Abstract
Autophagy, a cellular catabolic pathway, is evolutionarily conserved from yeast to mammals. Central to this process is the formation of autophagosomes, double-membrane vesicles responsible for delivering long-lived proteins and excess or damaged organelle into the lysosome for degradation and reuse of the resulting macromolecules. In addition to the hallmark discovery of core molecular machinery components involved in autophagosome formation, complex signaling cascades controlling autophagy have also begun to emerge, with mTOR as a central but far from exclusive player. Malfunction of autophagy has been linked to a wide range of human pathologies, including cancer, neurodegeneration and pathogen infection. Here we highlight recent advances in identifying and understanding the core molecular machinery and signaling pathways that are involved in mammalian autophagy.
Keywords: autophagy, lysosomes, mammalian cells, signal transduction, stress
Introduction
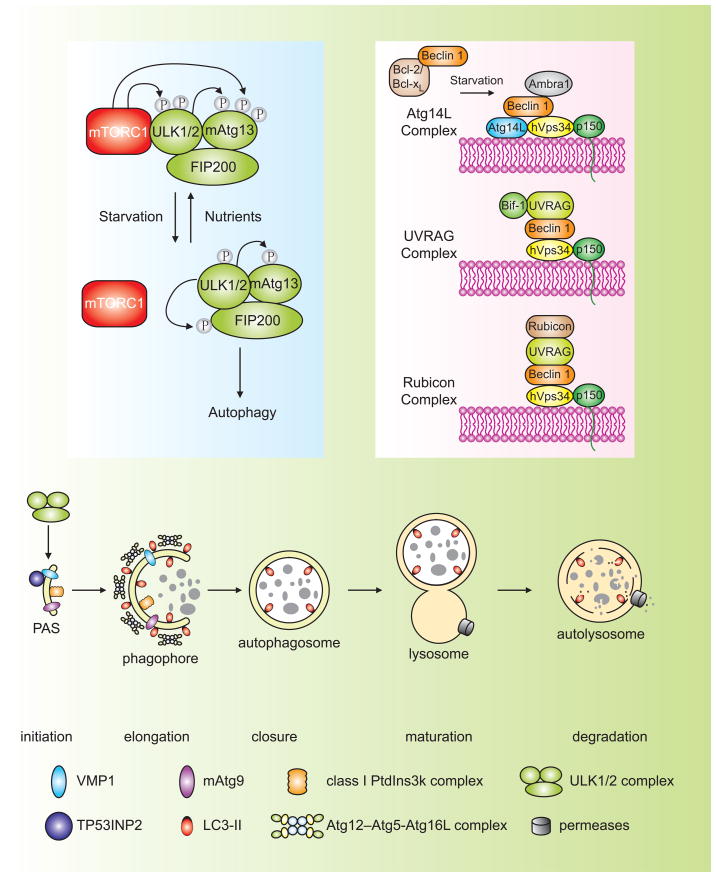
Autophagy, literally meaning “self-eating”, embraces three major intracellular pathways in eukaryotic cells, macroautophagy, microautophagy and chaperone-mediated autophagy (CMA), which share a common destiny of lysosomal degradation, but are mechanistically different from each other [1,2]. During macroautophagy, intact organelles (such as mitochondria) and portions of the cytosol are sequestered into a double-membrane vesicle, termed an autophagosome. Subsequently, the completed autophagosome matures by fusing with an endosome and/or lysosome, thereby forming an autolysosome. This latter step exposes the cargo to lysosomal hydrolases to allow its breakdown, and the resulting macromolecules are transported back into the cytosol through membrane permeases for reuse (Figure 1). By contrast, microautophagy involves the direct engulfment of cytoplasm at the lysosome surface, whereas CMA translocates unfolded, soluble proteins directly across the limiting membrane of the lysosome.
Figure 1. Schematic depiction of the autophagy pathway and its core molecular machinery in mammalian cells.
Mammalian autophagy proceeds through a series of steps, including initiation at the PAS (phagophore assembly site), elongation and expansion of the phagophore, closure and completion of the autophagosome, autophagosome maturation via docking and fusion with an endosome and/or lysosome, breakdown and degradation of the autophagosome inner membrane and cargo, and recycling of the resulting macromolecules. Regulatory components for autophagy induction include the ULK1 and ULK2 complexes that contain various Atg proteins (light blue box at left) that are required for autophagy. The association of mTORC1 with this complex and the activity of mTORC1 depend on the nutrient status. Under nutrient-rich conditions, mTORC1 is associated with the ULK1 and ULK2 complexes, and phosphorylates ULK1, ULK2, and mAtg13; upon inactivation of mTORC1 by nutrient starvation, mTORC1 disassociates, mAtg13, ULK1 and ULK2 are partially dephosphorylated, and activation of ULK1 and ULK2 promotes phosphosphorylation of FIP200. There are at least three class III PtdIns3K complexes (light red box at right), that are involved in autophagosome formation or clearance. The Atg14L (Atg14L-Beclin 1-hVps34-p150) and UVRAG (UVRAG-Beclin 1-hVps34-p150) complexes are required for autophagy, whereas the Rubicon complex (Rubicon-UVRAG-Beclin 1-hVps34-p150) negatively regulates autophagy. Ambra1 and Bif-1 are essential for induction of autophagy, through direct interaction with Beclin 1 and UVRAG, respectively, whereas Bcl-2 binds to Beclin 1 and disrupts the Beclin 1-associated hVps34 complex, thereby inhibiting autophagy.
