Summary
Autophagy is critical for maintaining cellular homeostasis, coping with metabolic stress, and limiting oxidative damage. Several autophagy-deficient or knockout models show increased tumor incidence, implicating autophagy as a tumor suppressor. Autophagy is involved in multiple processes which may curb transformation, including the control of oncogene-induced senescence (OIS), which can limit progression to full malignancy, and efficient antigen presentation, which is crucial for immune cell recognition and elimination of nascent cancer cells. Activation of the autophagy pathway may therefore hold promise as a chemoprevention strategy. Caloric restriction, bioactive dietary compounds, or specific pharmacological activators of the autophagy pathway are all possible avenues to explore in harnessing the autophagy pathway in cancer prevention.
Introduction
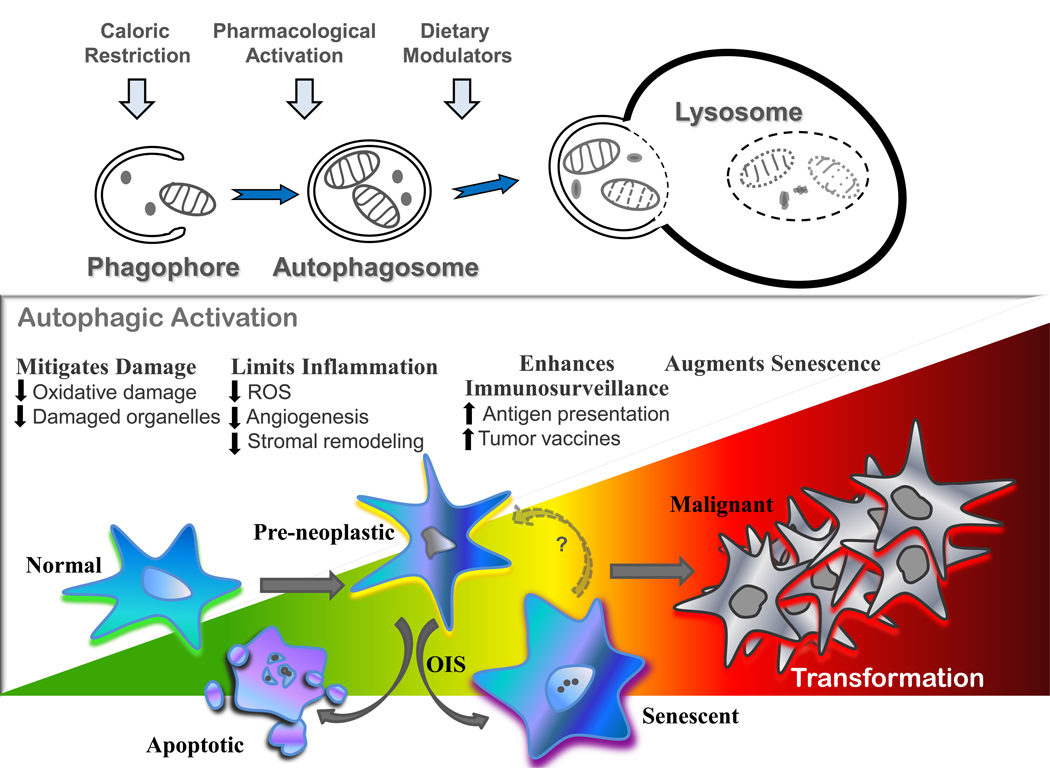
The macroautophagy (hereafter autophagy) pathway is the central recycling center of the cell. This pathway orchestrates the degradation of bulk cytoplasmic components and organelles, which are engulfed within double-membraned cytoplasmic vesicles coined autophagosomes that form following the fusion of isolation membranes called phagophores. The outer membrane of autophagosomes then fuse with the lysosome (Figure 1), delivering their cargo for degradation into monomers (e.g., amino acids, fatty acids, etc.) that are necessary to maintain the metabolic and bioenergetic demands of the cell under conditions of stress or nutrient deprivation. Indeed, the majority of long-lived proteins and organelles are degraded via autophagy, underscoring its essential role in cellular homeostasis. Further, autophagy is markedly upregulated by cellular stress, including nutrient or cytokine depletion, hypoxia or oxidative damage, and it is also required for removing protein aggregates and as an innate intracellular defense mechanism against several pathogens[1–3]. Finally, autophagy is induced by nearly every anti-cancer regimen, and this pathway therefore represents a major, tumor cell-intrinsic, resistance mechanism.
Figure 1. The role of autophagy in preventing neoplastic transformation.
The autophagy pathway can be activated through various mechanisms and performs essential cellular housekeeping and recycling functions that serve to limit the accumulation of cellular damage and inflammation. In addition, autophagic activation plays a key role in antigen presentation, immune system development, and homeostasis. Oncogene-induced senescence (OIS) has also been linked to autophagy and may provide an alternate cell cycle exit strategy when apoptotic pathways are disabled.
