Abstract
The mammalian ortholog of yeast Atg6/Vps30, Beclin 1, is an essential autophagy protein that has been linked to diverse biological processes, including immunity, development, tumor suppression, lifespan extension, and protection against certain cardiac and neurodegenerative diseases. In recent years, major advances have been made in identifying components of functionally distinct Beclin 1/class III phosphatidylinositol 3-kinase complexes, in characterizing the molecular regulation of interactions between Beclin 1 and the autophagy inhibitors, Bcl-2/BcL-XL, and in uncovering a role for viral antagonists of Beclin 1 in viral pathogenesis. The rapidly growing list of components of the ‘Beclin 1 interactome’ supports a model in which autophagy, and potentially other membrane trafficking functions of Beclin 1, are governed by differential interactions with different binding partners in different physiological or pathophysiological contexts.
Beclin 1: a conserved autophagy protein
Autophagy is a lysosomal degradation pathway that functions in a variety of stress conditions, in which long-lived or aggregated proteins, damaged organelles and pathogens are transported in double-membraned autophagosomes to lysosomes for destruction. So far, genetic screens have identified approximately 32 autophagy-related genes (known as ‘ATG’ genes) in the yeast S. cerevisiae, of which approximately 18 are essential for autophagy, and many of these are found in mammals and other higher eukaryotes [1]. Human Beclin 1 (coiled-coil, myosin-like BCL2-interacting protein) shares 24% sequence homology with yeast Atg6/Vps30 and restores autophagic activities in atg6 null yeast mutants, demonstrating that it is a functional homolog of Atg6/Vps30 [2••]. Like yeast Atg6/Vps30, mammalian Beclin 1 interacts with the class III phosphatidylinositol 3-kinase (PI3K), Vps34, and is involved in autophagic vesicle nucleation [3••]. Gene knockout/knockdown studies indicate a conserved requirement for ATG6/beclin 1 in autophagy in plants, slime molds, nematodes, fruit flies, mice, and human cells [4]. Decreases in Beclin 1 expression and/or functional activity have been linked to increased susceptibility to cancer, Alzheimer’s disease, Huntington’s disease, and desmin-related cardiomyopathy; alterations in microbial pathogenesis; defects in apoptotic corpse clearance and development; and aging [5]. An open question is whether these phenotypes are a direct consequence of deficient autophagy, or as-of-yet unidentified alternate functions of Beclin 1.
Beclin 1/class III PI3K complexes
The yeast ortholog of Beclin 1, Atg6/Vps30, was independently discovered in two different genetic screens, including one for proteins required for autophagy and one for proteins required for vacuolar protein sorting, a pathway that sorts hydrolases from the trans-Golgi network (TGN) to the yeast vacuole. Subsequently, two distinct Atg6/Vps30–class III PI3K complexes were described in yeast [6]. Atg6/Vps30, the class III PI3K Vps34, and the regulatory myristoylated kinase Vps15 are common elements of each complex, but Atg14 is uniquely present in the complex involved in autophagy and Vps38 is uniquely present in the complex involved in vacuolar protein sorting. Since the discovery of mammalian Beclin 1, an important question has been whether, Beclin 1, like yeast Atg6/Vps30, functions in distinct class III PI3K complexes that mediate different membrane trafficking events.
Three lines of evidence in early studies suggested that Beclin 1 may function specifically in autophagy, and not in vacuolar protein sorting. First, Beclin 1 rescued autophagy, but not vacuolar protein sorting, in atg6/vps30 null yeast [2••]. Second, the proteolytic processing of cathepsin D, which requires intact Vps34-dependent vacuolar protein sorting function, was found to be normal in autophagy-deficient, low Beclin 1-expressing mammalian cells [7]. Third, siRNA-mediated silencing of human Beclin 1 suppressed autophagy, but not other PI3K-dependent trafficking pathways such as the post-endocytic sorting of the epidermal growth factor (EGF) receptor or cathepsin D maturation [8]. Despite these negative data, suspicions remained that Beclin 1 may function in other membrane trafficking events given (1) the role of yeast Atg6/Vps30 in both autophagy and vacuolar protein sorting and (2) the different phenotype of beclin 1 null mouse embryos (which are early embryonically lethal) versus atg5 or atg7 null mouse embryos (which die during the early neonatal period) [5].
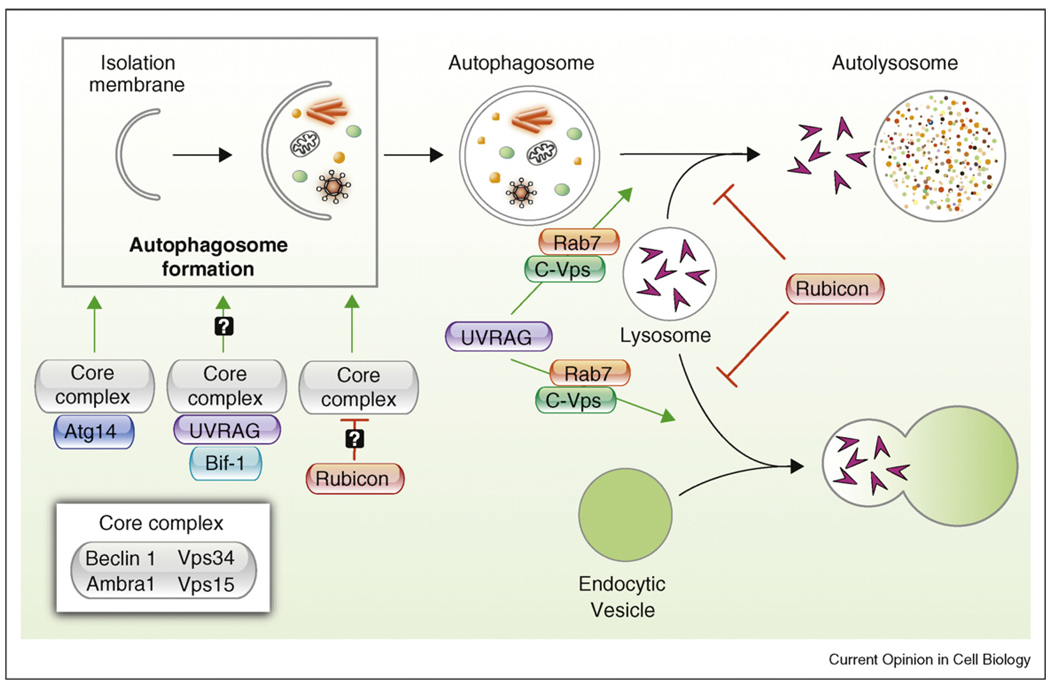
Recent findings provide strong biochemical evidence that mammalian Beclin 1 exists in distinct class III PI3K complexes. Like in yeast, each complex seems to consist of Beclin 1, Vps34, and Vps15 [9•,10•,11•], as well as possibly, a mammalian specific Beclin 1-interacting WD40 domain protein, Ambra1 [12••] (and personal communication, Francesco Cecconi); for purposes of this review, we designate Beclin 1, Vps34, Vps15, and Ambra1 as the core complex (Figure 1). Within the past year, four independent laboratories isolated Beclin 1-binding proteins that are part of biochemically distinct Beclin 1/class III PI3K core complexes and postulated to have distinct functions in membrane trafficking events, including human Atg14 (also known as Atg14L (Atg14-like protein) or Barkor (Beclin 1-associated autophagy-related key regulator)), UVRAG, and Rubicon [9•,10•,11•,13] (Figure 1).
