Summary
The generation of B lymphocytes from hematopoietic progenitors requires lineage-specific transcription factors that progressively direct cell fate choices. Differentiation of hematopoietic stem cells to lymphoid progenitors requires Ikaros-dependent lineage priming and graded levels of PU.1, which are controlled by Ikaros and Gfi1. E2A drives expression of EBF1, which initiates B lineage specification. EBF1, in addition to Pax5, is involved in commitment to the B cell lineage. As a model of gene activation in early B lymphopoiesis, mb-1 genes are activated sequentially by factors (e.g. EBF1) that initiate chromatin modifications prior to transcription. This review highlights the requisite interplay between transcription factors and epigenetic mechanisms in the context of B cell development.
Introduction
Pluripotent hematopoietic stem cells (HSCs) undergo successive rounds of lineage fate restrictions resulting in the specialized cells of the blood (Figure 1). HSCs yield multipotent progenitors (MPPs) that give rise to lymphoid-primed multipotent progenitors (LMPPs) and common lymphoid progenitors (CLPs). The transcription factors E box binding protein 2A (E2A) and Early B cell Factor-1 (EBF1) direct CLPs into the B cell developmental pathway. Together with Paired box protein 5 (Pax5) and Ikaros, these factors initiate the progressive steps of V(D)J recombination and expression of accessory proteins required for the display of pre- and mature B cell receptors (BCRs) [reviewed in 1]. Signaling via functional BCR complexes promotes the egress of immature B cells from the bone marrow. In the periphery, differentiation culminates with the production of antigen-experienced plasma and memory cells.
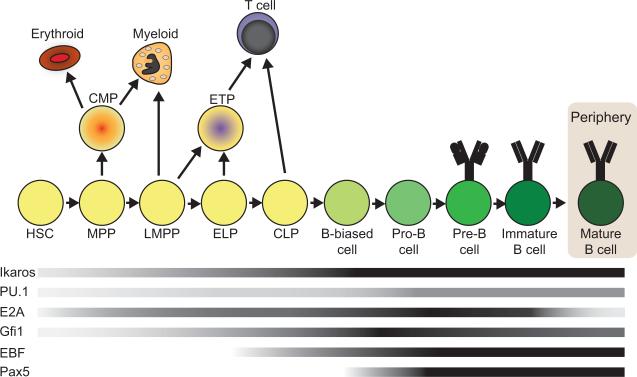
Figure 1.
Transcription factor expression during B lymphopoiesis. The progression of cells from hematopoietic stem cells through stages of B lymphopoiesis is shown. Shaded bars represent the levels of gene expression of transcription factors that are important in B cell development during the course of differentiation. HSCs (hematopoietic stem cells), MPPs (multipotent progenitors), LMPPs (lymphoid-primed MPPs), ELPs (early lymphoid progenitors), CLPs (common lymphoid progenitors), ETPs (early T lineage progenitors). Darker shading indicates increased gene expression. Important branch points during B lymphopoiesis are shown with arrows indicating alternative developmental pathways. Expression profiles were compiled using data from RefDIC (Reference Database of Immune Cells) http://refdic.rcai.riken.jp/profile.cgi and citations in the text.
In coining the term `epigenetics,' Conrad Waddington asserted the importance of regulatory mechanisms above and beyond the genes themselves [2]. Expression of the B cell program requires lineage-specific transcription factors which recruit proteins that mediate epigenetic changes in chromatin structure, including post-translational modifications of histones and DNA methylation. This review highlights recent advances concerning factors that govern early B cell development by modulating gene expression.
Regulation in progenitors prior to B lineage specification: Ikaros, PU.1 and E2A
HSC multipotency derives from low-level expression of lineage-affiliated genes associated with multiple hematopoietic backgrounds [3,4]. These genes possess `bivalent' chromatin in which both activating and repressive histone and DNA modifications are present simultaneously [5, *6, 7]. Primed regions of chromatin have been described at early developmental stages in many different systems, and are important in numerous developmental contexts for the rapid activation of poised genes after receiving appropriate cues [8]. Primed chromatin is characterized by greater accessibility, reduced nucleosome occupancy, low levels of associated RNAPII, and the presence of H3K4 mono-, di-, or trimethylation. In response to environmental stimuli, the expression of primed genes is modulated by a hierarchical network of transcription factors and epigenetic regulators. Consequently, dominant transcriptional programs emerge and direct differentiation of individual HSCs into discrete hematopoietic lineages. The transcription factors Ikaros, Purine box factor 1 (PU.1) and E2A are essential for priming lineage-associated genes and restricting fates to the B lineage [9–12].
Ikaros (encoded by Ikzf1) is a transcription factor containing variable numbers of Krüppel-like zinc fingers in two domains that mediate DNA binding and formation of dimers and multimeric complexes [13,14]. In Ikaros-deficient mice lymphoid cell development is arrested at the LMPP stage and myeloid development is perturbed [15,16]. Ikaros functions either as a transcriptional activator or repressor by recruiting various chromatin remodeling complexes (CRCs) including SWI/SNF (related to the yeast SWItch/Sucrose Non-Fermenter) or Mi-2/Nucleosome Remodeling and Deacetylase (Mi-2/NuRD) to DNA regulatory elements [17–19]. These associations are likely critical for Ikaros' roles in priming the expression of lineage-specific genes and regulating self-renewal in HSCs [**20,**21]. Subsequently, in LMPPs, Ikaros limits transcripts to the lymphoid and myeloid programs [**20,**22]. The lack of Ikaros in CLPs resulted in preferential development of NK cells at the expense of B and T cells [**21]. Other data suggest that Ikaros promotes B cell identity, because Ikaros-deficient pro-B cells expressing EBF1 and Pax5 were uncommitted [**23]. Following the expression of E2A and EBF1, Ikaros mediates chromatin accessibility necessary for V(D)J recombination. Ikaros also modulates the expression of early B cell-specific genes, including Igll1 (λ5), by competing with EBF1 for DNA binding [**24]. Thus, Ikaros restricts the self-renewal program in early hematopoiesis, while it advances the B lineage program at later stages of development.