In this review, we will focus on mammalian macroautophagy (hereafter referred to as autophagy), which plays important physiological roles in human health and disease. The basal, constitutive level of autophagy plays an important role in cellular homeostasis through the elimination of damaged/old organelles as well as the turnover of long-lived proteins and protein aggregates, and thus maintains quality control of essential cellular components. On the other hand, when cells encounter environmental stresses, such as nutrient starvation, hypoxia, oxidative stress, pathogen infection, radiation or anticancer drug treatment, the level of autophagy can be dramatically augmented as a cytoprotective response, resulting in adaptation and survival; however, dysregulated or excessive autophagy may lead to cell death. Thus, defective autophagy has been implicated in the pathogenesis of diverse diseases, such as certain types of neuronal degeneration and cancer, and also in aging [3].
Although autophagy was first identified in mammalian cells approximately 50 years ago, our molecular understanding of it only started in the past decade, largely based on the discovery of autophagy-related (ATG) genes initially in yeast followed by the identification of homologs in higher eukaryotes [4]. Among these Atg proteins, one subset is essential for autophagosome formation, and is referred to as the “core” molecular machinery [5]. These core Atg proteins are composed of four subgroups: (1) The Atg1/unc-51-like kinase (ULK) complex; (2) two ubiquitn-like protein (Atg12 and Atg8/LC3) conjugation systems; (3) the class III phosphatidylinositol 3-kinase (PtdIns3K)/Vps34 complex I; and (4) two transmembrane proteins, Atg9/mAtg9 (and associated proteins involved in its movement such as Atg18/WIPI-1) and VMP1. The proposed site for autophagosome formation, to which most of the core Atg proteins are recruited, is termed the phagophore assembly site (PAS).
In this review, we mainly highlight the recent advances in mammalian autophagy in terms of the molecular machinery involved in the formation and maturation of autophagosomes and the signaling cascades needed for the regulation of autophagy. The clarification of how autophagy is modulated in response to intracellular and extracellular stresses relies largely on the elucidation of the signaling network upstream of the Atg machinery.
Core molecular machinery
ULK complexes
The yeast serine/threonine kinase Atg1 plays a key role in the induction of autophagy, acting downstream of the target of rapamycin (TOR) complex 1 (TORC1). A family of mammalian Atg1 proteins has been identified; among these, unc-51-like kinase 1 (ULK1) and 2 have the highest similarity with yeast Atg1 and appear to be closely related. siRNA knockdown of ULK1 or ULK2 blocks autophagy in HEK293 cells [6]. However, ULK1−/− mice display normal autophagy in response to nutrient deprivation, but delay mitochondrial clearance during reticulocyte maturation [7]. The basis for these differences is not known. It is possible that in some tissues, ULK2 can compensate for the deficiency of ULK1. Furthermore, a role of ULK3 in autophagy induction in oncogene-induced cell senescence has been described recently [8]. Thus, at least three ULKs are involved in mammalian autophagy regulation and they have mechanistically different roles in vivo.
Yeast Atg1 exists in a complex with Atg13 and Atg17. Atg13 is phosphorylated in a TORC1-dependent manner and the phosphorylation state of Atg13 modulates its binding to Atg1 and Atg17; inactivation of TORC1 leads to dephosphorylation of Atg13, increasing Atg1-Atg13-Atg17 complex formation and activating autophagy [4,9]. ULK1 and ULK2 are also in a large complex that includes the mammalian homolog of Atg13 (mAtg13) and the scaffold protein FIP200 (an ortholog of yeast Atg17) [6,10,11]. mAtg13 is essential for autophagy, and it directly interacts with ULK1, ULK2 and FIP200 independent of its phosphorylation state [6,11]. FIP200 is also required for autophagy and binds to ULK1 and ULK2 independent of nutrient status [12], in contrast to the yeast Atg1-Atg17 interaction. In addition, under nutrient-rich conditions, the large ULK1-Atg13-FIP200 complex contains mammalian TORC1 (mTORC1); conversely, following nutrient deprivation, mTORC1 is quickly dissociated from the ULK1 complex [11]. There are several phosphorylation events within this complex, including phosphorylation of mAtg13 by ULK1, ULK2 and mTORC1, phosphorylation of FIP200 by ULK1 and ULK2, and phosphorylation of ULK1 and ULK2 by mTORC1 (Figure 1) [6,11]. Under conditions that induce autophagy, a decrease in mTORC1 activity leads to dephosphorylation of ULK1, ULK2 and mAtg13, activation of ULK1 and ULK2, and phosphorylation of mAtg13 and FIP200 by ULK1 and ULK2 [6,11]. Further studies are required to characterize the functional significance of these phosphorylation events. Recently, a new, mAtg13-interacting protein, Atg101, was found to interact with ULK1 in a mAtg13-dependent manner, and is essential for autophagy [13]. However, the role of the ULK1-Atg13-Atg101 complex in autophagy regulation remains unclear.
