Abstract
Plasmodium vivax merozoite surface protein-5 (PvMsp-5), a potential vaccine candidate, is encoded by a two-exon single copy gene. We have conducted a comprehensive analysis of PvMsp-5 by sequencing the entire gene of four parasite populations from northwestern Thailand (n = 73), southern Thailand (n = 53), Indonesia (n = 25) and Brazil (n = 24), and five isolates from other endemic areas. Results reveal that exon I exhibits a significantly higher level of nucleotide diversity at both synonymous and nonsynonymous sites than exon II (p < 0.01). Neutrality tests based on both intraspecific and interspecific nucleotide polymorphism have detected a signature of positive selection in exon I of all populations while substitutions in exon II mainly followed neutral expectation except that three residues in exon II of northwestern Thailand population appear to be positively selected using the Bayes Empirical Bayes method. Short imperfect repeats were identified in exon I at an equivalent region to its orthologue in P. knowlesi, supporting their close genetic relatedness. Significant levels of population subdivision were detected among most populations including those between northwestern and southern Thailand (p < 10−5), implying absent or minimal gene flow between these populations. Importantly, evidences for intragenic recombination in PvMsp-5 were found in most populations except that from southern Thailand in which haplotype diversity and nucleotide diversity were exceptionally low. Results from Fu and Li’s D*, F* and D and F tests suggested that PvMsp-5 of most P. vivax populations have been maintained by balancing selection whereas southern Thailand population could have gone through recent bottleneck events. These findings are concordant with a substantial reduction in number of P. vivax cases in southern Thailand during the past decade, followed by a very recent population expansion. Therefore, spatio-temporal monitoring of parasite population genetics provides important implications for disease control.
Keywords: malaria, Plasmodium vivax, merozoite surface protein-5, genetic diversity, genetic differentiation, natural selection, Plasmodium knowlesi
1. Introduction
Vaccination against pathogens has been proven a powerful means to eliminate and control a number of infectious diseases since the past two centuries (Andre, 2003). Although malaria can be controlled by integrated intervention such as early case detection, effective chemotherapy and vector control, these strategies have been complicated by inaccessibility of standard diagnostic tools, antimalarial resistant parasites and insecticide-resistant mosquitoes, thereby development of a malaria vaccine is essential. Priority has been justified for developing a vaccine against Plasmodium falciparum because it is the most prevalent and most virulent species. Despite the fact that P. vivax accounts for 130-145 million infections annually, progress in vaccine development against this parasite lags far behind vaccines against P. falciparum (Hay et al., 2004 Price et al., 2007). Recent studies have highlighted the importance of vivax malaria in causing severe disease manifestations ranging from severe anemia and respiratory distress to cerebral involvement akin to those found in severe malaria caused by P. falciparum (Baird et al., 2007; Price et al., 2007). Furthermore, the chronicity of vivax malaria due to the presence of hypnozoites and the emergence of chloroquine- and primaquine-resistant parasite strains from various endemic foci have underscored the burden of vivax malaria as an important global health problem (Galinski and Barnwell, 2008). One of the reasons underlying the ineffectiveness of malaria control is the lack of extensive knowledge on parasite population structure and population dynamics in each endemic area, which has theoretical and practical implications. Such knowledge will lead to understanding how genes of interest such as antigenic diversity, drug resistance and pathogenicity circulate in a given population while the interplay between selective forces on genes of interest and reproductive modes within populations will provide essential groundwork for theoretical assumptions, leading to a useful mathematical modeling to predict outcomes and consequences of certain control measures (Gauthier and Tibayrenc, 2005).
The complex anatomical niche of asexual blood stages of malaria parasites while living inside erythrocytes has secluded them from host destruction, probably by limiting direct accessibility of host macromolecule, especially antibodies. A critical event in the malaria life cycle in a human host is the process of release and reentry of merozoites into new erythrocytes because free merozoites in circulation are vulnerable to elimination by host humoral immunity. In fact, several proteins associated with merozoite surface or apical organelles are targets for naturally acquired antibodies and some of these antibodies, e.g. those to the polymorphic merozoite surface protein-1 (Msp-1) or apical membrane antigen-1 (AMA-1), are significantly correlated with lower clinical severity or reduced parasitemia than individuals incapable of producing specific antibodies (Egan et al., 1996; Conway et al., 2000; Polley et al., 2004). Therefore, a number of merozoite surface proteins are considered to be candidates for malaria vaccine development (Dooland and Steward, 2007).
The merozoite surface protein-5 of Plasmodium vivax (PvMsp-5) is a 41 kDa protein associated with micronemes or distributed over the polar cap, an apical organelle that is crucial for host cell entry by the merozoites. The gene encoding PvMsp-5 of the Thai-NYU strain is characterized by a two-exon structure of 861 bp and 300 bp separated by a 402-bp intron (Black et al., 2002). The PvMsp-5 locus and its paralogous gene, known as merozoite surface protein-4 (PvMsp-4), are arranged in tandem located on chromosome 4 (Carlton et al., 2008). The C-terminal part of PvMsp-5 possesses an epidermal growth factor (EGF)-like domain and glycosylphosphatidyl inositol (GPI) anchor sequences that are also found in Msp-1 of both P. falciparum and P. vivax. Importantly, experimental vaccination studies using either Msp-4/5 per se or in combination with the 19 kDa fragment of Msp-1 of P. yoelii have conferred significant protection against lethal parasite challenges (Kedzierski et al., 2002; Wang et al., 2004). Consistent results were obtained when Msp-4/5 of P. chabaudi adami was used as immunogens in an experimental murine model, highlighting the potential role of Msp-5 as a component in a malaria vaccine (Rainczuk et al., 2003).
A recent study of PvMsp-5 from 22 Colombian isolates has shown a remarkable sequence variation clustering in exon I while the intron and exon II contained relatively more conserved sequences (Gomez et al., 2006). Analysis of genetic diversity in these isolates has further suggested that intragenic recombination and probably positive selective pressure could cause sequence diversity at this locus. Meanwhile, malarial parasites from diverse geographic origins may possess different population genetic backgrounds that have practical significance for the development of strategic intervention of the disease, including a rational vaccine design (Putaporntip et al., 2002). The aims of the present study were to survey the extent of sequence diversity of this locus and to determine population history and genetic mechanisms underlying variation in PvMsp-5 of parasite populations from diverse disease endemic areas. Furthermore, to further elucidate the patterns of selection on this gene, we incorporated tests based on both intraspecific and interspecific polymorphisms by comparing with Msp-5 of P. knowlesi (PkMsp-5). Here we recruited P. vivax populations from Thailand, Indonesia and Brazil and isolates from Vietnam, Papua New Guinea, Solomon Island and India for population genetic analysis.
2. Materials and methods
Parasite populations
We recruited 180 blood samples from symptomatic P. vivax-infected patients in northwestern Thailand (51 males and 22 females from Tak Province), southern Thailand (30 males and 23 females from Yala and Narathiwat Provinces), Indonesia (15 males, 10 females from Lombok Island), Brazil (15 males, 9 females from the Acre State), Vietnam (2 males), Papua New Guinea (1 male), Solomon Island (1 male) and India (1 male). Blood samples were taken after informed consent. Venous blood samples obtained from Thai patients were preserved in EDTA anticoagulant while the remaining samples were from finger-prick blood spotted onto Whatman 3MM Chr filter. The parasite density was estimated for isolates from Thailand by examination of Giemsa-stained thick blood films for at least 200 leukocytes under x100 objectives. The mean parasite density from northwestern Thailand isolates and southern Thailand isolates was 12,610 parasites/μl (range 35 to 44,520 parasites/μl) and 11,980 parasites/μl (range 70 to 38,540 parasites/μl), respectively. Isolates from Thailand, Indonesia, Brazil and Vietnam were obtained during high transmission periods in 2006-2007, 2002, 1997 and 1995, respectively. The remaining isolates from elsewhere were collected in 1999. These isolates had been previously characterized for the presence of single clone infection of P. vivax using probes derived from the polymorphic blocks 2, 6 and 12 of PvMsp-1 by southern blot hybridization or direct sequencing of the PCR-amplified products encompassing these regions (Putaporntip et al., unpublished data). A blood sample from a Thai patient who was infected with P. knowlesi was also included as a closely related outgroup malaria for comparison (Escalante and Ayala, 1994; Escalante et al., 1998). The ethical aspects of this study have been approved by the Institutional Review Board of Faculty of Medicine, Chulalongkorn University.