Figure 1.
Function of proteins that interact with Beclin 1/class III PI3K complexes in different steps of autophagy. Beclin 1, Vps34, Vps15, and possibly Ambra 1, compose a class III PI3K core complex that binds either Atg14 or UVRAG. Atg14 activates the core complex and biogenesis of autophagosomes; however, there are conflicting reports as to whether UVRAG/Bif-1 facilitates the core complex and upregulates autophagosome formation. A better studied function of UVRAG is promoting the maturation of autophagosomes and endocytic vesicles by recruiting class C Vps and the Rab7 GTPase and inducing vesicle fusion. As an important negative regulator of autophagy, Rubicon inhibits multiple steps in autophagy, including the formation of autophagosomes and the fusion between lysosomes and autophagosomes/endocytic vesicles. However, it is not yet known whether Rubicon inhibits autophagosome formation directly through an inhibitory interaction with the core complex or whether Beclin 1 is involved in the inhibitory effects of Rubicon on autophagosome/endosome maturation. Green arrows, stimulatory action; red bars, inhibitory action.
These findings underscore certain common themes as well as unanswered questions regarding Beclin 1/class III PI3K complexes. One unequivocal finding is that human Atg14, which only has limited structural homology with yeast Atg14 (18% identity, 32% similarity), has functional parallels to yeast Atg14 [9•,10•,11•,13]. Human Atg14 is reported to localize to isolation membranes [9•,10•] as well as to the endoplasmic reticulum [9•] (which may be a source of autophagosomal membranes) and genetic deletion or siRNA silencing of Atg14 decreases Vps34 lipid kinase activity, autophagosome formation, and autophagic flux [9•,10•,11•,13]. The independent discovery of a similar function for human Atg14 in autophagosome formation by four independent laboratories provides strong evidence that it is a functional homolog of yeast Atg14 in a Beclin 1/class III PI3K autophagy-inducing complex (Figure 1).
The interrelationship between UVRAG (UV irradiation resistance-associated gene), Beclin 1/class III PI3K complexes, and different steps of autophagy is less clear. Three recent studies indicate that UVRAG and Atg14 are present in mutually exclusive Beclin 1/class III PI3K complexes [9•,10•,11•], and Itakura et al. provide evidence to support the hypothesis that UVRAG (which shares weak homology with yeast Vps38) is not involved in autophagosome formation [10•]. They showed that, at least in HeLa cells, UVRAG was primarily associated with Rab9-positive endosomes and UVRAG silencing did not suppress autophagosome formation or flux. It is not yet clear how to reconcile these data, and the biochemical evidence that Atg14 (which is required for autophagosome formation) and UVRAG exist in distinct biochemical complexes [9•,10•,11•], with previous reports that UVRAG and Bif-1/endophilin B1 are involved in the activation of Beclin 1/Vps34 complexes and autophagosome formation [14,15]. Perhaps cell type-specific differences exist in the composition and/or function of Beclin 1/UVRAG-containing complexes and in some contexts, UVRAG can function like Atg14 in autophagosome formation.
In addition to autophagosome formation, UVRAG has also been reported to function in autophagosome maturation and endocytic trafficking [16•] (Figure 1). The interaction between UVRAG and the class C Vps complex, which is known to be involved in endosomal fusion, triggers Rab7 GTPase activity and promotes the fusion of autophagosomes with late endosomes/lysosomes. UVRAG also accelerates endocytic trafficking of EGF to lysosomes and EGF-induced lysosomal degradation of EGF receptors. Of note, the function of UVRAG in autophagosome maturation does not require the Beclin 1-interacting coiled-coil domain of UVRAG, suggesting that it may be genetically separable from its interaction with Beclin 1. However, similar analyses have not been performed to evaluate whether UVRAG-dependent acceleration of the trafficking of endocytic cargo to late endosomes is also Beclin 1-independent. Thus, at present, a role for Beclin 1/class III PI3K complexes in UVRAG-dependent autophagosome maturation or endocytic trafficking is not established.
In contrast to Atg14, UVRAG and Bif-1, which are reported to be either positive regulators of autophagosome formation (e.g. Atg14 and more controversially, UVRAG and Bif-1), autophagosome maturation (e.g. UVRAG), or endosome maturation (e.g. UVRAG), another newly identified component of Beclin 1/class III PI3K complexes, Rubicon (a RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein), probably plays an important inhibitory role in some (or all) of these membrane trafficking events [9•,13]. Rubicon localizes to endosomes and lysosomes and its knockdown leads to an increase in numbers of autophagosomes and autolysosomes and an acceleration of the lysosomal degradation of endocytosed EGF receptor, demonstrating an inhibitory effect on autophagosome formation, autophagosomal maturation, and endosomal maturation. Although it is clearly established that there is a distinct Rubicon-containing Beclin 1/class III PI3K-containing complex that contains UVRAG (as well as a Rubicon-independent UVRAG Beclin 1/class III PI3K complex) [9•], it is not yet known whether Rubicon exerts its inhibitory effects on autophagosome maturation and endocytic trafficking through its interaction with the Beclin 1/class III PI3K core complex, or through Beclin 1-independent mechanisms. The ability of overexpressed Rubicon to decrease Vps34 kinase activity [13] raises the possibility that Rubicon may indeed interfere with autophagosome formation through inhibition of the Beclin 1/class III PI3K core complex. Similarly, Matsunaga et al. postulated that the Beclin 1/UVRAG/Rubicon complex may work in an opposing manner to the Beclin 1/UVRAG complex in the regulation of autophagosome maturation and endocytosis [9•]. However, as noted above, it is not yet clear that Beclin 1 is involved in UVRAG-dependent autophagosome or endocytic maturation.
