Abstract
The bHLH transcription factor Neurog1 (Ngn1, Neurod3, neurogenin 1) is involved in neuronal differentiation and cell-type specification in distinct regions of the developing nervous system. Here, transgenic mouse models were developed that use a Bacterial Artificial Chromosome (BAC) containing 208 kb flanking the Neurog1 gene to efficiently drive expression of GFP and Cre in all Neurog1 domains. Two characteristics of Neurog1 gene regulation were uncovered. First, a 4 kb region previously shown to be sufficient for driving expression of a reporter gene to a subset of the Neurog1 pattern in the developing midbrain, hindbrain, and spinal cord is required uniformly for high levels of expression in all Neurog1 domains, even those not originally identified as being regulated by this region. Second, a 0.8 kb enhancer was identified that is sufficient to drive Neurog1-like expression specifically in the ventral neural tube. Furthermore, Neurog1 progenitor cells in the ventral neural tube are largely fated to interneuron lineages rather than to motoneurons. These studies provide new tools for directing tissue specific expression in the developing neural tube, define Neurog1 lineages in the spinal cord, and further define the complex genomic structure required for obtaining the correct levels and spatial restriction of the neuronal differentiation gene Neurog1.
Keywords: gene regulation, bHLH transcription factor, Ngn1, neurogenin, BAC transgenic mice, spinal cord development, neurogenesis, neural tube enhancer, ventral spinal cord interneurons, Cre recombinase
Introduction
Proper neural function depends on development of the correct number of cells with the correct identity for accurate assembly of neuronal circuits. Neural basic helix-loop-helix (bHLH) transcription factors are known regulators of neuronal differentiation and neuronal sub-type specification. A subset of neural-bHLH transcription factors, including Neurog1, is transiently expressed in proliferating cells, and expression is lost as these cells become postmitotic and differentiate into more mature neural cell types (Cau et al., 2002; Fode et al., 2000; Gowan et al., 2001; Lee, 1997; Ma et al., 1996; Schuurmans et al., 2004). Overexpression studies have shown that Neurog1 is sufficient to induce neuronal differentiation in mouse embryonic carcinoma P19 cells, cortical progenitors in mouse, and neural tube in chick, Xenopus and zebrafish (Blader et al., 1997; Farah et al., 2000; Gowan et al., 2001; Ma et al., 1996; Sun et al., 2001). Furthermore, Neurog1 has been shown to play a role in specifying neuronal subtype in neural crest derivatives where ectopic expression induced sensory neuron-appropriate markers in non-sensory crest derivatives, and in chick dorsal neural tube where Neurog1 induced excess dI2 dorsal interneurons at the expense of neighboring dI1 and dI3 interneurons (Gowan et al., 2001; Perez et al., 1999). Loss-of-function studies in mouse have shown that Neurog1 is required for the formation of olfactory neurons and cranial sensory ganglia (Andermann et al., 2002; Cau et al., 2002; Ma et al., 1998; Ma et al., 1999), and along with the related factor Neurog2 (Ngn2, Math4A, neurogenin 2), is required for the proper development of dorsal root ganglia, dorsal interneuron population dI2 in the developing neural tube, and cerebral cortex (Gowan et al., 2001; Kriks et al., 2005; Ma et al., 1999; Nieto et al., 2001). Taken together these studies show that Neurog1 can induce general neuronal differentiation and specify neuronal subtype in the peripheral and central nervous systems. Hence, understanding how Neurog1 expression is regulated during neurogenesis is an important part of identifying the mechanisms involved in generating the correct numbers and types of neurons necessary for the accurate assembly of neuronal circuits.
Previous studies in mouse that tested regions of Neurog1 flanking sequence across a 15 kb region identified multiple intergenic regions sufficient to direct expression of reporter genes to a subset of the Neurog1 expression domain (Blader et al., 2004; Gowan et al., 2001; Murray et al., 2000; Nakada et al., 2004). However, these regulatory regions were not sufficient to recapitulate the entire Neurog1 pattern. Here we use a modified Bacterial Artificial Chromosome (BAC) and transgenic mice to demonstrate 208 kb flanking the Neurog1 gene is sufficient to direct expression to all Neurog1 domains. For efficient levels of Neurog1-like expression, the BAC requires sequences that fall within a previously identified enhancer and are conserved across multiple species including zebrafish (Blader et al., 2004; Gowan et al., 2001; Nakada et al., 2004). Even with the enhancer deleted, the BAC retains activity for low levels of tissue specific expression suggesting the presence of an autoregulatory element or a redundant secondary enhancer at a distinct location. Furthermore, we identify a 0.8 kb region that directs transgene expression specifically to the ventral Neurog1 domain, identifying an enhancer that is distinct from the previously defined dorsal neural tube enhancer for Neurog1 (Nakada et al., 2004), or the specific enhancers identified in zebrafish (Blader et al., 2004; Blader et al., 2003). Finally, we use the Neurog1 regulatory locus for in vivo genetic fate mapping using a Cre-flox system to demonstrate that a majority of Neurog1 progenitors in the ventral neural tube are preferentially fated to become ventral interneurons rather than motoneurons.
Materials and methods
Targeted Modification of Bacterial Artificial Chromsomes and Generation of Transgenic Mice
N1457-nGFP and N1457-Cre were developed using the RP23 457E22 BAC obtained from BACPAC Resources Center (BPRC) at Children’s Hospital Oakland Research Institute in Oakland, California. This BAC contains a genomic insert of 208 kb with the Neurog1 coding sequence located centrally. Homologous recombination in bacteria (Yang et al., 1997) was used to replace the Neurog1 coding region precisely with coding sequence for EGFP (Clontech) with a nuclear localization signal (Lumpkin et al., 2003) or for Cre recombinase. The N1457-nGFP BAC was further used to delete the regions for N1457-nGFPΔR1, N1457-nGFPΔR2, and N1457-nGFPΔR3 using BAC recombineering strategies (Lee et al., 2001). In each case the targeting constructs for the BAC recombineering contained 100–350 bp homology arms and deleted sequences from 2.5 kb to 4.0 kb (see Fig. 1 and Table 1). TgN1-16 was generated from N1457-nGFP using the BAC retrieval method (Liu et al., 2003). TgN1-15 and TgN1-15vnt were generated by cloning the region of interest by PCR into BgnGFP reporter cassette (Lumpkin et al., 2003). TgN1-15vntmgli is TgN1-15vnt with two candidate gli consensus sites mutated. The sequence TGGGTGTTCAGCCCCTGCTGGAAAAAGGCTCGGTGGGTGGG with the possible gli sites underlined was mutated from TGGGT in each case to GTATA. TgN1-2 and TgN1-13dnt have been described previously (Nakada et al., 2004, TgN1-13). Table 1 lists the positions on chromosome 13 for the transgene ends or deletion regions used in this study (using the mouse mm9 assembly July 2007).
Figure 1. Diagram and summary of activity of Neurog1-GFP transgenes.
Comparison of mouse and human genomes surrounding the Neurog1 coding sequence on mouse chromosome 13 reveals extensive conservation (shown 50–100%) in non-coding regions using ECR browser (Ovcharenko et al., 2004). Colors indicate over 70% conservation in sequence where blue is Neurog1 coding, yellow is UTR, and red is intergenic. Black blocks below the ECR diagram indicate sequence conserved to D. rerio that have been identified in functioning enhancers (LATE, ANPE, LSE) (Blader et al., 2004; Blader et al., 2003). The BAC transgene N1457-nGFP (modified BAC RP23 457E22) is shown with the location of deletions in ΔR1, ΔR2, ΔR3 indicated by brackets. The relative location of the deletions and the sequences tested in the transgenes are diagramed below the ECR browser image to highlight the conserved regions included in each. Precise coordinates in the genome are given in Table 1. Green boxes indicate the GFP reporter coding sequence. # Expressing indicates the number of independent transgenic founder embryos at E11.5 that had detectable GFP and were examined for expression in the tissues listed. Expression pattern was consistent across embryos with the same transgene and representative images are shown in Figs. 2 and 4. Black +/− indicate similarity to wildtype while red +/− highlight expression different from wildtype. The asterisks on TgN1-2 and TgN1-13dnt indicate they were reported in Nakada et al. 2004 and are shown here for comparison. ^indicates these transgenes aberrantly directed ventral telencephalon (vt) expression rather than dorsal. vnt, ventral neural tube; dnt, dorsal neural tube.
