Abstract
Rapid, accurate diagnosis of community-acquired pneumonia (CAP) due to Mycoplasma pneumoniae is compromised by low sensitivity of culture and serology. PCR has emerged as a sensitive method to detect M. pneumoniae DNA in clinical specimens. However, conventional real-time PCR is not cost-effective for routine out-patient or implementation. Here, we evaluate a novel microfluidic real-time PCR platform (Advanced Liquid Logic, Inc.) that is rapid, portable, and fully automated. We enrolled patients with CAP and extracted DNA from nasopharyngeal wash (NPW) specimens using a biotinylated capture probe and streptavidin-coupled magnetic beads. Each extract was tested for M. pneumoniae-specific DNA by real-time PCR on both conventional and microfluidic platforms using Taqman probe and primers. Three of 59 (5.0%) NPWs were positive, and agreement between the methods was 98%. The microfluidic platform was equally sensitive but three times faster and offers an inexpensive and convenient diagnostic test for microbial DNA.
Keywords: Mycoplasma pneumoniae, real-time PCR, DNA-based diagnostics, community-acquired pneumonia, diagnostic microbiology
Introduction
Despite numerous medical advances over the last several decades, pneumonia remains a leading, global cause of illness and mortality, and the majority of cases are community-acquired pneumonia (CAP) (Mandell et al., 2007;Venditti et al., 2009). CAP is difficult to diagnose because the clinical presentation and radiographic results are not distinctive or consistent (Fee and Weber, 2007;Hagaman et al., 2009;Lieberman et al., 2003;Plouffe and Martin, 2008). The progression, duration and degree of severity vary depending upon the etiology, as well as the underlying age, health and immunity of the patient (Jokinen et al., 2001;Luna et al., 2000;Mandell et al., 2007;Waites et al., 2005).
Mycoplasma pneumoniae causes up to a third of all cases of CAP and is particularly challenging to diagnose (Huang et al., 2006;Kashyap et al., 2008;Korppi et al., 2004;Kung and Wang, 2007;Luna et al., 2000;Ngeow et al., 2005;Reechaipichitkul et al., 2005;Thurman et al., 2009;Tsolia et al., 2004). Patients with mycoplasmal infections often lack the “classic” CAP-associated symptoms, which are characterized by acute, rapidly progressive lower respiratory infection with fever and purulent sputum. In contrast, mycoplasmal CAP usually has an insidious onset followed by several days or weeks of slowly worsening dry cough, fever, and malaise (Herold and Sailer, 2004;Luna et al., 2000;Plouffe, 2000). A radiographic diagnosis is challenging because most patients with mycoplasmal CAP rarely develop a focal pneumonia as seen with most other etiologies. Because a single gold standard for the diagnosis of mycoplasmal CAP is lacking, the usual approaches are a combination of culture, serology and PCR.
There are limitations to the diagnosis of mycoplasmal CAP by culture and serology. Successful culture of M. pneumoniae requires experienced technologists, specialized media, and prolonged incubation periods. Under optimal conditions, the sensitivity of recovery of M. pneumoniae by culture can vary from <30 to 60% (Ieven and Goossens, 1997;Kenny et al., 1990;Morozumi et al., 2006;Ozaki et al., 2007;Waris et al., 1998). Serology is a more reliable diagnostic tool, but since tests for anti-M. pneumoniae antibodies are usually outsourced to immunology laboratories, the results may pend for several weeks. In addition, it is often necessary to measure IgG, IgM and IgA titers and to compare acute and convalescent specimens to confirm or exclude the diagnosis. Specific IgA titers are the most helpful, but depending upon the serological reagents and methods, there is considerable variability in the reported specificity and sensitivity (Beersma et al., 2005;Daxboeck et al., 2003;Liu et al., 2008;Templeton et al., 2003;Thurman et al., 2009;Watkins-Riedel et al., 2001). Since a small percentage of healthy individuals benignly harbor M. pneumoniae in their respiratory tracts and/or have residual anti-M. pneumoniae IgG serum antibodies due to prior exposure or illness, serological tests and cultures may yield false-positive results. A major disadvantage to the lack of specific clinical criteria and a timely, reliable diagnostic test for mycoplasmal CAP is the frequent and unnecessary administration of empirical broad-spectrum antibiotics.
In recent years, PCR-based methods have offered the best potential for a specific, sensitive and rapid diagnosis of CAP due to M. pneumoniae as well as other agents. Several studies have evaluated methods for detection of M. pneumoniae DNA or RNA in respiratory specimens using various primers, regular or real-time thermocyclers, and different formats, such as nested, multiplex and hybridization methods (Daxboeck et al., 2003;Dumke et al., 2007;Dumke and Jacobs, 2009;Gullsby et al., 2008;Kashyap et al., 2008;Khanna et al., 2005;Kumar et al., 2008;Liu et al., 2007;Loens et al., 2003;Martínez et al., 2008;Ursi et al., 2003;Waites et al., 2005;Winchell et al., 2008). These and other reports convincingly demonstrate that PCR-based strategies are comparable or superior to culture and serology in specificity, sensitivity and predictive value. However, current PCR-based methods have not become routine diagnostic procedures because they suffer from multiple drawbacks, including high reagent costs, labor intensity, and inconvenience for rapid, convenient point-of-care testing.
This report introduces the initial version of a unique real-time PCR platform with potential to eliminate all of these disadvantages. Advanced Liquid Logic, Inc. (http://www.liquid-logic.com/) has developed an innovative real-time PCR lab-on-a-chip technology to detect microbial DNA in clinical specimens. The technology uses electrical fields to rapidly and precisely manipulate discrete nanoliter-sized droplets within an oil-filled chamber without the use of any pumps, valves or fixed channels. Each droplet manipulation is directly controlled within a software program enabling complex, multi-step protocols to be implemented and easily reconfigured without requiring modifications to the chip design.