Further studies are urgently needed to sort out the precise functions of Beclin 1-interacting partners, especially as they relate to the regulation of Beclin 1/class III PI3K complexes in mediating different membrane trafficking processes. Presently, we know that distinct Beclin 1/PI3K complexes exist; that Atg14 and other components of the Beclin 1/PI3K core complex (e.g. Beclin 1, Vps34, Ambra1) are involved in autophagosome formation; that UVRAG, a Beclin 1-interacting protein, may function in autophagosomal maturation, endosomal maturation, and autophagosome formation (at least in certain contexts); and that Rubicon, another Beclin 1-interacting protein, negatively regulates these three processes. However, aside from the Atg14/core complex that functions in autophagosome formation, the roles of Beclin 1/class III PI3K complexes in other membrane trafficking events (such as autophagosome maturation and endosome maturation) remain undefined, and it is not yet known whether UVRAG and Rubicon regulate these processes through their interactions with the Beclin 1/class III PI3K complex or through other mechanisms. While Atg14 and UVRAG seem to interact with Beclin 1 in a mutually exclusive manner through their coiled-coil domains, it is unclear which interactions in the core complex and which interactions between members of the core complex and other Beclin 1-binding partners are direct or indirect. Another crucial unanswered question is whether Beclin 1-binding partners alter the composition, subcellular localization, and/or function of the Beclin 1/class III PI3K complexes. Along these lines, there is growing research indicating that Atg14 and Ambra1 may help localize Beclin 1 to autophagosomes or the endoplasmic reticulum, which in the case of Ambra1 may involve interactions with the dynein motor complex regulated by autophagy-inducing stimuli [13] (and personal communication, Francesco Cecconi). These findings probably represent ‘the tip of the iceberg’ in understanding how the biochemical identification of distinct Beclin 1/class III PI3K complexes relates to the cellular processes of autophagosome formation, autophagosome maturation, and endocytic trafficking.
At a physiological level, it is noteworthy that Beclin 1, and three of its interacting partners, Ambra1, UVRAG, and Bif-1, play a role in negative growth control and/or tumor suppression. Loss of Ambra1 results in uncontrolled neural cell proliferation during embryogenesis [12••] and heterozygous deletion of Ambra1 results in a spectrum of spontaneous tumors (personal communication, Francesco Cecconi) similar to those observed in mice with monoallelic loss of beclin 1 [17,18]. UVRAG is frequently monoallelically deleted in human colon cancer, and its overexpression suppresses human colon cancer cell proliferation and tumorigenicity in nude mice [14]. Similarly, Bif-1 knockout mice display an increased incidence of spontaneous tumors [15]. Thus, while there are conflicting in vitro data regarding the role of UVRAG in autophagy (and by extension Bif-1, which functions through its interaction with UVRAG), it is tempting to speculate that the common roles of these different Beclin 1-interacting proteins in suppressing tumorigenesis may indicate common underlying functions in cell biology. The determination of whether Atg14, a Beclin 1-interacting partner that acts specifically in autophagosome formation, also has tumor suppressor function will be crucial to dissect whether the common underlying cell biology function is autophagy or other Beclin 1/class III PI3K complex-dependent processes.
Beclin 1–Bcl-2/Bcl-XL interactions
Beclin 1 was initially identified as a Bcl-2-interacting protein in a yeast two-hybrid screen [19], and subsequently, it was shown that Bcl-2 and Bcl-XL function as anti-autophagy proteins through their interactions with Beclin 1 [20••]. Structural and biochemical studies indicate that Beclin 1 contains a BH3 (Bcl-2-homology-3) domain that is necessary and sufficient to bind cellular and virally-encoded, anti-apoptotic Bcl-2 family members, such as Bcl-2, Bcl-XL, and murine γ-herpesvirus M11 [21,22,23•,24••,25]. Unlike other known BH3-only proteins, Beclin 1 does not function as a pro-apoptotic molecule [23•,26], suggesting that the BH3 domain may be a common structural motif by which Bcl-2 family members recognize and dually regulate both apoptosis and autophagy molecules rather than an invariant signature of a death-inducing protein [27]. Bcl-2 decreases Beclin 1 interactions with Vps34 and Beclin 1-associated Vps34 kinase activity [20••] and may therefore sequester Beclin 1 away from the autophagy-inducing class III PI3K ‘core complex’. Endoplasmic reticulum-localized Bcl-2, but not mitochondrial-localized Bcl-2, inhibits autophagy [20••], which is consistent with the growing evidence that ER-associated class III PI3K activity may be crucial in the initiation of autophagosome membrane formation. It has also been proposed that Bcl-XL inhibits Beclin 1 activity by stabilizing Beclin 1 homo-dimerization, which can be disrupted by UVRAG [28]. Further research is required to more fully elucidate the precise mechanisms by which Bcl-2/Bcl-XL inhibit autophagy.