Table 1.
Mouse chromosome positions for transgenes.
| Name | * mouse chromosome position (deleted regions in ΔR1, ΔR2, ΔR3) | Deletion or transgene size kb |
|---|---|---|
| Neurog 1 codingΨ | chr13:56,352,559-56,353,2943 | 0.735 |
| N1457-nGFP | chr13:56,245,586-56,454,4973 | 208 |
| N1457-nGFPΔR1 | chr13:56,354,675-56,357,1623 | 2.5 |
| N1457-nGFPΔR2 | chr13:56,357,364-56,361,3323 | 4.0 |
| N1457-nGFPΔR3 | chr13:56,361,620-56,365,3783 | 3.8 |
| TgN1-16 | chr13:56,345,580-56,362,0763 | 16.5 |
| TgN1-2 | chr13:56,356,824-56,364,4543 | 7.6 |
| TgN1-13dnt | chr13:56,359,269-56,360,0993 | 0.830 |
| TgN1-15 | chr13:56,361,901-56,364,4543 | 2.5 |
| TgN1-15vnt | chr13:56,363,312-56,364,1183 | 0.806 |
| ^ LATE | chr13:56,359,328-56,359,715 | 0.387 |
| ^ ANPE | chr13:56,360,975-56,361,111 | 0.136 |
| ^ LSE | chr13:56,361,733-56,361,970 | 0.237 |
Mouse mm9 assembly from July 2007 used to determine locations.
Transcribed in the reverse orientation.
Regulatory regions conserved with D. rerio (Blader et. al. 2003, 2004).
Transgenic mice were generated by pronuclear injection using fertilized eggs from B6SJLF1 (C57BL/6J x SJL/J) crosses using standard procedures (Hogan et al., 1986) in the UTSW Core Transgenic Facility. Qiagen purified BAC DNA (two independent clones per BAC deletion) was injected at 0.3–1ng/μl in 10mM Tris pH7.5, 0.1mM EDTA, 100mM NaCl. Non-BAC transgenes were isolated from the vector backbone and injected at 1–3ng/μl in the injection buffer above lacking the NaCl. Transgenic animals were identified by PCR using yolk sac DNA with primers to EGFP: 5′-TTACTTGTACAGCTCGTCCATGCC-3′; 5′-GTGAGCAAGGGCGAGGAGCTGTT-3′ or to Cre: 5′-GGACATGTTCAGGGATCGCCAGGCG-3′; 5′-GCATAACCAGTGAAACAGCATTGCTG-3′. The N1457-nGFP transgenic strain was used in studies of ear development as it marks Neurog1 expressing progenitors fated to be neurons in the VIIIth cranial ganglion (Raft et al., 2007), in studies of thalamus development (Vue et al., 2007), and it was contributed to the GENSAT project for their standard expression analysis (http://www.ncbi.nlm.nih.gov/projects/gensat/).
Cre reporter mouse strains R26R-stop-lacZ (Soriano, 1999), R26R-stop-YFP (Srinivas et al., 2001), and Z/EG (Novak et al., 2000) were genotyped by PCR using primers as previously published: for R26R; 5′-AAAGTCGCTCTGAGTTGTTAT-3′; 5′-GCGAAGAGTTTGTCCTCAACC-3′; 5′-GGAGCGGGAGAAATGGATATG-3′; and for Z/EG: 5′-TTACTTGTACAGCTCGTCCATGCC-3′; 5′-GTGAGCAAGGGCGAGGAGCTGTT-3′.
Tissue Preparation and Immunohistochemistry
E11.5 or E12.5 embryos were collected in Phosphate Buffer (PO4) at 4°C and imaged as whole embryos. Embryos were then fixed for 2 hrs in 4% paraformaldehyde-PO4 (pH 7.2), washed overnight with PO4-buffer, and sunk in 30% sucrose-PO4 all at 4°C. Embryos were embedded in OCT (Tissue Tek) and cryosectioned at 25 μm to 50 μm. Neural tube sections are from the forelimb region.
Immunohistochemistry was performed by incubating with the appropriate dilution of primary antibodies in PBS/1% goat serum/0.1% Triton X-100, followed by incubation with secondary antibodies goat-anti-rabbit or goat-anti-mouse IgG, conjugated to Alexa Fluors 488, 594, or 647 (Molecular Probes, Inc.). Primary antibodies include: mouse monoclonal antibodies anti-Ascl1 (Mash1) (1:100) (Lo et al., 1991), anti-Islet1/2 (1:100, 39.4D5, DSHB), anti-Lhx1/5 (1:100, 4F2, DSHB), and rabbit antibodies anti-Lhx2/9 (1:8000) (gift from T. Jessell), anti-Neurog1 (Ngn1) (1:500) (Gowan et al., 2001), anti-Atoh1 (Math1) (1:100)(Helms and Johnson, 1998), anti-Olig2 (1:1000, gift from C. Stiles and R. Lu), anti-Islet1/2 (1:500)(Tsuchida et al., 1994), anti-GFP (1:500, Molecular Probes, A6455), anti-HB9 (1:500, Abcam), and anti-Cre (1:500, Sigma), and guinea pig anti-Brn3a (1:250) (gift from E. Turner). Sections were imaged by confocal using Bio-Rad MRC1024 or Zeiss LSM510.
For β-galactosidase detection, embryos were fixed in 4% paraformaldehyde in PBS at room temperature for 30 minutes, washed with PBS twice for l0 min at room temperature, and incubated overnight in X-gal staining solution at 30°C (1 mg/ml 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal), 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, 5 mM MgCl2 in PBS). Stained embryos were rinsed in PBS and postfixed in 4% paraformaldehyde 3–6 hours at room temperature before preparing for cryosection as described above.
Results
208 kb surrounding Neurog1 contains sufficient cis-regulatory sequence for directing accurate Neurog1 expression
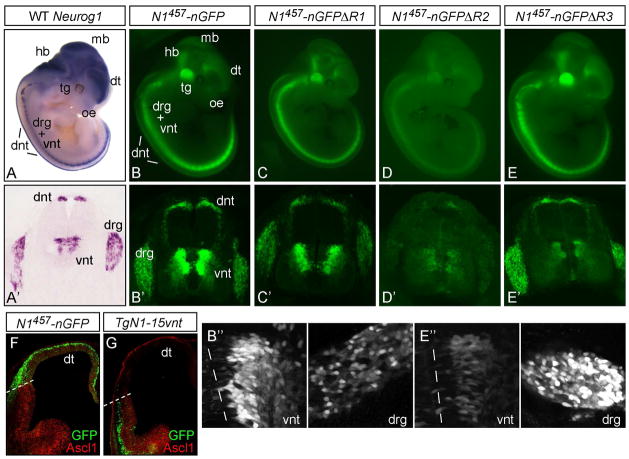
As no single regulatory region in the Neurog1 locus had been shown to direct expression to all Neurog1 domains, we tested a large genomic region using a modified BAC and transgenic mice. Using BAC recombineering techniques (Yang et al., 1997), the Neurog1 coding sequence, which is contained within one exon, was precisely replaced with a nuclear localized EGFP in BAC RP23 457E22. This modified BAC containing 101 kb 5′ and 107 kb 3′ genomic sequence flanking the Neurog1 coding region was injected into the pronuclei of single cell mouse embryos. Transgenic embryos were harvested at E11.5 and assayed for GFP expression. In N1457-nGFP transgenic embryos at E11.5, GFP expression was detected in all Neurog1 domains including dorsal and ventral neural tube (dnt and vnt), olfactory epithelium, cranial sensory ganglia, midbrain, and hindbrain (Figs. 1, 2A,A′B,B′). Notably, expression in the dorsal root ganglia (drg) and dorsal telencephalon were also detected (Fig. 1,2B,B′,F), and represent domains of Neurog1 expression where no cis-regulatory sequences had been previously identified in mouse.