DNA preparation, amplification and sequencing
DNA was extracted from either venous blood samples using QIAamp kit (Qiagen, Hilden, Germany) or finger-pricked blood spotted onto filter paper by the method as described by others (Sakihama et al., 2001). The complete PvMsp-5 sequence spanning ~1.5 kb was amplified using a forward primer (PvMsp-5-F0: 5′-TCTTCAATTTTCCGCTCAACC-3′, nucleotides −68 to −48 before the start codon of the Salvador I sequence, GenBank accession number XM-001612943) and a reverse primer (PvMsp-5-R0: 5′-CACAAGGTGAAGAGATCGAC-3′, nucleotides +35 to +54 after the stop codon of the Salvador I sequence). DNA amplification was carried out in a total volume of 30 μl of the reaction mixture containing template DNA, 2.5 mM MgCl2, 300 mM each deoxynucleoside triphosphate, 3 μl of 10X ExTaq PCR buffer, 0.3 μM of each primer and 1.25 units of ExTaq DNA polymerase (Takara, Seta, Japan). Thermal cycling profile included the preamplification denaturation at 94°C for 1 min followed by 35 cycles of 94°C for 30 s, 60°C for 30 s and 72°C for 2 min, and a final extension at 72°C for 5 min. DNA amplification was performed by using a GeneAmp 9700 PCR thermal cycler (Applied Biosystems, Foster City, CA). We used ExTaq DNA polymerase that possesses efficient 5′→3′ exonuclease activity to increase fidelity and no strand displacement (Takara, Japan). The size of PCR product was examined by electrophoresis in a 1% agarose gel and visualized under a UV transilluminator (Mupid Scope WD, Japan). The PCR product was purified by using QIAquick PCR purification kit (QIAGEN, Germany). DNA sequences were determined directly and bi-directionally from PCR-purified templates. Sequencing analysis was performed on an ABI3100 Genetic Analyzer using the Big Dye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, USA). Overlapping PvMsp-5 sequences were obtained by using sequencing primers (PvMsp-5-F1: 5′-CTTATGATGATTTATACAGC-3′, nucleotides −23 to −4 before the start codon; PvMsp-5-F2: 5′-TGACGACGACAGCAGACTTC-3′, nucleotides 837 to 856; PvMsp-5-R1: 5′-GTCAGCGGCACTACGCAG-3′, nucleotides +7 to +24 after the stop codon, and PvMsp-5-R2: 5′-TGATGTGGACATGTTTGA-3′, nucleotides 896 to 913). Whenever singleton substitution occurred, sequence was re-determined using PCR products from two independent amplifications from the same DNA template. Determination of the Msp-5 gene of P. knowlesi (PkMsp-5) was conducted essentially as described for PvMsp-5 except the PCR primers (PkMsp-5-F0: 5′-ATCGCATTAAATTACCTGTTC-3′, nucleotides −74 to −54 before the start codon of P. knowlesi H strain under the accession number AM910986, and PkMsp-5-R0: 5′-CGTGCGTGTTTGTTTAGTTGTA-3′, nucleotides +73 to +94 after the stop codon) and sequencing primers (PkMsp-5-F1: 5′-CATTTCCCCCCTTATGATGA-3′, nucleotides −33 to −14 from the start codon, PkMsp-5-F2: GCAAGGATTCGCCAGAAGG-3′, nucleotides 719 to 737, PkMsp-5-R1: 5′-ACGGGGCACTACTTAGAGGTT-3′, nucleotides +16 to +36 after the stop codon, and PkMsp-5-R2: 5′-AGGAGAATCAAGAAAATG-3′, nucleotides 776 to 793). Sequences in this study have been deposited in the GenBank™ Database under the accession numbers FJ874096 to FJ874276.
Statistical analyses
All PvMsp-5 sequences were aligned with the Thai NYU PvMsp-5 sequence (GenBank accession number AF403476) for intraspecific comparison using the default option of the Clustal X program (Thompson et al., 1997) and manually edited. In coding regions, indels were placed using information on multiple alignments of amino acid sequences to maintain the reading frame. Sequence of the Thai NYU strain (GenBank accession number AF403476) was used as reference sequence. We included the PvMsp-5 sequences of Colombian isolates available in the GenBank databases (accession numbers DQ341586 to DQ341601) for some comparative analyses (Gomez et al., 2006). For interspecific comparison, all P. vivax sequences were aligned with the two P. knowlesi sequences, one was from naturally acquired infection in a Thai patient that was newly determined in this study (Jongwutiwes et al., 2004) and the other was from a laboratory reared macaque, H strain (accession number AY573058) (Black et al., 2004). Tandem repeats are detected by scanning the sequence with a small window, determining the distance between exact matches and testing the statistical criteria as implemented in the Tandem Repeats Finder version 4.0 program (Benson, 1999). Tandem Repeats Explorer based on K-means algorithm in sequences (T-REKS) was deployed to reaffirm the results (Jorda and Kajava 2009). We also applied these algorithms to search for repeats in the orthologous genes in P. falciparum, designated PfMsp-4 and PfMsp-5 from our previous report (accession numbers AF447553 to AF447573) (Jongwutiwes et al., 2002).
Nucleotide diversity, Pi (π) or per-site heterozygosity, was calculated from the average number of pairwise sequence differences in the sample (Nei, 1987, equation 10.5) and its standard deviation (or standard error) is the square root of the variance (Nei, 1987, equation 10.7) using the MEGA 4.0 program (Tamura et al., 2007). Haplotype (gene) diversity and its sampling variance were estimated according to equations 8.4 and 8.12 but replacing 2n by n (Nei, 1987). Linkage disequilibrium between loci within each parasite population expressed in terms of the square of correlation coefficient (r2) between alleles (Hill and Robertson, 1968), the Lewontin’s standardized disequilibrium coefficient (D’) (Lewontin, 1964) was determined by the DnaSP 4.10.9 software (Rozas et al., 2003). Singleton polymorphic sites were not included in linkage disequilibrium analysis. The relationship between the r2 measure of linkage disequilibrium and molecular distance was tested under random permutation of the physical position of the single nucleotide polymorphism (SNP) using 1000 permutations as implemented in the LDhat 2.1 package (McVean et al., 2002). We estimated the parameter Rm which indicates the minimum number of recombination events in the history of the sample using the DnaSP 4.10.9 software (Hudson and Kaplan, 1985; Rozas et al., 2003).
Evidence for departure from the predictions of the neutral model of molecular evolution was determined by Tajima’s D, Fu and Li’s D* and F*, and Fu’s Fs tests that are based on measures of allele frequencies or intraspecific polymorphism. Tajima’s D statistic measures the difference between the average number of nucleotide differences and an estimate of θ = 4Neμ from the number of segregating sites (Tajima, 1983). Fu’s Fs test relies on the haplotype frequency distribution conditional the value of θ (Ewens, 1972; Fu, 1997). Fu and Li’s D* test is based on the differences between the number of singletons, and the total number of mutations while their F* test statistic takes into account the differences between the number of singletons and the average number of nucleotide differences between pairs of sequences (Fu and Li, 1993). Analysis based on interspecific nucleotide diversity was performed using Fu and Li’s D and F tests, Fay and Wu’s H statistic, and McDonald-Kreitman tests. Fu and Li’s D statistics rely on the differences between the total number of mutations in external branches of the genealogy and the total number of mutations while their F statistic compares the differences between the total number of mutations in external branches of the genealogy and the average number of nucleotide differences between pairs of sequences (Fu and Li, 1993). Fay and Wu’s H test statistic (Fay and Wu, 2000) is based on the frequency of the derived variants which measures departure from neutrality considering the differences between the average number of nucleotide differences between pairs of sequences and the frequency of the derived variants (Fay and Wu, 2000). Statistical significance of these parameters was estimated by coalescence simulation using 10,000 pseudoreplicates. All of these tests were performed in the DnaSP version 4.10.9 program (Rozas et al., 2003). The McDonald-Kreitman test compares the ratio of synonymous to nonsynonymous polymorphism within a species and the ration of synonymous to nonsynonymous polymorphism between closely related species. Statistical departure from neutral expectation was tested by Fischer’s exact test (McDonald and Kreitman, 1991).
Analysis of selective pressure on PvMsp-5 was performed by the maximum-likelihood method based on comparision of various site-specific models with different assumptions regarding the ratio of nonsynonymous to synonymous substitution rates (ω = dN/dS) as implemented in the CODEML program in PAML package version 4.0 (Yang, 1997; Yang, 2007). We computed dN/dS ratio average across all sites in exon I and exon II separately using model M0 with one ω for all sites. Signature of positive selection was determined by comparing the fit of the model pairs M1 (nearly neutral; considering two classes of sites, ω < 1 and ω = 1) vs. M2a (assuming an additional class of sites under positive selection, ω > 1) and between models M7 (following a beta distribution, ω varying across sites) vs. M8 (following a beta distribution and an additional class of sites under positive selection, ω > 1), assuming that twice the log likelihood difference between the two models has a χ2 distribution with a number of degrees of freedom equal to the difference in the number of free parameters (Whelan and Goldman, 1999). We searched for positively selected sites at which nonsynonymous substitutions occur at a higher rate than synonymous ones using the Bayes Empirical Bayes (BEB) estimation of posterior probabilities for site class implemented in CODEML models M2a and M8 (Yang et al., 2005).
Maximum-likelihood phylogeny based on PvMsp-5 from worldwide isolates was generated using the PHYML 3.0 software under the following search parameters: gamma distribution to account for site rate heterogeneity and the Hasegawa–Kishino–Yano two-parameter model variant for unequal base frequencies (HKY85 model) to account for transition-transversion and unequal nucleotide biases (Hasegawa et al., 1985). Initial trees were generated by an improved version of the neighbor-joining algorithm (BIONJ) (Gascuel, 1997). Reliability of branching patterns within trees was tested by the bootstrap method with 100 resamplings.
The genetic structure of population was investigated by analysis of molecular variance approach (AMOVA) implemented in the Arlequin 3.11 software which is similar to the method described by Weir and Cockerham but it takes into account the number of mutations between molecular haplotypes (Excoffier et al., 1992). The implemented algorithm calculates the fixation index FST identical to the weighted average F-statistic over loci, θw (Weir and Cockerham, 1984). The significance of the fixation indices was tested using a non-parametric permutation (Excoffier et al., 1992).
Characteristics of amino acid substitutions were based on charge and polarity of each residue. Charge property includes positive (H, R and K), negative (D and E) or neutral (the remainder). Polarity of amino acid residues is categorized as polar (S, Y, C, W, H, Q, T and N) or nonpolar (the remainder). Prediction of HLA-I binding peptides followed the method taken into account proteasomal C terminal cleavage and transporter associated with antigen processing (TAP) transport efficiency at http://www.cbs.dtu.dk/services/NetCTL (Larsen et al., 2005).