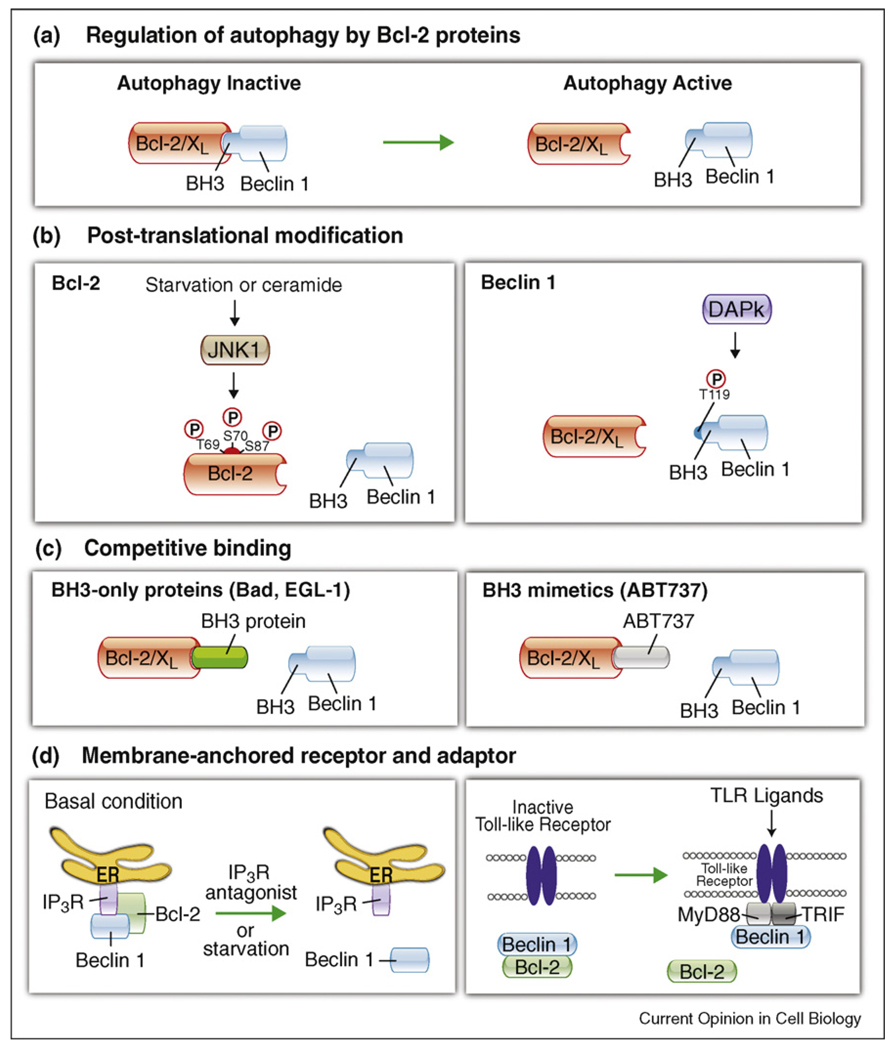
In the past few years, there has been increasing evidence that the regulation of Bcl-2/Bcl-XL interactions with Beclin 1 represents a central mechanism by which autophagy is turned on or off in response to diverse cellular stimuli [29]. There have also been numerous advances made in understanding the molecular mechanisms that govern such interactions: post-translational modifications of either Bcl-2 or Beclin 1 can alter Bcl-2/Bcl-XL–Beclin 1 interactions; BH3-only peptides or proteins can competitively disrupt Bcl-2/Bcl-XL–Beclin 1 interactions; and different membrane-anchored receptors or their adaptor proteins may also be able to regulate the interaction between Bcl-2 and Beclin 1 (Figure 2). Together, these and other regulatory mechanisms that have yet to be identified probably fine tune autophagy activities, at least partly, through the regulation of Bcl-2/Bcl-XL–Beclin 1 interactions. The disruption of Bcl-2/Bcl-XL–Beclin 1 interactions may be a mechanism that cells use to promote autophagy and cell survival during stress conditions, such as nutrient deprivation, hypoxia, or detachment from the extracellular matrix [30,31].
Figure 2.
Mechanisms underlying the regulation of Bcl-2/Bcl-XL interactions with Beclin 1. (a) When Bcl-2/Bcl-XL are bound to Beclin 1, they inhibit autophagy. The disruption of Bcl-2/Bcl-XL–Beclin 1 interactions is essential for autophagy activation. Mechanisms by which this occurs include post-translational modifications of either Bcl-2/Bcl-XL or Beclin 1 (b); competitive disruption of Bcl-2/Bcl-XL binding to Beclin 1 by BH3-only proteins (e.g. Bad, EGL-1) or BH3 mimetics (e.g. ABT737) (c), or potentially, effects of membrane-anchored receptors or their adaptors on Bcl-2/Beclin 1 interactions (d). In (b), starvation induces JNK1-dependent phosphorylation of residues T69, S70, and S87 of Bcl-2, resulting in the disruption of Bcl-2 from Beclin 1. The death-associated kinase, DAPk, phosphorylates residue T119 in the BH3 domain of Beclin 1, leading to the disruption of Bcl-2 from Beclin 1. In (d), Beclin 1 is released from ER-localized inositol-1,4,5-trisphosphate receptor (IP3R) and Bcl-2 after antagonist binding or during starvation, leading to activation of autophagy. The activation of Toll-like receptors recruits the adaptor proteins MyD88 and TRIF, which interact with Beclin 1 and decrease the inhibitory interaction of Bcl-2 with Beclin 1. Green arrows, autophagy induction; red bars, autophagy inhibition.
The most potent known physiological inducer of autophagy is starvation, and Pattingre et al. originally showed that nutrient status regulates the interaction of endogenous Bcl-2 and Beclin 1 [20••]. During nutrient rich conditions when autophagy is suppressed, Bcl-2 binding to Beclin 1 is maximal, whereas during nutrient deprivation when autophagy is stimulated, Bcl-2 binding to Beclin 1 is minimal. Wei et al. discovered that the underlying mechanism by which nutrient starvation disrupts the Bcl-2/Beclin 1 complex, leading to autophagy stimulation, is through JNK1-mediated multisite phosphorylation of residues Thr69, Ser70, and Ser97 of the non-structured loop of Bcl-2 [32•]. A similar mechanism has also been shown to be involved in ceramide-induced autophagy [33]. This mechanism of autophagy induction is likely to be physiologically relevant, as starvation fails to induce disruption of the Bcl-2/Beclin 1 complex and autophagy in MEFs that either lack JNK1 or that contain non-phosphorylatable knock-in mutations in Bcl-2 Thr69, Ser70, and Ser97 [32•].
The disruption of Bcl-2/Bcl-XL–Beclin 1 interactions (and subsequent autophagy activation) by phosphorylation is bidirectional. Not only does Bcl-2 phosphorylation disrupt Bcl-2/Beclin 1 interactions, the phosphorylation of Thr119 in the BH3 domain of Beclin 1 by enforced expression of the death-inducing kinase, DAP-kinase (DAPk), also promotes the dissociation of Beclin 1 from Bcl-XL (and Bcl-2) and induction of autophagy [34,35•]. It is not yet known whether endogenous DAPk is required for disruption of the Bcl-XL (or Bcl-2)-Beclin 1 complex, and if so, under which settings. It is also unknown whether this activity of DAPk contributes to its role in cell death and tumor suppression. For both Bcl-2 and Beclin 1 phosphorylation-mediated regulation of the Bcl-2/Bcl-XL–Beclin 1 complex, it is unknown whether adding a phosphate group sterically blocks the binding sites or induces conformational changes in Beclin 1 and Bcl-2/Bcl-XL proteins that prevent their interactions with each other. Another unanswered question is whether phosphorylation events on Bcl-2 proteins and Beclin 1 occur synergistically or independently under autophagy-inducing conditions.