Many other chip-based PCR technologies have been developed (Auroux et al., 2004;Chen et al., 2007;Sista et al., 2008;Zhang et al., 2006). They typically perform liquid movement either electrokinetically or with pneumatic pressure, and most of them utilize continuous streams of liquid. The Advanced Liquid Logic technology is based upon electrowetting, in which an electric field applied at the interface of a liquid and a hydrophobic insulator that can effectively modulate the surface tension of that interface (Pollack et al., 2000;Pollack et al., 2002). The microfluidics technology uses discrete unit-sized droplets that are individually positioned and manipulated by using an array of electrodes formed on a printed circuit board (PCB) substrate. The droplets are sandwiched between the PCB and a transparent glass cover plate with the surrounding space filled with an oil to prevent evaporation of the droplets. By applying different voltages to adjacent electrodes, a local and controllable surface energy gradient is generated, allowing droplets to be rapidly and programmatically transported along any path of contiguous electrodes.
This technique of droplet manipulation can be extended to other operations including merging, splitting, mixing and dispensing of microdroplets simply by applying different patterns of voltage activation. Large numbers of droplets can be simultaneously and independently manipulated allowing complex protocols to be flexibly implemented directly through software control, a concept sometimes referred to as “digital microfluidics” (Fair, 2007).
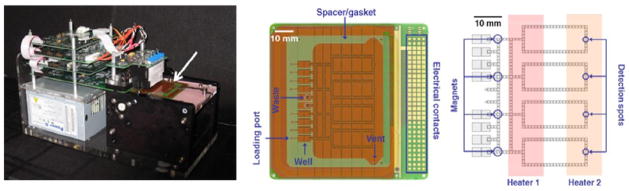
For on-chip real-time PCR, target DNA is loaded in chip well, nanodroplets are formed and mixed with PCR primers and reagents, and with each cycle, the droplet is moved from one on-chip heating sector to another (Paik et al., 2007;Sista et al., 2008). As depicted in Figure 1, the current prototype (86 ×86 mm) employs loops to circulate droplets through the thermal zones and can accommodate four simultaneous real-time PCRs. For detection of the amplicon, a miniature LED-photodiode fluorimeter measures fluorescence at the end of each extension cycle. The chips are intended for single use and are operated using a shoe-box sized instrument attached to a personal computer. In addition, the microfluidic real-time PCR platform has a user-friendly, fully automated interface and readout. Performing PCR on microfluidic chips offers obvious advantages. Multiple assays can be run in parallel on a self-contained, disposable chip that is protected from cross-contamination. The small droplet size permits the analyses of patient specimens that are too small for conventional real-time PCR, minimizes reagent costs and accelerates the cycle times because of the small thermal mass.
Figure 1.
Microfluidic PCR setup. Left: The self contained instrument including power supply, control electronics, fluorimeter module, heaters and cartridge deck, which is shown with the cartridge loaded (arrow). Center: Photograph of assembled microfluidic cartridge comprising an 86 × 86 mm PCB chip, polymer spacer/gasket and glass top-plate with drilled holes. Right: Schematic of PCR chip showing electrode positions relative to heaters, magnets and detectors.
The goal of this investigation was to evaluate this novel real-time platform. We optimized a real-time PCR protocol for the detection of M. pneumoniae DNA using a conventional real-time PCR thermocycler by developing methods of DNA extraction, recovery, amplification and detection that would be compatible with the microfluidics instrument. We then used simulated and actual respiratory specimens to compare PCR on the microfluidic platform with a conventional real-time thermocycler.
Materials and Methods
M. pneumoniae
For positive controls, standard curves and testing methods of DNA extraction and PCR, we used a reference strain of M. pneumoniae (ATCC # 15531), maintained by serial subculture in SP4 broth. To prepare stock cultures for PCRs, we pooled parallel broth cultures, enumerated the resulting bacterial concentration by viable plate counts on SP4 agar, adjusted the concentration to 2 × 105 CFU/ml in saline and stored aliquots at −80°C.
Healthy and simulated human nasopharyngeal wash (NPW) specimens
To test for non-specific amplification, NPWs were collected from healthy human volunteers by using a sterile syringe to spray at least 2 mL sterile saline (0.9% NaCl) into one nostril and asking the subject to expel the saline into a sterile tube. All control NPW specimens were stored at 4°C for future tests, while clinical NPW specimens were stored at −80C. Simulated NPW specimens were prepared by seeding sterile saline with dilutions of the stock culture of M. pneumoniae to create 10-fold concentrations of CFU/mL. We also seeded M. pneumoniae into NPWs from healthy volunteers to determine whether any components in normal nasopharyngeal wash fluid inhibited the PCR detection of target DNA using our protocols.
Clinical specimens
Beginning in March 2006, we enrolled patients who presented at the Duke University Hospital Emergency Department with symptoms and signs of community-acquired pneumonia (CAP) (DUMC IRB protocol 8368). Inclusion criteria were clinical diagnosis of pneumonia and fever (oral temperature >101.5°F), cough, leukocytosis (WBC≥12,000/mm3), compatible chest X-ray or physical examination suggestive of pneumonia. Patients were consented for enrollment, which included medical history and in some cases, pulmonary radiography. As described above, a NPW and blood were collected from each patient at the time of consent. All patients were seen in follow-up four weeks later for physical examination and the collection of convalescent respiratory and blood specimens. In this investigation, we evaluated the initial, acute NPW specimens on the first 59 patients for M. pneumoniae DNA.
