Abstract
Objective
RUNX1/AML1 is an essential regulator of hematopoiesis and has multiple isoforms arising from differential splicing and utilization of two promoters. We hypothesized that the rare Runx1c isoform has a distinct role in hematopoietic stem cells (HSCs).
Methods
We have characterized the expression pattern of Runx1c in mouse embryos and human embryonic stem cell (hESC)-derived embryoid bodies using in situ hybridization, and expression levels in mouse and human HSCs by real-time PCR. We then determined the functional effects of Runx1c using enforced retroviral over-expression in mouse HSCs.
Results
We observed differential expression profiles of RUNX1 isoforms during hematopoietic differentiation of hESCs. The RUNX1a and RUNX1b isoforms were expressed consistently throughout hematopoietic differentiation whereas the RUNX1c isoform was only expressed at the time of emergence of definitive HSCs. RUNX1c was also expressed in the AGM region of E10.5–11.5 mouse embryos, the region where definitive HSCs arise. These observations suggested that the RUNX1c isoform may be important for the specification or function of definitive HSCs. However, using retroviral over-expression to study the effect of RUNX1 isoforms on HSCs in a gain-of-function system, no discernable functional difference could be identified between RUNX1 isoforms in mouse HSCs. Over-expression of both RUNX1b and RUNX1c induced quiescence in mouse HSCs in vitro and in vivo.
Conclusions
Although the divergent expression profiles of Runx1 isoforms during development suggest specific roles for these proteins at different stages of HSC maturation, we could not detect an important functional distinction in adult mouse HSCs using our assay systems.
INTRODUCTION
Runt-related transcription factor 1 (RUNX1), also known as acute myeloid leukemia 1 (AML1), is a powerful and pivotal regulator of hematopoiesis. RUNX1 is the alpha subunit of the core binding factor (CBF) complex and is the most frequent chromosomal translocation associated with human leukemia [1,2]. The major physiological function of RUNX1 was revealed by gene targeting studies showing that RUNX1 is required for definitive hematopoiesis [3–5]. Although the absence of RUNX1 does not affect primitive hematopoiesis or development of the yolk sac vasculature, RUNX1−/− embryos die between E11.5–E12.5 due to extensive hemorrhaging and complete effacement of definitive hematopoiesis [3,4]. In the embryo, RUNX1 is expressed in all sites from which hematopoietic cells emerge, and all definitive hematopoietic stem cells (HSCs) in the embryo express RUNX1 [6]. RUNX1 appears to regulate the specification of definitive HSCs in developing mouse embryos as the intra-aortic hematopoietic clusters associated with the hemogenic endothelium from which definitive HSCs emerge are absent in RUNX1−/− embryos [7,8]. In contrast, RUNX1 appears to be dispensable for HSC function in the adult as mice which have RUNX1 conditionally-deleted in the bone marrow show mild defects including a decrease in platelets (due to a maturational defect of the megakaryocytes), a block of lymphocyte development, and an expansion of hematopoietic progenitors, but no significant impairment to HSC function [9–11].
RUNX1 has been recognized to have multiple isoforms due to differential splicing and promoter utilization (Figure 1A). The c-isoform is transcribed from the distal P1 promoter, which results in a transcript encoding for 32 amino acids unique to this isoform. The major isoform in terms of relative abundance is the Runx1b isoform, which contains 5 unique N-terminal amino acids and emanates from the proximal P2 promoter. The c-isoform is rare, and only a few reports have explored its expression in mouse [12] or man [13,14]. The P1 promoter element is much more complex than the P2, containing binding sites of several key hematopoietic transcription factors, while the P2 promoter is much more “generic” [13]. All vertebrates have three Runx genes, and all of the three genes has a distal P1 and a proximal P2 promoter [15]. Thus, this striking dual-promoter structure is conserved through 250 million years of evolution, consistent with an important function. Promoter-reporter transfection experiments have shown some differential specificity of expression derived from the promoters [13]. Experiments with the Runx3 P1 and P2 promoter regions showed that a small (around 600 bp) P1 element could direct cell-type-specific expression with fidelity [16]. There is also evidence for functional significance of Runx isoform paralogs as deletion of both Runx2-I and Runx2-II isoforms results in complete arrest of bone development, whereas selective loss of Runx2-II is sufficient to form a grossly intact skeleton with impaired endochondral bone development [17].
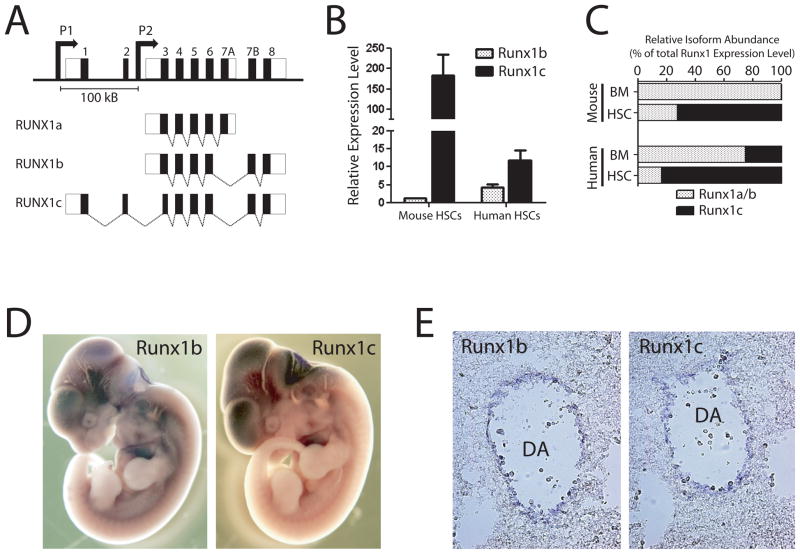
Figure 1.
RUNX1 genomic locus and isoform expression patterns. (A) Genomic organization of human RUNX1 locus. The RUNX1c isoform is transcribed from the distal P1 promoter and contains the unique N-terminal amino acids encoded by exons 1 and 2. The RUNX1a and predominant RUNX1b isoforms are transcribed from the proximal P2 promoter. The promoters are separated by over 100 Kb in the genome. (B) Expression of RUNX1 isoforms in hematopoietic stem cells. Real-time PCR analysis showed that the RUNX1c isoform is much more highly expressed in HSCs compared to RUNX1b relative to whole bone marrow in both human and mouse. (C) Relative abundance of RUNX1c versus RUNX1a and RUNX1b isoforms in mouse and human bone marrow (BM) and HSCs calculated as a proportional ratio of total RUNX1 expression level. These data show that the RUNX1c isoform is much more abundant in HSCs compared to whole BM. (D) Wholemount in situ hybridization analysis of Runx1 isoform expression patterns in E11.5 mouse embryos. On a gross level, the expression patterns of Runx1 isoforms at this stage of mouse development were highly overlapping. (E) Sectioning of stained embryos showed that Runx1b and Runx1c were both expressed in the endothelium lining the dorsal aorta in the AGM region where definitive HSCs are born.
Most strikingly, the N-terminal amino acids that are unique to the P1 isoform (RUNX1c) are 100% identical between the mouse and human proteins, suggesting an important function for this isoform that has been conserved throughout evolution. A recent study of hematopoietic differentiation from human embryonic stem cells (hESCs) revealed that the RUNX1c isoform was expressed exclusively at the time of emergence of definitive hematopoietic cells whereas the RUNX1b isoform was expressed broadly [18]. As the design of the Runx1 knock-out and Runx-lacZ-reporter mice result in knock-out of all isoforms [7,12] and given the highly specific expression pattern of the rare RUNX1c isoform during hematopoietic differentiation of ES cells, we hypothesized that RUNX1c may be essential for the specification of definitive HSCs during embryonic development and may be a marker of definitive HSCs.