3. Results
Genetic diversity in PvMsp-5
Analysis of the entire PvMsp-5 gene of 180 P. vivax isolates from seven countries in this study has identified 107 haplotypes. All of these haplotypes were novel and differed from recently reported sequences from Colombia (Gomez et al., 2006). Size variation was observed in exon I (range = 849 to 873 bp) and in intron (311 to 498 bp) while exon II of all isolates contained 100 codons. We observed 104 amino acid changes and insertion/deletion of 13 codons in exon I while 6 amino acid substitutions were detected in exon II comparing with the PvMsp-5 sequence of the Thai NYU strain (GenBank accession number AF403476)(Supplementary Fig. 1). The nucleotide diversity (π) of PvMsp-5 coding regions from Thailand (including both northwestern and southern isolates), Indonesia and Brazil did not show significantly different values, ranging from 0.03723 to 0.04210 (p > 0.05) (Table 1). However, when P. vivax populations from northwestern and southern Thailand were considered separately, the latter population exhibited significantly less diversity than the former (π = 0.01058 and 0.04405, respectively) (p < 0.001). Nucleotide diversity, synonymous and nonsynonymous nucleotide polymorphisms were significantly more pronounced in exon I than in exon II of PvMsp-5 and this pattern occurred in all populations examined (p < 0.001) (Table 1). Despite a low level of nucleotide diversity observed in exon II, all 6 amino acid exchanges were located in the EGF-like domain and GPI anchor sequences (supplementary Fig. 1). The haplotype diversity was high across P. vivax populations from northwestern Thailand, Indonesia and Brazil while a remarkably low value was observed in the population from southern Thailand (h = 0.365) (Table 1). Like PvMsp-5, the orthologous gene of P. knowlesi accumulated more nucleotide substitutions in exon I than in exon II, and most of the substitutions were nonsynonymous replacements (data not shown).
Table 1.
Haplotype and nucleotide diversity of the Msp-5 gene from Thai, Indonesian and Brazilian populations of Plasmodium vivax
| Population | H | M | S | h±S.D. | π±S.E. | πS±S.E | πN±S.E |
|---|---|---|---|---|---|---|---|
| Thailand (n=126) | |||||||
| Exon I | 50 | 158 | 148 | 0.873±0.027 | 0.05014±0.00472***,††† | 0.02149±0.00526*** | 0.06175±0.00656***,### |
| Exon II | 4 | 3 | 3 | 0.519±0.013 | 0.00179±0.00159 | 0.00000±0.00000 | 0.00253±0.00227 |
| Exons I & II | 54 | 161 | 151 | 0.884±0.025 | 0.03723±0.00348††† | 0.01567±0.00398 | 0.04603±0.00506### |
| Intron | 2 | 1 | 1 | 0.077±0.032 | 0.00028±0.00012 | - | - |
| Northwestern (n=73) | |||||||
| Exon I | 48 | 156 | 146 | 0.983±0.006 | 0.05970±0.00538***,††† | 0.02611±0.00643*** | 0.07331±0.00734***,### |
| Exon II | 4 | 3 | 3 | 0.341±0.061 | 0.00123±0.00100 | 0.00000±0.00000 | 0.00175±0.00150 |
| Exons I & II | 50 | 159 | 149 | 0.984±0.006 | 0.04405±0.00391††† | 0.01901±0.00464 | 0.05426±0.00539### |
| Intron | 2 | 1 | 1 | 0.129±0.051 | 0.00047±0.00019 | - | - |
| Southern (n=53) | |||||||
| Exon I | 3 | 72 | 71 | 0.365±0.068 | 0.01431±0.00234***,††† | 0.00847±0.00304** | 0.01665±0.00320*** |
| Exon II | 2 | 1 | 1 | 0.038±0.036 | 0.00018±0.00018 | 0.00000±0.00000 | 0.00018±0.00017 |
| Exons I & II | 3 | 73 | 72 | 0.365±0.068 | 0.01058±0.00175††† | 0.00620±0.00226 | 0.01235±0.00225 |
| Intron | 2 | 1 | 1 | 0.038±0.036 | 0.00009±0.00009 | - | - |
| Indonesia (n=25) | |||||||
| Exon I | 20 | 138 | 132 | 0.977±0.019 | 0.05747±0.00523***,††† | 0.02723±0.00659*** | 0.06961±0.00739***,### |
| Exon II | 2 | 1 | 1 | 0.080±0.072 | 0.00027±0.00025 | 0.00000±0.00000 | 0.00038±0.00037 |
| Exons I & II | 20 | 139 | 133 | 0.977±0.019 | 0.04210±0.00402††† | 0.01978±0.00514 | 0.05112±0.00563### |
| Intron | 2 | 1 | 1 | 0.220±0.100 | 0.00079±0.00036 | - | - |
| Brazil (n=24) | |||||||
| Exon I | 13 | 115 | 111 | 0.924±0.032 | 0.05043±0.00515***,††† | 0.02387±0.00642*** | 0.06112±0.00703***,### |
| Exon II | 5 | 4 | 4 | 0.627±0.093 | 0.00250±0.00133 | 0.00183±0.00187 | 0.00278±0.00177 |
| Exons I & II | 14 | 119 | 115 | 0.931±0.033 | 0.03764±0.00383††† | 0.01789±0.00482 | 0.04564±0.00526### |
| Intron | 3 | 2 | 2 | 0.359±0.110 | 0.00143±0.00049 | - | - |
H = the number of haplotypes. M = the number of mutations. S = the number of segregating sites.
Tests of the hypothesis that mean πS equals that for πN:
p<0.05
p<0.01
p<0.001
Tests of the hypothesis that mean π, πS or πN of exon 1 equals the corresponding value of exon 2:
p<0.05
p<0.01
p<0.001
Tests of the hypothesis that mean π equals that for π of intron:
p<0.05
p<0.01
p<0.001
Both Tandem Repeats Finder program and T-REKS program yielded identical results for repeats identification in PvMsp-5 and PkMsp-5 (Benson, 1999; Jorda and Kajava 2009). Although no apparent amino acid repeats occurred in PvMsp-5, three copies of imperfect repeats characterized by a consensus sequence ‘AAGGAGATGGAGGAG’, occurred in exon I of 10 out of 180 isolates (5.6%) analyzed in this study, i.e. TF127, TFF20, TFF23, TC137, T1031 and T1192 from Thailand, DLB13a from Indonesia, and PC36, AD86 and BP29 from Brazil. The other imperfect repeats with a consensus sequence ‘GGAG(or gap)AAGA(or gap)GAAAAAG(or gap)GAAC(or gap)GGT’ were also found in a Colombian isolate (accession number DQ341589) (Gomez et al., 2006). It is noteworthy that the orthologous gene in P. knowlesi, possessed three copies of 24-bp repeat motifs at both nucleotide and amino acid levels, characterized by ‘AGAAAATGG(C/T)AAAAAAAAGTGATGA’ motif also found to be located at a similar region in exon I, spanning nucleotides 351 - 434 (positions after the H strain sequence). Likewise, two copies of imperfect repeat motif were noted in PfMsp-4 but none in PfMsp-5 (Fig. 1).
Figure 1.
Imperfect repeats in exon I of PvMsp-5 of (A) isolates Thai-TF127, Thai-TFF20, Thai-TFF23, Thai-TC137, Thai-1031, Thai-1192, Indonesia-DLB13A, Brazil-PC36, Brazil-BP29 and Brazil-AD86 and (B) isolate Colombia-DQ341589. Repeats in Msp-5 of P. knowlesi (C) and in Msp-4 of P. falciparum (D).
The intron in PvMsp-5 of worldwide isolates displayed considerable size variation, ranging from 279 to 498 bp. Polymorphism in the intron is due to insertion/deletion of single nucleotides and variation in numbers of the repeat arrays (Supplementary Fig. 2). Of all isolates examined including 16 Colombian isolates from the GenBank database, 41 haplotypes were observed. Most worldwide isolates (41.6%) shared identical intron sequence with the NYU strain from Thailand (Gomez et al., 2006.) Nucleotide diversity in intron was significantly lower than those in exon I (p < 0.001) while statistical difference was not observed between nucleotide diversity in intron and in exon II (Table 1).
Recombination and linkage disequilibrium
Analysis of linkage disequilibrium between polymorphic sites across the PvMsp-5 gene was performed for each population. A decline in numbers of significant linkage disequilibrium between pairs of polymorphic loci (both r2 and D’) over molecular distance was observed (data not shown). The correlation between the linkage disequilibrium expressed by the r2 values and nucleotide distance estimated from the LDhat version 2.1 package yielded a significant negative correlation (corr r2, d = −0.24045, df = 71, p < 10−4) for the northwestern Thai isolates, indicating intragenic recombination in this gene. Likewise, evidence for intragenic recombination in PvMsp-5 occurred in parasite populations from Indonesia (corr r2, d = −0.26992, df = 23, p < 10−4) and Brazil (corr r2, d = −0.17068, df = 22, p < 10−4). Consistently, the minimum number of recombination events estimated by means of the four-gamete test revealed 30, 24 and 15 recombination sites in the P. vivax populations from northwestern Thailand, Indonesia and Brazil, respectively, reaffirming the results of linkage disequilibrium analysis. It is noteworthy that Rm occurred mainly within exon I of PvMsp-5. On the other hand, significant inverse relationship between the r2 values and nucleotide distance along the PvMsp-5 gene was not observed in the parasite population from southern Thailand (corr r2, d = ∞, df = 51, p = 1.0). Consistently, no detectable Rm value was generated by means of the four gamete test (Hudson and Kaplan 1985), implying no recombination in the PvMsp-5 locus between P. vivax isolates in southern Thailand.
