Abstract
The C. elegans the 4-cell stage blastomere EMS is an endomesodermal precursor. Its anterior daughter, MS, makes primarily mesodermal cells, while its posterior daughter E generates the entire intestine. The gene regulatory network underlying specification of MS and E has been the subject of study for more than 15 years. A key component of the specification of the two cells is the involvement of the Wnt/β-catenin asymmetry pathway, which through its nuclear effector POP-1, specifies MS and E as different from each other. Loss of pop-1 function results in the misspecification of MS as an E-like cell, because POP-1 directly represses the end-1 and end-3 genes in MS, which would otherwise promote an endoderm fate. A long-standing question has been whether POP-1 plays a role in specifying MS fate beyond repression of endoderm fate. This question has been difficult to ask because the only chromosomal lesions that remove both end-1 and end-3 are large deletions removing hundreds of genes. Here, we report the construction of bona fide end-1 end-3 double mutants. In embryos lacking activity of end-1, end-3 and pop-1 together, we find that MS fate is partially restored, while E expresses early markers of MS fate and adopts characteristics of both MS and C. Our results suggest that POP-1 is not critical for MS specification beyond repression of endoderm specification, and reveal that Wnt-modified POP-1 and END-1/3 further reinforce E specification by repressing MS fate in E. By comparison, previous work suggested that in the related nematode C. briggsae, Cb-POP-1 is not required to repress endoderm specification in MS, in direct contrast with Ce-POP-1, but is critical for repression of MS fate in E. The findings reported here shed new light on the flexibility of combinatorial control mechanisms in endomesoderm specification in Caenorhabditis.
Keywords: C. elegans, endomesoderm, specification, POP-1, gene network
Introduction
Combinatorial control, achieved through spatiotemporally precise activation of genes in regulatory networks, is a central mechanism by which cells choose appropriate pathways of cell specification in metazoan development. Elucidation of such networks allows an appreciation for the complexity of the instructions encoded in the genome that bring about embryogenesis from the zygote. The endomesoderm specification network in the nematode C. elegans is a model for such networks in general. While an exhaustive compendium of protein-DNA interactions has yet to be made, decades of forward and reverse genetics, transcriptome analysis and protein-DNA studies have revealed a core network that includes motifs found in many other similar networks, such as feed-forward loops and autoregulation (Maduro, 2006). Evolutionary comparisons of this network have begun to be performed in related nematodes, and important similarities and differences have been identified (Coroian et al., 2005; Lin et al., 2009).
In C. elegans, the 4-cell stage blastomere EMS is an endomesoderm precursor: Its anterior daughter, MS, generates some 80 cells that are primarily mesodermal, which includes a portion of the body muscles, many posterior cells of the pharynx, and four embryonically-derived coelomocytes (Fig. 1A) (Sulston et al., 1983). Its posterior sister E is the sole endoderm progenitor and generates 20 intestinal cells. Specification of MS and E involves the participation of two pathways that work in parallel (Fig. 1B): The SKN-1 pathway assigns endomesodermal identity to the EMS daughters, while the Wnt/β-catenin asymmetry pathway directs the two cells to adopt different fates (Bowerman et al., 1992; Lin et al., 1995; Maduro and Rothman, 2002; Mizumoto and Sawa, 2007; Rocheleau et al., 1997; Rocheleau et al., 1999; Thorpe et al., 1997).
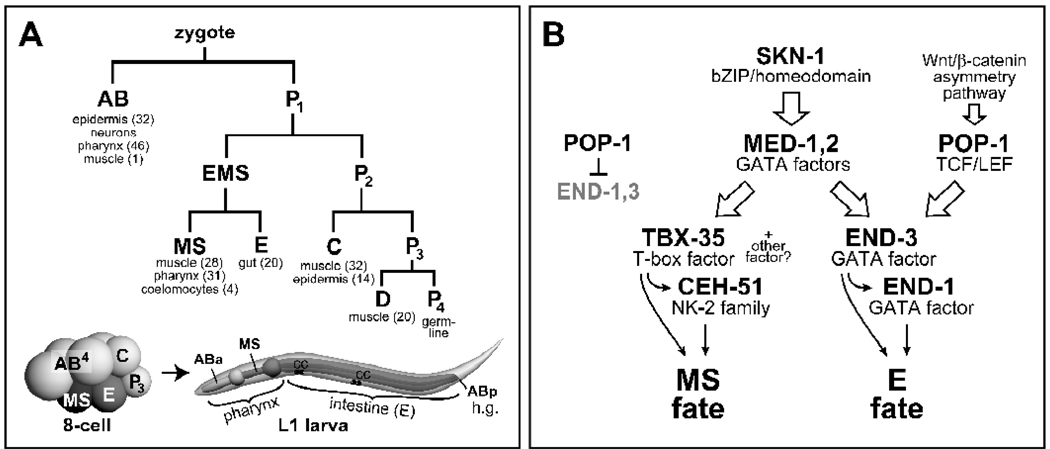
Fig. 1.
The early C. elegans lineage and an abbreviated version of the endomesoderm gene regulatory network. (A) An abbreviated lineage shows the origin of the six founder cells and tissues made by each, with numbers of cells for gut, pharynx, muscle and coelomocytes in brackets (Sulston et al., 1983). Diagrams of an 8-cell stage embryo and larva show the lineal origins of the components of the digestive tract: Foregut/pharynx, midgut, and hindgut/rectum (h.g.). The assignment of ABa to ‘anterior’ pharynx and MS to ‘posterior’ pharynx is convenient but not strictly correct (Priess et al., 1987). On the larva, coelomocytes are indicated by ‘CC’. (B) A flow diagram depicts a simplified version of the endomesoderm gene regulatory network, showing combinatorial control through the SKN-1 and Wnt/β-catenin pathways (Broitman-Maduro et al., 2009; Maduro, 2008).
Endomesoderm specification begins with maternal SKN-1, a bZIP/homeodomain transcription factor that is present in the nuclei of EMS and its sister P2 at the 4-cell stage (Bowerman et al., 1993). Loss of SKN-1 leads to misspecification of MS all the time, and E most (~70–80%) of the time (Bowerman et al., 1992). In addition to the MS-derived posterior portion of the pharynx, mutants for skn-1 also lack the anterior portion of the pharynx, which is specified by a GLP-1/Notch-dependent cell-cell interaction between MS and descendants of the AB founder cell (Mello et al., 1994; Priess et al., 1987). Within EMS, SKN-1 activates expression of a presumptive ligand for this interaction and the med-1,2 divergent GATA factor gene pair (Lowry et al., 2009; Maduro et al., 2001). Loss of med-1,2 together results in a penetrant mis-specification of MS, and low-penetrance mis-specification of E, owing to parallel inputs into E specification (Broitman-Maduro et al., 2009; Goszczynski and McGhee, 2005; Maduro et al., 2007; Maduro et al., 2001). Within the early MS lineage, the MEDs activate the T-box gene tbx-35, and TBX-35 (perhaps in combination with another factor) activates the NK-2 homeobox gene ceh-51 (Broitman-Maduro et al., 2009). Loss of tbx-35 and ceh-51 together results in a penetrant mis-specification of MS that resembles the MS phenotype of med-1,2 mutants (Broitman-Maduro et al., 2009). TBX-35 and CEH-51 are hypothesized to activate further pathways that lead to specification of pharynx, muscle and other tissues made by MS. Pharynx specification involves activation of a gene network with PHA-4/FoxA at the top (Gaudet and Mango, 2002), and which includes the pharynx muscle-specific regulator CEH-22/Nkx2.5 (Okkema and Fire, 1994). Muscle specification occurs via a three-way collaboration of HND-1/Hand, HLH-1/MyoD and UNC-120/Srf (Fukushige et al., 2006) and activation of a muscle gene network (Roy et al., 2002).
Within the early E lineage, the MEDs, SKN-1, the Wnt effector POP-1/TCF, and PAL-1/Caudal contribute to endoderm specification through activation of the GATA factor genes end-1 and end-3. Expression of end-3 occurs slightly earlier than end-1 and there is evidence that END-3 also contributes to end-1 activation (Baugh et al., 2003; Maduro et al., 2007). Downstream of end-1,3, the principal target is the GATA factor gene elt-2 (Fukushige et al., 1998), which activates a network of targets for intestinal development (McGhee et al., 2009; McGhee et al., 2007; Pauli et al., 2006).
For both MS and E, loss of function of SKN-1, the MEDs, ENDs or TBX-35/CEH-51 causes adoption of a C-like fate by the mis-specified blastomeres. In contrast, mutations in the Wnt/β-catenin asymmetry pathway, which makes MS and E different, result in either an MS to E transformation (a ‘Pop’ phenotype) or the reverse, an E to MS transformation (a ‘Mom’ phenotype). Specification of E requires a cell-cell interaction between EMS and its sister P2 (Goldstein, 1992). This interaction involves overlapping Wnt/MAPK/Src pathways and ultimately results in differential modification of the nuclear effector POP-1/TCF (Bei et al., 2002; Lin et al., 1998; Lin et al., 1995; Meneghini et al., 1999; Rocheleau et al., 1997; Rocheleau et al., 1999; Shin et al., 1999; Thorpe et al., 1997). This modification results in the nuclear export of POP-1 in E (a phenomenon called ‘POP-1 asymmetry’), lowering its concentrations relative to the divergent β-catenin SYS-1, and permitting it to function as an activator in E (Huang et al., 2007; Maduro et al., 2002; Phillips et al., 2007). Within MS, the high nuclear levels of POP-1 allow it to repress end-1 and end-3, while in E, POP-1 contributes to activation of (at least) end-1 (Maduro et al., 2005b; Maduro et al., 2002; Shetty et al., 2005). Mutations in the upstream Wnt/MAPK/Src components result in a Mom phenotype, while loss of pop-1 is epistatic to Mom mutants and results in a Pop phenotype. POP-1 and the Wnt/β-catenin asymmetry pathway form a switch that is used multiple times in C. elegans development (Kaletta et al., 1997; Lin et al., 1998; Mizumoto and Sawa, 2007).
Outside of the EMS lineage, multiple parallel pathways block activity of SKN-1 or promote its degradation (Bei et al., 2002; Lin, 2003; Maduro et al., 2001; Mello et al., 1992; Page et al., 2007; Shirayama et al., 2006). For example, the CCCH zinc finger protein PIE-1 blocks activity of SKN-1 in P2: Loss of pie-1 function results in an ectopic mis-specification of the P2 daughters as MS- and E-like (Mello et al., 1992). Similarly, a gain-of-function mutation in oma-1, which encodes a zinc finger protein similar to PIE-1, results in ectopic mis-specification of the C blastomere as an EMS-like cell, due to increased concentrations of SKN-1 in C (Lin, 2003).
The POP-1 switching system in C. elegans participates in the MS/E decision primarily by repressing endoderm specification in MS, although it makes a weaker contribution to endoderm specification in E (Lin et al., 1995; Shetty et al., 2005). In the related nematode, C. briggsae, analysis of the Cb-pop-1 and Cb-skn-1 RNAi phenotypes led to the conclusion that Cb-POP-1 contributes to MS and E specification primarily as an essential activator of the Cb-end genes in E, with a parallel function in MS specification with Cb-SKN-1 (Lin et al., 2009). This observation, along with other observations regarding expression of MS factors in pop-1-depleted embryos (Broitman-Maduro et al., 2006; Broitman-Maduro et al., 2009), prompted us to examine the role of C. elegans POP-1 in MS outside of repression of endoderm specification.
Here, we describe the construction of chromosomal end-1 end-3 double mutants, and from a fully penetrant gut defect, conclude that these two genes are completely essential for endoderm specification in C. elegans. We test the requirement for POP-1 in MS specification outside of repression of the ends, and find that loss of pop-1 function in end-3 single mutants, and end-1,3 double mutants, results in an apparent restoration of some aspects of MS specification to the MS cell, and a transformation of the E lineage to one that makes both MS- and C-like tissues. Loss of end-1,3, or loss of pop-1, results in derepression of MS specification genes in the early E lineage, implying other functions for these regulators in E. These findings are integrated into an updated model for the C. elegans endomesoderm gene specification network, and are discussed in terms of a hierarchy of competing cell specification pathways and evolutionary flexibility in combinatorial control.
Materials and methods
Mutations and transgenic strains
Mutants: LG X: med-1(ok804). I: pop-1(zu189), dpy-5(e61). II: tbx-35(tm1789). III: med-2(cx9744), unc-119(ed3), unc-119(ed4). IV: him-8(e1498). V: end-1(ok558), end-3(ok1448), end-3(zu247), dpy-11(e224), unc-76(e911), him-5(e1490), ceh-51(tm2123). Rearrangements: hT1 [I;V]. Mapped reporters: wIs84 [elt-2::GFP] X, gvIs402 [unc-120::GFP] I, cdIs42 [cup-4::GFP] I, cdIs41 [cup-4::GFP] II, ruIs37 [myo-2::GFP] III, pxIs6 [pha-4::GFP] IV, cuIs1 [ceh-22::GFP] V. Unmapped: qtIs9 [nhr-25::YFP], wIs117 [med-1::GFP::POP-1]. Extrachromosomal arrays: teEx226 [tbx-35::GFP], teEx420 [med-1::GFP::SYS-1]. All pop-1(zu189) chromosomes also carried dpy-5(e61), permitting identification of homozygous zu189 mothers by their Dpy phenotype.