METHODS
Mouse Hematopoietic Stem Cell Purification
All animal procedures were IUCAC-approved and conducted in accordance with the Baylor College of Medicine (Houston, Texas, USA) institutional guidelines. All mice were housed in a specific pathogen-free barrier facility and fed autoclaved acidified water and mouse chow ad libitum. Whole bone marrow was isolated from femurs and tibias, and SP cell staining was performed with the vital dye Hoechst 33342 (Sigma-Aldrich, St Louis, MO) as previously reported [19]. Briefly, whole bone marrow was resuspended in staining media at 106 cells/mL and incubated with 5 μg/mL Hoechst 33342 for 90 minutes at 37°C. For antibody staining, cells were suspended at a concentration of 108 cells/mL and incubated on ice for 20 minutes with the following antibodies (all 1:100 dilution); PeCy5–conjugated Mac-1, Gr-1, CD4, CD8, B220 and Ter119 (eBioscience); Sca-1-PE (BD Pharmingen, Franklin Lakes, NJ) and c-Kit-FITC (BD Pharmingen). Cell sorting and analysis were performed on a MoFlo cell sorter (Dako North America, Carpinteria, CA).
Human Hematopoietic Stem Cell Purification
Human bone marrow samples were obtained from the clinic with prior approval and resuspended at 108 cells/mL and incubated on ice for 20 minutes with the following antibodies (1:100 dilutions); CD34-PE and CD38-APC (BD Pharmingen). Cells were washed, spun down, resuspended in propidium iodide and cell sorting and analysis were performed on a MoFlo cell sorter (Dako North America, Carpinteria, CA).
Real-Time PCR Analysis
RNA was isolated using the RNAqueous kit (Ambion) and reverse transcribed with random hexamer primers using Super Script II (Invitrogen). For isoform-specific PCRs, primers were designed to be specific to unique sequences of the isoform transcripts. Primer sequences can be found in Table 1. cDNA input was standardized and RT-PCRs were performed with SYBR Green PCR master mix (Applied Biosystems) for 50 cycles using an AbiPrism 7900HT (Applied Biosystems). Samples were normalized to mouse or human GAPDH and fold change determined by the ΔΔCt method. For Notch pathway PCR, samples were analyzed using Taqman master Mix (Applied Biosystems), 18 s-rRNA probe (VIC-MGB; Applied Biosystems), and a gene-specific probe (FAM-MGB; Applied Biosystems).
Table 1.
Sequences of primers used in real-time PCR.
| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| Runx1b | CCTCCGGTAGTAATAAAGGCTTCTG | CCGATTGAGTAAGGACCCTGAA |
| Runx1c | GTGTGCTGGAATTCGGCTTAG | AAGCCATCGTTTCCTTTCGA |
| mGAPDH | AGAACATCATCCCTGCATCCA | CAGATCCACGACGGACACATT |
| OCT4 | CTGCAGCAGATCAGCCACAT | GACCCAGCAGCCTCAAAATC |
| NANOG | TGAAGCATCCGACTGTAAAGAATC | GCAGATCCATGGAGGAAGGA |
| SOX2 | CGCCGAGTGGAAACTTTTGT | CCTTCTTCATGAGCGTCTTGGT |
| T | CCACCTGCAAATCCTCATCCT | AGAATTGTTCCGATGAGCATAGG |
| MIXL1 | GCGTCAGAGTGGGAAATCCTT | GGCAGTTCACATCTACCTCAAGAG |
| CD34 | CAATGAGGCCACAACAAACATC | GGTGGTGAACACTGTGCTGATT |
| CD43 | CCTTTACCTGAGCCAACAACCT | TCACGGTGTGGGATCCTAGAG |
| TAL1 | CCACCAACAATCGAGTGAAGAG | AGGCCCCGTTCACATTCTG |
| RUNX1 | ACTCGGCTGAGCTGAGAAATG | GACTTGCGGTGGGTTTGTG |
| RUNX1b | TGCATGATAAAAGTGGCCTTGT | CGAAGAGTAAAACGATCAGCAAAC |
| RUNX1c | TGGTTTTCGCTCCGAAGGT | CATGAAGCACTGTGGGTACGA |
| hGAPDH | CCTGCACCACCAACTGCTTA | CCATCACGCCACAGTTTCC |
RNA in situ Hybridization
Expression patterns of RUNX1 isoforms were analyzed by RNA in situ hybridization using isoform-specific digoxigenin-labelled sense and antisense riboprobes. The following primers were used to amplify unique 5′ sequences for each isoform from bone marrow cDNA pools: mouse RUNX1b-forward; ATACTCGCTGATACCTCACTAAAGG, mouse RUNX1b-reverse ACGGGGATACGCATCACA, mouse RUNX1c-forward; AGCCGCGCTGATTACCCT, mouse RUNX1c-reverse; CTCATGAAGCACTGTGGATATGA, human RUNX1B-forward; TCAACTCCCAACAAACCAGAG, human RUNX1B-reverse ATCATGCAAAACTGGCTTCAG, human RUNX1c-forward GGCCTCATAAACAACCACAGA, human RUNX1c-reverse; CTGTGGGTACGAAGGAAATGA. Probes were synthesized as described previously [20]. Whole mount in situ hybridizations were performed as described [21] with minor modifications. All probes were hybridized at 65°C. Images were captured with a Zeiss Stemi SV11 microscope equipped with a Zeiss Axiocam color camera.
Human Embryonic Stem Cell Culture and Immunofluorescence
H9 hES cells (NIH registry WA09; obtained from WiCell Research Institute, Madison, WI, USA) were grown on gamma-irradiated mouse embryonic fibroblast feeders in 80% Dulbecco’s modified Eagle’s medium-F12 (Gibco, Grand Island, NY, USA), 20% Knockout Serum Replacement (Gibco), 1 mM glutamine (Gibco), 0.1 mM βmercapto-ethanol (Sigma-Aldrich), 1% nonessential amino acids (Gibco), and 4 ng/ml human recombinant basic fibroblast growth factor (bFGF) (Invitrogen) as described [22]. Hematopoietic differentiation of hEBs was performed exactly as previously described [18]. Wholemount in situ hybridization was performed on hEBs as described above. Stained hEBs were then briefly fixed by immersion in 4% paraformaldehyde/PBS and mounted in OCT and snap frozen. Sections were cut on a cryostat and transferred to slides which were air-dried and stored at −80°C until use. For immunofluorescence, slides were washed in PBS, permeabilized for 10 minutes in PBTX and blocked for 30 minutes in PBS + 1% BSA. Slides were incubated in PBS + 1% BSA with mouse anti-human CD34 (BD Pharmingen) and goat anti-human VE-CADHERIN (R&D Systems) overnight at 4°C (both 1:100 dilution). Slides were then washed in PBS and incubated with anti-mouse-Alexa594 and anti-goat-Alexa488 for 45 minutes at room temperature. Slides were mounted in Vectashield + DAPI (Vector Laboratories) and images captured on a Zeiss Axioplan 2 microscope equipped with Photometrics Coolsnap HQ camera.
Plasmids and Retroviral Over-Expression
Full-length human RUNX1b (Accession NM_001001890) and RUNX1c (Accession NM_001754) cDNAs were purchased from Genscript (Piscataway, NJ, USA). Coding sequences were subcloned into MSCV-RFB-IRES-GFP vector using Gateway recombination. Briefly, RUNX1 isoforms were PCR amplified using the following primers; RUNX1b-forward; CACCGATGCGTATCCCCGTAGATGCCAGC, RUNX1c-forward; CACCGATGGCTTCAGACAGCATATTTGAGTCATTT, RUNX1-reverse; GTCAGTAGGGCCTCCACACGGCCT and inserted into the pENTR-D/TOPO vector (Invitrogen, K2400-20) by TOPO cloning. Correct clones were sequence verified and recombined into an MSCV-RFB-IRES-GFP vector (containing attR recombination sites) using LR clonase enzyme mixture (Gateway LR ClonaseII Enzyme Mix, 11791-020, Invitrogen) to produce retroviral vectors. MSCV-IRES-GFP containing no open reading frame was used as a control vector in all experiments. Viruses were packaged by cotransfection with pCL-Eco [23] into 293T cells. Viral supernatants were collected 48-hours post-transfection and viral titers determined using 3T3 cells. All experiments were carried out using an MOI of 1.
