Abstract
Vitamin E (VE) in soybean seed has value for foods, medicines, cosmetics, and animal husbandry. Selection for higher VE contents in seeds along with agronomic traits was an important goal for many soybean breeders. In order to map the loci controlling the VE content, F5-derived F6 recombinant inbred lines (RILs) were advanced through single-seed-descent (SSD) to generate a population including 144 RILs. The population was derived from a cross between ‘OAC Bayfield’, a soybean cultivar with high VE content, and ‘Hefeng 25’, a soybean cultivar with low VE content. A total of 107 polymorphic simple sequence repeat markers were used to construct a genetic linkage map. Seed VE contents were analyzed by high performance liquid chromatography for multiple years and locations (Harbin in 2007 and 2008, Hulan in 2008 and Suihua in 2008). Four QTL associated with α-Toc (on four linkage groups, LGs), eight QTL associated with γ-Toc (on eight LGs), four QTL associated with δ-Toc (on four LGs) and five QTL associated with total VE (on four LGs) were identified. A major QTL was detected by marker Satt376 on linkage group C2 and associated with α-Toc (0.0012 > P > 0.0001, 5.0% < R 2 < 17.0%, 25.1 < α-Toc < 30.1 μg g−1), total VE (P < 0.0001, 7.0% < R 2 < 10.0%, 118.2 < total VE < 478.3 μg g−1). A second QTL detected by marker Satt286 on LG C2 was associated with γ-Toc (0.0003 > P > 0.0001, 6.0% < R 2 < 13.0%, 141.5 < γ-Toc < 342.4 μg g−1) and total VE (P < 0.0001, 2.0% < R 2 < 9.0%, 353.9 < total VE < 404.0 μg g−1). Another major QTL was detected by marker Satt266 on LG D1b that was associated with α-Toc (0.0002 > P > 0.0001, 4.0% < R 2 < 6.0%, 27.7 < α-Toc < 43.7 μg g−1) and γ-Toc (0.0032 > P > 0.0001, 3.0% < R 2 < 10.0%, 69.7 < γ-Toc < 345.7 μg g−1). Since beneficial alleles were all from ‘OAC Bayfield’, it was concluded that these three QTL would have great potential value for marker assisted selection for high VE content.
Introduction
Vitamin E (VE) is composed of a group of compounds known as α-(α-Toc), β-(β-Toc), γ-(γ-Toc), δ-(δ-Toc) tocopherol and four corresponding unsaturated derivatives, α-, β-, γ-, and δ-tocotrienol (Van Eenennaam et al. 2003). All tocopherols are amphipathic molecules consisting of a polar chromanol head group derived from homogentisic acid and a lipophilic tail derived from phytyl-diphosphate (Bramley et al. 2000; Savidge et al. 2002). Tocotrienols differ from tocopherols only in having a lipophilic tail derived from geranylgeranyl diphosphate with double bonds at carbon positions 3′, 7′ and 11′ (Cahoon et al. 2003; Munne-Bosch and Alegre 2002). Four tocopherols, α-Toc, β-Toc, γ-Toc and δ-Toc, differ from one another in the number and position of methyl groups on the chromanol head group (Tavva et al. 2007). The VE activities of β-Toc, γ-Toc and δ-Toc are 50, 10 and 3% equivalent to α-Toc (Kamal-Eldin and Appelqvist 1996; Sheppard et al. 1993; Shintani and DellaPenna 1998). Tocopherols are synthesized from precursors derived from two pathways. The methylerythritol phosphate pathway contributes to the side chain of tocopherol through the synthesis of phytyldiphosphate (Bramley et al. 2000). The shikimate pathway produces homogentisic acid, which contributes the aromatic ring of tocopherol. The first committed step in the tocopherol biosynthetic pathway is the prenylation of homogentisic acid with phytyldiphosphate to form 2-methyl-6-phytylbenzoquinol (Van Eenennaam et al. 2003). Many studies showed VE has a number of vital functions in plants, including the protection of chloroplasts from photooxidative damage (Munne-Bosch and Alegre 2002). Moreover, human nutritional studies have suggested that VE might play an important role in enhancing the function of the immune system (Adachi et al. 1997), and the treatment or prevention of a number of serious diseases including cardiovascular disease (Pryor 2000) and cancer (Prasad et al. 1999).