Construction of end-1 end-3 double mutant strains
We constructed a dpy-11 end-1 unc-76 triple mutant using standard methods (Fay, 2006) and confirmed homozygosity of ok558 by PCR. We obtained dpy-11 end-1 unc-76 / end-3(ok1448) heterozygotes after crossing with end-3(ok1448) males, from which 120 Dpy non-Unc self progeny were singled. Two of these segregated ~25% dead eggs lacking endoderm. We injected one of these with end-3(+) (pMM214) and unc-119::CFP (pMM809) to generate a dpy-11 end-1,3; Ex[CFP] line. PCR was used to confirm the presence of ok558 and ok1448 and absence of wild-type chromosomal end-1 and end-3 sequences (primer sequences are available on request). A similar strategy was used to recover a dpy-11 end-1 end-3(zu247) strain from 93 Dpy non-Unc progeny of dpy-11 end-1 unc-76 / end-3(zu247) heterozygotes. Strains were backcrossed to N2 to remove dpy-11, and derivative strains marked with unc-119::YFP (pMM531), unc-119::mCherry (pMM824) or sur-5::dsRed (pAS152) in the rescuing arrays were made by microinjection to replace the CFP-expressing array. Such strains are superficially wild-type and segregate arrested embryos and larvae that lack the transgene array. In total, three independent end-1,3 double mutants were isolated at a frequency of 1.4% (n=213), comparable to the predicted frequency of 1.1% obtained by subtracting the end-1 and end-3 interpolated genetic positions in WormBase (WS200).
To introduce the end-1(ok558) end-3(ok1448) chromosome into a pop-1(zu189) strain, males from MS749 [pop-1 dpy-5 / hT1[I;V] I; ruIs37 unc-119 III; him-5 / him-5 hT1[I;V] V] were crossed to end-1,3; Ex[unc-119::CFP] hermaphrodites. F1 CFP(+) males were crossed to MS749 to generate strain MS1172 [pop-1 dpy-5 / hTI I; ruIs37 III; end-1,3 / him-5 hT1 V]. From 50 singled wild-type CFP(+) MS1172 hermaphrodites, we obtained three isolates of genotype pop-1 dpy-5 / hT1 I; end-1,3 / end-1,3 hT1 V; ruIs37 III; Ex[unc-119::CFP] as confirmed by absence of CFP(−) progeny that contained gut.
A med-1,2; end-1,3 mutant strain was made as follows. med-1; med-2 males rescued by an unc-119::CFP array carrying med-1(+) were crossed to a dpy-11 end-1 end-3; pha-4::GFP strain rescued by an array carrying end-3(+) and unc-119::YFP. Dpy F2 hermaphrodites expressing YFP, CFP and GFP were singled to identify those showing complete transgene dependence on the med (+) and end (+) arrays, and which showed 100% transmission of pha-4::GFP. The two arrays were replaced by a single array expressing unc-119::mCherry and which carried cosmids T24D3 (med-1(+)) and F58E10 (end-1,3(+)). A cup-4::GFP version was made using a similar strategy.
To make a tbx-35; end-1 end-3 ceh-51 strain, we first constructed + + ceh-51 / end-1 end-3 + hermaphrodites carrying an unc-119::CFP array rescuing ceh-51 and an unc-119::YFP array rescuing end-1,3. YFP(+), CFP(+) animals were identified in which all viable progeny expressed CFP and which segregated gutless embryos (hence end-1 end-3 ceh-51 / + + ceh-51). YFP(+), CFP(+) animals were picked in the next generation and examined for expression of both arrays by all viable animals. These animals were crossed to MS1291 males to generate a quadruple mutant tbx-35; end-1 end-3 ceh-51 strain rescued by an unc-119::mCherry array (tbx-35(+), ceh-51(+)) and an unc-119::YFP array (end-3(+)). The cup-4::GFP and pha-4::GFP markers were introduced and the quadruple mutant genotypes re-segregated.
Microscopy, imaging and laser ablations
Embryos were imaged on agar pads using an Olympus BX-61 equipped with a Canon 350D camera. Images were processed using Adobe Photoshop 7. In images for birefringent granules and fluorescent reporter expression, images from multiple focal planes were digitally combined. Laser ablations were performed at the UC Riverside Microscopy and Imaging Core Facility as described (Lin et al. 2009).
Results
Chromosomal loss of end-1 and end-3 blocks endoderm specification
END-1 and END-3 are GATA factors that contain a single zinc finger and associated basic domain (Lowry and Atchley, 2000; Maduro et al., 2005a; Zhu et al., 1997). The end-1 and end-3 genes are located ~30kbp apart on chromosome V, apparently the products of an ancestral gene duplication (Gillis et al., 2007). Deletions that remove both end-1 and end-3 and a large number of neighboring genes, defining the Endoderm Determining Region (EDR), result in a onefold embryonic arrest and a complete absence of gut (Zhu et al., 1997). In contrast, single mutants of end-1 and end-3 have only minor effects on endoderm. The ok558 lesion of end-1, which removes the DNA-binding domain, results in no apparent phenotype, while the zu247 allele, a missense mutation causing a P202L change in the END-3 DNA-binding domain, and ok1448, which deletes the domain, both result in mild endoderm defects (5%-9% lacking endoderm; Table 1 and Maduro et al., 2005a; Maduro et al., 2005b). The single end mutants can enhance the penetrance of other mutants that themselves have only a partial endoderm specification defect (Maduro et al., 2005a; Maduro et al., 2005b). RNAi of end-3 in an end-1 mutant blocked endoderm specification in 89–93% of progeny embryos (Maduro et al., 2005a), but it was not resolved whether the incomplete penetrance reflected the efficiency of RNAi targeted to end-3 or if there are other genes in the EDR that can contribute to endoderm specification. Indeed, a nuclear hormone receptor gene located in the EDR, R10D12.2/dpr-1 (previously named end-2) could apparently rescue endoderm specification in a small proportion of transgenic embryos homozygous for a deficiency that removes end-1 and end-3 (Zhu et al., 1997).
Table 1.
Endoderm specification in wild-type and mutant strains
| Genotype | % of embryos containing endoderm1 |
|---|---|
| wild type (N2) | 100% (>500) |
| end-1(ok558) | 100% (322) |
| end-3(ok1448) | 95% (155) |
| end-3(zu247) | 91% (247) |
| end-1(ok558) end-3(ok1448) | 0% (190) |
| end-1(ok558) end-3(zu247) | 0% (377) |
| end-1(ok558) end-3(ok1448); Ex[end-1,3(+)] | 99% (134) |
| end-1(ok558) end-3(zu247); Ex[end-1,3(+)] | 96% (83) |
| end-1(ok558) end-3(ok1448); pie-1(RNAi) | 0% (49) |
| end-1(ok558) end-3(zu247); pie-1(RNAi) | 0% (72) |
| end-3(zu247); pop-1(RNAi) | 3% (175) |
| end-3(ok1448); pop-1(zu189)2 | 9% (274) |
| end-3(ok1448); pop-1(RNAi) | 1% (262) |
| end-1(ok558) end-3(zu247); pop-1(RNAi) | 0% (51) |
| end-1(ok558) end-3(ok1448); pop-1(RNAi) | 0% (127) |
| end-1(ok558) end-3(ok1448); pop-1(zu189)2 | 0% (34) |
Some data were previously reported in Maduro et al. 2005b.
embryos also homozygous for dpy-5(e61).
To directly assess the requirement for end-1 and end-3 in endoderm specification, we constructed double mutants of end-1(ok558) with end-3(ok1448) and end-3(zu247) (see Materials and Methods). As the same end-1 allele was used for all analyses presented here, the end-1(ok558) end-3(ok1448) genotype will be referred to as end-1,3(ok1448) and the end-1(ok558) end-3(zu247) genotype as end-1,3(zu247). Both end-1,3(ok1448) and end-1,3(zu247) double mutants failed to make endoderm 100% of the time (n=377 and n=190) as scored by morphology with DIC microscopy and using polarized light to detect birefringent gut granules (Fig. 2A–D) (Laufer et al., 1980). Hence, although the zu247 allele is only a missense mutation, all endoderm specification in end-3(zu247) is dependent on end-1. Ectopic endoderm made in pie-1(RNAi) embryos also showed a requirement for end-1,3 in the two strains (Table 1) (Mello et al., 1992). The majority of end-1,3 embryos (83%, n=124 for end-1,3(ok1448)) underwent a developmental arrest after completing elongation, with many of these eventually hatching as arrested L1s; the remainder arrested as embryos at varying stages of elongation. The gut, elongation and viability defects were restored to 96–99% of end-1,3 double mutants by an array carrying end-1(+) and end-3(+) or end-3(+) alone (Table 1, Fig. 2 and data not shown).
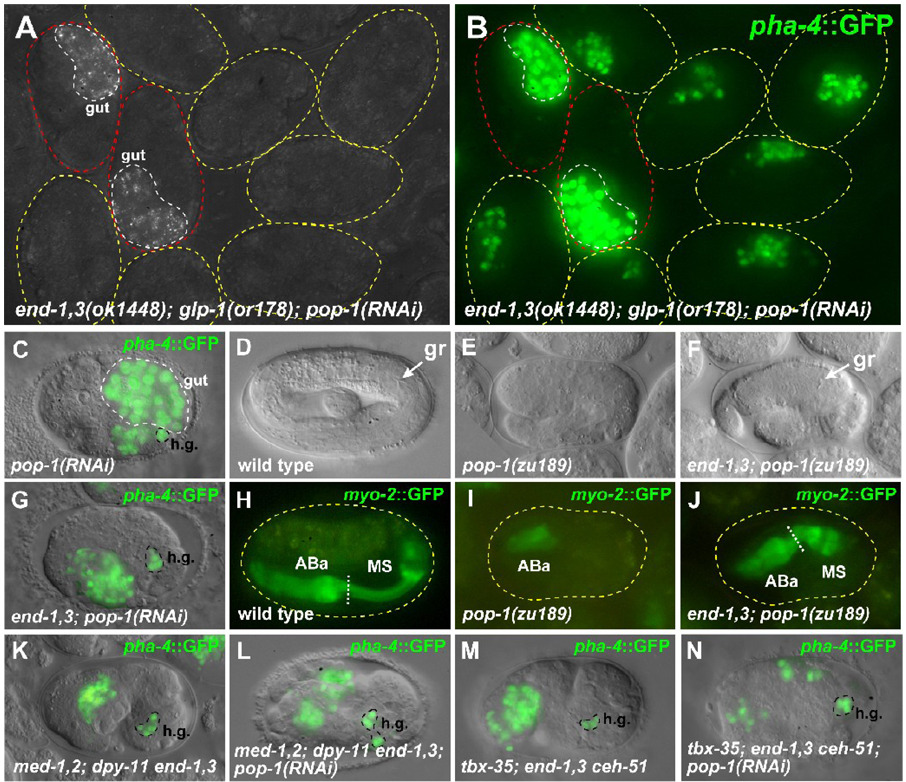
Fig. 2.
Chromosomal end-1,3 double mutants lack endoderm. (A) DIC appearance of end-1(ok558) end-3(ok1448) embryos. The embryo on the left is rescued by an extrachromosomal transgene array carrying end-1(+) and end-3(+) (referred to as an end-1,3(+) array). On the middle embryo, internal cavities are indicated by arrowheads. (B) Polarized light micrograph showing appearance of birefringent gut granules in the rescued embryo (left, outlined in red) but not in the mutant embryos (outlined in yellow). (C) end-1(ok558) end-3(zu247) embryos. The embryo on the left carries an end-1,3(+) array. (D) Gut granules present only in the rescued embryo (red outline). (E) Newly-hatched wild-type L1 (left) and arrested end-1(ok558) end-3(ok1448) double mutant. The gut in the wild-type has been shaded blue, and the pharynx and hindgut (rectum) have been shaded in red. The wild-type is approximately 240µm long. (F) Expression of elt-2::GFP (Fukushige et al., 1998) in a wild-type elongated embryo. DIC and fluorescence image have been digitally combined in this panel and panels G, H, J and K. (G,H) Absence of elt-2 expression in end-1(ok558) end-3(ok1448) and end-1(ok558) end-3(zu247) embryos. (I) Accumulation of gut granule-like material in part of the excretory cell (arrowheads) of an end-1(ok558) end-3(zu247) arrested embryo. (J) Expression of pha-4::GFP (Horner et al., 1998; Kalb et al., 1998) in a developing end-1,3 embryo carrying an end-1,3(+)array. The pharynx and hindgut (h.g.) components of pha-4 expression are indicated; the remaining expressing cells are from the developing intestine. (K) Absence of the intestinal component of pha-4 expression in an end-1(ok558) end-3(ok1448) embryo at a similar stage as panel J. (L) end-1(ok558) end-3(ok1448) embryos arrested with a one-fold appearance following heat shock overexpression of elt-2. Two embryos that carry an end-1,3(+) array are indicated. (M) Accumulation of large amounts of gut granules in rescued (red outline) and non-rescued (yellow outline) embryos. A C. elegans embryo is approximately 50µm long. In panels J and K, anterior is to the left and dorsal is up.
Two additional phenotypes were noted among end-1,3 mutant embryos. First, ectopic gut granule-like material was observed in the excretory cell in 11% (n=90) of end-1,3(zu247) and 13% (n=106) of end-1,3(ok1448) embryos (Fig. 2I). Second, while wild-type L1s were measured to be 242 ± 4.4 µm (n=12) long, hatched end-1(ok558) end-3(ok1448) double mutants were only about 75% as long, with an average length of 175 ± 7µm (n=19, p<10−8) (Fig. 2E). Such hatched end-1,3 embryos frequently contained apparent hypodermal abnormalities (not shown). Excretory-cell granules and a contraction of embryo length were also reported for mutants in the glo genes, which have defects in gut granule biogenesis but nonetheless make an intestine (Hermann et al., 2005). At least one of these, glo-1, is expressed in the embryonic E lineage and the intestine through adulthood, and is proposed to be a target of END-1,3 (Hermann et al., 2005). By in situ hybridization, we detected transcripts for glo-1 in the E lineage in 86% (n=49) of wild-type embryos at gastrulation stage or later. In contrast, expression was detected in only 33% (n=42) of embryos from an end-1,3(ok1448) strain rescued by an array with a transmission frequency of ~40%, consistent with the absence of glo-1 expression in all end-1,3 mutant embryos (data not shown). These results suggest that later embryonic phenotypes of end-1,3 double mutants result from the absence of functions that are normally provided to the embryo by the gut, such as osmoregulation or lysosomal trafficking (Hermann et al., 2005).