Population differentiation
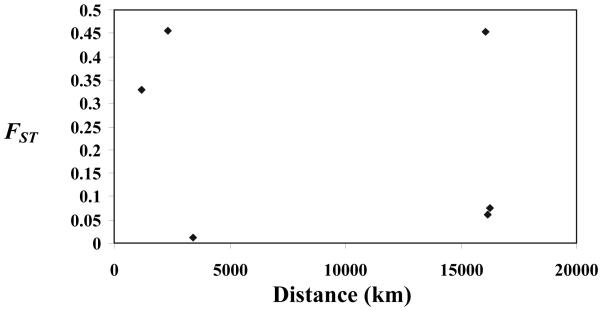
We measured the FST statistics for all pairwise comparisons between populations to assess whether there was evidence of genetic differentiation between P. vivax populations in this study. All pairwise FST values among P. vivax populations from different geographic regions were significantly greater than zero, indicating genetic differentiation between populations (p < 10−5) (Table 2, upper diagonal values). Because P. vivax samples from northwestern and southern Thailand were from different endemic areas with a distance of more than 1,000 km apart and had a remarkable difference in both haplotype and nucleotide diversities at the PvMsp-5 locus, these parasite populations had FST values significantly different from zero (p < 10−5) and thus were considered to be distinct populations with minimum or absence of gene flow. However, the FST value between P. vivax populations from northwestern Thailand and Indonesia did not show significant departure from zero, indicating a lack of genetic differentiation between these populations (Table 2, lower diagonal values). When the FST values were plotted against the geographic distance where samples were collected, no significant correlation was observed (r = −0.22829; p = 0.66) (Fig. 2).
Table 2.
Genetic differentiation (FST indices) of P. vivax populations from northwestern and southern regions of Thailand, Indonesia and Brazil
| Northwestern Thailand |
Southern Thailand |
Indonesia | Brazil | |
|---|---|---|---|---|
| Thailand (all) | 0.07225* | 0.09479* | ||
| Northwestern | ||||
| Southern | 0.32855* | |||
| Indonesia | 0.01084# | 0.45430* | 0.03801* | |
| Brazil | 0.06139* | 0.45380* | 0.07529* |
Upper diagonal and lower diagonal FST values were computed considering parasites from northwestern and southern Thailand as single and distinct populations, respectively. Test of hypothesis that FST does not depart from zero by permutation:
p < 10−5
p = 0.18919.
Figure 2.
Lack of association analysis between FST and geographic distance for P. vivax populations from northwestern and southern Thailand, Indonesia and Brazil (r = −0.22829; p = 0.66).
Phylogenetic relationship
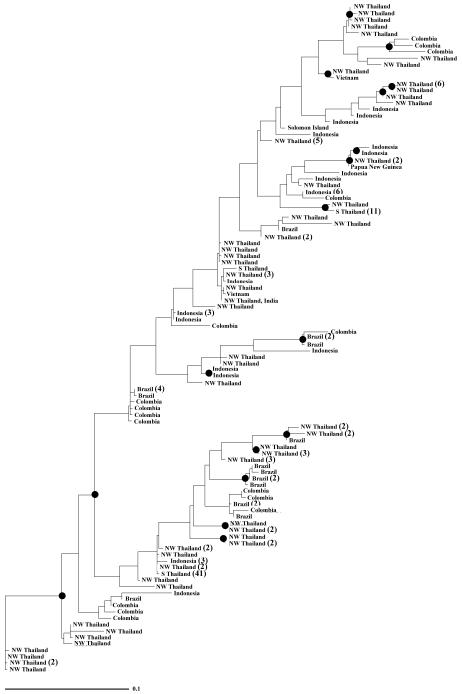
In total of 197 isolates of P. vivax from diverse geographic origins (180 isolates in this study, 16 isolates from Colombia and the Thai NYU strain) contained 105 unique coding sequences. Maximum likelihood phylogeny revealed that the PvMsp-5 alleles could not be assigned into any particular lineage, indicating that no distinctive parental types could be discerned at this locus. The distribution of the PvMsp-5 alleles in the phylogenetic tree did not exhibit geographic clustering but rather that parasites from diverse geographic origins could be found throughout the tree (Fig. 3). Phylogenetic trees generated separately from exon I, exon II or intron yielded different topologies, implying no apparent association between these regions (data not shown). High nucleotide diversity and the presence of recombination in exon I could be attributable to difference in these phylogenetic relationships. Despite significant genetic differentiation between P. vivax populations from northwestern and southern Thailand, the genealogy of all 3 haplotypes found in southern region were closely related to the isolates from northwestern area (Fig. 3).
Figure 3.
Maximum likelihood tree inferred from coding regions of PvMsp-5 from diverse geographic origins. Numbers of identical sequences are in parentheses. Bootstrap values > 70% from 100 pseudoreplicates are shown as filled circles at the branches. NW and S Thailand denotes isolates from northwestern and southern Thailand, respectively.
Tests of neutrality
Maximum-likelihood estimates of rates of evolution at synonymous and nonsynonymous sites in the PvMsp-5 gene have shown that exon I of each population had average dN/dS ratio (ω) across sites significantly greater than 1 (p < 0.00001) (Table 3). Comparison of nested site models assuming neutral evolution or null models (M1 and M7) to models containing positively evolving sites (M2a and M8) has revealed that the models M2a and M8 fitted the data of exon I significantly better than the models M1 and M7 by likelihood-ratio tests (p < 0.00001), supporting the presence of positive selection in this region. On the other hand, application of similar comparison of these nested models for exon II did not yield significant difference; thereby no evidence of positive selection was observed (Table 3). The BEB analysis under the models M2a and M8 identified a number of sites in exon I to be under positive selection (posterior probability > 0.95 to > 0.99). No positively selected sites were observed in exon II of parasite populations from southern Thailand, Indonesia and Brazil. However, three sites in exon II of P. vivax population from northwestern Thailand had posterior probability under the model M8 reaching significant level (> 0.95), suggesting micro-scale signature of positive selection in this segment (Table 2). In the presence of recombination, identification of sites under positive selection by the BEB method has shown to be more robust than the likelihood ratio test (Anisimova et al., 2003).
Table 3.
Estimates of selection parameters on the Msp-5 gene from Thai, Indonesian and Brazilian populations of Plasmodium vivax
|
P values |
No. positively selected sites* |
|||||||
|---|---|---|---|---|---|---|---|---|
| Population | L | Lnonsyn | Lsyn | dN/dS | M1 vs. M2a | M7 vs. M8 | M2a | M8 |
| Thailand (n=126) | ||||||||
| Exon I | 2.302 | 0.968 | 0.265 | 3.654 | <0.00001 | <0.00001 | 53 | 60 |
| Exon II | 0.031 | 0.013 | 0.000 | NA | 0.999 | 0.291 | 0 | 3 |
| Northwestern (n=73) | ||||||||
| Exon I | 2.004 | 0.849 | 0.219 | 3.879 | <0.00001 | <0.00001 | 52 | 56 |
| Exon II | 0.031 | 0.013 | 0.000 | NA | 0.521 | 0.521 | 0 | 3 |
| Southern (n=53) | ||||||||
| Exon I | 0.267 | 0.111 | 0.033 | 3.385 | <0.00001 | <0.00001 | 10 | 10 |
| Exon II | 0.011 | 0.005 | 0.000 | NA | 0.708 | 0.709 | 0 | 0 |
| Indonesia (n=25) | ||||||||
| Exon I | 1.223 | 0.509 | 0.160 | 3.189 | <0.00001 | <0.00001 | 61 | 63 |
| Exon II | 0.010 | 0.005 | 0.000 | NA | 0.711 | 0.709 | 0 | 0 |
| Brazil (n=24) | ||||||||
| Exon I | 0.725 | 0.300 | 0.099 | 3.019 | <0.00001 | <0.00001 | 43 | 48 |
| Exon II | 0.041 | 0.014 | 0.012 | 1.233 | 0.999 | 0.984 | 0 | 0 |
L, tree length in substitutions per codon under PAML M0 model with one ω for all sites; Lnonsyn, tree length for dN; Lsyn, tree length for dS; dN/dS, ratio average across all sites under PAML model M0; P values, probability values of the likelihood-ratio tests between specified models of PAML.
NA, values could not be estimated because denominator is zero.
estimates from Bayes Empirical Bayes analysis implemented in the PAML program (p < 0.05).
Application of the Tajima’s D statistics and Fu’s Fs test to each population revealed that none of the population showed significant departure from neutral expectation. Likewise, no significant results were obtained when exon I and exon II of PvMsp-5 were analyzed separately. When Fu and Li’s D* and F* tests were applied to P. vivax population from northwestern Thailand, significant positive values were obtained for exon I and the entire coding region (p < 0.05) while exon II yielded a non-significant negative value, suggesting that balancing or diversifying selection maintains polymorphism in exon I. It is noteworthy that the parasite population from southern Thailand gave high negative Fu and Li’s D* and F* values for exon I and the entire coding region of PvMsp-5 along with a negative value of Tajima’s D test, implying that directional or purifying selection might have acted against less frequent genotypes or a recent population bottleneck might have occurred, thus eliminating less frequent alleles. On the contrary, no significant departure from neutrality was observed when these tests were applied to the parasite populations from Brazil and Indonesia. When Fu and Li’s D and F tests were applied to each population using two PkMsp-5 sequences as closely related outgroups, significant departure from neutrality could be found in exon I and the entire coding region of PvMsp-5 in populations from northwestern Thailand and Indonesia (p < 0.05). Meanwhile, Fay and Wu’s H test yielded highly significant negative values in P. vivax population from southern Thailand (p < 0.01), reflecting a relative excess of high-frequency derived variant alleles as expected to occur immediately after a selective sweep (Table 4).