Twenty to thirty percent of VE in the American diet is derived from oil-based products such as margarines, dressings, and mayonnaise (Sheppard et al. 1993; Eitenmiller 1997). In many developed countries, the recommended daily intake of VE has been increased from 7–10 to 15 mg (Cho et al. 2005). Soybean oil accounts for 30% of the worldwide oil consumption (Van Eenennaam et al. 2003). Soybean oils has a much higher VE content than other oil seed crops such as safflower and sunflower (Dwiyanti et al. 2007). However, the relatively inactive γ-Toc is the most abundant component (about 70% of VE) in soybean, while in safflower and sunflower, highly active α-Toc was the most abundant component (about 80% of VE content; Dwiyanti et al. 2007). VE activity in soybean is actually lower than that of safflower or sunflower (Bramley et al. 2000). Therefore, increase of VE activity in soybean was necessary through increase of total VE or α-Toc contents.
VE biosynthesis occurs in the plastids of higher plants (Soll and Schultz 1980; Soll et al. 1985). The biosynthetic enzymes are associated with the chloroplast envelope and have proven difficult to isolate and purify. Some studies have focused on increasing VE content through transgenic techniques using proteins predicted to encode genes in the VE synthesis pathways (Cho et al. 2005; Van Eenennaam et al. 2003). However, the difficulties of soybean transformation and legal obstacles to transgenic soybean in China limited applications. Hence, selecting cultivars or breeding lines with higher VE and α-Toc content may be most effective.
Traditionally, plant improvement has relied on phenotypic selection of populations from crosses between commercial cultivars or experimental lines (Stuber et al. 1992). Selection for high VE and α-Toc contents in seeds based on phenotype is complicated by genotype × environment interaction (GE interaction) that significantly influences seed VE accumulation (Ujiie et al. 2005; Almonor et al. 1998; Dolde et al. 1999). Hence, selection for soybean cultivars with high VE and α-Toc contents in seeds requires evaluation in multiple environments over several years, which is expensive, time consuming, and labor intensive. Molecular markers offer a faster and more accurate approach to breeding, since selection could be based on genotype rather than solely on phenotype. The use of molecular markers for indirect selection of important agronomic traits, or marker-assisted selection (MAS) could improve the efficiency of traditional plant breeding. Some aspects of plant breeding that can be improved by MAS include; identification and elimination of undesirable individuals in the early stages of selection; identification of individuals prior to flowering when backcrossing genes governing the favorable expression of quantitative traits into adapted genotypes, and facilitation of selection for several traits simultaneously (Allen 1994). MAS could potentially improve selection of traits that have low heritability by using markers with high heritability.
Cregan et al. (1999) and Song et al. (2004) developed an integrated genetic linkage map of soybean containing 1,849 markers, including 1,015 SSR markers polymorphic in one or more of five different populations. Markers were aligned into molecular linkage groups (LGs) into a consensus map of 20 LGs which correspond to the 11 of the 20 pairs of soybean chromosomes (Zou et al. 2003). The remaining nine LGs were assigned to chromosomes based on size (Soybase; unpublished).
Currently, few studies attempted to map quantitative trait loci (QTL) associated with individual and total VE in soybean seed. Dwiyanti et al. (2007) analyzed individual and total VE using a set of F2 seeds and F3 seeds from a cross between ‘Keszthelyi A.S.’ and ‘Ichihime’ soybean. The results showed SSR markers Sat_243 and Sat167 in LG K were significantly associated with α-Toc. By 2009 the authors could find no report of QTL associated with individual tocopherols and total VE of soybean based on RIL populations tested in multiple environments.
The objective of the present study was to identify QTL associated with individual tocopherols and total VE using ‘Hefeng 25’ × ‘OAC Bayfield’ RILs in multiple environments.
Materials and methods
Plant materials
The mapping population contains 144 F5:6 recombinant inbred lines (RILs) which were advanced by single-seed-descent (SSD) in 2003–2008 from the cross between ‘Hefeng 25’ (developed by the Heilongjiang Academy of Agriculture, Jiamushi, China) and ‘OAC Bayfield’ (developed by University of Guelph, Guelph, Canada) (Tanner et al. 1998). ‘OAC Bayfield’ showed consistently high seed VE content (mean α-Toc 49.1 μg g−1; γ-Toc 503.3 μg g−1; δ-Toc 254.2 μg g−1; total VE 853.9 μg g−1). ‘Hefeng 25’ had low seed VE content (mean α-Toc 8.8 μg g−1; γ-Toc 84.4 μg g−1; δ-Toc 44.0 μg g−1; total VE 176.6 μg g−1).