For retroviral transduction of hematopoietic progenitors, donor CD45.2 mice were treated with 5-fluorouracil (150 mg/kg, American Pharmaceutical Partners) six days prior to bone marrow harvest. Whole bone marrow was enriched for Sca-1+ cells using magnetic enrichment (AutoMACS, Miltenyi) and adjusted to a concentration of 5 × 105 cells/ml in transduction medium, containing Stempro 34 (GIBCO), nutrient supplement, penicillin/streptomycin, L-glutamine (2 mM), mSCF (10 ng/ml, R&D Systems), mTPO (100 ng/ml, R&D Systems), and polybrene (4 ug/ml, Sigma). The suspension was spin-infected at 250 × g at room temperature for two hours [24], and cells were incubated for a further three hours at 37°C. For in vitro assays, transduced cells were cultured in fresh transduction medium for a further two days. For clonal colony-forming analysis, single HSCs (Sca-1+GFP+) were sorted into 96-well plates containing Methocult 3434 medium (Vancouver, BC, Canada) supplemented with penicillin/streptomycin and cultured in vitro at 37°C for two-weeks unless stated otherwise.
Transplantation and Peripheral Blood Analysis
For in vivo assay of transduced cells, recipient CD45.1 C57Bl/6 mice were given a split dose of 10.5 Gy of whole-body irradiation. 5 × 104 transduced cells per recipient were transplanted by retro-orbital intravenous injection. For peripheral blood analysis, mice were bled retro-orbitally at 4, 8, 12 and 16 weeks after transplantation, the red blood cells were lysed, and each sample was incubated with the following antibodies (all 1:100 dilution; eBioscience) on ice for 20 minutes – CD45.2-APC, CD4-Pacific Blue, CD8-Pacific Blue, B220-Pacific Blue, B220-PeCy7, Mac1-PeCy7, and Gr-1-PeCy7 as previously described [25]. Cells were then spun down, resuspended in a propidium iodide solution, and analysis was accomplished on live cells with an LSRII (Becton Dickinson).
HSC Analysis of Transplanted Mice
>18 weeks after transplantation, transplanted mice were killed and bone marrow was isolated. Bone marrow was prepared for HSC isolation using the SPKLS scheme discussed above with the following antibodies; Lineage markers-PeCy5, CD45.2-PE, Sca-1-PE-Cy7 and c-Kit-APC (all 1:100). Samples were washed, resuspended in propidium iodide solution and HSCs analyzed using a MoFlo cell sorter as above.
Homing Assay
CD45.1 C57Bl/6 mice were given a single dose of 10.5 Gy of whole-body irradiation and 24-hours later were transplanted with 2 × 106 transduced bone marrow cells. Recipient mice were then sacrificed 24-hours post-transplant for bone marrow collection. Bone marrow was stained with CD45.1-PE, CD45.2-APC, Lineage Markers-PeCy5, c-Kit-APC-Cy7 and Sca-1-PeCy7 and analyzed on an LSRII (Becton Dickinson).
Spleen Colony Assay
Lethally irradiated recipient mice were transplanted with 2.5 × 104 transduced Sca-1+ cells as above. Recipient mice were sacrificed 12-days post-transplant for spleen collection. The total number of spleen colonies formed was visually counted, and GFP+ spleen colonies were identified under a fluorescent inverted microscope.
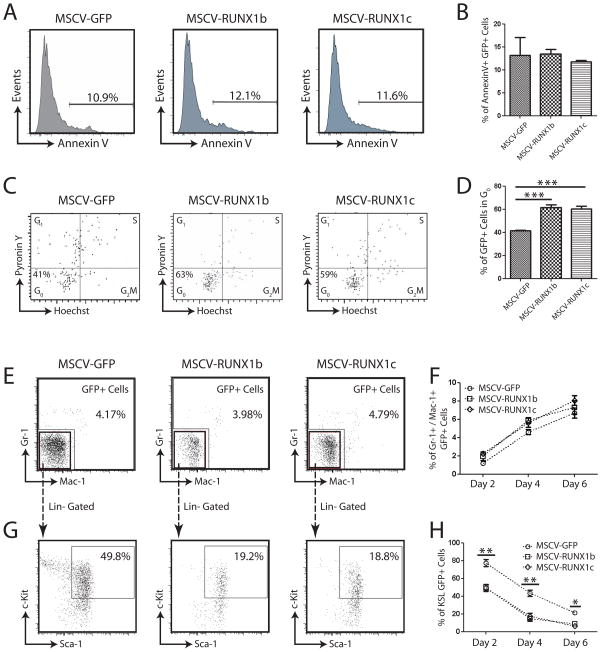
AnnexinV Staining
Annexin V and PI staining was used to assess cell death and apoptosis. Briefly, cells were washed twice with cold PBS and incubated at room temperature in 1x binding buffer (10 mM HEPES, 140 mM NaCl, 2.5 mM CaCl2) containing Annexin V-APC (BD-Pharmingen) and PI. Cells were analyzed by flow cytometry within one hour of staining.
Pyronin Y Staining
200,000 B220-FITC+ splenocytes were pre-sorted into collection tubes. Sca-1+GFP+ cells (more than 1000) were then sorted into the same tube. The sorted cells were then incubated for 45 minutes with 20μg/mL Hoechst 33342 and 50 μg/mL Verapamil (Sigma-Aldrich) in PBS supplemented with 3% fetal bovine serum. Pyronin Y (Sigma-Aldrich) was then added at 1 μg/ml, and the cells were incubated for another 15 min at 37°C, washed, and immediately analyzed on a BD LSRII. During flow analysis, both Hoechst 33342 and Pyronin Y signal were displayed under linear mode. The carrier B-cells, were then a control to define the G0/G1 DNA content (2N).
Statistics
Student t test and 1-way ANOVA’s were used for statistical comparisons where appropriate. Significance is indicated on the figures using the following convention: *p<0.05, **p<0.01, ***p<0.001. Error bars on all graphs represent the SEM.
RESULTS
The Rare RUNX1c Isoform is Highly Expressed in Mouse and Human Adult HSCs
We decided to first characterize the endogenous expression patterns of the RUNX1 isoforms in developing mouse embryos and mouse and human HSCs. The relative expression levels of RUNX1 isoforms was examined in adult HSCs using isoform-specific real-time PCR primers. Mouse HSCs were purified from the bone marrow as side-population+Lineage−Sca-1+c-Kit+ cells [25] while human HSCs were isolated by the CD34+CD38− phenotype. While the RUNX1b isoform was detected at a slightly higher level in both mouse and human HSCs, relative to whole bone marrow, RUNX1c was much more significantly increased in both mouse and human HSCs (182-fold and 11.7-fold respectively; Figure 1B). Thus, although the RUNX1c isoform is relatively rare in whole bone marrow, it is highly expressed in adult HSCs from both mouse and human, which are present in the marrow at around 1 in 10,000 cells. In addition, we calculated the relative abundance of RUNX1c versus RUNXla and RUNX1b isoforms in mouse and human bone marrow and HSCs calculated as a proportional ratio of total RUNX1 expression level and show that the RUNX1c isoform is the major isoform in terms of expression level in both mouse and human HSCs (Fig 1C).
RUNX1c is Expressed During the Emergence of Definitive HSCs in Embryonic Development
The endogenous expression patterns of Runx1 isoforms was determined by in situ hybridization of mid-gestation mouse embryos (the timepoint where definitive HSCs are generated) utilizing isoform-specific riboprobes. Wholemount in situ hybridization revealed no discernable differences in expression patterns of Runx1 isoforms in E9.5–E11.5 mouse embryos (Figure 1D). Sectioning of the AGM region (the intra-embryonic site of definitive HSC generation) of the stained E11.5 embryos showed Runx1b was expressed specifically in the endothelial cell layer lining the dorsal aorta (Figure 1E), consistent with existing data on embryonic Runx1 expression pattern. However, we also noted that the Runx1c isoform was also expressed in these cells in a highly similar pattern. This result demonstrates for the first time visually that Runx1c is also expressed in the region from which definitive HSCs arise during mouse embryogenesis. As both Runx1b and Runx1c are expressed in the same endothelial cells, it appears that both isoforms are expressed in HSC-generating and non-HSC-generating endothelial cells.