PU.1, an Ets family transcription factor encoded by Sfpi1, regulates the bifurcation between myeloid and B lymphoid development [25]. Low concentrations of PU.1 favor the B cell fate, while higher concentrations promote myeloid differentiation. Recent work illuminated pathways that modulate the expression of Sfpi1. Mice lacking the Growth factor independent 1 transcriptional repressor Gfi1 exhibited significant reductions of B lineage cells in the bone marrow and elevated levels of PU.1 in MPPs [26,**27]. The B cell potential of MPPs in Gfi1−/− mice is reduced substantially, but can be reversed by enforced expression of Sfpi1-specific shRNAs. Gfi1 thus represses Sfpi1 expression by displacing PU.1 from its upstream autoregulatory element. Interestingly, the expression of Gfi1 is up-regulated by Ikaros in a subset of MPPs. Thus, Ikaros drives the production of B cells at the expense of granulocytes and macrophages.
In contrast, enforced expression of the C/EBPα transcription factor converted a progenitor B cell line into macrophages in vitro [**28]. Activation of a C/EBPα-estrogen receptor fusion protein rapidly stimulated dramatic alterations in transcription, including up-regulation of myeloid-associated genes and down-regulation of B cell-specific genes such as E2A, EBF1, and Pax5. Several chromatin modifiers were down-regulated, including Hdac11, Polycomb group protein Ezh2 and the DNA methyltransferase DNMT3b. This finding underscores the importance of chromatin modifications in maintaining lineage commitment. Similar to PU.1, C/EBPα must be restrained to allow B cell development.
E2A (encoded by Tcfe2a) is a basic helix-loop-helix transcription factor required for B cell development beyond the pre-pro stage [29,30]. It occurs in two splice variants, E12 and E47 [reviewed in 31]. E2A contributes to the maintenance of the HSC pool [32–34]. Additionally, it is necessary for lymphoid-lineage priming in MPPs, as evidenced by severely reduced numbers of LMPPs and CLPs in the absence of both E2A proteins and reduced expression of lymphoid-specific genes in Tcfe2a−/− LMPPs [**33,**34]. E2A is not necessary for the expression of Ikaros, PU.1 and Gfi1; however, it activates genes in LMPPs synergistically with these factors. E2A is required for initiating and maintaining expression of EBF, Pax5 and the B cell-specific program in pro-B cells, as well as directing B cell maturation in germinal centers [*35]. Although both E12 and E47 are present in LMPPs and CLPs, E47 alone is required for B cell lineage specification [*36]. Ikaros, PU.1 and E2A are therefore essential for transcriptional priming that lays the foundation for commitment to the B cell lineage.
The role of EBF in B cell commitment
EBF1 (encoded by Ebf1) comprises novel DNA-binding and helix-loop-helix domains and is essential for the development of functional B cells [37–39]. EBF1 is expressed first in CLPs and regulates many genes involved in B cell development including mb-1 (Cd79a) and Pax5 [40,41]. Ebf1 transcription is mediated by two promoters that are differentially regulated in B cells [42,*43]. The distal (α) promoter is controlled by interleukin-7 signaling (STAT5), E47 and EBF1 autoregulation. The proximal (β) promoter appears to be the stronger of the two modules and is up-regulated by Pax5, Ets1 and PU.1. Consequently, activation of the Pax5 gene by EBF1 up-regulates Ebf1 transcription. Although EBF1 is not critical for the development of CLPs, EBF1-deficient CLPs displayed reduced expression of B lineage genes in both conventional and single cell expression analyses [*44]. In CLPs, expression of Ebf1 initiates a pre-commitment process in which B cell-specific genes are primed for later expression.
Pax5 is essential for maintaining commitment to the B lineage program [45,**46]. Recently, a requisite role of EBF1 in this process was elucidated. MPPs cultured under B-lymphoid-promoting conditions retained T lymphoid and myeloid potential in the absence of EBF1 [**47]. Accordingly, enforced expression of EBF1 in wild-type MPPs generated B cells while suppressing myeloid cell development by attenuating expression of both PU.1 and C/EBPα. Significantly, EBF1 expression rescued the generation of B cells from Pax5−/− progenitors, while blocking promiscuous differentiation. Thus, the developmental plasticity previously demonstrated in Pax5−/− cells may result, in part, from reduced levels of EBF1.
EBF1 plays an additional role in maintaining B lineage development by preventing Id2- and Id3-mediated inhibition of E47 [**47,*48]. Exogenous expression of E47 in pre-pro B cells induced Id2 and Id3 expression. EBF1 was found to bind the Id2 and Id3 promoters and dramatically down-regulated Id2 and Id3 mRNAs. These results indicate that E47 mediates a negative feedback loop by activating Id2 and Id3 expression, while EBF1 maintains E47 activity through down-regulation of Id2 and Id3. Overexpression of either Id2 or Id3 in wild-type murine bone marrow cells resulted in developmental arrest at the pre-pro B cell stage, confirming that down-regulation of these factors is essential for appropriate B lymphopoiesis. Together, these findings demonstrate the importance of EBF1 in maintaining B lineage commitment.