These responses are most consistent with the well-established pro-survival roles of the autophagy pathway, yet aberrantly high levels of autophagy can also trigger macroautophagic (type II) cell death. However, under most physiological conditions, for example, following long-term starvation, cells can undergo a complete recovery when returned to nutrient-replete conditions [4]. Thus, cell death via autophagy likely reflects the finite ability to survive prolonged famine rather than an active form of cell death. Indeed, bioenergetic failure leads to apoptosis or necrosis, and these bona fide cell death programs and autophagy are interconnected at many levels. For example, apoptotic proteins such as Bcl2 and Bcl-xL also regulate autophagy through their interactions with Beclin1, a required cofactor for the class III PI3-kinase Vps34[5,6] that generates phosphoinositides essential for autophagosome formation. Further, blocking both autophagy and apoptosis leads to death by necrosis[4], which provokes an inflammatory response that drives tumor progression.[1]
Dualism of autophagy in the development and maintenance of cancer
Links between autophagy and cancer are evident on many levels. First, loss of heterozygosity (LOH) of some autophagic regulators such as Beclin1 or bif, or components of the pathway such as Atg4c or UVRAG, generates a tumor -prone phenotype in mice[7–10]. Second, beclin1 heterozygosity accelerates Myc-driven tumorigenesis in the Eµ-Myc transgenic model of human B cell lymphoma (F. Dorsey and J. Cleveland, unpublished data). Third, Beclin1 or Atg5 knockdown augments transformation of cell lines[11,12]. Finally, LOH of Beclin1 appears common in ovarian and breast cancers, yet complete loss of Beclin1 is not observed, suggesting essential roles of autophagy in the maintenance of the malignant state[13,14].
Collectively, these findings are consistent with a tumor suppressor role for the autophagy pathway in blocking cancer development. Indeed, given that autophagy limits the accumulation of cellular damage to proteins, organelles, and DNA, all of which can contribute to mutation and/ or metabolic dysfunction and initiate transformation, this pathway should intuitively suppress tumorigenesis [1,12,15]. Accordingly, one would predict that oncogenes suppress this pathway, and indeed our recent studies have established that the Myc oncoprotein suppresses the expression of autophagic components. However, the loss of other essential components of the autophagy pathway such as Atg7 or Atg5, which are required for autophagosome formation, is not sufficient to trigger a tumor-prone phenotype and Atg7 loss does not affect the onset of Eµ-Myc-driven lymphoma (F. Dorsey and J. Cleveland, unpublished data). Thus, the autophagy pathway may only curb tumorigenesis in select contexts.
Despite its rather confusing roles in tumor development, autophagy provides a definitive advantage to tumors in the management of metabolic stress. For example, autophagy is specifically induced in hypoxic regions of tumors[1], where it provides a metabolic safety net for oxygen- and nutrient-starved cells. Furthermore, Atg7 functions contribute to the maintenance of Myc-induced lymphoma. Thus, though reductions of the autophagy pathway may favor mutations that promote transformation, as seen in Beclin1 haploinsufficiency, autophagy likely plays essential survival roles in the maintenance of extant tumors.
Impact of the autophagy pathway on transformation
Oncogene activation triggers a series of DNA-damage, proliferative, and apoptotic checkpoints that are disabled during tumor progression and autophagy may impact these responses at several levels (Figure 1). First, autophagy functions in a cell-intrinsic fashion as a guardian that removes damaged and/ or damaging materials, thereby limiting mutational insults to the cell. Thus, one prediction is that autophagy may dampen activation of these checkpoints. Second, autophagy appears to play an important role in oncogene-induced senescence, which may serve to limit tumor progression. Finally, there are likely several non-tumor cell autonomous effects of the autophagy pathway on tumorigenesis. For example, autophagy has clear effects in monitoring and controlling damage invoked by the inflammatory response, which can drive tumorigenesis, and autophagy is necessary for aspects of immune cell function and for immune surveillance. Specifically, autophagy is necessary for the processing of MHC peptides in antigen-presenting cells (APC), which is necessary for loading these peptides on APC and for presentation and activation of effector T cells. Accordingly, the autophagy pathway has a direct impact on all cancer vaccine development. However, the early role of autophagy in preventing transformation contrasts with its roles in promoting survival of tumor cells undergoing metabolic stress. Thus, while autophagy provides an attractive target for cancer prevention and therapeutics, selective preneoplastic- and/ or tumor-specific anti-autophagy agents are clearly desired to maximize anti-tumor effects without compromising immune surveillance.
Minimizing cellular damage
Autophagy functions as a sentinel against cellular damage, a byproduct of metabolism and aging that must be constantly curtailed. This is largely accomplished by the steady - state turnover of proteins and organelles by autophagy. Accordingly, deficiencies in autophagy trigger increases in protein dysfunction and aggregation, damage to mitochondria, the induction of reactive oxygen species (ROS), and genomic instability. Further, autophagy plays clear roles in age-related and chronic disease and also in the response to acute stress, including hypoxia, oxidation, and heat shock, all of which contribute to protein dysfunction and genotoxic damage[1,11,12,16,17].
Autophagy-related disruption of normal protein/ organelle turnover is implicated in several pathologies, including cancer, heart disease, and neurodegeneration, and chronic liver disease. Improper clearance of injurious protein aggregates and increases in levels of oxidized proteins are hallmarks of neuronal cells in Huntington ’s, Parkinson ’s, and Alzheimer’s diseases, and similar phenotypes are manifest in autophagy-deficient mice[18,19]. Accumulation of p62 in autophagy-deficient tumor cells leads to DNA and organelle damage, and increases in ROS[17]. Similarly, defects in removal of mutant fibrinogen, which leads to chronic liver disease and cirrhosis in humans, have been recapitulated in atg5-deficient cells[20], and alpha-1-antitrypsin (AT) deficiency, which can lead to hepatocellular carcinomas, is harnessed by autophagy-mediated clearance of mutant AT protein[21].