Figure 2. GFP expression from Neurog1-GFP BAC transgenic mice.
mRNA in situ hybridization for Neurog1 in an E11.5 mouse embryo in whole mount (A) or in a cross section of the neural tube (A′). (BE) GFP expression in representative BAC transgenic embryos is shown in whole mount at E11.5. (B′-E′) cross sections show the Neurog1-like expression in the dorsal neural tube (dnt), the ventral neural tube (vnt), and the dorsal root ganglia (drg). (B″,E″) show a higher magnification of the GFP expression in the vnt and drg from N1457-nGFP compared to N1457-nGFPΔR3 to highlight the specific reduction in vnt activity with this deletion. (F,G) telencephalon showing GFP relative to Ascl1 immunofluorescence (red). GFP from N1457-nGFP reflects the endogenous Neurog1 in the dorsal telencephalon while TgN1-15vnt has aberrant expression in the ventral telencephalon. Transgenes are diagramed and results summarized in Fig. 1. dt, dorsal telencephalon; hb, hindbrain; mb, midbrain; oe, olfactory epithelium; tg, trigeminal ganglion.
To determine if N1457-nGFP directs expression precisely to Neurog1 domains, we examined whether GFP respected the endogenous boundaries of Neurog1 expression in the telencephalon and neural tube. In the telencephalon, Neurog1 is restricted to dorsal regions with its ventral boundary marked by the bHLH factor Ascl1 (Mash1) (Ma et al., 1997). GFP from N1457-nGFP mimics this pattern and shares the same ventral boundary (Fig. 2F). In the E11.5 neural tube, co-expression of GFP with Neurog1 demonstrates expression from the BAC is restricted to the Neurog1 domain in these regions as well (Fig. 3A–D). In the dorsal neural tube, Neurog1 is present precisely in progenitors to the dorsal interneuron population 2 (dI2), non-overlapping with the closely neighboring progenitors marked by the bHLH factors Ascl1 (Mash1) and Atoh1 (Math1) (Fig. 3 upper diagram). We demonstrate that GFP overlaps with Neurog1 (Fig. 3E) but not Ascl1 or Atoh1 (Fig. 3F,G). Furthermore, GFP persists into differentiating interneurons, and the GFP overlaps with the dI2 markers Lhx1/5 but not dI1 markers Lhx2/9, consistent with GFP expression precisely in dI2 but not dI1 neurons (Fig. 3H,I). In the ventral neural tube, the Neurog1 domain is bounded by Ascl1 dorsally and Olig2 ventrally (Fig. 3 lower diagram). Likewise, GFP is present in the ventral progenitor domain overlapping with Neurog1 (Fig. 3J) but not with either Ascl1 or Olig2 (Fig. 3K,L). Although GFP is at lower levels in Neurog1 cells closest to the ventricle, essentially all Neurog1 positive cells have detectable GFP. Again, since the GFP persists into differentiating neurons, we detect extensive overlap with Lhx1/5 that marks interneurons dI6, V0, V1 and V2, and Chox10 that also marks V2 (Fig. 3M,N). GFP positive cells are bounded by dI5 neurons marked by Brn3a at the dorsal boundary (Fig. 3M) and motoneurons marked by Isl1/2 at the ventral boundary (Fig. 3O). Overall, the BAC transgene, N1457-nGFP, reliably reports Neurog1 expression domains at E11.5. A stable transgenic strain with this BAC was contributed to the GENSAT project and the standard GENSAT characterization at E15.5, P7 and adult is available through that project at (http://www.ncbi.nlm.nih.gov/projects/gensat/). This transgenic strain was also shown to express in a Neurog1-like pattern in the VIIIth cranial ganglia (Raft et al., 2007), developing thalamus (Vue et al., 2007), and olfactory epithelium (data not shown).
Figure 3. N1457-nGFP directs GFP expression precisely to the Neurog1 lineage.
(A,B) show a cross section through the neural tube of an E11.5 N1457-nGFP embryo labeled by immunoflorescence for Neurog1 (A) or direct fluorescence of GFP (B) and indicate GFP expression is restricted to Neurog1 domains in the dnt, vnt, and drg at this level. Arrows indicate the dorsal and ventral boundaries of expression for Neurog1 and GFP match. (C,D) are high magnification of views of the dnt and vnt demonstrating the co-expression of Neurog1 and GFP. (E-I) focuses on the dnt and compares GFP with immunofluorescence for Neurog1, Ascl1, Atoh1, Lhx1/5 or Lhx2/9 from N1457-nGFP in E11.5 dnt. Overlap of the GFP signal with Neurog1 (yellow) and Lhx1/5 (turquoise) but none of the other factors illustrates the precision with which the BAC transgene is controlling GFP expression. (J-O) focuses on the vnt and compares GFP with immunofluorescence for Neurog1, Ascl1, Olig2, Lhx1/5, Brn3a, Chx10 and Isl1 from N1457-nGFP in E11.5 vnt. Dashed line indicates position of the ventricular zone. Overlap of the GFP signal with Neurog1 (yellow), Lhx1/5 (turquoise), and Chox10 (yellow) but none of the other factors suggests the Neurog1 lineage in the ventral neural tube comprises interneurons rather than motoneurons. Diagrams for progenitor/neuron relationships and markers are shown for both dnt and vnt. dI5 and dI6, dorsal interneurons 5 and 6; MN, motoneuron; V0-V2, ventral interneurons.
Redundant regulatory information for tissue specific expression
The reliable expression of N1457-nGFP in transgenic mice provided a tool to test whether the enhancer sequences previously described as sufficient for directing expression to specific subsets of the Neurog1 pattern are also required, a test rarely performed for identified enhancers. Three deletions were made in the N1457-nGFP BAC using homologous recombination in bacteria (Lee et al., 2001). Each mutated BAC was used to generate transgenic embryos that were assayed at E11.5. To control for unplanned, undetected rearrangements in the BAC sequences, two independently derived BAC constructs for each deletion were used to generate multiple transgenic embryos. One deletion, N1457-nGFPΔR1, that deleted a largely non-conserved genomic region, had no detectable alteration in the expression of the reporter gene at E11.5 (Fig. 1, 2C,C′). This is consistent with early studies that showed this region worked only inefficiently at directing expression of a reporter to Neurog1 domains in transgenic mice (Murray et al., 2000; Nakada et al., 2004).
Two additional deletions were designed to test the requirement for highly conserved sequences shown previously to direct expression of reporter transgenes in a subset of the Neurog1 pattern in transgenic mice (Gowan et al., 2001) (also see Figs. 1,4B, TgN1-2). The deletion in N1457-nGFPΔR2 includes 4 kb of the 7.6 kb enhancer (TgN1-2) shown previously to be sufficient to drive reporter expression to many, but not all, Neurog1 domains (Gowan et al., 2001; Nakada et al., 2004), and contains two highly conserved sequences, LATE and ANPE, shown to be important in zebrafish neurogenin 1 expression (Blader et al., 2004; Blader et al., 2003). This sequence also contains an element specific for dorsal neural tube expression (TgN1-13dnt), but lacks information sufficient to direct expression to the dorsal root ganglia and the dorsal telencephalon (Nakada et al., 2004) (also see Figs. 1,4B,F). Thus, we predicted that N1457-nGFPΔR2 would lack expression in multiple domains particularly the dorsal neural tube, but expression in dorsal root ganglia and dorsal telencephalon would be spared. Surprisingly, in N1457-nGFPΔR2 there was no specific loss of activity in one tissue over another. Rather, there was a dramatic decrease in overall reporter expression levels across all domains when compared to N1457-nGFP (Fig. 1,2 compare B,B′ with D,D′). Although signal amplification was required to detect the GFP consistently in all N1457-nGFPΔR2 embryos, expression in all Neurog1 domains was detected and the precise dI2 progenitor boundaries were maintained (data not shown). This dramatic decrease in overall expression demonstrates the presence of a general enhancer component, or locus control region, within the sequence deleted. This broad activity across all Neurog1 domains of the 4 kb sequence was not detected previously since it is not sufficient to direct expression to all Neurog1 domains on its own. Furthermore, the fact that low expression remains precisely in a Neurog1-like pattern suggests either the existence of an inefficient autoregulatory enhancer, or a secondary redundant enhancer elsewhere within the 208 kb BAC. Secondary enhancers have been seen in multiple developmental genes in Drosophila and mouse, and may be a common strategy to ensure reproducible expression of essential genes (Hong et al., 2008; Jeong et al., 2006; Markstein et al., 2002; Zeitlinger et al., 2007).