To study the roles of RUNX1 isoforms in human hematopoiesis, we utilized hESCs cells as an in vitro model system. We chose to use the hematopoietic differentiation protocol of Zambidis et al. because it generates two waves of hematopoietic development in vitro [18], mimicking the primitive and definitive waves of hematopoiesis that occur in the embryo (Figure 2A). The differentiation process was analyzed by real-time PCR to confirm the events occurring in vitro accurately reflected in vivo development. Human embryoid bodies (hEBs) were collected at four day intervals for gene expression analysis (Figure 2B). Expression of the pluripotency factors OCT-4, SOX2 and NANOG decreased in expression over the timecourse as expected as hESCs differentiated into more restricted cell types. The mesoderm markers T and MIXL1 became rapidly upregulated, peaking in expression between days 4–8 of hEB development, before decreasing in expression again. This suggested that mesoderm was generated in this system during culture days 4–8. The human HSC markers CD34, CD43 and TAL1 show a dynamic expression pattern, peaking in expression between days 12–16 in culture before switching off. This suggests that definitive HSCs are generated in this culture system during the day 12–16 day time period. Analysis of RUNX1 isoform expression in this experiment revealed that the total level of RUNX1 and the RUNX1b isoform were expressed at consistent levels throughout the entire differentiation process. However, the RUNX1c isoform was dynamically expressed in a similar profile to the HSC markers, indicating that this isoform may be important in the specification or function of definitive human HSCs.
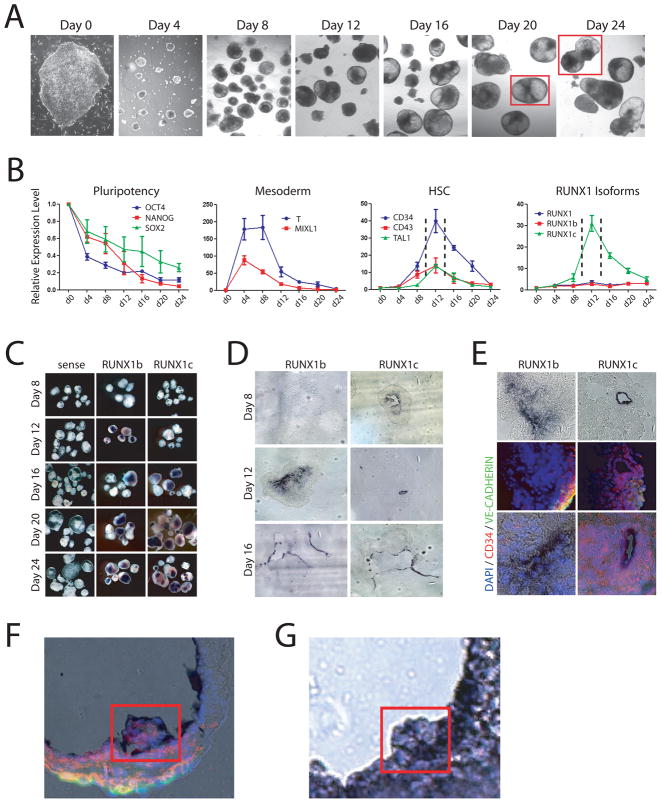
Figure 2.
Modeling human hematopoiesis in vitro with human embryonic stem cell culture. (A) Hematopoietic development in embryoid bodies. Bright field images showing development of hEBs from undifferentiated hESCs (day 0) over time at 4-day intervals. Red squares outline merged inset images. (B) Real-time PCR analysis of hEB differentiation. The pluripotency markers OCT4, NANOG and SOX2 decrease in expression over time as pluripotent hESCs differentiate. The mesoderm markers T and MIXL1 show a dramatic spike in expression between days 4–8 before downregulating, implying that mesoderm is generated between these timepoints in this culture system. The human HSC markers CD34, CD43 and TAL1 peak in expression at day 12 (enclosed by dashed lines) of culture indicating definitive HSCs are being generated during this time window. Analysis of the expression of RUNX1 isoforms during hematopoietic development show that total RUNX1 expression and the RUNX1b isoform are consistent throughout the differentiation process but expression of RUNX1c is dynamic, peaking at day 12 in a similar pattern to that of HSC markers. (C) Wholemount in situ hybridization analysis of RUNX1 isoforms in hEBs. Sense controls show no background staining, but on a gross level no discernable difference can be discriminated between RUNX1b and RUNX1c. (D) Section analysis of stained embryoid bodies showed that at day 12 when there is the most differential expression, RUNX1b is broadly expressed at low levels while RUNX1c is specifically expressed in cells lining the developing cavities in the hEBs. By day 16, the expression profiles of RUNX1b and RUNX1c become highly overlapping. (E) Co-immunofluorescence analysis of stained day 12 hEBs for vascular (VE-CADHERIN) and HSC (CD34) markers. While RUNX1b was broadly expressed throughout the day 12 hEBs, and its expression did not generally correlate with VE-CADHERIN or CD34 expression. RUNX1c however was expressed in a subpopulation of CD34+ cells. (F,G) hEB differentiation in vitro mimics mouse hematopoietic development in vivo. (F) Merged RUNX1b in situ hybridization (purple staining) and immunofluorescence (CD34 – red; VE-CADHERIN – green; DAPI – blue) images of day 20 hEB showing clusters of hematopoietics cells budding off into the lumen. (G) Section of RUNX1b-stained in situ hybridization of AGM region of E11.5 mouse embryo showing presumptive HSCs budding off into the dorsal aorta.
A similar approach was used as with mouse embryos to analyze the expression patterns of RUNX1 isoforms in hEBs using in situ hybridization. There appeared to be minimal discernable differences between isoform expression patterns at a gross level (Figure 2C). To obtain a more detailed view, the stained hEBs were sectioned (Figure 2D). The most distinct differences in expression profiles were observed at the earliest stages of hEB development. At day 8, little expression of RUNX1b could be detected by in situ staining but RUNX1c was highly expressed by small pockets of cells, which could possibly represent emerging definitive HSCs. The fact that RUNX1b could be detected by real-time PCR at this timepoint suggests that this isoform may be expressed at very low levels by most if not all cells of the hEB while RUNX1c is highly expressed by few specific cells. By day 12, RUNX1b was broadly expressed by many cells in the interior of the hEB but the restricted expression pattern of RUNX1c remained being expressed by cells lining forming cavities in the hEBs. By day 16 onwards the expression profiles of the isoforms began to overlap more with all isoforms expressed by cells lining the cavities that were forming within the hEBs. It appeared that these cavities were analogous to the formation of large vessels in embryos which give rise to intra-embryonic HSCs in vivo. The outermost cells lining the cavities expressed the endothelial marker VE-CADHERIN, and the inner layers expressed CD45 (Figure 2F,G). So it appeared that the structure of these cavities included an endothelial cell layer, overlayed by a layer of hematopoietic cells, which then gave rise to budding clumps of hematopoietic cells which filled the lumens.
Immunofluorescence analysis was performed to overlay markers of hematopoietic and endothelial cells on top of the in situ hybridization staining to define what cell types were expressing the different RUNX1 isoforms (Figure 2E). While a broad expression of RUNX1b was observed in day 12 hEBs, none of these cells appeared to express either the endothelial marker VE-CADHERIN or the human HSC marker CD34. In contrast, RUNX1c expression was restricted to a very specific population of cells. All of these cells were CD34+, consistent with the emergence of primitive hemato-endothelial progenitors. However, it should be noted that while it appeared all RUNX1c+ cells were CD34+, not all CD34+ cells were RUNX1c+. This could suggest that RUNX1c may mark a distinct subpopulation of human hemato-endothelial progenitors.
Over-Expression of RUNX1 Isoforms in Adult and Embryonic Mouse HSPCs Induces Quiescence
To examine whether the RUNX1 isoforms had distinct roles in the adult, we decided to perform isoform-specific over-expression in hematopoietic stem/progenitor cells (HSPCs). The full-length human RUNX1b and RUNX1c coding sequences were cloned into a murine stem cell virus (MSCV) vector containing GFP which was then used to generate retrovirus. For experimentation, MSCV-RUNX1b and MSCV-RUNX1c retroviruses (along with a empty vector control MSCV-GFP virus) were used to transduce Sca-1+ bone marrow cells from mice injected with 5-flurouracil six days prior to bring the HSPCs into cycle. The transduced HSPCs were then cultured for in vitro assays or transplanted into lethally irradiated recipient mice for in vivo analysis with the effect of RUNX1 isoform over-expression monitored in terms of peripheral blood engraftment (by way of donor CD45.2 allele) and hematopoietic lineage distribution (by lineage analysis of GFP+ cells).