Table 4.
Substitutions in predicted T cell epitopes in PvMsp-5 among isolates from Thailand, Indonesia, Brazil and Colombia that alter HLA-binding
| Domain | HLA Supertype |
Predicted epitope* | Predicted score** |
Geographic distribution (n) |
|||
|---|---|---|---|---|---|---|---|
| Thailand | Indonesia | Brazil | Colombia | ||||
| Exon I | A1 | KTDNAAAGK | 0.80 | 18 | 8 | - | 1 |
| ETDNAAAGK | 0.78 | 7 | 2 | 8 | 4 | ||
| KTGNAAAGK | 0.44 | 4 | 2 | - | - | ||
| ETKKEATGK | 0.40 | 16 | 4 | 3 | 3 | ||
| GKDNAAAGK | 0.39 | 67 | 7 | 10 | 8 | ||
| GKDNADAGK | 0.38 | 5 | 1 | - | - | ||
| GKDKAAAGK | 0.38 | 2 | - | - | - | ||
| ETKKEAAGK | 0.36 | 1 | - | 1 | - | ||
| GKNNAAAGK | 0.35 | 6 | 1 | 1 | - | ||
| A3 | GSAGESGSK | 1.07 | 43 | 11 | 6 | 4 | |
| GSDGESGSK | 0.67 | 6 | - | 3 | 1 | ||
| GSDGESGEK | 0.49 | 12 | 6 | 5 | 5 | ||
| GSEGESGEK | 0.43 | 65 | 8 | 10 | 6 | ||
| A3 | RLKPLDSEK | 1.10 | 8 | - | 1 | - | |
| KMKTLDSEK | 1.07 | 3 | - | - | - | ||
| RLKTLISGE | 0.18 | 45 | 3 | - | - | ||
| RLKTLNSRE | 0.18 | - | - | 5 | - | ||
| RLKTLNSGE | 0.14 | 16 | 1 | - | 4 | ||
| RLKTLNSEE | 0.13 | 2 | 2 | 3 | 1 | ||
| RLKPLNSGE | 0.13 | 9 | - | - | - | ||
| KMKTLGSGE | 0.10 | 13 | 1 | 1 | - | ||
| KMKTLNSEE | 0.10 | - | - | 5 | - | ||
| KMKTLGSEE | 0.10 | - | 1 | - | - | ||
| QMKTLDSGD | 0.05 | - | - | - | 3 | ||
| RLKTLDSGE | 0.04 | - | - | - | 2 | ||
| RLKPLDSGE | 0.04 | 3 | 2 | 1 | 1 | ||
| QMKTLGSGD | 0.03 | - | 1 | 8 | 3 | ||
| KMKTLGSGD | 0.03 | - | - | - | 1 | ||
| KMKTLDSGD | 0.02 | 9 | 3 | - | - | ||
| KMKTLDSGE | 0.02 | - | 1 | - | - | ||
| QMKTLGSGE | 0.01 | 18 | 10 | - | 1 | ||
| B7 | RAKDKQASV | 0.86 | 11 | 2 | 1 | 1 | |
| RAKEENASV | 0.75 | 1 | 1 | - | - | ||
| RAKNEQASV | 0.75 | 6 | 2 | - | - | ||
| RAKDENASV | 0.72 | 13 | 9 | 8 | 5 | ||
| RAKEEQASV | 0.70 | 59 | 3 | - | - | ||
| RAKDEQASV | 0.67 | 24 | 6 | 14 | 7 | ||
| RAEGENASV | 0.54 | 12 | 2 | 1 | 3 | ||
| B8 | NLKKTDNAA | 1.14 | 18 | 9 | - | 1 | |
| NLKKTGNAA | 1.26 | 4 | 2 | - | - | ||
| NLKETDNAA | 0.76 | 7 | 2 | 8 | 4 | ||
| NLEGKNNAA | 0.48 | 6 | 1 | 1 | - | ||
| NLEGKDNAA | 0.47 | 68 | 6 | 9 | 8 | ||
| NLEGKDKAA | 0.43 | 2 | - | - | - | ||
| NLEETKKEA | 0.27 | 16 | 4 | 4 | 3 | ||
| NLEGKDNAD | 0.13 | 5 | 1 | - | - | ||
| B39 | SEEEQMKTL | 1.03 | 18 | 10 | - | 1 | |
| SEEDRLKTL | 0.86 | 2 | 3 | 8 | 7 | ||
| PEEDKMKTL | 0.56 | - | - | - | 1 | ||
| PEEEQMKTL | 0.55 | - | 1 | 8 | 6 | ||
| PEEEKMKTL | 0.53 | 13 | 2 | 6 | - | ||
| PEEDRLKTL | 0.50 | 61 | 4 | - | - | ||
| SEKDRLKPL | 0.49 | 20 | 1 | 1 | - | ||
| PEEKKMKTL | 0.47 | 3 | 2 | - | - | ||
| PEKEKMKTL | 0.36 | 9 | 2 | - | - | ||
| B58 | STSGEHTNL | 0.94 | - | 1 | - | - | |
| STSEEAPNL | 0.84 | 1 | 1 | - | - | ||
| STSGEDTNL | 0.78 | 30 | 10 | 14 | 9 | ||
| STSGKDTNL | 0.56 | 2 | 4 | - | - | ||
| SRSGEHTNL | 0.28 | 2 | 1 | - | - | ||
| PQTEGDTNL | 0.21 | 68 | 4 | 5 | 4 | ||
| PPSGEDTNL | 0.20 | 12 | 1 | 5 | - | ||
| PPSEEAPNL | 0.20 | 1 | 3 | - | 3 | ||
| B62 | KQSGDVHPA | 0.98 | 1 | - | - | - | |
| KQAEEVNST | 0.91 | 20 | 4 | 1 | - | ||
| KQAEGVDST | 0.80 | 5 | - | - | - | ||
| KQSGDVEPT | 0.71 | - | 1 | - | - | ||
| KQAEEVDPT | 0.70 | 2 | 1 | - | - | ||
| KQAEQVDPA | 0.67 | - | 1 | - | - | ||
| KQAEEVDST | 0.63 | - | 2 | 9 | 4 | ||
| KQSGDVDPA | 0.62 | 2 | 2 | 8 | 5 | ||
| KQAEKVDST | 0.60 | 9 | - | 4 | - | ||
| KQSGYVDPT | 0.59 | 2 | 2 | - | - | ||
| KQAKEVDST | 0.41 | - | - | - | 3 | ||
| KKAEQIDPA | 0.10 | 9 | - | - | - | ||
| KKSGDVDPA | 0.09 | 11 | 7 | - | - | ||
| KKAEQVDPA | 0.08 | 4 | - | 2 | 3 | ||
| KNAEEVDST | 0.07 | 1 | - | - | - | ||
| KKAEEVDST | 0.06 | 59 | 5 | - | - | ||
| KKAEQVDPT | 0.05 | 1 | - | - | - | ||
| Exon II | A2 | YLFGDKCIL | 1.14 | 1 | - | - | - |
| HLFGGKCIL | 0.68 | 60 | 24 | 5 | 2 | ||
| HLFGDKCIL | 0.67 | 67 | 1 | 19 | 14 | ||
Underlined residues were positively selected sites based on the model M8 estimated using the BEB method implemented in PAML package (Yang et al, 2005).
Threshold for epitope identification was set at ≥ 0.75 using weight on C terminal cleavage at ≥ 0.15 and weight on transporter associated with antigen processing transport efficiency at ≥ 0.05 (Larsen et al, 2005).
Comparison of synonymous and nonsynonymous polymorphic changes within P. vivax and between P. vivax and P. knowlesi following the MacDonald-Kreitman test has shown that exon I and the entire coding region of PvMsp-5 in each parasite population had significant deviation in positive direction from neutral expectation (p < 0.01), a signature of positive selection (balancing selection). Consistent with other tests, none of these populations generated significant values of the MacDonald-Kreitman test for exon II (Table 5).
Table 5.
Tests for neutrality in the PvMsp-5 gene from P. vivax populations in Thailand, Indonesia and Brazil
| Intraspecific polymorphism |
Interspecific polymorphism# |
||||||
|---|---|---|---|---|---|---|---|
| Tajima’s D | Fu&Li’s D* | Fu&Li’s F* | Fu’s Fs | Fu&Li’s D | Fu&Li’s F | Fay&Wu’s H | |
| Thailand (n=126) | |||||||
| Exon I | 1.115 | 1.581* | 1.647 | 2.606 | 2.108** | 1.996* | −19.222* |
| Exon II | −0.062 | −2.082 | −1.691 | −0.100 | −2.107 | −1.713 | −0.019 |
| All | 1.091 | 1.418 | 1.525 | 1.602 | 1.799* | 1.768 | −19.241* |
| Northwestern (n=73) | |||||||
| Exon I | 1.636 | 1.500* | 1.866* | −0.833 | 2.019** | 2.261** | −11.546 |
| Exon II | −0.766 | −1.877 | −1.792 | −1.294 | −1.918 | −1.831 | 0.294 |
| All | 1.576 | 1.368 | 1.743* | −1.662 | 1.751* | 2.019* | −11.252 |
| Southern (n=53) | |||||||
| Exon I | −1.055 | −3.687** | −3.236** | 20.715 | −0.772 | −0.878 | −20.427*** |
| Exon II | −1.097 | −1.858 | −1.896 | −1.685 | −1.918 | −1.831 | −0.956 |
| All | −1.085 | −3.747** | −3.294** | 20.780 | −0.693 | −0.849 | −22.352*** |
| Indonesia (n=25) | |||||||
| Exon I | 1.075 | 0.991 | 1.198 | 0.295 | 1.787** | 1.900* | −14.624 |
| Exon II | −1.158 | −1.620 | −1.716 | −1.061 | −1.666 | −1.771 | 0.077 |
| All | 1.045 | 0.944 | 1.149 | 0.301 | 1.675* | 1.787* | −14.547 |
| Brazil (n=24) | |||||||
| Exon I | 1.312 | 0.961 | 1.259 | 6.565 | 1.228 | 1.499 | −12.034 |
| Exon II | −0.829 | 0.114 | −0.178 | −1.651 | 1.105 | 0.646 | −2.507* |
| All | 1.220 | 0.934 | 1.204 | 5.293 | 1.297 | 1.485 | −14.544 |
The Msp-5 gene of P. knowlesi strain H and isolate A1 were used as out group sequences.
p < 0.05
p < 0.02
p < 0.01.
