
- Munroe RJ, Prabhu V, Acland GM, Johnson KR, Harris BS, O’Brien TP, Welsh IC, Noden DM, Schimenti JC. Mouse H6 Homeobox 1 (Hmx1) mutations cause cranial abnormalities and reduced body mass. BMC Dev Biol. 2009;9:27. doi: 10.1186/1471-213X-9-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohuchi H, Yasue A, Ono K, Sasaoka S, Tomonari S, Takagi A, Itakura M, Moriyama K, Noji S, Nohno T. Identification of cis-element regulating expression of the mouse Fgf10 gene during inner ear development. Dev Dyn. 2005;233:177–87. doi: 10.1002/dvdy.20319. [DOI] [PubMed] [Google Scholar]
- Ohyama T. Unraveling inner ear induction by gene manipulation using Pax2-Cre BAC transgenic mice. Brain Res. 2009;1277:84–9. doi: 10.1016/j.brainres.2009.02.036. [DOI] [PubMed] [Google Scholar]
- Ohyama T, Groves AK. Expression of mouse Foxi class genes in early craniofacial development. Dev Dyn. 2004;231:640–6. doi: 10.1002/dvdy.20160. [DOI] [PubMed] [Google Scholar]
- Ohyama T, Groves AK, Martin K. The first steps towards hearing: mechanisms of otic placode induction. Int J Dev Biol. 2007;51:463–72. doi: 10.1387/ijdb.072320to. [DOI] [PubMed] [Google Scholar]
- Ohyama T, Mohamed OA, Taketo MM, Dufort D, Groves AK. Wnt signals mediate a fate decision between otic placode and epidermis. Development. 2006;133:865–75. doi: 10.1242/dev.02271. [DOI] [PubMed] [Google Scholar]
- Park BY, Saint-Jeannet JP. Hindbrain-derived Wnt and Fgf signals cooperate to specify the otic placode in Xenopus. Dev Biol. 2008;324:108–21. doi: 10.1016/j.ydbio.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauley S, Lai E, Fritzsch B. Foxg1 is required for morphogenesis and histogenesis of the mammalian inner ear. Dev Dyn. 2006;235:2470–82. doi: 10.1002/dvdy.20839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauley S, Wright TJ, Pirvola U, Ornitz D, Beisel K, Fritzsch B. Expression and function of FGF10 in mammalian inner ear development. Dev Dyn. 2003;227:203–15. doi: 10.1002/dvdy.10297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian D, Jones C, Rzadzinska A, Mark S, Zhang X, Steel KP, Dai X, Chen P. Wnt5a functions in planar cell polarity regulation in mice. Dev Biol. 2007;306:121–33. doi: 10.1016/j.ydbio.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccomagno MM, Takada S, Epstein DJ. Wnt-dependent regulation of inner ear morphogenesis is balanced by the opposing and supporting roles of Shh. Genes Dev. 2005;19:1612–23. doi: 10.1101/gad.1303905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Germain N, Lee YH, Zhang Y, Sargent TD, Saint-Jeannet JP. Specification of the otic placode depends on Sox9 function in Xenopus. Development. 2004;131:1755–63. doi: 10.1242/dev.01066. [DOI] [PubMed] [Google Scholar]
- Schimmang T. Expression and functions of FGF ligands during early otic development. Int J Dev Biol. 2007;51:473–81. doi: 10.1387/ijdb.072334ts. [DOI] [PubMed] [Google Scholar]
- Schlosser G. Induction and specification of cranial placodes. Dev Biol. 2006;294:303–51. doi: 10.1016/j.ydbio.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Schlosser G, Ahrens K. Molecular anatomy of placode development in Xenopus laevis. Dev Biol. 2004;271:439–66. doi: 10.1016/j.ydbio.2004.04.013. [DOI] [PubMed] [Google Scholar]
- Schwenk F, Baron U, Rajewsky K. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 1995;23:5080–1. doi: 10.1093/nar/23.24.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sechrist J, Nieto MA, Zamanian RT, Bronner-Fraser M. Regulative response of the cranial neural tube after neural fold ablation: spatiotemporal nature of neural crest regeneration and up-regulation of Slug. Development. 1995;121:4103–15. doi: 10.1242/dev.121.12.4103. [DOI] [PubMed] [Google Scholar]
- Sienknecht UJ, Fekete DM. Mapping of Wnt, frizzled, and Wnt inhibitor gene expression domains in the avian otic primordium. J Comp Neurol. 2009;517:751–64. doi: 10.1002/cne.22169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon KS, Kwak SJ, Fritz A. Genetic interactions underlying otic placode induction and formation. Dev Dyn. 2004;230:419–33. doi: 10.1002/dvdy.20067. [DOI] [PubMed] [Google Scholar]
- Stark MR, Biggs JJ, Schoenwolf GC, Rao MS. Characterization of avian frizzled genes in cranial placode development. Mech Dev. 2000;93:195–200. doi: 10.1016/s0925-4773(00)00263-x. [DOI] [PubMed] [Google Scholar]
- Streit A. The preplacodal region: an ectodermal domain with multipotential progenitors that contribute to sense organs and cranial sensory ganglia. Int J Dev Biol. 2007;51:447–61. doi: 10.1387/ijdb.072327as. [DOI] [PubMed] [Google Scholar]