Optimization of conventional real-time PCR
Using standardized cultures and purified DNA of M. pneumoniae, we conducted extensive preliminary experiments on the conventional real-time PCR thermocycler (ABI) to determine the optimal protocol. (i) Comparisons of filtration (Qiagen QIAmp DNA blood mini kit) vs. magnetic bead-based DNA extraction methods revealed that the commercial filtration and centrifugation technique consistently provided higher yields of DNA (data not shown). However, extraction with magnetic beads was faster, did not require a microcentrifuge and was more amenable to direct loading onto the microfluidic chips. In addition, the beads offer the potential for future development of on-chip sample processing of DNA. As a capture probe, we designed the oligonucleotide given below. (ii) We tested several M. pneumoniae-specific primers pairs that amplified sequences of the single copy P1 adhesin gene (Kong et al., 2000) and the multicopy genes, MP5 (Bernet et al., 1989), repMP1 (Dumke et al., 2007), and Mp181 (Winchell et al., 2008). The Mp181 primers amplify specific sequences of the mycoplasmal community-acquired respiratory distress syndrome (CARDS) toxin gene sequences, which proved to be the most sensitive and generated an optimal 73-bp amplicon (data not shown). A thorough search of GenBank sequences found no variation among strains at the location of either the capture probe or the PCR probe and primers.
Extraction of DNA from simulated and actual NPW specimens
As noted above, exhaustive optimization studies were used to develop protocols for the concentration of target DNA in simulated or actual NPW specimens, as well as the design of our probes and primers and Streptavidin-coated magnetic bead extraction method. These reagents and conditions were compatible with both conventional and microfluidic real-time PCR platforms.
DNA extraction was carried out as follows: 200 μL of each real or simulated NPW specimen was treated directly with 20 μL proteinase K (Qiagen, Inc.) and 200 μL AL buffer (Qiagen, Inc.). These specimens were vortexed for 5 s, briefly minifuged to clear caps, and incubated at 56°C for 15 min. Then, 4.5 μL oligonucleotide capture probe (5′-Biotin-AGAGTGGATCTTCTGACACTTCCGGGTCTAAC-3′, Sigma-Aldrich Co.) was added to each specimen, and the mixture was vortexed for 5 s, briefly minifuged, and incubated at 95°C for 15 min and 56°C for 20 min to denature the target DNA and allow the probe to hybridize. We then added 5.0 μL pre-washed m-270 Streptavidin Dynabeads (Invitrogen, Inc.). Tubes were incubated at room temperature for 30 min, during which time they were mixed by inversion 5 times every 5 minutes. The specimens were then minifuged for 5 s and placed on the magnetized rack for 10 min, after which the liquid was withdrawn from each specimen, leaving the beads with attached target DNA. Each specimen was then treated with 200 μL TE buffer, gently mixed and returned to the magnetized rack for 2 min. The beads were washed once more and gently resuspended in 10 μL TE buffer with 50 mM NaCl and 0.075% (w/v) Tween 20. Magnetic beads with attached DNA either were used immediately or stored at −80°C. Preliminary experiments confirmed that DNA was not degraded by storage. For real-time PCR testing, each 10 μL bead-DNA preparation was split: 6 μL were used for conventional real-time PCR and 4 μL for the microfluidic platform.
Conventional real-time PCR
Each real-time PCR assay plate included a positive control of simulated NPW containing M. pneumoniae at 104 CFU/mL and two non-template controls (NTC). One NTC consisted of sterile saline that had been concurrently subjected to the extraction protocol, and a second NTC consisted of PCR grade water used in place of extracted specimen template. All controls and simulated or real NPW specimens were processed in parallel from DNA extraction to assay, and all were assayed in triplicate.
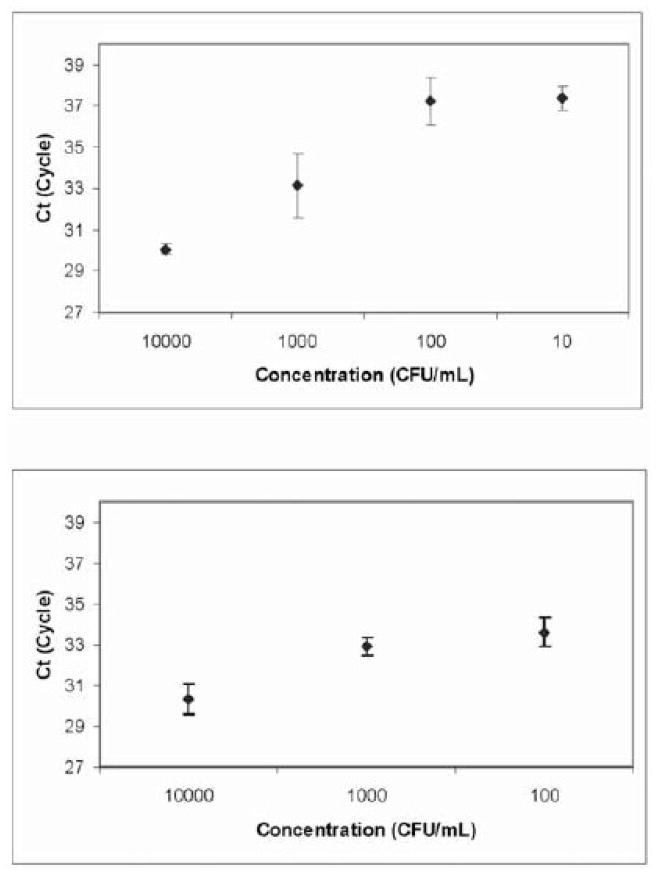
The Mp181 primer sequences were as follows (Winchell et al., 2008): Mp181-F 5′-TTTGGTAGCTGGTTACGGGAAT-3′; Mp181-R 5′-GGTCGGCACGAATTTCATATAAG-3′; Mp181 [FAM]-TGTACCAGAGCACCCCAGAAGGGCT-[BHQ]). Each PCR well had a final volume of 20 μL and contained 250 nM of each primer, 10 μL Taqman Fast Universal Real-Time Master Mix, 0.58 U uracil-DNA glycosylase (Applied Biosystems), and 2.0 μL DNA-bead extract. The master mix included UNG (New England Biolabs) and UTP to prevent amplification of any previously made amplicons. The following program was used with the thermocycler (7900 HT, Applied Biosystems): Initial amplification at 95°C for 120 s, followed by 45 cycles each at 95°C for 15 s and 57.8°C for 30 s, and concluding with a final cycle at 95°C for 60 s and 55 °C for 60 s. The thermocycler was located in a separate building from that in which the specimens were prepared, and the PCR plates were never returned to the preparatory laboratory. Evaluation of magnetic bead DNA extraction and real-time PCR precision was assessed by repeated extractions and multiple reactions of simulated clinical samples. These samples were created by serial dilution of the M. pneumoniae reference culture to 104, 103, 102, and 101 CFU/mL saline. Two sets of samples underwent magnetic bead DNA extraction on consecutive days, and samples were reacted in quadruplicate and triplicate, respectively (Figure 2A). These data also served to determine assay sensitivity.