Field experiments
RILs were grown together with the parents; at Harbin (N45°, fine-mesic chernozen soil) in 2007 and 2008; at Hulan (46°, fine-mesic chernozen soil) in 2008 and Suihua (N47°, fine-mesic chernozen soil) in 2008. Seeds were planted with rows 3 m long, 0.65 m apart and with a space of 6 cm between two plants. Three replicates were used with a randomized complete block design. Each plot of a single genotype provided 20 plants as seed donors that were later used to analyze individual and total VE content.
Sample preparation and high performance liquid chromatography (HPLC) analysis
High performance liquid chromatography was performed following the procedure of Ujiie et al. (2005). Soybean seed samples (about 5 g) were ground to a fine powder. A total of 0.1 g of soybean flour and 0.05 g of ascorbic acid were mixed in a 5 ml tube, and stirred with 3 ml of 80% (w/v) ethanol for 10 s. After super-sonication for 15 min at room temperature exactly 2 ml of hexyl-hydride was added and stirred for 10 min. The slurry was centrifuged at 13,000×g for 15 min. The clear aliquot was filtered through a 0.45-μm PTFE filter. The resulting supernatant was analyzed for VE contents by HPLC (Dionex ASI-100, USA) with a C18 reverse-phase column (250 mm length and 4.6 μm particle size). The condition used was as follows: mobile phases were solvent A (100% methanol), flow rate 1.5 ml min−1, the temperature of the column was 40°C; and the injection volume was 20 μl. A dionex fluorescent light detector was used with excitation at 295 nm and emission at 330 nm (Abbasi et al. 2007). The α-Toc, γ -Toc and δ-Toc standards, obtained from Sigma Inc (St. Louis, MO, USA), were dissolved and serially diluted in ethanol. Standard concentrations ranged from 5 to 100 μM, and 10 μl volumes (5–1,000 pM) were injected (Tavva et al. 2007). FR spectra were recorded and its responses were integrated by software Dionex 2.0. Quantification of the VE was done by the external standard method.
SSR analyses
Total DNA of parents and each RIL were isolated from freeze-dried leaf tissue by the CTAB method (Doyle and Doyle 1990). SSR analysis was performed with the primers developed by Cregan et al. (1999). PCR was performed in 20 μl reaction containing 2 μl genomic DNA (25 ng μl−1), 1.5 μl MgCl2 (25 mM), 0.3 μl dNTP mixtures (10 mM), 2 μl 10 × PCR buffer, 2 μl each primer (2 μM), 0.2 μl Taq polymerase enzyme (10 units μl−1), 12 μl double distilled water. The amplification profiles were; 2 min at 94°C, followed by 35 cycles of 30 s at 94°C; 30 s at 47°C, 30 s at 72°C; then 5 min at 72°C. After amplification, the PCR products was mixed with loading buffer [2.5 mg ml−1 bromophenol blue, 2.5 mg ml−1 diphenylamine blue, 10 mM EDTA, 95% (v/v) formamide], denatured for 5 min at 94°C and put on ice for 5 min. The denatured PCR products were separated on 6% (w/v) denaturing polyacrylamide gel and visualized by silver straining (Trigizano and Caetano-Anolles 1998).
Data analysis
Broad-sense heritability and statistic parameters for RILs were analyzed using the SAS procedure (line regression and GLM.SAS). A linkage map was constructed by MAPMAKER/EXP version 3.0b (Lander et al. 1987) as described by Primomo et al. (2005). The commands “group,” “map,” “sequence,” “lod table,” “try,” and “compare” were used for building the linkage groups. The error detection ratio was set at 1%. The Haldane mapping function was used with a minimum LOD score of 3.0 and a maximum distance of 50 cM.
QTL were identified by single-factor analysis of variance (PROC. GLM. SAS) as described by Primomo et al. (2005), based on individual environment value. Locus main effects were considered for linear models if they were significant at P ≤ 0.01. Significant loci on the same LG were tested by two-factor analysis of variance without interactions. If both loci were significant at P ≤ 0.05 in the two-factor model, they would both be considered for linear models. Otherwise, the locus with the larger individual R 2 value was chosen to represent the effect of the putative QTL on the LG. The nomenclature of QTL included four parts. For example QαC2_1, Q, α, C2 and 1 represent QTL, trait (α-Toc), linkage group name and QTL order in the linkage group, respectively.
