Abstract
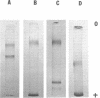
Two isoenzymes of malate dehydrogenase (MDH) were demonstrated in plasmodia of Physarum polycephalum by polyacrylamide-gel electrophoresis. The more “cathodal” form was uniquely associated with mitochondria (M-MDH) and the other form was found in the soluble cytoplasm (S-MDH). The isoenzymes were separated by acetone fractionation of soluble plasmodial homogenates acidified to pH 5.0. The M-MDH was purified 201-fold by cetylpyridinium chloride treatment, fractionation with ammonium sulfate, gradient elution from sulfoethyl cellulose at pH 6.0, and Sephadex G-100 chromatography. The S-MDH was purified 155-fold by ammonium sulfate fractionation, diethylaminoethyl cellulose chromatography, gradient elution from sulfoethyl cellulose at pH 5.5, and Sephadex G-100 chromatography. The optimal cis-oxalacetate concentrations were 0.35 mM for M-MDH and 0.25 mM for S-MDH, and the optimal pH for both isoenzymes was 7.6 for oxalacetate reduction. The optimal l-malate concentrations were 5 mM for S-MDH and 6 mM for M-MDH, and both isoenzymes exhibited an optimal pH of 10.0 for L-malate oxidation. The Michaelis constants of S-MDH and M-MDH served to discriminate between the isoenzymes. The S-MDH was more heat-stable than the M-MDH. High concentrations of oxalacetate and malate inhibited S-MDH more than M-MDH. The isoenzymes were further distinguished by their utilization of analogues of nicotinamide adenine dinucleotide. Many properties of the Physarum isoenzymes were similar to those of more complex organisms, especially vertebrates.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abou-Zamzam A. M., Wallace A. Some characteristics of the mitochondrial and soluble forms of malate dehydrogenase in lemon fruits. Biochim Biophys Acta. 1970 Dec 16;220(3):396–409. doi: 10.1016/0005-2744(70)90271-8. [DOI] [PubMed] [Google Scholar]
- Atzpodien W., Gancedo J. M., Duntze W., Holzer H. Isoenzymes of malate dehydrogenase in Saccharomyces cerevisiae. Eur J Biochem. 1968 Dec;7(1):58–62. doi: 10.1111/j.1432-1033.1968.tb19573.x. [DOI] [PubMed] [Google Scholar]
- BOXER G. E., DEVLIN T. M. Pathways of intracellular hydrogen transport. Science. 1961 Nov 10;134(3489):1495–1501. doi: 10.1126/science.134.3489.1495. [DOI] [PubMed] [Google Scholar]
- Bailey G. S., Wilson A. C., Halver J. E., Johnson C. L. Multiple forms of supernatant malate dehydrogenase in salmonid fishes. J Biol Chem. 1970 Nov 25;245(22):5927–5940. [PubMed] [Google Scholar]
- Benveniste K., Munkres K. D. Cytoplasmic and mitochondrial malate dehydrogenases of Neurospora. Regulatory and enzymic properties. Biochim Biophys Acta. 1970 Nov 11;220(2):161–177. doi: 10.1016/0005-2744(70)90003-3. [DOI] [PubMed] [Google Scholar]
- Dupourque D., Kun E. Malate dehydrogenases of ox kidney. 2. Two substrate kinetic and inhibition analyses. Eur J Biochem. 1969 Jan;7(2):247–252. doi: 10.1111/j.1432-1033.1969.tb19599.x. [DOI] [PubMed] [Google Scholar]