To initially check that over-expression of RUNX1 isoforms was not toxic to HSPCs, Sca-1+ cells were transduced with retrovirus and cultured in vitro for 48-hours. Single Sca-1+GFP+ cells were then sorted into individual wells of 96-well plates containing Methocult medium to observe hematopoietic colony-forming potential. After two weeks culture in Methocult, colony numbers and phenotypes were scored (Figure 3A). Over-expression of RUNX1 isoforms caused a reduction in colony-forming potential of transduced HSPCs, particularly in the number of multi-lineage CFU-GEMM colonies (data not shown). Moreover, colonies arising from HSPCs transduced with RUNX1 isoforms tended to be smaller, suggesting that although over-expression of RUNX1 isoforms did not appear to be toxic, it had substantial effects on HSPC proliferation.
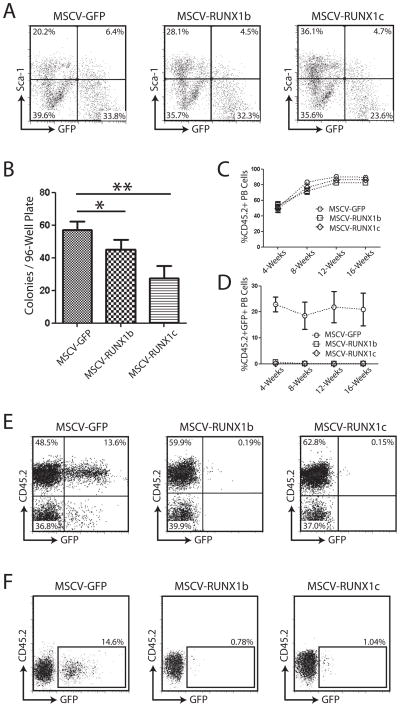
Figure 3.
Over-expression of RUNX1 isoforms in mouse hematopoietic progenitor cells. (A) Purification of transduced HSPCs following two days of in vitro culture. All viruses had comparable levels of transduction at this stage. (B) Clonal colony-forming assay for hematopoietic progenitors transduced with RUNX1 isoform retroviruses. Following transduction and culture in vitro for 48-hours, single Sca-1+GFP+ cells were sorted into individual wells of 96-well plates containing Methocult medium. Over-expression of RUNX1 isoforms caused a significant decrease in the number of colonies formed in comparison to cells transduced with MSCV-GFP control virus. (C–E) Peripheral blood analysis of mice transplanted with HSPCs transduced with control (MSCV-GFP) or RUNX1 isoform retroviruses. There was no difference in the total number of donor-derived (CD45.2+) peripheral blood cells between the three transplant groups (C), but there was a marked difference in the number of GFP+ cells generated from transduced cells (D) with virtually no generation of progeny from stem/progenitor cells over-expressing RUNX1 isoforms. (E) Representative examples of peripheral blood analysis of mice transplanted with HSPCs transduced with retroviruses 4-weeks post-transplant. While robust GFP expression was detected in peripheral blood cells of control mice (MSCV-GFP), virtually no GFP+ cells were detected in mice transplanted with cells over-expressing RUNX1 isoforms. (F) Analysis of the HSC compartment of long-term transplanted mice. Analysis of the SPKLS fraction from the bone marrow of mice transplanted with hematopoietic progenitors transduced with retrovirus showed that some GFP+ RUNX1 over-expressing HSCs could be detected >16-weeks post-transplant. Although the percentage was greatly reduced compared to control animals, this suggests that over-expression of RUNX1 isoforms in mouse HSCs was not toxic but induced extreme quiescence.
We then examined the potentials of RUNX1-isoform over-expressing HSPCs in vivo by transplantation of transduced HSPCs. The level of hematopoietic chimerism from donor cells was monitored by analysis of CD45.2+ cells in recipient mice peripheral blood at four-week intervals post-transplant. The overall level of donor cell engraftment was not significantly different between groups (Figure 3B), however the percentage of GFP+ cells in the peripheral blood showed a striking trend (Figure 3C). Virtually no GFP+ cells could be detected in the peripheral blood of mice transplanted with HSPCs over-expressing RUNX1b and RUNX1c (Figure 3D,E). There are several possibilities to account for this including, (1) over-expression of RUNX1 isoforms caused selective apoptosis in HSPCs, (2) over-expression of RUNX1 isoforms forced HSPCs to remain in quiescence, rather than differentiate, or (3) our retroviruses were not effective. However, we routinely achieved titers of MSCV-RUNX1b and MSCV-RUNX1c comparable to control MSCV-GFP virus and abundant GFP+ HSPCs could be detected after transduction and two days of in vitro culture which suggests our viruses were of good functional quality despite the large size of the RUNX1 coding sequences. We also performed Western blots on transfected 293T cells and observed RUNX1 protein levels dramatically upregulated with the retroviral vectors (data not shown).
We then decided to analyze the bone marrow of transplanted mice long-term post-transplant (>16-weeks) to determine if any GFP+ donor HSCs could still be detected (Figure 3F). Although there was a significant reduction in the number compared to control GFP-transplanted mice (13.9 ± 0.70%), GFP+ HSCs could be identified in the bone marrow of mice transplanted with HSPCs transduced with MSCV-RUNX1b (0.94 ± 0.14%) and –RUNX1c (1.03 ± 0.29%). These data indicate that RUNX1-transduced HSCs persisted in the bone marrow, with the limited number being consistent with maintenance of quiescent HSCs that are blocked from differentiating into mature cells of the peripheral blood.
Over-Expression of RUNX1 Isoforms in Mouse HSPCs Does Not Affect Homing to the Bone Marrow
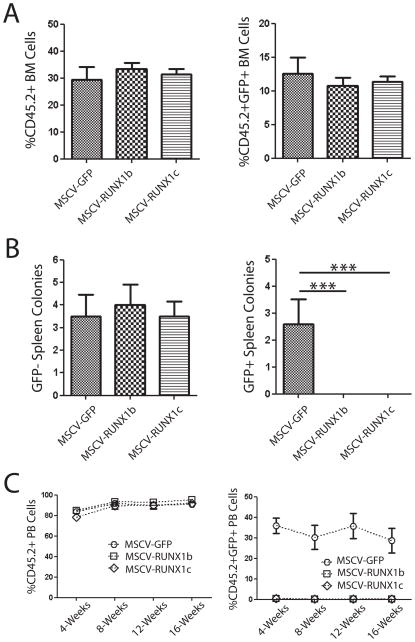
Another possible explanation as to why very few GFP+ RUNX1-transduced cells were found in the blood and bone marrow of recipient mice could be that over-expression inhibited HSPC homing to the bone marrow. To exclude this possibility, a homing assay was performed in which lethally irradiated CD45.1 mice were transplanted with 2 × 106 CD45.2 Sca-1+ RUNX1-transduced or control-transduced cells and then sacrificed 24-hours later for bone marrow collection. The bone marrow of recipient mice was then analyzed to determine the number of total CD45.2+ donor cells and CD45.2+GFP+ transduced donor cells (Figure 4A) that had successfully homed to the new niche. There was no difference between the total number of donor cells or transduced cells in the bone marrow of recipients post-transplant of transduced HSPCs, indicating over-expression of RUNX1 isoforms did not inhibit homing to their new niche.
Figure 4.
