Abstract
The basic helix-loop-helix DNA binding protein Hand2 has critical functions in cardiac development both in neural crest-derived and mesoderm-derived structures. Targeted deletion of Hand2 in the neural crest has allowed us to genetically dissect Hand2-dependent defects specifically in outflow tract and cardiac cushion independent of Hand2 functions in mesoderm-derived structures. Targeted deletion of Hand2 in the neural crest results in misalignment of the aortic arch arteries and outflow tract, contributing to development of double outlet right ventricle (DORV) and ventricular septal defects (VSD). These neural crest-derived developmental anomalies are associated with altered expression of Hand2-target genes we have identified by gene profiling. A number of Hand2 direct target genes have been identified using ChIP and ChIP-on-chip analyses. We have identified and validated a number of genes related to cell migration, proliferation/cell cycle and intracellular signaling whose expression is affected by Hand2 deletion in the neural crest and which are associated with development of VSD and DORV. Our data suggest that Hand2 is a multifunctional DNA binding protein affecting expression of target genes associated with a number of functional interactions in neural crest-derived cells required for proper patterning of the outflow tract, generation of the appropriate number of neural crest-derived cells for elongation of the conotruncus and cardiac cushion organization. Our genetic model has made it possible to investigate the molecular genetics of neural crest contributions to outflow tract morphogenesis and cell differentiation.
Keywords: Hand2(dHand), neural crest, bHLH, heart, outflow tract, cardiac cushion, cell type-specific gene regulation, gene microarray
Introduction
Proper function of the four-chambered mammalian heart requires that oxygenated blood and deoxygenated blood be routed, respectively, to the systemic and pulmonic circulatory systems. The elegantly structured cardiac outflow tract (OFT) fulfills this task. The OFT is composed of cells derived from both neural crest and splanchnic mesoderm that collectively generate a properly septated ascending aorta and pulmonary artery, which is required for postnatal survival. Cardiac morphogenesis initiates at E7.5 in the mouse where anterior lateral plate mesoderm migrates towards the midline forming a linear heart tube. This early tube gives rise to both right and left atria, the left ventricle, and is defined as the primary heart field. During looping of the primary heart tube, a second distinct population of pharyngeal mesoderm-derived cells, termed the secondary heart field, contribute to both myocardial and endocardial components of the right ventricle, interventricular septum, venous pole and base of the OFT where it connects to the great vessels. In addition to the mesoderm-derived cells, the OFT acquires a substantial contribution from the cardiac neural crest. A more complete understanding of OFT formation might therefore be based in determining the function of regulatory molecules expressed in both mesoderm-derived and neural crest-derived components. The basic helix-loop-helix (bHLH) DNA biding protein Hand2 is of particular interest as it has critical functions in both mesoderm-derived and neural crest-derived components of the developing heart.
Hand2 is expressed within cells of both the primary and secondary heart fields, as well as cardiac neural crest-derived cells. Cells from the primary heart field contribute to heart formation beginning at the cardiac crescent stage (E7). Transcripts encoding Hand2 are present in the cardiac crescent but as cardiac looping initiates (E8) Hand2 expression within the primary heart field down-regulates to be replaced by expression within the forming right ventricle and OFT (McFadden et al., 2000; Firulli, 2003; Barnes and Firulli, 2009). Although systemic knock-out of Hand2 does not directly affect development of primary heart field-derived structures, phenotypic anomalies are observed within the OFT and right ventricle (Srivastava et al., 1995; Srivastava et al., 1997; Thomas et al., 1998) suggesting a requirement of Hand2 for proper development of these structures. Expression of Hand2 is also crucial for the formation of several neural crest-derived structures including sympathetic chain ganglia (Howard, 2005; Hendershot et al., 2008; Schmidt et al., 2009), cranio-facial elements (Funato et al., 2009) and the enteric nervous system (Wu and Howard, 2002; Hendershot et al., 2007; Morikawa et al., 2007).
A constellation of congenital heart defects (CHDs) including ventricular septal defects, aortic arch artery patterning defects, and aortic and pulmonary valvular defects, is attributed to cardiac neural crest dysfunction; these disorders represent a substantial proportion of all observed CHDs (reviewed in Srivastava and Olson, 2000; Snider et al., 2007; and Waldo et al., 1998; Stoller and Epstein, 2005; Lie-Venema et al., 2007; Mitchell et al., 2007; Obler et al., 2008; Snarr et al., 2008). These late-stage phenotypes cannot be assessed in systemic Hand2 knockout mice due to early developmental lethality. To better dissect the molecular mechanisms of Hand2 function specifically in neural crest-derived cells important for OFT morphogenesis and independent from its function in mesoderm-derived components, we took advantage of our floxed-Hand2 mice (Hendershot et al., 2007; Hendershot et al., 2008). We targeted deletion of Hand2 in the neural crest using the Wnt1-Cre driver line of mice (Danielian et al., 1998; Jiang et al., 2002). Our results demonstrate that targeted deletion of Hand2 in the cardiac neural crest results in embryos with defects in OFT and aortic arch artery development, which phenocopy documented neural crest-dependent defects (reviewed in Stoller and Epstein, 2005; Mitchell et al., 2007; Hutson and Kirby, 2007). Our studies confirm and extend previously published aortic arch artery defects in a similar but independently generated Hand2 conditional allele (Morikawa and Cserjesi, 2008), by additional analysis of OFT phenotypic anomalies and analyses of differential patterns of gene expression. We show that targeted deletion of Hand2 results in double outlet right ventricle (DORV) and accompanying ventricular septal defects (VSD). By combining targeted deletion of Hand2 with microarray and ChIP-on-chip analyses, we have identified a number of transcriptional regulators and signaling molecules within the neural crest whose expression is modulated by loss of Hand2. Moreover, we show that Hand2 directly regulates a number of genes expressed within the neural crest suggesting a role for Hand2 in mediating transcriptional programs that orchestrate cell-cell interactions within the forming OFT and which affect cell cycle regulation.
Materials and Methods
Targeting Strategy and Mouse Strains
All breeding procedures, animal care and experimental protocols were approved by the Medical University of Ohio (renamed University of Toledo Health Sciences Campus) animal care and use committee prior to initiation of this work. Our strategy for targeting Hand2 is published elsewhere (Hendershot et al., 2007). Our analysis of Hand2fl/fl mice demonstrates that the mice are fertile, viable and phenotypically normal; introduction of loxP sites in the 5′ UTR therefore does not negatively impact transcription at the targeted locus. Normal levels and patterns of Hand2 expression have been confirmed by in-situ hybridization and qRT-PCR (Hendershot et al., 2007; Hendershot et al., 2008). Both systemic and targeted deletion of Hand2 is embryonic lethal (Srivastava et al., 1997; Hendershot et al., 2008). To generate embryos older than E11 (Hendershot et al., 2008) pregnant dams are fed water containing a cocktail of catecholamines (100 μg/ml L-phenylephrine, 100 μg/ml isoproterenol, and 2 mg/ml ascorbic acid) beginning at embryonic day (E) 8. This pharmacological approach allows us to rescue Hand2flneo/flneo and Hand2fl/fl;Wnt1-Cre embryos from pre-term death, a phenotype observed in other mouse models in which norepinephrine is absent or expression curtailed (Hendershot et al., 2007; Lim et al., 2000; Thomas et al., 1995; Zhou and Palmiter, 1995). Reporter mice were generated as previously described (Hendershot et al., 2007). Homozygous R26RYFP females were mated to hemizygous Wnt1-Cre males and Hand2fl/+;Wnt1-Cre males were mated to Hand2fl/fl;R262YFP females.
Immunocytochemistry and Histology
The antibodies used for the current studies are detailed in Table 1. Embryos were prepared and treated as previously described (Hendershot et al., 2007; Hendershot et al., 2008). Briefly, embryos or tissues are emersion fixed in 4% paraformaldehyde overnight at 4°C, extensively washed in PBS and stored in 30% sucrose unless otherwise stated. For analysis of heart development, hearts were removed in ice-cold PBS, fixed in 10% neutral buffered formalin, dehydrated through a graded series of ethanol and embedded in paraffin. Paraffin sections were prepared and stained with hematoxylin and eosin (University of Texas, Southwestern Medical center, Molecular Pathology Core Laboratory) using standard procedures. Frozen sections of Hand2wt/wt and Hand2flneo/flneo E11 hearts were stained using a modified hematoxylin and eosin stain (Sanderson, 1994). Briefly, tissue sections were post-fixed in 4% paraformaldehyde (10 minutes), rinsed in PBS (2 × 5 minutes) and stained in hematoxylin (1 minute), followed by washing in running water, differentiation in 1% HCl in 70% EtOH, bluing in Scott’s tap water (3.6 g sodium bicarbonate, 20g magnesium sulphate/litre) with counterstaining in Eosin Y for 10–20 seconds, followed by dehydration and mounting in Permount (Fisher Scientific, Fair Lawn, NJ) or EUKITT (O. Kindler, Germany). Immunostaining was done according to our established procedures (Howard et al., 1999, Wu and Howard, 2002, Liu et al., 2005, Hendershot et al., 2007). Tissue sections were blocked in a solution containing 0.1M Tris, pH. 7.5, 1.5% NaCl, 0.3% Triton-X 100 and 10% horse serum for 3 × 5 minutes. Primary antibody is applied in this same solution but containing 4% horse serum and incubated overnight at 4°C. Following washing in 0.1M Tris pH 7.5, 15% NaCl, secondary antibody was applied in this same solution with 4% horse serum for three hours at room temperature. Sections are then washed 3 × 5 minutes in 0.1M Tris, pH 7.5, 15% NaCl and mounted in Vectashield (Vector Laboratories, Burlingame, CA) or Fluoromount-G (Southern Biotech, Birmingham, Al). To view sites of antibody binding in whole embryos (Young et al., 1999; Hendershot et al., 2007; Hendershot et al., 2008) samples were incubated overnight in PBS containing 0.3% Triton-X 100 with gentle shaking at 4°C. We used our standard blocking step and primary antibody was applied as described above and incubated for two days at 4°C with gentle shaking. Following extensive washing secondary antibody was applied and embryos incubated overnight at 4°C with shaking. Whole embryos were mounted on depression slides in Fluoromount-G (Southern Biotech, Birmingham, AL).
Table 1.
Antibodies used for immunostaining in this study are listed. The antigen, dilution, and secondary antibodies are listed. The source for each antibody is listed.
| Antigen/Antibody | Dilution | Source | Secondary Antibody | Dilution | Source |
|---|---|---|---|---|---|
| Connexin-40 (rabbit) | 1:50 | Zymed, Carlsbad, CA | Donkey anti-rabbit TRITC Donkey anti-rabbit Alexa 647 | 1:200 1:500 |
Jackson ImmunoReserach, West Grove, PA Inivitrogen, Carlsbad, CA |
| GFP (chicken) | 1:300 | Aves Labs, Tigard, OR | Donkey anti-chicken FITC | 1:100 | Jackson ImmunoReserach West Grove, PA |
| Ki-67 (rabbit) | 1:200 | AbCam, Cambridge, MA | Donkey anti-rabbit TRITC | 1:200 | JacksonImmuno Reserach West Grove, PA |
Confocal Microscopy
All confocal images were acquired using a Leica Microsystems multiphoton confocal microscope (TCS SP5). Confocal Z-stack images were captured using either a 10X objective (n.a.= 0.3) or 63X (oil immersion) objective (n.a.= 1.4) with 4X software magnification at 0.5 μm (tissue sections) or 1.0 μm (whole-mount) steps. To acquire images, fluorescent FITC–coupled, TRITC–coupled and AlexaFlour 647-coupled secondary antibodies were excited using argon, HeNE or 633 laser lines, respectively.
Gene Array Analysis
For this array (Affymetrix 430 2.0, mouse array) total cellular RNA was isolated from Hand2wtwt and Hand2fl/del;Wnt1-Cre E10.5 hearts using RNeasy (Qiagen, Valencia, CA) according to manufacturer’s directions. The RNA samples were submitted to the NIH Neurosciences Microarray Consortium (Phoenix, AZ, USA; http://arrayconsortium.tgen.org/np2/home.do;) for array preparation and analysis. The screen included three biological replicates for each genotype (3 chips/genotype). Expression and statistical analyses were compiled using GeneSpring (GX V 7.3.1 (Agilent Technologies, Santa Clara CA). The signal intensities from each probe set were normalized at the 50th percentile for each chip; the expression level for each gene was normalized to the median level across the arrays. The linear regression for all replicates was greater than 0.9 indicating that replicates correlated very well. A list of significantly expressed genes was generated based on a comparison of 3 arrays derived from control and three arrays derived form mutant hearts. The list was filtered first for the absent genes, secondly for a fold change cutoff of 2, and thirdly for p value of ≤ 0.05 using Welch’s t-test. Genes whose expression was significantly altered by Hand2 deletion in the neural crest were subjected to network analysis using GeneGo (St. Joseph, MI), MetaCore and amiGO to group genes based on function and/or pathway analysis. We chose genes for further analysis based on their fold change, relationship to Hand2 and cardiac development.
Real-Time RT-PCR
Expression of transcripts encoding proteins identified in the gene array analysis were verified using quantitative reverse transcription-real time PCR as previously described (Zhou et al., 2004; Liu et al., 2005b; Hendershot et al., 2008). Briefly, we calculated the levels of transcript from the Ct values and normalized them based on expression of GAPDH. A detailed description of the protocol and equations can be found in Zhou et al., (2004) or Liu et al., (2005b). Each triplicate sample was analyzed (in triplicate) using cDNA corresponding to 10 ng of input RNA isolated from either Hand2wt/fl or Hand2fl/del;Wnt1-Cre E10.5 hearts. cDNA was mixed with Taqman Gene Expression master mix (Applied Biosystems, Foster City, CA, USA) and premixed (20X) Taqman probes (5 μM) and primers (18 μm). Forward and reverse primers (1.1 μM final concentration) with 6-FAM (carboxyfluorescein reporter dye) and a non-fluorescent quencher dye incorporated at 5′ and 3′ ends, respectively were used for all assays. Primers and probes were purchased from Applied Biosystems (Foster City, CA, USA) from their inventory of pre-developed Taqman® Gene Expression Assays. Triplicate PCR reactions were run and analyzed using a 7500 Fast Real-Time PCR sequence detection system (Applied Biosystems); the increase in product is monitored directly and reflects the threshold number of cycles (C) required for a detectable change in fluorescence based on release of probe. The efficiency of each primer and probe set was determined in separate reactions and based on the slope of C versus input cDNA dilution. Efficiency values were ≥ 1.875 for all transcripts.