- ENGLARD S., BREIGER H. H. Beef-heartmalic dehydrogenases. II. Preparation and properties of crystalline supernatant malic dehydrogenase. Biochim Biophys Acta. 1962 Jan 29;56:571–583. doi: 10.1016/0006-3002(62)90609-1. [DOI] [PubMed] [Google Scholar]
- GRIMM F. C., DOHERTY D. G. Properties of the two forms of malic dehydrogenase from beef heart. J Biol Chem. 1961 Jul;236:1980–1985. [PubMed] [Google Scholar]
- Hayden D. B., Cook F. S. Malate dehydrogenase in maize endosperm: the intracellular location and characterization of the two major particulate isozymes. Can J Biochem. 1972 Jun;50(6):663–671. doi: 10.1139/o72-091. [DOI] [PubMed] [Google Scholar]
- Henney H. R., Jr, Lynch T. Growth of Physarum flavicomum and Physarum rigidum in chemically defined minimal media. J Bacteriol. 1969 Aug;99(2):531–534. doi: 10.1128/jb.99.2.531-534.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyde E., Ainsworth S. Kinetic studies on the mechanism of the malate dehydrogenase reaction. J Biol Chem. 1968 May 10;243(9):2413–2423. [PubMed] [Google Scholar]
- KAPLAN N. O. Symposium on multiple forms of enzymes and control mechanisms. I. Multiple forms of enzymes. Bacteriol Rev. 1963 Jun;27:155–169. doi: 10.1128/br.27.2.155-169.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KORN E. D., GREENBLATT C. L., LEES A. M. SYNTHESIS OF UNSATURATED FATTY ACIDS IN THE SLIME MOLD PHYSARUM POLYCEPHALUM AND THE ZOOFLAGELLATES LEISHMANIA TARENTOLAE, TRYPANOSOMA LEWISI, AND CRITHIDIA SP.: A COMPARATIVE STUDY. J Lipid Res. 1965 Jan;6:43–50. [PubMed] [Google Scholar]
- Kitto G. B., Kottke M. E., Bertland L. H., Murphey W. H., Kaplan N. O. Studies on malate dehydrogenases and aspartate aminotransferases from Neurospora crassa. Arch Biochem Biophys. 1967 Jul;121(1):224–232. doi: 10.1016/0003-9861(67)90028-8. [DOI] [PubMed] [Google Scholar]
- Kitto G. B., Lewis R. G. Purification and properties of tuna supernatant and mitochondrial malate dehydrogenases. Biochim Biophys Acta. 1967 May 16;139(1):1–15. doi: 10.1016/0005-2744(67)90107-6. [DOI] [PubMed] [Google Scholar]
- LaNoue K. F., Williamson J. R. Interrelationships between malate-aspartate shuttle and citric acid cycle in rat heart mitochondria. Metabolism. 1971 Feb;20(2):119–140. doi: 10.1016/0026-0495(71)90087-4. [DOI] [PubMed] [Google Scholar]
- Lowenstein J. M. Citrate and the conversion of carbohydrate into fat. Biochem Soc Symp. 1968;27:61–86. [PubMed] [Google Scholar]
- MUNKRES K. D., RICHARDS F. M. GENETIC ALTERATION OF NEUROSPORA MALATE DEHYDROGENASE. Arch Biochem Biophys. 1965 Mar;109:457–465. doi: 10.1016/0003-9861(65)90390-5. [DOI] [PubMed] [Google Scholar]
- Markert C. L., Møller F. MULTIPLE FORMS OF ENZYMES: TISSUE, ONTOGENETIC, AND SPECIES SPECIFIC PATTERNS. Proc Natl Acad Sci U S A. 1959 May;45(5):753–763. doi: 10.1073/pnas.45.5.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElroy F. A., Wong G. S., Williams G. R. Distribution of malate across the mitochondrial membrane as a signifacant factor in respiratory control. Arch Biochem Biophys. 1968 Nov;128(2):563–565. doi: 10.1016/0003-9861(68)90063-5. [DOI] [PubMed] [Google Scholar]