(A) Over-expression of RUNX1 isoforms in mouse hematopoietic stem/progenitor cells does not affect homing to the bone marrow. Analysis of the bone marrow of recipient mice 24-hours post-transplant of transduced hematopoietic progenitors showed that there was no difference in the total number of donor cells (CD45.2+) or transduced donor cells (CD45.2+GFP+) in the bone marrow of recipient mice. (B) The spleen CFU-S colony assay demonstrated that although there was no difference in the number of GFP− spleen colonies in mice transplanted with transduced cells, no GFP+ colonies were detected from cells over-expressing RUNX1 isoforms. (C) Over-expression of RUNX1 isoforms in fetal liver HSCs. Transplantation of transduced fetal liver HSCs showed that while there was no difference in total number of donor-derived peripheral blood cells (CD45.2+), essentially no peripheral blood progeny (CD45.2+GFP+) could be detected from HSCs over-expressing RUNX1 isoforms.
Absence of GFP+ CFU-S from RUNX1 Over-Expressing HSPCs
Because of the discrepancy between in vitro assays and the long-term transplant assays, we decided to perform spleen colony assays, which are a short-term semi-quantitative in vivo assay for early hematopoietic progenitors. Early progenitors give rise to discrete nodules in the spleens 8–12 days post-transplant which are the progeny of primitive progenitors (termed CFU-S) with multi-lineage reconstituting capacity [26–32]. Again we transduced HSPCs with retroviruses and transplanted 2.5 × 104 cells into lethally irradiated recipient mice. 12-days post-transplant, recipients were sacrificed and spleens analyzed for CFU-S. Total colonies were visually counted and GFP+ colonies were easily identified by viewing spleens under a fluorescent microscope. There was no difference in the total number of GFP− colonies that were formed in the spleens of mice transplanted with HSPCs transduced by MSCV-GFP, -RUNX1b and –RUNX1c, but there was a striking difference in the number of GFP+ colonies formed (Figure 4B). No GFP+ colonies could be detected in the spleens of mice transplanted with MSCV-RUNX1b and –RUNX1c transduced HSPCs. So while RUNX1 over-expressing HSPCs homed to the bone marrow post-transplantation, none seemed to migrate to the spleen, or if they migrated, they were unable to give rise to colonies.
Over-Expression of RUNX1 Isoforms in Mouse Fetal Liver HSPCs Also Induces Quiescence
Because over-expression of both RUNX1b and RUNX1c in adult HSPCs produced the effect of inducing quiescence, we wondered if over-expression of the isoforms in more developmentally primitive HSPCs might produce different results. This was assessed by repeating the transplant experiments using fetal liver HSPCs as donor material. Analysis of the peripheral blood of transplant recipients showed the same trend as mice transplanted with adult HSPCs over-expressing RUNX1 isoforms. While there was no difference in donor cell engraftment (%CD45.2+ peripheral blood cells) between control MSCV-GFP and MSCV-RUNX1b and –RUNX1c transduced HSPCs, there was a dearth of GFP+ cells in the peripheral blood of mice transplanted with RUNX1 over-expressing cells (Figure 4C).
Over-Expression of RUNX1 in HSPCs Does Not Increase Apoptosis or Differentiation, But Induces Quiescence
The lack of GFP+ RUNX1 over-expressing cells in the peripheral blood of transplanted mice could be due to selective apoptosis. Although this did not seem likely due to the fact that Methocult colonies were still formed in a clonal assay, we performed an apoptosis assay to exclude this possibility. HSPCs were transduced with MSCV-GFP, -RUNX1b and –RUNX1c retroviruses and cultured in vitro for two days. Sca-1+GFP+ cells were then purified and re-stained for AnnexinV to assess the percentage of transduced cells which were undergoing apoptosis (Figure 5A). There was no statistical difference between the numbers of AnnexinV+ transduced cells (Figure 5B), indicating that over-expression of RUNX1 does not affect HSC apoptosis, at least in vitro.
Figure 5.
(A) Examples of AnnexinV staining for apoptosis in retrovirally transduced hematopoietic stem/progenitor cells. (B) No difference was detected in the number of RUNX1 over-expressing cells undergoing apoptosis compared to control cells (MSCV-GFP). (C) Examples of Hoechst/Pyronin Y staining of retrovirally transduced HSPCs following two days of in vitro culture. (D) Over-expression of RUNX1 isoforms in HSPCs caused an inhibition of proliferation in vitro with a significantly higher proportion of cells in G0 or quiescence. (E) Examples of flow cytometric analysis of transduced HSPCs after four days culture for the differentiation markers Mac-1 and Gr-1. (F) Over-expression of RUNX1-isoforms in HSPCs did not increase the rate of differentiation compared to MSCV-GFP transduced control. (G) Examples of flow cytometric analysis of transduced HSPCs after four days culture for the hematopoietic progenitor compartment (KSL). (H) Over-expression of RUNX1-isoforms resulted in a significant reduction in the primitive progenitor compartment from transduced cells.
To confirm that over-expression of RUNX1 isoforms induced HSPC quiescence, a cell cycle analysis of transduced HSPCs was performed. HSPCs were transduced with MSCV-GFP, -RUNX1b and –RUNX1c retroviruses and cultured in vitro for two days. Sca-1+GFP+ cells were then purified and re-stained with Hoechst and Pyronin Y to obtain a cell cycle profile for the transduced cells (Figure 5C). This confirmed that over-expression of RUNX1 isoforms in HSPCs induced a state of quiescence as there was a significantly higher proportion of GFP+ cells in G0 (Figure 5D) resulting from transduction with MSCV-RUNX1b (61.5 ± 2.24%) and –RUNX1c (60.4 ± 2.15%) than MSCV-GFP transduced controls (41.6 ± 1.26%).
To exclude the possibility that over-expression of RUNX1-isoforms in HSPCs induced rapid differentiation, preventing engraftment and slowing growth, we examined transduced cells at two-day intervals in liquid culture. We observed no differences in cell morphology or viable cell counts over the timecourse (data not shown). We analyzed the GFP+ cells by flow cytometry (Figure 5E–H) and determined that over-expression of RUNX1-isoforms did not increase the rate of HSPC differentiation as there was no difference in the percentage of Mac-1+/Gr-1+ cells (Figure 5F). We did observe a significant decrease in the percentage of GFP+ progenitor cells (c-Kit+Sca-1+Lineage−; KSL) resulting from over-expression of RUNX1 isoforms. As we observed no differences in apoptosis or differentiation of this population, and we noted a higher proportion of HSPCs in G0 following RUNX1 over-expression, we concluded that the observed reduction in transduced progenitors was due to their increased state of quiescence or lack of proliferation.
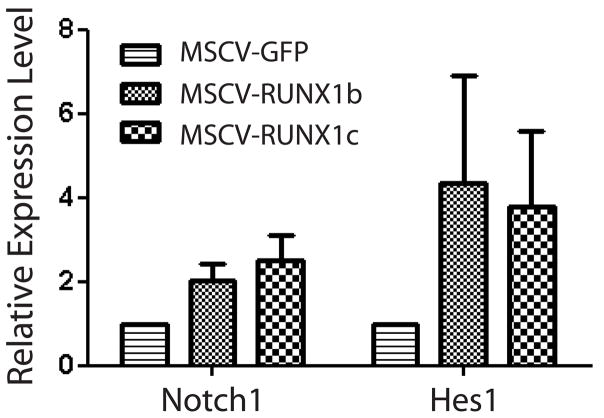
Over-Expression of RUNX1 in HSPCs Upregulates the Notch Signaling Pathway
We tried to determine the mechanism by which RUNX1 over-expression leads to increased HPSC quiescence. RUNX1 interacts with the Notch signaling pathway during the developmental specification of HSCs and subsequent homeostasis of HSC number [33], and perhaps the RUNX1 over-expression induced HSPC quiescence we observed was being mediated by the Notch pathway. To analyze this, HSPCs were transduced with retroviruses, cultured in vitro for two days, and then sorted to purify Sca-1+GFP+ cells. Real-time PCR analysis for Notch1 signaling pathway members showed RUNX1 over-expression in HSPCs caused an upregulation of the receptor Notch1 and the downstream target gene Hes1 (Figure 6). Upregulation of Hes1 in HSCs has been shown to inhibit cell proliferation [34] and dysregulation of Hes1 leads to increased HSC proliferation [35]. These data suggest that over-expression of RUNX1 in HSPCs induces quiescence which is mediated through the Notch1 signaling pathway leading to upregulation of Hes1.
Figure 6.