Chromatin Immunoprecipitation (ChIP) Assay
To determine whether Cx40 is a direct Hand2 target, we employed Chromatin Immunoprecipitation (ChIP) on genomic DNA cross-linked, immunoprecipitated and purified from H92c cells, using a protocol modified from manufacturer’s guidelines (Millipore Corp., Billerica, MA). Briefly, ~1 × 106 H9c2 cells (~75% confluent, 10 cm dish) were cross-linked in 1% formaldehyde for 10 minutes at RT. Cross linking was quenched in 1 ml of 1.25 M glycine in PBS with shaking for 5 minutes at RT. Cells were rinsed twice with ice-cold PBS, collected to 1.5 ml Eppendorf tubes, and pelleted at 2,000× g for 4 minutes at 4°C. Cell pellets were washed with 1 ml of PBS containing protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 1 μg/ml aprotinin, 1 μg/ml pepstatin, Roche Inc., Madison WI) then resuspended in 200 μl of lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-Cl, pH 8.1, 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml aprotinin, 1 μg/ml pepstatin). The cell suspensions were incubated on ice for 10 minutes and sonicated on ice 4 times, 30 seconds each, at 1 minute intervals using a setting of 3 (Fisher Scientific Model: 550 Sonic Dismembrator). The appropriate sonication protocol was determined empirically. Lysates were centrifuged (1000 rpm, 7 minutes) and supernatants were added to 1.8 ml of dilution buffer (1.1% Triton X-100, 0.01% SDS, 1.2 mM EDTA, 167 mM NaCl, 16.7 mM Tris-HCl, pH 8.1, 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml aprotinin, 1 μg/ml pepstatin), then pre-cleared with 50 μl of Protein A/G PLUSAgarose (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) for 1 hour with end-over-end rotation at 4°C. Protein A/G PLUSAgarose was removed by brief centrifugation and immunoprecipitation was performed overnight at 4°C with 5 μg of Hand2 antibody.
The following day, an additional 50 μl of Protein A/G PLUSAgarose was added and the incubation continued for 2 hours at 4°C. Immuno-complexes were collected by centrifugation at 1000 rpm for 1 minute at 4°C and subjected to sequential 5 minute washes in 1 ml of each of the following buffers: low salt immune complex wash buffer (1% Triton X-100, 0.1% SDS, 2 mM EDTA, 20 mM Tris-HCl, pH 8.1, 150 mM NaCl), high salt immune complex wash buffer (1% Triton X-100, 0.1% SDS, 2 mM EDTA, 20 mM Tris-HCl, pH 8.1, 500 mM NaCl), and LiCl immune complex wash buffer (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris-HCl, pH 8.1). Precipitates were washed twice with TE buffer, then eluted twice in 250 μl of elution buffer (1% SDS, 0.1M NaHCO3) for 15 min at RT. The eluates were pooled and 20 μl of 5M NaCl was added; the eluates were subsequently heated at 65°C for 4 hours. Protein was removed from the samples by addition of 10 μl 0.5 M EDTA, 20 μl 1M Tris-HCl, pH 6.5 and 1 μl of 20 mg/ml Proteinase K for 1 hour at 45°C. DNA fragments were recovered by phenol/chloroform extraction and ethanol precipitation; 10 μg of yeast tRNA was added to each eluate to aid in recovery of DNA pellets. Following centrifugation (12,000 × g, 20 minutes at 4°C), the pellets were resuspended in 50 μl of TE; 1 μl was used per PCR reaction. PCR primers were as follows: Cx40(F)ChIP[−1144]: 5′-ACTGTCCCTCAGTTTCCCTG-3′, Cx40(R)ChIP[−814]: 5′-ACAGAGGGTCAAGGACATGG-3′, Cx40(F)ChIP[−655]: 5′-GAACACTCTGATTGGTGGGG-3′, Cx40(R)ChIP[−283]: 5′-CCAGTGGCTGTCCTTGTTTT-3′, GAPDH: 5′-TCCCACTCTTCCACCTTC-3′, GAPDH: 5′-CTGTAGCCGTATTCATTGTC-3′. PCR was conducted using GoTaq polymerase (Promega Corp., Madison, WI) with a 58°C, 1 min annealing step over 40 cycles of amplification.
ChIP-on-chip
Neural crest-derived cells were FACsorted from 20 E10.5 hearts isolated from wild-type Wnt1-reporter embryos and samples were prepared according to the manufacturer’s ChIP-chip user’s guide (Roche NimbleGen, Inc. Madison, WI). Briefly, hearts were extensively washed in ice-cold PBS, incubated in 0.5% trypsin in CMF-PBS for 15 minutes and dissociated into single cells using a fire-polished Pasteur pipette. The cells were washed one time in PBS, pelleted, and cross linked in 1% formaldehyde for 8 minutes at RT. The cross linking was quenched in 125 mM glycine. The washed cells were filtered through a 70 μm nylon cell strainer (Falcon #352350, Bedford, MA) and subjected to FACsorting (BDFacs Aria, BD Biosciences, San Jose, CA); the non-neural crest cells from the non-sorted sample were used for the controls. The sorted neural crest cells were lysed and sonicated (Fisher Scientific Model: 550 Sonic Dismembrator) for 4 rounds for 15 seconds on a setting of 3 with a one minute incubation on ice in between each round. The appropriate settings for sonication were determined on genomic DNA and the fragment size verified in a 1% agarose gel. For IP, we used our Hand2 antibody (raised in rabbit) coupled to Dynabeads (M-20, Invitrogen, Carlsbad, CA). Complexes were allowed to form for 2 hours at 4° C with rotation and isolated on a magnetic table. Sample DNA was amplified according to manufacturer’s directions (WGA3, Sigma-Aldrich, St. Louis, MO) and sent to Roach for processing (2.1×106 Deluxe Promoter Array).
Transfection and Luciferase Reporter Assay
HEK293a cells were grown in DMEM plus 10% FBS supplemented with l-glutamine, Na-pyruvate and 100 mg/ml Pen/Stryp (Invitrogen, Carlsbad, CA); cells were transfected when 50% confluent using CaPO4 according to our published procedures (Firulli et al., 1994). In each well, 2.5 μg of pcDNA-Hand2 and/or pCS2-MT+E12 (plus empty pcDNA vector to make a total of 5 μg of plasmid DNA) was co-transfected with 5 μg of Cx40 luciferase reporter (Cx40+pXP2) and 150 ng of Renilla reporter (pRL-CMV) as a transfection control. Cultures were harvested 48 hours after transfection and diluted ~10 fold in lysis buffer; 50 μl of this lysate was assayed. Luciferase assays were performed as previously reported (Xu et al., 2003) using the dual luciferase assay kit (Promega Corp., Madison, WI) according to the manufacturer’s protocol. Luciferase activities were read using a 96-well microtiter plate luminometer (Thermo Labsystems, Franklin, MA). Assays were done in triplicate.
Statistics
Data are presented as the mean ± S.E.M unless otherwise stated. Statistical significance was determined using Student’s unpaired t test or ANOVA and Bonferroni post hoc test, unless otherwise stated. At least four embryos were examined for each condition from at least three different matings unless otherwise stated. For microarray analysis, three biological replicates were run and each in triplicate.
Results
Hand2 Misexpression Affects Heart Development
In order to define the molecular basis for defects in heart development associated with loss of Hand2 function, we generated a conditional targeted mutation of Hand2 (Hendershot et al., 2007; Hendershot et al., 2008). For the current studies, we targeted deletion of Hand2 specifically within neural crest-derived cells by intercrossing our compound heterozygous floxed-Hand2 (Hand2f/ldel) mice with mice expressing Cre recombinase under control of the Wnt1 promoter. The Wnt1-Cre transgene allows floxed genes to be targeted in the neural crest (Danielian et al., 1998; Jiang et al., 2000; Brewer et al., 2004; Hendershot et al., 2008). Because loss of Hand2 expression within neural crest-derived cells is embryonic lethal between E10-E11 due to insufficient levels of norepinephrine (Lim et al., 2000; Hendershot et al., 2008), we routinely feed pregnant dams a cocktail of catecholamine intermediates beginning at E8 (Lim et al 2000; Thomas et al., 1995; Zhou and Palmiter, 1995; Kaufman et al., 2003; Hendershot et al., 2008). This pharmacological rescue makes it possible to bring embryos to term, allowing assessment of Hand2 loss-of-function within neural crest-derived cells and tissues, throughout late stage OFT morphogenesis and up to birth. Wnt1-mediated deletion of Hand2 results in developmental defects in all neural crest-derived structures dependent upon Hand2 for early aspects of their development (Hendershot et al., 2007; Hendershot et al., 2008). Briefly, there is significant loss of sympathetic and enteric neurons, alterations in neurotransmitter expression and regulation, as well as a number of defects in cranio-facial development, consequent to the loss of Hand2.
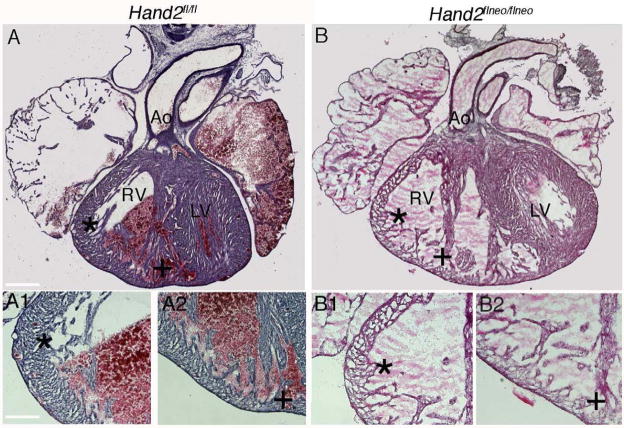
We previously reported that embryos harboring the neomycin cassette (Hand2flneo/flneo) in their genome misexpress Hand2 resulting in a Hand2 hypomorph (Hendershot et al., 2008). Interestingly, the impact of Hand2 under-expression was observed to be tissue dependent demonstrating both gene dosage and penetrance effects. The importance of Hand gene dosage for cardiac development is well documented (McFadden et al., 2005) and this current study confirms and extends these previous findings. At E18 we observe differential effects on heart development in Hand2flneo/flneo embryos compared to Hand2fl/fl embryos (Fig. 1). In Hand2 hypomorphic (Hand2flneo/flneo) embryos, under-expression of Hand2 could be affecting neural crest-derived structures, primary heart field myocardium, and/or intraventricular septal myocardium derived from the second heart field. In the cardiovascular system, the neural crest makes contributions to the aortic arch arteries and OFT. In Hand2flneo/flneo embryos, there were no obvious developmental defects within the aortic arch arteries (one embryo had patent ductus arteriosus); ~50% of hearts examined showed membranous VSDs (not shown). Development of the right ventricle is dependent upon Hand2 but independent of the neural crest (Srivastava et al., 1997; Thomas et al., 1998; Srivastava, 1999). Development of the right ventricle was affected in all hypomorphic hearts examined (Fig. 1B). In these hearts ventricular hypoplasia and disorganized trabeculations indicates likely effects on proliferation and/or apoptosis of cardiomyocytes. We have previously reported that Hand2 regulates proliferation of neural crest-derived noradrenergic neural precursor cells and neuroblasts (Hendershot et al., 2008) suggesting a potential function for Hand2 in the heart (see data below). The hypomorph phenotype thus recapitulates some of the defects observed in Hand2 systemic null embryos (Thomas et al., 1998; McFadden et al., 2005). Since we were interested in dissecting the function of Hand2 in cardiac neural crest-derived structures and given this potential complication, we employed SYCP-Cre mice to remove the neomycin-resistance cassette within our Hand2 conditional allele and completely rescue these hypomorphic defects (Hendershot et al., 2008).
Fig. 1. Cardiac development is affected in Hand2 hypomorph embryos.
Whole hearts were removed from Hand2fl/fl [A] and Hand2flneo/flneo [B] E18.5 embryos and 20 μm frozen transverse sections were visualized following staining with Hematoxylin and Eosin. Misexpression of Hand2 [B] consistently results in enlarged hypoplastic right ventricles and disorganized trabeculations. Abbreviations: right ventricle (RV), left ventricle (LV), aorta (Ao). Scale bars indicate 100 μm [A,B] and 25 m [A1,A2,B1,B2].
Targeted Deletion of Hand2 Results in Outflow Tract Defects
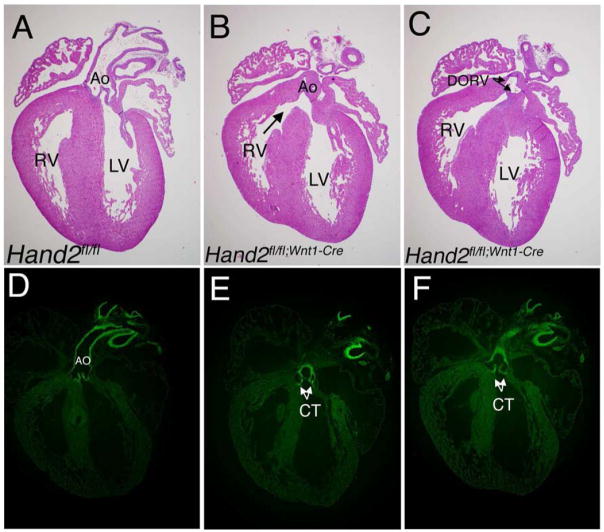
Based on the expression pattern of Hand2 within the cardiac neural crest, we predicted that deletion of Hand2 specifically within this cell population would adversely effect proper development of outflow tract structures. Analysis of Hand2 loss-of-function within the neural crest consistently demonstrated effects on ventricular septation and alignment of the great vessels (Fig. 2). We consistently observed double outlet right ventricle (DORV) with an associated membranous VSD in all Hand2fl/fl;Wnt1-Cre embryos (Fig. 2B & C) examined. This neural crest-derived anomaly (DORV) was never observed in Hand2 hypomorphic embryos. Analysis of Hand2wt/wt;Wnt1-Cre;R26R/YFP mice (Fig. 2D) and Hand2fl/fl;Wnt1-Cre/R26R/YFP mutant reporter mice (Fig. 2E) demonstrates that Hand2 will be recombined in the neural crest-derived cells which contribute to the conotruncus and aortic arch arteries. The effect of misexpression of Hand2 within the heart therefore depends heavily upon Hand2 expression levels as well as the identity of the cells where expression of Hand2 is affected.
Fig 2. Conditional deletion of Hand2 effects outflow tract development.
Whole hearts were removed from Hand2fl/fl (WT) [A] and Hand2fl/fl;Wnt1-Cre (−/−) [B, C] embryos, embedded in paraffin and 20 μm transverse sections stained with Hematoxylin and Eosin. In all mutant embryos examined (5) we observed membranous VSDs (B, black arrow) and double outlet right ventricle (C, DORV, arrows). Based on the expression pattern of GFP in comparable cryosections of E18.5 Hand2wt/wt;Wnt1-Cre/R26RYFP reporter [D] and Hand2fl/fl;Wnt1-Cre/R26RYFP mutant reporter [E–F] embryo hearts, the defects that we observe following deletion of Hand2 can be ascribed to neural crest-derived anomalies. Abbreviations: conotruncus (CT), aorta (Ao).
Defects in Neural Crest Migration and Proliferation
Between days E10 and E13 in the mouse, the neural crest not only remodels the pharyngeal arches, contributing to the smooth muscle of the forming arch arteries, but also invades the OFT to form the septum that divides the aorta and pulmonary trunk (Conway et al., 1997; Conway et al., 2000; Snarr et al., 2008). We considered that aberrant migration of neural crest-derived cells into the OFT might contribute to the VSDs observed in all embryos deficient in expression of Hand2 in neural crest-derived cells. At E10.5 neural crest-derived cells have migrated into the OFT traversing approximately 50% of this developing structure (Fig. 3). The patterns of migration observed in Hand2wt/wt;R26R/Wnt1-Cre and Hand2fl/del;R26R/Wnt1-Cre E10.5 reporter embryos is strikingly affected by deletion of Hand2. At this stage of development it is possible to observe the “prongs” of neural crest-derived cells that will migrate into the truncal cushions (arrows in Fig. 3A, C) and which contribute to septation of the aorta and pulmonary arteries. Visual inspection suggests that loss of Hand2 negatively impacts cell migration and/or cell numbers. There is a dramatic reduction in the number of neural crest-derived cells within the OFT (compare Fig. 3B and D). The migration of neural crest-derived cells into the aortic sac and truncus is not only required for septation but elongation of the conotruncus as well (Waldo, 1999; Epstein et al., 2000; Snider et al., 2007). Examination of neural crest-derived cells (Fig. 3B and 3D) actively migrating into this region suggested that cell-cell interactions are important. It appears that neural crest-derived cells migrate as a coherent sheet of cells with the majority of cells being in contact with their neighbors. In the Hand2fl/del;Wnt1-Cre;R262RYFP hearts we observe not only fewer cells that have reached the truncus but in addition they appear to migrate more as individual cells with fewer contacts with neighboring cells.