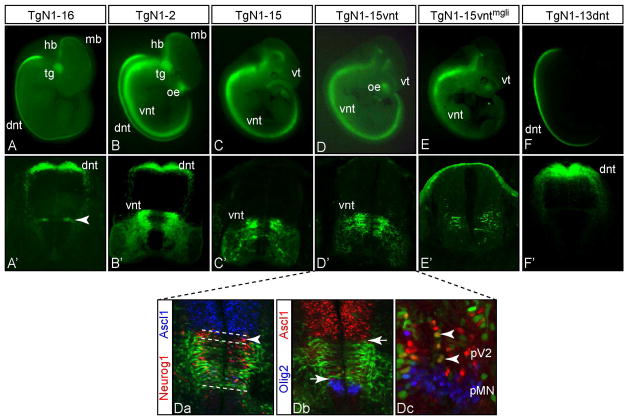
Figure 4. An enhancer for Neurog1-like expression in the ventral neural tube is identified.
(A–F) GFP expression in representative transgenic embryos is shown in whole mount at E11.5. (A′–F′) cross sections show the activity of each transgene in the neural tube. Transgenes are diagramed and results summarized in Fig. 1. Within the spinal neural tube, TgN1-2 directs expression to both dorsal neural tube (dnt) and ventral neural tube (vnt) whereas TgN1-16 and TgN1-13dnt are largely restricted to the dnt (A′, arrowhead indicates the narrow strip of ventral neural tube expression consistently seen with TgN1-16), and TgN1-15 and TgN1-15vnt are restricted to the vnt. Activity for expression in dorsal root ganglia (drg), dorsal telencephalon, olfactory epithelium (oe), trigeminal ganglion (tg), and subsets of hindbrain (hb) and midbrain (mb) is not present in every transgene. For example, drg and dt are largely absent in all transgenes, whereas oe is only seen in TgN1-2, TgN1-15 (tail obscuring oe in this image), and TgN1-15vnt, and tg is only in TgN1-16 and TgN1-2. (Da-Dc) immunofluorescence on E11.5 cross sections of TgN1-15vnt neural tube. (Da) Dashed lines delineate the dorsoventral boundaries of the ventral Neurog1 (red) domain is shared by GFP but also highlight the more dorsal part of this domain is lacking GFP (arrowhead). (Db) Arrows highlight the boundaries of non-overlap of GFP with Ascl1 (red) and Olig2 (blue). (Dc) is a higher magnification of the progenitor domains for V2 interneurons and motoneurons (MN). In the pV2 region there are a few cells that co-express Ascl1 and Neurog1 (Da) and GFP (Dc, arrowheads). Ventral telencephalon (vt) expression, non-Neurog1 pattern, appears in TgN1-15 and its derivatives (see Fig. 2G for a cross section).
A ventral neural tube specific enhancer is identified
In contrast to the phenotype in the first two deletions, N1457-nGFPΔR3 resulted in a dramatic loss of expression in a specific subset of the Neurog1 pattern (Fig. 1,2E,E′). The region deleted in N1457-nGFPΔR3 contains the other half of TgN1-2, sequence that includes the LSE conserved region important for neurogenin 1 expression in primary neurogenesis and telencephalon in zebrafish (Blader et al., 2003) (Fig. 1). With this deletion, there was a dramatic decrease in GFP levels specifically in the ventral neural tube relative to the GFP levels in the other Neurog1 domains. This is highlighted by comparing the levels in the dorsal root ganglia to the levels in the ventral neural tube in the wild type N1457-nGFP versus the N1457-nGFPΔR3 transgenic embryos (Fig. 2B′, B″, E′,E″). From these data, it is clear that the sequence deleted is required for efficient expression in the ventral neural tube but not other domains. The low level expression remaining in the ventral neural tube is consistent with the existence of an autoregulatory component, or secondary enhancer as suggested above with N1457-nGFPΔR2 (see previous section).
Analyzing ventral neural tube expression from additional transgenic constructs identified the location of the ventral neural tube enhancer. First, a 16 kb sequence including 9 kb 5′ and 7 kb 3′ of Neurog1 (including LATE, ANPE, and LSE conserved regions) was retrieved from N1457-nGFP using recombination in bacteria (Liu et al., 2003), and tested for activity in transgenic mice (TgN1-16). This transgene was designed to test the most highly conserved sequence from the locus that might reveal efficient expression in such domains as the drg and telencephalon that had so far only been seen with the 208 kb BAC. However, the 16 kb was not sufficient to recapitulate all Neurog1 domains. GFP expression from this construct mimicked Neurog1 expression in the dorsal neural tube, a narrow strip in the ventral neural tube, trigeminal ganglia, and at least a subset of the hindbrain and midbrain domains. Strikingly, expression in the majority of the ventral neural tube, dorsal root ganglia, dorsal telencephalon, and olfactory epithelium was lost (Figs. 1,4A). This clearly places required enhancer sequences for these last four domains outside the 16 kb tested. For the ventral neural tube, comparing the 5′ end of TgN1-16 with the previously reported TgN1-2 that does contain ventral neural tube enhancer activity (Nakada et al., 2004), suggested the required information for at least this domain was coded in a 2.5 kb region (Figs. 1,4B,B′). In transgenic mice, we tested this 2.5 kb (TgN1-15) as well as a more restricted 0.8 kb region (TgN1-15vnt) that contained the sequence with the highest conservation between mammalian species. Both transgenes were sufficient to direct GFP expression to the ventral neural tube from spinal regions to the hindbrain, the olfactory epithelium, and a subset of the dorsal telencephalon, but not other Neurog1 expression domains (Figs. 1, 4C–D′). Unexpectedly GFP was also detected in the telencephalon. A closer examination of sections from these embryos revealed that this expression did not mimic Neurog1 but rather was restricted to the ventral telencephalon (Fig. 2, compare F,G). These later data suggest the presence of a repressor sequence in the TgN1-2 that is not present in TgN1-15.
To characterize the expression pattern of the 0.8 kb vnt enhancer, we used immunofluorescence to delineate boundaries and overlap of GFP with other markers. First we compared the GFP signal to endogenous Neurog1 and found that the domains largely overlap (Fig. 4Da). The exception is the most dorsal boundary of the ventral Neurog1 domain where GFP is missing (Fig. 4Da, arrowhead). The regulatory element for this subset of the Neurog1 pattern is likely located at a distinct site within TgN1-16 (Fig. 4A′, arrowhead). The ventral boundary of the GFP pattern abuts the Olig2 domain that marks progenitors to motoneurons (Fig. 4Db,Dc), just as was demonstrated for the N1457-GFP BAC transgenic embryos (Fig. 3L). Thus, the 0.8 kb vnt enhancer directs expression to the ventral Neurog1 domain except for a small subset of progenitors at the dorsal boundary that likely give rise to the dI6 interneurons.
Examination of the 0.8 kb vnt region revealed two possible gli consensus binding sites (Vokes et al., 2007) in sequence conserved across multiple mammalian species. Gli factors are transcription factors downstream of Shh signaling. Since Shh is active in the developing ventral neural tube, this possible direct connection of Shh signaling to Neurog1 regulation through the vnt enhancer was an attractive hypothesis to link patterning signals to this transcription factor regulating differentiation. We mutated the two candidate gli sites within the context of the TgN1-15vnt reporter and tested it in transgenic embryos. Mutating the gli sites (TgN1-15vntmgli) did not alter the pattern of GFP expression. However, as a group these embryos had lower expression that required longer exposure times to image the pattern (Fig. 4E,E′). These results suggest a possible role for Shh signaling through gli for enhancer activity but not for the pattern.