Two ubiquitin-like proteins, Atg12 and Atg8/LC3, and their conjugation systems
Studies in yeast and mammals have identified two ubiquitin-like proteins, Atg12 and Atg8/LC3, and their respective, partially overlapping, conjugation systems, which are proposed to act during elongation and expansion of the phagophore membrane. Atg12 is conjugated to Atg5 in a reaction that requires Atg7 and Atg10 (E1 and E2-like enzymes, respectively). The Atg12–Atg5 conjugate then interacts non-covalently with Atg16L, which oligomerizes to form a large multimeric complex called the Atg16L complex. Atg8/LC3 is cleaved at its C terminus by Atg4 to generate the cytosolic LC3-I with a C-terminal glycine residue, which is conjugated to phosphatidylethanolamine (PE) in a reaction that requires Atg7 and the E2-like enzyme Atg3. The lipidated form of LC3 (LC3-II) is attached to both faces of the phagophore membrane, but is ultimately removed from the autophagosome outer membrane, which is followed by fusion of the autophagosome with a late endosome/lysosome [4].
Recent work suggests that these two ubiquitination-like systems are closely connected. On the one hand, the Atg16L complex is localized to the phagophore and it can act as a novel E3-like enzyme, determining the sites of Atg8/LC3 lipidation [14,15]. On the other hand, the Atg8/LC3 conjugation machinery seems to be essential for formation of the Atg16L complex. In Atg3-deficient mice, where no LC3-II can be detected, Atg12–Atg5 conjugation is markedly reduced, and dissociation of the Atg16L complex from the phagophore is delayed; autophagosomes are smaller than in the wild type and appear either open-ended or multi-lamellar [16], indicating a role for the Atg16L complex and LC3-lipidation for the elongation and closure of the phagophore. This hypothesis is futher supported by the observation that overexpression of an inactive mutant of Atg4 inhibits the lipidation of LC3, and in these cells a significant number of nearly complete autophagosomes are not closed [17].
Class III phosphatidylinositol 3-kinase complex
In yeast, the only phosphatidylinositol 3-kinase (PtdIns3K) is Vps34, and it exists in two different complexes, complex I and II. Complex I, consisting of Vps34, Vps15, Atg6 and Atg14, is required for the induction of autophagy, and the lipid kinase activity of Vps34 is essential for generating phosphatidylinositol (3)-phosphate (PtdIns(3)P) at the PAS to allow recruitment of other Atg proteins. Complex II, consisting of Vps34, Vps15, Atg6 and Vps38, is required for the vacuolar sorting of carboxypeptidase Y. In mammals, there are two types of PtdIns3K: class I and III. Formation of the mammalian class III PtdIns3K complex, including hVps34, Beclin 1 (a homolog of Atg6), and p150 (a homolog of Vps15), is conserved. The orthologs of Atg14 and Vps38 have recently been identified and are called Atg14-like protein (Atg14L, or Barkor) and ultraviolet irradiation resistant-associated gene (UVRAG), respectively [18–20].
Atg14L plays an important role in mammalian autophagy. Under nutrient-rich conditions, a subpopulation of Atg14L localizes to the ER; upon starvation, Atg14L localizes to Atg16L- and LC3-positive structures, indicating the phagophore and autophagosome, respectively, independently of the interaction of Atg14L with hVps34 and Beclin 1 [18,21]. Importantly, depletion of Atg14L reduces Atg16L and LC3 puncta formation [21]. Overexpression of Atg14L stimulates the kinase activity of hVps34, and induces autophagy, whereas Atg14L knockdown reduces PtdIns(3)P production, and inhibits autophagy [19,22]. Thus, a possible role of Atg14L is to direct the class III PtdIns3K complex to the phagophore to initiate the recruitment of Atg machinery.
Recent studies suggest that UVRAG participates in at least four different mechanisms to regulate autophagy. First, UVRAG competes with Atg14L for binding to Beclin 1; the interactions of Atg14L and UVRAG with the Beclin 1-hVps34-p150 complex are mutually exclusive [18,19]. Second, UVRAG interacts with Bif-1 (Bax-interacting factor 1); Bif-1 is required for autophagy and colocalizes with Atg5, LC3 and mAtg9 during starvation [23]. It is proposed that recruitment of Bif-1 via UVRAG may provide the machinery to deform membranes, as Bif-1 has an N-BAR domain and shows membrane binding and bending activities [24]. Third, UVRAG interacts with the class C Vps/HOPS proteins, promoting autophagosome fusion with the late endosome/lysosome, thereby accelerating delivery and degradation of autophagic cargo [25]. Fourth, the recently identified Rubicon (RUN domain and cysteine-rich domain containing, Beclin 1-interacting) protein forms a complex with UVRAG-Beclin 1-hVps34-p150; this complex localizes to the late endosome/lysosome and negatively regulates autophagosome maturation [21,22]. Rubicon reduces hVps34 activity and inhibits autophagy.
In addition to hVps34, Atg14L and UVRAG, Beclin 1 also interacts with Ambra 1 (activating molecule in Beclin 1-regulated autophagy). Ambra 1 functions as a positive regulator of autophagy and the mechanism remains unclear [26]. Collectively, there exist multiple mammalian hVps34-Beclin 1 complexes that may participate in distinct steps of autophagy regulation (Figure 1), either at the early stage to promote autophagosome formation or at the later stage to promote autophagosome maturation.
Transmembrane proteins in mammalian autophagy
Mammalian Atg9 (mAtg9) and vacuole membrane protein 1 (VMP1) are the two transmembrane proteins so far identified that are required for mammalian autophagy. mAtg9, with both the N and C termini in the cytosol, spans the membrane six times. It is located in the trans-Golgi network and late endosomes, and upon starvation or rapamycin treatment, redistributes to peripheral sites, overlapping with GFP-LC3-positive autophagosomes. The cycling of mAtg9 after starvation is ULK1-dependent, and also requires the kinase activity of hVps34 [27], which is similar to the yeast protein [28]. Although its functions remain unclear, based on the existing data from yeast Atg9, mAtg9 potentially contributes to the delivery of membrane to the forming autophagosome, an attrative model that needs to be experimentally tested in mammalian cells.