Autophagy also plays critical roles following exposure to DNA damage. For example, immortalized Atg5−/− or beclin1+/− Bcl-2-expressing kidney epithelial cells are hypersensitive to DNA damage and manifest increased centrosome numbers, microtubule abnormalities, and aneuploidy[12]. Similar results have been reported in autophagy-deficient immortalized mammary epithelial cells[11]. Curiously, these cell lines have increased tumorigenic potential, supporting the role of autophagy in limiting both the DNA damage and resultant genomic instability, which ultimately leads to malignant transformation.
Autophagy and senescence
Cellular senescence is an important tumor suppression mechanism whereby cells undergo a terminal proliferative arrest but retain normal metabolic capacity. While replicative senescence is defined as compulsory mitotic arrest imposed after a finite number of divisions[22] (i.e., the classic Hayflick limit), senescence can also be induced by DNA or organelle damage, oxidative insults, loss of tumor suppressors, or following the activation of oncogenes. Unlike replicative senescence, oncogene-induced senescence (OIS) is a rapid and acute process triggered by oncogenes (i.e., Ras) that has been implicated in the control of benign tumors by impeding frank malignancy[23]. Interestingly, autophagic flux and expression of components of the autophagy pathway are increased in senescent cells, where autophagy appears to provide essential monomers for these highly anabolic cells[24]. Modulation of key autophagic components such as Ulk3, Atg5, or Atg7 has been shown to control senescence, possibly through feedback control of the Pi3K-Akt-mTOR pathway[25], which acts to limit oncogene signaling and enable cell cycle exit.
Autophagy and immune surveillance
Immune detection and elimination of nascent cancer cells, first posited over a century ago, is now recognized as a major barrier to clinical disease[26]. Human transplant patients who undergo long-term immunosuppression have a greatly increased cancer incidence[27]. Further, increased levels of select immune cells within tumors are positive prognostic indicators, as is immunity to various tumor antigens. Finally, several strains of immunodeficient mice are highly susceptible to chemically-induced tumors, and can also develop early-onset spontaneous cancers[28].
Immunosurveillance provides a fundamental defense against cancer through cell-mediated, humoral, and innate mechanisms. Cytotoxic CD8+ T lymphocytes (CTLs) recognize tumor-associated antigens (TAA)[29–31], including the cancer/ testis (CT) antigens, cyclin B1, and mucin 1 (Muc1), and correspondingly, CTLs within tumor tissue connote favorable patient prognosis[32,33]. Antibodies to oncoproteins correlate with positive outcome in ovarian cancer[34]. Additionally, innate immune effectors such as natural killer (NK), natural killer T (NKT), and γδ T cells are important effectors in eradicating malignancies[28,35]. Conversely, chronic inflammation leads to infiltration of tumor-associated macrophages and mast cells that promote tumorigenesis and presage poor outcome. Predictably, tumors evolve means of immune avoidance, including tolerizing T cells and secreting pro-inflammatory and/or immunosuppressive cytokines and chemokines[36]. Ironically, over time the immune response may sculpt the tumor repertoire by negatively selecting highly immunogenic tumor cells whilst sparing the poorly immunogenic, enabling persistence and the acquisition of additional mutations[28].
A classic example of tumor immunosurveillance and its interplay with autophagy is found with Epstein-Barr virus (EBV). EBV infections are associated with Burkitt and Hodgkin’s lymphomas, nasopharyngeal carcinomas, and gastric cancers, and immunocompromised individuals have a much higher frequency of EBV-transformed B cell lymphoma[37]. With an infection rate estimated >90% of the general population, the efficacy of this immunosurveillance is clear. Importantly, autophagy plays critical roles in processing and presentation of EBNA-1 peptides with MHC II molecules on EBV-transformed cells, as presentation is blocked by knockdown of Atg12[38]. Antigen presentation of other TAAs, such as Muc1, also requires autophagy[39], and indeed, proteins fused to Atg8/ LC3 can enhance MHC class II presentation[40].
Autophagy also plays roles in innate and adaptive immunity that may be exploited for cancer prevention[3]. For example, autophagy is critical for normal T and B cell development[41,42], B cell receptor (BCR) signaling and co-stimulation[43], macrophage function[44], apoptotic cell clearance[45], and antimicrobial function of Paneth cells[46] in the small intestine. Interestingly, two recent studies highlight a T cell-intrinsic role for mTOR inhibition in promoting memory CD8+ T cell differentiation[47,48]. As mTOR is a negative regulator of autophagy, activation of autophagy may play a role in the generation of effective memory.
Thus, agents that selectively augment autophagy functions may increase the efficacy of immune surveillance, the potency of tumor vaccines, and ameliorate the inflammatory phenotype manifest during chronic infections that are associated with increased cancer risk.
Activating autophagy as a cancer prevention strategy
Cancer prevention strategies must be readily accessible, safe over the long-term, easily implemented, and have no major side effects. There are several avenues available to pursue in targeting autophagy for cancer prevention, but all need to be optimized to meet the above criteria.