Figure 2.
Limit of detection and precision testing performed with simulated clinical samples on (A) the conventional real-time PCR platform and (B) the microfluidic real-time PCR platform. Concentration versus mean CT with standard deviations shown.
Microfluidic real-time PCR
Prior to processing patient specimens, validation of the precision of on-chip microfluidic PCR was performed. Five simulated clinical specimens were prepared at a concentration of 104 CFU/mL sterile saline and extracted with magnetic beads, and the final 10 μL of each were pooled. Twenty real-time PCR reactions were run on the microfluidic platform, consisting of four reactions each on five separate, four-loop chips (Figure 1). As described above, 10-fold dilutions of M. pneumoniae were used to generate a standard curve comparing CFU/ml with the real-time PCR cycle threshold of detection (Figure 2B). Three real-time PCRs of each concentration were run on the microfluidic platform, and each run included a NTC consisting of PCR grade water in place of extracted specimen template. In addition, separate studies using beads added to spin column-purified M. pneumoniae DNA were undertaken to confirm that the presence of beads in the chip loops would not inhibit the PCR (data not shown).
As noted above, DNA-extracts of NPW specimens were stored at −20°C for subsequent testing on the microfluidic platform. Two 2-μL aliquots of each extracted NPW were tested. Each 2 μL extract was diluted 1:1 in TE buffer (pH 8.0, 50 mM NaCl, 0.075% Tween 20), which was necessary because the chip wells were designed to accept a minimum input volume of 3 μL. The two aliquots of each NPW specimen underwent real-time PCR on-chip as close to the same time as possible, and always on separate chips. Three patient specimen aliquots and one positive control, consisting of 200 fg/μL of M. pneumoniae genomic DNA, were run on each chip. Negative controls were included only periodically to maximize throughput and because previous tests of NTCs were invariably negative. The master mix was prepared daily and included, per reaction, 1.2 μL PCR buffer with final concentrations after 1:1 mixing of sample and master mix droplets of 3 mM MgCl2, 1 μM Mp181 primers, 1 μM Taqman Mp181 probe, and 0.5 U/μLKAPATaq. For on-chip real-time PCR, the heaters were preheated to 95°C and 58°C. The chip was then removed from its vacuum-sealed package, filled with ca. 1.5 mL degassed 10 cSt silicone oil, and inserted into the real-time instrument. Four microliters of magnetic bead-extracted DNA were added to the appropriate wells, and 3 μL master mix were added to the appropriate wells. The magnetic Dynabeads are concentrated by permanent magnets embedded in the deck of the instrument and situated directly underneath particular locations of the chip when in place on the real-time instrument. A 330-nL droplet containing all the beads concentrated from the 4 μL sample was dispensed from the loading zone and mixed with a 330 nL droplet of master mix. The combined droplet was physically cycled between the denaturation and annealing zones corresponding to the cycle temperatures of 95°C and 58°C, respectively. The dwell times were 10 s at 95°C and 45 s at 58°C with 4 s of transport time between the two zones. Fluorescence readings were taken at the end of each annealing extension cycle. At the conclusion of the run, the software program computed the earliest cycle threshold (CT) of detection of amplicon above the baseline, that is, a positive test for the target DNA. The CT is inversely related to the amount of template DNA. Patient specimens that were discrepant between the two platforms were re-extracted and re-tested on both platforms.
Results
Validation of conventional real-time PCR and microfluidic platform
Positive control samples for the conventional qPCR were created by extracting a 104 CFU/mL suspension of M. pneumoniae in sterile normal saline. All 29 conventional qPCR positive control samples tested alongside patient samples amplified with a mean cycle threshold (CT) of 29.7 +/− 0.50 SD. The efficiency of conventional qPCR runs varied from 91.6 to 101.3%. All but one NTC tested by conventional qPCR were negative. After finding amplification of one NTC sample on the conventional real-time PCR, all reagents were replaced, and the tests on that run were successfully repeated.