In contrast to mAtg9, VMP1 has no known homologs in yeast. The localization of VMP1 is controversial: in mammalian cells it is localized to the plasma membrane and also colocalizes with LC3 and Beclin 1 upon autophagy induction [29], whereas the VMP1 homolog in Dictyostelium discoideum localizes to the ER [30]. In mammalian cells, ectopical overexpression of VMP1 triggers autophagy even under nutrient-rich conditions, whereas depletion of VMP1 blocks starvation- and rapamycin-induced autophagy [29]. Importantly, VMP1 interacts with Beclin 1, and this interaction is essential for autophagy induced by VMP1 overexpression [29]. VMP1 might function as a transmembrane protein that recruits Beclin 1 and other components in the class III PtdIns3K complex to the phagophore. This is supported by a recent finding that a novel VMP1-interacting protein, TP53INP2 (tumor protein 53-induced nuclear protein 2), is essential for translocation of Beclin 1 and LC3 to autophagosomes upon autophagy stimulation, potentially through its interaction with VMP1 [31]. TP53INP2 is essential for autophagy. It translocates from the nucleus to autophagosomes upon autophagy induction, where it interacts with LC3 as well as VMP1, but not Beclin 1.
Signaling pathways regulating autophagy
PtdIns3K-Akt-mTORC1
The target of rapamycin (TOR) is a highly conserved serine/threonine protein kinase that acts as a central sensor of growth factors, nutrient signals and engery status. TOR serves as a master regulator of autophagy [32]. TOR exists in two distinct complexes, TORC1 and TORC2 that are conserved from yeast to mammals, and TORC1 has a primary function in regulating autophagy. In yeast, inhibiting the TORC1 complex during nitrogen starvation or by rapamycin stimulates autophagy [4]. The mammalian TORC1 (mTORC1) is also sensitive to rapamycin, which in many settings stimulates autophagy. However, a recent report challenged this view by showing that rapamycin and siRNA knockdown of one of the key downstream effectors of mTORC1, S6 kinase 1 (S6K1), inhibit autophagy in cancer cells [33], and a more recent finding shows that mTORC1 regulates autophagy through an unknown mechanism that is essentially insensitive to rapamycin [34].
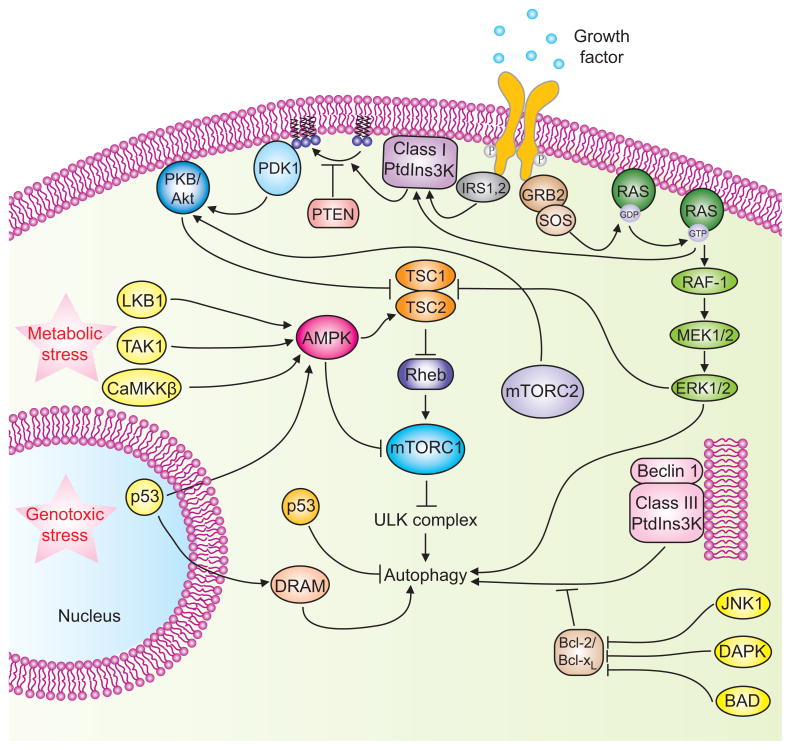
mTORC1 integrates upstream activating signals that inhibit autophagy through the class I PtdIns3K-protein kinase B (PKB, also known as Akt) pathway (Figure 2). Upon association with growth factor, receptor tyrosine kinases undergo autophosphorylation and become activated, leading to stimulation of two key signal-transducing components: the small GTPase Ras and class I PtdIns3K. Class I PtdIns3K catalyzes production of PtdIns(3)P at the plasma membrane, which increases membrane recruitment of both PKB and its activator PDK1 (phosphoinositide-dependent protein kinase 1), leading to activation of PKB. PtdIns3K kinase activity can be opposed by PTEN, a 3′-phosphoinositide phosphatase, subsequently decreasing PKB activity, and inhibiting mTOR. PtdIns3K-PKB activation suppresses autophagy in mammalian cells. PKB further activates mTORC1 through inhibiting a downstream protein complex, the tuberous sclerosis complex 1/2 (TSC1/TSC2). The TSC1/TSC2 heterodimer, which is a stable complex, senses the upstream inputs from various kinases, including PKB and ERK1/2 [35,36]. Phosphorylation of TSC2 by PKB or ERK1/2 leads to the disruption of its complex with TSC1, and results in mTOR activation. TSC1/TSC2 acts as the GTPase-activating protein for Rheb, a small GTP-binding protein that binds to and activates mTOR in its GTP-bound form. Ras has opposing roles in autophagy regulation: It inhibits autophagy by activating the PtdIns3K-PKB-mTORC1 pathway, and at the same time, it may induce autophagy via the Raf-1-MEK1/2-ERK1/2 pathway [37,38]. Finally, the mTORC2 complex is also involed in autophagy regulation. Full activation of PKB requires mTORC2 [39], and inhibition of PKB, caused by mTORC2 depletion, reduces the phosphorylation of, and therefore activates, the forkhead box O (FoxO3) transcription factor, which stimulates autophagy in muscle cells independent of the activity of mTORC1 [40].