In prior studies that examined the fate of E descendants in EDR deficiency embryos or following RNAi of end-1 and end-3, E was observed to adopt a C-like fate, generating epidermis and body muscle cells instead of gut (Maduro et al., 2005a; Zhu et al., 1997). A small number of end-3(zu247) mutant embryos appeared to show evidence of partial transformation to an MS-like fate, suggesting that at least part of the E lineage can produce ectopic MS-like cells when endoderm specification is compromised (Maduro et al., 2005a). The chromosomal double end-1,3 mutant embryos were examined for evidence of the tissue types made by E descendants. First, the formation of internal cavities has previously been associated with internalization of ectopic C-like cells in skn-1, med-1,2 or tbx-35 mutants, resulting in ectopic epidermis in the descendants (Bowerman et al., 1992; Broitman-Maduro et al., 2006). We observed such cavities in 13% (n=176) of end-1(ok558) end-3(zu247) and 15% (n=124) of end-1(ok558) end-3(ok1448) embryos (Fig. 2A and data not shown). By comparison, cavities were observed in approximately 30% of med-1,2 and tbx-35 mutants (Broitman-Maduro et al., 2006; Maduro et al., 2001).
To more precisely evaluate the fate of E descendants in end-1,3 embryos, a laser microbeam was used to isolate early blastomeres in end-1,3(ok1448) embryos carrying various tissue-specific reporters. Expression of nhr-25::YFP, a reporter that is normally expressed in epidermis (Baugh et al., 2005), did not occur in the embryonic intestine of intact embryos (n=80), while descendants of isolated C blastomeres showed expression as expected (5/5 partial embryos). However, 10/10 isolated E blastomeres from end-1,3(ok1448) embryos displayed nhr-25::YFP (Table 3). Production of body muscle cells from isolated E blastomeres was observed in end-1,3(ok1448) mutant embryos (n=3) as assessed by expression of unc-120::GFP (Table 3) (Fukushige et al., 2006). Moreover, in intact end-1,3(ok1448) and end-1,3(zu247) embryos, neither ectopic pharynx (scored by pha-4::GFP) nor ectopic coelomocytes (cup-4::GFP) were apparent (Table 2) (Horner et al., 1998; Kalb et al., 1998; Patton et al., 2005). These results confirm that chromosomal loss of end-1 and end-3 results causes E to adopt a C-like fate, making body muscles and epidermal cells. Furthermore, we could find no evidence that such embryos produce ectopic pharynx or coelomocyte cells as would be consistent with ectopic MS specification.
Table 3.
Production of tissues in laser-operated embryos
| genotype | blastomeres ablated or isolated |
partial embryos making tissues (# of cells) | |||
|---|---|---|---|---|---|
| coelomocytes (cup-4::GFP) |
pharynx muscle (myo-2::GFP)1 |
body muscle (unc-120::GFP) |
epidermis (nhr-25::YFP)1 |
||
| wild type | MS abl. | 0/3 (0.0 ± 0.0) | - | - | - |
| MS iso. | 3/3 (4.0 ± 0.0) | - | - | 0/1 | |
| E iso. | 0/6 (0.0 ± 0.0) | - | 0/4 (0.0 ± 0.0) | 0/6 | |
| C iso. | - | - | - | 5/5 | |
| skn-1(RNAi) | MS iso. | - | - | - | 3/3 |
| end-3(ok1448) | MS abl. | 0/5 (0.0 ± 0.0) | - | - | - |
| E abl. | 2/2 (4.0 ± 0.0) | - | - | - | |
| end-3(ok1448); pop-1(zu189)2 | ABa+MS abl. | - | 3/3 (weak) | - | - |
| ABa+E abl. | - | 2/2 | - | - | |
| ABa+MS+E abl. | - | 0/3 | - | - | |
| end-3(ok1448); pop-1(RNAi) | MS abl. | 4/4 (3.0 ± 0.6) | - | - | - |
| E abl. | 5/5 (3.0 ± 0.5) | - | - | - | |
| MS+E abl. | 1/4 (0.3 ± 0.2) | - | - | - | |
| end-1(ok558) end-3(ok1448) | MS abl. | 1/5 (0.2 ± 0.2) | - | - | - |
| E abl. | 5/5 (3.6 ± 0.2) | - | - | - | |
| MS+E abl. | 0/3 (0.0 ± 0.0) | - | - | - | |
| MS iso. | 5/5 (4.0 ± 0.0)3 | - | 3/3 (18.7 ± 2.0) | 0/53 | |
| E iso. | 0/4 (0.0 ± 0.0)3 | - | 3/3 (15.3 ± 0.7) | 10/103 | |
| pop-1(zu189)2 | MS abl. | - | 1/1 | - | - |
| ABa abl. | - | 0/3 | - | - | |
| pop-1(RNAi) | MS iso. | - | - | - | 0/3 |
| E iso. | - | - | - | 0/4 | |
| end-1(ok558) end-3(ok1448); | ABa+MS abl. | - | 5/5 | - | - |
| pop-1(zu189)2 | ABa+E abl. | - | 4/4 | - | - |
| ABa+MS+E | - | 0/3 | - | - | |
| end-1(ok558) end-3(ok1448); | MS abl. | 3/3 (6.7 ± 0.5) | - | - | - |
| pop-1(RNAi) | E abl. | 3/3 (4.7 ± 0.7) | - | - | - |
| MS+E abl. | 0/4 (0.0 ± 0.0) | - | - | - | |
| MS iso. | 4/4 (5.0 ± 0.4) | - | - | 0/8 | |
| E iso. | 6/6 (5.5 ± 0.3) | - | 6/6 (10.2 ± 1.2) | 9/11 | |
| ABx+MS abl. | - | 5/8 | - | - | |
| ABx+E abl. | - | 4/6 | - | - | |
Numbers of cells were not counted because individual cells could not be distinguished.
Embryos from homozygous pop-1(zu189) dpy-5(e61) mothers (with other mutations as indicated).
Numbers from experiments on a strain carrying both nhr-25::YFP and cup-4::GFP were included.
Abbreviations: abl, ablated; iso, isolated (all other blastomeres ablated); ABx, both daughters of AB were ablated.
Table 2.
Coelomocytes and pharynx cells made in wild-type, mutant and RNAi backgrounds
| Genotype | coelomocytes cup-4::GFP1 |
pharynx cells pha-4::GFP1,2 |
|---|---|---|
| Wild type (N2) | 3.7 ± 0.2 (105) | 50.0 ± 0.9 (21) |
| pop-1(RNAi) | 0.0 ± 0.0 (50) | 21.3 ± 0.8 (18) |
| glp-1(RNAi) | nd | 23.1 ± 0.6 (15) |
| end-1(ok558) end-3(ok1448) | 4.5 ± 0.1 (65) | 49.1 ± 1.0 (10) |
| end-1(ok558) end-3(zu247) | 4.6 ± 0.1 (77) | 51.1 ± 1.3 (15) |
| end-3(ok1448); pop-1(RNAi) | 6.1 ± 0.2 (77) | 28.5 ± 7.0 (22) |
| end-1(ok558) end-3(zu247); pop-1(RNAi) | 9.3 ± 0.2 (80) | 43.5 ± 1.3 (23) |
| end-1(ok558) end-3(ok1448); pop-1(RNAi) | 10.7 ± 0.3 (77) | 44.8 ± 2.2 (17) |
| end-3(ok1448); glp-1(or178) | nd | 21.2 ± 0.8 (18) |
| end-3(ok1448); glp-1(or178); pop-1(RNAi) | nd | 9.5 ± 0.8 (75) |
| end-1(ok558) end-3(ok1448); glp-1(or178) | nd | 25.2 ± 1.0 (17) |
| end-1(ok558) end-3(ok1448); glp-1(or178); | nd | 16.8 ± 1.1 (93) |
| pop-1(RNAi) | ||
| end-1(ok558) end-3(ok1448); glp-1(or178); | nd | 1.4 ± 0.3 (40) |
| pop-1(RNAi); Ex[end-1,3(+)] | ||
| med-1(ok804); med-2(cx9744) | 0.1 ± 0.0 (34) | 31.3 ± 0.6 (26) |
| ceh-51(tm2123); tbx-35(tm1789) | 0.2 ± 0.0 (124) | 30.2 ± 0.5 (44) |
| med-1(ok804); med-2(cx9744); end-1(ok558) | 0.0 ± 0.0 (52) | 29.9 ± 1.1 (12) |
| end-3(ok1448)3 | ||
| med-1(ok804); med-2(cx9744); end-1(ok558) | 1.0 ± 0.2 (57) | 22.4 ± 0.9 (24) |
| end-3(ok1448); pop-1(RNAi)3 | ||
| end-1(ok558) end-3(ok1448) ceh-51(tm2123); | 0.1 ± 0.0 (122) | 30.3 ± 1.0 (21) |
| tbx-35(tm1789) | ||
| end-1(ok558) end-3(ok1448) ceh-51(tm2123); | 0.1 ± 0.1 (35) | 22.1 ± 1.4 (19) |
| tbx-35(tm1789); pop-1(RNAi) |
Some data are from Broitman-Maduro et al. 2009.
Expression of pha-4 in gut and/or rectum cells (if present) was not included.
Embryos were also homozygous for dpy-11(e224).
The apparent target of end-1 and end-3 is the GATA factor gene elt-2 (Fukushige et al., 1998; Maduro et al., 2005a; Maduro and Rothman, 2002). ELT-2 is not essential for embryonic gut specification, but is required for maintenance of the gut fate (Fukushige et al., 1998). A chromosomally integrated elt-2::GFP reporter was introduced into the end-1,3 double mutants. Expression of elt-2 was absent in 100% (n=112) of end-1,3(ok1448) and 100% (n=142) of end-1,3(zu247) embryos (Fig. 2F–H). Similarly, expression of a reporter for the gut esterase gene ges-1, a direct target of ELT-2 (Egan et al., 1995), and the intestine expression of pha-4 (Horner et al., 1998), were both absent in end-1,3(ok1448) double mutants (n=238 and n=30, respectively; Fig. 2J,K). Hence, end-1 and end-3 together are necessary for activation of elt-2 and its downstream targets.
To test if overexpression of ELT-2 in an end-1 end-3 double mutant background was sufficient to restore endoderm, we introduced a heat shock (hs) elt-2 integrated transgene (Fukushige et al., 1998) into the end-1,3(ok1448) background rescued by a transgenic end-1,3(+) extrachromosomal array that can be scored for its presence by sur-5::dsRed expression (Yochem et al., 1998). Among heat shocked embryos that contained excess gut, 36% (16/44) carried the end-1,3(+) rescuing array, while the remainder (64%, 28/44) did not, confirming that overexpression of ELT-2 is sufficient to specify intestinal fate in the absence of end-1 and end-3 (Fig. 2L,M).
Taken together, these results confirm that end-1 and end-3 together are essential for endoderm specification, that E adopts a C-like fate in end-1,3 double mutants, and that a key contribution of END-1,3 to intestine fate is the activation of elt-2. Furthermore, the ability of end-1,3 mutants to elongate and hatch demonstrates the capacity of the embryo to undergo morphogenesis in the complete absence of the endoderm.
MS-derived tissues are made in pop-1; end-3 and pop-1; end-1,3 embryos
The nuclear Wnt effector TCF/POP-1 plays a central role in specification of MS. Loss of maternal pop-1 function through the maternal-specific allele zu189 or by RNAi results in a penetrant mis-specification of MS as an E-like precursor (Lin et al., 1998; Lin et al., 1995). Consistent with negative regulation of the end genes by POP-1 in MS, end-1 and end-3 are expressed ectopically in both MS and E in pop-1 mutant embryos (Maduro et al., 2005a; Maduro et al., 2007; Shetty et al., 2005). This repression is direct, as GFP::POP-1 interacts in vivo with extrachromosomal arrays containing either promoter (Maduro et al., 2002), and a POP-1 binding site is required for repression of an end-1 transgene reporter in MS (Shetty et al., 2005). What has not been clear from prior studies with pop-1 and MS fate is whether there is a requirement for POP-1 in MS in addition to repression of endoderm fate. Indeed, the MS specification genes tbx-35 and ceh-51 are still expressed in MS in pop-1 mutants (Broitman-Maduro et al., 2006; Broitman-Maduro et al., 2009), suggesting that the normal pathways for MS specification might still be active.
A direct assessment of the involvement of POP-1 in MS specification could be made by evaluating the phenotype of pop-1 mutant embryos in which endoderm specification is blocked by reduction or elimination of end activity. Interpretation of these results may be complicated by the fact that POP-1 and the Wnt/β-catenin asymmetry pathway function in many asymmetric cell divisions, including those that follow the MS-E division within the endomesoderm (Huang et al., 2007; Kaletta et al., 1997; Lin et al., 1998; Maduro et al., 2002; Mizumoto and Sawa, 2007; Schroeder and McGhee, 1998). In the embryo, pop-1 function in MS and E is contributed maternally, as revealed by the maternal-effect zu189 allele of pop-1 (Lin et al., 1995). In contrast, pop-1(RNAi) abolishes both the zygotic and maternal contributions of pop-1, resulting in a more severe phenotype in pop-1(RNAi) embryos compared to pop-1(zu189) (Lin et al., 1998). Hence, to make the best assessment of MS specification, we have evaluated both pop-1(zu189) and pop-1(RNAi) backgrounds, and used multiple criteria to evaluate production of MS-derived tissues.