A third mechanism for regulating the interaction between Bcl-2/Bcl-XL and activating autophagy involves competitive disruption by either BH3-only proteins (e.g. Bad in mammalian cells and EGL-1 in C. elegans) or pharmacological BH3 peptidomimetic agents (e.g. ABT737) [23•] (Figure 2) or, by a less-defined mechanism, the binding of the ARF tumor suppressor to mitochondrial Bcl-XL [36]. Bcl-2 and Bcl-XL bind to Beclin 1 with a relatively low affinity; therefore, this interaction can be readily disrupted by BH3 proteins or peptides that have higher affinity interactions with Bcl-2/Bcl-XL. This mechanism is likely to be physiologically relevant, since the knockdown of Bad or deletion of EGL-1 impairs starvation-induced autophagy in mammalian cells and C. elegans, respectively [23•]. However, virtually nothing is known about the relative contributions of BH3-only proteins versus post-translational modifications of Bcl-2/Bcl-XL and Beclin 1 in the regulation of Bcl-2/Bcl-XL–Beclin 1 interactions and autophagy. Another important question is to what extent BH3 mimetics exert their anti-cancer effects through autophagy induction (in addition to their established role in apoptosis induction [37–40]) and whether more selective BH3 mimetics can be designed that inhibit the anti-autophagy activity of Bcl-2 family members without inhibiting the anti-apoptosis activity of Bcl-2 family members. Such reagents could be useful in the treatment of conditions other than cancer where selective autophagy activation may be beneficial, such as infectious diseases, neurodegenerative diseases, and aging.
A newly emerging area of research relates to the effects of different membrane-anchored proteins or their adaptor proteins on Bcl-2/Bcl-XL–Beclin 1 interactions. For example, the endoplasmic reticulum-localized inositol-1,4,5-triphosphate receptor (IP3R), which is a major regulator of apoptotic signaling, may also regulate autophagy through its effects on Bcl-2/Beclin 1 interactions [41]. Treatment with the IP3R antagonist or physiological induction of autophagy by nutrient starvation disrupts the interaction between IP3R and Beclin 1, which is an interaction that is abolished by Bcl-2 knockdown or enhanced by Bcl-2 overexpression. Another study suggested that Toll-like receptor (TLR) signaling may indirectly regulate Bcl-2/Beclin 1 interactions and thereby, induce autophagy; the TLR adaptor proteins, MyD88 and Trif interact with Beclin 1, which results in the reduced binding of Beclin 1 to Bcl-2 [42]. However, for both the IP3R antagonist and MyD88/Trif-dependent induction of autophagy, further studies are required to determine whether disruption of the Beclin 1/Bcl-2 interaction is mechanistically involved in autophagy induction, and if so, how the IP3R and TLR adaptors regulate the interaction.
Although not yet related to Bcl-2/Bcl-XL–Beclin 1 interactions, it is worth noting in this section that other membrane-anchored and nuclear receptors are also part of the ‘Beclin 1 interactome’. This includes the membrane-anchored delta 2 glutamate receptor that is aberrantly activated in mice with cerebellar degeneration and linked to Beclin 1 through an interaction with a PDZ domain-containing protein, nPIST [43], the pathogen receptor CD46 that is linked to Beclin 1 through an isoform of nPIST, GOPC (also known as PIST) [44], and the estrogen receptor, ERα [45]. It will be interesting to explore in future studies whether these receptors regulate autophagy either through altering Beclin 1/class III PI3K complexes and/or through altering Bcl-2/Bcl-XL–Beclin 1 interactions.
The Beclin 1 viral protein interactome
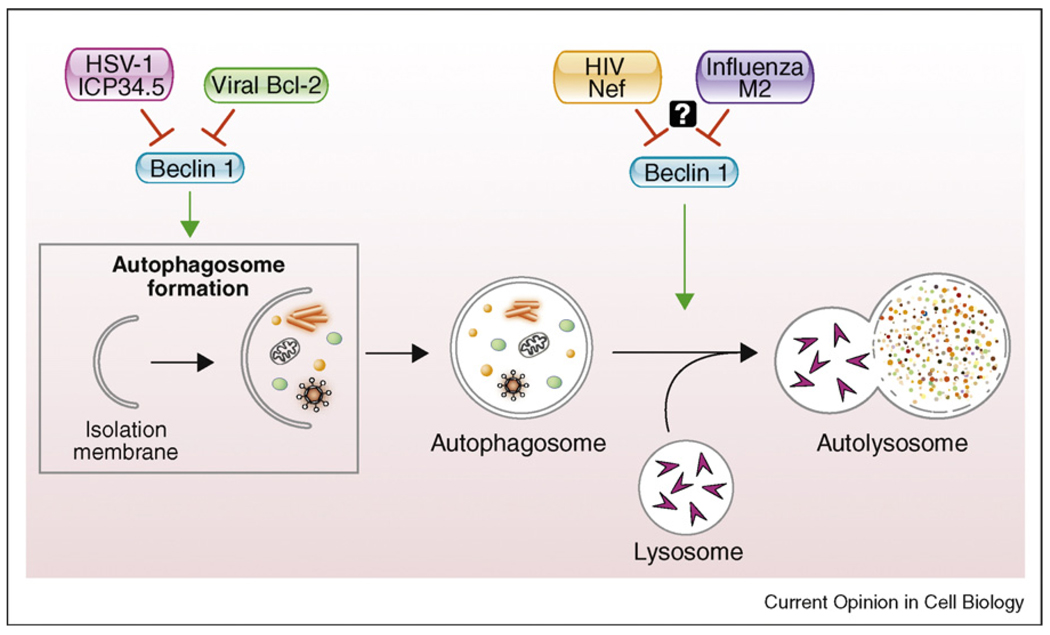
Perhaps the best evidence that Beclin 1 is an important autophagy protein stems from the diversity of the expanding known components of its ‘viral protein interactome’. Autophagy plays a crucial role in antiviral host defense and functions to limit viral replication, activate Type I interferon signaling, and enhance MHC class II presentation of endogenously synthesized viral antigens [46,47]. Consequently, there is probably a strong selection pressure for viruses to evolve mechanisms to counteract host autophagy; the most common molecular strategy identified thus far by which viruses accomplish this goal is through encoding virulence proteins that bind to Beclin 1 and inhibit its autophagy function. The list of viral proteins that interact with Beclin 1 and modulate its autophagy activity includes viral Bcl-2-like proteins encoded by the oncogenic γ-herpesviruses (e.g. KSHV vBcl-2, murine γHV68 M11) [22,24••,25,32•,48]; a neurovirulence protein, ICP34.5 encoded by the α-herpesvirus, HSV-1 [49•,50]; a pathogenic factor, Nef, encoded by HIV-1 [51•]; and possibly the M2 protein of influenza virus [52]. The herpesvirus-encoded Beclin 1 antagonists, including the viral Bcl-2 molecules and HSV-1 ICP34.5, inhibit autophagosome formation, whereas HIV-1 Nef and influenza M2 inhibit autophagosome maturation (Figure 3).
Figure 3.