Neurog1 progenitor cells in the ventral neural tube preferentially give rise to interneurons and a limited contribution to motoneurons
The transient expression of the Neurog1 gene and the relatively fast degradation of its protein product makes it difficult to determine which neural cell types arise from Neurog1 expressing cells. Based on its expression pattern in the neural tube, it has been inferred that progenitors expressing this protein will largely give rise to ventral interneurons and motoneurons. To determine precisely which ventral neurons are in the Neurog1 lineage, we exploited the GFP stability in the N1457-nGFP mice to perform short-term lineage tracing by co-labeling with markers specific for various neuronal populations as described above and shown in Fig. 3. Analysis of the N1457-nGFP revealed that a majority of ventral interneurons are derived from Neurog1 progenitors, with much less GFP detected in the motoneuron domain (MN) (Fig. 3). Indeed, the lack of overlap with Olig2 in the progenitor domain predicts exclusion of the Neurog1 lineage in the motor pools since Olig2 progenitors are known to become motor neurons (Mukouyama et al., 2006; Park et al., 2002; Zhou and Anderson, 2002). However, since GFP from N1457-nGFP is transient it is possible that the earlier formed MN populations lost their GFP by E11.5. Examination of the embryos at E10.5 still did not reveal extensive GFP in the MN population (data not shown).
To determine whether the Neurog1 progenitor cells do preferentially give rise to ventral interneurons over motoneurons, we used in vivo genetic fate mapping with Cre recombinase. We generated N1457-Cre mice with the same strategy used to generate the N1457-GFP mice. Just as the GFP in the N1457-GFP mice, expression of Cre recombinase in the N1457-Cre mice closely mimics the pattern of endogenous Neurog1 (Fig. 5A–C). Immunofluoresence detecting Cre in E11.5 embryos revealed the same pattern of expression as Neurog1 (compare Fig. 3A and 5B) including the dorsal root ganglia, and the dorsal and ventral neural tubes. Embryos from N1457-Cre;R26RYFP or N1457-Cre;R26RlacZ transgenic mice revealed a pattern of reporter gene expression in the neural tube and the sensory ganglia consistent with labeling of Neurog1 lineages at E11.5 (Fig. 5A,C,C′). A cross section through the neural tube region of a β-gal stained embryo shows expression in differentiated neurons (Fig. 5C′). By adult stages the Neurog1 lineage cells are mature neurons that have settled in the intermediate and ventral gray matter of the spinal cord (Fig. 5D).
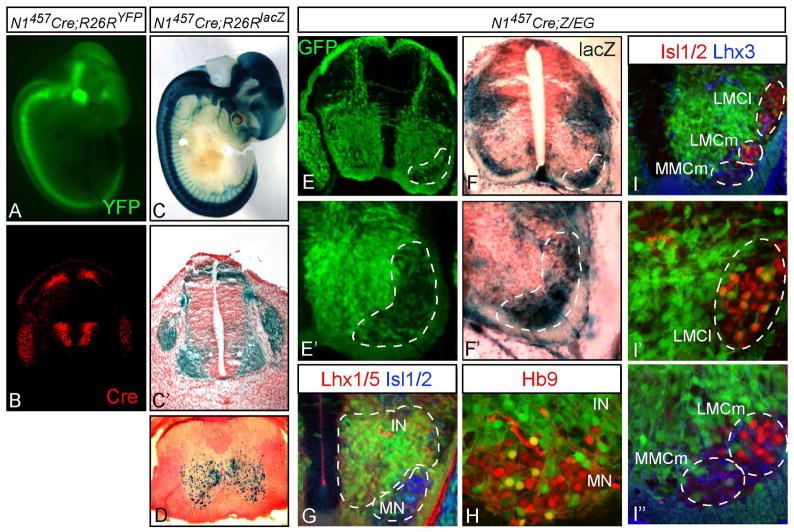
Figure 5. In vivo analysis of the Neurog1 lineage in the neural tube using Cre recombinase.
(A,C) E11.5 embryos from N1457-Cre crossed with Cre reporter strains are shown in whole mount (A, R26RYFP; C, R26RlacZ), or cross section (C′). (B) Cre immunofluorescence mimics the Neurog1 pattern in the neural tube at E11.5 (compare to Fig. 3A). (D) spinal cord from a 30-day old N1457-Cre;R26RlacZ mouse showing the location of Neurog1-lineage neurons. (E-I) Cross sections through the neural tube of N1457-Cre;Z/EG E12.5 embryos. (E,E′) GFP shows Neurog1 lineages where Cre activity has recombined the reporter and includes the drg, a minor dorsal population, and a major ventral population in the neural tube. (F,F′) X-gal staining indicates non-Neurog1 lineages that lack Cre activity. The dashed area highlights motoneuron populations that are mostly not included in the Neurog1 lineages. (G) extensive co-expression of the GFP is detected with the broad interneuron (IN) marker Lhx1/5. (H,I) co-expression with motoneuron (MN) markers Hb9, Isl1/2, and Lhx3 is low. LMCm, lateral motor column medial; LMCl, lateral motor column lateral; MMCm, medial motor column medial.
We used the N1457-Cre mice with the Z/EG reporter line that expresses GFP in cells where Cre recombinase is active, and expresses LacZ in cells in the absence of recombination (Novak et al., 2000) to assess the fate of the Neurog1 progenitors in the ventral neural tube. In E12.5 neural tubes of N1457-Cre;Z/EG mice, GFP expression was predominant in ventral interneuron domains, but were notably sparse in motoneuron domains (Fig. 5E,E′). Indeed, the LacZ expression pattern in this region was complementary to that of GFP, where LacZ was found predominantly in the motoneuron domain (compare Fig. 5E,E′ with F,F′). These results show that Neurog1 lineage cells largely contribute to the ventral interneuron populations, and to a much lesser extent to motoneurons. This finding was verified by co-labeling GFP from the Z/EG locus with homeodomain factors Hb9, Isl1/2, Lhx1/5, and Lhx3. As suspected, Neurog1 lineage cells have broad overlap with the interneuron marker Lhx1/5 (Fig. 5G; yellow IN), while only a small subset overlapped with the motoneuron markers Hb9 and Isl1/2 (Fig. 5G-I). To determine if the Neurog1 lineage motoneurons belonged to a specific motoneuron pool, we compared the overlap of GFP with Lhx3 and Isl1/2. A small subset of Lhx3+ cells in the MMCm (Fig. 5I,I″) and a small subset of Isl1/2 in the LMCl (Fig. 5I,I′) co-localize with GFP. However, GFP did not co-localize with Isl1/2 in the LMCm. Thus, only a subset of motoneurons is derived from the Neurog1 lineage, and even within this subset, there is a bias towards specific neuronal subtypes.
Discussion
Cis-regulation of Neurog1 expression
We demonstrate that cis-regulatory regions for Neurog1 are contained within 208 kb flanking the coding region of Neurog1 with the N1457-nGFP transgenic mice reliably reporting domains of Neurog1 expression throughout development. Neurog1 is expressed in neuronal progenitor cells in many different regions throughout the central and peripheral nervous systems in a pattern largely non-overlapping to another bHLH factor Ascl1 (Ma et al., 1997; Sommer et al., 1996). As such, the temporal and spatial control of Neurog1 expression is complex and includes multiple discrete regulatory regions spread over more than 20 kb surrounding the coding exon. Over the past decade, work in zebrafish and mouse has identified multiple cis-regulatory sequences for Neurog1 using sufficiency assays in transgenic animals (Blader et al., 2004; Blader et al., 2003; Gowan et al., 2001; Murray et al., 2000; Nakada et al., 2004). Here, using BAC transgenic mice to test the requirement of these sequences in directing neural specific expression in vivo, we find additional complexities including an apparent redundancy in enhancer activity, new activities for a previously defined enhancer, and localization of an enhancer for ventral neural tube expression. The current understanding of how Neurog1 is regulated is summarized below.