Epigenetic regulation of the Pax5 gene and maintenance of B cell identity
Pax5 has been extensively characterized as a commitment factor that maintains B cell identity through activation of B cell-specific genes and repression of genes associated with other lineages [45,49–**50]. Strikingly, conditional inactivation of Pax5 in mature B cells resulted in de-differentiation to lymphoid progenitors, which gave rise to functional T cells [46]. These effects were largely due to the loss of Pax5 repressive activities, which include recruiting histone deacetylases (HDACs) and Groucho family corepressors to modulate chromatin structure [51,52].
The potent activities of Pax5 suggest that its expression must be tightly governed throughout lymphopoiesis. However, little was known about the control of Pax5 expression until recently. Decker and colleagues defined a tissue-specific enhancer in intron 5 of the Pax5 gene that reconstitutes Pax5 expression in concert with the Pax5 promoter [**53]. Both the Pax5 promoter and intron 5 enhancer are contained in regions of active chromatin in pro-B cells, as indicated by the presence of active histone marks (acetylated H3K9, di- and trimethylated H3K4) and absence of the repressive histone mark trimethylated H3K27. This active motif also was detected within hypersensitive regions of Pax5 genes in E2A- or EBF1-deficient progenitor cells. These data suggest that an active chromatin configuration is present at Pax5 enhancers in the absence of E2A and EBF1, though mechanisms that establish this epigenetic state have not yet been determined. CpGs in the Pax5 enhancer are demethylated in MPPs during early hematopoiesis. In contrast, the Pax5 promoter region possessed marks of inactive chromatin in E2A- or EBF1-deficient progenitor cells, and DNA methylation appeared to be less important for epigenetic regulation of the promoter. E2A binding was not detected at the Pax5 promoter or enhancer in pro-B cells, suggesting that E2A regulates Pax5 expression indirectly by inducing EBF1 expression. Thus, EBF1 is critical for initiating modifications that result in an active chromatin configuration at the Pax5 promoter, but not at the enhancer.
Maintenance of Pax5 expression at later stages of development requires multiple factors. ChIP analyses demonstrated binding of PU.1 and the interferon regulatory factors (IRF) 4 and IRF8 to the Pax5 enhancer in pro-B and mature B cell lines. IRF4 and 8 are each directly regulated by Pax5. Therefore, their activities at the Pax5 enhancer constitute an indirect positive feedback loop. Together these data suggest that activation of the Pax5 enhancer and promoter regions occurs in a stepwise fashion: PU.1 likely contributes to early remodeling of the Pax5 enhancer [**53], where low levels of IRF4/8 may aid its activity. At the onset of B cell specification, the expression of EBF1 overcomes repression of the Pax5 promoter by Polycomb group proteins, resulting in Pax5 transcription.
Epigenetic regulation of mb-1 and Cd19 transcription by EBF and Pax5
The mb-1 (Cd79a) gene encodes Igα, a signaling component of the pre- and mature B cell receptors that directs their assembly at the plasma membrane. Multiple transcription factors including E2A, EBF, Runx1/CBFβ, Pax5, and Ets family proteins are required for transcriptional activation of mb-1 [40,54]. The mb-1 promoter is hypermethylated at CpGs in HSCs, and becomes progressively demethylated and accessible to transcription factors over the course of B cell development [54]. EBF1 initiates chromatin remodeling necessary for the binding of Pax5 and its Ets protein partners to the mb-1 promoter. Additionally, transcriptional activation of the mb-1 promoter requires the recruitment of SWI/SNF chromatin remodeling complexes (CRCs) by EBF and Pax5 [**55]. Brahma (Brm)-related gene-1 (Brg1), a catalytic (ATPase) component of SWI/SNF, was detected at mb-1 promoters following their activation by EBF1. Furthermore, knock-down of Brm and Brg1 prevented increased chromatin accessibility mediated by EBF alone or in conjunction with Pax5. This suggests that SWI/SNF complexes directly remodel the mb-1 promoter during activation. Enforced expression of EBF1 in Ebf1-deficient fetal liver cells activated endogenous Pax5 expression and promoted demethylation, increased accessibility and transcriptional activation of mb-1 genes. However, reconstitution of Ebf1−/−Pax5−/− progenitors with EBF1 did not enable demethylation of the mb-1 promoter or achieve transcriptional activation of the gene. These results identified mechanisms necessary for the observed synergy between EBF1 and Pax5.
Notably, demethylation of mb-1 promoter DNA and efficient transcriptional activation (>1700-fold) were achieved only following knock-down of Mi-2β, a catalytic (ATPase) component of the Mi-2/NuRD CRCs. The dramatic increase in mb-1 transcription corresponded with histone displacement/eviction and increased chromatin accessibility. These data suggest that Mi-2/NuRD CRCs function as a barrier to the activation of mb-1 transcription. EBF, Pax5 and SWI/SNF were proposed to assist with the exclusion of Mi-2/NuRD. Thus, at mb-1 promoters, SWI/SNF and Mi-2 NuRD function in opposition by shifting the balance between activating and repressive chromatin architecture (Figure 2).