Caloric restriction
Cancer risk rises dramatically with age. Thus, attempts to extend the collective human lifespan will have to reckon with cancer. Further, the ability to induce autophagy appears to decline with age[49]. Interestingly, the only consistent strategy documented to extend life and inhibit tumorigenesis involves limitation of food intake. Discovered in rodents over 70 years ago[50], this principle is widespread and robust. For example, Caenorhabditis elegans GLD-1 mutants, which develop lethal germ cell tumors, are rescued by longevity-conferring mutations that impair insulin receptor signaling, prevent food consumption, or disrupt optimal mitochondrial function [51], and this has been linked to induction of autophagy[52]. Further, caloric restriction (CR) nearly doubles the lifespan of a mouse and delays spontaneous tumorigenesis in p53−/− mice[53,54]. Recently, CR has been shown to increase longevity in rhesus monkeys[55]. Extension of lifespan is also circumstantially linked to autophagy in yeast [56], C. elegans[57], and Drosophila[58], where cumulative cellular damage is associated with an accelerated aging phenotype. Therefore, activation of autophagy through CR or periodic fasting would appear to be a logical avenue to pursue in cancer prevention. Conversely, obesity (i.e., excessive caloric intake) is a strong risk factor for cancer, and overweight cancer patients have consistently poorer prognoses than those having normal weight. These effects may be largely due to inflammation, as adipocytes secrete a number of pro-inflammatory cytokines that increase both cancer risk and tumor progression[59]. Interestingly, autophagy has been shown to be critical for rapid clearance of apoptotic cells, which is known to limit the inflammation associated with cell death.[45] Thus, autophagy induced by caloric restriction may reduce both inflammation and cancer incidence.
Activation of autophagy by nutrient limitation is also tied to other anti-aging pathways. Sirtuins, some of which have NAD-dependent deacetylase function, control lifespan in eukaryotes and Sirtuin-1 (Sirt1) is induced in tissues of rodents subjected to CR[60,61]. Sirt1 activation following CR activates autophagy in a deacetylase-dependent manner, and Sirt1-deficient mice have defects in mobilizing the autophagic machinery following nutrient deprivation, and this has been linked to Sirt1-dependent deacetylation of Atg7 and LC3[62]. Conversely, the acetyltransferase p300 appears to acetylate Atg5, Atg7, LC3 and Atg12 and, accordingly, p300 knockdown induces autophagic activation[63]. Therefore, the lifespan extension seen with CR in many model systems may involve the activation of multiple anti-aging pathways that target the autophagy pathway.
Dietary modulation
In addition to CR, other diet modifications hold promise in preventing cancer through activation of autophagy. The high-fat, low-fiber diet of modern, industrialized societies is implicated in a third of all cancers; thus, cancer -protective compounds in food are receiving much attention. These include the carotenoids, lycopene (watermelon, tomatoes), β-carotene (carrots, leafy greens) and lutein (kale family), the polyphenols quercetin (onions, apples), resveratrol (grapes), and curcumin (turmeric), and epigallocatechin-3-gallate (green tea). All inhibit ROS and reactive nitrogen species, and have anti-angiogenic and anti-inflammatory properties. Likewise, polyunsaturated fats have anti-inflammatory effects, and these compounds, and other phytochemicals (e.g., indole-3-carbinol, proanthocyanin) and vitamin D3, are thought to have cancer prevention effects[64,65]. Interestingly, many of these dietary compounds also modulate autophagy[66,67]. For example, several phytochemicals reverse okadaic acid-induced inhibition of autophagy, and vitamins D3 and K2, and resveratrol, a known activator of Sirt1, also appear to augment autophagy. Unfortunately, experiments are lacking that clearly demonstrate that dietary compounds act to prevent transformation via their effects on autophagy.
There are several hurdles that must be addressed in using dietary compounds to modulate autophagy [65]. First, concentrations of these compounds in foods are rarely high enough to have physiological effects and data on their bioavailability, absorption, and metabolism are lacking. Second, the synergistic effects associated with many food - related compounds is lost if they are taken in isolation. Conversely, some compounds demonstrate antagonistic effects with each other or with traditional chemotherapeutics. Finally, the scope of assessing diet in cancer chemoprevention is large. For example, while thousands of phytochemicals have been identified, few have been rigorously assessed for their ability to modulate autophagy.
Pharmacologic activation of autophagy
Autophagy has apparent conflicting roles in the development and maintenance of cancer. Agonists of the autophagy pathway will likely inhibit transformation and prevent cancer by limiting damage. In contrast, agents that inhibit the pathway are desired for extant malignancies, where autophagy plays critical roles in tumor cell survival. Thus, both agonists and inhibitors of the pathway are desired.
There are several drugs that alter autophagy and impair tumorigenesis. Agents that mimic metabolic or environmental stresses, or those that disrupt mitogenic signaling cascades induce autophagy[68]. For example, 2-deoxyglucose impairs glycolysis, thus mimicking starvation, and inhibitors of growth factor factors receptors, the PI3K-mTOR pathway and histone deacetylases are all potent activators of the autophagy pathway[69]. Indeed, the rapamycin family (rapamycin/ sirolimus, Rad001/ Everolimus, CCI-779/ Temsirolimus, and AP23573) of macrolide antibiotics activate autophagy through mTOR inhibition and can extend lifespan in mice[70], though this could not be directly attributed to reduced cancer incidence. In addition, inhibitors that selectively target class I PI3K are also potent agonists of the autophagy pathway, as are agents that target 1,4,5-inositol triphosphate (IP3) uptake[7]. Collectively, such compounds have promise as chemoprevention drugs, and their efficacy as therapeutics will be markedly augmented by the derivation of new autophagy inhibitors.