Studies in zebrafish uncovered three enhancers within an 8.4 kb 5′ proximal genomic region, LSE, ANPE, and LATE, which are conserved to mouse and have distinct activities when assayed as reporter transgenes in vivo (for review see Strahle and Rastegar, 2008). The LSE directs expression to the lateral stripes of the neural plate during primary neurogenesis in cells that give rise to the Rohan Beard neurons, with later activity directing expression to the telencephalon. In contrast, the ANPE directs expression to the anterior neural plate, and the LATE directs expression after completion of primary neurogenesis to the neural tube, hindbrain, and diencephalon. When tested in mouse the zebrafish LATE also had activity in the lateral telencephalon. None of these enhancers were reported to direct expression to the dorsal root ganglia or to the motor neuron domain in the ventral neural tube—two other areas of neurogenin 1 expression, although this information is within the 8.4 kb 5′ genomic region tested.
Early studies on Neurog1 regulation in mouse showed that the proximal 4.5 kb 5′ of Neurog1 was inefficient at directing expression to Neurog1 domains in transgenic mice (Gowan et al., 2001; Murray et al., 2000). This proximal 5′ region was also recently reported to undergo chromatin remodeling during activation of Neurog1 in retinoic acid induced differentiation of P19 cells (Wu et al., 2009). In contrast, a more efficient regulatory region found further 5′ (TgN1-2) that contains sequence conserved to the zebrafish LSE, ANPE, and LATE could direct expression of reporter transgenes efficiently to both dorsal and ventral neural tube, more anterior domains in the hindbrain and midbrain, some cranial sensory ganglia, and the olfactory epithelium. Notably, enhancer activity for directing expression to the telencephalon and dorsal root ganglia was absent (Gowan et al., 2001). A subsequent study further delineated a 0.8 kb region that includes homology to the zebrafish LATE element that directs expression specifically to the dorsal neural tube (here called TgN1-13dnt) (Nakada et al., 2004). In the current study, we delineate another enhancer, distinct from any of those described above, that is sufficient to direct expression to Neurog1 cells in the ventral neural tube. The ventral neural tube enhancer has less conservation between species than sequence for the other enhancers since it is restricted to mammals. Taking these studies together, it is clear that there is a cassette-like organization of discrete regulatory regions that work together to direct the diverse pattern of Neurog1-specific expression.
The regulatory regions identified to date are only part of the story, since we cannot recapitulate the full Neurog1 pattern without using a large genomic region as tested in the BAC. One promising approach in the literature is to identify tissue specific enhancers using ChIP-Seq to localize co-activators such as p300 (Visel et al., 2009). Since Visel et al used E11.5 forebrain and midbrain, tissues where Neurog1 is actively expressed, we examined the published data set for p300 occupied sites within the 208 kb in the BAC RP23 457E22. With forebrain tissue there was a p300 binding region about 40 kb 3′ of Neurog1 (chr13: 56,309,425-56,310,401), a region not specifically tested in the transgenic mice here. In contrast, p300 in midbrain tissue occupies a region (chr13: 56,359,200-56,359,676) that is included within the identified dorsal neural tube enhancer tested in TgN1-13dnt, a region with homology to fish. Paradoxically, TgN1-13dnt alone did not show activity in the midbrain suggesting other sequences and complexes lacking p300 are required for efficient expression in midbrain. So while the use of ChIP-Seq with co-activators like p300 to identify tissue specific enhancers is powerful in identifying some important enhancer regions, using the current technology it is unlikely to identify all enhancers.
The studies described above define activity of regulatory sequences by determining whether they are sufficient to direct reporter gene expression to a particular tissue. In the current study we asked whether the identified enhancers are required for expression in particular tissues. We predicted that deletion of sequence that includes the dnt element as well as homology to the ANPE from the 208 kb Neurog1 BAC transgene would have lost GFP expression in the dnt but not lose expression in other tissues like dorsal root ganglia. Unexpectedly we found a uniform attenuation of enhancer activity in all domains. This suggests the sequence deleted, while not sufficient to direct expression to all Neurog1 domains (see TgN1-2), is required generally for expression of the locus (see N1457-nGFPΔR2). Furthermore, deletion of this region revealed the existence of a weaker tissue specific secondary enhancer within the 208 kb BAC since the complete Neurog1 pattern was detected but just at much lower levels. The existence of secondary enhancers in developmental genes has recently been reported and is suggested to be an important mechanism driving animal diversity (Hong et al., 2008; Jeong et al., 2006; Markstein et al., 2002; Zeitlinger et al., 2007). Indeed, a recent study tested the requirement for ultraconserved elements by knocking them out in the mouse genome (Ahituv et al., 2007). Of the four elements tested, none resulted in notable abnormalities in expression of the locus or in negative consequences for viability. It remains to be determined if the decrease in expression resulting from deletion of R2 or R3 (Fig. 2) would be sufficient to disrupt nervous system development.
There has been little advance in identifying factors that regulate through the Neurog1 enhancers. Pax6 is a good candidate for an upstream regulator since its expression in the telencephalon and neural tube in mouse overlaps that of Neurog1. Indeed, Pax6 can bind zebrafish LATE in vitro, and LATE transgene activity in mouse telencephalon and zebrafish diencephalon requires Pax6 (Blader et al., 2004). However, the sequence in the mouse Neurog1 gene that is conserved with zebrafish LATE does not have activity in mouse telencephalon, but rather is restricted to directing expression to the dorsal neural tube (TgN1-13dnt) where Pax6 is not present. Thus, although we cannot rule out a role for Pax6 in regulating mouse Neurog1, it does not appear to be functioning through the same enhancer in zebrafish and mice, nor does there appear to be a Pax6 site within the newly localized ventral neural tube enhancer. Gli factors are also candidates for regulating Neurog1 expression, particularly in the ventral neural tube since they are downstream effectors of Shh signaling and are active in patterning the ventral neural tube (Vokes et al., 2007). Mutating two gli consensus sites within the ventral neural tube enhancer appeared to attenuate enhancer activity, although the overall pattern of expression remained. Identifying the full complement of upstream signaling events and transcription factors that regulate through the distinct Neurog1 enhancers remains an area where little is known.
Neurog1 lineage in the developing spinal cord
The bHLH proteins such as Neurog1 act in balance with the Notch pathway to control the timing of differentiation of neural progenitors (Bertrand et al., 2002; Lee and Pfaff, 2001). In addition, in combination with homeodomain factors, bHLH factors define distinct progenitor cells in the developing spinal cord, and act to confer diversity to the emerging nervous system (Bertrand et al., 2002; Briscoe et al., 2000; Helms and Johnson, 2003; Shirasaki and Pfaff, 2002). Defining the fate of Neurog1 progenitor cells in the ventral spinal cord has been complicated by multiple factors including 1) its transient expression in progenitor cells, 2) its broad expression in ventral progenitor domains, and 3) presumptive redundancy with the closely related Neurog2 making phenotypic analysis of mutants more difficult. In particular, although Neurog1 mutants have defects in proximal cranial ganglia, olfactory neurons, and inner ear (Cau et al., 2002; Ma et al., 2000; Ma et al., 1998), defects in other neural regions such as dorsal root ganglia, spinal cord, and dorsal telencephalon require Neurog1/Neurog2 double mutants to detect a phenotype (Fode et al., 2000; Gowan et al., 2001; Ma et al., 1999; Scardigli et al., 2001). Our data using in vivo genetic fate mapping places Neurog1 preferentially in progenitors to interneurons including dI2, dI6, V0, V1, and V2, with a minor contribution to motoneurons. Although Neurog1 is present in these progenitor cells, no obvious phenotype has been seen in the ventral spinal cord in Neurog1 mutant mice (Scardigli et al., 2001)(HIQ and JEJ, unpublished data) possibly due to compensation by Neurog2. In contrast, although Neurog1 can compensate for the loss of Neurog2 in the initiation of neurogenesis, it does not compensate for the regulation of homeodomain markers like Hb9 and Isl1 in motoneuron development (Scardigli et al., 2001).