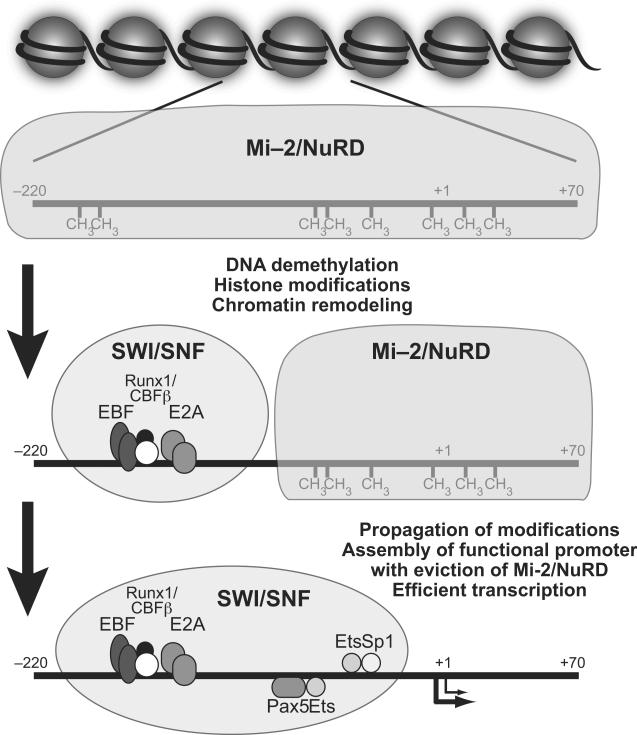
Figure 2.
Model of epigenetic regulation of the mb-1 promoter. Sequential steps involved in mb-1 transcriptional activation are shown. Association of chromatin remodeling complexes with promoter elements is indicated by shaded regions representing hypothesized Mi-2/NuRD and SWI/SNF occupancy. We propose that Mi-2/NuRD complexes are required to maintain hypermethylated, transcriptionally silent mb-1 promoters (top). The mb-1 promoter is primed for expression prior to transcriptional activation (middle). Priming following DNA binding of EBF, E2A and Runx1/CBFβ involves demethylation of CpGs at distal sites and chromatin remodeling following recruitment (by EBF1) of SWI/SNF. Efficient transcriptional activation of the mb-1 gene (bottom) requires 5' to 3' propagation of DNA demethylation, which allows appropriate binding of Pax5, Ets and Sp1, displacement of Mi-2/NuRD, and further modifications of chromatin by SWI/SNF.
Regulation of the Cd19 locus was examined as a model for epigenetic priming of lymphoid genes in early progenitors [**56]. Chromatin remodeling was associated with E2A binding in MPPs at a newly-identified upstream enhancer of Cd19. The enhancer exhibited low levels of monomethylated histone H3K9 in the absence of Pax5, indicating chromatin that is poised for transcription before Pax5 expression. Interestingly, RNA polymerase II (RNAP II) was enriched at the enhancer even in the absence of EBF1 and Pax5. However, binding of Pax5 to the Cd19 promoter was required for efficient transcription. The transient presence of RNAP II prior to transcriptional activation may enable rapid activation of the gene. Notably, demethylation of CpG dinucleotides at the Cd19 enhancer in very early progenitors preceded demethylation of CpGs in the promoter. E2A remodels the enhancer prior to binding of the promoter by EBF and Pax5. These observations suggest that CpG demethylation initiates at discrete sites and spreads to proximal areas as differentiation proceeds. Thus, similar to the Pax5 gene, activation of Cd19 genes is characterized by “primed” chromatin at the enhancer prior to gene expression.
Conclusions and Perspectives
Although many of the details regarding the hierarchical regulation of B cell development by transcription factors have been elucidated, the epigenetic topography that regulates this process remains largely unknown. In the near future, mechanisms of epigenetic change will be revealed by global analysis of modifications using techniques involving deep sequencing technologies, such as ChIP-seq and genomic bisulfite sequencing. Resulting data will address regulation of gene targets by transcription factors described in this review, epigenetic consequences of their binding and changes in binding patterns throughout B cell development. Other areas of analysis include identification of proteins recruited to mediate epigenetic changes and the order of events necessary for propagation of DNA demethylation, histone modifications, and nucleosome eviction/displacement in various contexts. Ultimately, it will be important to address how expression of transcription factors and ensuing changes in the epigenetic landscape contribute to aberrant lymphocyte development, resulting in diseases such as cancer. As the answers to these and many other questions are revealed, a more detailed framework for understanding the control of lymphocyte development and the significance of regulatory steps will emerge.
Acknowledgements
J.R. was supported by NIH Postdoctoral Training Grant T32 AI07405. J.H. is generously supported by NIH grants R01 AI54661, P01 AI22295 and R21 AI075177.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the last two years, have been highlighted as:
*of special interest
**of outstanding interest
- 1.Hardy RR, Kincade PW, Dorshkind K. The protean nature of cells in the B lymphocyte lineage. Immunity. 2007;26:703–714. doi: 10.1016/j.immuni.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 2.Waddington CH. The epigenotype. Endeavour. 1942;1:18–20. [Google Scholar]
- 3.Hu M, Krause D, Greaves M, Sharkis S, Dexter M, Heyworth C, Enver T. Multilineage gene expression precedes commitment in the hemopoietic system. Genes Dev. 1997;11:774–785. doi: 10.1101/gad.11.6.774. [DOI] [PubMed] [Google Scholar]
- 4.Ye M, Iwasaki H, Laiosa CV, Stadfeld M, Xie H, Heck S, Clausen B, Akashi K, Graf T. Hematopoietic stem cells expressing the myeloid lysozyme gene retain long-term, multilineage repopulation potential. Immunity. 2003;19:689–699. doi: 10.1016/s1074-7613(03)00299-1. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein B, Mikkelsen T, Xie X, Kamal M, Huebert D, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- *6.Weishaupt H, Sigvardsson M, Attema J. Epigenetic chromatin states uniquely define the developmental plasticity of murine hematopoietic stem cells. Blood. 2009 doi: 10.1182/blood-2009-07-235176. in press. [DOI] [PubMed] [Google Scholar]; Genome-wide miniChIP was used to investigate chromatin modifications in murine HSCs. Lineage-restricted promoters, including those of regulatory genes, are associated with bivalent and combinatorial histone modifications that control their expression patterns during development.