Amino acid substitutions
The majority of amino acid substitutions in PvMsp-5 were conservative with respect to polarity as 63.6% of these remained unchanged while the reverse was true with respect to charge property as 67.3% of these changes altered their charge profiles. Closer look into the substituted residues in exons I and II of PvMsp-5 has identified a number of potential HLA class I-binding peptides in exon I and a few in exon II as predicted by the high scores for the C-terminal cleavage and the transporter associated with antigen processing efficiency (Larsen et al., 2005) (data not shown). Importantly, amino acid substitutions in some of these peptides abolished the predicted property of the epitopes (Table 4). It is noteworthy that substituted residues in some of these epitopes in both exons I and II were shown to be positively selected based on the BEB analysis (Table 3).
4. Discussion
Both PfMsp-5 and PvMsp-5 are frequently recognized by species-specific antibodies induced by natural infections (Woodberry et al., 2009). Analysis of polymorphism at the Msp-5 locus of P. vivax populations from Thailand, Indonesia and Brazil, and isolates from four other endemic areas in Asia revealed a substantial number of nucleotide substitutions in the coding region with a nonrandom distribution. Polymorphism in PvMsp-5 is characterized by both nucleotide substitutions and indels that mainly clustered in the middle portion of exon I while a limited number of codon changes was observed in exon II. Variation in the intron of PvMsp-5 was largely due to deletion or insertion of repeats or partial repeat units, following those previously observed among P. vivax population in Colombia (Gomez et al., 2006). Although no repeats were observed in previous study by others (Gomez et al., 2006), our analysis has identified two types of imperfect repeats in exon I of PvMsp-5 of some isolates (Fig. 1). Indels in these imperfect repeats that could be generated by slipped-strand mispairing or related mechanisms have disrupted repeat characteristics at amino acid level by frameshift mutations (Levinson and Gutman, 1987). Likewise, one of two orthologues in P. falciparum (PfMsp-4) contained two copies of imperfect repeats akin to those found in PvMsp-5 (Figure 1) (Jongwutiwes et al, 2002). Furthermore, three copies of 8-codon repeats occurred in exon I of the orthologous gene in P. knowlesi that was not noted previously (Black et al., 2004). Because both P. vivax and P. knowlesi are closely related in terms of mitochondrial genome-based and small subunit ribosomal RNA-based phylogenies (Escalante et al., 1994; Escalante et al., 1998; Jongwutiwes et al., 2005), the imperfect repeats or remnant of repeat motifs in PvMsp-5 found in some P. vivax isolates could further support their close genetic relatedness, thereby the generation of repeats in exon I plausibly preceded speciation, followed by subsequent loss in most P. vivax lineages.
We observed a strong population structure in P. vivax isolates from Thailand (considering northwestern and southern isolates as a single population), Indonesia and Brazil as evidenced by the FST values among parasites from different geographic origins being significantly greater than zero. However, when P. vivax from northwestern and southern Thailand were considered as separate populations, a low and not significant FST value was found between populations from northwestern Thailand and Indonesia, suggesting indistinguishable or meager genetic differentiation between them. Likewise, analysis of the apical membrane antigen-1 (AMA-1) locus of P. vivax has revealed similar trends of genetic differentiation, i.e. populations from northwestern Thailand and Indonesia were not differentiated while a striking geographical differentiation occurred between populations from Thailand and Brazil (Grynberg et al., 2008). Although gene flow between these populations could have been ongoing, no evidence of frequent historical migration of hosts or vectors between northwestern Thailand and Indonesia could support such a process. Alternatively, it might be suggested that ancient balanced polymorphism could have maintained the genetic make-up of these populations after the expansion of P. vivax from this region (Jongwutiwes et al., 2005). Plots of FST values against geographical distance did not yield significant correlation, further purporting an ancient selectively maintained polymorphism at the PvMsp-5 locus. Nevertheless, a closely related genealogy of all PvMsp-5 haplotypes from southern Thailand to those from northern area could suggest that they might originate from a common ancestral haplotypes preceding current population subdivision rather than being introduced from elsewhere.
Intragenic recombination has reportedly been implicated in sequence diversity in other P. vivax surface proteins such as genes encoding the merozoite surface protein-1, thrombospondin-related adhesive protein and apical membrane antigen-1 (Putaporntip et al., 2001; Putaporntip et al., 2002; Putaporntip et al., 2009). Analysis of linkage disequilibrium between parsimony informative sites and estimation of the minimum number of recombination events in the history of the PvMsp-5 gene have supported previous study that intragenic recombination enhances sequence diversity at this locus (Gomez et al., 2006). It is noteworthy that P. vivax populations from northwestern Thailand and Indonesia had higher Rm than that from Brazil, implying that recombination events occurred more frequently in the former populations. More importantly, the zero value of Rm and no correlation between r2 and molecular distance in the PvMsp-5 locus by linkage disequilibrium analysis indicate no intragenic recombination in parasite population from southern Thailand, conforming to clonal expansion. Meanwhile, it is noteworthy that no mixed infections of PvMsp-1 haplotypes were detected in southern P. vivax isolates in this study. Taken together, the paucity of haplotypes (n= 3) in southern P. vivax population and extremely low prevalence of mixed infections of P. vivax could compromise interallelic recombination at this locus.
In the present study, we used various tests based on either intraspecific or interspecific comparisons to investigate the pattern of selection on the PvMsp-5 locus for each population because each test may have different statistical powers to detect departure from neutrality (Zhai et al., 2008). The results of several maximum likelihood approaches provide strong evidence that exon I has experienced positive selection. Although no evidence of positive selection was observed in exon II, BEB analysis using the model M8 has identified 3 positively selected sites in this region. The MacDonald-Kreitman tests, excluding sites equivalent to repeat regions because of uncertainty in sequence alignment, reaffirmed a significant departure from neutral expectation in positive direction for exon I and the entire coding region but not in exon II of all populations. Meanwhile, a number of HLA-class I binding peptides were predicted to locate mainly in exon I and a few in exon II. Several of these peptides contained amino acid substitutions at positively selected sites and a number of these exchanges resulted in alteration of peptide-binding scores (Table 4). Despite the presence of 6 amino acid substitutions in exon II, all cysteine residues forming the EGF-like domain were perfectly conserved in all isolates. Therefore, limited diversity in exon II of PvMsp-5 could partly stem from some functional or structural constraints. Taken together, these results support previous analysis using P. vivax isolates from Colombia that polymorphism in exon I is greater than that in exon II (Gomez et al., 2006) while exon I and a few residues in exon II have been maintained by positive selection, probably from host immune pressure.
Meanwhile, polymorphism in PvMsp-5 of populations from northwestern Thailand and Indonesia seems to be under balancing selection as evidenced by a significant positive tests based on Fu and Li’s D and F statistics as well as equivalent tests without an outgroup for population from Indonesia. However, P. vivax from southern Thailand exhibits significantly negative departure from neutrality by Fu and Li’s D* and F* statistics and a negative value for Tajima’s D test, albeit not significant, which could be resulted from directional or purifying selection or a recent population bottleneck. Consistently, a large positive value of Fu’s Fs test may further suggest population bottleneck or overdominant selection at the PvMsp-5 locus of P. vivax from southern Thailand. Although the negative Tajima’s D value may also suggest recent population growth, this skew will be caused by low-frequency-derived variants (Fu, 1997). As a result, population growth per se will always generate a positive skew in Fay and Wu’s H statistic. On the contrary, the effect of a recent bottleneck or a selective sweep (a locus-specific bottleneck followed by population expansion) could fit the results of significantly negative value of Fay and Wu’s H test while older bottlenecks will result in a more negative Tajima’s D and a positive H statistics when new mutations emerge in the population (Haddrill et al., 2005). It is noteworthy that genetic differentiation seems to be most pronounced between P. vivax population of southern Thailand and all others (northwestern Thailand, Brazil and Indonesia) (Table 2) although the FST values between other pairs of populations are very low but their p values reached a significant level. Meanwhile, the phylogenetic relationships in samples from southern Thailand were not intermixed as much as other samples. Although the reasons underlying such contradictive findings are not well understood, an interplay among factors such as variability in selective pressure generated by host immune responses to PvMsp-5, difference in frequency of intragenic recombination in mosquito vectors, genetic drift that occurs as a result of a drastic reduction in population by an event having little to do with the usual forces of natural selection and introduction of new variants into the local gene pool by population migration could shape genetic profiles of these populations differently. Further studies related to these issues would be required to address this phenomenon.