Figure 2. Signaling cascades involved in the regulation of mammalian autophagy.
Autophagy is regulated by a complex signaling network of various stimulatory (arrowheads) and inhibitory (bars) inputs. Activation of growth factor receptors stimulates the class I PtdIns3K complex and small GTPase Ras, which leads to activation of the PtdIns3K-PKB-mTORC1 pathway and the Raf-1-MEK1/2-ERK1/2 pathway, respectively. PKB and ERK1/2 phosphorylate and inhibit the GTPase-activating protein complex TSC1/TSC2, leading to the stabilization of Rheb-GTPase, which, in turn, activates mTORC1, causing inhibition of autophagy. Activated ERK1/2 also stimulates autophagy. mTORC2 inhibits autophagy through the phosphorylation and activation of PKB. Metabolic stress, such as high AMP/ATP ratios resulting from energy depletion, or an increase in the cytosolic free Ca2+ concentration or cytokines, cause the AMP-activated protein kinase (AMPK) to be phosphorylated and activated by LKB1, CaMKKβ and TAK1, respectively. AMPK phosphorylates and activates TSC1/TSC2, leading to inactivation of mTORC1 and autophagy induction. Genotoxic and oncogenic stresses result in nuclear p53 stabilization and activation, which stimulates autophagy through activation of AMPK or upregulation of DRAM. In contrast, cytosolic p53 has an inhibitory effect on autophagy. Anti-apoptotic proteins, Bcl-2 or Bcl-XL, associate with Beclin 1 and inhibit the Beclin 1-associated class III PtdIns3K complex, causing inhibition of autophagy. For additional details, see the main text.
AMPK
The AMP-activated protein kinase (AMPK) is another sensor of cellular bioenergetics, specifically in response to energy stress. During nutrient and energy depletion, AMPK is activated by a decreased ATP/AMP ratio through the upstream LKB1 kinase (encoded by the Peutz-Jeghers syndrome gene). Active AMPK leads to phosphorylation and activation of TSC1/2 and inhibition of mTORC1 activity. Thus, the phosphorylation of TSC1/TSC2 by AMPK and PKB has opposite effects on mTORC1 and connects mTORC1 with energy and growth factor signaling, respectively (Figure 2). Recently, it is reported that AMPK regulates mTORC1 signaling through an alternative mechanism, whereby AMPK directly phosphorylates Raptor, a subunit of mTORC1, and this Raptor phosphorylation is important for the inhibition of mTORC1 signaling by AMPK [41]. Thus, AMPK serves as a positive regulator of autophagy. Under stress conditions, the LKB1-AMPK pathway phosphorylates and stabilizes p27kip1, a cell cycle inhibitor, and stabilized p27kip1 induces autophagy [42]. An increase in the cytosolic free Ca2+ concentration and cytokines (such as TRAIL) activates AMPK via activation of the Ca2+/calmodulin-dependent kinase kinase-β (CaMKKβ) and transforming growth factor-β-activating kinase 1 (TAK1), respectively, and these pathways are required for Ca2+- or TRAIL-induced autophagy [43,44]. Moreover, AMPK activity contributes to the induction of autophagy during hypoxia [45].
p53
The p53 tumor suppressor, the “guardian of the cellular genome”, has dual positive and negative regulatory roles in autophagy induction (Figure 2) [46]. Upon genotoxic stress or oncogenic activation, the activation of p53 induces autophagy; p53 activates AMPK, which in turn, activates the TSC1/2 complex, leading to the inhibition of the mTORC1 pathway [47]. p53 can also induce autophagy through upregulation of DRAM (damage-regulated modulator of autophagy) [48].
Remarkably, chemical inhibition of p53, knockdown of p53 with siRNA, or deletion of the p53 gene can trigger the onset of autophagy [49]. Several stimuli, including starvation or ER stress, can induce HDM2-dependent proteasomal degradation of p53 to favor autophagy induction, positioning p53 as a negative regulator of autophagy. HDM2, the p53-specific E3 ubiquitin ligase, targets p53 to proteasome-mediated destruction. Inhibition of HDM2 blocks the depletion of p53 and also prevents the activation of autophagy [49]. More importantly, it is the cytoplasmic p53 that exerts its inhibitory function towards autophagy, in contrast to the transcriptionally active nuclear p53 that promotes autophagy. Upon reintroduction into p53−/− cancer cells, mutants of p53 that are restricted to the cytosol effectively inhibit autophagy, whereas mutants of p53 that accumulate within the nucleus fail to block autophagy [49]. The inhibitory role of cytosolic p53 in autophagy may contribute to the strong oncogenic action of certain p53 mutants that are preferentially localized to the cytosol [50].