Neurog1 functions in many regions of the developing nervous system. In the CNS, its two accepted roles include 1) a general role in initiating neuronal differentiation while suppressing glial lineages, and 2) its particular role in specifying glutamatergic type neurons. We note that Neurog1 lineage cells are almost exclusively neurons, consistent with its function in neurogenesis (HIQ and JEJ, unpublished). In contrast, with respect to the second role of Neurog1, it is worth noting that it is not restricted to glutamatergic neuronal lineages. For example, using the same N1457-Cre transgenic line reported here, it was recently shown that in the cerebellum the Neurog1 lineage maps to a subset of Purkinje cells which are GABAergic (Lundell et al., 2009). Future studies using these mice will aid in identifying the full complement of Neurog1 derived cell types in the animal.
With this study we extended our understanding of the complex transcriptional mechanisms that have evolved to direct the precise temporal and spatial expression of Neurog1, and defined the cell types within the neural tube that belong to the Neurog1 lineage. Our understanding of these mechanisms is far from complete, particularly with respect to identifying the upstream factors functioning through the identified enhancers. Controlling the precise levels, timing, and pattern of Neurog1 expression is critical for generating a normal nervous system as its function is necessary for generating the correct numbers and diversity of neurons required for the accurate assembly of neuronal circuits.
Acknowledgments
We acknowledge Nathaniel Heintz and Neil Copeland for providing their respective BAC recombination system reagents used extensively to generate the modified BACs in this study. We acknowledge the outstanding services of the UTSW Transgenic Facility and expert advice from Robert Hammer on generating transgenic mice with BACs. We thank E. J. Kim for critical comments on this manuscript and Dongbo Yu for generating TgN1-15. This work was supported by grants from NIH R01 HD037932 to JEJ, and F31 GM066454 to HIQ.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References Cited
- Ahituv N, Zhu Y, Visel A, Holt A, Afzal V, Pennacchio LA, Rubin EM. Deletion of ultraconserved elements yields viable mice. PLoS Biol. 2007;5:e234. doi: 10.1371/journal.pbio.0050234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andermann P, Ungos J, Raible DW. Neurogenin1 defines zebrafish cranial sensory ganglia precursors. Dev Biol. 2002;251:45–58. doi: 10.1006/dbio.2002.0820. [DOI] [PubMed] [Google Scholar]
- Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neuroscience. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- Blader P, Fischer N, Gradwohl G, Guillemot F, Strahle U. The activity of neurogenin1 is controlled by local cues in the zebrafish embryo. Development. 1997;124:4557–4569. doi: 10.1242/dev.124.22.4557. [DOI] [PubMed] [Google Scholar]
- Blader P, Lam CS, Rastegar S, Scardigli R, Nicod JC, Simplicio N, Plessy C, Fischer N, Schuurmans C, Guillemot F, Strahle U. Conserved and acquired features of neurogenin1 regulation. Development. 2004;131:5627–37. doi: 10.1242/dev.01455. [DOI] [PubMed] [Google Scholar]
- Blader P, Plessy C, Strähle U. Multiple regulatory elements with spatially and temporally distinct activities control neurogenin1 expression in primary neurons of the zebrafish embryo. Mech Dev. 2003;120:211–218. doi: 10.1016/s0925-4773(02)00413-6. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Alessandra P, Jessell TM, Ericson J. A homeodomain protein code specifies progeitor cell identity and neuronal fate in the ventral neural tube. Cell. 2000;101:435–445. doi: 10.1016/s0092-8674(00)80853-3. [DOI] [PubMed] [Google Scholar]
- Cau E, Casarosa S, Guillemot F. Mash1 and Ngn1 control distinct steps of determination and differentiation in the olfactory sensory neuron lineage. Development. 2002;129:1871–1880. doi: 10.1242/dev.129.8.1871. [DOI] [PubMed] [Google Scholar]
- Farah MH, Olson JM, Sucic HB, Hume RI, Tapscott SJ, Turner DL. Generation of neurons by transient expression of neural bHLH proteins in mammalian cells. Development. 2000;127:693–702. doi: 10.1242/dev.127.4.693. [DOI] [PubMed] [Google Scholar]
- Fode C, Ma Q, Casarosa S, Ang SL, Anderson DJ, Guillemot F. A role for neural determination genes in specifying the dorsoventral identity of telencephalic neurons. Genes Dev. 2000;14:67–80. [PMC free article] [PubMed] [Google Scholar]
- Gowan K, Helms AW, Hunsaker TL, Collisson T, Ebert PJ, Odom R, Johnson JE. Crossinhibitory activities of Ngn1 and Math1 allow specification of distinct dorsal interneurons. Neuron. 2001;31:219–232. doi: 10.1016/s0896-6273(01)00367-1. [DOI] [PubMed] [Google Scholar]
- Helms AW, Johnson JE. Progenitors of dorsal commissural interneurons are defined by MATH1 expression. Development. 1998;125:919–925. doi: 10.1242/dev.125.5.919. [DOI] [PubMed] [Google Scholar]
- Helms AW, Johnson JE. Specification of dorsal spinal cord interneurons. Curr op in Neurobiology. 2003;13:42–49. doi: 10.1016/s0959-4388(03)00010-2. [DOI] [PubMed] [Google Scholar]
- Hogan B, Costantini F, Lacy E. Manipulating the mouse embryo: A laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y: 1986. [Google Scholar]
- Hong JW, Hendrix DA, Levine MS. Shadow enhancers as a source of evolutionary novelty. Science. 2008;321:1314. doi: 10.1126/science.1160631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong Y, El-Jaick K, Roessler E, Muenke M, Epstein DJ. A functional screen for sonic hedgehog regulatory elements across a 1 Mb interval identifies long-range ventral forebrain enhancers. Development. 2006;133:761–72. doi: 10.1242/dev.02239. [DOI] [PubMed] [Google Scholar]
- Kriks S, Lanuza GM, Mizuguchi R, Nakafuku M, Goulding M. Gsh2 is required for the repression of Ngn1 and specification of dorsal interneuron fate in the spinal cord. Development. 2005;132:2991–3002. doi: 10.1242/dev.01878. [DOI] [PubMed] [Google Scholar]
- Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- Lee JE. Basic helix-loop-helix genes in neural development. Curr Opin Neurobiol. 1997;7:13–20. doi: 10.1016/s0959-4388(97)80115-8. [DOI] [PubMed] [Google Scholar]
- Lee SK, Pfaff SL. Transcriptional networks regulating neuronal identity in the developing spinal cord. Nat Neurosci. 2001;4(Suppl):1183–91. doi: 10.1038/nn750. [DOI] [PubMed] [Google Scholar]
- Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–484. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo L-C, Johnson JE, Wuenschell CW, Saito T, Anderson DJ. Mammalian achaete-scute homolog 1 is transiently expressed by spatially-restricted subsets of early neuroepithelial and neural crest cells. Genes Dev. 1991;5:1524–1537. doi: 10.1101/gad.5.9.1524. [DOI] [PubMed] [Google Scholar]
- Lumpkin EA, Collisson T, Parab P, Omar-Abdalla A, Haeberle H, Chen P, Doetzlhofer A, White P, Groves A, Segil N, Johnson JE. Math1-driven GFP expression in the developing nervous system of transgenic mice. Gene Expression Patterns. 2003;3:389–395. doi: 10.1016/s1567-133x(03)00089-9. [DOI] [PubMed] [Google Scholar]
- Lundell TG, Zhou Q, Doughty ML. Neurogenin1 expression in cell lineages of the cerebellar cortex in embryonic and postnatal mice. Dev Dyn. 2009;238:3310–25. doi: 10.1002/dvdy.22165. [DOI] [PubMed] [Google Scholar]