Finally, there is mounting evidence that current FDA-approved agents for other indications may affect the autophagy pathway and cancer therapy. Chloroquine (CQ), an anti-malarial compound, has been shown to prevent lymphoma development by impacting autophagy and the lysosomal pathway in mouse cancer models of ataxia telangiectasia mutated (Atm −/− mice) and human B lymphoma (Eµ-Myc transgenics)[71]. These findings suggest CQ as a chemoprevention agent, and indeed this notion is supported by a clinical trial assessing the efficacy of CQ as an anti-malarial in Tanzania, where CQ reduced the incidence of Burkitt lymphoma by over 80 percent[72]. The mechanism of CQ chemoprevention is not entirely clear, as measurements of autophagic flux indicate that it can activate the pathway. However, CQ is weak base and, as a lysosomotropic agent, it also disrupts lysosomal functions and impairs the destruction of cargo delivered by the autophagosomes. Importantly, however, CQ has synergistic anti-cancer effects in combination with conventional therapeutic agents in treating otherwise refractory malignancies. Accordingly, CQ is currently being tested in several clinical trials in combination therapies[68].
Additionally, several independent studies have shown reduced cancer mortality in type 2 diabetes patients treated with metformin, a biguanide that lowers blood glucose levels by inducing glucose uptake into muscle tissue and by inhibiting its production in the liver[73–76]. As type-2 diabetes patients have an overall increased cancer risk, this implicates metformin as a potential chemoprevention agent. Interestingly, metformin can inhibit p53-deficient tumor growth through the induction of AMPK kinase, which in turn inhibits mTOR and induces autophagy[77], suggesting an autophagy-related mechanism is responsible for metformin’s anti-cancer effects.
Conclusions and future directions
Future emphasis must be placed on specificity, selectivity and clearly defined mechanisms of action when identifying agonists of the autophagy pathway, which should be effective chemoprevention agents. Indeed, most current prevention strategies rely on non-specific modes of action. Moving forward, the oncology field must consider taking a specific approach by targeting key components of the autophagy pathway, yet how these agents impact immunosurveillance as well as other aspects of host physiology must be considered. Indeed, given the likely roles of autophagy in maintenance of the malignant state, the use of such chemoprevention strategies must be carefully tailored.
Acknowledgements
The authors thank members of the Cleveland laboratory for their numerous contributions to autophagy projects. This work was supported in part by a NRSA grant 5F32 CA123777-03 (to F.C.D), by monies from the Lehman Brothers Foundation, and by start up funds provided to Scripps Florida from the State of Florida.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
The authors declare no conflicts of interest.
References and annotations
- 1. Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gelinas C, Fan Y, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. This is one of the first papers to mechanistically explain how autophagy can act as a tumor suppressor. This work was expanded in references [11•; and 12•] to additional model systems.
- 2.Amaravadi RK, Yu D, Lum JJ, Bui T, Christophorou MA, Evan GI, Thomas-Tikhonenko A, Thompson CB. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest. 2007;117:326–336. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levine B, Deretic V. Unveiling the roles of autophagy in innate and adaptive immunity. Nat Rev Immunol. 2007;7:767–777. doi: 10.1038/nri2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lum JJ, Bauer DE, Kong M, Harris MH, Li C, Lindsten T, Thompson CB. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–248. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 5.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 6.Tracy K, Dibling BC, Spike BT, Knabb JR, Schumacker P, Macleod KF. BNIP3 is an RB/E2F target gene required for hypoxia-induced autophagy. Mol Cell Biol. 2007;27:6229–6242. doi: 10.1128/MCB.02246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, Jung JU. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8:688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- 9.Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, Liang C, Jung JU, Cheng JQ, Mule JJ, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9:1142–1151. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marino G, Salvador-Montoliu N, Fueyo A, Knecht E, Mizushima N, Lopez-Otin C. Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in Atg4C/autophagin-3. J Biol Chem. 2007;282:18573–18583. doi: 10.1074/jbc.M701194200. [DOI] [PubMed] [Google Scholar]
- 11. Karantza-Wadsworth V, Patel S, Kravchuk O, Chen G, Mathew R, Jin S, White E. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 2007;21:1621–1635. doi: 10.1101/gad.1565707. Along with [1•], this paper and [12•] provide further mechanistic evidence of autophagy as a tumor suppressor.
- 12. Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, Degenhardt K, Chen G, Jin S, White E. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007;21:1367–1381. doi: 10.1101/gad.1545107. Along with [1•], this paper and [11•] provide further mechanistic evidence of autophagy as a tumor suppressor.