Bcl-2 protein family
In mammals, the Bcl-2 protein family plays a dual role in autophagy regulation. Anti-apoptotic proteins, such as Bcl-2, Bcl-XL, Bcl-w and Mcl-1, can inhibit autophagy, whereas pro-apoptotic BH3-only proteins, such as BNIP3, Bad, Bik, Noxa, Puma and BimEL, can induce autophagy [51]. The binding of Bcl-2 to Beclin 1 disrupts the association of Beclin 1 with hVps34, decreases Beclin 1-associated hVps34 PtdIns3K activity and thereby inhibits autophagy. There are at least three distinct mechanisms that may account for the release of Beclin 1 from its inhibitory interaction with Bcl-2/Bcl-XL (Figure 2). One model depicts that the BH3 domain of BH3-only proteins such as Bad, may competitively disrupt the inhibitory interaction of Beclin 1 and Bcl-2/Bcl-XL [52]. A second mechanism for the dissociation of Beclin 1 from its inhibitory interaction with Bcl-2 involves the phosphorylation of Bcl-2 by the stress-activated c-Jun N-terminal Kinase 1 (JNK1). Starvation induces phosphorylation of Bcl-2 at residues T69, S70 and S87 of the non-structured loop; expression of a non-phosphorylatable Bcl-2 mutant (T69A, S70A, S87A) or inhibition of JNK1 abolishes the starvation-triggered dissociation of Bcl-2 from Beclin 1, and inhibits autophagy; expression of a constitutively active JNK1 results in constitutive Bcl-2 multisite phosphorylation, dissociation of Bcl-2 from Beclin 1 and stimulation of autophagy [53]. Third, a recent finding shows that the activation of Beclin 1 to induce autophagy involves the phosphorylation of Beclin 1 by the death-associated protein kinase (DAPK). DAPK physically interacts with Beclin 1, and phosphorylates Beclin 1 on Thr119 located at a crucial position within the BH3 domain of Beclin 1, and thus promotes the dissociation of Beclin 1 from its inhibitor, Bcl-XL, and autophagy induction [54].
Concluding remarks
In the past decade there has been a tremendous advance in our understanding of the molecular machinery involved in mammalian autophagy. Nonetheless, many outstanding questions remain to be answered, including the mystery of the membrane source for autophagosome formation. By comparison, our knowledge about the signaling regulation of autophagy is relatively limited, in particular, with regard to the complex coordination between autophagy machinery and signaling inputs. As an intracellular self-destructive system, autophagy must be tightly regulated in order to adapt to different intracellular and extracellular stresses. This raises a fundamental question: How does the cell determine the specificity and magnitude of autophagy based on the inputs from a variety of signaling mechanisms? Mammalian autophagy has gained tremendous attention due to its implications in a wide range of physiological processes and diseases in humans. Our current understanding of this process and continued examination of its mechanism and regulation hold the potential for practical modulation of autophagy and its use as a therapeutic intervention.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Klionsky DJ. The molecular machinery of autophagy: unanswered questions. J Cell Sci. 2005;118:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Massey AC, Zhang C, Cuervo AM. Chaperone-mediated autophagy in aging and disease. Curr Top Dev Biol. 2006;73:205–235. doi: 10.1016/S0070-2153(05)73007-6. [DOI] [PubMed] [Google Scholar]
- 3.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang Z, Klionsky DJ. An overview of the molecular mechanism of autophagy. Curr Top Microbiol Immunol. 2009;335:1–32. doi: 10.1007/978-3-642-00302-8_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 6 ●.Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. This paper, along with reference 11, shows that ULK1 and ULK2 are in a large protein complex with Atg13 and FIP200, and that complex formation is independent of nutrient status. mTOR directly regulates the complex by phosphorylating Atg13, ULK1 and ULK2; ULK1 and ULK2 phosphorylate Atg13 and FIP200. Inactivation of mTOR by rapamycin treatment or starvation, leads to dephosphorylation of Atg13, ULK1 and ULK2, and hence activates ULK1 and ULK2 to phosphorylate FIP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kundu M, Lindsten T, Yang CY, Wu J, Zhao F, Zhang J, Selak MA, Ney PA, Thompson CB. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood. 2008;112:1493–1502. doi: 10.1182/blood-2008-02-137398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Young ARJ, Narita M, Ferreira M, Kirschner K, Sadaie M, Darot JF, Tavare S, Arakawa S, Shimizu S, Watt FM. Autophagy mediates the mitotic senescence transition. Genes Dev. 2009;23:798–803. doi: 10.1101/gad.519709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ganley IG, Lam du H, Wang J, Ding X, Chen S, Jiang X. ULK1·ATG13·FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284:12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11 ●.Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. See the annotation to reference 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12 ●.Hara T, Takamura A, Kishi C, Iemura S-i, Natsume T, Guan J-L, Mizushima N. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. This paper demonstrates the identification of a novel ULK-interacting protein, FIP200, which is an ortholog of yeast Atg17. FIP200 is required for autophagy, and is important for the kinase activity of ULK1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mercer CA, Kaliappan A, Dennis PB. A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy. Autophagy. 2009;5:649–662. doi: 10.4161/auto.5.5.8249. [DOI] [PubMed] [Google Scholar]