- Ma Q, Anderson DJ, Fritzsch B. Neurogenin 1 null mutant ears develop fewer, morphologically normal hair cells in smaller sensory epithelia devoid of innervation. J Assoc Res Otolaryngol. 2000;1:129–43. doi: 10.1007/s101620010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Chen Z, del Barco Barrantes I, de la Pompa JL, Anderson DJ. neurogenin1 is essential for the determination of neuronal precursors for proximal cranial sensory ganglia. Neuron. 1998;120:469–482. doi: 10.1016/s0896-6273(00)80988-5. [DOI] [PubMed] [Google Scholar]
- Ma Q, Fode C, Guillemot F, Anderson DJ. Neurogenin1 and neurogenin2 control two distinct waves of neurogenesis in developing dorsal root ganglia. Genes & Development. 1999;13:1717–28. doi: 10.1101/gad.13.13.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Kintner C, Anderson DJ. Identification of neurogenin, a vertebrate neuronal determination gene. Cell. 1996;87:43–52. doi: 10.1016/s0092-8674(00)81321-5. [DOI] [PubMed] [Google Scholar]
- Ma Q, Sommer L, Cserjesi P, Anderson DJ. Mash1 and neurogenin1 expression patterns define complementary domains of neuroepithelium in the developing CNS and are correlated with regions expressing notch ligands. J Neurosci. 1997;17:3644–3652. doi: 10.1523/JNEUROSCI.17-10-03644.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M, Markstein P, Markstein V, Levine MS. Genome-wide analysis of clustered Dorsal binding sites identifies putative target genes in the Drosophila embryo. Proc Natl Acad Sci U S A. 2002;99:763–8. doi: 10.1073/pnas.012591199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukouyama YS, Deneen B, Lukaszewicz A, Novitch BG, Wichterle H, Jessell TM, Anderson DJ. Olig2+ neuroepithelial motoneuron progenitors are not multipotent stem cells in vivo. Proc Natl Acad Sci U S A. 2006;103:1551–6. doi: 10.1073/pnas.0510658103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray RC, Tapscott SJ, Petersen JW, Calof AL, McCormick MB. A fragment of the neurogenin1 gene confers regulated expression of a reporter gene in vitro and in vivo. Dev Dyn. 2000;218:189–194. doi: 10.1002/(SICI)1097-0177(200005)218:1<189::AID-DVDY16>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Nakada Y, Parab P, Simmons A, Omer-Abdalla A, Johnson JE. Separable enhancer sequences regulate the expression of the neural bHLH transcription factor Ngn1. Dev Biol. 2004;271:479–487. doi: 10.1016/j.ydbio.2004.04.021. [DOI] [PubMed] [Google Scholar]
- Nieto M, Schuurmans C, Britz O, Guillemot F. Neural bHLH genes control the neuronal versus glial fate decision in cortical progenitors. Neuron. 2001;29:401–413. doi: 10.1016/s0896-6273(01)00214-8. [DOI] [PubMed] [Google Scholar]
- Novak A, Guo C, Yang W, Nagy A, Lobe CG. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon cre-mediated excision. Genesis. 2000;28:147–155. [PubMed] [Google Scholar]
- Ovcharenko I, Nobrega MA, Loots GG, Stubbs L. ECR Browser: a tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res. 2004;32:W280–6. doi: 10.1093/nar/gkh355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HC, Mehta A, Richardson JS, Appel B. olig2 is required for zebrafish primary motor neuron and oligodendrocyte development. Dev Biol. 2002;248:356–68. doi: 10.1006/dbio.2002.0738. [DOI] [PubMed] [Google Scholar]
- Perez SE, Rebelo S, Anderson DJ. Early specification of sensory neuron fate revealed by expression and function of neurogenins in the chick embryo. Development. 1999;126:1715–28. doi: 10.1242/dev.126.8.1715. [DOI] [PubMed] [Google Scholar]
- Raft S, Koundakjian EJ, Quinones H, Jayasena CS, Goodrich LV, Johnson JE, Segil N, Groves AK. Cross-regulation of Ngn1 and Math1 coordinates the production of neurons and sensory hair cells during inner ear development. Development. 2007;134:4405–15. doi: 10.1242/dev.009118. [DOI] [PubMed] [Google Scholar]
- Scardigli R, Schuurmans C, Gradwohl G, Guillemot F. Crossregulation between neurogenin2 and pathways specifying neuronal identity in the spinal cord. Neuron. 2001;31:203–217. doi: 10.1016/s0896-6273(01)00358-0. [DOI] [PubMed] [Google Scholar]
- Schuurmans C, Armant O, Nieto M, Stenman JM, Britz O, Klenin N, Brown C, Langevin LM, Seibt J, Tang H, Cunningham JM, Dyck R, Walsh C, Campbell K, Polleux F, Guillemot F. Sequential phases of cortical specification involve Neurogenin-dependent and -independent pathways. Embo J. 2004;23:2892–902. doi: 10.1038/sj.emboj.7600278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasaki R, Pfaff SL. Transcriptional codes and the control of neuronal identity. Annu Rev Neurosci. 2002;25:251–81. doi: 10.1146/annurev.neuro.25.112701.142916. [DOI] [PubMed] [Google Scholar]
- Sommer L, Ma Q, Anderson DJ. Neurogenins, a novel family of atonal-related bHLH transcription factors, are putative mammalian neuronal determination genes that reveal progenitor cell heterogenity in the developing CNS and PNS. Mol Cell Neurosci. 1996;8:221–241. doi: 10.1006/mcne.1996.0060. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genetics. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahle U, Rastegar S. Conserved non-coding sequences and transcriptional regulation. Brain Res Bull. 2008;75:225–30. doi: 10.1016/j.brainresbull.2007.11.010. [DOI] [PubMed] [Google Scholar]
- Sun Y, Nadal-Vicens M, Misono S, Lin MZ, Zubiaga A, Hua X, Fan G, Greenberg ME. Neurogenin promotes neurogenesis and inhibits glial differentiation by independent mechanisms. Cell. 2001;104:365–376. doi: 10.1016/s0092-8674(01)00224-0. [DOI] [PubMed] [Google Scholar]
- Tsuchida T, Ensini M, Morton SB, Baldassare M, Edlund T, Jessell TM, Pfaff SL. Topographic organization of embryonic motor neurons defined by expression of LIM homeobox genes. Cell. 1994;79:957–970. doi: 10.1016/0092-8674(94)90027-2. [DOI] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, Afzal V, Ren B, Rubin EM, Pennacchio LA. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–8. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vokes SA, Ji H, McCuine S, Tenzen T, Giles S, Zhong S, Longabaugh WJ, Davidson EH, Wong WH, McMahon AP. Genomic characterization of Gli-activator targets in sonic hedgehog-mediated neural patterning. Development. 2007;134:1977–89. doi: 10.1242/dev.001966. [DOI] [PubMed] [Google Scholar]
- Vue TY, Aaker J, Taniguchi A, Kazemzadeh C, Skidmore JM, Martin DM, Martin JF, Treier M, Nakagawa Y. Characterization of progenitor domains in the developing mouse thalamus. J Comp Neurol. 2007;505:73–91. doi: 10.1002/cne.21467. [DOI] [PubMed] [Google Scholar]
- Wu M, Zhang Y, Wu NH, Shen YF. Histone marks and chromatin remodelers on the regulation of neurogenin1 gene in RA induced neuronal differentiation of P19 cells. J Cell Biochem. 2009;107:264–71. doi: 10.1002/jcb.22122. [DOI] [PubMed] [Google Scholar]
- Yang XW, Model P, Heintz N. Homologous recombination based modification in Escherichia coli and germline transmission in transgenic mice of a bacterial artificial chromosome. Nat Biotechnol. 1997;15:859–865. doi: 10.1038/nbt0997-859. [DOI] [PubMed] [Google Scholar]
- Zeitlinger J, Zinzen RP, Stark A, Kellis M, Zhang H, Young RA, Levine M. Whole-genome ChIP-chip analysis of Dorsal, Twist, and Snail suggests integration of diverse patterning processes in the Drosophila embryo. Genes Dev. 2007;21:385–90. doi: 10.1101/gad.1509607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Anderson DJ. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109:61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]