Nineteen of the 20 aliquots of 104 CFU/mL that underwent real-time PCR on the microfluidic platform amplified, and the mean CT +/− SD for these runs was 29.6 +/− 0.96 (Table 1). Positive controls for the microfluidic qPCR platform consisted of 67 fg commercial M. pneumoniae DNA. Thirty-seven of forty positive controls were amplified with a mean CT of 31.28 +/− 0.98 SD. The CTs for these controls was comparable to the results obtained by Winchell et al. (Winchell et al., 2008), who also amplified the Mp181 target.
Table 1.
Precision of the microfluidic real-time PCR platform using simulated clinical samples at 104 CFU/mL
| Loop 1 | Loop 2 | Loop 3 | Loop 4 | Mean ± SD | |
|---|---|---|---|---|---|
| Chip 1 | 31.1 | 30.3 | 30.1 | 29.5 | 30.25 ± 0.61 |
| Chip 2 | 29.7 | 29 | 28.5 | 29.7 | 29.24 ± 0.59 |
| Chip 3 | 29.4 | 28.6 | 29.6 | 28.9 | 29.14 ± 0.45 |
| Chip 4 | 29.1 | 28.2 | 28.9 | 28.4 | 28.68 ± 0.42 |
| Chip 5 | 30 | 31 | 31.7 | ND | 30.94 ± 0.85 |
| Mean | 29.86 ±0.69 | 29.42 ±1.06 | 29.76 ±1.12 | 29.13 ±0.51 | 29.58 ± 0.96 |
SD, Standard Deviation; ND, not done
Comparison of conventional real-time PCR and the microfluidic platform
The limit of detection of M. pneumoniae in simulated specimens extracted with magnetic beads was 10 CFU/mL for the conventional real-time PCR platform. The microfluidic platform was not tested at lower concentrations but did detect 100 CFU/mL. The limit of detection was more sensitive on both platforms for specimens extracted via QIAamp DNA blood mini kit (data not shown), which is consistent with a study comparing five DNA extraction methods, including the QIAamp DNA mini kit and magnetic bead extraction with Dynabeads (Cler et al., 2006). That study showed the QIAamp mini kit produced a higher yield of extracted DNA than did magnetic bead extraction, which should lead to increased sensitivity.
Six of the 59 patient NPWs initially were positive on at least one real-time PCR platform. Two NPWs tested positive for M. pneumoniae DNA on both the conventional and the microfluidic platforms. Two NPWs were positive only on the conventional platform, and two others were positive only on the microfluidic platform. When additional aliquots of the four discrepant NPW specimens were extracted and tested on both platforms, one specimen was positive on the conventional platform, and none was positive on the microfluidic platform. Thus, we concluded that a total of three specimens were positive (Table 2), and the agreement between platforms for the 59 samples was 98%. None of the NPWs from healthy individuals was positive on either instrument, and the spiked non-patient NPWs did not inhibit the PCR; that is, there was no inhibition or significant increase in the CT of detection (data not shown). The time required to run 50 PCR cycles on the microfluidic platform was 60 min versus 165 min on the conventional platform.
Table 2.
Comparison of real-time PCR results of acute patient NPWs on conventional and microfluidic real-time PCR platforms
| Conventional real-time PCR | |||
|---|---|---|---|
| Positive | Negative | ||
| Microfluidic real-time PCR | Positive | 2 | 0 |
| Negative | 1 | 56 | |
Discussion
This report describes the first direct comparison of digital microfluidics to conventional bench methods for the detection of microbial DNA in clinical specimens. In recent years, real-time PCR has emerged as a rapid and sensitive diagnostic technique for respiratory infections caused by M. pneumoniae, an organism whose detection by culture and serology has long been recognized as arduous and ineffectual. Consequently, many variations on DNA-based detection of M. pneumoniae in clinical specimens have been developed, and most have demonstrated significantly greater sensitivity and specificity than culture or serology (Daxboeck et al., 2003;Loens et al., 2003). These methods have not become routine in hospital clinical laboratories or outpatient clinics because they require significant training, and most of the protocols and instruments are not expandable to multiple pathogens, fully automated or portable.
The microfluidic PCR platform developed by Advanced Liquid Logic obviates all of these disadvantages of conventional PCR thermocycling. This investigation compared the microfluidic platform with conventional real-time PCR for the capability to detect M. pneumoniae DNA in respiratory specimens from patients with CAP, using the same protocol for extraction, probe and primers. The microfluidic real-time PCR performed as well as conventional PCR. Furthermore, the microfluidic real-time PCR was easier to use and at least three times faster. It is versatile, portable and potentially inexpensive and amenable to point-of-care use in the hospital or outpatient setting. Consequently, we are currently conducting a large scale evaluation on more than 200 patients with CAP, including additional laboratory data, such as cultures, serologies, and tests for viral pathogens.
We observed that the bead-based extraction was less efficient than the column filtration method and confirmed these results by conventional real-time PCR. However, we chose to use a magnetic bead extraction protocol because microcentrifugation steps are not required. The next version of the microfluidic platform, currently under development, will include an automated specimen preparation step using on-chip, bead-based extraction of DNA. Magnetic beads also provide a mechanism for concentrating DNA from a 10 μL specimen volume into a 1–2 μL volume, which is ideal for a single microfluidic real-time PCR. The sensitivity of the microfluidic platform was equivalent to conventional real-time over the range that was tested (≥100 CFU/mL). Based on estimates of the census of M. pneumoniae in respiratory specimens of infected patients, this level of sensitivity appears to be adequate for diagnosis (Kenny et al., 1990;Skakni et al., 1992).