Real-time PCR analysis of retrovirally transduced hematopoietic stem/progenitor cells for Notch pathway genes. Over-expression of RUNX1 isoforms induced an upregulation of the surface receptor Notch1 and the downstream target gene Hes1 compared to control cells (MSCV-GFP).
DISCUSSION
RUNX1 is a pivotal regulator of definitive hematopoiesis. Expression of RUNX1 is regulated by two distinct promoters which creates diversity in distribution and protein-coding potential of the mRNA transcripts. We show here that the RUNX1c isoform is highly expressed in mouse and human HSCs, and this isoform is expressed in a spatio-temporal pattern which coincides with the emergence of definitive HSCs in both mice (E11.5 AGM region) and human (day 12 hEBs). A previous study performed a systematic spatio-temporal analysis of RUNX1 P1 and P2 promoter usage during developmental hematopoiesis and found that RUNX1 expression in hemogenic endothelial cells of the dorsal aorta commenced with the P2 (RUNX1b), while later endothelial cells and the emerging HSCs contained both P1- and P2-derived transcripts [36]. Another group that used mice bearing a hypomorphic RUNX1 allele with largely diminished P2 activity found that contrary to RUNX1 knockout mice in which embryonic lethality occurs, P2neo/neo mice develop to term, implying that during early development the activity of the P1 promoter prevails [37]. These data, combined with the expression profile of RUNX1 isoforms we report here, suggested a potentially novel or unexplored role for the RUNX1c isoform in hematopoiesis or HSC function.
Despite their divergent expression profiles, when the roles of RUNX1 isoforms were examined in mouse HSPCs using a gain-of-function model with enforced retroviral over-expression, no significant differences in function were observed. Over-expression of both RUNX1b and RUNX1c in fetal liver and adult bone marrow HSPCs induced a state of quiescence. Another study which utilized over-expression to study the actions of RUNX1 isoforms in HSCs also noted that enforced expression of RUNX1b in HSCs abrogated engraftment potential [38]. However, this group also found that over-expression of the RUNX1a isoform, which encodes a truncated molecule with DNA-binding but no transactivation capacity, increased the competitive engraftment potential of mouse HSCs [38]. Thus, there appears to be distinct roles for RUNX1 isoforms in HSCs, and given the appropriate context, a specific role for the RUNX1c isoform may yet be elucidated. It should be noted that we transduced only the Sca-1+ population of cycling HSCs, different results may be obtained from over-expression of RUNX1 isoforms in a cell population where they are essential such as VE-Cadherin+ AGM HSCs or adult niche cells.
We show that over-expression of RUNX1b and –c in mouse HSPCs induces quiescence in vivo and inhibits proliferation in vitro. There is evidence to suggest a role for RUNX1 regulating the quiescence or cell cycle of HSCs. RUNX1 levels vary during the cell cycle, and RUNX1 regulates G1 to S cell cycle progression [39]. Mouse RUNX1-deficient bone marrow contains an increased total number of HSCs [40], which from our results might be interpreted as resulting from a loss of RUNX1-mediated HSC quiescence. One of the signaling pathways thought to contribute to HSC quiescence is the Notch pathway [41],[42]. We show here that over-expression of RUNX1 isoforms in HSPCs causes upregulation of Notch1 and the downstream Notch target gene Hes1, which inhibits HSC proliferation [34]. This suggests that the Notch-RUNX1-Hes1 axis plays an important role in regulating HSC quiescence. Another study also demonstrated that Notch1 expands the HSPC compartment in zebrafish by causing a G0 population to upregulate RUNX1-dependent gene expression [33]. In combination with our results, these data identify the Notch-RUNX1 interaction as critical for the homeostasis of HSC number.
Although the roles of RUNX1b and RUNX1c seem to be largely redundant in adult HSPCs, the evidence suggests that the RUNX1c isoform may play a role in the initial specification of definitive HSCs during development. It is possible that the P1 promoter is required for epigenetic opening (activation) of the locus. Thus, conditional deletion of the individual RUNX1 promoters in the mouse E10.5–11.5 AGM region may produce a more striking result or reveal a specific function for the RUNX1c isoform at this stage of hematopoiesis. It should be noted we also tried to investigate the functions of RUNX1 isoforms in HSCs with isoform-specific gene expression knockdown. However, we were unable to generate a system which specifically ablated expression of the RUNX1b or RUNX1c isoform without affecting the expression of the other using miRNA or shRNA assays.
Given the intriguing expression pattern of RUNX1c, we sought to determine if this isoform might be a marker of definitive HSCs and if it has a specific role in HSC function. If RUNX1c was a specific marker of the first definitive HSCs, theoretically it could be used to purify powerful HSCs during mouse development, or as a readout for more efficient strategies for generating HSCs from hESC culture systems. P1-reporter hES cell lines could be used to screen for small molecules that induced hematopoietic differentiation and efficient differentiation of hES cells into hematopoietic cells could be utilized to make universal donor cells for blood or bone marrow transplantation. While the alternative promoter usage of P1 and P2 likely plays an important role in RUNX1 biology, much remains to be determined about the function of the P1/P2 switch in mediating tissue- and stage-specific expression of RUNX1.
Acknowledgments
The authors would like to thank all members of the Goodell lab for scientific advice, and Chris Threeton for flow cytometric sorting and analysis. G.A.C. was supported by an Australian National Health and Medical Research Council (NHMRC) CJ Martin Fellowship. This work was also supported by NIH grants HL081007, EB005173, DK075355 and DK58192.
Footnotes
Conflict of interest disclosure
The authors have no financial conflicts of interest to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Look AT. Oncogenic transcription factors in the human acute leukemias. Science. 1997;278:1059–1064. doi: 10.1126/science.278.5340.1059. [DOI] [PubMed] [Google Scholar]
- 2.Speck NA, Gilliland DG. Core-binding factors in haematopoiesis and leukaemia. Nat Rev Cancer. 2002;2:502–513. doi: 10.1038/nrc840. [DOI] [PubMed] [Google Scholar]
- 3.Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 4.Wang Q, Stacy T, Binder M, Marin-Padilla M, Sharpe AH, Speck NA. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc Natl Acad Sci U S A. 1996;93:3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okada H, Watanabe T, Niki M, et al. AML1(−/−) embryos do not express certain hematopoiesis-related gene transcripts including those of the PU.1 gene. Oncogene. 1998;17:2287–2293. doi: 10.1038/sj.onc.1202151. [DOI] [PubMed] [Google Scholar]
- 6.North TE, de Bruijn MF, Stacy T, et al. Runx1 expression marks long-term repopulating hematopoietic stem cells in the midgestation mouse embryo. Immunity. 2002;16:661–672. doi: 10.1016/s1074-7613(02)00296-0. [DOI] [PubMed] [Google Scholar]
- 7.North T, Gu TL, Stacy T, et al. Cbfa2 is required for the formation of intra-aortic hematopoietic clusters. Development. 1999;126:2563–2575. doi: 10.1242/dev.126.11.2563. [DOI] [PubMed] [Google Scholar]
- 8.Yokomizo T, Ogawa M, Osato M, Kanno T, Yoshida H, Fujimoto T, Fraser S, Nishikawa S, Okada H, Satake M, Noda T, Nishikawa S, Ito Y. Requirement of Runx1/AML1/PEBP2alphaB for the generation of haematopoietic cells from endothelial cells. Genes Cells. 2001;6:13–23. doi: 10.1046/j.1365-2443.2001.00393.x. [DOI] [PubMed] [Google Scholar]