- Tang SH, Silva FJ, Tsark WM, Mann JR. A Cre/loxP-deleter transgenic line in mouse strain 129S1/SvImJ. Genesis. 2002;32:199–202. doi: 10.1002/gene.10030. [DOI] [PubMed] [Google Scholar]
- Tien JY, Spicer AP. Three vertebrate hyaluronan synthases are expressed during mouse development in distinct spatial and temporal patterns. Dev Dyn. 2005;233:130–41. doi: 10.1002/dvdy.20328. [DOI] [PubMed] [Google Scholar]
- van Amerongen R, Nusse R. Towards an integrated view of Wnt signaling in development. Development. 2009;136:3205–14. doi: 10.1242/dev.033910. [DOI] [PubMed] [Google Scholar]
- Wang W, Chan EK, Baron S, Van de Water T, Lufkin T. Hmx2 homeobox gene control of murine vestibular morphogenesis. Development. 2001;128:5017–29. doi: 10.1242/dev.128.24.5017. [DOI] [PubMed] [Google Scholar]
- Wang W, Grimmer JF, Van De Water TR, Lufkin T. Hmx2 and Hmx3 homeobox genes direct development of the murine inner ear and hypothalamus and can be functionally replaced by Drosophila Hmx. Dev Cell. 2004;7:439–53. doi: 10.1016/j.devcel.2004.06.016. [DOI] [PubMed] [Google Scholar]
- Wang W, Lufkin T. Hmx homeobox gene function in inner ear and nervous system cell-type specification and development. Exp Cell Res. 2005;306:373–9. doi: 10.1016/j.yexcr.2005.03.016. [DOI] [PubMed] [Google Scholar]
- Wang W, Van De Water T, Lufkin T. Inner ear and maternal reproductive defects in mice lacking the Hmx3 homeobox gene. Development. 1998;125:621–34. doi: 10.1242/dev.125.4.621. [DOI] [PubMed] [Google Scholar]
- Whitfield TT, Hammond KL. Axial patterning in the developing vertebrate inner ear. Int J Dev Biol. 2007;51:507–20. doi: 10.1387/ijdb.072380tw. [DOI] [PubMed] [Google Scholar]
- Wilkinson DG. Whole mount in situ hybridization of vertebrate embryos. In: Wilkinson DG, editor. In Situ Hybridization: A Practical Approach. IRL Press; Oxford: 1992. pp. 75–83. [Google Scholar]
- Wright E, Hargrave MR, Christiansen J, Cooper L, Kun J, Evans T, Gangadharan U, Greenfield A, Koopman P. The Sry-related gene Sox9 is expressed during chondrogenesis in mouse embryos. Nat Genet. 1995;9:15–20. doi: 10.1038/ng0195-15. [DOI] [PubMed] [Google Scholar]
- Wright TJ, Hatch EP, Karabagli H, Karabagli P, Schoenwolf GC, Mansour SL. Expression of mouse fibroblast growth factor and fibroblast growth factor receptor genes during early inner ear development. Dev Dyn. 2003;228:267–72. doi: 10.1002/dvdy.10362. [DOI] [PubMed] [Google Scholar]
- Wright TJ, Ladher R, McWhirter J, Murre C, Schoenwolf GC, Mansour SL. Mouse FGF15 is the ortholog of human and chick FGF19, but is not uniquely required for otic induction. Dev Biol. 2004;269:264–75. doi: 10.1016/j.ydbio.2004.02.003. [DOI] [PubMed] [Google Scholar]
- Wright TJ, Mansour SL. Fgf3 and Fgf10 are required for mouse otic placode induction. Development. 2003a;130:3379–90. doi: 10.1242/dev.00555. [DOI] [PubMed] [Google Scholar]
- Wright TJ, Mansour SL. FGF signaling in ear development and innervation. Curr Top Dev Biol. 2003b;57:225–59. doi: 10.1016/s0070-2153(03)57008-9. [DOI] [PubMed] [Google Scholar]
- Xu J, Song P, Miller ML, Borgese F, Barone S, Riederer B, Wang Z, Alper SL, Forte JG, Shull GE, Ehrenfeld J, Seidler U, Soleimani M. Deletion of the chloride transporter Slc26a9 causes loss of tubulovesicles in parietal cells and impairs acid secretion in the stomach. Proc Natl Acad Sci U S A. 2008;105:17955–60. doi: 10.1073/pnas.0800616105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S, Nishimura O, Misaki K, Nishita M, Minami Y, Yonemura S, Tarui H, Sasaki H. Cthrc1 selectively activates the planar cell polarity pathway of Wnt signaling by stabilizing the Wnt-receptor complex. Dev Cell. 2008;15:23–36. doi: 10.1016/j.devcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Yoshiura K, Leysens NJ, Reiter RS, Murray JC. Cloning, characterization, and mapping of the mouse homeobox gene Hmx1. Genomics. 1998;50:61–8. doi: 10.1006/geno.1998.5284. [DOI] [PubMed] [Google Scholar]
- Zelarayan LC, Vendrell V, Alvarez Y, Dominguez-Frutos E, Theil T, Alonso MT, Maconochie M, Schimmang T. Differential requirements for FGF3, FGF8 and FGF10 during inner ear development. Dev Biol. 2007;308:379–91. doi: 10.1016/j.ydbio.2007.05.033. [DOI] [PubMed] [Google Scholar]
- Zhang P, Li MZ, Elledge SJ. Towards genetic genome projects: genomic library screening and gene-targeting vector construction in a single step. Nat Genet. 2002;30:31–9. doi: 10.1038/ng797. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.