Fig 3. Conditional Deletion of Hand2 effects cell-cell interactions and migration of cardiac neural crest-derived cells.
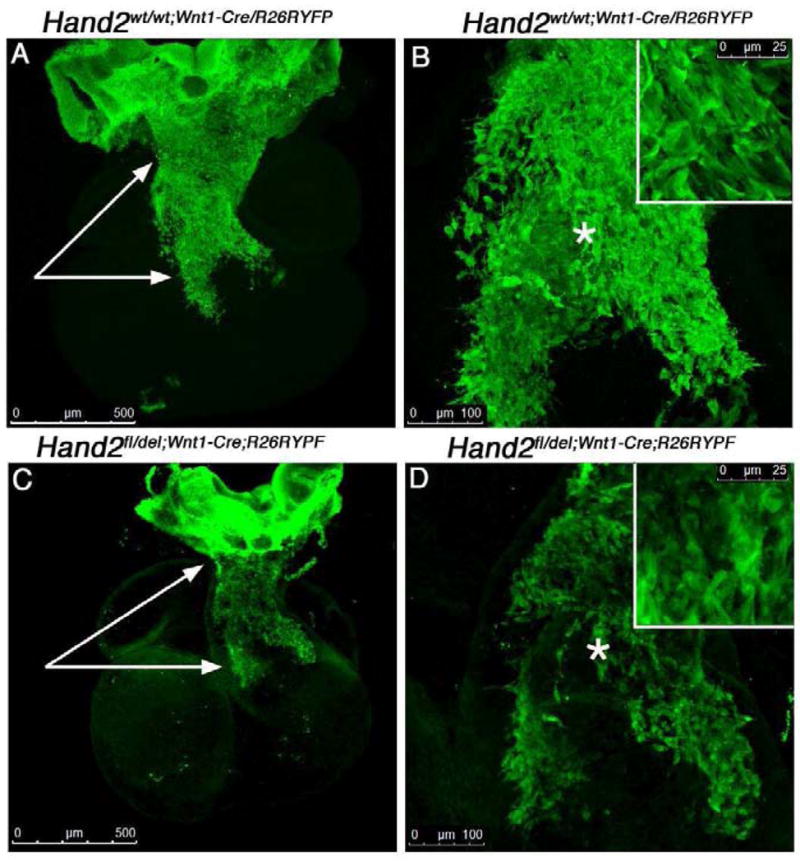
Migrating neural crest-derived cells were visualized in E10.5 Hand2wt/wt;Wnt1-cre/R26RYFP reporter [A–B] and Hand2fl/del;Wnt1-cre/R26RYFP mutant reporter hearts [C–D] immunostained for YFP expression in whole-mount. Confocal optical sections (1μm) were compressed into a z-stack (~400 sections) and analyzed. The pattern of migrating neural crest-derived cells observed within the outflow tract of mutant embryos was strikingly affected by loss of Hand2 expression. Neural crest-derived cells failed to migrate as a sheet and individual cells appear to have lost apparent contact with their neighbors routinely observed in control migrating cells (compare panels B and D). The number of neural crest-derived cells localized within the conotruncus (white arrows) appears reduced (see Figure 4) and the patterning of the AP Prongs is defective in the mutant compared to control hearts. The insets show high power views of the area indicated by the (*).
Based on our previous findings indicating that Hand2 is required to generate the appropriate number of neuronal precursor cells and neuroblasts in developing noradrenergic sympathetic ganglia (Hendershot et al., 2008) we counted the number of cells expressing the proliferation marker Ki-67 at E10.5 in Hand2wt/wt;Wnt1-Cre;R26RYFP and Hand2fl/del;Wnt1-CreR262RYFP hearts. The number of GFP (YFP) marked neural crest cells that labeled for immunoreactivity to Ki-67 was determined on 20 μm frozen cross sections at the level of the OFT (Fig. 4). Cells were counted in the entire outflow tract of control and mutant hearts. In control embryos, the mean number of proliferating neural crest-derived cells per tissue section in the OFT was 47 ± 6 compared to 10 ± 2 (P < 0.01) in the mutant OFT. Based on the number of neural crest-derived cells (GFP) migrating into and within the OFT, we observed a significant reduction in the proportion of proliferating cells based on the total neural crest cell population (Fig. 4G); in control embryos 41% + 4 of neural crest-derived cells were proliferating compared to 24% ± 3 (P < 0.05) in the mutant outflow tract. This result suggested possible impact of loss of Hand2 on mitosis. To determine if the number of cells in mitosis was affected by loss of Hand2, we counted the number of mitotic figures in the OFT of Hand2wt/wt;Wnt1-Cre;R26RYFP and Hand2fl/del;Wnt1-CreR262RYFP E10.5 embryos labeled with Ki-67 (Fig. 4E, F, H). The percentage of mitotic figures in Hand2fl/del;Wnt1-CreR262RYFP OFT was not significantly different from control indicating that equivalent numbers of neural crest-derived cells initiated mitosis; this likely results from problems in either G1 or S phases of the cell cycle. This suggests that the effects we observe on proliferation likely reflect alterations in the regulatory mechanisms controlling transit through the various stages of mitosis.
Fig 4. Hand2 affects proliferation of cardiac neural crest-derived cells.
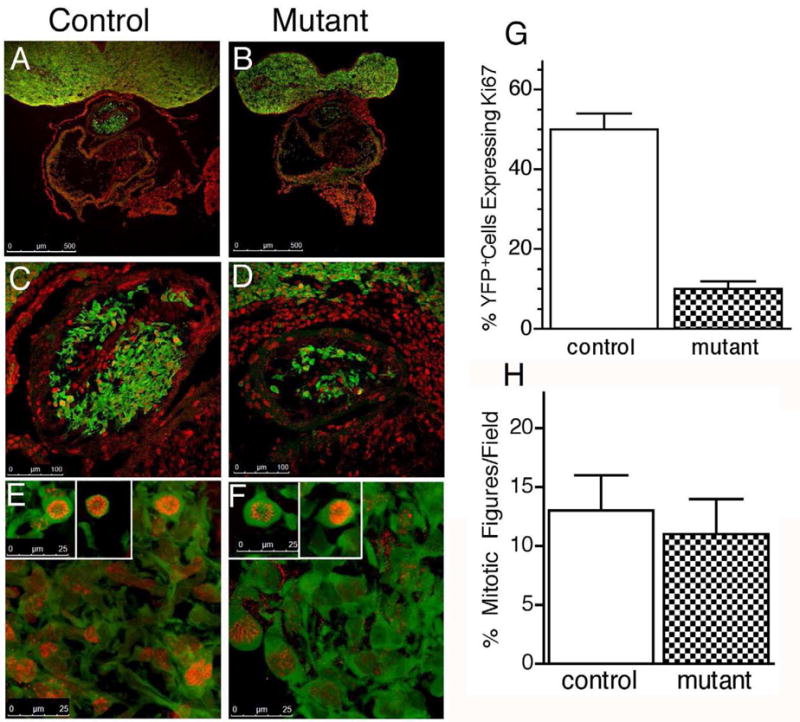
To determine if the apparent decrease in neural crest-derived cells migrating within the outflow tract of Hand2fl/del;Wnt1-Cre;R26R/YFP hearts was due to a defect in cell proliferation, we determined the number of neural crest-derived cells expressing the proliferation marker, Ki-67 in E10.5 outflow tract of Hand2wt/wt;Wnt1-cre/R26RYFP reporter [A, C, E] and Hand2fl/del;Wnt1-cre;R26RYFP mutant reporter hearts [B, D, F]. Cells were visualized, following immunostaining for Ki-67 (red) and GFP (green; marks YFP expressing neural crest-derived cells) in 20 μm frozen cross sections. There was no apparent difference in the number of cells expressing Ki-67 localized to second heart field. Visual inspection of the entire set of sections encompassing the developing outflow tract indicated a substantial decrease in the number of neural crest-derived cells (green) and neural crest-derived cells proliferating (green + red) within the outflow tract of mutant [B, D] compared to control [A, C] hearts. This result was quantified [G] by counting the number of cells co-expressing GFP and Ki-67 over the entire length of the outflow tract (N=3 for each genotype). There was a significant (P < 0.001) decrease in the number of proliferating neural crest-derived cells in response to targeted deletion of Hand2. [H] The percentage of mitotic figures did not differ significantly in control compared to mutant OFT.
Outflow Tract Defects Associated with Altered Gene Expression
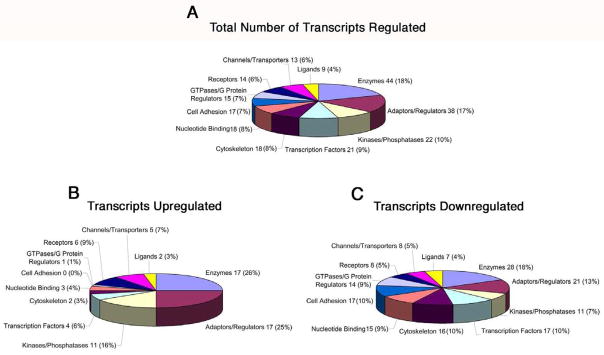
In order to directly address the genetic basis of the OFT phenotype we observe in Hand2fl/del;Wnt1-Cre embryos, we compared the panel of transcripts whose expression was altered in mutant compared to wild-type embryos. We screened gene arrays using RNA isolated from E10.5 Hand2wt/wt and Hand2fl/del;Wnt1-Cre whole hearts. The screen included three biological replicates for each genotype (9 chips/genotype). Our results demonstrate that targeted deletion of Hand2 in neural crest-derived cells in the heart significantly (P < 0.05) affected expression of 309 identified transcripts and 56 unidentified transcripts regulated minimally by greater than or equal to a 2-fold change (Fig. S1). The identified transcripts were associated with 11 functional classes (Fig. 5A; GeneGo Inc., St. Joseph, MI). Of the identified regulated genes, 68 were up-regulated (Fig. 5B) and 162 were down-regulated (Fig. 5C). Of the identified genes (309) 79 proteins were identified but which have no known function. Of the genes regulated, a notable fraction are known to affect heart development and/or cardiac function (Table 2). We validated a set of genes whose apparent function is associated with some aspect of the morphological or functional consequences of Hand2 deletion; a subset of these genes were validated using RNA purified from cardiac neural crest-derived cells FACsorted from E10.5 heart showing that our transcript analysis yielded alterations in expression of neural crest-specific genes (* in Table 2.).
Fig 5. Functional classes of differentially regulated genes in the heats of Hand2wt/wt and Hand2fl/del;wnt1-cre embryos.
Regulated genes were grouped into 11 functional categories and graphed as a percentage of the total based on their GeneGo designation. 309 genes were differentially regulated based on analysis of the array data; 79 of these regulated genes did not have any known function and were eliminated from this analysis [A]. Of the regulated genes, 68 were up-regulated [B] and 162 were down-regulated [C]. A number of down-regulated genes (17) are associated with cell adhesion; none of the genes in this category were up-regulated. The cytoskeletal-related transcript category contained 16 down-regulated genes and 2 up-regulated genes. In the transcription factor category, a notable difference in the number of transcripts down-regulated (17) and up-regulated (4) related to Hand2 was observed. In the kinases/phosphatases category a significant number of transcripts (11 up-regulated, 11 down-regulated) were affected by Hand2 deletion. We have identified a number of Hand2-target genes in each of these categories that are associated with the developmental anomalies that we observe in embryos with targeted deletion of Hand2 in the neural crest.
Table 2.
Hand2 Target Genes
| Biological Function | Gene Network | Gene Symbol | Expression | ChIP-on-chip (FDR) | Classification |
|---|---|---|---|---|---|
| Migration | |||||
| Migration | Cap1 | ↑7.78 | Cytoskeleton | ||
| Itga4 | ↑3.83 | Receptor | |||
| Clasp2 | ↑2.59 | .06031 | Cytoskeleton | ||
| Mmp14 | ↓2.02 | .0201 | Enzyme | ||
| Nr4a2 | ↓2.11 | 0 | Transcription Factor | ||
| Pard3 | ↓2.11 | .003 | Cell Adhesion | ||
| Ankrd6 | ↓2.19 | .00076 | Cytoskeleton | ||
| Nav1 | ↓2.43 | .0488 | Cytoskeleton | ||
| FoxC1* | ↓3.78 | .0256 | Transcription Factor | ||
| Focal Adhesion | Itga9* | ↑2.94 | Receptor | ||
| Col11a1* | ↓2.1 | .0671 | Cell Adhesion | ||
| PdgfD* | ↓3.89 | Ligand | |||
| Sox11* | ↓2.5 | .0134 | Transcription Factor | ||
| Insm1* | ↓20.7 | .009 | Transcription Factor | ||
| Gja5* | ↓23.87 | Channel/Transporter | |||
| Cell Cycle/Proliferation | |||||
| Cytoskeleton | Ccnb1ip | ↑32.47 | .19718 | Enzymes | |
| Cdk6 | ↑2.53 | Kinase/Phosphatase | |||
| Eif2ak1 | ↑2.16 | .10235 | Kinase/Phosphatase | ||
| Pttg1* | ↑2.11 | Transcription Factor | |||
| Stat5b | ↑2.05 | .00464 | Transcription Factor | ||
| Mmp14 | ↓2.02 | .021 | Enzyme | ||
| Rps6 | ↓2.03 | .17212 | Adaptor/Regulator | ||
| Hoxa3 | ↓2.15 | .15815 | Transcription Factor | ||
| Camk2a | ↓2.17 | .07472 | Kinase/Phosphatase | ||
| Tial1 | ↓2.22 | .0609 | Nucleotide Binding | ||
| Foxp2* | ↓2.45 | Transcription Factor | |||
| Sox11* | ↓2.5 | .0134 | Transcription Factor | ||
| H3f3b | ↓2.55 | NucleotideBinding | |||
| Anxa6 | ↓2.6 | .03856 | Cytoskeleton | ||
| Lgals7 | ↓3.01 | .01206 | Cell Adhesion | ||
| Gli3 | ↓3.54 | .03856 | Transcription Factor | ||
| FoxC1* | ↓3.73 | .0256 | Transcription Factor | ||
| RpL17 | ↓5 | .10235 | Adaptors/Regulator | ||
| Khdrbs1 | ↓8.85 | .17212 | Nucleotide Binding | ||
| Heart Morphogenesis | |||||
| Itga4 | ↑3.83 | Kinase/Phosphatase | |||
| Col11a1 | ↓2.01 | .0671 | Cell Adhesion | ||
| Adam19* | ↓2.16 | Enzyme | |||
| NF-ATc2 | ↓2.29 | .003 | Transcription Factor | ||
| Gli3 | ↓3.54 | .03856 | Transcription Factor | ||
| Foxc1* | ↓3.73 | .0256 | Transcription Factor | ||
| Gja5* | ↓23.87 | Channels/Transporter | |||
| Neural Related Genes | |||||
| Differentiation | Ywhag | ↑2.1 | .06031 | Adaptor/Regulator | |
| Nr4a2 | ↓2.11 | 0 | Transcription Factor | ||
| Hey1 | ↓2.0 | .1772 | Transcription Factor | ||
| Nav1 | ↓2.43 | .0488 | Cytoskeleton | ||
| Gli3 | ↓2.54 | .03856 | Transcription Factor | ||
| Tgif2 | ↓2.8 | 0 | Transcription Factor | ||
| Prph | ↓6.41 | 0 | Cytoskeleton | ||
Expression Verified by qRT-PCR of FACS-isolated ROSA-YFP cardiac neural crest. For the ChIP-on-chip the false discovery rate (FDR) is presented. Each reported interval is considered significant and indicative of a potential binding site. Within the three confidence levels, FDR ≤ 0.05 represents the highest confidence level; FDR >.05 ≤ 0.1 is the second highest confidence level and FDR > 0.1 ≤ 0.2 is the lowest confidence level where binding remains statistically significant. The primary data for the heart micro-arrays is available at the NIH microarray consortium website http://np2.ctrl.ucla.edu/np2/home.do. To navigate to the data, click on the “navigate repository” button and write Howard under investigator and click go. The project will come up and the data can be accessed by clicking on the magnifying glass.