- 9.Ichikawa M, Asai T, Saito T, et al. AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nat Med. 2004;10:299–304. doi: 10.1038/nm997. [DOI] [PubMed] [Google Scholar]
- 10.Growney JD, Shigematsu H, Li Z, et al. Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood. 2005;106:494–504. doi: 10.1182/blood-2004-08-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Putz G, Rosner A, Nuesslein I, Schmitz N, Buchholz F. AML1 deletion in adult mice causes splenomegaly and lymphomas. Oncogene. 2006;25:929–939. doi: 10.1038/sj.onc.1209136. [DOI] [PubMed] [Google Scholar]
- 12.Fujita Y, Nishimura M, Taniwaki M, Abe T, Okuda T. Identification of an alternatively spliced form of the mouse AML1/RUNX1 gene transcript AML1c and its expression in early hematopoietic development. Biochem Biophys Res Commun. 2001;281:1248–1255. doi: 10.1006/bbrc.2001.4513. [DOI] [PubMed] [Google Scholar]
- 13.Ghozi MC, Bernstein Y, Negreanu V, Levanon D, Groner Y. Expression of the human acute myeloid leukemia gene AML1 is regulated by two promoter regions. Proc Natl Acad Sci U S A. 1996;93:1935–1940. doi: 10.1073/pnas.93.5.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miyoshi H, Ohira M, Shimizu K, et al. Alternative splicing and genomic structure of the AML1 gene involved in acute myeloid leukemia. Nucleic Acids Res. 1995;23:2762–2769. doi: 10.1093/nar/23.14.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rennert J, Coffman JA, Mushegian AR, Robertson AJ. The evolution of Runx genes I. A comparative study of sequences from phylogenetically diverse model organisms. BMC Evol Biol. 2003;3:4. doi: 10.1186/1471-2148-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bangsow C, Rubins N, Glusman G, et al. The RUNX3 gene--sequence, structure and regulated expression. Gene. 2001;279:221–232. doi: 10.1016/s0378-1119(01)00760-0. [DOI] [PubMed] [Google Scholar]
- 17.Xiao ZS, Hjelmeland AB, Quarles LD. Selective deficiency of the “bone-related” Runx2-II unexpectedly preserves osteoblast-mediated skeletogenesis. J Biol Chem. 2004;279:20307–20313. doi: 10.1074/jbc.M401109200. [DOI] [PubMed] [Google Scholar]
- 18.Zambidis ET, Peault B, Park TS, Bunz F, Civin CI. Hematopoietic differentiation of human embryonic stem cells progresses through sequential hematoendothelial, primitive, and definitive stages resembling human yolk sac development. Blood. 2005;106:860–870. doi: 10.1182/blood-2004-11-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodell MA, Brose K, Paradis G, Conner AS, Mulligan RC. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J Exp Med. 1996;183:1797–1806. doi: 10.1084/jem.183.4.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Challen GA, Martinez G, Davis MJ, et al. Identifying the molecular phenotype of renal progenitor cells. J Am Soc Nephrol. 2004;15:2344–2357. doi: 10.1097/01.ASN.0000136779.17837.8F. [DOI] [PubMed] [Google Scholar]
- 21.Challen G, Gardiner B, Caruana G, et al. Temporal and spatial transcriptional programs in murine kidney development. Physiol Genomics. 2005;23:159–171. doi: 10.1152/physiolgenomics.00043.2005. [DOI] [PubMed] [Google Scholar]
- 22.Thomson JA, Itskovitz-Eldor J, Shapiro SS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 23.Naviaux RK, Costanzi E, Haas M, Verma IM. The pCL vector system: rapid production of helper-free, high-titer, recombinant retroviruses. J Virol. 1996;70:5701–5705. doi: 10.1128/jvi.70.8.5701-5705.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kotani H, Newton PB, 3rd, Zhang S, et al. Improved methods of retroviral vector transduction and production for gene therapy. Hum Gene Ther. 1994;5:19–28. doi: 10.1089/hum.1994.5.1-19. [DOI] [PubMed] [Google Scholar]
- 25.Challen GA, Boles N, Lin KK, Goodell MA. Mouse hematopoietic stem cell identification and analysis. Cytometry A. 2009;75:14–24. doi: 10.1002/cyto.a.20674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baines P, Bol SJ, Rosendaal M. Physical and kinetic properties of haemopoietic progenitor cell populations from mouse marrow detected in five different assay systems. Leuk Res. 1982;6:81–88. doi: 10.1016/0145-2126(82)90046-7. [DOI] [PubMed] [Google Scholar]
- 27.Bartelmez SH, Andrews RG, Bernstein ID. Uncovering the heterogeneity of hematopoietic repopulating cells. Exp Hematol. 1991;19:861–862. [PubMed] [Google Scholar]
- 28.Bearpark AD, Gordon MY. Adhesive properties distinguish sub-populations of haemopoietic stem cells with different spleen colony-forming and marrow repopulating capacities. Bone Marrow Transplant. 1989;4:625–628. [PubMed] [Google Scholar]
- 29.Curry JL, Trentin JJ. Hemopoietic spleen colony studie s. I. Growth and differentiation. Dev Biol. 1967;15:395–413. doi: 10.1016/0012-1606(67)90034-6. [DOI] [PubMed] [Google Scholar]
- 30.Harris RA, Hogarth PM, Wadeson LJ, Collins P, McKenzie IF, Penington DG. An antigenic difference between cells forming early and late haematopoietic spleen colonies (CFU-S) Nature. 1984;307:638–641. doi: 10.1038/307638a0. [DOI] [PubMed] [Google Scholar]
- 31.Molineux G, Schofield R, Testa NG. Development of spleen CFU-S colonies from day 8 to day 11: relationship to self-renewal capacity. Exp Hematol. 1986;14:710–713. [PubMed] [Google Scholar]
- 32.Ploemacher RE, Brons RH. Separation of CFU-S from primitive cells responsible for reconstitution of the bone marrow hemopoietic stem cell compartment following irradiation: evidence for a pre-CFU-S cell. Exp Hematol. 1989;17:263–266. [PubMed] [Google Scholar]
- 33.Burns CE, Traver D, Mayhall E, Shepard JL, Zon LI. Hematopoietic stem cell fate is established by the Notch-Runx pathway. Genes Dev. 2005;19:2331–2342. doi: 10.1101/gad.1337005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu X, Alder JK, Chun JH, et al. HES1 inhibits cycling of hematopoietic progenitor cells via DNA binding. Stem Cells. 2006;24:876–888. doi: 10.1634/stemcells.2005-0598. [DOI] [PubMed] [Google Scholar]
- 35.Santaguida M, Schepers K, King B, et al. JunB protects against myeloid malignancies by limiting hematopoietic stem cell proliferation and differentiation without affecting self-renewal. Cancer Cell. 2009;15:341–352. doi: 10.1016/j.ccr.2009.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bee T, Liddiard K, Swiers G, et al. Alternative Runx1 promoter usage in mouse developmental hematopoiesis. Blood Cells Mol Dis. 2009;43:35–42. doi: 10.1016/j.bcmd.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 37.Pozner A, Lotem J, Xiao C, et al. Developmentally regulated promoter-switch transcriptionally controls Runx1 function during embryonic hematopoiesis. BMC Dev Biol. 2007;7:84. doi: 10.1186/1471-213X-7-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsuzuki S, Hong D, Gupta R, Matsuo K, Seto M, Enver T. Isoform-specific potentiation of stem and progenitor cell engraftment by AML1/RUNX1. PLoS Med. 2007;4:e172. doi: 10.1371/journal.pmed.0040172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cao W, Britos-Bray M, Claxton DF, et al. CBF beta-SMMHC, expressed in M4Eo AML, reduced CBF DNA-binding and inhibited the G1 to S cell cycle transition at the restriction point in myeloid and lymphoid cells. Oncogene. 1997;15:1315–1327. doi: 10.1038/sj.onc.1201305. [DOI] [PubMed] [Google Scholar]
- 40.Ichikawa M, Goyama S, Asai T, et al. AML1/Runx1 negatively regulates quiescent hematopoietic stem cells in adult hematopoiesis. J Immunol. 2008;180:4402–4408. doi: 10.4049/jimmunol.180.7.4402. [DOI] [PubMed] [Google Scholar]
- 41.Maillard I, Koch U, Dumortier A, et al. Canonical notch signaling is dispensable for the maintenance of adult hematopoietic stem cells. Cell Stem Cell. 2008;2:356–366. doi: 10.1016/j.stem.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stier S, Cheng T, Dombkowski D, Carlesso N, Scadden DT. Notch1 activation increases hematopoietic stem cell self-renewal in vivo and favors lymphoid over myeloid lineage outcome. Blood. 2002;99:2369–2378. doi: 10.1182/blood.v99.7.2369. [DOI] [PubMed] [Google Scholar]