A further complication could arise if mutation of end-1,3 affected ratios of POP-1::SYS-1, the primary determinant of POP-1 function as a repressor or activator (Huang et al., 2007; Phillips et al., 2007). However, this seems highly unlikely, as zygotic expression of the end genes is determined by, and occurs downstream of, the activity of maternal POP-1 and SYS-1 proteins (Huang et al., 2007; Maduro et al., 2007; Shetty et al., 2005). Nonetheless, we examined med-1-driven GFP::SYS-1 and GFP::POP-1 reporter strains in an end-1,3(ok1448) mutant background and saw no apparent effect on SYS-1 or POP-1 protein asymmetries (data not shown) (Huang et al., 2007; Maduro et al., 2002).
Previous results suggested that a potent knockdown of endoderm specification occurs in end-3; pop-1 mutant embryos. END-3 appears to contribute to end-1 activation (Maduro et al., 2007), and POP-1 makes a positive contribution to end expression in E (Phillips et al., 2007; Shetty et al., 2005). As a result, although there is also input into endoderm specification by SKN-1, MED-1,2 and PAL-1, loss of pop-1 and end-3 compromises end-1 activation enough that endoderm fails to be specified in 91–99% of embryos (Table 1) (Maduro et al., 2007; Maduro et al., 2005b; Maduro et al., 2001). Because end-3 homozygotes are viable, we performed our first studies with end-3; pop-1 embryos, then subsequently with end-1,3; pop-1 embryos when the double end-1,3 mutants became available. The results of both sets of experiments, collectively referred to as end; pop-1, are described below and summarized in Table 2 and Table 3.
We examined end-3(ok1448); pop-1(RNAi) and end-1(ok558) end-3(ok1448); pop-1(RNAi) embryos for production of MS-derived tissues (Table 2). Using pha-4::GFP to score pharynx cells (Horner et al., 1998; Kalb et al., 1998), control pop-1(RNAi) embryos displayed an average of 21.3 ± 0.8 (n=18) pharynx cells (compared with wild-type, 50.0 ± 0.9, n=21), consistent with the loss of MS-derived pharynx resulting from an MS-to-E transformation (Fig. 2J and Fig. 3C). In contrast, end-3(ok1448); pop-1(RNAi) embryos contained 28.5 ± 7.0 (n=22) pharynx cells (p<0.0002 compared with pop-1(RNAi) alone), while end-1,3(ok1448); pop-1(RNAi) and end-1,3(zu247); pop-1(RNAi) embryos had 44.8 ± 2.2 (n=17) and 43.5 ± 1.3 (n=23) pharynx cells each (p<10−14 compared with pop-1(RNAi) alone) (Fig. 3G and data not shown). From the absence of gut, and the presence of significantly more pharynx cells in end-1,3; pop-1 embryos over pop-1 alone, the most straightforward interpretation is that MS and/or E produced pharynx in end; pop-1 embryos.
Fig. 3.
Embryos lacking end and pop-1 function make MS-type pharynx. (A) Polarized light image of a field of end-1(ok558) end-3(ok1448); glp-1(or178ts) embryos raised at the non-permissive temperature for or178 and treated with pop-1(RNAi). The two embryos on the left (outlined in red) carry an end-1,3(+) transgene and are the only embryos making gut. (B) Fluorescence image of the same embryos in panel A showing pha-4::GFP expression (Horner et al., 1998; Kalb et al., 1998). Expression in the two rescued embryos (red outline) is from gut cells, while in the remaining embryos expression is from GLP-1-independent pharynx cells (an average of 16.8 ± 1.1 cells, n=93). We do not observe the rectum component of pha-4::GFP expression in our glp-1(or178) strain when it is raised at 25°C (data not shown). (C) GFP overlay of pha-4::GFP expression in a pop-1(RNAi) embryo. The bright signals in large nuclei are gut nuclei (confirmed by polarized light birefringence), while the remaining expression is from hindgut cells (h.g.) and ABa-derived pharynx (an average of 21.3 ± 0.8 cells, n=18). (D) Elongated wild-type embryo with grinder (gr) indicated. (E) Arrested pop-1(zu189) dpy-5(e61) embryo from pop-1 dpy-5 mother. (F) Arrested end-1(ok558) end-3(ok1448); pop-1(zu189) dpy-5(e61) embryo from an end-1,3; pop-1 dpy-5 mother rescued by an end-1,3(+) transgene. The grinder (gr) within a posterior pharynx-like structure is indicated. (G) Expression of pha-4::GFP in an end-1(ok558) end-3(ok1448); pop-1(RNAi) embryo in hindgut (h.g.) and pharynx (average of 44.8 ± 2.2 pharynx cells, n=17). (H–J) myo-2::GFP expression (Okkema et al., 1993) showing presence of ABa- and MS-derived portions of pharynx in wild-type (H) and end-1,3; pop-1 (J) embryos, but lacking MS-derived pharynx in a pop-1 mutant (I). (K–N) pha-4::GFP expression in chromosomal mutant backgrounds that block MS specification, with and without pop-1(RNAi). (K,L) med-1(ok804); med-2(cx9744); dpy-11(e224) end-1(ok558) end-3(ok1448) background. (M,N) tbx-35(tm1789); end-1(ok558) end-3(ok1448) ceh-51(tm2123) background. In these mutant backgrounds, the number of pha-4::GFP-expressing cells is actually reduced from ~30 to ~22 when pop-1(RNAi) is added, owing to an apparent requirement for pop-1 in ABa-derived pharynx (Table 2). An additional requirement for pop-1 in morphogenesis (Lin et al., 1998) results in dispersal of pha-4::GFP-expressing cells in panels L and N. Embryos in most panels have been oriented with anterior to the left and dorsal up.
As MS-derived pharynx is produced independently of maternal GLP-1 (Priess et al., 1987), we examined whether glp-1 was required for production of extra pharynx cells in end; pop-1 embryos. First, end-1,3(ok1448); glp-1(or178); pop-1(RNAi); Ex[end-1,3(+)] embryos, rescued for endoderm specification by a transgene array, made almost no pharynx cells (1.4 ± 0.3 cells, n=40) (Fig. 3A,B). This control result is consistent with the loss of both AB- and MS-derived pharynx in glp-1; pop-1 mutant embryos (Lin et al., 1995). In contrast, non-rescued end-1,3(ok1448); glp-1(or178); pop-1(RNAi) embryos made 16.8 ± 1.1 (n=93) pharynx cells (p<10−24) while end-3(ok1448); glp-1(or178); pop-1(RNAi) embryos made 9.5 ± 0.8 (n=75) cells (p<10−13) (Fig. 3A,B). The complement of GLP-1-independent pharynx was 21.3 ± 0.8 (n=18) in glp-1(RNAi) embryos, 21.2 ± 0.8 (n=18) in end-3(ok1448); glp-1(or178) embryos, and 25.2 ± 1.0 (n=17) in end-1,3(ok1448); glp-1(or178) embryos (Table 2). Hence, ~45% and ~67% of the number of GLP-1-independent pharynx cells are restored in end-3(ok1448); pop-1(RNAi) and end-1,3(ok1448); pop-1(RNAi) embryos, respectively.
To look for evidence that these pharynx cells were capable of undergoing organogenesis, we looked for a posterior pharynx in end-1,3(ok1448); pop-1(zu189) embryos that also carried a myo-2::GFP pharynx muscle reporter (Okkema et al., 1993). At least a partial grinder was visible in 23% (n=74) of end-1,3(ok1448); pop-1(zu189) embryos, but this was never seen in sibling embryos that were rescued for end-1,3 by a transgene array (n=43) (Fig. 3D–F, H–J). This suggests that the additional pharynx cells made in end-1,3; pop-1 embryos can sometimes undergo organogenesis to generate part of the posterior pharynx.
To examine the origin of the additional pharynx cells made in end; pop-1(RNAi) embryos, we performed laser ablations on embryos using end-3(ok1448); pop-1(zu189) and end-1(ok558) end-3(ok1448); pop-1(zu189) strains carrying a cytoplasmic myo-2::GFP reporter, which marks pharynx muscle (Okkema et al., 1993). While ablation of ABa, MS and E together led to production of no pharynx muscle (n=3 for each genotype), ablation of only ABa+MS or ABa+E resulted in production of pharynx muscle in partial embryos of each genotype, although myo-2 expression in ABa+MS ablated end-3(ok1448); pop-1(zu189) embryos was somewhat weaker (Table 3). In a separate experiment, ablation of the AB daughters plus either MS or E in end-1,3(ok1448); pop-1(RNAi) embryos resulted in myo-2 expression in 5/8 and 4/6 partial embryos, respectively. These results suggest that both MS and E can produce MS-like tissues in end-3; pop-1 and end-1,3; pop-1 embryos.
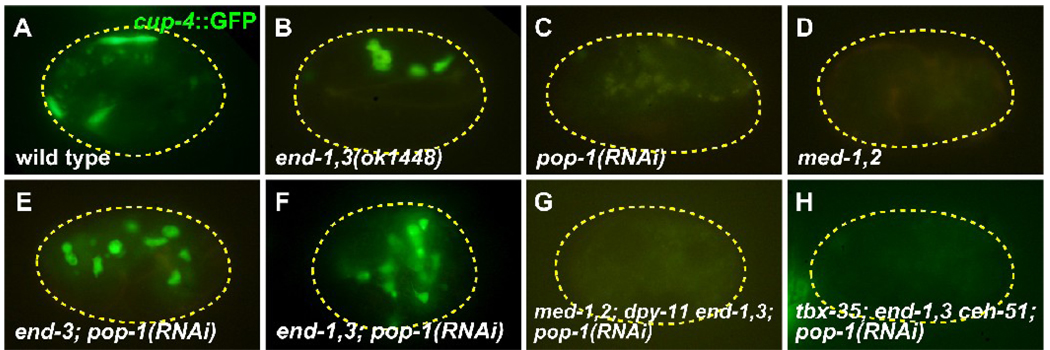
Next, we tested for embryonic production of coelomocytes, which comprise four cells derived exclusively from MS (Sulston et al., 1983). As scored by a cup-4::GFP marker (Patton et al., 2005), pop-1(RNAi) embryos made no coelomocytes (n=50) compared with wild-types, which showed an average of 3.7 ± 0.2 (n=105), and end-1,3 mutants, which showed an average of ~4.5 cells (Table 2, Fig. 4A–C). In contrast, end-3(ok1448); pop-1(RNAi) embryos made an average of 6.1 ± 0.2 (n=77) coelomocytes, end-1,3(ok1448); pop-1(RNAi) embryos made an average of 9.3 ± 0.2 (n=80), and end-1,3(zu247); pop-1(RNAi) embryos made an average of 10.7 ± 0.3 (n=77) (Fig. 4E,F and data not shown). Using a laser microbeam, we ablated MS, E, or both in end-3(ok1448) and end-1,3(ok1448) backgrounds with or without pop-1(RNAi). The end-3 and end-1,3 backgrounds showed production of coelomocytes only from MS, while end-3; pop-1 and end-1,3; pop-1 showed production of coelomocytes from both MS and E (results summarized in Table 3). Taken together, these results suggest that coelomocytes and GLP-1-independent pharynx are made by both MS and E in end; pop-1 embryos. This suggests that POP-1 is not essential for production of mesoderm tissue from MS, and furthermore, that POP-1 and/or END-1,3 may repress MS fate in the E cell. The failure to obtain a high number of completely restored MS-derived pharynxes implies additional requirements for POP-1 function in MS lineage development; this is consistent with known involvement of the Wnt/β-catenin asymmetry pathway in pharynx lineages in MS (Kaletta et al., 1997).
Fig. 4.
Coelomocytes are made in end-1,3; pop-1 mutant embryos. All embryos carry a cup-4::GFP coelomocyte-specific marker (Patton et al., 2005). (A) Wild-type embryo showing accumulation of cup-4::GFP expression in four coelomocytes (the top two are adjacent). (B) Normal coelomocytes in an end-1(ok558) end-3(ok1448) mutant. (C,D) Loss of coelomocytes in pop-1(RNAi) and med-1(ok804); med-2(cx9744) embryos, in which a penetrant mis-specification of MS occurs (Broitman-Maduro et al., 2009; Lin et al., 1995; Maduro et al., 2001). (E,F) Supernumerary coelomocytes are made in end-3(ok1448); pop-1(RNAi) embryos (average of 6.1 ± 0.2 cup-4::GFP expressing cells, n=77) and end-1(ok558) end-3(ok1448); pop-1(RNAi) embryos (average of 10.7 ± 0.3 cells, n=77). (G,H) Loss of coelomocytes in med-1(ok804); med-2(cx9744); dpy-11(e224) end-1(ok558) end-3(ok1448); pop-1(RNAi) and tbx-35(tm1789); end-1(ok558) end-3(ok1448) ceh-51(tm2123); pop-1(RNAi) embryos. Faint yellow signal in some panels corresponds to gut granules.