Genes associated with neural crest cell migration
In our initial analysis of the consequences on heart development due to targeted deletion of Hand2 in the neural crest, we observed that population of the conotruncus by neural crest-derived cells was aberrant (Fig. 3). Several genes associated with migration were differentially expressed in the Hand2fl/del;Wnt-Cre mice. These regulated genes fall into at least three functional networks (Table 2). We validated genes from each group using qRT-PCR and/or immunostaining. We observed a decrease in transcript encoding Pdgf (↓3.89); Pdgf has been associated with neural crest cell migration (Orr-Urtreger and Lonai, 1992). Interestingly, defects in the Pdgf receptor results in developmental defects in the OFT as well as other neural crest-derived structures (Morrison-Graham et al., 1992). We validated integrin α9 (Itga9; ↑2.94) because it has an important function in angiogenesis (Brooks et al., 1994), which is adversely affected in Hand2 mutant embryos (and see Morikawa et al., 2008) as well as a potential role in cell migration (Young et al., 2001; Huang et al., 2005). The integrin subunit α4 is a component of the α4b1 fibronectin-binding domain, which is one important interaction regulating cell migration (Sheppard et al., 1994; Pinco et al., 2001), and was up-regulated (↑3.83) in response to deletion of Hand2. Interestingly, Adam19 (↓2.16) inhibits migration mediated by α4b1 integrins (Huang et al., 2005). Deletion of α4 integrin is embryonic lethal in part due to defective development of the epicardium and coronary vessels (Yang et al., 1995).
The regulation of connexin 40 (Cx40; Gja5) was of particular interest because it is heart-specific (Dahl et al., 1995; Kirchhoff et al., 2000) and associated with both adhesion and cell-cell communication. Based on the array results, transcripts encoding Cx40 were down-regulated 24-fold in Hand2fl/del;Wnt1-Cre hearts compared to control; this transcript was not detectable by qRT-PCR in the linear range of amplification (transcript became detectable at 40 cycles of amplification). We used immunostaining and confocal microscopy to visualize sites of Cx40 expression on tissue sections of E10.5 OFT (Fig. 6). Interestingly, although we could not detect transcripts encoding Cx40 there remains a small number of cells (Fig. 6B, D, F, arrows) that express Cx40 protein in the mutant OFT. The majority of neural crest-derived cells express Cx40 in the wild-type OFT, a novel finding in itself (Fig. 6A, C, E). Connexin 40 is also expressed in some cells in the second heart field-derived component of the conotruncus in both the control and mutant hearts (white arrows Fig. 6A, B) validating that neural crest specific deletion of Hand2 is revealing a cell autonomous effect. We further assessed regulation of Cx40 because it has not previously been reported in the neural crest and is affected cell autonomously by Hand2 loss-of-function.
Fig 6. Connexin 40 expression is decreased by targeted deletion of Hand2 in the neural crest.

Expression of Connexin 40 was examined in the outflow tract of Hand2wt/wt;Wnt1-Cre;R26R/YFP [A,C,E] and Hand2fl/del;Wnt1-Cre;R26R/YFP [B,D,F] E10.5 cryostat sections encompassing the entire outflow tract. Tissue sections (20 μm) were immuno-labeled for expression of YFP (reporter, green) and Connexin 40 (red). The entire outflow tract was imaged at 20X and 63X; 0.5 μm optical sections were compressed in the z-plane and assessed. Ten pairs of matched sections were examined. The majority of neural crest-derived cells migrating within the outflow tract in control embryos [A,C,E] express connexin 40 at the cell surface and in contact with neighboring cells. In Hand2 mutant embryos, only a few neural crest-derived cells maintain expression of Connexin 40 [B,D,E]; the expression pattern appears normal in those few cells that maintain Connexin 40. The arrows point to connexin 40 expressed in the myocardium. Abbreviation: Cx40 (Connexin 40).
Hand2 binds to and trans-activates the Cx40 promoter
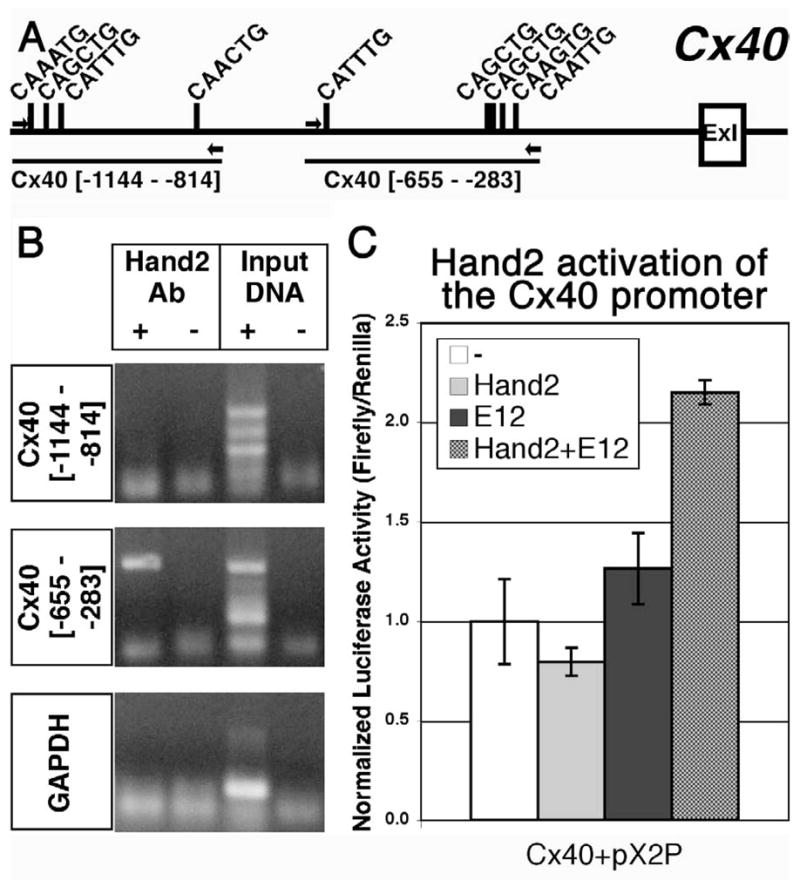
We sought to validate that a subset of the genes identified in our microarray screen represent direct Hand2 transcriptional targets (Table 2 (+); Fig. 7). Based on ChIP (Cx40) or ChIP-on-chip we identified a number of putative Hand2-target genes. We analyzed Cx40 in some detail because Cx40 null mice display membranous VSDs phenotypically similar to those occurring in our Hand2-loss-of-function embryos (Kirchhoff, 2000) and we found that Cx40 is expressed in cardiac neural crest cells (Fig. 6). A putative cis-regulatory region for the Cx40 gene has been previously identified (Seul et al., 1997) and spans bases from −1190 to +121. Initially, we analyzed this region for E-box (bHLH consensus) binding sites (CANNTG) and identified a total of 9 sites 5′ to the Cx40 transcription start site 5′ to exon 1 (Fig. 7A). For ChIP analysis we used H9c2 cells, a rat cardiomyocyte cell line which endogenously expresses Hand2 (Zang et al., 2004) and which upregulates Cx40 expression in response to Hand2 overexpression (Togi et al., 2006). PCR amplification of DNA fragments immunoprecipitated using our Hand2 antibody revealed that DNA fragments overlapping the proximal (−655 to −283 bp), but not distal (−1144 to −814 bp) E-box-containing DNA sequences co-immunoprecipitated with Hand2 (Fig. 7B); the two sets of primers we designed and tested flank all 9 of the E-boxes in the Cx40 5′ putative cis-regulatory region. The PCR product band was TOPO®cloned (InVitrogen, Carslbad, CA) and the sequence verified as the appropriate region of Cx40 (not shown). These data suggest that Hand2 directly binds to the putative Cx40 promoter at E-box sites located between −655 and −283 bp relative to the transcription start site. In order to directly demonstrate that this binding could have functional relevance, we conducted trans-activation studies to confirm that Hand2 can regulate Cx40 transcriptional activity. We used a reporter construct (generously provided by Benoit Bruneau, Gladstone Institute of Cardiovascular Disease) containing sequence corresponding to bases −1068 to +121 of the Cx40 gene, that drives expression of a luciferase cassette (Bruneau et al., 2001); this was co-transfected with Hand2 and/or E12 expression constructs into HEK 293 cells (Fig. 7C). While expression of neither Hand2 nor E12 alone was sufficient to modulate Cx40-luc reporter expression, expression of both factors together up-regulated Cx40-luciferase activity (~2.2 fold) over baseline (p < 0.001). These results indicate that Hand2 and its bHLH dimer partner, E12, can positively regulate gene expression from the Cx40 promoter. Together, these results indicate that Cx40 is a direct target of Hand2 within the cardiac neural crest.
Fig. 7. Connexin 40 is a direct Hand2 target gene.
[A] Schematic representation of the E-boxes (consensus: CANNTG) located within genomic region 5′ to Exon 1 (ExI) of the Cx40 gene. Amplicons of the distal (Cx40 [−1144 – −814]) and proximal (Cx40 [−655 – −283]) primer sets (arrows) used for ChIP assays are denoted. [B] ChIP using a Hand2-specific antibody co-immunoprecipitates DNA fragments overlapping the proximal (Cx40 [−655 – −283]), but not distal (Cx40 [−1144 – −814]), primer sets, indicating that Hand2 binds to the Cx40 promoter within this region. [C] Luciferase-reporter transactivation assays in HEK 293 cells show that Hand2 and E12 cooperatively up-regulate expression of a Cx40-luciferase reporter (Cx40-pX2P) ~2.2 fold over baseline (n = 3, p < 0.001). Individually, Hand2 and E12 do not significantly influence Cx40-pX2P expression.
We validated (qRT-PCR) an additional subset of genes, identified in the array screen, that appear in one or more of the functional pathways associated with cell-cell interaction and/or adhesion (Table 2; validated genes are shown in blue). In addition to qRT-PCR and ChIP, we also performed a ChIP-on-chip screen on FACsorted cardiac neural crest-derived cells (Table 2; direct Hand2 target genes indicated by a +) to determine which of the validated genes are potentially neural crest-specific Hand2 direct transcriptional targets.
Genes associated with neural crest cell cycle
We validated a subset of genes associated with cell cycle because of the significant reduction in proliferation that we observe (Fig. 4) when Hand2 is deleted in neural crest-derived cells (Hendershot et al., 2008). The genes that we validated, based on qRT-PCR and ChIP-on-chip, include cdk6 (↑2.53), Insm1 (↓20.7 array Vs undetectable qRT-PCR); each of these genes is associated with some aspect of cell proliferation and/or known Hand2-related genes. Several classical “cell cycle” genes are differentially regulated in the absence of Hand2. The most highly regulated gene in the array was cyclin B1 interacting protein 1 (Ccnb1ip1; ↑37.42). The protein coded for by this gene is a Ring finger protein. It is a member of a group of B-type cyclin E3 ubiquitin ligases that also has cyclin-dependent kinase and cyclin-dependent phosphorylation sites (Toby et al., 2003). This protein interacts with cyclin B1, an important checkpoint regulator of cell cycle progression, and causes its degradation (Toby et al., 2003). Overexpression of Ccnb1ip1 substantially reduces expression of cyclin B1 resulting in decreased proliferation (Toby et al, 2003). This protein also has been shown to affect migration and metastasis (Singh et al., 2007) potentially linking cell cycle with migration; functions which are both effected by deletion of Hand2. It is interesting to note that none of the cyclin proteins were identified in our array but several cyclin-dependent kinases or interacting proteins were. Cyclin dependent kinase 6 (↑2.53) was increased. Cyclin dependent kinase 6 is a serine/threonine protein kinase that is activated by cyclin D1, D2 and D3, and which is required for the G0 to G1 transition (Malumbres and Barbacid, 2005). Additional genes coding proteins that have a role in cell cycle regulation were also identified. Insm1 (Insulinoma associated 1) was of particular interest because it has been identified as a member of a transcription factor network that regulates development of neural crest-derived noradrenergic sympathetic ganglion neurons (Howard, 2005; Hendershot et al., 2008; Wildner et al., 2008; Parlier et al., 2008). Hand2 has a central role in this pathway and likely functions upstream or in parallel with Insm1 (Hendershot et al., 2008; Lan and Breslin, 2009). Targeted deletion of Hand2 or Insm1 results in a marked decrease in proliferation of the pool of neural precursor cells (Hendershot et al., 2008; Wildner et al., 2008). A potential site of Insm1 activity in the heart is at cyclin D1. Insm1 binds to cyclin D1 (Liu et al., 2006) which competes for the interaction of cyclin D1 with Cdk4. The Cdk4/cyclin D1 interaction is required for proper phosphorylation of retinoblastoma protein Prb necessary for cell cycle progression from G0 to G1 (Zhang et al., 2009). The putative Insm1 interacting protein, CAP1 (adenylate cyclase associated protein; Xie et al., 2002) transcript level was also increased (↑7.21) in the mutant hearts. Interestingly, transcripts encoding peripherin, a neuron specific gene and associated with Hand2 and Insm1 was down-regulated 6.41 fold in the mutant hearts. Expression of several histones was also affected by deletion of Hand2 including: 1) histone cluster 1 (↓2.1; Hist1h2bb); 2) H2a histone family member V (↓2.27); 3) H3 histone family 3B (↓2.55); and 4) histone cluster 2 H2be (↓2.67). Although the nucleosome (core histone complex) is associated with multiple aspects of DNA replication, repair and chromatin structure, one of these histones, H2be, is required for Cdk6 to bind its substrate, retinoblastoma protein Prb, required for initiation of cell cycle progression from G to S phase (Zhao et al., 2000; Marzluff et al., 2002; Jaeger et al., 2005; Ewen, 2009). Although we found a significant reduction in cell proliferation (Fig. 4G), there is no significant reduction in the percentage of cells that initiate mitosis (Fig. 4H) suggesting a potential role for Hand2 in regulation of cell cycle progression.