- 14.Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 2008;19:2092–2100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, Inagaki F, Ohsumi Y. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem. 2007;282:37298–37302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- 16.Sou YS, Waguri S, Iwata J, Ueno T, Fujimura T, Hara T, Sawada N, Yamada A, Mizushima N, Uchiyama Y, et al. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol Biol Cell. 2008;19:4762–4775. doi: 10.1091/mbc.E08-03-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujita N, Hayashi-Nishino M, Fukumoto H, Omori H, Yamamoto A, Noda T, Yoshimori T. An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol Biol Cell. 2008;19:4651–4659. doi: 10.1091/mbc.E08-03-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun Q, Fan W, Chen K, Ding X, Chen S, Zhong Q. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci U S A. 2008;105:19211–19216. doi: 10.1073/pnas.0810452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, Jung JU. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8:688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- 21 ●●.Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, Maejima I, Shirahama-Noda K, Ichimura T, Isobe T, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11:385–396. doi: 10.1038/ncb1846. This paper, along with reference 22, demonstrates the existence of three Beclin 1 complexes: Atg14L-Beclin 1- hVps34-p150, UVRAG-Beclin 1-hVps34-p150 and Rubicon-UVRAG-Beclin 1-hVps34-p150, which regulate autophagy at multiple steps. Atg14L is required for autophagy, whereas Rubicon is negatively involved in the maturation of autophagosomes. Atg14L localizes to the phagophore, and autophagosome, as well as the ER, whereas Rubicon resides on the endosome/lysosome. [DOI] [PubMed] [Google Scholar]
- 22 ●●.Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, Heintz N, Yue Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11:468–476. doi: 10.1038/ncb1854. See the annotation to reference 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, Liang C, Jung JU, Cheng JQ, Mule JJ, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9:1142–1151. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takahashi Y, Meyerkord CL, Wang HG. Bif-1/endophilin B1: a candidate for crescent driving force in autophagy. Cell Death Differ. 2009;16:947–955. doi: 10.1038/cdd.2009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25 ●.Liang C, Lee JS, Inn KS, Gack MU, Li Q, Roberts EA, Vergne I, Deretic V, Feng P, Akazawa C, et al. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol. 2008;10:776–787. doi: 10.1038/ncb1740. This study demonstrates that UVRAG interacts with the class C Vps complex, and this interaction promotes autophagosome maturation as well as endocytic vesicle trafficking, which is independent of Beclin 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, Corazzari M, Fuoco C, Ucar A, Schwartz P, et al. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–1125. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- 27.Young ARJ, Chan EYW, Hu XW, Köchl R, Crawshaw SG, High S, Hailey DW, Lippincott-Schwartz J, Tooze SA. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci. 2006;119:3888–3900. doi: 10.1242/jcs.03172. [DOI] [PubMed] [Google Scholar]
- 28.Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell. 2004;6:79–90. doi: 10.1016/s1534-5807(03)00402-7. [DOI] [PubMed] [Google Scholar]
- 29.Ropolo A, Grasso D, Pardo R, Sacchetti ML, Archange C, Lo Re A, Seux M, Nowak J, Gonzalez CD, Iovanna JL, et al. The pancreatitis-induced vacuole membrane protein 1 triggers autophagy in mammalian cells. J Biol Chem. 2007;282:37124–37133. doi: 10.1074/jbc.M706956200. [DOI] [PubMed] [Google Scholar]
- 30.Calvo-Garrido J, Carilla-Latorre S, Lazaro-Dieguez F, Egea G, Escalante R. Vacuole membrane protein 1 is an endoplasmic reticulum protein required for organelle biogenesis, protein secretion, and development. Mol Biol Cell. 2008;19:3442–3453. doi: 10.1091/mbc.E08-01-0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31 ●.Nowak J, Archange C, Tardivel-Lacombe J, Pontarotti P, Pebusque MJ, Vaccaro MI, Velasco G, Dagorn JC, Iovanna JL. The TP53INP2 protein is required for autophagy in mammalian cells. Mol Biol Cell. 2009;20:870–881. doi: 10.1091/mbc.E08-07-0671. This study describes the identification of a novel protein, TP53INP2, essential for mammalian autophagy. Moreover, TP53INP2 functions as a scaffold protein that recruits LC3 and Beclin 1 to the autophagosome through interaction with the transmembrane protein VMP1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Codogno P, Meijer AJ. Autophagy and signaling: their role in cell survival and cell death. Cell Death Differ. 2005;12 Suppl 2:1509–1518. doi: 10.1038/sj.cdd.4401751. [DOI] [PubMed] [Google Scholar]
- 33.Zeng X, Kinsella TJ. Mammalian target of rapamycin and S6 kinase 1 positively regulate 6-thioguanine-induced autophagy. Cancer Res. 2008;68:2384–2390. doi: 10.1158/0008-5472.CAN-07-6163. [DOI] [PubMed] [Google Scholar]