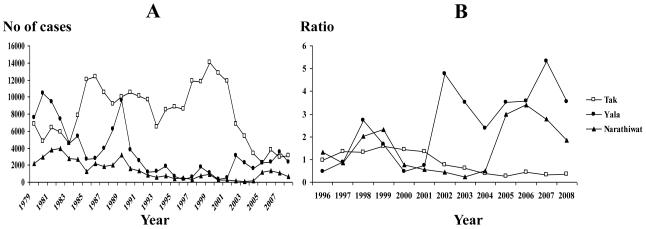
The annual parasite incidence (API) or the number of malaria cases positive by microscopy per 1,000 persons per year in Thailand has an overall decline since 1982 because of early case detection, timely change of national antimalarial drug policy and active implementation of vector control measures (Zhou et al., 2005; Annual Statistics, Division of Vector-Borne Diseases, Ministry of Public Health, Thailand 2009). However, the number of P. vivax cases in Tak, Yala and Narathiwat Provinces has been fluctuating over the past 3 decades (Fig. 4A). Vivax malaria in Tak Province reached a maximum peak in the year 1999 (n=14,037), followed by a rapid decline in the total number of cases in the following years and remained relatively stable during 2004-2008. A striking reduction in total number of vivax cases in Yala Province occurred twice in the year 1996 (n=308) and in 2000 (n=299), followed by a rapid increase in the number of cases in 2001 (n=3,142). A similar trend was found in P. vivax in Narathiwat Province with the lowest number of cases observed in the year 2003 (n=86), followed by a relatively substantial increase in the number of cases during the past 4 years, coinciding with a reduction or absence of vector control activities (Fig. 4B). Therefore, it seems that P. vivax populations in all of these endemic areas are subject to bottleneck effects on a different time scale. It is noteworthy that our recent sequence analysis of the same parasite populations in Thailand has shown that the merozoite surface protein-4 (PvMsp-4) locus of southern population (n=53) exhibited exceptionally lower nucleotide diversity than that of northwestern population (n=77), i.e. π±S.D. for southern population = 0.00056±0.00013 and for northwestern population = 0.00214±0.00035 (Putaporntip et al., 2009). Consistently, the low nucleotide polymorphism and the negatively significant values of parameters Fu and Li’s D* and F* at the PvMsp-5 locus of population from southern Thailand would suggest the population bottleneck effect. However, the extent of nucleotide diversity, significant positive direction of Fu and Li’s D*, D, F and F*, and recombination parameters at the PvMsp-5 locus of Tak population rather indicate balancing selection. This paradox can be explained by the fact that vivax malaria epidemiology in Tak Province has been influenced by migrants and transmigrants carrying malaria from Myanmar that could contribute to and maintain variants of PvMsp-5 in this endemic area while almost all malaria cases in Yala and Narathiwat Provinces were indigenous cases and transmigrants carrying malaria from Malaysia seems to be trivial (Chareonviriyaphap et al., 2000). Importantly, our study has depicted a correlation between P. vivax population dynamics and the results predicted from molecular data based on the PvMsp-5 locus. Undoubtedly, the effectiveness of a malaria vaccine requires knowledge about parasite evolutionary dynamics, population history and social factors.
Figure 4.
Annual number of microscopy-positive cases caused by P. vivax in Tak, Yala and Narathiwat Provinces from 1979 to 2008 (A) and ratio of P. vivax-infected cases to P. vivax-infected cases detected in 1995. (source: Division of Vector-Borne Diseases, Ministry of Public Health, Thailand).
Supplementary Material
Table 6.
McDonald-Kreitman tests on Msp-5 of Plasmodium vivax from diverse geographic origins with P. knowlesi orthologue as outgroup species
| Population | Domain | Polymorphic changes within P.vivax |
Fixed differences between species* |
Neutrality index |
Fisher’s exact test (p) |
||
|---|---|---|---|---|---|---|---|
| Synonymous | Nonsynonymous | Synonymous | Nonsynonymous | ||||
| Thailand | |||||||
| Northwestern (n=73) | |||||||
| Exon I | 15 | 86 | 58 | 119 | 2.79 | 0.00107 | |
| Exon II | 1 | 5 | 20 | 20 | 5.00 | 0.19813 | |
| All | 16 | 91 | 78 | 139 | 3.19 | 0.00008 | |
| Southern (n=53) | |||||||
| Exon I | 6 | 49 | 62 | 133 | 3.81 | 0.00182 | |
| Exon II | 1 | 3 | 20 | 20 | 3.00 | 0.60862 | |
| All | 7 | 52 | 82 | 153 | 3.98 | 0.00042 | |
| All (n=126) | |||||||
| Exon I | 16 | 87 | 58 | 119 | 2.65 | 0.00187 | |
| Exon II | 1 | 5 | 20 | 20 | 5.00 | 0.19813 | |
| All | 17 | 92 | 78 | 139 | 3.04 | 0.00010 | |
| Indonesia (n=25) | |||||||
| Exon I | 12 | 74 | 59 | 118 | 3.08 | 0.00102 | |
| Exon II | 1 | 3 | 20 | 20 | 3.00 | 0.60862 | |
| All | 13 | 77 | 79 | 138 | 3.39 | 0.00011 | |
| Brazil (n=24) | |||||||
| Exon I | 9 | 71 | 61 | 129 | 3.73 | 0.00024 | |
| Exon II | 2 | 5 | 20 | 19 | 2.63 | 0.41766 | |
| All | 11 | 76 | 81 | 148 | 3.78 | 0.00005 | |
Differences between PvMsp-5 and PkMsp-5 excluding repeat regions.
Acknowledgements
We are grateful to all patients who donated their blood samples for this study; to Professor Hiroji Kanbara, Department of Protozoology, Institute of Tropical Medicine, Nagasaki University, Nagasaki, Japan for parasite samples from Indonesia, Papua New Guinea, Solomon Island and India; and to Professor Marcelo U. Ferreira, Department of Parasitology, Institute of Biomedical Sciences, University of Sao Paulo, Sao Paulo, Brazil, for samples from Brazil. We thank Pannadhat Areekul, Rattiporn Kosuwin, Thongchai Hongsrimueng and the staff of the Bureau of Vector Borne Disease, Department of Disease Control, Ministry of Public Health, Thailand, for assistance in field work. C.P. was supported by The Thailand Research Fund (RMU5080002). This research was supported by grant from the National Research Council of Thailand and the Thai Government Research Budget to S.J. and C.P. and grant from The Fogarty International Center, NIH (grant D43TW006571) to C.P. and L.C.
Abbreviations
- PvMsp-5
Plasmodium vivax merozoite surface protein-5
- PkMsp-5
Plasmodium knowlesi merozoite surface protein-5
- π
nucleotide diversity
- πS
synonymous nucleotide diversity
- h
haplotype diversity
- πN
nonsynonymous nucleotide diversity
- dS
number of synonymous substitutions per synonymous site
- dN
number of nonsynonymous substitutions per nonsynonymous site
- FST
fixation index
- GPI
glycosylphosphatidyl inositol
- EGF
epidermal growth factor
- API
annual parasite incidence
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andre FE. Vaccinology: past achievements, present roadblocks and future promises. Vaccine. 2003;21:593–595. doi: 10.1016/s0264-410x(02)00702-8. [DOI] [PubMed] [Google Scholar]
- Anisimova M, Nielsen R, Yang Z. Effect of recombination on the accuracy of the likelihood method for detecting positive selection at amino acid sites. Genetics. 2003;164:1229–1236. doi: 10.1093/genetics/164.3.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annual Statistics. Division of Vector-Borne Diseases, Ministry of Public Health; Thailand: [Accessed 3 March 2009]. Available at: http://www.thaivbd.org. [Google Scholar]
- Baird JK. Neglect of Plasmodium vivax malaria. Trends Parasitol. 2007;23:533–539. doi: 10.1016/j.pt.2007.08.011. [DOI] [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black CG, Barnwell JW, Huber CS, Galinski MR, Coppel RL. The Plasmodium vivax homologues of merozoite surface proteins 4 and 5 from Plasmodium falciparum are expressed at different locations in the merozoite. Mol. Biochem. Parasitol. 2002;120:215–224. doi: 10.1016/s0166-6851(01)00458-3. [DOI] [PubMed] [Google Scholar]
- Black CG, Wang L, Topolska AE, Finkelstein DI, Horne MK, Thomas AW, Mohandas N, Coppel RL. Merozoite surface proteins 4 and 5 of Plasmodium knowlesi have differing cellular localisation and association with lipid rafts. Mol. Biochem. Parasitol. 2004;138:153–158. doi: 10.1016/j.molbiopara.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Carlton JM, Adams JH, Silva JC, et al. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature. 2008;455:757–763. doi: 10.1038/nature07327. 40 co-authors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chareonviriyaphap T, Bangs MJ, Ratanatham S. Status of malaria in Thailand. Southeast Asian J. Trop. Med. Pub. Health. 2000;31:225–237. [PubMed] [Google Scholar]
- Conway DJ, Cavanagh DR, Tanabe K, et al. A principal target of human immunity to malaria identified by molecular population genetic and immunological analyses. Nat. Med. 2000;6:689–692. doi: 10.1038/76272. 12 co-authors. [DOI] [PubMed] [Google Scholar]
- Dooland DL, Steward VN. Status of malaria vaccine R&D in 2007. Expert. Rev. Vaccines. 2007;6:755–766. doi: 10.1586/14760584.6.6.903. [DOI] [PubMed] [Google Scholar]
- Egan AF, Morris J, Barnish G, Allen S, Greenwood BM, Kaslow DC, Holder AA, Riley EM. Clinical immunity to Plasmodium falciparum malaria is associated with serum antibodies to the 19-kDa C-terminal fragment of the merozoite surface antigen, PfMSP-1. J. Infect. Dis. 1996;173:765–769. doi: 10.1093/infdis/173.3.765. [DOI] [PubMed] [Google Scholar]