Restored MS-like fates in end-1,3; pop-1 mutants require normal pathways for MS specification
The simplest explanation for the origin of MS-derived pharynx and coelomocytes in end; pop-1 mutants is that these arise through activation of the same genes that specify MS in wild-type embryos. However, it is possible that mesoderm specification is activated later by some default mechanism in response to the absence of POP-1 and END-1,3. We have shown that med-1,2 together, and tbx-35; ceh-51 together are essential for production of coelomocytes and GLP-1-independent pharynx (Fig. 3K, 3M and Fig. 4D) (Broitman-Maduro et al., 2009). To confirm that coelomocytes and pharynx cells made in end; pop-1 mutant embryos are the result of normal MS specification, we constructed quadruple med-1(ok804); med-2(cx9744); end-1(ok558) end-3(ok1448) and tbx-35(tm1789); end-1(ok558) end-3(ok1448) ceh-51(tm2123) mutant strains carrying pha-4::GFP or cup-4::GFP (see Materials and Methods). As summarized in Table 2, both quadruple-mutant backgrounds resulted in loss of MS-derived coelomocytes (none, for med-1,2; end-1,3 (n=52), and 0.1 ± 0.0 (n=122) for tbx-35; end-1,3 ceh-51), similar to med-1,2 or tbx-35; ceh-51 alone. However, even though end-1,3; pop-1 embryos made an average of 10.7 ± 0.3 (n=77) coelomocytes as reported above, addition of pop-1(RNAi) resulted in very few coelomocytes (1.0 ± 0.2 in the case of med-1,2; end-1,3; pop-1(RNAi) or nearly none (0.1 ± 0.1, n=35) in the case of tbx-35; end-1,3 ceh-51 (Fig. 4G,H). A similar pattern was observed for expression of pha-4::GFP: While med-1,2; end-1,3 and tbx-35; end-1,3 ceh-51 quadruple mutants made 29.9 ± 1.1 (n=12) and 30.3 ± 1.0 (n=21) pharynx cells each, addition of pop-1(RNAi) did not result in an increase in pharynx cells as was seen with end-1,3; pop-1 (Fig. 3C,L,N). Indeed, the quadruple mutant + pop-1(RNAi) backgrounds produced even fewer pharynx cells (22.4 ± 0.9 (n=24) and 22.1 ± 1.4 (n=19) than the ~30 cells in the med-1,2 or tbx-35; ceh-51 backgrounds, comparable to pop-1(RNAi) alone (21.3 ± 0.8, n=18), and demonstrating a requirement for the Wnt/β-catenin asymmetry pathway in production of pharynx cells from the AB lineage (Kaletta et al., 1997; Lin et al., 1998). These results show that the MS-derived tissues made in end-1,3; pop-1 embryos require the genes that normally specify MS. The detection of greater than 8 coelomocytes (as would be expected from 2xMS cells in the embryo) further suggests that POP-1 plays a role in MS lineage asymmetries that give rise to coelomocyte precursors, a phenomenon that has been reported for postembryonically-specified coelomocytes (Amin et al., 2009).
E produces epidermal tissue as well as MS tissues in end; pop-1 embryos
It is possible that in addition to MS-type cells, MS and/or E in end; pop-1 mutant embryos could make cells that are normally derived from C. The Caudal-like regulator PAL-1, which specifies the C and D fates, is present in the early MS and E lineages, and is responsible for the C transformations that result when MS and E specification is compromised by mutation in the SKN-1/MED-1,2 pathway (Broitman-Maduro et al., 2006; Broitman-Maduro et al., 2009; Hunter and Kenyon, 1996; Maduro et al., 2005a). To evaluate this possibility, we constructed an end-1,3(ok1448) strain carrying both a cup-4::GFP reporter, to mark coelomocytes, and an nhr-25::YFP reporter to mark epidermal cells (Baugh et al., 2005; Patton et al., 2005); the two reporters can be distinguished with appropriate filter sets (Miller et al., 1999). We used a laser microbeam to isolate MS or E and evaluated whether the resultant partial embryos produced epidermal cells, coelomocytes or both. MS blastomeres isolated from end-1,3(ok1448); pop-1(RNAi) embryos generated coelomocytes but not epidermal cells (n=4), while isolated E blastomeres made coelomocytes all the time (6/6 embryos) and, simultaneously, epidermal cells part of the time (4/6 embryos). Therefore, in end; pop-1 mutant embryos, E, but not MS, frequently adopts a mixed fate that includes production of tissues normally made by MS and C.
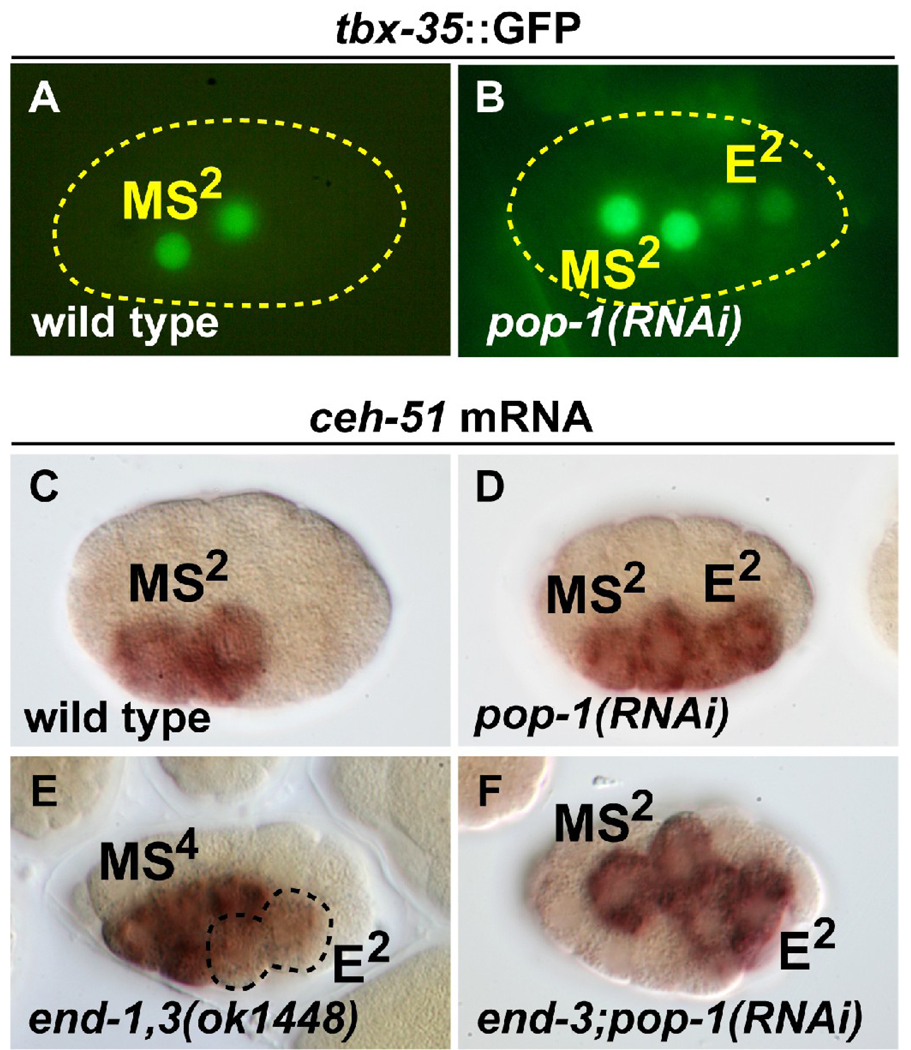
Early MS markers are expressed in E in pop-1 and end-1,3 mutant embryos
The production of pharynx cells from E in end-1,3; pop-1(RNAi) embryos suggests that POP-1 contributes to repression of MS fate in E. A role for POP-1 in repressing MS fate in E would explain why Wnt/β-catenin asymmetry mutants, which act upstream of POP-1, cause a transformation of E to MS, while loss of end-1,3 results in an E to C transformation (Kaletta et al., 1997; Maduro et al., 2005a; Rocheleau et al., 1997; Rocheleau et al., 1999; Thorpe et al., 1997; Zhu et al., 1997). We previously failed to detect ectopic expression of the MS specification gene tbx-35 in the E lineage in pop-1 mutant embryos, either by in situ hybridization or a tbx-35::GFP reporter with 605bp of putative tbx-35 promoter (Broitman-Maduro et al., 2006). However, such ectopic expression was observed with a tbx-35::GFP reporter with 734bp of promoter (Premnath Shetty and Rueyling Lin, personal communication; we reproduced this result with their reporter as shown in Fig. 5A,B). It was recently reported that pop-1(RNAi) embryos frequently showed expression of endogenous ceh-51 and a ceh-51::GFP reporter in the early E lineage (Broitman-Maduro et al., 2009) (Fig. 5C,D). The ability of E to consistently produce MS tissues in end-1,3; pop-1 mutant embryos suggested to us that loss of end-1,3 may synergize with loss of pop-1 to result in activation of MS genes. Indeed, we detected low levels of ceh-51 transcripts in the E lineage in end-1,3(ok1448) mutants, consistent with a parallel role for END-1,3 in repressing MS fate (Fig. 5E). Furthermore, expression of ceh-51 was strong in both the E and MS lineages in end-3; pop-1(RNAi) embryos, though no further enhancement of the expression seen in pop-1(RNAi) alone was noted (Fig. 5F). Taken together, these results confirm that the MS tissues made by E in end; pop-1 embryos correlate with ectopic activation of MS specification in E, and they also provide evidence that POP-1 and END-1,3 contribute to the repression of early MS genes in the E lineage.
Fig. 5.
Expression of MS specification factors in pop-1(RNAi) and end-1,3 mutant backgrounds. (A,B) Expression of a tbx-35::GFP reporter in wild-type and pop-1(RNAi) embryos. 100% (n=40) of expressing embryos showed MS lineage-only expression in wild-type, while 12% (n=67) of tbx-35::GFP-expressing pop-1(RNAi) embryos showed ectopic expression in the E lineage as shown; 75% of embryos overall had weak E lineage expression. (C–F) in situ hybridization with a ceh-51 probe. (C) Wild-type staining pattern observed in 91% of embryos in the early MS lineage (MS2 as shown here, or MS4; n=101). (D) Ectopic expression of ceh-51 in the early E lineage was observed in 86% (n=44) of pop-1(RNAi) embryos (the remainder showed either normal expression or did not stain). Results in C and D were previously reported (Broitman-Maduro et al., 2009). (E) Weak ectopic expression of ceh-51 in the E lineage (at E2, as shown here; ectopic expression was also observed at E4), as seen in 41% (n=209) of progeny from end-1(ok558) end-3(ok1448) mothers carrying an end-1,3(+) rescuing array. As this array is transmitted to ~50% of the progeny, ~80% of end-1,3 mutant embryos showed activation of ceh-51 in the E lineage. The remaining embryos showed wild-type expression (MS only). (F) Expression of ceh-51 in MS and E was seen in 88% (n=45) of end-3(ok1448); pop-1(RNAi) embryos. The remaining embryos were either unstained or appeared normal.