- 7.Maes J, Maleszewska M, Guillemin C, Pflumio F, Six E, André-Schmutz I, Cavazzana-Calvo M, Charron D, Francastel C, Goodhardt M. Lymphoid-affiliated genes are associated with active histone modifications in human hematopoietic stem cells. Blood. 2008;112:2722–2729. doi: 10.1182/blood-2008-02-140806. [DOI] [PubMed] [Google Scholar]
- 8.Wang Z, Zang C, Cui K, Schones D, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ikawa T, Kawamoto H, Wright LY, Murre C. Long-term cultured E2A-deficient hematopoietic progenitor cells are pluripotent. Immunity. 2004;20:349–360. doi: 10.1016/s1074-7613(04)00049-4. [DOI] [PubMed] [Google Scholar]
- 10.Stopka T, Amanatullah DF, Papetti M, Skoultchi AI. PU.1 inhibits the erythroid program by binding to GATA-1 on DNA and creating a repressive chromatin structure. EMBO J. 2005;24:3712–3723. doi: 10.1038/sj.emboj.7600834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dakic A, Metcalf D, Di Rago L, Mifsud S, Wu L, Nutt SL. PU.1 regulates the commitment of adult hematopoietic progenitors and restricts granulopoiesis. J. Exp. Med. 2005;201:1487–1502. doi: 10.1084/jem.20050075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshida T, Ng SY, Zuniga-Pflucker JC, Georgopoulos K. Early hematopoietic lineage restrictions directed by Ikaros. Nat. Immunol. 2006;7:382–391. doi: 10.1038/ni1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Georgopoulos K, Moore DD, Derfler B. Ikaros: an early lymphoid-specific transcription factor and a putative mediator for T cell commitment. Science. 1992;258:808–812. doi: 10.1126/science.1439790. [DOI] [PubMed] [Google Scholar]
- 14.Hahm K, Ernst P, Lo K, Kim GS, Turck C, Smale ST. The lymphoid transcription factor LyF-1 is encoded by specific, alternatively-spliced mRNAs derived from the Ikaros gene. Mol. Cell. Biol. 1994;14:7111–7123. doi: 10.1128/mcb.14.11.7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Georgopoulos K, Bigby M, Wang JH, Molnar A, Wu P, Winandy S, Sharpe A. The Ikaros gene is required for the development of all lymphoid lineages. Cell. 1994;79:143–156. doi: 10.1016/0092-8674(94)90407-3. [DOI] [PubMed] [Google Scholar]
- 16.Wang JH, Nichogiannopoulou A, Wu L, Sun L, Sharpe AH, Bigby M, Georgopoulos K. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity. 1996;5:537–549. doi: 10.1016/s1074-7613(00)80269-1. [DOI] [PubMed] [Google Scholar]
- 17.O'Neill DW, Schoetz SS, Lopez RA, Castle M, Rabinowitz L, Shor E, Krawchuk D, Goll MG, Renz M, Seelig HP, et al. An ikaros-containing chromatin-remodeling complex in adult-type erythroid cells. Mol. Cell. Biol. 2000;20:7572–7582. doi: 10.1128/mcb.20.20.7572-7582.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim J, Sif S, Jones B, Jackson A, Koipally J, Heller E, Winandy S, Viel A, Sawyer A, Ikeda T, et al. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. 1999;10:345–355. doi: 10.1016/s1074-7613(00)80034-5. [DOI] [PubMed] [Google Scholar]
- 19.Sridharan R, Smale ST. Predominant interaction of both Ikaros and Helios with the NuRD complex in immature thymocytes. J. Biol. Chem. 2007;282:30227–30238. doi: 10.1074/jbc.M702541200. [DOI] [PubMed] [Google Scholar]
- **20.Ng SY, Yoshida T, Zhang J, Georgopoulos K. Genome-wide lineage-specific transcriptional networks underscore Ikaros-dependent lymphoid priming in hematopoietic stem cells. Immunity. 2009;30:493–507. doi: 10.1016/j.immuni.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]; Multilineage priming was demonstrated in HSCs that contain lymphoid, myeloid, and erythroid-specific transcripts. Ikaros is required for lymphoid-lineage priming in HSCs as well as repression of self-renewal and genetic programs downstream of HSCs.
- **21.Papathanasiou P, Attema JL, Karsunky H, Hosen N, Sontani Y, Hoyne GF, Tunningley R, Smale ST, Weissman IL. Self-renewal of the long-term reconstituting subset of hematopoietic stem cells is regulated by Ikaros. Stem Cells. 2009;27:3082–3092. doi: 10.1002/stem.232. [DOI] [PMC free article] [PubMed] [Google Scholar]; HSCs of Ikaros mutant mice lost the capacity for self-renewal and did not support erythrocyte/megakaryocyte development. CLPs from Ikaros mutant mice preferentially committed to the NK cell lineage.