- 13.Aita VM, Liang XH, Murty VV, Pincus DL, Yu W, Cayanis E, Kalachikov S, Gilliam TC, Levine B. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics. 1999;59:59–65. doi: 10.1006/geno.1999.5851. [DOI] [PubMed] [Google Scholar]
- 14.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sato K, Tsuchihara K, Fujii S, Sugiyama M, Goya T, Atomi Y, Ueno T, Ochiai A, Esumi H. Autophagy is activated in colorectal cancer cells and contributes to the tolerance to nutrient deprivation. Cancer Res. 2007;67:9677–9684. doi: 10.1158/0008-5472.CAN-07-1462. [DOI] [PubMed] [Google Scholar]
- 16.Jin S, White E. Tumor suppression by autophagy through the management of metabolic stress. Autophagy. 2008;4:563–566. [PMC free article] [PubMed] [Google Scholar]
- 17. Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–1075. doi: 10.1016/j.cell.2009.03.048. This elegant paper provides the first evidence linking autophagy to the NF-κB signaling pathway. This also ties in nicely with references [11• and 12•] in providing mechanistic support that autophagy works to inhibit transformation.
- 18.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 19.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 20.Kruse KB, Dear A, Kaltenbrun ER, Crum BE, George PM, Brennan SO, McCracken AA. Mutant fibrinogen cleared from the endoplasmic reticulum via endoplasmic reticulum-associated protein degradation and autophagy: an explanation for liver disease. Am J Pathol. 2006;168:1299–1308. doi: 10.2353/ajpath.2006.051097. quiz 1404-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamimoto T, Shoji S, Hidvegi T, Mizushima N, Umebayashi K, Perlmutter DH, Yoshimori T. Intracellular inclusions containing mutant alpha1-antitrypsin Z are propagated in the absence of autophagic activity. J Biol Chem. 2006;281:4467–4476. doi: 10.1074/jbc.M509409200. [DOI] [PubMed] [Google Scholar]
- 22.Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Exp Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- 23.Collado M, Serrano M. The power and the promise of oncogene-induced senescence markers. Nat Rev Cancer. 2006;6:472–476. doi: 10.1038/nrc1884. [DOI] [PubMed] [Google Scholar]
- 24. Young AR, Narita M, Ferreira M, Kirschner K, Sadaie M, Darot JF, Tavare S, Arakawa S, Shimizu S, Watt FM, et al. Autophagy mediates the mitotic senescence transition. Genes Dev. 2009;23:798–803. doi: 10.1101/gad.519709. This novel paper implicates autophagy in the induction of senescence. Overexpression of the ATG1 family member, ULK3, both upregulated autophagy and induced a premature senescence phenotype.
- 25.Courtois-Cox S, Genther Williams SM, Reczek EE, Johnson BW, McGillicuddy LT, Johannessen CM, Hollstein PE, MacCollin M, Cichowski K. A negative feedback signaling network underlies oncogene-induced senescence. Cancer Cell. 2006;10:459–472. doi: 10.1016/j.ccr.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 27.Buell JF, Gross TG, Woodle ES. Malignancy after transplantation. Transplantation. 2005;80:S254–S264. doi: 10.1097/01.tp.0000186382.81130.ba. [DOI] [PubMed] [Google Scholar]
- 28.Reiman JM, Kmieciak M, Manjili MH, Knutson KL. Tumor immunoediting and immunosculpting pathways to cancer progression. Semin Cancer Biol. 2007;17:275–287. doi: 10.1016/j.semcancer.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu B, Finn OJ. T-cell death and cancer immune tolerance. Cell Death Differ. 2008;15:70–79. doi: 10.1038/sj.cdd.4402274. [DOI] [PubMed] [Google Scholar]
- 30.Rentzsch C, Kayser S, Stumm S, Watermann I, Walter S, Stevanovic S, Wallwiener D, Guckel B. Evaluation of pre-existent immunity in patients with primary breast cancer: molecular and cellular assays to quantify antigen-specific T lymphocytes in peripheral blood mononuclear cells. Clin Cancer Res. 2003;9:4376–4386. [PubMed] [Google Scholar]
- 31.Beckhove P, Feuerer M, Dolenc M, Schuetz F, Choi C, Sommerfeldt N, Schwendemann J, Ehlert K, Altevogt P, Bastert G, et al. Specifically activated memory T cell subsets from cancer patients recognize and reject xenotransplanted autologous tumors. J Clin Invest. 2004;114:67–76. doi: 10.1172/JCI20278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caballero OL, Chen YT. Cancer/testis (CT) antigens: potential targets for immunotherapy. Cancer Sci. 2009;100:2014–2021. doi: 10.1111/j.1349-7006.2009.01303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vella LA, Yu M, Fuhrmann SR, El-Amine M, Epperson DE, Finn OJ. Healthy individuals have T-cell and antibody responses to the tumor antigen cyclin B1 that when elicited in mice protect from cancer. Proc Natl Acad Sci U S A. 2009;106:14010–14015. doi: 10.1073/pnas.0903225106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goodell V, Salazar LG, Urban N, Drescher CW, Gray H, Swensen RE, McIntosh MW, Disis ML. Antibody immunity to the p53 oncogenic protein is a prognostic indicator in ovarian cancer. J Clin Oncol. 2006;24:762–768. doi: 10.1200/JCO.2005.03.2813. [DOI] [PubMed] [Google Scholar]
- 35.Ishigami S, Natsugoe S, Tokuda K, Nakajo A, Che X, Iwashige H, Aridome K, Hokita S, Aikou T. Prognostic value of intratumoral natural killer cells in gastric carcinoma. Cancer. 2000;88:577–583. [PubMed] [Google Scholar]
- 36.Shafer-Weaver K, Anderson M, Malyguine A, Hurwitz AA. T cell tolerance to tumors and cancer immunotherapy. Adv Exp Med Biol. 2007;601:357–368. doi: 10.1007/978-0-387-72005-0_38. [DOI] [PubMed] [Google Scholar]
- 37.Crawford DH. Biology and disease associations of Epstein-Barr virus. Philos Trans R Soc Lond B Biol Sci. 2001;356:461–473. doi: 10.1098/rstb.2000.0783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paludan C, Schmid D, Landthaler M, Vockerodt M, Kube D, Tuschl T, Munz C. Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science. 2005;307:593–596. doi: 10.1126/science.1104904. [DOI] [PubMed] [Google Scholar]
- 39.Dorfel D, Appel S, Grunebach F, Weck MM, Muller MR, Heine A, Brossart P. Processing and presentation of HLA class I and II epitopes by dendritic cells after transfection with in vitro-transcribed MUC1 RNA. Blood. 2005;105:3199–3205. doi: 10.1182/blood-2004-09-3556. [DOI] [PubMed] [Google Scholar]
- 40. Schmid D, Pypaert M, Munz C. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity. 2007;26:79–92. doi: 10.1016/j.immuni.2006.10.018. This is an elegant paper describing MHC class II presentation of cytosolic antigens as a function of autophagy. The authors also report that direct targeting of a viral protein to autophagosomes increased its effective presentation and recognition by CD4+ T cells.