- 34.Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS. An ATP-competitive Mammalian Target of Rapamycin Inhibitor Reveals Rapamycin-resistant Functions of mTORC1. J Biol Chem. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- 36.Ma L, Chen Z, Erdjument-Bromage H, Tempst P, Pandolfi PP. Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell. 2005;121:179–193. doi: 10.1016/j.cell.2005.02.031. [DOI] [PubMed] [Google Scholar]
- 37.Furuta S, Hidaka E, Ogata A, Yokota S, Kamata T. Ras is involved in the negative control of autophagy through the class I PI3-kinase. Oncogene. 2004;23:3898–3904. doi: 10.1038/sj.onc.1207539. [DOI] [PubMed] [Google Scholar]
- 38.Pattingre S, Bauvy C, Codogno P. Amino acids interfere with the ERK1/2-dependent control of macroautophagy by controlling the activation of Raf-1 in human colon cancer HT-29 cells. J Biol Chem. 2003;278:16667–16674. doi: 10.1074/jbc.M210998200. [DOI] [PubMed] [Google Scholar]
- 39.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 40.Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, Burden SJ, Di Lisi R, Sandri C, Zhao J, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 41.Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liang J, Shao SH, Xu ZX, Hennessy B, Ding Z, Larrea M, Kondo S, Dumont DJ, Gutterman JU, Walker CL, et al. The energy sensing LKB1-AMPK pathway regulates p27(kip1) phosphorylation mediating the decision to enter autophagy or apoptosis. Nat Cell Biol. 2007;9:218–224. doi: 10.1038/ncb1537. [DOI] [PubMed] [Google Scholar]
- 43.Høyer-Hansen M, Bastholm L, Szyniarowski P, Campanella M, Szabadkai G, Farkas T, Bianchi K, Fehrenbacher N, Elling F, Rizzuto R, et al. Control of macroautophagy by calcium, calmodulin-dependent kinase kinase-beta, and Bcl-2. Mol Cell. 2007;25:193–205. doi: 10.1016/j.molcel.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 44 ●.Herrero-Martin G, Høyer-Hansen M, Garcia-Garcia C, Fumarola C, Farkas T, Lopez-Rivas A, Jäättelä M. TAK1 activates AMPK-dependent cytoprotective autophagy in TRAIL-treated epithelial cells. EMBO J. 2009;28:677–685. doi: 10.1038/emboj.2009.8. This study shows that TRAIL-induced autophagy involves a novel signaling pathway in which the activation of AMPK is mediated by TAK1, but independent of LKB1 and CaMKKβ. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Papandreou I, Lim AL, Laderoute K, Denko NC. Hypoxia signals autophagy in tumor cells via AMPK activity, independent of HIF-1, BNIP3, and BNIP3L. Cell Death Differ. 2008;15:1572–1581. doi: 10.1038/cdd.2008.84. [DOI] [PubMed] [Google Scholar]
- 46.Levine B, Abrams J. p53: The Janus of autophagy? Nat Cell Biol. 2008;10:637–639. doi: 10.1038/ncb0608-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feng Z, Zhang H, Levine AJ, Jin S. The coordinate regulation of the p53 and mTOR pathways in cells. Proc Natl Acad Sci U S A. 2005;102:8204–8209. doi: 10.1073/pnas.0502857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Crighton D, Wilkinson S, O’Prey J, Syed N, Smith P, Harrison PR, Gasco M, Garrone O, Crook T, Ryan KM. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 49 ●●.Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D’Amelio M, Criollo A, Morselli E, Zhu C, Harper F, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008;10:676–687. doi: 10.1038/ncb1730. This paper, along with reference 50, reveals that p53 has dual positive and negative regulatory roles in autophagy induction, with cytosolic p53 acting as an endogenous repressor of autophagy, in contrast to nuclear p53. These hallmark studies provide evidences to link autophagy to the cancer-associated dysregulation of p53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50 ●.Morselli E, Tasdemir E, Maiuri MC, Galluzzi L, Kepp O, Criollo A, Vicencio JM, Soussi T, Kroemer G. Mutant p53 protein localized in the cytoplasm inhibits autophagy. Cell Cycle. 2008;7:3056–3061. doi: 10.4161/cc.7.19.6751. See the annotation to reference 49. [DOI] [PubMed] [Google Scholar]
- 51.Levine B, Sinha S, Kroemer G. Bcl-2 family members: dual regulators of apoptosis and autophagy. Autophagy. 2008;4:600–606. doi: 10.4161/auto.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52 ●.Maiuri MC, Le Toumelin G, Criollo A, Rain JC, Gautier F, Juin P, Tasdemir E, Pierron G, Troulinaki K, Tavernarakis N, et al. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J. 2007;26:2527–2539. doi: 10.1038/sj.emboj.7601689. This study demonstrates that the interaction between Beclin 1 and its inhibitors, Bcl-2 and Bcl-XL, involves the BH3 domain of Beclin 1. Importantly, the pharmacological BH3 mimics ABT737 and the BH3-only protein Bad, competitively disrupt the interaction between Beclin 1 and Bcl-2 or Bcl-XL, and hence stimulate autophagy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53 ●.Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008;30:678–688. doi: 10.1016/j.molcel.2008.06.001. This study establishes that JNK1 mediates starvation-induced multisite phosphorylation of Bcl-2, leading to dissociation of Bcl-2 from Beclin 1, and hence activation of autophagy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54 ●.Zalckvar E, Berissi H, Mizrachy L, Idelchuk Y, Koren I, Eisenstein M, Sabanay H, Pinkas-Kramarski R, Kimchi A. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep. 2009;10:285–292. doi: 10.1038/embor.2008.246. This report demonstrates that DAPK phosphorylates Beclin 1, and this phosphorylation promotes dissociation of Beclin 1 from its inhibitor, Bcl-XL, and hence leads to activation of autophagy. [DOI] [PMC free article] [PubMed] [Google Scholar]