After resolving three discrepant test results, approximately 5% of patient specimens were positive on at least one platform. These results are within the ranges of reported prevalence of mycoplasmal CAP. Explanation for the discrepant specimens is uncertain. There was no indication of contamination or thermal cycler failure. Most likely, the discrepant specimens had marginal levels of M. pneumoniae DNA as the CT’s on the initially positive tests were higher. Overall, these initial results of the diagnostic potential of digital microfluidics are most promising.
Acknowledgments
We greatly appreciate the invaluable assistance of the clinical coordinators, Gisselle Mani, Debra Freeman and Denise Beaver, who helped to enroll patients, arrange follow-ups, collect specimens, record data and store specimens. This project was supported by US PHS grants from the NIH, U01 AI 066590 and K24 AI 072522 (BDA).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Auroux PA, Koc Y, deMello A, Manz A, Day PJ. Miniaturised nucleic acid analysis. Lab Chip. 2004;4:534–546. doi: 10.1039/b408850f. [DOI] [PubMed] [Google Scholar]
- Beersma MFC, Dirven K, van Dam AP, Templeton KE, Claas ECJ, Goossens H. Evaluation of 12 commercial tests and the complement fixation test for Mycoplasma pneumoniae-specific immunoglobulin G (IgG) and IgM antibodies, with PCR used as the “gold standard”. J Clin Microbiol. 2005;43:2277–2285. doi: 10.1128/JCM.43.5.2277-2285.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernet C, Garret M, de Barbeyrac B, Bébéar CM, Bonnet J. Detection of Mycoplasma pneumoniae by using the polymerase chain reaction. J Clin Microbiol. 1989;27:2492–2496. doi: 10.1128/jcm.27.11.2492-2496.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Manz A, Day PJ. Total nucleic acid analysis integrated on microfluidic devices. Lab Chip. 2007;7:1413–1423. doi: 10.1039/b708362a. [DOI] [PubMed] [Google Scholar]
- Cler L, Bu D, Lewis C, Euhus D. A comparison of five methods for extracting DNA from paucicellular clinical samples. Mol Cell Probes. 2006;20:191–196. doi: 10.1016/j.mcp.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Daxboeck F, Krause R, Wenisch C. Laboratory diagnosis of Mycoplasma pneumoniae infection. Clin Microbiol Infect. 2003;9:263–273. doi: 10.1046/j.1469-0691.2003.00590.x. [DOI] [PubMed] [Google Scholar]
- Dumke R, Jacobs E. Comparison of commercial and in-house real-time PCR assays used for detection of Mycoplasma pneumoniae. J Clin Microbiol. 2009;47:441–444. doi: 10.1128/JCM.01989-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumke R, Schurwanz N, Lenz M, Schuppler M, Luck C, Jacobs E. Sensitive detection of Mycoplasma pneumoniae in human respiratory tract samples by optimized real-time PCR approach. J Clin Microbiol. 2007;45:2726–2730. doi: 10.1128/JCM.00321-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fair RB. Digital microfluidics: is a true lab-on-a-chip possible? Microfluidics and Nanofluidics. 2007;3:245–281. [Google Scholar]
- Fee C, Weber EJ. Identification of 90% of patients ultimately diagnosed with community-acquired pneumonia within four hours of emergency department arrival may not be feasible. Ann Emerg Med. 2007;49:553–559. doi: 10.1016/j.annemergmed.2006.11.008. [DOI] [PubMed] [Google Scholar]
- Gullsby K, Storm M, Bondeson K. Simultaneous detection of Chlamydophila pneumoniae and Mycoplasma pneumoniae by use of molecular beacons in a duplex real-time PCR. J Clin Microbiol. 2008;46:727–731. doi: 10.1128/JCM.01540-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagaman JT, Rouan GW, Shipley RT, Panos RJ. Admission chest radiograph lacks sensitivity in the diagnosis of community-acquired pneumonia. Am J Med Sci. 2009;337:236–240. doi: 10.1097/MAJ.0b013e31818ad805. [DOI] [PubMed] [Google Scholar]
- Herold CJ, Sailer JG. Community-acquired and nosocomial pneumonia. Eur Radiol. 2004;14(Suppl 3):E2–20. doi: 10.1007/s00330-003-2162-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HH, Zhang YY, Xiu QY, Zhou X, Huang SG, Lu Q, Wang DM, Wang F. Community-acquired pneumonia in Shanghai, China: microbial etiology and implications for empirical therapy in a prospective study of 389 patients. Eur J Clin Microbiol Infect Dis. 2006;25:369–374. doi: 10.1007/s10096-006-0146-7. [DOI] [PubMed] [Google Scholar]
- Ieven M, Goossens H. Relevance of nucleic acid amplification techniques for diagnosis of respiratory tract infections in the clinical laboratory. Clin Microbiol Rev. 1997;10:242–256. doi: 10.1128/cmr.10.2.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jokinen C, Heiskanen L, Juvonen H, Kallinen S, Kleemola M, Koskela M, Leinonen M, Rönnberg PR, Saikku P, Sten M, Tarkiainen A, Tukiainen H, Pyorala K, Makela PH. Microbial etiology of community-acquired pneumonia in the adult population of 4 municipalities in eastern Finland. Clin Infect Dis. 2001;32:1141–1154. doi: 10.1086/319746. [DOI] [PubMed] [Google Scholar]