The Beclin 1 viral protein interactome and their presumed sites of anti-autophagy action. HSV-1 ICP34.5 and γ-herpesvirus-encoded viral Bcl-2 molecules inhibit autophagosome formation through their interactions with Beclin 1, whereas HIV-1 Nef and influenza M2 block the maturation step of autophagy. The anti-autophagic maturation activity of HIV-1 Nef is thought to be mediated by interactions with Beclin 1 (see text), whereas it is not yet known whether the anti-autophagic maturation activity of influenza M2 is mediated by its interaction with Beclin 1.
For both HSV-1 and murine γHV68, there is strong in vitro evidence that viral proteins inhibit autophagosome formation through their interaction with Beclin 1 and strong in vivo evidence that viral inhibition of Beclin 1 is important in viral pathogenesis. With respect to HSV-1, Orvedahl et al. originally showed that a mutant virus containing a small deletion in ICP34.5 that blocks its ability to interact with Beclin 1 is highly attenuated following intracerebral inoculation in mice, with significantly reduced CNS viral replication and animal lethality [49•]. In addition, a more recent study demonstrated that this mutant virus stimulates a significantly stronger CD4 T cell response and is more rapidly cleared in a corneal infection model [50]. With respect to murine γHV68, a virus that contains a deletion mutation in the α1 helix of the viral Bcl-2, which blocks its interaction with Beclin 1 and anti-autophagy activity but not its interaction with pro-apoptotic BH3 proteins or its anti-apoptotic activity, is deficient in the maintenance of viral latent infection [48].
The precise mechanisms by which herpesvirus-encoded viral proteins inhibit autophagosome formation are unknown. On the basis of crystallographic and NMR studies of the viral Bcl-2 bound to the BH3 domain of Beclin 1, the analysis of binding affinities, and mutational analyses [27], the structural determinants of viral Bcl-2/Beclin 1 interactions generally resemble those observed for cellular Bcl-2/Bcl-XL interactions with Beclin 1. However, of note, it appears that viral Bcl-2s may have evolved mechanisms to be more successful than their cellular counterparts in inhibiting autophagy. The γHV68-encoded Bcl-2 molecule, M11, binds to Beclin 1 with significantly higher affinity than do cellular Bcl-2 or Bcl-XL [22,25]. In addition, KSHV vBcl-2, unlike cellular Bcl-2, lacks potential JNK1 target phosphorylation sites, escapes physiological regulation of binding to Beclin 1, and constitutively inhibits autophagy [32•]. Given the importance of herpesvirus antagonism of Beclin 1 function in viral pathogenesis in mouse models, pharmacological agents that can disrupt HSV-1 ICP34.5/Beclin 1 or viral Bcl-2/Beclin 1 interactions may be beneficial in treating HSV-1 and KSHV infections in patients.
Recent studies also provide strong evidence that HIV-1 Nef and the influenza M2 proteins interact with Beclin 1 and block autophagosome maturation [51•,52]. Yet, it is not yet definitively known whether the mechanism by which these viral proteins inhibit autophagosome maturation is through their interactions with Beclin 1. Kyei et al. propose that this is the case for HIV-1 Nef, since they observed a correlation between the effects of mutations in HIV-1 Nef on Beclin 1-binding activity and the ability of HIV-1 Nef to block autophagosome maturation [51•]. However, the 174DD175 → 174AA175 mutation in the diacidic motif that blocks Nef’s interaction with Beclin 1 and Nef’s inhibition of autophagosome maturation also blocks its interaction with the V1 domain of vacuolar H+ ATPase. Thus, an interesting question is whether the interaction with the vacuolar H+ ATPase may also contribute to Nef’s ability to block autophagosome maturation. With respect to influenza, Gannage et al. demonstrate that the viral M2 protein, including a fragment containing the N-terminal 60 amino acids that are sufficient to block autophagosome fusion with lysosomes, also interacts with Beclin 1 [52]. They speculate that the N-terminal 60 amino acids of M2, independent of its proton channel function (which requires the C-terminal membrane spanning residues), may block autolysosome formation through interfering with Beclin 1 and UVRAG-containing PI3K complexes. However, further studies are required to test this hypothesis.
If HIV-1 Nef and influenza virus M2 inhibit autolysosome formation through their interaction with Beclin 1, this will be an important indication that Beclin 1 functions not only in autophagosome formation, but also in autophagosome maturation. Moreover, such a finding will underscore the crucial importance of viral antagonism of multiple stages of autophagy. Perhaps, a dual role of Beclin 1 in both autophagosome formation and autophagosome maturation renders it a prime target for viral inhibitors of autophagy, that is, the virus can ‘kill two birds with one stone’.
Conclusion
In the past few years, the repertoire of the Beclin 1 interactome has vastly expanded, with the identification of new components of Beclin 1/PI3K complexes, the identification of new mechanisms for regulating Bcl-2/Bcl-XL–Beclin 1 interactions, and the identification of new viral inhibitors of Beclin 1, and many of these interactions have been shown between endogenous proteins (Table 1). Together, these findings suggest that Beclin 1 may not only function in autophagosome formation, but also in autophagosome/endosome maturation, and that temporally-modulated or spatially-modulated interactions between Beclin 1 and its positive regulators and negative regulators may govern these activities. Within the next few years, there will probably be a dramatic further expansion of known proteins in the Beclin 1 interactome, as a result of ongoing genetic and proteomic screens in diverse laboratories, as well as enhanced efforts to detect proteins that form unstable or transient complexes with Beclin 1 under specific conditions. Dissecting the Beclin 1 interactome in further detail will help elucidate the mechanisms by which complex regulatory networks integrate diverse environmental cues with this core component of the autophagy machinery to fine tune autophagy levels, coordinate distinct steps in autophagy, and maintain cellular homeostasis.
Table 1.
Summary of studies that have confirmed endogenous interactions in mammalian cells between Beclin 1 and the indicated Beclin 1-binding partners
| Beclin 1-binding partner | References |
|---|---|
| A. Class III PI3K complex components | |
| Autophagy inducers: | |
| Vps34 | [3••,9•,10•] |
| Vps15 | [9•] |
| Ambra1 | [12••] |
| Atg14 | [9•,10•,11•] |
| UVRAG | [9•,10•,14] |
| Bif | [15] |
| Autophagy inhibitors: | |
| Rubicon | [9•] |
| B. Bcl-2 family members | |
| Bcl-2 | [20••,23•,32•,33] |
| Bcl-XL | [36] |
| C. Receptor and adaptor proteins | |
| Inositol 1,4,5-trisphosphate receptor (IP3R) | [41] |
| Estrogen receptor (ERα) | [45] |
| MyD88 and TRIF | [42] |
| nPIST | [43] |
Acknowledgements
The work in the authors’ own laboratory was supported by NIH grants ROI AI151267, ROI CA84254, and ROI CA109618 to B.L. The authors thank Angela Diehl for expert medical illustration. We apologize to those authors whose work could not be cited due to space limitations.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. This work was the first to show that beclin 1 is an autophagy gene and tumor suppressor molecule.