- **22.Mansson R, Hultquist A, Luc S, Yang L, Anderson K, Kharazi S, Al-Hashmi S, Liuba K, Thorén L, Adolfsson J, et al. Molecular evidence for hierarchical transcriptional lineage priming in fetal and adult stem cells and multipotent progenitors. Immunity. 2007;26:407–419. doi: 10.1016/j.immuni.2007.02.013. [DOI] [PubMed] [Google Scholar]; Analysis of global and single-cell gene expression of early hematopoietic progenitors revealed a bifurcation between lymphoid and erythroid lineages during early development. In comparison with HSCs, megakaryocyte/erythroid lineage genes were repressed, common lymphoid genes were up-regulated, and granulocyte/monocyte gene expression were maintained in LMPPs.
- **23.Reynaud D, Demarco IA, Reddy KL, Schjerven H, Bertolino E, Chen Z, Smale ST, Winandy S, Singh H. Regulation of B cell fate commitment and immunoglobulin heavy-chain gene rearrangements by Ikaros. Nat. Immunol. 2008;9:927–936. doi: 10.1038/ni.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ikzf1−/− promotes B cell identity. Ikzf1−/− pro-B cells were unable to perform V(D)J recombination due to deficiencies of Rag1 and Rag2. Enforced expression of EBF1 reconstituted B cell development in Ikaros-null (Ikzf1−/−) MPPs, but the resulting cells were uncommitted. Expression of the Ik-1 isoform of Ikaros in these cells reconstituted normal B cell differentiation and commitment.
- **24.Thompson EC, Cobb BS, Sabbattini P, Meixlsperger S, Parelho V, Liberg D, Taylor B, Dillon N, Georgopoulos K, Jumaa H, et al. Ikaros DNA-binding proteins as integral components of B cell developmental-stage-specific regulatory circuits. Immunity. 2007;26:335–344. doi: 10.1016/j.immuni.2007.02.010. [DOI] [PubMed] [Google Scholar]; Ikaros was found to repress λ5 (Igll1) expression in pre-B cells independently of pre-BCR signaling. Ikaros and EBF1 compete for binding to the Igll1 promoter. These factors function in opposition to repress or activate Igll1 transcription, respectively. Appropriate expression of Aiolos was required for Ikaros-mediated silencing of Igll1.
- 25.DeKoter RP, Singh H. Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science. 2000;288:1439–1441. doi: 10.1126/science.288.5470.1439. [DOI] [PubMed] [Google Scholar]
- 26.Hock H, Hamblen MJ, Rooke HM, Traver D, Bronson RT, Cameron S, Orkin SH. Intrinsic requirement for zinc finger transcription factor Gfi-1 in neutrophil differentiation. Immunity. 2003;18:109–120. doi: 10.1016/s1074-7613(02)00501-0. [DOI] [PubMed] [Google Scholar]
- **27.Spooner C, Cheng J, Pujadas E, Laslo P, Singh H. A recurrent network involving the transcription factors PU.1 and Gfi1 orchestrates innate and adaptive immune cell fates. Immunity. 2009;31:576–586. doi: 10.1016/j.immuni.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gfi1 promotes B cell differentiation via repression of Sfpi1 transcription by displacing PU.1 from auto-regulatory elements in the Sfpi1 promoter. Ikaros up-regulates Gfi1 expression to antagonize PU.1 expression in MPPs, thus favoring B cell development.
- **28.Bussmann LH, Schubert A, Vu Manh TP, De Andres L, Desbordes SC, Parra M, Zimmermann T, Rapino F, Rodriguez-Ubreva J, Ballestar E, et al. A robust and highly efficient immune cell reprogramming system. Cell Stem Cell. 2009;5:554–566. doi: 10.1016/j.stem.2009.10.004. [DOI] [PubMed] [Google Scholar]; Inducible in vitro expression of C/EBPα converted a B cell progenitor line to macrophage-like cells within days. This lineage conversion was accompanied by striking changes in gene expression, including the up-regulation of factors associated with macrophage development and down-regulation of transcription factors associated with B cell development.
- 29.Bain G, Maandag ECR, Izon DJ, Amsen D, Kruisbeek AM, Weintraub BC, Krop I, Schlissel MS, Feeney AJ, van Roon M, et al. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell. 1994;79:885–892. doi: 10.1016/0092-8674(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 30.Zhuang Y, Soriano P, Weintraub H. The helix-loop-helix gene E2A is required for B cell formation. Cell. 1994;79:875–884. doi: 10.1016/0092-8674(94)90076-0. [DOI] [PubMed] [Google Scholar]
- 31.Murre C. Helix-loop-helix proteins and lymphocyte development. Nat. Immunol. 2005;6:1079–1086. doi: 10.1038/ni1260. [DOI] [PubMed] [Google Scholar]
- 32.Yang Q, Kardava L, St Leger A, Martincic K, Varnum-Finney B, Bernstein ID, Milcarek C, Borghesi L. E47 controls the developmental integrity and cell cycle quiescence of multipotential hematopoietic progenitors. J. Immunol. 2008;181:5885–5894. doi: 10.4049/jimmunol.181.9.5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **33.Dias S, Mansson R, Gurbuxani S, Sigvardsson M, Kee B. E2A proteins promote development of lymphoid-primed multipotent progenitors. Immunity. 2008;29:217–227. doi: 10.1016/j.immuni.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]; The investigators established that E2A proteins play a crucial role in early lymphocyte development. E2A proteins were required for LMPP development, including the primed expression of lymphoid-associated genes. E2A opposed differentiation to the myeloid fate in LMPPs and restricted proliferation of early hematopoietic cells.
- **34.Semerad C, Mercer E, Inlay M, Weissman I, Murre C. E2A proteins maintain the hematopoietic stem cell pool and promote the maturation of myelolymphoid and myeloerythroid progenitors. Proc. Natl. Acad. Sci. USA. 2009;106:1930–1935. doi: 10.1073/pnas.0808866106. [DOI] [PMC free article] [PubMed] [Google Scholar]; E2A proteins promote the development of all early hematopoietic progenitors and repress erythroid-specific gene programs in other cell lineages. They also regulate the expression of p21, p27, and thrombopoietin receptor in HSCs, thus controlling cell cycle progression.