- 41.Pua HH, Dzhagalov I, Chuck M, Mizushima N, He YW. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J Exp Med. 2007;204:25–31. doi: 10.1084/jem.20061303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nedjic J, Aichinger M, Emmerich J, Mizushima N, Klein L. Autophagy in thymic epithelium shapes the T-cell repertoire and is essential for tolerance. Nature. 2008;455:396–400. doi: 10.1038/nature07208. [DOI] [PubMed] [Google Scholar]
- 43.Watanabe K, Ichinose S, Hayashizaki K, Tsubata T. Induction of autophagy by B cell antigen receptor stimulation and its inhibition by costimulation. Biochem Biophys Res Commun. 2008;374:274–281. doi: 10.1016/j.bbrc.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 44.Sanjuan MA, Dillon CP, Tait SW, Moshiach S, Dorsey F, Connell S, Komatsu M, Tanaka K, Cleveland JL, Withoff S, et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–1257. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- 45.Qu X, Zou Z, Sun Q, Luby-Phelps K, Cheng P, Hogan RN, Gilpin C, Levine B. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 2007;128:931–946. doi: 10.1016/j.cell.2006.12.044. [DOI] [PubMed] [Google Scholar]
- 46.Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pearce EL, Walsh MC, Cejas PJ, Harms GM, Shen H, Wang LS, Jones RG, Choi Y. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature. 2009;460:103–107. doi: 10.1038/nature08097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Araki K, Turner AP, Shaffer VO, Gangappa S, Keller SA, Bachmann MF, Larsen CP, Ahmed R. mTOR regulates memory CD8 T-cell differentiation. Nature. 2009;460:108–112. doi: 10.1038/nature08155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cuervo AM, Bergamini E, Brunk UT, Droge W, Ffrench M, Terman A. Autophagy and aging: the importance of maintaining "clean" cells. Autophagy. 2005;1:131–140. doi: 10.4161/auto.1.3.2017. [DOI] [PubMed] [Google Scholar]
- 50.McCay CM, Crowell MF, Maynard LA. The effect of retarded growth upon length of lifespan and upon ultimate body size. Journal of Nutrition. 1935;10:63–79. [PubMed] [Google Scholar]
- 51.Pinkston JM, Garigan D, Hansen M, Kenyon C. Mutations that increase the life span of C. elegans inhibit tumor growth. Science. 2006;313:971–975. doi: 10.1126/science.1121908. [DOI] [PubMed] [Google Scholar]
- 52.Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, Kenyon C. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet. 2008;4:e24. doi: 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bartke A, Wright JC, Mattison JA, Ingram DK, Miller RA, Roth GS. Extending the lifespan of long-lived mice. Nature. 2001;414:412. doi: 10.1038/35106646. [DOI] [PubMed] [Google Scholar]
- 54.Berrigan D, Perkins SN, Haines DC, Hursting SD. Adult-onset calorie restriction and fasting delay spontaneous tumorigenesis in p53-deficient mice. Carcinogenesis. 2002;23:817–822. doi: 10.1093/carcin/23.5.817. [DOI] [PubMed] [Google Scholar]
- 55. Colman RJ, Anderson RM, Johnson SC, Kastman EK, Kosmatka KJ, Beasley TM, Allison DB, Cruzen C, Simmons HA, Kemnitz JW, et al. Caloric restriction delays disease onset and mortality in rhesus monkeys. Science. 2009;325:201–204. doi: 10.1126/science.1173635. This report summarizes a 20 year longtitudinal study of the effects of CR in a primate model. As well as increasing longevity, CR resulted in a decrease in cancer incidence.