- McReynolds M. S., Kitto G. B. Purification and properties of Drosophila malate dehydrogenases. Biochim Biophys Acta. 1970 Feb 11;198(2):165–175. doi: 10.1016/0005-2744(70)90048-3. [DOI] [PubMed] [Google Scholar]
- Mukerji S. K., Ting I. P. Malic dehydrogenase isoenzymes in green stem tissue of Opuntia: isolation and characterization. Arch Biochem Biophys. 1969 May;131(2):336–351. doi: 10.1016/0003-9861(69)90406-8. [DOI] [PubMed] [Google Scholar]
- Ozaki H., Whiteley A. H. L-malate dehydrogenase in the development of the sea urchin Strongylocentrotus purpuratus. Dev Biol. 1970 Feb;21(1):196–215. doi: 10.1016/0012-1606(70)90068-0. [DOI] [PubMed] [Google Scholar]
- PURVIS J. L., LOWENSTEIN J. M. The relation between intra- and extramitochondrial pyridine nucleotides. J Biol Chem. 1961 Oct;236:2794–2803. [PubMed] [Google Scholar]
- RAVAL D. N., WOLFE R. G. Malic dehydrogenase. IV. pH dependence of the kinetic parametrs. Biochemistry. 1962 Nov;1:1118–1123. doi: 10.1021/bi00912a024. [DOI] [PubMed] [Google Scholar]
- Rocha V., Ting I. P. Malate dehydrogenases of leaf tissue from Spinacia oleracea: properties of three isoenzymes. Arch Biochem Biophys. 1971 Nov;147(1):114–122. doi: 10.1016/0003-9861(71)90316-x. [DOI] [PubMed] [Google Scholar]
- SHRAGO E., FALCONE A. B. Purification and properties of human-erythrocyte malic dehydrogenase. Biochim Biophys Acta. 1963 May 7;73:7–16. doi: 10.1016/0006-3002(63)90354-8. [DOI] [PubMed] [Google Scholar]
- SIEGEL L., ENGLARD S. Beef-heart malic dehydrogenases. III. Comparative studies of some properties of M-malic dehydrogenase and S-malic dehydrogenase. Biochim Biophys Acta. 1962 Oct 8;64:101–110. doi: 10.1016/0006-3002(62)90763-1. [DOI] [PubMed] [Google Scholar]
- Simon H. L., Henney H. R. Chemical composition of slime from three species of myxomycetes. FEBS Lett. 1970 Mar 16;7(1):80–82. doi: 10.1016/0014-5793(70)80623-8. [DOI] [PubMed] [Google Scholar]
- Stromeyer C. T., Cole F. E., Arquembourg P. C. Purification and properties of malate dehydrogenase from Chlorella pyrenoidosa. Catalytic mechanism of the particulate form. Biochemistry. 1971 Mar 2;10(5):729–735. doi: 10.1021/bi00781a002. [DOI] [PubMed] [Google Scholar]
- Sulebele G., Silverstein E. Malate dehydrogenase and aspartate aminotransferase of Phycomyces blakesleeanus. Arch Biochem Biophys. 1969 Sep;133(2):425–435. doi: 10.1016/0003-9861(69)90472-x. [DOI] [PubMed] [Google Scholar]
- WIGGERT B. O., VILLEE C. A. MULTIPLE MOLECULAR FORMS OF MALIC AND LACTIC DEHYDROGENASES DURING DEVELOPMENT. J Biol Chem. 1964 Feb;239:444–451. [PubMed] [Google Scholar]
- Whitt G. S. Genetic variation of supernatant and mitochondrial malate dehydrogenase isozymes in the teleost Fundulus heteroclitus. Experientia. 1970;26(7):734–736. doi: 10.1007/BF02232514. [DOI] [PubMed] [Google Scholar]
- Zee D. S., Zinkham W. H. Malate dehydrogenase in Ascaris suum: characterization, ontogeny, and genetic control. Arch Biochem Biophys. 1968 Aug;126(2):574–584. doi: 10.1016/0003-9861(68)90444-x. [DOI] [PubMed] [Google Scholar]