Discussion
Regulatory interactions in the C. elegans endomesoderm gene regulatory network
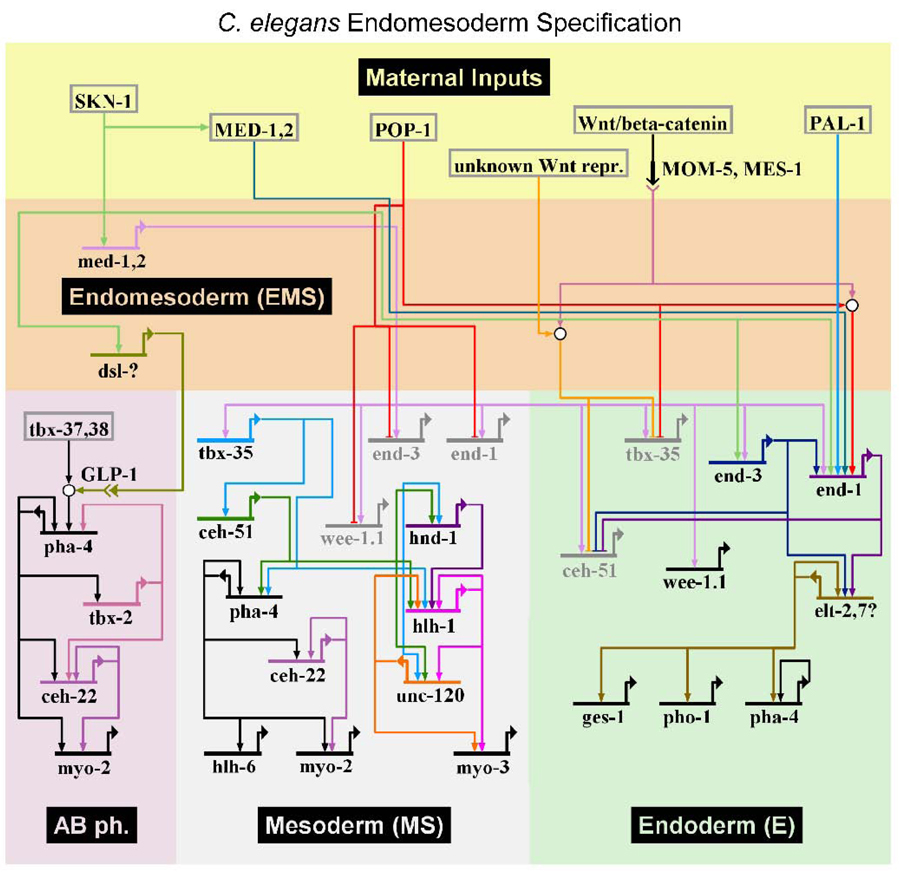
An updated gene network for C. elegans endomesoderm specification is shown in Fig. 7. Although this network focuses on EMS and its daughters, secondary mesoderm generated by ABa is included because it is induced by MS and involves downstream target genes that are shared with MS (Priess et al., 1987; Roy Chowdhuri et al., 2006; Smith and Mango, 2007). The network features early maternal factors that act combinatorially to direct blastomere-specific activation of transiently-expressed cell specification factors, which ultimately results in activation of stably-expressed tissue/organ identify factors in their descendants (Labouesse and Mango, 1999; Maduro, 2006; Maduro, 2008; Maduro and Rothman, 2002). The maternal inputs include SKN-1 (Bowerman et al., 1992; Maduro et al., 2001), POP-1 (Huang et al., 2007; Lin et al., 1995; Shetty et al., 2005), the Wnt/β-catenin asymmetry pathway that acts downstream of the P2-EMS interaction (Bei et al., 2002; Huang et al., 2007; Phillips et al., 2007; Rocheleau et al., 1997; Rocheleau et al., 1999; Thorpe et al., 1997), and the weak contribution by PAL-1 (Maduro et al., 2005b). Also included is the weak maternal contribution of MED-1,2 (Maduro et al., 2007), the evidence for which has been disputed by others (Captan et al., 2007). Within EMS, the only genes in the network are zygotic med-1 and med-2 (Maduro et al., 2001), and an unknown Delta-Serrate-Lag (DSL) gene that mediates the GLP-1/Notch-dependent cell-cell interaction that specifies pharynx from ABa (Mello et al., 1992; Priess et al., 1987). The MS/E zygotic factors as discussed above are activated combinatorially by the SKN-1 pathway and Wnt/β-catenin asymmetry pathway: These are end-1 and end-3, which specify E, and tbx-35 and ceh-51, which specify MS (Broitman-Maduro et al., 2009; Maduro et al., 2005a). As tissue/organogenesis factors, we have included the muscle regulatory network consisting of HND-1/Hand, HLH-1/MyoD, and UNC-120/SRF (Fukushige et al., 2006), the pharynx identity factor PHA-4/FoxA (Gaudet and Mango, 2002; Kaltenbach et al., 2005), genes that direct specification of pharynx cell subtypes (CEH-22/Nkx2.5 and HLH-6) (Okkema and Fire, 1994; Okkema et al., 1993; Raharjo and Gaudet, 2007; Vilimas et al., 2004), and factors required for specification of AB-derived pharynx (TBX-37/38) (Good et al., 2004) and AB-derived pharynx muscle (TBX-2) (Roy Chowdhuri et al., 2006; Smith and Mango, 2007). Based on genetic evidence that TBX-35 and CEH-51 define MS-derived muscle and pharynx, we have connected these regulators directly to the muscle network and pha-4, although evidence for direct interaction has not yet been obtained. Rather, the ability of PAL-1 to directly activate hlh-1 in the C lineage strongly suggests that TBX-35 and/or CEH-51 may do the same in the MS lineage (Broitman-Maduro et al., 2009; Lei et al., 2009). As markers of terminal differentiation, the network includes as examples the pharynx muscle myosin gene myo-2, the body muscle myosin gene myo-3, the gut esterase gene ges-1, and the gut-expressed phosphatase gene pho-1 (Fukushige et al., 2005; Kennedy et al., 1993; Okkema et al., 1993). The elt-7 GATA factor is included, as it is activated at the same time as elt-2 and may be involved in parallel functions with ELT-2 (Maduro and Rothman, 2002), and mutation of elt-7 was reported to result in a decrease of intestinal expression of pha-4 (Murray et al., 2008). The elt-2 paralog elt-4 appears to be nonfunctional and has not been included (Fukushige et al., 2003). As we have focused on early specification events, the model does not include many other regulators known to be involved in pharynx and intestine development; these have been described in recent reviews (Mango, 2007; McGhee, 2007).
Fig. 7.
Updated gene regulatory network for C. elegans endomesoderm generated with BioTapestry (Longabaugh et al., 2009) and modified in Adobe Photoshop. A subset of the connections shown have been established at the protein-DNA level, while others have been inferred from extensive genetic analysis. See Discussion for further details.
From the studies presented here, we have included the apparent roles of END-1,3 in repression of ceh-51 and the role of POP-1 in repression of tbx-35. As derepression of tbx-35 was not complete in a pop-1 mutant background, the model retains repression of tbx-35 by an unknown Wnt-dependent repressor, accounting for persistent differences between the two EMS daughters in the absence of pop-1 (discussed below). As the results presented here also do not require a positive input of POP-1 in specification of MS, it is not included. We note that this differs from the related nematode C. briggsae, in which an apparent positive role for POP-1, in parallel with SKN-1, is implied by RNAi knockdown experiments (also discussed below) (Lin et al., 2009).
A relatively mild phenotype of mutants lacking endoderm
We have reported the construction of a bona fide end-1 end-3 double mutant strain and shown that it results in a complete failure of endoderm specification, as predicted by prior experiments (Maduro et al., 2005a; Zhu et al., 1997). It might be expected that the complete mis-specification of a germ layer would result in profound changes to the remainder of embryonic development, but end-1,3 mutants demonstrated a surprisingly mild phenotype, sometimes hatching into (inviable) larvae. By comparison, in Drosophila, loss of the endoderm (midgut) through mutation of the GATA factor serpent resulted in embryos that began gastrulation normally but eventually arrested, with midgut progenitors adopting ectodermal (hindgut and foregut) fates (Reuter, 1994). The mild phenotype of C. elegans end-1,3 mutants explains why smaller deletions removing end-1,3 alone (as opposed to large deletions removing hundreds of genes) were not isolated in screens for penetrant zygotic endoderm-defective mutants, if the design of the screens assumed that misspecification of endoderm would result in a strict embryonic lethality (Maduro et al., 2005a; Zhu et al., 1997).
In C. elegans end-1,3 mutant embryos, the transformed descendants of E are capable of an otherwise fairly normal morphogenesis of the embryo. In normal development, the E daughter cells are the first to ingress during gastrulation (Knight and Wood, 1998; Nance et al., 2005). The ability of most end-1,3(−) embryos to elongate suggests that there are mechanisms that can direct early gastrulation movements within the early E lineage independently of E specification, or that errors made during early E gastrulation are compensated by the embryo, or a combination of both mechanisms. Previous studies have used Wnt/β-catenin pathway mutants to perturb E specification and gastrulation (Lee et al., 2006; Rohrschneider and Nance, 2009), but these mutants may have effects outside of the E lineage. Hence, it may be that the chromosomal end-1,3 double mutant will provide a new resource to further dissect the connection between gut specification and morphogenesis. It is also possible that the double mutant will permit evaluation of contributions that the intestine makes to development, as end-1,3 mutants had later phenotypes that are also seen in lysosomal trafficking mutants (Hermann et al., 2005).
A hierarchy of specification pathways and reinforcement of fate choices
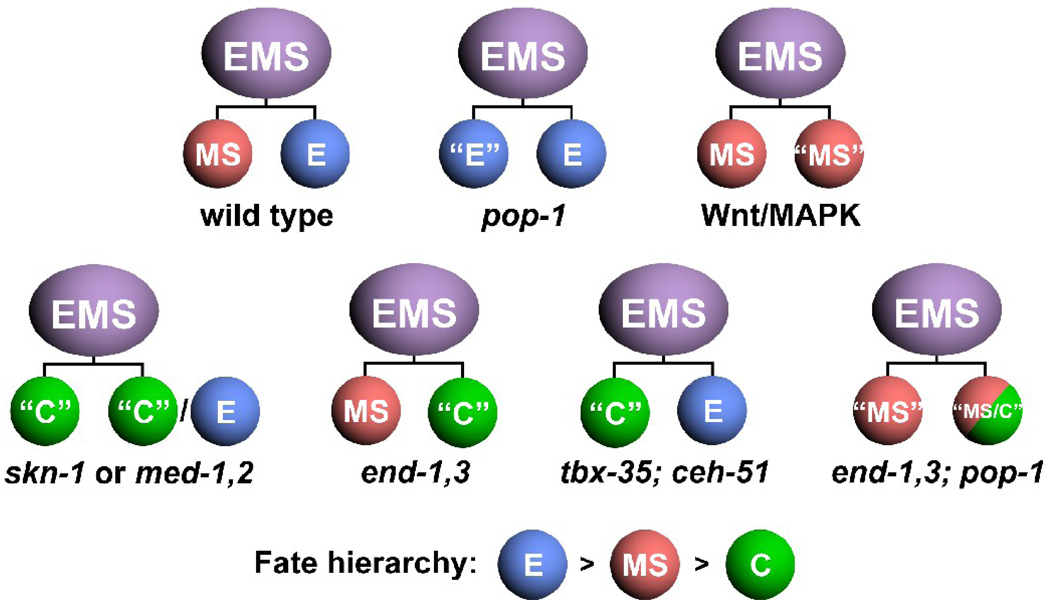
Prior work on MS specification, and the analysis reported here, highlight the ability of blastomere specification pathways to exhibit phenotype prevalence. Normally, specification of MS and E is driven by activation of tbx-35/ceh-51 in MS, and end-1,3 in E (Broitman-Maduro et al., 2006; Broitman-Maduro et al., 2009; Maduro et al., 2005a). In pop-1 mutant embryos, end-1 and end-3 become activated in MS, but tbx-35 and ceh-51 continue to be activated in MS as well (Broitman-Maduro et al., 2009; Maduro et al., 2007; Shetty et al., 2005). As well, maternal PAL-1 protein, which specifies the C lineage, is present in the early MS and E lineages (Hunter and Kenyon, 1996). Hence, in pop-1 mutant embryos, the factors that specify E, MS and C all coexist within MS, and yet a high-penetrance MS to E transformation results in such embryos (Lin et al., 1995). Here we have shown that when end-1 and end-3 are removed in pop-1 mutant embryos, MS can now be specified by TBX-35 and CEH-51. In mutants for skn-1, the meds, or tbx-35/ceh-51, MS adopts a C-like fate (Bowerman et al., 1992; Broitman-Maduro et al., 2006; Broitman-Maduro et al., 2009; Maduro et al., 2001). Taken together, this reveals a hierarchy of E > MS > C fates. This is reminiscent of the posterior prevalence of Hox genes, in which the most posteriorly expressed Hox gene controls regional identity when multiple Hox genes are forcibly coexpressed, though it is almost certainly not the result of a similar mechanism (Capovilla and Botas, 1998; Yekta et al., 2008).
Unlike MS, which adopts an apparently strict MS, E or C fate in various mutant backgrounds (Bowerman et al., 1992; Broitman-Maduro et al., 2009; Lin et al., 1995), the E lineage can apparently adopt a mixed fate. We found that in an end-1,3; pop-1 mutant background, E makes MS tissues (coelomocytes and pharynx) in all embryos, and epidermal cells, characteristic of C, some of the time (4/6 embryos). There are other examples of mixed E fates from prior studies. In end-3 mutants, a small number of embryos showed apparent “half-guts” in which only one half of the intestine was present (Maduro et al., 2005a). In at least one lineage recording of an end-3(zu247) embryo, an E grand-daughter followed a lineage pattern that resembled an MS grand-daughter (Maduro et al., 2005a). The end genes are still sensitive to POP-1 repression in Ea (Maduro et al., 2007), suggesting that it is possible for Ea and Ep to adopt different fates if end activity is partly compromised, but this mechanism does not explain how E can make both MS and C tissue types in a single embryo. Our analysis did not find evidence of production of C tissues from MS in end-1,3; pop-1 embryos. While we detected similar levels of ceh-51 expression in the MS and E lineages in pop-1(RNAi) embryos (Fig. 5C,D), this was not the case for tbx-35, in which a tbx-35::GFP reporter was activated only weakly (Fig. 5A,B) and which was not detected ectopically in E at all with a different reporter and in situ hybridization (Broitman-Maduro et al., 2006). The MS and E expression of ceh-51::GFP in pop-1(RNAi) embryos was found to be dependent on tbx-35 (Broitman-Maduro et al., 2009), suggesting that some amount of TBX-35 is ectopically expressed in E in pop-1(RNAi) embryos, but it is at lower levels in E than in MS. The conclusion is that some differences between MS and E are still present in pop-1-depleted embryos.
These results nonetheless provide further evidence that Wnt-signaled POP-1 and END-1,3 also actively repress MS specification in E, perhaps as another Wnt-dependent mechanism to enforce an endodermal fate. Within MS, POP-1 is not modified by the Wnt/β-catenin asymmetry pathway, and end-1,3 are repressed by POP-1, permitting the MEDs to activate tbx-35 and the TBX-35 target ceh-51 (Broitman-Maduro et al., 2006; Broitman-Maduro et al., 2009). We recently reported that while ceh-51 activation is completely dependent on the MEDs for activation, tbx-35 mutants still activate ceh-51 at reduced levels, suggesting that another activator works in parallel with TBX-35 (Broitman-Maduro et al., 2009). Knockdown of pop-1 by RNAi in a tbx-35 mutant background resulted in abrogation of ceh-51::GFP expression, suggesting that this second factor is dependent on POP-1 (Broitman-Maduro et al., 2009). Hence, there is some evidence that unmodified POP-1 in MS contributes to MS fate, but this contribution is not at all evident from the phenotype data (as MS fate is restored to end-1,3; pop-1 mutant embryos), and it is similarly not clear how POP-1 could activate expression of ceh-51 in the MS daughters at equal levels, given that POP-1 asymmetry exists in MSa/p (Lin et al., 1998; Maduro et al., 2002).