Genes associated with VSD and DORV
A number of genes have been identified as risk loci for VSDs and DORV. We were very interested in these genes since targeted deletion of Hand2 in the neural crest results in these defects in 100% of embryos examined (Fig. 2). Sox11 (↓2.5), a member of the SRY-related HMG-box family of transcriptional regulatory genes is an important regulatory gene affecting neural, skeletal and cardiac development. Sox11 null embryos develop VSDs with 100% penetrance (Sock et al., 2004). These embryos also display DORV and other OFT defects similar to that which we observe in the Hand2 mutant embryos. Gli3 (↓2.54; C2H2-type zinc finger protein) was identified as a VSD susceptibility gene in a study focused on 12q13 chromosomal region previously identified as associated with occurrence of VSD in humans (Qui et al., 2006). Although the molecular mechanism of Gli function in the cardiac neural crest is not yet known, Gli-family members mediate sonic hedgehog (Shh) signaling (te Welscher et al., 2002a; te Welscher et al., 2002b; Stamataki et al., 2005) and Zic3 translocation to the nucleus appears dependent upon Gli3 (Bedard et al., 2007). Disruption of Zic3 expression results in a number of developmental defects related to left-right asymmetry including conotruncal malformations (Mégarbané et al., 2000; Ware et al., 2006). Mutations in Foxc1 (↓3.73; forkhead-winged helix transcription factor family) are associated with abnormal vasculogenesis, coarctation of the aortic arch, and VSDs, related to Notch signaling (Kume et al., 2001; Seo et al., 2006); the only linkage to Notch signaling in our array is Hey1 (↓2.0). Notch signaling has been implicated as important for septation and Hey 1 (hairy and enhancer of split-related family of bHLH-type transcriptional repressor) is a Notch-downstream target gene (Sakata et al., 2002). In conjunction with Hey 2 (not regulated) Hey 1 has been implicated in Notch-mediated epithelial-mesenchymal transition (EMT) (Fischer et al., 2004; Niessen and Karsan, 2008). This occurs via a decrease in expression of several metalloproteinases (MMP; Brou et al., 2000). Two MMPs, MMP14 (↓2.02) and Adam 19 (↓2.16) were down-regulated in response to deletion of Hand2 in the cardiac neural crest suggesting that the reduction in migration and invasion of the cardiac cushions could be related to decreased expression of these genes.
Valvulogenesis
The cardiac cushions develop in the distal portion of the common OFT and between the common atria and ventricle (reviewed in Barnett and Desgrosellier, 2003; Goodwin et al., 2005). Cushion formation requires production of extracellular matrix material that becomes situated between the endocardial and myocardial layers of the developing heart. Neural crest-derived cells contribute to the truncal cushions that eventually form the aorticopulmonary septum and semilunar valves (de Lange et al., 2004). A number of genes associated with aspects of cardiac cushion formation, valvulogenesis and/or septation have been identified and which are affected by targeted deletion of Hand2 in the neural crest. In the absence of Nf-ATc (nuclear factor of activated T-cells) heart valves fail to form and embryos die (de la Pompa et al., 1998, Zhou et al., 2002). Expression of NF-ATc1 is induced by NF-ATc2 (↓2.29) that is down-regulated in response to Hand2-deletion in the neural crest. Collagen type XI a1 (Col11a1, ↓2.1 array Vs. ↓1.43 PCR) is expressed in the non-cartilaginous tissue of the heart. Mutations in the Col11a1 gene have been implicated in valve defects. Adam 19 (↓2.16 array Vs. ↓2.63 qRT-PCR) is a metalloprotease that is expressed in the endocardial cushion. Adam 19 null mice die within a week of birth due to thickened and misshapen semilunar valves, tricuspid valves, and membranous VSD (Horiuchi et al., 2005; Komatsu et al., 2007). EMT is required for proper cardiac cushion transformation (Barnett and Desgrosellier, 2003) and is stimulated by members of the transforming growth factor-β family of proteins. Deletion of latent transforming growth factor beta binding protein 1 (Ltbp1, ↓2.13) results in multiple heart defects including improper septation of the OFT, patent truncus arteriosus (PTA), and interrupted aortic arch (IAA) (Todorovic et al., 2007).
Discussion
The consequences to loss of Hand2 in the heart have been a focus of much investigation (Srivastava et al., 1997; Yamagishi et al., 2001; McFadden et al., 2005; Morikawa and Cserjesi, 2008; Reviewed in Firulli and Thattaliyath, 2002; Firulli, 2003; Bhattacharya et al., 2006; Olson, 2006; Srivastava, 2006). Hand2 contributes to normal development of both the OFT and right ventricular myocardium. Cells from the secondary heart field and cardiac neural crest contribute to the OFT by migrating into the formed heart tube, whereas only secondary heart field cells contribute to right ventricle myocardium development (reviewed in Kelly and Buckingham, 2002; Garry and Olson, 2006; Hutson and Kirby, 2007). Since Hand2 is expressed within the second heart field myocardium and cardiac neural crest it has been difficult to separate effects of Hand2 deletion in neural crest-derived or mesoderm-derived components of the heart because of the close interaction of cells from these two lineages. This problem has been addressed in the current studies. We employed an intercross of our mice harboring a Hand2 conditional allele with the Wnt1-Cre transgenic mouse line to interrogate the consequences to loss of Hand2 specifically in the neural crest. Our analysis of OFT development combined with gene profiling data demonstrate that Hand2-target genes fall into a variety of functional categories including cell adhesion, cell cycle regulation, proliferation control and intracellular signaling. We have identified a large number of genes as likely or identified loci for neural crest-dependent cardiac developmental anomalies, including VSD, DORV, and valve malformation.
Hand2 Gene Dosage and Cardiac Development
Conditional inactivation of Hand2 in secondary heart field cells presages malformation of the right ventricle manifesting as ventricular hypoplasia and in the conotruncus resulting in membranous ventricular septal defects (VSDs. Specific deletion of Hand2 in neural crest-derived cells that contribute to the conotruncus results in developmental abnormalities expected based on expression pattern. Interestingly, occurrence of OFT anomalies and VSDs was not predicted to be prevalent in Hand2fl/fl;Wnt1-Cre embryos because of potential redundant function with Hand1, which is co-expressed with Hand2 in a subset of cardiac neural crest cells (McFadden et al., 2005). Deletion of Hand1 in neural crest-derived cells does not affect phenotypically, embryonic development. However, in these same mice, Hand2 haploinsufficiency, due to deletion in the ventrolateral branchial arch, shows novel defects in craniofacial neural crest-derived structures (Yanagisawa et al., 2003; Barbosa et al., 2007). We observe clear defects in the cardiac neural crest of Hand2fl/fl;Wnt1-Cre embryos, where Hand1 is coexpressed, suggesting that Hand2 function would be dominant to that of Hand1; these closely related transcriptional regulators do not appear to have redundant functions related to migration or differentiation of cardiac neural crest. Our data suggest that Hand1 and Hand2 have unique functions in the cardiac neural crest.
The defect we observe in right ventricle development in the Hand2 hypomorph embryos, disorganized trabeculations and thin ventricular wall, was predictable given that Hand1 expression is low (albeit undetectable) within the right ventricle. The phenotypic characteristics we describe in both Hand2fl/fl;Wnt1-Cre and Hand2flneo/flneo embryos supports previous data suggesting that Hand2 protein function is dosage dependent (McFadden et al., 2005; Firulli et al., 2005; Hendershot et al., 2008). The phenotype observed in the Hand2 hypomorph embryos compared to loss of Hand2 in neural crest-derived cardiac structures underscores the complex functional interactions that the Twist family of bHLH proteins exhibits in-vivo (Firulli et al, 2003; Firulli et al., 2005; Barnes and Firulli, 2009); our data indicate a complex pattern of interaction between different cell types that express Hand2. The nature of the defects we observe in both the Hand2 hypomorph embryos and Hand2 neural crest-specific knockout embryos attests to the utility of our genetic model. Because of the acknowledged importance of gene dosage in Hand factor function, the Hand2 hypomorph allele will provide a valuable tool for future examination of phenotypic response to misexpression of Hand2 as well as genetic and functional interaction of Hand2 with other transcriptional regulators.
Outflow Tract Defects
In the mouse, formation of the septum and valves initiates between E9.5 and E10 (Webb et al., 1998; Jiang et al., 2000). Cells in the OFT undergo an EMT and migrate into the cardiac jelly (Song et al., 2000; Camenisch et al., 2002; Gitler et al., 2003; Person et al., 2005); these cells derive from the neural crest and the endocardium (Runyan and Markwald, 1983; Camenisch et al., 2000; Kisanuki et al., 2001). In conjunction with cells located in the AV canal (endocardial origin), neural crest-derived cells will form the membranous septum and valves (Runyan and Markwald, 1983, Poelmann et al., 1998; De Lange et al., 2004). Formation of the aorticopulmonary septation complex involves neural crest-derived cells and non-neural crest-derived cells in the conotruncal ridges (Waldo et al., 1998). Neural crest-derived cells form the prongs of the aorticopulmonary septum at around E9.5 (Waldo et al., 1998; Waldo et al., 1999; Kisanuki et al., 2001); these prongs of cells eventually form parts of both the aorta and pulmonary structures, valves and arterial smooth muscle (Waldo et al., 1998). Our results indicate that formation of the AP prongs is affected by loss of Hand2 in the neural crest-derived cells that migrate into the OFT. The reduced cell density and apparent decrease of cell-cell contact between the migrating cells is reflected in altered expression of a number of genes associated with these functions.
Formation of the OFT septum begins at around E10.5 (Jiang et al., 2002). The defects that we observe in septation and OFT vessel formation likely result from a substantial reduction in the number of neural crest-derived cells that form the AP prongs. At E9.5 neural crest-derived cells are located proximal to the aortic sac and initiating migration through it to the truncus arteriosus (Jiang et al., 2002). Later in development, the OFT will form the aortic and pulmonary OFT vessels as the aorticopulmonary septum forms at the distal pole and the conotruncal septum forms at the proximal end. The outflow valves form concurrently. Our results demonstrate that Hand2 is required for these morphogenetic events to proceed properly. As described previously (Morikawa and Cserjesi, 2008), the absence of Hand2 in cardiac neural crest causes misalignment of the aortic arch arteries and OFT. Our data suggest that cell-cell communication, mediated by gap-junctions, constitutes a critical component of signaling in the cardiac neural crest; anomalies in vessel alignment and septation appear to occur, in part, as a result of decreased expression of Cx40 in neural crest-derived cells. The reduction in Cx40 expression appears to be a direct result of Hand2 loss-of-function since our data indicate that Cx40 is a direct target of Hand2.
Cell-cell contact and intercellular signaling via gap junctions has been associated with cell cycle control (Lowenstein, 1988) and neural crest migration (Huang et al., 1998; Lo et al., 1997). Both gain-of-function of connexin 43 in the neural crest and systemic loss-of-function of connexin 43 results in defective development of the conotruncus as well as the pulmonary outflow vessel (Reaume et al., 1995; Ewart et al., 1997; Huang et al., 1998; Sullivan et al., 1998; Simon et al., 2004). It is clear from these previous studies that expression levels of connexin proteins in the neural crest is critical and contributes to development of the OFT; Cx40 has not previously been reported in the neural crest. Importantly, it appears that Hand2 directly affects expression of Cx40 in the developing OFT. It will be interesting in future studies to validate other Hand2 targets or interacting molecules identified in our mRNA screen and validated as potential direct Hand2 targets in out ChIP-on-chip screen that are involved in cell-cell contact and migration in OFT formation. Based on our gene profiling, expression of connexin 43 was not altered by deletion of Hand2 in the neural crest. We provide evidence that in the absence of Hand2, neural crest-derived cells migrating within the OFT do not migrate as a coherent sheet of cells as they normally do (Bancroft and Bellairs, 1976; Davis and Trinkaus, 1981; Raible and Eisen, 1996; Gammill et al., 2006; Kuriyama and Mayor; 2008). It is interesting to note however, that the extent of migration is not adversely affected by Hand2 deletion. Although we did not detect any notable difference in the length of the OFT in Hand2 mutant embryos, the apparent reduction in the numbers of neural crest-derived cells coupled with the decreased adhesion that we observe in the AP prongs suggests that it is aberrant remodeling affecting proper generation of the aorticopulmonary septation complex that contributes to generation of DORV and VSD.
Interestingly, in about 20% of the hearts that we examined, we found neural crest-derived cells that had tracked incorrectly and migrated into the ventricles. Although we did not observe any apparent effect of these mis-located neural crest-derived cells on ventricle development, abnormal ventricle development has been reported previously using an independently derived line of floxed Hand2 mice (Morikawa and Cserjesi, 2008). These authors concluded that loss-of-Hand2 function in cardiac neural crest induced proliferation of cardiomyocytes resulting in enlarged right ventricles (Morikawa and Cserjesi, 2008). This was an intriguing finding because neural crest-derived cells do not normally contribute to myocardial development although expression of Hand2 in cardiomyocytes is necessary for right ventricle development (Srivastava, 1997). The hypertrabeculations observed in these mutant hearts is reminiscent of what we observe in our Hand2 hypomorphic embryos. It is likely that this reported aberrant right ventricle development was in fact a primary hypomorphic phenotype within the cardiomyocytes, as this group observed hypomorphic phenotypes from their floxed Hand2 mice (Morikawa et al., 2007); in these embryos, although targeted Hand2 deletion in the neural crest would give rise to the expected defects in OFT development, misexpression of Hand2 in the right venrtricle would be expected to affect trabeculations and cell cycle.
Neural Crest Cell Cycle Regulation
In the peripheral nervous system Hand2 is required for neurogenesis and neurotransmitter specification and expression in noradrenergic sympathetic ganglion neurons (Howard et al., 1999; Howard et al., 2000; Wu and Howard, 2002; Xu et al., 2003; Hendershot et al., 2007; Hendershot et al., 2008; Morikawa et al., 2008; Schmidt et al., 2009). One important function of Hand2 is in cell cycle regulation; in the absence of Hand2 neural precursor cells cease to divide (Hendershot et al., 2008). We identified a number of cell cycle and proliferation genes regulated downstream of Hand2 in the heart. The number of cells that populate the OFT is reduced following targeted deletion of Hand2 in the neural crest; this could occur as a result of aberrant migration, decreased proliferation and/or abnormal cell cycle regulation; gene profiling suggests that all of these events are affected in the mutant hearts. The appropriate number of cells in the AP prongs of neural crest-derived cells that move into the truncal cushions is required for elongation of the conotruncus and septation into the aortic and pulmonary trunks. The abnormal pattern of migration, decreased cell numbers and loss of cell-cell signaling likely contribute to the formation of VSDs observed in all mutant embryos. Normal closure of the membranous septum involves three coordinated morphogenetic events: the muscular septum must grow upwards, the endocardial cushions must form the inlet septum (AV canal) and the formation of outlet septum by growth of the conotruncal ridges must occur (Webb et al., 1998; Lamers and Moorman, 2002; Raid et al., 2009). As part of this fusion, the valves are also formed. We observed in 100% of mutant embryos DORV that may have developed as a consequence of the development of VSD (Yelbuz et al., 2002; Xu et al., 2004) or due to the misalignment of the great vessels (described in Morikawa and Cserjesi, 2008) observed in embryos with neural crest targeted deletions of Hand2. Abnormal development of the cardiac cushions could contribute to the generation of these defects. Mis-regulation of transit through the cell cycle, resulting in a decreased number of neural crest-derived cells populating the AP prongs likely affects truncal cushion development and septation. Based on the profile of Hand2-trancriptioanl targets and the array of proliferation/cell cycle genes modulated by loss of Hand2, we suggest that the cell cycle is arrested/delayed in G1 or S phase, a situation that would account for a reduction in cells while maintaining a stable proportion of cells in M phase.