- *35.Kwon K, Hutter C, Sun Q, Bilic I, Cobaleda C, Malin S, Busslinger M. Instructive role of the transcription factor E2A in early B lymphopoiesis and germinal center B cell development. Immunity. 2008;28:751–762. doi: 10.1016/j.immuni.2008.04.014. [DOI] [PubMed] [Google Scholar]; Conditional mutagenesis in mice was used to demonstrate that E2A is necessary for the development of pro-B, pre-B, and immature B cells, but not mature B cells or plasma cells in the periphery. E2A is also necessary for maintaining the expression of EBF1 and the B cell program in pro-B cells.
- *36.Beck K, Peak M, Ota T, Nemazee D, Murre C. Distinct roles for E12 and E47 in B cell specification and the sequential rearrangement of immunoglobulin light chain loci. J. Exp. Med. 2009;206:2271–2284. doi: 10.1084/jem.20090756. [DOI] [PMC free article] [PubMed] [Google Scholar]; Using mice deficient in E47 but not E12, the investigators determined that E47 is necessary for developmental progression beyond the pre-pro-B cell stage. However, both E12 and E47 are essential for Ig λ gene transcription and VJ rearrangement in pre-B and immature B cells.
- 37.Hagman J, Belanger C, Travis A, Turck CW, Grosschedl R. Cloning and functional characterization of early B-cell factor, a regulator of lymphocyte-specific gene expression. Genes Dev. 1993;7:760–773. doi: 10.1101/gad.7.5.760. [DOI] [PubMed] [Google Scholar]
- 38.Hagman J, Gutch MJ, Lin H, Grosschedl R. EBF contains a novel zinc coordination motif and multiple dimerization and transcriptional activation domains. EMBO J. 1995;14:2907–2916. doi: 10.1002/j.1460-2075.1995.tb07290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin H, Grosschedl R. Failure of B cell differentiation in mice lacking the transcription factor EBF. Nature. 1995;376:263–267. doi: 10.1038/376263a0. [DOI] [PubMed] [Google Scholar]
- 40.Sigvardsson M, Clark DR, Fitzsimmons D, Doyle M, Akerblad P, Breslin T, Bilke S, Li R, Yeamans C, Zhang G, et al. Early B-cell factor, E2A, and Pax5 cooperate to activate the early B-cell specific mb-1 promoter. Mol. Cell. Biol. 2002;22:8539–8551. doi: 10.1128/MCB.22.24.8539-8551.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Riordan M, Grosschedl R. Coordinate regulation of B cell differentiation by the transcription factors EBF and E2A. Immunity. 1999;11:21–31. doi: 10.1016/s1074-7613(00)80078-3. [DOI] [PubMed] [Google Scholar]
- 42.Smith EM, Gisler R, Sigvardsson M. Cloning and characterization of a promoter flanking the early B cell factor (EBF) gene indicates roles for E-proteins and autoregulation in the control of EBF expression. J. Immunol. 2002;169:261–270. doi: 10.4049/jimmunol.169.1.261. [DOI] [PubMed] [Google Scholar]
- *43.Roessler S, Györy I, Imhof S, Spivakov M, Williams RR, Busslinger M, Fisher AG, Grosschedl R. Distinct promoters mediate the regulation of Ebf1 gene expression by interleukin-7 and Pax5. Mol. Cell. Biol. 2007;27:579–594. doi: 10.1128/MCB.01192-06. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ebf1 expression is controlled by differentially regulated distal and proximal promoters, which generate different isoforms of EBF1. IL-7 signaling, E2A, and EBF1 regulate the distal promoter, while Pax5, Ets1, and PU.1 regulate the stronger proximal promoter.
- *44.Zandi S, Mansson R, Tsapogas P, Zetterblad J, Bryder D, Sigvardsson M. EBF1 is essential for B-lineage priming and establishment of a transcription factor network in common lymphoid progenitors. J. Immunol. 2008;181:3364–3371. doi: 10.4049/jimmunol.181.5.3364. [DOI] [PubMed] [Google Scholar]; EBF1-deficient fetal liver cells generate CLPs, though these cells lack transcription of B-lineage associated genes. EBF1-deficient CLPs also supported development of T cells. The investigators described two novel direct targets of EBF1, Foxo1 and Pou2af1.
- 45.Nutt SL, Heavey B, Rolink AG, Busslinger M. Commitment to the B lymphoid lineage depends on the transcription factor Pax5. Nature. 1999;401:556–562. doi: 10.1038/44076. [DOI] [PubMed] [Google Scholar]
- **46.Cobaleda C, Jochum W, Busslinger M. Conversion of mature B cells into T cells by dedifferentiation to uncommitted progenitors. Nature. 2007;449:473–477. doi: 10.1038/nature06159. [DOI] [PubMed] [Google Scholar]; Conditional ablation of Pax5 in mature peripheral B cells resulted in their dedifferentiation to uncommitted progenitor cells and rescued T cell development in Rag-deficient mice. The re-differentiated T cells contained rearranged Ig heavy- and light-chain genes and, remarkably, were functional in vivo.