Comparisons between C. briggsae and C. elegans: Network evolution
In contrast to C. elegans, a positive contribution of POP-1 to MS fate was found in the related nematode C. briggsae. Although the embryonic lineages of C. elegans and C. briggsae are highly similar (Zhao et al., 2008), there are notable differences in the mechanisms of endomesoderm specification as determined by knockdown phenotypes. C. elegans skn-1 mutants show a loss of MS and E tissues (Bowerman et al., 1992). In C. briggsae, knockdown of Cb-skn-1 resulted in a more penetrant loss of endoderm, while GLP-1-independent pharynx was still made from MS (Lin et al., 2009). Knockdown of Cb-pop-1 resulted in production of GLP-1-independent pharynx from both MS and E, and when Cb-skn-1(RNAi) and Cb-pop-1(RNAi) were performed simultaneously, both MS and E failed to make pharynx (Lin et al., 2009). Hence, there is a positive function for Cb-POP-1 in specification of MS fate in parallel with Cb-SKN-1, and this mechanism is very different from C. elegans. Here we have confirmed that the primary contribution of Ce-POP-1 to MS specification is by repression of Ce-end-1,3, as loss of Ce-pop-1 is compatible with MS specification if the Ce-ends are also mutated. While the mechanistic basis for these interspecific phenotype differences has yet to be elucidated, comparisons between the two species have already revealed flexibility in how the same genes can function differently in the same combinatorial specification events, and showing that for at least some apparently hardwired cell fate decisions, there is more than one way of accomplishing the same developmental endpoint. To this end, the weaker ‘reinforcing’ functions that have been identified in C. elegans may, in other species, be the primary mechanisms that determine blastomere fates.
Fig. 6.
Summary of fate hierarchies revealed by EMS daughter cell fate transformations in various mutant backgrounds. Loss of pop-1 results in an MS to E transformation (Lin et al., 1995); loss of Wnt/MAPK function causes an E to MS transformation (Rocheleau et al., 1997; Thorpe et al., 1997); for skn-1 or med-1,2 mutant embryos, MS adopts a C-like fate, while E appears to adopt either an E or C fate (Bowerman et al., 1992; Maduro et al., 2001); loss of end-1,3 results in an E to C transformation (Maduro et al., 2005a; Zhu et al., 1997); loss of tbx-35 and ceh-51 results in a transformation of MS to C (Broitman-Maduro et al., 2009); and loss of end-1,3 and pop-1 together results in a restored partial MS lineage, and an apparent mixed fate from E that is a combination of MS and C fates (this work).
Acknowledgements
We thank Attila Stetak, Christian Frøkjær-Jensen, Erik Jorgensen and Andy Fire for plasmids, and Joel Rothman, Michael Krause, Premnath Shetty, Shuyi Huang, Rueyling Lin, Johnny Fares and Jim McGhee for transgenic strains. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR). The ok558 and ok1448 mutations were generated by the C. elegans Knockout Consortium. H.R. was an undergraduate trainee with the MARCU* program at UC Riverside. This work was supported by grants from the NSF (IBN#0416922 and IOS#0643325) and NIH (1R03HD054589-01) to M.M., and in part by NIH award T34GM062756 to UC Riverside.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amin NM, Lim SE, Shi H, Chan TL, Liu J. A conserved Six-Eya cassette acts downstream of Wnt signaling to direct non-myogenic versus myogenic fates in the C. elegans postembryonic mesoderm. Dev Biol. 2009;331:350–360. doi: 10.1016/j.ydbio.2009.05.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh LR, Hill AA, Claggett JM, Hill-Harfe K, Wen JC, Slonim DK, Brown EL, Hunter CP. The homeodomain protein PAL-1 specifies a lineage-specific regulatory network in the C elegans embryo. Development. 2005;132:1843–1854. doi: 10.1242/dev.01782. [DOI] [PubMed] [Google Scholar]
- Baugh LR, Hill AA, Slonim DK, Brown EL, Hunter CP. Composition and dynamics of the Caenorhabditis elegans early embryonic transcriptome. Development. 2003;130:889–900. doi: 10.1242/dev.00302. [DOI] [PubMed] [Google Scholar]
- Bei Y, Hogan J, Berkowitz LA, Soto M, Rocheleau CE, Pang KM, Collins J, Mello CC. SRC-1 and Wnt signaling act together to specify endoderm and to control cleavage orientation in early C. elegans embryos. Dev Cell. 2002;3:113–125. doi: 10.1016/s1534-5807(02)00185-5. [DOI] [PubMed] [Google Scholar]
- Bowerman B, Draper BW, Mello CC, Priess JR. The maternal gene skn-1 encodes a protein that is distributed unequally in early C. elegans embryos. Cell. 1993;74:443–452. doi: 10.1016/0092-8674(93)80046-h. [DOI] [PubMed] [Google Scholar]
- Bowerman B, Eaton BA, Priess JR. skn-1, a maternally expressed gene required to specify the fate of ventral blastomeres in the early C. elegans embryo. Cell. 1992;68:1061–1075. doi: 10.1016/0092-8674(92)90078-q. [DOI] [PubMed] [Google Scholar]
- Broitman-Maduro G, Lin KT-H, Hung W, Maduro M. Specification of the C. elegans MS blastomere by the T-box factor TBX-35. Development. 2006;133:3097–3106. doi: 10.1242/dev.02475. [DOI] [PubMed] [Google Scholar]
- Broitman-Maduro G, Owraghi M, Hung W, Kuntz S, Sternberg PW, Maduro M. The NK-2 class homeodomain factor CEH-51 and the T-box factor TBX-35 have overlapping function in C. elegans mesoderm development. Development. 2009;176:2735–2746. doi: 10.1242/dev.038307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capovilla M, Botas J. Functional dominance among Hox genes: repression dominates activation in the regulation of Dpp. Development. 1998;125:4949–4957. doi: 10.1242/dev.125.24.4949. [DOI] [PubMed] [Google Scholar]
- Captan VV, Goszczynski B, McGhee JD. Neither maternal nor zygotic med-1/med-2 genes play a major role in specifying the Caenorhabditis elegans endoderm. Genetics. 2007;175:969–974. doi: 10.1534/genetics.106.066662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coroian C, Broitman-Maduro G, Maduro MF. Med-type GATA factors and the evolution of mesendoderm specification in nematodes. Dev Biol. 2005;289:444–455. doi: 10.1016/j.ydbio.2005.10.024. [DOI] [PubMed] [Google Scholar]
- Egan CR, Chung MA, Allen FL, Heschl MF, Van Buskirk CL, McGhee JD. A gut-to-pharynx/tail switch in embryonic expression of the Caenorhabditis elegans ges-1 gene centers on two GATA sequences. Dev Biol. 1995;170:397–419. doi: 10.1006/dbio.1995.1225. [DOI] [PubMed] [Google Scholar]
- Fay D. Genetic mapping and manipulation: chapter 7--Making compound mutants. WormBook. 2006:1–4. doi: 10.1895/wormbook.1.96.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushige T, Brodigan TM, Schriefer LA, Waterston RH, Krause M. Defining the transcriptional redundancy of early bodywall muscle development in C. elegans: evidence for a unified theory of animal muscle development. Genes Dev. 2006;20:3395–3406. doi: 10.1101/gad.1481706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushige T, Goszczynski B, Tian H, McGhee JD. The Evolutionary Duplication and Probable Demise of an Endodermal GATA Factor in Caenorhabditis elegans. Genetics. 2003;165:575–588. doi: 10.1093/genetics/165.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushige T, Goszczynski B, Yan J, McGhee JD. Transcriptional control and patterning of the pho-1 gene, an essential acid phosphatase expressed in the C. elegans intestine. Dev Biol. 2005;279:446–461. doi: 10.1016/j.ydbio.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Fukushige T, Hawkins MG, McGhee JD. The GATA-factor elt-2 is essential for formation of the Caenorhabditis elegans intestine. Dev Biol. 1998;198:286–302. [PubMed] [Google Scholar]
- Gaudet J, Mango SE. Regulation of organogenesis by the Caenorhabditis elegans FoxA protein PHA-4. Science. 2002;295:821–825. doi: 10.1126/science.1065175. [DOI] [PubMed] [Google Scholar]
- Gillis WJ, Bowerman B, Schneider SQ. Ectoderm- and endomesoderm-specific GATA transcription factors in the marine annelid Platynereis dumerilli. Evol Dev. 2007;9:39–50. doi: 10.1111/j.1525-142X.2006.00136.x. [DOI] [PubMed] [Google Scholar]
- Goldstein B. Induction of gut in Caenorhabditis elegans embryos. Nature. 1992;357:255–257. doi: 10.1038/357255a0. [DOI] [PubMed] [Google Scholar]
- Good K, Ciosk R, Nance J, Neves A, Hill RJ, Priess JR. The T-box transcription factors TBX-37 and TBX-38 link GLP-1/Notch signaling to mesoderm induction in C. elegans embryos. Development. 2004;131:1967–1978. doi: 10.1242/dev.01088. [DOI] [PubMed] [Google Scholar]
- Goszczynski B, McGhee JD. Re-evaluation of the role of the med-1 and med-2 genes in specifying the C. elegans endoderm. Genetics. 2005 doi: 10.1534/genetics.105.044909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann GJ, Schroeder LK, Hieb CA, Kershner AM, Rabbitts BM, Fonarev P, Grant BD, Priess JR. Genetic analysis of lysosomal trafficking in Caenorhabditis elegans. Mol Biol Cell. 2005;16:3273–3288. doi: 10.1091/mbc.E05-01-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner MA, Quintin S, Domeier ME, Kimble J, Labouesse M, Mango SE. pha-4, an HNF-3 homolog, specifies pharyngeal organ identity in Caenorhabditis elegans. Genes Dev. 1998;12:1947–1952. doi: 10.1101/gad.12.13.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Shetty P, Robertson SM, Lin R. Binary cell fate specification during C. elegans embryogenesis driven by reiterated reciprocal asymmetry of TCF POP-1 and its coactivator beta-catenin SYS-1. Development. 2007;134:2685–2695. doi: 10.1242/dev.008268. [DOI] [PubMed] [Google Scholar]
- Hunter CP, Kenyon C. Spatial and temporal controls target pal-1 blastomere-specification activity to a single blastomere lineage in C. elegans embryos. Cell. 1996;87:217–226. doi: 10.1016/s0092-8674(00)81340-9. [DOI] [PubMed] [Google Scholar]
- Kalb JM, Lau KK, Goszczynski B, Fukushige T, Moons D, Okkema PG, McGhee JD. pha-4 is Ce-fkh-1, a fork head/HNF-3alpha,beta,gamma homolog that functions in organogenesis of the C. elegans pharynx. Development. 1998;125:2171–2180. doi: 10.1242/dev.125.12.2171. [DOI] [PubMed] [Google Scholar]
- Kaletta T, Schnabel H, Schnabel R. Binary specification of the embryonic lineage in Caenorhabditis elegans. Nature. 1997;390:294–298. doi: 10.1038/36869. [DOI] [PubMed] [Google Scholar]
- Kaltenbach LS, Updike DL, Mango SE. Contribution of the amino and carboxyl termini for PHA-4/FoxA function in Caenorhabditis elegans. Dev Dyn. 2005;234:346–354. doi: 10.1002/dvdy.20550. [DOI] [PubMed] [Google Scholar]
- Kennedy BP, Aamodt EJ, Allen FL, Chung MA, Heschl MF, McGhee JD. The gut esterase gene (ges-1) from the nematodes Caenorhabditis elegans and Caenorhabditis briggsae. J Mol Biol. 1993;229:890–908. doi: 10.1006/jmbi.1993.1094. [DOI] [PubMed] [Google Scholar]
- Knight JK, Wood WB. Gastrulation initiation in Caenorhabditis elegans requires the function of gad-1, which encodes a protein with WD repeats. Dev Biol. 1998;198:253–265. [PubMed] [Google Scholar]
- Labouesse M, Mango SE. Patterning the C. elegans embryo: moving beyond the cell lineage. Trends Genet. 1999;15:307–313. doi: 10.1016/s0168-9525(99)01750-3. [DOI] [PubMed] [Google Scholar]
- Laufer JS, Bazzicalupo P, Wood WB. Segregation of developmental potential in early embryos of Caenorhabditis elegans. Cell. 1980;19:569–577. doi: 10.1016/s0092-8674(80)80033-x. [DOI] [PubMed] [Google Scholar]
- Lee JY, Marston DJ, Walston T, Hardin J, Halberstadt A, Goldstein B. Wnt/Frizzled signaling controls C. elegans gastrulation by activating actomyosin contractility. Curr Biol. 2006;16:1986–1997. doi: 10.1016/j.cub.2006.08.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei H, Liu J, Fukushige T, Fire A, Krause M. Caudal-like PAL-1 directly activates the bodywall muscle module regulator hlh-1 in C. elegans to initiate the embryonic muscle gene regulatory network. Development. 2009 doi: 10.1242/dev.030668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin KT, Broitman-Maduro G, Hung WW, Cervantes S, Maduro MF. Knockdown of SKN-1 and the Wnt effector TCF/POP-1 reveals differences in endomesoderm specification in C. briggsae as compared with C. elegans. Dev Biol. 2009;325:296–306. doi: 10.1016/j.ydbio.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R. A gain-of-function mutation in oma-1, a C. elegans gene required for oocyte maturation, results in delayed degradation of maternal proteins and embryonic lethality. Dev Biol. 2003;258:226–239. doi: 10.1016/s0012-1606(03)00119-2. [DOI] [PubMed] [Google Scholar]