- Kashyap B, Kumar S, Sethi GR, Das BC, Saigal SR. Comparison of PCR, culture & serological tests for the diagnosis of Mycoplasma pneumoniae in community-acquired lower respiratory tract infections in children. Indian J Med Res. 2008;128:134–139. [PubMed] [Google Scholar]
- Kenny GE, Kaiser GG, Cooney MK, Foy HM. Diagnosis of Mycoplasma pneumoniae pneumonia: sensitivities and specificities of serology with lipid antigen and isolation of the organism on soy peptone medium for identification of infections. J Clin Microbiol. 1990;28:2087–2093. doi: 10.1128/jcm.28.9.2087-2093.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna M, Fan J, Pehler-Harrington K, Waters C, Douglass P, Stallock J, Henrickson KJ, Kehl S. The Pneumoplex assays, a multiplex PCR-enzyme hybridization assay that allows simultaneous detection of five organisms, Mycoplasma pneumoniae, Chlamydia (Chlamydophila) pneumoniae, Legionella pneumophila, Legionella micdadei, and Bordetella pertussis, and its real-time counterpart. J Clin Microbiol. 2005;43:565–571. doi: 10.1128/JCM.43.2.565-571.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong F, Gordon S, Gilbert GL. Rapid-cycle PCR for detection and typing of Mycoplasma pneumoniae in clinical specimens. J Clin Microbiol. 2000;38:4256–4259. doi: 10.1128/jcm.38.11.4256-4259.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korppi M, Heiskanen-Kosma T, Kleemola M. Incidence of community-acquired pneumonia in children caused by Mycoplasma pneumoniae: serological results of a prospective, population-based study in primary health care. Respirology. 2004;9:109–114. doi: 10.1111/j.1440-1843.2003.00522.x. [DOI] [PubMed] [Google Scholar]
- Kumar S, Wang L, Fan J, Kraft A, Bose ME, Tiwari S, Van Dyke M, Haigis R, Luo T, Ghosh M, Tang H, Haghnia M, Mather EL, Weisburg WG, Henrickson KJ. Detection of 11 common viral and bacterial pathogens causing community-acquired pneumonia or sepsis in asymptomatic patients by using a multiplex reverse transcription-PCR assay with manual (enzyme hybridization) or automated (electronic microarray) detection. J Clin Microbiol. 2008;46:3063–3072. doi: 10.1128/JCM.00625-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung CM, Wang HL. Seroprevalence of Mycoplasma pneumoniae in healthy adolescents in Taiwan. Jpn J Infect Dis. 2007;60:352–354. [PubMed] [Google Scholar]
- Lieberman D, Shvartzman P, Korsonsky I, Lieberman D. Diagnosis of ambulatory community-acquired pneumonia. Comparison of clinical assessment versus chest X-ray. Scand J Prim Health Care. 2003;21:57–60. doi: 10.1080/02813430310000582. [DOI] [PubMed] [Google Scholar]
- Liu F-C, Chen P-Y, Huang F-L, Tsai C-R, Lee C-Y, Lin CF. Do serological tests provide adequate rapid diagnosis of Mycoplasma pneumoniae infection? Jpn J Infect Dis. 2008;61:397–399. [PubMed] [Google Scholar]
- Liu F-C, Chen P-Y, Huang F-L, Tsai C-R, Lee C-Y, Wang L-C. Rapid diagnosis of Mycoplasma pneumoniae infection in children by polymerase chain reaction. J Microbiol Immunol Infect. 2007;40:507–512. [PubMed] [Google Scholar]
- Loens K, Ursi D, Goossens H, Ieven M. Molecular diagnosis of Mycoplasma pneumoniae respiratory tract infections. J Clin Microbiol. 2003;41:4915–4923. doi: 10.1128/JCM.41.11.4915-4923.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna CM, Famiglietti A, Absi R, Videla AJ, Nogueira FJ, Fuenzalida AD, Gené RJ. Community-acquired pneumonia: etiology, epidemiology, and outcome at a teaching hospital in Argentina. Chest. 2000;118:1344–1354. doi: 10.1378/chest.118.5.1344. [DOI] [PubMed] [Google Scholar]
- Mandell LA, Wunderink RG, Anzueto A, Bartlett JG, Campbell GD, Dean NC, Dowell SF, File TM, Jr, Musher DM, Niederman MS, Torres A, Whitney CG. Infectious Diseases Society of America/American Thoracic Society consensus guidelines on the management of community-acquired pneumonia in adults. Clin Infect Dis. 2007;44(Suppl 2):S27–S72. doi: 10.1086/511159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez MA, Ruiz M, Zunino E, Luchsinger V, Avendaño LF. Detection of Mycoplasma pneumoniae in adult community-acquired pneumonia by PCR and serology. J Med Microbiol. 2008;57:1491–1495. doi: 10.1099/jmm.0.2008/003814-0. [DOI] [PubMed] [Google Scholar]
- Morozumi M, Ito A, Murayama SY, Hasegawa K, Kobayashi R, Iwata S, Kawamura N, Kuroki H, Nakayama E, Tajima T, Ubukata K. Assessment of real-time PCR for diagnosis of Mycoplasma pneumoniae pneumonia in pediatric patients. Can J Microbiol. 2006;52:125–129. doi: 10.1139/w05-118. [DOI] [PubMed] [Google Scholar]
- Ngeow YF, Suwanjutha S, Chantarojanasriri T, Wang F, Saniel M, Alejandria M, Hsueh P-R, Ping-Ing L, Park SC, Sohn JW, Aziah AM, Liu Y, Seto WH, Ngan CC, Hadiarto M, Hood A, Cheong YM. An Asian study on the prevalence of atypical respiratory pathogens in community-acquired pneumonia. Int J Infect Dis. 2005;9:144–153. doi: 10.1016/j.ijid.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Ozaki T, Nishimura N, Ahn J, Watanabe N, Muto T, Saito A, Koyama N, Nakane K, Funahashi K. Utility of a rapid diagnosis kit for Mycoplasma pneumoniae pneumonia in children, and the antimicrobial susceptibility of the isolates. J Infect Chemother. 2007;13:204–207. doi: 10.1007/s10156-007-0519-6. [DOI] [PubMed] [Google Scholar]