- **47.Pongubala J, Northrup D, Lancki D, Medina K, Treiber T, Bertolino E, Thomas M, Grosschedl R, Allman D, Singh H. Transcription factor EBF restricts alternative lineage options and promotes B cell fate commitment independently of Pax5. Nat. Immunol. 2008;9:203–215. doi: 10.1038/ni1555. [DOI] [PubMed] [Google Scholar]; EBF1 plays an important role in B cell lineage commitment. Expression of Ebf1 in Pax5-deficient pro-B cells was sufficient to block their differentiation to T cells or myeloid cells. EBF1 repressed genes associated with other lineages, indicating that reduced EBF1 expression is, in part, responsible for promiscuous expression of non-B-lineage genes in the absence of Pax5.
- *48.Thal MA, Carvalho TL, He T, Kim HG, Gao H, Hagman J, Klug CA. Ebf1-mediated down-regulation of Id2 and Id3 is essential for specification of the B cell lineage. Proc. Natl. Acad. Sci. USA. 2009;106:552–557. doi: 10.1073/pnas.0802550106. [DOI] [PMC free article] [PubMed] [Google Scholar]; Enforced EBF1, but not E47, expression restored B cell differentiation in IL7Rα−/− mice. In the absence of EBF1, E47 up-regulates the E2A inhibitory factors Id2 and Id3. This mechanism is repressed by EBF1. EBF1 binds and represses the Id3 promoter. Expression of either Id2 or Id3 in wild-type murine bone marrow blocked B cell development at the pre-pro B cell stage.
- 49.Delogu A, Schebesta A, Sun Q, Aschenbrenner K, Perlot T, Busslinger M. Gene repression by Pax5 in B cells is essential for blood cell homeostasis and is reversed in plasma cells. Immunity. 2006;24:269–281. doi: 10.1016/j.immuni.2006.01.012. [DOI] [PubMed] [Google Scholar]
- **50.Schebesta A, McManus S, Salvagiotto G, Delogu A, Busslinger GA, Busslinger M. Transcription factor Pax5 activates the chromatin of key genes involved in B cell signaling, adhesion, migration and immune function. Immunity. 2007;27:49–63. doi: 10.1016/j.immuni.2007.05.019. [DOI] [PubMed] [Google Scholar]; The investigators identified 170 genes activated by Pax5. Conditional deletion of Pax5 in pro-B and mature B cells reveled that these genes require continuous Pax5 activity to support normal expression. ChIP-on-chip analysis revealed that Pax5 directly binds and facilitates changes in chromatin structure at regulatory elements of its target genes.
- 51.Eberhard D, Jiménez G, Heavey B, Busslinger M. Transcriptional repression by Pax5 (BSAP) through interaction with corepressors of the Groucho family. EMBO J. 2000;19:2292–2303. doi: 10.1093/emboj/19.10.2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Linderson Y, Eberhard D, Malin S, Johansson A, Busslinger M, Pettersson S. Corecruitment of the Grg4 repressor by PU.1 is critical for Pax5-mediated repression of B-cell specific genes. EMBO Rep. 2004;5:291–296. doi: 10.1038/sj.embor.7400089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **53.Decker T, Pasca di Magliano M, McManus S, Sun Q, Bonifer C, Tagoh H, Busslinger M. Stepwise activation of enhancer and promoter regions of the B cell commitment genePax5in early lymphopoiesis. Immunity. 2009;30:508–520. doi: 10.1016/j.immuni.2009.01.012. [DOI] [PubMed] [Google Scholar]; Pax5 transgene deletion mapping and DNase I hypersensitivity were used to identify an enhancer in intron 5 of the Pax5 locus that contained functional binding sites for PU.1, IRF4, IRF8 and NF-κB. CpG dinucleotides at the enhancer were methylated in HSCs, but were demethylated at the MPP stage. In contrast, the promoter remained demethylated throughout development and is regulated by Polycomb-group proteins and EBF1.
- 54.Maier H, Ostraat R, Gao H, Fields S, Shinton SA, Medina KL, Ikawa T, Murre C, Singh H, Hardy RR, et al. Early B cell factor cooperates with Runx1 and mediates epigenetic changes associated withmb-1transcription. Nat. Immunol. 2004;5:1069–1077. doi: 10.1038/ni1119. [DOI] [PubMed] [Google Scholar]
- **55.Gao H, Lukin K, Ramírez J, Fields S, Lopez D, Hagman J. Opposing effects of SWI/SNF and Mi-2/NuRD chromatin remodeling complexes on epigenetic reprogramming by EBF and Pax5. Proc. Natl. Acad. Sci. USA. 2009;106:11258–63. doi: 10.1073/pnas.0809485106. [DOI] [PMC free article] [PubMed] [Google Scholar]; EBF and Pax5 synergize to promote transcription of mb-1 (Cd79a). SWI/SNF and Mi-2/NuRD CRCs function in opposition: SWI/SNF mediates transcriptional activation by EBF and Pax5, while Mi-2/NuRD limits activation. The SWI/SNF complex directly binds to the mb-1 promoter and enables changes in chromatin including mobilization of nucleosomes and increased accessibility. Displacement of the Mi-2/NuRD complex is necessary for DNA demethylation required for efficient transcriptional activation.
- **56.Walter K, Bonifer C, Tagoh H. Stem cell-specific epigenetic priming and B cell-specific transcriptional activation at the mouseCd19locus. Blood. 2008;112:1673–1682. doi: 10.1182/blood-2008-02-142786. [DOI] [PubMed] [Google Scholar]; An enhancer was identified upstream of the Cd19 gene. Stepwise activation of Cd19 expression begins in MPPs with E2A-associated demethylation of enhancer CpGs. However, Cd19 is not expressed until later stages when EBF and Pax5 bind the promoter.