- Lin R, Hill RJ, Priess JR. POP-1 and anterior-posterior fate decisions in C. elegans embryos. Cell. 1998;92:229–239. doi: 10.1016/s0092-8674(00)80917-4. [DOI] [PubMed] [Google Scholar]
- Lin R, Thompson S, Priess JR. pop-1 encodes an HMG box protein required for the specification of a mesoderm precursor in early C. elegans embryos. Cell. 1995;83:599–609. doi: 10.1016/0092-8674(95)90100-0. [DOI] [PubMed] [Google Scholar]
- Longabaugh WJ, Davidson EH, Bolouri H. Visualization, documentation, analysis, and communication of large-scale gene regulatory networks. Biochim Biophys Acta. 2009;1789:363–374. doi: 10.1016/j.bbagrm.2008.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry JA, Atchley WR. Molecular evolution of the GATA family of transcription factors: conservation within the DNA-binding domain. J Mol Evol. 2000;50:103–115. doi: 10.1007/s002399910012. [DOI] [PubMed] [Google Scholar]
- Lowry JA, Gamsjaeger R, Thong SY, Hung W, Kwan AH, Broitman-Maduro G, Matthews JM, Maduro M, Mackay JP. Structural analysis of MED-1 reveals unexpected diversity in the mechanism of DNA recognition by GATA-type zinc finger domains. J Biol Chem. 2009;284:5827–5835. doi: 10.1074/jbc.M808712200. [DOI] [PubMed] [Google Scholar]
- Maduro M, Hill RJ, Heid PJ, Newman-Smith ED, Zhu J, Priess J, Rothman J. Genetic redundancy in endoderm specification within the genus Caenorhabditis. Dev Biol. 2005a;284:509–522. doi: 10.1016/j.ydbio.2005.05.016. [DOI] [PubMed] [Google Scholar]
- Maduro MF. Endomesoderm specification in Caenorhabditis elegans and other nematodes. Bioessays. 2006;28:1010–1022. doi: 10.1002/bies.20480. [DOI] [PubMed] [Google Scholar]
- Maduro MF. Structure and evolution of the C. elegans embryonic endomesoderm network. Biochim Biophys Acta. 2008 doi: 10.1016/j.bbagrm.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduro MF, Broitman-Maduro G, Mengarelli I, Rothman JH. Maternal deployment of the embryonic SKN-1-->MED-1,2 cell specification pathway in C. elegans. Dev Biol. 2007;301:590–601. doi: 10.1016/j.ydbio.2006.08.029. [DOI] [PubMed] [Google Scholar]
- Maduro MF, Kasmir JJ, Zhu J, Rothman JH. The Wnt effector POP-1 and the PAL-1/Caudal homeoprotein collaborate with SKN-1 to activate C. elegans endoderm development. Dev Biol. 2005b;285:510–523. doi: 10.1016/j.ydbio.2005.06.022. [DOI] [PubMed] [Google Scholar]
- Maduro MF, Lin R, Rothman JH. Dynamics of a developmental switch: recursive intracellular and intranuclear redistribution of Caenorhabditis elegans POP-1 parallels Wnt-inhibited transcriptional repression. Dev Biol. 2002;248:128–142. doi: 10.1006/dbio.2002.0721. [DOI] [PubMed] [Google Scholar]
- Maduro MF, Meneghini MD, Bowerman B, Broitman-Maduro G, Rothman JH. Restriction of mesendoderm to a single blastomere by the combined action of SKN-1 and a GSK-3beta homolog is mediated by MED-1 and -2 in C. elegans. Mol Cell. 2001;7:475–485. doi: 10.1016/s1097-2765(01)00195-2. [DOI] [PubMed] [Google Scholar]
- Maduro MF, Rothman JH. Making worm guts: the gene regulatory network of the Caenorhabditis elegans endoderm. Dev Biol. 2002;246:68–85. doi: 10.1006/dbio.2002.0655. [DOI] [PubMed] [Google Scholar]
- Mango SE. The C. elegans pharynx: a model for organogenesis. WormBook. 2007:1–26. doi: 10.1895/wormbook.1.129.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee JD. The C. elegans intestine. WormBook. 2007:1–36. doi: 10.1895/wormbook.1.133.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee JD, Fukushige T, Krause MW, Minnema SE, Goszczynski B, Gaudet J, Kohara Y, Bossinger O, Zhao Y, Khattra J, Hirst M, Jones SJ, Marra MA, Ruzanov P, Warner A, Zapf R, Moerman DG, Kalb JM. ELT-2 is the predominant transcription factor controlling differentiation and function of the C. elegans intestine, from embryo to adult. Dev Biol. 2009;327:551–565. doi: 10.1016/j.ydbio.2008.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee JD, Sleumer MC, Bilenky M, Wong K, McKay SJ, Goszczynski B, Tian H, Krich ND, Khattra J, Holt RA, Baillie DL, Kohara Y, Marra MA, Jones SJ, Moerman DG, Robertson AG. The ELT-2 GATA-factor and the global regulation of transcription in the C. elegans intestine. Dev Biol. 2007;302:627–645. doi: 10.1016/j.ydbio.2006.10.024. [DOI] [PubMed] [Google Scholar]
- Mello CC, Draper BW, Krause M, Weintraub H, Priess JR. The pie-1 and mex-1 genes and maternal control of blastomere identity in early C. elegans embryos. Cell. 1992;70:163–176. doi: 10.1016/0092-8674(92)90542-k. [DOI] [PubMed] [Google Scholar]
- Mello CC, Draper BW, Priess JR. The maternal genes apx-1 and glp-1 and establishment of dorsal-ventral polarity in the early C. elegans embryo. Cell. 1994;77:95–106. doi: 10.1016/0092-8674(94)90238-0. [DOI] [PubMed] [Google Scholar]
- Meneghini MD, Ishitani T, Carter JC, Hisamoto N, Ninomiya-Tsuji J, Thorpe CJ, Hamill DR, Matsumoto K, Bowerman B. MAP kinase and Wnt pathways converge to downregulate an HMG-domain repressor in Caenorhabditis elegans. Nature. 1999;399:793–797. doi: 10.1038/21666. [DOI] [PubMed] [Google Scholar]
- Miller DM, 3rd, Desai NS, Hardin DC, Piston DW, Patterson GH, Fleenor J, Xu S, Fire A. Two-color GFP expression system for C. elegans. Biotechniques. 1999;26:914–918. 920–921. doi: 10.2144/99265rr01. [DOI] [PubMed] [Google Scholar]
- Mizumoto K, Sawa H. Two betas or not two betas: regulation of asymmetric division by beta-catenin. Trends Cell Biol. 2007;17:465–473. doi: 10.1016/j.tcb.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Murray JI, Bao Z, Boyle TJ, Boeck ME, Mericle BL, Nicholas TJ, Zhao Z, Sandel MJ, Waterston RH. Automated analysis of embryonic gene expression with cellular resolution in C. elegans. Nat Methods. 2008;5:703–709. doi: 10.1038/nmeth.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nance J, Lee JY, Goldstein B. Gastrulation in C. elegans. WormBook. 2005:1–13. doi: 10.1895/wormbook.1.23.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okkema PG, Fire A. The Caenorhabditis elegans NK-2 class homeoprotein CEH-22 is involved in combinatorial activation of gene expression in pharyngeal muscle. Development. 1994;120:2175–2186. doi: 10.1242/dev.120.8.2175. [DOI] [PubMed] [Google Scholar]
- Okkema PG, Harrison SW, Plunger V, Aryana A, Fire A. Sequence requirements for myosin gene expression and regulation in Caenorhabditis elegans. Genetics. 1993;135:385–404. doi: 10.1093/genetics/135.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page BD, Diede SJ, Tenlen JR, Ferguson EL. EEL-1, a Hect E3 ubiquitin ligase, controls asymmetry and persistence of the SKN-1 transcription factor in the early C. elegans embryo. Development. 2007;134:2303–2314. doi: 10.1242/dev.02855. [DOI] [PubMed] [Google Scholar]
- Patton A, Knuth S, Schaheen B, Dang H, Greenwald I, Fares H. Endocytosis function of a ligand-gated ion channel homolog in Caenorhabditis elegans. Curr Biol. 2005;15:1045–1050. doi: 10.1016/j.cub.2005.04.057. [DOI] [PubMed] [Google Scholar]
- Pauli F, Liu Y, Kim YA, Chen PJ, Kim SK. Chromosomal clustering and GATA transcriptional regulation of intestine-expressed genes in C. elegans. Development. 2006;133:287–295. doi: 10.1242/dev.02185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips BT, Kidd AR, 3rd, King R, Hardin J, Kimble J. Reciprocal asymmetry of SYS-1/beta-catenin and POP-1/TCF controls asymmetric divisions in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2007;104:3231–3236. doi: 10.1073/pnas.0611507104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priess JR, Schnabel H, Schnabel R. The glp-1 locus and cellular interactions in early C. elegans embryos. Cell. 1987;51:601–611. doi: 10.1016/0092-8674(87)90129-2. [DOI] [PubMed] [Google Scholar]
- Raharjo I, Gaudet J. Gland-specific expression of C. elegans hlh-6 requires the combinatorial action of three distinct promoter elements. Dev Biol. 2007;302:295–308. doi: 10.1016/j.ydbio.2006.09.036. [DOI] [PubMed] [Google Scholar]
- Reuter R. The gene serpent has homeotic properties and specifies endoderm versus ectoderm within the Drosophila gut. Development. 1994;120:1123–1135. doi: 10.1242/dev.120.5.1123. [DOI] [PubMed] [Google Scholar]
- Rocheleau CE, Downs WD, Lin R, Wittmann C, Bei Y, Cha YH, Ali M, Priess JR, Mello CC. Wnt signaling and an APC-related gene specify endoderm in early C. elegans embryos. Cell. 1997;90:707–716. doi: 10.1016/s0092-8674(00)80531-0. [DOI] [PubMed] [Google Scholar]
- Rocheleau CE, Yasuda J, Shin TH, Lin R, Sawa H, Okano H, Priess JR, Davis RJ, Mello CC. WRM-1 activates the LIT-1 protein kinase to transduce anterior/posterior polarity signals in C. elegans. Cell. 1999;97:717–726. doi: 10.1016/s0092-8674(00)80784-9. [DOI] [PubMed] [Google Scholar]
- Rohrschneider MR, Nance J. Polarity and cell fate specification in the control of Caenorhabditis elegans gastrulation. Dev Dyn. 2009;238:789–796. doi: 10.1002/dvdy.21893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy Chowdhuri S, Crum T, Woollard A, Aslam S, Okkema PG. The T-box factor TBX-2 and the SUMO conjugating enzyme UBC-9 are required for ABa-derived pharyngeal muscle in C. elegans. Dev Biol. 2006 doi: 10.1016/j.ydbio.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Roy PJ, Stuart JM, Lund J, Kim SK. Chromosomal clustering of muscle-expressed genes in Caenorhabditis elegans. Nature. 2002;418:975–979. doi: 10.1038/nature01012. [DOI] [PubMed] [Google Scholar]
- Schroeder DF, McGhee JD. Anterior-posterior patterning within the Caenorhabditis elegans endoderm. Development. 1998;125:4877–4887. doi: 10.1242/dev.125.24.4877. [DOI] [PubMed] [Google Scholar]
- Shetty P, Lo MC, Robertson SM, Lin R. C. elegans TCF protein, POP-1, converts from repressor to activator as a result of Wnt-induced lowering of nuclear levels. Dev Biol. 2005;285:584–592. doi: 10.1016/j.ydbio.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Shin TH, Yasuda J, Rocheleau CE, Lin R, Soto M, Bei Y, Davis RJ, Mello CC. MOM-4, a MAP kinase kinase kinase-related protein, activates WRM-1/LIT-1 kinase to transduce anterior/posterior polarity signals in C. elegans. Mol Cell. 1999;4:275–280. doi: 10.1016/s1097-2765(00)80375-5. [DOI] [PubMed] [Google Scholar]
- Shirayama M, Soto MC, Ishidate T, Kim S, Nakamura K, Bei Y, van den Heuvel S, Mello CC. The Conserved Kinases CDK-1, GSK-3, KIN-19, and MBK-2 Promote OMA-1 Destruction to Regulate the Oocyte-to-Embryo Transition in C. elegans. Curr Biol. 2006;16:47–55. doi: 10.1016/j.cub.2005.11.070. [DOI] [PubMed] [Google Scholar]
- Smith PA, Mango SE. Role of T-box gene tbx-2 for anterior foregut muscle development in C. elegans. Dev Biol. 2007;302:25–39. doi: 10.1016/j.ydbio.2006.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Thorpe CJ, Schlesinger A, Carter JC, Bowerman B. Wnt signaling polarizes an early C. elegans blastomere to distinguish endoderm from mesoderm. Cell. 1997;90:695–705. doi: 10.1016/s0092-8674(00)80530-9. [DOI] [PubMed] [Google Scholar]
- Vilimas T, Abraham A, Okkema PG. An early pharyngeal muscle enhancer from the Caenorhabditis elegans ceh-22 gene is targeted by the Forkhead factor PHA-4. Dev Biol. 2004;266:388–398. doi: 10.1016/j.ydbio.2003.10.015. [DOI] [PubMed] [Google Scholar]
- Yekta S, Tabin CJ, Bartel DP. MicroRNAs in the Hox network: an apparent link to posterior prevalence. Nat Rev Genet. 2008;9:789–796. doi: 10.1038/nrg2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yochem J, Gu T, Han M. A new marker for mosaic analysis in Caenorhabditis elegans indicates a fusion between hyp6 and hyp7, two major components of the hypodermis. Genetics. 1998;149:1323–1334. doi: 10.1093/genetics/149.3.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Boyle TJ, Bao Z, Murray JI, Mericle B, Waterston RH. Comparative analysis of embryonic cell lineage between Caenorhabditis briggsae and Caenorhabditis elegans. Dev Biol. 2008;314:93–99. doi: 10.1016/j.ydbio.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Hill RJ, Heid PJ, Fukuyama M, Sugimoto A, Priess JR, Rothman JH. end-1 encodes an apparent GATA factor that specifies the endoderm precursor in Caenorhabditis elegans embryos. Genes Dev. 1997;11:2883–2896. doi: 10.1101/gad.11.21.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]