- Escalante AA, Ayala FJ. Phylogeny of the malarial genus Plasmodium, derived from rRNA gene sequences. Proc. Natl. Acad. Sci. USA. 1994;91:11373–11377. doi: 10.1073/pnas.91.24.11373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante AA, Freeland DE, Collins WE, Lal AA. The evolution of primate malaria parasites based on the gene encoding cytochrome b from the linear mitochondrial genome. Proc. Natl. Acad. Sci. USA. 1998;95:8124–8129. doi: 10.1073/pnas.95.14.8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewens WJ. The sampling theory of selectively neutral alleles. Theor. Popul. Biol. 1972;3:87–112. doi: 10.1016/0040-5809(72)90035-4. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JC, Wu CI. Hitchhiking under positive Darwinian selection. Genetics. 2000;155:1405–1413. doi: 10.1093/genetics/155.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YX. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YX, Li WH. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galinski MR, Barnwell JW. Plasmodium vivax: who cares? Malar. J. 2008;7(Suppl 1):S9. doi: 10.1186/1475-2875-7-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascuel O. BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol. Biol. Evol. 1997;14:685–695. doi: 10.1093/oxfordjournals.molbev.a025808. [DOI] [PubMed] [Google Scholar]
- Gauthier C, Tibayrenc M. Population structure of malaria parasites: the driving epidemiological forces. Acta Trop. 2005;94:241–50. doi: 10.1016/j.actatropica.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Gomez A, Suarez CF, Martinez P, Saravia C, Patarroyo MA. High polymorphism in Plasmodium vivax merozoite surface protein-5 (MSP5) Parasitology. 2006;133:661–672. doi: 10.1017/S0031182006001168. [DOI] [PubMed] [Google Scholar]
- Grynberg P, Fontes CJ, Hughes AL, Braga EM. Polymorphism at the apical membrane antigen 1 locus reflects the world population history of Plasmodium vivax. BMC Evol. Biol. 2008;8:123. doi: 10.1186/1471-2148-8-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddrill PR, Thornton KR, Charlesworth B, Andolfatto P. Multilocus patterns of nucleotide variability and the demographic and selection history of Drosophila melanogaster populations. Genome Res. 2005;15:790–799. doi: 10.1101/gr.3541005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M, Kishino H, Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- Hay SI, Guerra CA, Tatem AJ, Noor AM, Snow RW. The global distribution and population at risk of malaria: past, present, and future. Lancet Infect. Dis. 2004;4:327–336. doi: 10.1016/S1473-3099(04)01043-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill WG, Robertson A. Linkage disequilibrium in finite populations. Theor. Appl. Genet. 1968;38:226–231. doi: 10.1007/BF01245622. [DOI] [PubMed] [Google Scholar]
- Hudson RR, Kaplan NL. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics. 1985;111:147–164. doi: 10.1093/genetics/111.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongwutiwes S, Putaporntip C, Iwasaki T, Ferreira MU, Kanbara H, Hughes AL. Mitochondrial genome sequences support ancient population expansion in Plasmodium vivax. Mol. Biol. Evol. 2005;22:1733–1739. doi: 10.1093/molbev/msi168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongwutiwes S, Putaporntip C, Friedman R, Hughes AL. The extent of nucleotide polymorphism is highly variable across a 3-kb region on Plasmodium falciparum chromosome 2. Mol. Biol. Evol. 2002;19:1585–1590. doi: 10.1093/oxfordjournals.molbev.a004220. [DOI] [PubMed] [Google Scholar]
- Jongwutiwes S, Putaporntip C, Iwasaki T, Sata T, Kanbara H. Naturally acquired Plasmodium knowlesi malaria in human, Thailand. Emerg. Infect. Dis. 2004;10:2211–2213. doi: 10.3201/eid1012.040293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorda J, Kajava AV. T-REKS: identification of Tandem REpeats in sequences with a K-mean S based algorithm. Bioinformatics. 2009;25:2632–2638. doi: 10.1093/bioinformatics/btp482. [DOI] [PubMed] [Google Scholar]
- Kedzierski L, Black CG, Goschnick MW, Stowers AW, Coppel RL. Immunization with a combination of merozoite surface proteins 4/5 and 1 enhances protection against lethal challenge with Plasmodium yoelii. Infect. Immun. 2002;70:6606–6613. doi: 10.1128/IAI.70.12.6606-6613.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen MV, Lundegaard C, Lamberth K, Buus S, Brunak S, Lund O, Nielsen M. An integrative approach to CTL epitope prediction: a combined algorithm integrating MHC class I binding, TAP transport efficiency, and proteasomal cleavage predictions. Eur. J. Immunol. 2005;35:2295–2303. doi: 10.1002/eji.200425811. [DOI] [PubMed] [Google Scholar]
- Levinson G, Gutman GA. Slipped-strand mispairing: A major mechanism for DNA sequence evolution. Mol. Biol. Evol. 1987;4:203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- Lewontin RC. The interaction of selection and linkage. I. General considerations: heterotic models. Genetics. 1964;49:49–69. doi: 10.1093/genetics/49.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- McVean G, Awadalla P, Fearnhead P. A coalescent-based method for detecting and estimating recombination rates from gene sequences. Genetics. 2002;160:1231–1241. doi: 10.1093/genetics/160.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Molecular evolutionary genetics. Columbia University Press; New York: 1987. [Google Scholar]
- Polley SD, Mwangi T, Kocken CH, et al. Human antibodies to recombinant protein constructs of Plasmodium falciparum apical membrane antigen 1 (AMA1) and their associations with protection from malaria. Vaccine. 2004;23:718–728. doi: 10.1016/j.vaccine.2004.05.031. 13 co-authors. [DOI] [PubMed] [Google Scholar]
- Price RN, Tjitra E, Guerra CA, Yeung S, White NJ, Anstey NM. Vivax malaria: neglected and not benign. Am. J. Trop. Med. Hyg. 2007;77:79–87. [PMC free article] [PubMed] [Google Scholar]
- Putaporntip C, Jongwutiwes S, Ferreira MU, Kanbara H, Udomsangpetch R, Cui L. Limited global diversity of the Plasmodium vivax merozoite surface protein 4 gene. Infect. Genet. Evol. 2009;9:821–826. doi: 10.1016/j.meegid.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putaporntip C, Jongwutiwes S, Grynberg P, Cui L, Hughes AL. Nucleotide sequence polymorphism at the apical membrane antigen-1 locus reveals population history of Plasmodium vivax in Thailand. Infect. Genet. Evol. 2009;9:1295–1300. doi: 10.1016/j.meegid.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putaporntip C, Jongwutiwes S, Sakihama N, Ferreira MU, Kho WG, Kaneko A, Kanbara H, Hattori T, Tanabe K. Mosaic organization and heterogeneity in frequency of allelic recombination of the Plasmodium vivax merozoite surface protein-1 locus. Proc. Natl. Acad. Sci. USA. 2002;99:16348–16353. doi: 10.1073/pnas.252348999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putaporntip C, Jongwutiwes S, Tia T, Ferreira MU, Kanbara H, Tanabe K. Diversity in the thrombospondin-related adhesive protein gene (TRAP) of Plasmodium vivax. Gene. 2001;268:97–104. doi: 10.1016/s0378-1119(01)00425-5. [DOI] [PubMed] [Google Scholar]
- Rainczuk A, Smooker PM, Kedzierski L, Black CG, Coppel RL, Spithill TW. The protective efficacy of MSP4/5 against lethal Plasmodium chabaudi adami challenge is dependent on the type of DNA vaccine vector and vaccination protocol. Vaccine. 2003;21:3030–3042. doi: 10.1016/s0264-410x(03)00116-6. [DOI] [PubMed] [Google Scholar]
- Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Sakihama N, Mitamura T, Kaneko A, Horii T, Tanabe K. Long PCR amplification of Plasmodium falciparum DNA extracted from filter paper blots. Exp. Parasitol. 2001;97:50–54. doi: 10.1006/expr.2000.4591. [DOI] [PubMed] [Google Scholar]
- Tajima F. Evolutionary relationship of DNA sequences in finite populations. Genetics. 1983;105:437–460. doi: 10.1093/genetics/105.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Goschnick MW, Coppel RL. Oral immunization with a combination of Plasmodium yoelii merozoite surface proteins 1 and 4/5 enhances protection against lethal malaria challenge. Infect. Immun. 2004;72:6172–6175. doi: 10.1128/IAI.72.10.6172-6175.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan S, Goldman N. Distributions of statistics used for the comparison of models of sequence evolution in phylogenetics. Mol. Biol. Evol. 1999;16:1292–1299. [Google Scholar]
- Weir BS, Cockerham CC. Estimating F statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Woodberry T, Minigo G, Piera KA, Hanley JC, de Silva HD, Salwati E, Kenangalem E, Tjitra E, Coppel RL, Price RN, Anstey NM, Plebanski M. Antibodies to Plasmodium falciparum and Plasmodium vivax merozoite surface protein 5 in Indonesia: species-specific and cross-reactive responses. J. Infect. Dis. 2008;198:134–142. doi: 10.1086/588711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comp Appl BioSci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yang Z, Wong WS, Nielsen R. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 2005;22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Zhai W, Nielsen R, Slatkin M. An investigation of the statistical power of neutrality tests based on comparative and population genetic data. Mol. Biol. Evol. 2008;26:273–283. doi: 10.1093/molbev/msn231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, Sirichaisinthop J, Sattabongkot J, Jones J, Bjørnstad ON, Yan G, Cui L. Spatio-temporal distribution of Plasmodium falciparum and P. vivax malaria in Thailand. Am. J. Trop. Med. Hyg. 2005;72:256–262. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.