Cardiac Cushion and Valve Development
Proper patterning and differentiation of the cardiac cushions is required for formation of the semilunar valves and aorticopulomnary septation. In order for the cardiac cushions to develop extracellular matrix must be deposited into the acellular cardiac jelly. In order for valves to form, EMT must occur to remodel the cushions (Wang et al., 2004). Genes associated with transcriptional control (Nf-ATc; Foxc1, Foxp1), extracellular matrix composition (collagen XI a1; Ltbp1) and protein modifying enzymes (metalloproteases), all associated with aspects of cardiac cushion development, were affected by deletion of Hand2; a number of these genes are likely direct Hand2 targets. The cardiac phenotype of mouse embryos deficient in Nf-ATc, FoxP1, and Sox11 show nearly identical defects resulting from problems in cardiac cushion development (De la Pompa, 1998, Ranger et al., 1998; Kume et al., 2001; Wang et al., 2004; Sock et al., 2004). Previously none of these genes has been associated with Hand2. Hand2 is a BMP-downstream target (Howard et al., 2000; Liu et al., 2005b). BMP is an important signaling molecule regulating aspects of OFT development including cushion transition and valvulogenesis (reviewed in Délot, 2003; Jia et al., 2007).
Generation of VSD and DORV
Conditional targeted deletion of Hand2 in the neural crest resulted in DORV with associated VSD with 100% penetrance. Interestingly, neither DORV nor VSD is reported in a similar line of mice where Hand2 was conditionally deleted in the neural crest (Morikawa and Cserjesi, 2008). Although this could reflect phenotypic differences between these two Hand2 conditional alleles, our examination of the mice generated in the Cserjesi laboratory (kindly provided by Peter Cserjesi) demonstrated that these mice also present DORV and associated VSD (Firulli and Vincentz, unpublished observations). Taken together, studies reporting on OFT development (current paper) and aortic arch development (Morikawa and Cserjesi, 2008) define fairly completely consequences on OFT and aortic arch artery development due to targeted deletion of Hand2. Additionally, we identified a number of Hand2-target genes correlated with or identified as susceptibility loci for VSD and/or DORV (Zhang et al., 2006). We identified Gli3 as a direct Hand2 target and whose expression is significantly reduced as a consequence to loss of Hand2 in cardiac neural crest. The decreased expression of Gli3 is of interest as Gli3 has been identified as a susceptibility locus for VSD (Qui et al., 2006) and is associated with Hand2 expression and function in the developing limb bud (te Welscher et al., 2002a; te Welscher et al., 2002b; Liu et al., 2005a; Barnes and Firulli, 2009). Gli3 represses sonic hedgehog (SHH), a signaling molecule associated with second heart field development but not associated with OFT morphogenesis (Dyer and Kirby, 2009). Sox11 is another gene associated with DORV and VSD (Sock et al., 2004) that we identified as a direct Hand2 target and which is downregulated in the neural crest in response to deletion of Hand2. A number of genes regulated in response to deletion of Hand2, which we showed to be direct Hand2 targets and regulated in the neural crest, (Hey1, Foxc1, Mmp14) are related to the Notch signaling pathway. Notch signaling is implicated in migration and development of the cardiac cushions. Expression of Hand2 in neural crest-derived neural precursor cells down-regulates expression of Hes1 (Howard unpublished result); Hey 1 is regulated in the cardiac neural crest. This intriguing result raises the possibility of identifying networks of Hand2 target genes that affect cell cycle and cell-cell contact in a number of differentiating cell types but which together mediate morphogenetic movements, proliferation and differentiation.
The constellation of birth defects associated with disrupted development of the OFT appears to represent a common phenotype susceptible to multiple sites of regulation. The results of our gene profiling have identified Hand2 as a critical transcriptional regulatory factor that directly or indirectly impacts expression of genes associated with all aspects of OFT development. Given the complexity of OFT morphogenesis and the role Hand2 plays in multiple cell lineages that contribute to its formation, our studies add insight into the cell autonomous neural crest contributions to OFT morphogenesis.
Supplementary Material
Acknowledgments
Grant Sponsor: National Institutes of Health DK067064, NS040644 (to MJH); IP30-AG13319; 9730181N (to ABF)
Acknowledgements: The authors would like to thank Janet Lambert, Jennifer Duerr and Sean Eringer for excellent technical assistance. We thank Dr. Phyllis Pugh for her assistance with the initial analysis of the gene microarray data. We appreciate the invaluable assistance of Dr. Andrea Kalinoski with confocal microscopy. All confocal images were acquired in the Marci Kaptor Advanced Imaging Center at the University of Toledo. We would like to thank Dr. Benoit Bruneau for the Cx40-pX2P reporter construct. We thank Simon Conway for helpful comments and suggestions.
Footnotes
The Medical University of Ohio changed its name to the Health Sciences Campus, University of Toledo following a merger in 2007. 419-383-5438 (Tel.), 419-383-3008 (Fax)
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bancroft M, Bellairs R. The development of the notochord in the chick embryo, studied by scanning and transmission electron microscopy. J Embryol Exp Morphol. 1976;35:383–401. [PubMed] [Google Scholar]
- Barbosa AC, Funato N, Chapman S, McKee MD, Richardson JA, Olson EN, Yanagisawa H. Hand transcription factors cooperatively regulate development of the distal midline mesenchyme. Dev Biol. 2007;310:154–168. doi: 10.1016/j.ydbio.2007.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes RM, Firulli AB. A twist of insight-the role of Twist-family bHLH factors in development. Int J Dev Biol. 2009 doi: 10.1387/ijdb.082747rb. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett JV, Desgrosellier JS. Early events in valvulogenesis: a signaling perspective. Birth Defects Res C Embryo Today. 2003;69:58–72. doi: 10.1002/bdrc.10006. [DOI] [PubMed] [Google Scholar]
- Bedard JE, Purnell JD, Ware SM. Nuclear import and export signals are essential for proper cellular trafficking and function of ZIC3. Hum Mol Genet. 2007;16:187–198. doi: 10.1093/hmg/ddl461. [DOI] [PubMed] [Google Scholar]
- Bhattacharya S, MacDonald ST, Farthing CR. Molecular mechanisms controlling the coupled development of myocardium and coronary vasculature. Clin Sci. 2006;111:35–46. doi: 10.1042/CS20060003. [DOI] [PubMed] [Google Scholar]
- Brewer S, Feng W, Huang J, Sullivan S, Williams T. Wnt1-Cre-mediated deletion of AP-2alpha causes multiple neural crest-related defects. Dev Biol. 2004;267:135–152. doi: 10.1016/j.ydbio.2003.10.039. [DOI] [PubMed] [Google Scholar]
- Brooks PC, Clark RA, Cheresh DA. Requirement of vascular integrin alpha v beta 3 for angiogenesis. Science. 1994;264:569–571. doi: 10.1126/science.7512751. [DOI] [PubMed] [Google Scholar]
- Brou C, Logeat F, Gupta N, Bessia C, LeBail O, Doedens JR, Cumano A, Roux P, Black RA, Israel A. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol Cell. 2000;5:207–216. doi: 10.1016/s1097-2765(00)80417-7. [DOI] [PubMed] [Google Scholar]
- Bruneau BG, Nemer G, Schmitt JP, Charron F, Robitaille L, Caron S, Conner DA, Gessler M, Nemer M, Seidman CE, Seidman JG. A murine model of the Holt-Oram syndrome defines role of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell. 2001;106:709–721. doi: 10.1016/s0092-8674(01)00493-7. [DOI] [PubMed] [Google Scholar]
- Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, Evans S. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5:877–889. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camenisch TD, Spicer AP, Brehm-Gibson T, Biesterfeldt J, Augustine ML, Calabro A, Jr, Kubalak S, Klewer SE, McDonald JA. Disruption of hyaluronan synthase-2 abrogates normal cardiac morphogenesis and hyaluronan-mediated transformation of epithelium to mesenchyme. J Clin Invest. 2000;106:349–360. doi: 10.1172/JCI10272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camenisch TD, Molin DG, Person A, Runyan RB, Gittenberger-de Groot AC, McDonald JA, Klewer SE. Temporal and distinct TGFbeta ligand requirements during mouse and avian endocardial cushion morphogenesis. Dev Biol. 2002;248:170–181. doi: 10.1006/dbio.2002.0731. [DOI] [PubMed] [Google Scholar]
- Conway SJ, Henderson DJ, Copp AJ. Pax3 is required for cardiac neural crest migration in the mouse: evidence from the splotch (Sp2H) mutant. Development. 1997;124:505–514. doi: 10.1242/dev.124.2.505. [DOI] [PubMed] [Google Scholar]
- Conway SJ, Bundy J, Chen J, Dickman E, Rogers R, Will BM. Decreased neural crest stem cell expansion is responsible for the conotruncal heart defects within the splotch (Sp(2H))/Pax3 mouse mutant. Cardiovasc Res. 2000;47:314–328. doi: 10.1016/s0008-6363(00)00098-5. [DOI] [PubMed] [Google Scholar]
- Dahl E, Winterhager E, Traub O, Willecke K. Expression of gap junction genes, connexin40 and connexin43, during fetal mouse development. Anat Embryol (Berl) 1995;191:267–278. doi: 10.1007/BF00187825. [DOI] [PubMed] [Google Scholar]
- Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol. 1998;8:1323–1326. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- Davis EM, Trinkaus JP. Significance of cell-to cell contacts for the directional movement of neural crest cells within a hydrated collagen lattice. J Embryol Exp Morphol. 1981;63:29–51. [PubMed] [Google Scholar]
- de la Pompa JL, Timmerman LA, Takimoto H, Yoshida H, Elia AJ, Samper E, Potter J, Wakeham A, Marengere L, Langille BL, Crabtree GR, Mak TW. Role of the NF-ATc transcription factor in morphogenesis of cardiac valves and septum. Nature. 1998;392:182–186. doi: 10.1038/32419. [DOI] [PubMed] [Google Scholar]
- de Lange FJ, Moorman AF, Anderson RH, Manner J, Soufan AT, Gier-de Vries C, Schneider MD, Webb S, van den Hoff MJ, Christoffels VM. Lineage and morphogenetic analysis of the cardiac valves. Circ Res. 2004;95:645–654. doi: 10.1161/01.RES.0000141429.13560.cb. [DOI] [PubMed] [Google Scholar]
- Délot EC. Control of endocardial cushion and cardiac valve maturation by BMP signaling pathways. Mol Gen and Metab. 2003;80:27–35. doi: 10.1016/j.ymgme.2003.07.004. [DOI] [PubMed] [Google Scholar]
- Dodou E, Verzi MP, Anderson JP, Xu SM, Black BL. Mef2c is a direct transcriptional target of ISL1 and GATA factors in the anterior heart field during mouse embryonic development. Development. 2004;131:3931–3942. doi: 10.1242/dev.01256. [DOI] [PubMed] [Google Scholar]
- Dyer LA, Kirby ML. Sonic hedgehog maintains proliferation in secondary heart field progenitors and is required for normal arterial pole formation. Dev Biol. 2009 doi: 10.1016/j.ydbio.2009.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein JA, Li J, Lang D, Chen F, Brown CB, Jin F, Lu MM, Thomas M, Liu E, Wessels A, Lo CW. Migration of cardiac neural crest cells in Splotch embryos. Development. 2000;127:1869–1878. doi: 10.1242/dev.127.9.1869. [DOI] [PubMed] [Google Scholar]
- Ewart JL, Cohen MF, Meyer RA, Huang GY, Wessels A, Gourdie RG, Chin AJ, Park SM, Lazatin BO, Villabon S, Lo CW. Heart and neural tube defects in transgenic mice overexpressing the Cx43 gap junction gene. Development. 1997;124:1281–1292. doi: 10.1242/dev.124.7.1281. [DOI] [PubMed] [Google Scholar]
- Ewen ME. Where the cell cycle and histones meet. Genes and Dev. 2009;14:2265–2270. doi: 10.1101/gad.842100. [DOI] [PubMed] [Google Scholar]
- Firulli AB. A Handful of questions: the molecular biology of the heart and neural crest derivatives (Hand)-subclass of basic helix-loop-helix transcription factors. Gene. 2003;312:27–40. doi: 10.1016/s0378-1119(03)00669-3. [DOI] [PubMed] [Google Scholar]
- Firulli AB, Thattaliyath BD. Transcription factors in cardiogenesis: the combinations that unlock the mysteries of the heart. Int Rev Cytol. 2002;214:1–62. doi: 10.1016/s0074-7696(02)14002-2. [DOI] [PubMed] [Google Scholar]
- Firulli BA, Howard MJ, McDaid JR, McIlreavey L, Dionne KM, Centonze VE, Cserjesi P, Virshup DM, Firulli AB. PKA, PKC, and the protein phosphatase 2A influence HAND factor function: a mechanism for tissue-specific transcriptional regulation. Mol Cell. 2003;12:1225–1237. doi: 10.1016/s1097-2765(03)00425-8. [DOI] [PubMed] [Google Scholar]
- Firulli BA, Krawchuk D, Centonze VE, Vargesson N, Virshup DM, Conway SJ, Cserjesi P, Laufer E, Firulli AB. Altered Twist1 and Hand2 dimerization is associated with Saethre-Chotzen syndrome and limb abnormalities. Nat Genet. 2005;37:373–381. doi: 10.1038/ng1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funato N, Chapman SL, McKee MD, Funato H, Morris JA, Shelton JM, Richardson JA, Yanagisawa H. Hand2 controls osteoblast differentiation in the branchial arch by inhibiting DNA binding of Runx2. Develop. 2009;136:615–625. doi: 10.1242/dev.029355. [DOI] [PubMed] [Google Scholar]
- Fischer A, Schumacher N, Maier M, Sendtner M, Gessler M. The Notch target genes Hey1 and Hey2 are required for embryonic vascular development. Genes Dev. 2004;18:901–911. doi: 10.1101/gad.291004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammill LS, Gonzalez C, Gu C, Bronner-Fraser M. Guidance of trunk neural crest migration requires neuropilin 2/semaphorin 3F signaling. Development. 2006;133:99–106. doi: 10.1242/dev.02187. [DOI] [PubMed] [Google Scholar]