- 56.Alvers AL, Fishwick LK, Wood MS, Hu D, Chung HS, Dunn WA, Jr, Aris JP. Autophagy and amino acid homeostasis are required for chronological longevity in Saccharomyces cerevisiae. Aging Cell. 2009;8:353–369. doi: 10.1111/j.1474-9726.2009.00469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hars ES, Qi H, Ryazanov AG, Jin S, Cai L, Hu C, Liu LF. Autophagy regulates ageing in C. elegans. Autophagy. 2007;3:93–95. doi: 10.4161/auto.3636. [DOI] [PubMed] [Google Scholar]
- 58.Juhasz G, Erdi B, Sass M, Neufeld TP. Atg7-dependent autophagy promotes neuronal health, stress tolerance, and longevity but is dispensable for metamorphosis in Drosophila. Genes Dev. 2007;21:3061–3066. doi: 10.1101/gad.1600707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Colotta F, Allavena P, Sica A, Garlanda C, Mantovani A. Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability. Carcinogenesis. 2009;30:1073–1081. doi: 10.1093/carcin/bgp127. [DOI] [PubMed] [Google Scholar]
- 60.Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, de Cabo R, Sinclair DA. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 61.Nemoto S, Fergusson MM, Finkel T. Nutrient availability regulates SIRT1 through a forkhead-dependent pathway. Science. 2004;306:2105–2108. doi: 10.1126/science.1101731. [DOI] [PubMed] [Google Scholar]
- 62.Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, Tsokos M, Alt FW, Finkel T. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci U S A. 2008;105:3374–3379. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee IH, Finkel T. Regulation of autophagy by the p300 acetyltransferase. J Biol Chem. 2009;284:6322–6328. doi: 10.1074/jbc.M807135200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pan MH, Ho CT. Chemopreventive effects of natural dietary compounds on cancer development. Chem Soc Rev. 2008;37:2558–2574. doi: 10.1039/b801558a. [DOI] [PubMed] [Google Scholar]
- 65.Davis CD. Nutritional Interactions: Credentialing of moleular targets for cancer prevention. Soc Exp Biol Med. 2007;232:176–183. [PubMed] [Google Scholar]
- 66.Hannigan AM, Gorski SM. Macroautophagy: the key ingredient to a healthy diet? Autophagy. 2009;5:140–151. doi: 10.4161/auto.5.2.7529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Singletary K, Milner J. Diet, autophagy, and cancer: a review. Cancer Epidemiol Biomarkers Prev. 2008;17:1596–1610. doi: 10.1158/1055-9965.EPI-07-2917. This review provides an in-depth and comprehensive examination of the effects of diet on autophagy and its role in cancer.
- 68.White E, DiPaola RS. The double-edged sword of autophagy modulation in cancer. Clin Cancer Res. 2009;15:5308–5316. doi: 10.1158/1078-0432.CCR-07-5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Park MA, Zhang G, Martin AP, Hamed H, Mitchell C, Hylemon PB, Graf M, Rahmani M, Ryan K, Liu X, et al. Vorinostat and sorafenib increase ER stress, autophagy and apoptosis via ceramide-dependent CD95 and PERK activation. Cancer Biol Ther. 2008;7:1648–1662. doi: 10.4161/cbt.7.10.6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. This report is one of the first demonstrations that targeting the mTOR pathway can increase longevity in mammals. Though not directly attributed to autophagy, rapamycin is a well known activator of autophagy.
- 71. Maclean KH, Dorsey FC, Cleveland JL, Kastan MB. Targeting lysosomal degradation induces p53-dependent cell death and prevents cancer in mouse models of lymphomagenesis. J Clin Invest. 2008;118:79–88. doi: 10.1172/JCI33700. This is the first report that chloroquine-mediated modulation of the autophagolysosomal system delays tumor development in two genetically distinct mouse models of cancer. It is one of few reports which document a chemopreventative effect.
- 72.Geser A, Brubaker G, Draper CC. Effect of a malaria suppression program on the incidence of African Burkitt's lymphoma. Am J Epidemiol. 1989;129:740–752. doi: 10.1093/oxfordjournals.aje.a115189. [DOI] [PubMed] [Google Scholar]
- 73.Evans JM, Donnelly LA, Emslie-Smith AM, Alessi DR, Morris AD. Metformin and reduced risk of cancer in diabetic patients. Bmj. 2005;330:1304–1305. doi: 10.1136/bmj.38415.708634.F7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bowker SL, Majumdar SR, Veugelers P, Johnson JA. Increased cancer-related mortality for patients with type 2 diabetes who use sulfonylureas or insulin. Diabetes Care. 2006;29:254–258. doi: 10.2337/diacare.29.02.06.dc05-1558. [DOI] [PubMed] [Google Scholar]
- 75.Landman GW, Kleefstra N, van Hateren KJ, Groenier KH, Gans RO, Bilo HJ. Metformin associated with lower cancer mortality in type 2 diabetes (ZODIAC-16) Diabetes Care. 2009 doi: 10.2337/dc09-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stumvoll M, Nurjhan N, Perriello G, Dailey G, Gerich JE. Metabolic effects of metformin in non-insulin-dependent diabetes mellitus. N Engl J Med. 1995;333:550–554. doi: 10.1056/NEJM199508313330903. [DOI] [PubMed] [Google Scholar]
- 77.Buzzai M, Jones RG, Amaravadi RK, Lum JJ, DeBerardinis RJ, Zhao F, Viollet B, Thompson CB. Systemic treatment with the antidiabetic drug metformin selectively impairs p53-deficient tumor cell growth. Cancer Res. 2007;67:6745–6752. doi: 10.1158/0008-5472.CAN-06-4447. [DOI] [PubMed] [Google Scholar]