- 3. Kihara A, Kabeya Y, Ohsumi Y, Yoshimori T. Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep. 2001;2:330–335. doi: 10.1093/embo-reports/kve061. This work was the first to show that mammalian beclin 1, like yeast ATG6/VPS30, controls autophagy as part of a class III PI3K complex.
- 4.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 5.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kihara A, Noda T, Ishihara N, Ohsumi Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol. 2001;152:519–530. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Furuya N, Yu J, Byfield M, Pattingre S, Levine B. The evolutionarily conserved domain of Beclin 1 is required for Vps34 binding, autophagy and tumor suppressor function. Autophagy. 2005;1:46–52. doi: 10.4161/auto.1.1.1542. [DOI] [PubMed] [Google Scholar]
- 8.Zeng X, Overmeyer JH, Maltese WA. Functional specificity of the mammalian Beclin-Vps34 PI 3-kinase complex in macroautophagy versus endocytosis and lysosomal enzyme trafficking. J Cell Sci. 2006;119:259–270. doi: 10.1242/jcs.02735. [DOI] [PubMed] [Google Scholar]
- 9. Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, Maejima I, Shirahama-Noda K, Ichimura T, Isobe T, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11:385–396. doi: 10.1038/ncb1846. Together with Ref. [13], using affinity chromatography/mass spectrometry approaches, the authors identified Atg14 and Rubicon in Beclin 1-containing protein complexes. They present evidence that Atg14 is required for autophagosome formation and that Rubicon inhibits the formation of autophagosomes/autolysosomes and inhibits endocytic trafficking.
- 10. Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. This paper first demonstrated mammalian Atg14 and UVRAG as candidate functional homologs of yeast Atg14 and Vps38 and components of mutually exclusive Beclin 1/class III PI3K complexes. The authors show an essential role for mammalian Atg14 in autophagosome formation.
- 11. Sun Q, Fan W, Chen K, Ding X, Chen S, Zhong Q. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci U S A. 2008;105:19211–19216. doi: 10.1073/pnas.0810452105. This paper demonstrated that Barkor (an identical protein as Atg14) is a Beclin 1-interacting and Vps34-interacting protein that functions in autophagosome formation.
- 12. Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, Corazzari M, Fuoco C, Ucar A, Schwartz P, et al. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–1125. doi: 10.1038/nature05925. This study identified Ambra1 as a novel binding partner of Beclin 1 and Vps34 that is essential for autophagy and the control of cell proliferation and cell death in the developing embryonic nervous system.
- 13.Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, Heintz N, Yue Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11:468–476. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, Jung JU. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8:688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- 15.Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, Liang C, Jung JU, Cheng JQ, Mule JJ, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9:1142–1151. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liang C, Lee JS, Inn KS, Gack MU, Li Q, Roberts EA, Vergne I, Deretic V, Feng P, Akazawa C, et al. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol. 2008;10:776–787. doi: 10.1038/ncb1740. This study identified UVRAG as a positive regulator in both autophagosome and endocytic vesicle maturation, through a mechanism that involves interactions with class C Vps and activation of Rab7 GTPase.
- 17.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang XH, Kleeman LK, Jiang HH, Gordon G, Goldman JE, Berry G, Herman B, Levine B. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J Virol. 1998;72:8586–8596. doi: 10.1128/jvi.72.11.8586-8596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. This work provided the first demonstration that cellular and viral Bcl-2 function as anti-autophagy proteins.
- 21.Feng W, Huang S, Wu H, Zhang M. Molecular basis of Bcl-xL’s target recognition versatility revealed by the structure of Bcl-xL in complex with the BH3 domain of Beclin-1. J Mol Biol. 2007;372:223–235. doi: 10.1016/j.jmb.2007.06.069. [DOI] [PubMed] [Google Scholar]
- 22.Ku B, Woo JS, Liang C, Lee KH, Hong HS, Xiaofei E, Kim KS, Jung JU, Oh BH. Structural and biochemical bases for the inhibition of autophagy and apoptosis by viral BCL-2 of murine gamma-herpesvirus 68. PLoS Pathog. 2008;4:e25. doi: 10.1371/journal.ppat.0040025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Maiuri MC, Le Toumelin G, Criollo A, Rain JC, Gautier F, Juin P, Tasdemir E, Pierron G, Troulinaki K, Tavernarakis N, et al. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J. 2007;26:2527–2539. doi: 10.1038/sj.emboj.7601689. This paper demonstrated that BH3-only proteins or BH3 mimetics can disrupt Bcl-2/Bcl-XL–Beclin 1 interactions and induce autophagy.
- 24. Oberstein A, Jeffrey PD, Shi Y. Crystal structure of the Bcl-XL–Beclin 1 peptide complex: Beclin 1 is a novel BH3-only protein. J Biol Chem. 2007;282:13123–13132. doi: 10.1074/jbc.M700492200. This paper solved the crystal structure of a fragment of Beclin 1 bound to Bcl-XL and provided the first evidence that Beclin 1 contains a BH3 domain that mediates its interactions with Bcl-2/Bcl-XL.
- 25.Sinha S, Colbert CL, Becker N, Wei Y, Levine B. Molecular basis of the regulation of Beclin 1-dependent autophagy by the gamma-herpesvirus 68 Bcl-2 homolog M11. Autophagy. 2008;4:989–997. doi: 10.4161/auto.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ciechomska IA, Goemans GC, Skepper JN, Tolkovsky AM. Bcl-2 complexed with Beclin-1 maintains full anti-apoptotic function. Oncogene. 2009;28:2128–2141. doi: 10.1038/onc.2009.60. [DOI] [PubMed] [Google Scholar]
- 27.Sinha S, Levine B. The autophagy effector Beclin 1: a novel BH3-only protein. Oncogene. 2008;27 Suppl. 1:S137–S148. doi: 10.1038/onc.2009.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noble CG, Dong JM, Manser E, Song H. Bcl-xL and UVRAG cause a monomer–dimer switch in Beclin1. J Biol Chem. 2008;283:26274–26282. doi: 10.1074/jbc.M804723200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levine B, Sinha S, Kroemer G. Bcl-2 family members: dual regulators of apoptosis and autophagy. Autophagy. 2008;4:600–606. doi: 10.4161/auto.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gelinas C, Fan Y, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fung C, Lock R, Gao S, Salas E, Debnath J. Induction of autophagy during extracellular matrix detachment promotes cell survival. Mol Biol Cell. 2008;19:797–806. doi: 10.1091/mbc.E07-10-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008;30:678–688. doi: 10.1016/j.molcel.2008.06.001. This study identified a biochemical mechanism by which starvation regulates autophagy that involves disruption of the Bcl-2–Beclin 1 complex by JNK1-mediated multisite phosphorylation of Bcl-2.