- Garry DJ, Olson EN. A common progenitor at the heart of development. Cell. 2006;127:1101–1104. doi: 10.1016/j.cell.2006.11.031. [DOI] [PubMed] [Google Scholar]
- Gitler AD, Lu MM, Jiang YQ, Epstein JA, Gruber PJ. Molecular markers of cardiac endocardial cushion development. Dev Dyn. 2003;228:643–650. doi: 10.1002/dvdy.10418. [DOI] [PubMed] [Google Scholar]
- Goodwin RL, Nesbitt T, Price RL, Wells JC, Yost MJ, Potts JD. Three-dimensional model system of valvulogenesis. Dev Dyn. 2005;233:122–129. doi: 10.1002/dvdy.20326. [DOI] [PubMed] [Google Scholar]
- Hendershot TJ, Liu H, Sarkar AA, Giovannucci DR, Clouthier DE, Abe M, Howard MJ. Expression of Hand2 is sufficient for neurogenesis and cell type-specific gene expression in the enteric nervous system. Dev Dyn. 2007;236:93–105. doi: 10.1002/dvdy.20989. [DOI] [PubMed] [Google Scholar]
- Hendershot TJ, Liu H, Clouthier DE, Shepherd IT, Coppola E, Studer M, Firulli AB, Pittman DL, Howard MJ. Conditional deletion of Hand2 reveals critical functions in neurogenesis and cell type-specific gene expression for development of neural crest-derived noradrenergic sympathetic ganglion neurons. Dev Biol. 2008;319:179–191. doi: 10.1016/j.ydbio.2008.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi K, Zhou HM, Kelly K, Manova K, Blobel CP. Evaluation of the contributions of ADAMs 9, 12, 15, 17, and 19 to heart development and ectodomain shedding of neuregulins beta1 and beta2. Dev Biol. 2005;283:459–471. doi: 10.1016/j.ydbio.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Howard MJ, Foster DN, Cserjesi P. Expression of HAND gene products may be sufficient for the differentiation of avian neural crest-derived cells into catecholaminergic neurons in culture. Dev Biol. 1999;215:62–77. doi: 10.1006/dbio.1999.9450. [DOI] [PubMed] [Google Scholar]
- Howard MJ, Stanke M, Schneider C, Wu X, Rohrer H. The transcription factor dHAND is a downstream effector of BMPs in sympathetic neuron specification. Development. 2000;127(18):4073–4081. doi: 10.1242/dev.127.18.4073. [DOI] [PubMed] [Google Scholar]
- Howard MJ. Mechanisms and perspectives on differentiation of autonomic neurons. Dev Biol. 2005;277:271–286. doi: 10.1016/j.ydbio.2004.09.034. [DOI] [PubMed] [Google Scholar]
- Huang GY, Cooper ES, Waldo K, Kirby ML, Gilula NB, Lo CW. Gap junction-mediated cell-cell communication modulates mouse neural crest migration. J Cell Biol. 1998;143:1725–1734. doi: 10.1083/jcb.143.6.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Bridges LC, White JM. Selective modulation of integrin-mediated cell migration by distinct ADAM family members. Mol Biol Cell. 2005;16:4982–4991. doi: 10.1091/mbc.E05-03-0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutson MR, Kirby ML. Model systems for the study of heart development and disease. Cardiac neural crest and conotruncal malformations. Semin Cell Dev Biol. 2007;18:101–110. doi: 10.1016/j.semcdb.2006.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger S, Barends S, Giege R, Eriani G, Martin F. Expression of metazoan replication-dependent histone genes. Biochimie. 2005;87:827–834. doi: 10.1016/j.biochi.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Jia Q, McDIll BW, Li SZ, Deng C, Chang CP, Chen F. Smad signaling in the neural crest regulates cardiac outflow tract remodeling through cell autonomous and non-cell autonomous effects. Dev Biol. 2007;311:172–184. doi: 10.1016/j.ydbio.2007.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cardiac neural crest. Development. 2000;127:1607–1616. doi: 10.1242/dev.127.8.1607. [DOI] [PubMed] [Google Scholar]
- Jiang X, Choudhary B, Merki E, Chien KR, Maxson RE, Sucov HM. Normal fate and altered function of the cardiac neural crest cell lineage in retinoic acid receptor mutant embryos. Mech Dev. 2002;117:115–122. doi: 10.1016/s0925-4773(02)00206-x. [DOI] [PubMed] [Google Scholar]
- Kaartinen V, Dudas M, Nagy A, Sridurongrit S, Lu MM, Epstein JA. Cardiac outflow tract defects in mice lacking ALK2 in neural crest cells. Development. 2004;131:3481–3490. doi: 10.1242/dev.01214. [DOI] [PubMed] [Google Scholar]
- Kaufman CK, Zhou P, Pasolli HA, Rendl M, Bolotin D, Lim KC, Dai X, Alegre ML, Fuchs E. GATA-3: an unexpected regulator of cell lineage determination in skin. Genes Dev. 2003;17:2108–2122. doi: 10.1101/gad.1115203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly RG, Buckingham ME. The anterior heart-forming field: voyage to the arterial pole of the heart. Trends Genet. 2002;18:210–216. doi: 10.1016/s0168-9525(02)02642-2. [DOI] [PubMed] [Google Scholar]
- Kim RY, Robertson EJ, Solloway MJ. Bmp6 and Bmp7 are required for cushion formation and septation in the developing mouse heart. Dev Biol. 2001;235:449–466. doi: 10.1006/dbio.2001.0284. [DOI] [PubMed] [Google Scholar]
- Kirchhoff S, Kim JS, Hagendorff A, Thonnissen E, Kruger O, Lamers WH, Willecke K. Abnormal cardiac conduction and morphogenesis in connexin40 and connexin43 double deficient mice. Circ Res. 2000;87:399–405. doi: 10.1161/01.res.87.5.399. [DOI] [PubMed] [Google Scholar]
- Kisanuki YY, Hammer RE, Miyazaki J, Williams SC, Richardson JA, Yanagisawa M. Tie2-Cre transgenic mice: a new model for endothelial cell-lineage analysis in vivo. Dev Biol. 2001;230:230–242. doi: 10.1006/dbio.2000.0106. [DOI] [PubMed] [Google Scholar]
- Komatsu K, Wakatsuki S, Yamada S, Yamamura K, Miyazaki J, Sehara-Fujisawa A. Meltrin beta expressed in cardiac neural crest cells is required for ventricular septum formation of the heart. Dev Biol. 2007;303:82–92. doi: 10.1016/j.ydbio.2006.10.037. [DOI] [PubMed] [Google Scholar]
- Kume T, Jiang H, Topczewska JM, Hogan BL. The murine winged helix transcription factors, Foxc1 and Foxc2, are both required for cardiovascular development and somitogenesis. Genes Dev. 2001;15:2470–2482. doi: 10.1101/gad.907301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriyama S, Mayor R. Molecular analysis of neural crest migration. Philos Trans R Soc Lond B Biol Sci. 2008;363(1495):1349–1362. doi: 10.1098/rstb.2007.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers WH, Mooreman AF. Cardiac septation: a late contribution of the embryonic primary myocardium to heart morphogenesis. Circ Res. 2002;91:93–103. doi: 10.1161/01.res.0000027135.63141.89. [DOI] [PubMed] [Google Scholar]
- Lan MS, Breslin MB. Structure, expression, and biological function of INSM1 transcription factor in neuroendocrine differentiation. FASEB J. 2009 doi: 10.1096/fj.08-125971. fj.08-125971. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lie-Venema H, van den Akker NM, Bax NA, Winter EM, Maas S, Kekarainen T, Hoeben RC, deRuiter MC, Poelmann RE, Gittenberger-de Groot AC. Origin, fate, and function of epicardium-derived cells (EPDCs) in normal and abnormal cardiac development. Scientific World Journal. 2007;12:1777–1798. doi: 10.1100/tsw.2007.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KC, Lakshmanan G, Crawford SE, Gu Y, Grosveld F, Engel JD. Gata3 loss leads to embryonic lethality due to noradrenaline deficiency of the sympathetic nervous system. Nat Genet. 2000;25:209–212. doi: 10.1038/76080. [DOI] [PubMed] [Google Scholar]
- Liu A, Wang B, Niswander LA. Mouse intraflagellar transport proteins regulate both the activator and repressor functions of Gli transcription factors. Development. 2005a;132:3103–3111. doi: 10.1242/dev.01894. [DOI] [PubMed] [Google Scholar]
- Liu H, Margiotta JF, Howard MJ. BMP4 supports noradrenergic differentiation by a PKA-dependent mechanism. Dev Biol. 2005b;286:521–536. doi: 10.1016/j.ydbio.2005.08.022. [DOI] [PubMed] [Google Scholar]
- Liu WD, Wang HW, Muguira M, Breslin MB, Law MS. INSM1 functions as a transcriptional repressor of the neuroD/b2 gene through the recruitment of cyclin D1 and histone deacetylases. Biochem J. 2006;397:169–177. doi: 10.1042/BJ20051669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo L, Sommer L, Anderson DJ. MASH1 maintains competence for BMP2-induced neuronal differentiation in post-migratory neural crest cells. Curr Biol. 1997;7:440–450. doi: 10.1016/s0960-9822(06)00191-6. [DOI] [PubMed] [Google Scholar]
- Lowenstein WR. Regulation of cell-to-cell communication by phosphorylation. Biochem Soc Symp. 1985;50:43–58. [PubMed] [Google Scholar]
- Lowenstein WR. Genetic regulation of cell-to-cell communication. Braz J Med Biol Res. 1988;21:1213–1223. [PubMed] [Google Scholar]
- Maeda J, Yamagishi H, McAnally J, Yamagishi C, Srivastava D. Tbx1 is regulated by forkhead proteins in the secondary heart field. Dev Dyn. 2006;235:701–710. doi: 10.1002/dvdy.20686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malumbres M, Barbacid M. Mammalian cyclin-dependent kinases. Trends Biochem Sci. 2005;30:630–641. doi: 10.1016/j.tibs.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Marzluff WF, Gongidi P, Woods KR, Jin J, Maltais LJ. The human and mouse replication-dependent histone genes. Genomics. 2002;80:487–498. [PubMed] [Google Scholar]
- McFadden DG, Charite J, Richardson JA, Srivastava D, Firulli AB, Olson EN. A GATA-dependent right ventricular enhancer controls dHAND transcription in the developing heart. Development. 2000;127:5331–5341. doi: 10.1242/dev.127.24.5331. [DOI] [PubMed] [Google Scholar]
- McFadden DG, Barbosa AC, Richardson JA, Schneider MD, Srivastava D, Olson EN. The Hand1 and Hand2 transcription factors regulate expansion of the embryonic cardiac ventricles in a gene dosage-dependent manner. Development. 2005;132:189–201. doi: 10.1242/dev.01562. [DOI] [PubMed] [Google Scholar]
- Mégarbané A, Salem N, Stephan E, Ashooush R, Lenoir D, Delague V, Kassab R, Loiselet J, Bouvagnet P. X-linked transposition of the great arteries and incomplete penetrance among males with a nonsense mutation in ZIC3. Eur J Hum Gen. 2000;8:704–708. doi: 10.1038/sj.ejhg.5200526. [DOI] [PubMed] [Google Scholar]
- Mitchell ME, Sander TL, Klinker DB, Tomita-Mitchell A. The molecular basis of congenital heart disease. Semin Thorac Cardiovasc Surg. 2007;19:228–37. doi: 10.1053/j.semtcvs.2007.07.013. [DOI] [PubMed] [Google Scholar]
- Morikawa Y, D’Autreaux F, Gershon MD, Cserjesi P. Hand2 determines the noradrenergic phenotype in the mouse sympathetic nervous system. Dev Biol. 2007;307:114–126. doi: 10.1016/j.ydbio.2007.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa Y, Cserjesi P. Cardiac neural crest expression of Hand2 regulates outflow and second heart field development. Circ Res. 2008;103:1422–1429. doi: 10.1161/CIRCRESAHA.108.180083. [DOI] [PubMed] [Google Scholar]
- Morrison-Graham K, Schatteman GC, Bork T, Bowen-Pope DF, Weston JA. A PDGF receptor mutation in the mouse (Patch) perturbs the development of a non-neuronal subset of neural crest-derived cells. Development. 1992;115:133–142. doi: 10.1242/dev.115.1.133. [DOI] [PubMed] [Google Scholar]
- Niessen K, Karsan A. Notch signaling in cardiac development. Circ Res. 2008;102:1169–1181. doi: 10.1161/CIRCRESAHA.108.174318. [DOI] [PubMed] [Google Scholar]
- Obler D, Juraszek AL, Smoot LB, Natowicz MR. Double outlet right ventricle: aetiologies and associations. J Med Genet. 2008;45:481–97. doi: 10.1136/jmg.2008.057984. [DOI] [PubMed] [Google Scholar]
- Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313:1922–7. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Urtreger A, Lonai P. Platelet-derived growth factor-A and its receptor are expressed in separate, but adjacent cell layers of the mouse embryo. Development. 1992;115:1045–1058. doi: 10.1242/dev.115.4.1045. [DOI] [PubMed] [Google Scholar]
- Parlier D, Ariza A, Christulia F, Geneco F, Vanhomwegen J, Kricha S, Souopgui J, Bellefroid EJ. Xenopus zinc finger transcription factor IA1 (Insm1) expression marks anteroventral noradrenergic neuron progenitors in Xenopus embryos. Dev Dyn. 2008;237:2147–2157. doi: 10.1002/dvdy.21621. [DOI] [PubMed] [Google Scholar]
- Person AD, Klewer SE, Runyan RB. Cell biology of cardiac cushion development. Int Rev Cytol. 2005;243:287–335. doi: 10.1016/S0074-7696(05)43005-3. [DOI] [PubMed] [Google Scholar]
- Pinco KA, Liu S, Yang JT. Alpha4 integrin is expressed in a subset of cranial neural crest cells and in epicardial progenitor cells during early mouse development. Mech Dev. 2001;100:99–103. doi: 10.1016/s0925-4773(00)00503-7. [DOI] [PubMed] [Google Scholar]
- Poelmann RE, Mikawa T, Gittenberger-de Groot AC. Neural crest cells in outflow tract septation of the embryonic chicken heart: differentiation and apoptosis. Dev Dyn. 1998;212:373–384. doi: 10.1002/(SICI)1097-0177(199807)212:3<373::AID-AJA5>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Qui GR, Gong LG, He G, Xu XY, Xin N, Sun GF, Yuan YH, Sun KL. Association of the GLI gene with ventricular septal defect after the susceptibility gene being narrowed to 3.56 cM in 12q13. Chin Med J (Engl) 2006;119:267–274. [PubMed] [Google Scholar]
- Raible DW, Eisen JS. Regulative interactions in zebrafish neural crest. Development. 1996;122:501–507. doi: 10.1242/dev.122.2.501. [DOI] [PubMed] [Google Scholar]
- Raid R, Krinka D, Bakhoff L, Abdelwahid E, Jokin E, Kärner M, Malva M, Meier R, Pelliniemi LJ, Ploom M, Sizarov A, Pooga M, Karis A. Lack of Gata3 results in conotruncal heart anomalies in mouse. Mech Dev. 2009;126:80–89. doi: 10.1016/j.mod.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Ranger AM, Grusby MJ, Hodge MR, Gravallese EM, Charles de la Brousse F, Hoey T, Mickanin C, Baldwin HS, Glimcher LH. The transcription factor NF-ATc is essential for cardiac valve formation. Nature. 1998;392:186–190. doi: 10.1038/32426. [DOI] [PubMed] [Google Scholar]