- Paik PY, Allen DJ, Eckhardt AE, Pamula VK, Pollack MG. Programmable flow-through real-time PCR using digital microfluidics. Proc Micro TAS. 2007:1559–1561. [Google Scholar]
- Plouffe JF. Importance of atypical pathogens of community-acquired pneumonia. Clin Infect Dis. 2000;31(Suppl 2):S35–S39. doi: 10.1086/314058. [DOI] [PubMed] [Google Scholar]
- Plouffe JF, Martin DR. Pneumonia in the emergency department. Emerg Med Clin North Am. 2008;26:389–411. doi: 10.1016/j.emc.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack MG, Fair RB, Shenderov AD. Electrowetting-based actuation of liquid droplets for microfulidic applications. Applied Physics Letters. 2000;77:1725–1726. [Google Scholar]
- Pollack MG, Shenderov AD, Fair RB. Electrowetting-based actuation of droplets for integrated microfluidics. Lab Chip. 2002;2:96–101. doi: 10.1039/b110474h. [DOI] [PubMed] [Google Scholar]
- Reechaipichitkul W, Lulitanond V, Tantiwong P, Saelee R, Pisprasert V. Etiologies and treatment outcomes in patients hospitalized with community-acquired pneumonia (CAP) at Srinagarind Hospital, Khon Kaen, Thailand. Southeast Asian J Trop Med Public Health. 2005;36:156–161. [PubMed] [Google Scholar]
- Sista RS, Hua Z, Thwar P, Sudarsan A, Srinivasan V, Eckhardt AE, Pollack MG, Pamula VK. Development of a digital microfluidic platform for point of care testing. Lab Chip. 2008;8:2091–2104. doi: 10.1039/b814922d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skakni L, Sardet A, Just J, Landman-Parker J, Costil J, Moniot-Ville N, Bricout F, Garbarg-Chenon A. Detection of Mycoplasma pneumoniae in clinical samples from pediatric patients by polymerase chain reaction. J Clin Microbiol. 1992;30:2638–2643. doi: 10.1128/jcm.30.10.2638-2643.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton KE, Scheltinga SA, Graffelman AW, Van Schie JM, Crielaard JW, Sillekens P, van den Broek PJ, Goossens H, Beersma MFC, Claas ECJ. Comparison and evaluation of real-time PCR, real-time nucleic acid sequence-based amplification, conventional PCR, and serology for diagnosis of Mycoplasma pneumoniae. J Clin Microbiol. 2003;41:4366–4371. doi: 10.1128/JCM.41.9.4366-4371.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman KA, Walter ND, Schwartz SB, Mitchell SL, Dillon MT, Baughman AL, Deutscher MP, Fulton JP, Tongren JE, Hicks LA, Winchell JM. Comparison of laboratory diagnostic procedures for detection of Mycoplasma pneumoniae in community outbreaks. Clin Infect Dis. 2009;48:1244–1249. doi: 10.1086/597775. [DOI] [PubMed] [Google Scholar]
- Tsolia MN, Psarras S, Bossios A, Audi H, Paldanius M, Gourgiotis D, Kallergi K, Kafetzis DA, Constantopoulos A, Papadopoulos NG. Etiology of community-acquired pneumonia in hospitalized school-age children: evidence for high prevalence of viral infections. Clin Infect Dis. 2004;39:681–686. doi: 10.1086/422996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursi D, Dirven K, Loens K, Ieven M, Goossens H. Detection of Mycoplasma pneumoniae in respiratory samples by real-time PCR using an inhibition control. J Microbiol Meth. 2003;55:149–153. doi: 10.1016/s0167-7012(03)00131-3. [DOI] [PubMed] [Google Scholar]
- Venditti M, Falcone M, Corrao S, Licata G, Serra P. Outcomes of patients hospitalized with community-acquired, health care-associated, and hospital-acquired pneumonia. Ann Intern Med. 2009;150:19–26. doi: 10.7326/0003-4819-150-1-200901060-00005. [DOI] [PubMed] [Google Scholar]
- Waites KB, Katz B, Schelonka RL. Mycoplasmas and ureaplasmas as neonatal pathogens. Clin Microbiol Rev. 2005;18:757–789. doi: 10.1128/CMR.18.4.757-789.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waris ME, Toikka P, Saarinen T, Nikkari S, Meurman O, Vainionpaa R, Mertsola J, Ruuskanen O. Diagnosis of Mycoplasma pneumoniae pneumonia in children. J Clin Microbiol. 1998;36:3155–3159. doi: 10.1128/jcm.36.11.3155-3159.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins-Riedel T, Stanek G, Daxboeck F. Comparison of SeroMP IgA with four other commercial assays for serodiagnosis of Mycoplasma pneumoniae pneumonia. Diagn Microbiol Infect Dis. 2001;40:21–25. doi: 10.1016/s0732-8893(01)00250-4. [DOI] [PubMed] [Google Scholar]
- Winchell JM, Thurman KA, Mitchell SL, Thacker WL, Fields BS. Evaluation of three real-time PCR assays for detection of Mycoplasma pneumoniae in an outbreak investigation. J Clin Microbiol. 2008;46:3116–3118. doi: 10.1128/JCM.00440-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Xu J, Ma W, Zheng W. PCR microfluidic devices for DNA amplification. Biotechnol Adv. 2006;24:243–284. doi: 10.1016/j.biotechadv.2005.10.002. [DOI] [PubMed] [Google Scholar]