- 33.Pattingre S, Bauvy C, Carpentier S, Levade T, Levine B, Codogno P. Role of JNK1-dependent Bcl-2 phosphorylation in ceramide-induced macroautophagy. J Biol Chem. 2009;284:2719–2728. doi: 10.1074/jbc.M805920200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zalckvar E, Berissi H, Eisenstein M, Kimchi A. Phosphorylation of Beclin 1 by DAP-kinase promotes autophagy by weakening its interactions with Bcl-2 and Bcl-XL. Autophagy. 2009;5:720–722. doi: 10.4161/auto.5.5.8625. [DOI] [PubMed] [Google Scholar]
- 35. Zalckvar E, Berissi H, Mizrachy L, Idelchuk Y, Koren I, Eisenstein M, Sabanay H, Pinkas-Kramarski R, Kimchi A. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep. 2009;10:285–292. doi: 10.1038/embor.2008.246. This paper identified another mechanism by which the Bcl-2/Bcl-XL–Beclin 1 interaction and autophagy can be regulated that involves death-associated protein kinase (DAPk)-mediated phosphorylation of the BH3 domain of Beclin 1.
- 36.Pimkina J, Humbey O, Zilfou JT, Jarnik M, Murphy ME. ARF induces autophagy by virtue of interaction with Bcl-xl. J Biol Chem. 2009;284:2803–2810. doi: 10.1074/jbc.M804705200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Konopleva M, Contractor R, Tsao T, Samudio I, Ruvolo PP, Kitada S, Deng X, Zhai D, Shi YX, Sneed T, et al. Mechanisms of apoptosis sensitivity and resistance to the BH3 mimetic ABT-737 in acute myeloid leukemia. Cancer Cell. 2006;10:375–388. doi: 10.1016/j.ccr.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 38.van Delft MF, Wei AH, Mason KD, Vandenberg CJ, Chen L, Czabotar PE, Willis SN, Scott CL, Day CL, Cory S, et al. The BH3 mimetic ABT-737 targets selective Bcl-2 proteins and efficiently induces apoptosis via Bak/Bax if Mcl-1 is neutralized. Cancer Cell. 2006;10:389–399. doi: 10.1016/j.ccr.2006.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Del Gaizo Moore V, Brown JR, Certo M, Love TM, Novina CD, Letai A. Chronic lymphocytic leukemia requires BCL2 to sequester prodeath BIM, explaining sensitivity to BCL2 antagonist ABT-737. J Clin Invest. 2007;117:112–121. doi: 10.1172/JCI28281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, Bruncko M, Deckwerth TL, Dinges J, Hajduk PJ, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 41.Vicencio JM, Ortiz C, Criollo A, Jones AW, Kepp O, Galluzzi L, Joza N, Vitale I, Morselli E, Tailler M, et al. The inositol 1,4,5-trisphosphate receptor regulates autophagy through its interaction with Beclin 1. Cell Death Differ. 2009;16:1006–1017. doi: 10.1038/cdd.2009.34. [DOI] [PubMed] [Google Scholar]
- 42.Shi CS, Kehrl JH. MyD88 and Trif target Beclin 1 to trigger autophagy in macrophages. J Biol Chem. 2008;283:33175–33182. doi: 10.1074/jbc.M804478200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yue Z, Horton A, Bravin M, DeJager PL, Selimi F, Heintz N. A novel protein complex linking the delta 2 glutamate receptor and autophagy: implications for neurodegeneration in lurcher mice. Neuron. 2002;35:921–933. doi: 10.1016/s0896-6273(02)00861-9. [DOI] [PubMed] [Google Scholar]
- 44.Joubert PE, Meiffren G, Grégoire IP, Pontini G, Richetta C, Flacher M, Azocar O, Vidalain PO, Vidal M, Lotteau V, et al. Autophagy induction by the pathogen receptor CD46. Cell Host Microbe. 2009;6:354–366. doi: 10.1016/j.chom.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 45.John S, Nayvelt I, Hsu HC, Yang P, Liu W, Das GM, Thomas T, Thomas TJ. Regulation of estrogenic effects by Beclin 1 in breast cancer cells. Cancer Res. 2008;68:7855–7863. doi: 10.1158/0008-5472.CAN-07-5875. [DOI] [PubMed] [Google Scholar]
- 46.Deretic V, Levine B. Autophagy, immunity, and microbial adaptations. Cell Host Microbe. 2009;5:527–549. doi: 10.1016/j.chom.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Virgin HW, Levine B. Autophagy genes in immunity. Nat Immunol. 2009;10:461–470. doi: 10.1038/ni.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.E X, Hwang S, Oh S, Lee JS, Jeong JH, Gwack Y, Kowalik TF, Sun R, Jung JU, Liang C. Viral Bcl-2-mediated evasion of autophagy aids chronic infection of gammaherpesvirus 68. PLoS Pathog. 2009;5:e1000609. doi: 10.1371/journal.ppat.1000609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Orvedahl A, Alexander D, Talloczy Z, Sun Q, Wei Y, Zhang W, Burns D, Leib DA, Levine B. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe. 2007;1:23–35. doi: 10.1016/j.chom.2006.12.001. This paper provided the first demonstration that viral evasion of autophagy is essential for viral pathogenesis, and established the importance of molecular targeting of Beclin 1 by viral virulence proteins.
- 50.Leib DA, Alexander DE, Cox D, Yin J, Ferguson TA. Interaction of ICP34.5 with Beclin 1 modulates herpes simplex virus type 1 pathogenesis through control of CD4+ T-cell responses. J Virol. 2009;83:12164–12171. doi: 10.1128/JVI.01676-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kyei GB, Dinkins C, Davis AS, Roberts E, Singh SB, Dong C, Wu L, Kominami E, Ueno T, Yamamoto A, et al. Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages. J Cell Biol. 2009;186:255–268. doi: 10.1083/jcb.200903070. This paper described a role for early autophagosomes in enhancing viral yields and HIV Gag processing, and a role for the HIV pathogenic factor Nef as an anti-autophagic maturation factor through its interactions with Beclin 1.
- 52.Gannage M, Dormann D, Albrecht R, Dengjel J, Torossi T, Ramer PC, Lee M, Strowig T, Arrey F, Conenello G, et al. Matrix protein 2 of influenza A virus blocks autophagosome fusion with lysosomes. Cell Host Microbe. 2009;6:367–380. doi: 10.1016/j.chom.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]