- Reaume AG, de Sousa PA, Kulkarni S, Langille BL, Zhu D, Davies TC, Juneja SC, Kidder GM, Rossant J. Cardiac malformation in neonatal mice lacking connexin43. Science. 1995;267:1831–1834. doi: 10.1126/science.7892609. [DOI] [PubMed] [Google Scholar]
- Runyan RB, Markwald RR. Invasion of mesenchyme into three-dimensional collagen gels: a regional and temporal analysis of interaction in embryonic heart tissue. Dev Biol. 1983;95:108–114. doi: 10.1016/0012-1606(83)90010-6. [DOI] [PubMed] [Google Scholar]
- Sakata Y, Kamei CN, Nakagami H, Bronson R, Liao JK, Chin MT. Ventricular septal defect and cardiomyopathy in mice lacking the transcription factor CHF1/Hey2. Proc Natl, Acad Sci. 2002;99:16197–16202. doi: 10.1073/pnas.252648999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson J. Biological Microtechnique (Royal Microscopical Society Microscopy Handbooks) London: Garland Science; 1994. pp. 163–170. [Google Scholar]
- Sanford LP, Ormsby I, Gittenberger-de Groot AC, Sariola H, Friedman R, Boivin GP, Cardell EL, Doetschman T. TGFbeta2 knockout mice have multiple developmental defects that are non-overlapping with other TGFbeta knockout phenotypes. Development. 1997;124:2659–2670. doi: 10.1242/dev.124.13.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M, Lin S, Pape M, Ernsberger U, Howard MJ, Rohrer H. The bHLH transcription factor Hand2 is essential for the maintenance of noradrenergic properties in differentiated sympathetic neurons. Dev Biol. 2009;329:191–200. doi: 10.1016/j.ydbio.2009.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S, Fujita H, Nakano A, Kang M, Duarte A, Kume T. The forkhead transcription factors, Foxc1 and Foxc2, are required for arterial specification and lymphatic sprouting during vascular development. Dev Biol. 2006;294:458–470. doi: 10.1016/j.ydbio.2006.03.035. [DOI] [PubMed] [Google Scholar]
- Seul KH, Tadros PN, Beyer EC. Mouse connexin40: gene structure and promoter analysis. Genomics. 1997;46:120–126. doi: 10.1006/geno.1997.5025. [DOI] [PubMed] [Google Scholar]
- Sheppard AM, Onken MD, Rosen GD, Noakes PG, Dean DC. Expanding roles for alpha 4 integrin and its ligands in development. Cell Adhes Commun. 1994;2:27–43. doi: 10.3109/15419069409014200. [DOI] [PubMed] [Google Scholar]
- Simon AM, McWhorter AR, Dones JA, Jackson CL, Chen HDR. Heart and head defects in mice lacking pairs of connexins. Dev Biol. 2004;265:369–383. doi: 10.1016/j.ydbio.2003.09.036. [DOI] [PubMed] [Google Scholar]
- Singh MK, Nicolas E, Gherraby W, Dadke D, Lessin S, Golemis EA. HEI10 negatively regulates cell invasion by inhibiting cyclin B/Cdk1 and other promotility proteins. Oncogene. 2007;26:4825–4832. doi: 10.1038/sj.onc.1210282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snarr BS, Kern C, Wessels A. Origins and fate of cardiac mesenchyme. Dev Dyn. 2008;237:2804–2819. doi: 10.1002/dvdy.21725. [DOI] [PubMed] [Google Scholar]
- Snider P, Olaopa M, Firulli AB, Conway SJ. Cardiovascular development and the colonizing cardiac neural crest lineage. ScientificWorldJournal. 2007;7:1090–1113. doi: 10.1100/tsw.2007.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sock E, Rettig SD, Enderich J, Bosl MR, Tamm ER, Wegner M. Gene targeting reveals a widespread role for the high-mobility-group transcription factor Sox11 in tissue remodeling. Mol Cell Biol. 2004;24:6635–6644. doi: 10.1128/MCB.24.15.6635-6644.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Jackson K, McGuire PG. Degradation of type IV collagen by matrix metalloproteinases is an important step in the epithelial-mesenchymal transformation of the endocardial cushions. Dev Bio. 2000;222:606–617. doi: 10.1006/dbio.2000.9919. [DOI] [PubMed] [Google Scholar]
- Srivastava D. HAND proteins: molecular mediators of cardiac development and congenital heart disease. Trends Cardiovasc Med. 1999;9:11–18. doi: 10.1016/s1050-1738(98)00033-4. [DOI] [PubMed] [Google Scholar]
- Srivastava D. Making or breaking the heart: form lineage determination to morphogenesis. Cell. 2006;126:1037–1048. doi: 10.1016/j.cell.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Srivastava D, Cserjesi P, Olson EN. A subclass of bHLH proteins required for cardiac morphogenesis. Science. 1995;270:1995–1999. doi: 10.1126/science.270.5244.1995. [DOI] [PubMed] [Google Scholar]
- Srivastava D, Thomas T, Lin Q, Kirby ML, Brown D, Olson EN. Regulation of cardiac mesodermal and neural crest development by the bHLH transcription factor, dHAND. Nat Genet. 1997;16:154–160. doi: 10.1038/ng0697-154. [DOI] [PubMed] [Google Scholar]
- Srivastava D, Olson EN. A genetic blueprint for cardiac development. Nature. 2000;407:221–226. doi: 10.1038/35025190. [DOI] [PubMed] [Google Scholar]
- Stamataki D, Ulloa F, Tsoni SV, Mynett A, Briscoe J. A gradient of Gli activity mediates graded Sonic Hedgehog signaling in the neural tube. Genes Dev. 2005;19:626–641. doi: 10.1101/gad.325905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoller JZ, Epstein JA. Cardiac neural crest. Sem Cell and Dev Biol. 2005;16:704–715. doi: 10.1016/j.semcdb.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Sullivan R, Huang GY, Meyer RA, Wessels A, Linask KK, Lo CW. Heart malformations in transgenic mice exhibiting dominant negative inhibition of gap junctional communication in neural crest cells. Dev Biol. 1998;204:224–234. doi: 10.1006/dbio.1998.9089. [DOI] [PubMed] [Google Scholar]
- te Welscher P, Zuniga A, Kuijper S, Drenth T, Goedemans HJ, Meijlink F, Zeller R. Progression of vertebrate limb development through SHH-mediated counteraction of gli3. Science. 2002a;298:827–830. doi: 10.1126/science.1075620. [DOI] [PubMed] [Google Scholar]
- te Welscher P, Fernandez-Terran M, Ros MA, Zeller R. Mutual genetic antagonism involving GLI3 and dHAND prepatterns the vertebrate limb bud mesenchyme prioir to SHH signaling. Genes and Dev. 2002b;16:421–426. doi: 10.1101/gad.219202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas SA, Matsumoto AM, Palmiter RD. Noradrenaline is essential for mouse fetal development. Nature. 1995;374:643–646. doi: 10.1038/374643a0. [DOI] [PubMed] [Google Scholar]
- Thomas T, Yamagishi H, Overbeek PA, Olson EN, Srivastava D. The bHLH factors, dHAND and eHAND, specify pulmonary and systemic cardiac ventricles independent of left-right sidedness. Dev Biol. 1998;196:228–236. doi: 10.1006/dbio.1998.8849. [DOI] [PubMed] [Google Scholar]
- Toby GG, Gherraby W, Coleman TR, Golemis EA. A novel RING finger protein, human enhancer of invasion 10, alters mitotic progression through regulation of cyclin B levels. Mol Cell Biol. 2003;23:2109–2122. doi: 10.1128/MCB.23.6.2109-2122.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todorovic V, Frendewey D, Gutstein DE, Cehn Y, Freyer L, Finnegan E, Liu F, Murphy A, Valenzuela D, Yancopoilos G, Rifkin DB. Long form of latent TGF-b binding protein 1(Ltbp1L) is essential for cardiac outflow tract septation and remodeling. Development. 2007;134:3723–3732. doi: 10.1242/dev.008599. [DOI] [PubMed] [Google Scholar]
- Togi K, Yoshida Y, Matsumae H, Nakashima Y, Kita T, Tanaka M. Essential role of Hand2 in interventricular septum formation and trabeculation during cardiac development. Biochem Biophys Res Commun. 2006;343:144–151. doi: 10.1016/j.bbrc.2006.02.122. [DOI] [PubMed] [Google Scholar]
- von Both I, Silvestri C, Erdemir T, Lickert H, Walls JR, Henkelman RM, Rossant J, Harvey RP, Attisano L, Wrana JL. Foxh1 is essential for development of the anterior heart field. Dev Cell. 2004;7:331–345. doi: 10.1016/j.devcel.2004.07.023. [DOI] [PubMed] [Google Scholar]
- Waldo K, Miyagawa-Tomita S, Kumiski D, Kirby ML. Cardiac neural crest cells provide new insight into septation of the cardiac outflow tract: aortic sac to ventricular septal closure. Dev Biol. 1998;196:129–144. doi: 10.1006/dbio.1998.8860. [DOI] [PubMed] [Google Scholar]
- Waldo KL, Lo CW, Kirby ML. Connexin 43 expression reflects neural crest patterns during cardiovascular development. Dev Biol. 1999;208:307–323. doi: 10.1006/dbio.1999.9219. [DOI] [PubMed] [Google Scholar]
- Wang B, Weidenfeld J, Lu MM, Maika S, Kuziel WA, Morrisey EE, Tucker PW. Foxp1 regulates cardiac outflow tract, endocardial cushion morphogenesis and myocyte proliferation and maturation. Development. 2004;131:4477–4487. doi: 10.1242/dev.01287. [DOI] [PubMed] [Google Scholar]
- Ware SM, Harutyunyan KG, Belmont JW. Heart defects in X-linked heterotaxy: evidence for a genetic interaction of Zic3 with the nodal signaling pathway. Dev Dyn. 2006;235:1631–1637. doi: 10.1002/dvdy.20719. [DOI] [PubMed] [Google Scholar]
- Webb S, Brown NA, Anderson RH. Formation of the atrioventricular septal structures in the normal mouse. Circ Res. 1998;82:645–656. doi: 10.1161/01.res.82.6.645. [DOI] [PubMed] [Google Scholar]
- Wildner H, Gierl MS, Strehle M, Pla P, Birchmeier C. Insm1 (IA-1) is a crucial component of the transcriptional network that controls differentiation of the sympatho-adrenal lineage. Development. 2008;135:473–481. doi: 10.1242/dev.011783. [DOI] [PubMed] [Google Scholar]
- Wu X, Howard MJ. Transcripts encoding HAND genes are differentially expressed and regulated by BMP4 and GDNF in developing avian gut. Gene Expr. 2002;10:279–293. doi: 10.3727/000000002783992361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Cai T, Zhang H, Lan MS, Notkins AL. The zinc-finger transcription factor INSM1 is expressed dring embryo development and interacts with the Cbl-associated protein. Genomics. 2002;80:54–61. doi: 10.1006/geno.2002.6800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Firulli AB, Zhang X, Howard MJ. HAND2 Synergistically Enhances Transcription of Dopamine-b-hydroxylase in the Presence of Phox2a. Dev Biol. 2003;262:183–193. doi: 10.1016/s0012-1606(03)00361-0. [DOI] [PubMed] [Google Scholar]
- Xu H, Morishima M, Wylie JN, Schwartz RJ, Burneau BG, Lindsay EA, Baldini A. Tbx1 has a dual role in the morphogenesis of the cardiac outflow tract. Development. 2004;131:3217–3227. doi: 10.1242/dev.01174. [DOI] [PubMed] [Google Scholar]
- Yamagishi H, Yamagishi C, Nakagawa O, Harvey RP, Olson EN, Srivastava D. The combinatorial activities of Nkx2.5 and dHAND are essential for cardiac ventricle formation. Dev Biol. 2001;239:190–203. doi: 10.1006/dbio.2001.0417. [DOI] [PubMed] [Google Scholar]
- Yanagisawa H, Clouthier DE, Richardson JA, Charite’ J, Olson EN. Targeted deletion of a branchial arch-specific enhancer reveals a role of dHAND in craniofacial development. Development. 2003;130:1069–1078. doi: 10.1242/dev.00337. [DOI] [PubMed] [Google Scholar]
- Yang JT, Rayburn H, Hynes RO. Cell adhesion evenets mediated by a4 integrins are essential in placental and cardiac development. Development. 1995;121:549–560. doi: 10.1242/dev.121.2.549. [DOI] [PubMed] [Google Scholar]
- Yelbuz TM, Waldo KL, Kumiski DH, Stadt HA, Wolfe RR, Leatherbury L, Kirby ML. Circulation. 2002;106:504–510. doi: 10.1161/01.cir.0000023044.44974.8a. [DOI] [PubMed] [Google Scholar]
- Young BA, Taooka Y, Liu S, Askins KJ, Yokosaki Y, Thomas SM, Sheppard D. The cytoplasmic domain of the integrin alpha9 subunit requires the adaptor protein paxillin to inhibit cell spreading but promotes cell migration in a paxillin-independent manner. Mol Biol Cell. 2001;12:3214–3225. doi: 10.1091/mbc.12.10.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young HM, Ciampoli D, Hsuan J, Canty AJ. Expression of Ret-, p75(NTR)-, Phox2a-, Phox2b-, and tyrosine hydroxylase-immunoreactivity by undifferentiated neural crest-derived cells and different classes of enteric neurons in the embryonic mouse gut. Dev Dyn. 1999;216:137–152. doi: 10.1002/(SICI)1097-0177(199910)216:2<137::AID-DVDY5>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Zang MX, Li Y, Wang H, Wang JB, Jia HT. Cooperative interactions between the basic helix-loop-helix transcription factor dHand and myocyte enhancer factor 2C regulates mycoradial gene expression. J Biol Chem. 2004;279 (52):54258–54263. doi: 10.1074/jbc.M408502200. [DOI] [PubMed] [Google Scholar]
- Zhang H, Zhou L, Yang R, Sheng Y, Sum W, Kong X, Cao K. Identification of differentially expressed genes in human heart with ventricular septal defect using suppression subtractive hybridization. BBRC. 2006;342:135–144. doi: 10.1016/j.bbrc.2006.01.113. [DOI] [PubMed] [Google Scholar]
- Zhang T, Liu WD, Saunee NA, Breslin MB, Lan MS. Zinc finger transcription factor INSM1 interrupts cyclin D1 and CDK4 binding and induces cell cycle arrest. J Biol Chem. 2009;284:5574–5581. doi: 10.1074/jbc.M808843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Kennedy BK, Lawrence BD, Barbie DA, Matera AG, Fletcher JA, Harlow E. NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev. 2000;14:2283–2297. [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Cron RQ, Wu B, Genin A, Wang Z, Liu S, Robson P, Baldwin HS. Regulation of the murine Nfatc1 gene by NFATc2. J Biol Chem. 2002;277:10704–10711. doi: 10.1074/jbc.M107068200. [DOI] [PubMed] [Google Scholar]
- Zhou QY, Palmiter RD. Dopamine-deficient mice are severely hypoactive, adipsic, and aphagic. Cell. 1995;83:1197–1209. doi: 10.1016/0092-8674(95)90145-0. [DOI] [PubMed] [Google Scholar]
- Zhou X, Nai Q, Chen M, Dittus JD, Howard MJ, Margiotta JF. Brain-derived neurotrophic factor and trkB signaling in parasympathetic neurons: relevance to regulating alpha7-containing nicotinic receptors and synaptic function. J Neurosci. 2004;24:4340–4350. doi: 10.1523/JNEUROSCI.0055-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.