Genotype by trait (GT) biplot methodology (Yan 2001) was employed to analyze the interactions between QTL and environments or individual and total VE means, based on the formula; T ij − T j/S j = λ 1 ζ i1 τ j1 + λ 2 ζ i2 τ j2 + ε ij; where T ij was the mean value of QTL i for environment j; T j was the mean value of environment j over all QTL; S j was the standard deviation of environment j among QTL mean; ζ i1 and ζ i2 were the PC1 (first principle component) and PC2 (second principle component) scores, respectively, for QTL mean i; τ j1 and τ j2 were the PC1 and PC2 scores, respectively, for environment j; and ε ij was the residual of the model associated with QTL i, challenged with environment j.
Results
Phenotypic analysis of individual and total VE content
The results of α-Toc, γ-Toc, δ-Toc and total VE contents from RILs and parents across four different environments were shown in Table 1. The mean values of individual and total VE for ‘OAC Bayfield’ were significantly higher than those of ‘Hefeng 25’ across four environments. In contrast, the variation of these traits for 144 RILs across various environments, and individual or total VE means were not significant (P > 0.05). Both Skewness and Kurtosis values for these traits and their means were less than 1.0 in the different environments, suggesting that the segregation of these traits approximatively fit a normal distribution model. Broad-sense heritability for 144 RILs across various environments and individual or total VE means were listed in Table 1, most of them showed relatively low heritability.
Table 1.
Range, average, standard deviation, coefficient of variation, Skewness, Kurtosis, and broad-sense heritability for vitamin E contents of RILs under multiple environments
| Trait | Environment | Parents | RIL | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Hefeng 25a | OAC Bayfielda | Range | Mean | SDb | CVc | Skewness | Kurtosis | BSHd | ||
| α-Toce | 07 Harbin | 10.36 | 37.78 | 9.91–62.23 | 26.97 | 7.58 | 0.28 | 0.52 | 0.21 | 0.37 |
| 08 Harbin | 14.52 | 80.26 | 2.53–95.295 | 34.42 | 16.62 | 0.48 | 0.67 | 0.37 | 0.32 | |
| 08 Hulan | 5.99 | 40.73 | 1.92–42.26 | 19.48 | 8.54 | 0.43 | 0.22 | −0.16 | 0.28 | |
| 08 Suihua | 4.32 | 37.79 | 0.98–39.85 | 18.45 | 7.28 | 0.39 | 0.27 | 0.26 | 0.33 | |
| α-Toce,f | 8.79 | 49.14 | 3.84–59.91 | 24.83 | 10.01 | 0.39 | 0.42 | 0.25 | 0.30 | |
| γ-Tocg,f | 07 Harbin | 100.54 | 158.49 | 85.18–196.95 | 139.73 | 118.24 | 0.13 | −0.17 | 0.32 | 0.40 |
| 08 Harbin | 43.87 | 372.08 | 8.22–533.80 | 145.18 | 107.08 | 0.50 | 0.996 | 0.97 | 0.40 | |
| 08 Hulan | 87.54 | 682.52 | 34.84–808.84 | 450.51 | 155.995 | 0.34 | −0.345 | −0.16 | 0.33 | |
| 08 Suihua | 105.64 | 800.05 | 29.93–991.12 | 515.83 | 165.61 | 0.32 | −0.28 | 0.58 | 0.42 | |
| γ-Tocg,f | 84.39 | 503.28 | 39.54–632.68 | 312.81 | 111.73 | 0.38 | 0.45 | 0.51 | 0.33 | |
| δ-Toc | 07 Harbin | 14.62 | 60.01 | 27.39–82.72 | 57.97 | 10.57 | 0.18 | −0.01 | −0.59 | 0.22 |
| 08 Harbin | 53.65 | 486.17 | 27.43–522.89 | 244.30 | 88.48 | 0.36 | 0.35 | 0.18 | 0.31 | |
| 08 Hulan | 32.87 | 277.71 | 14.28–312.64 | 175.75 | 59.99 | 0.34 | −0.499 | −0.06 | 0.40 | |
| 08 Suihua | 74.75 | 193.07 | 15.13–293.03 | 187.11 | 56.03 | 0.29 | −0.54 | 0.25 | 0.36 | |
| δ-Toch,f | 43.97 | 254.24 | 21.06–302.82 | 166.28 | 53.77 | 0.29 | 0.35 | 0.27 | 0.29 | |
| TE | 07 Harbin | 157.69 | 294.68 | 139.85–306.20 | 224.23 | 27.53 | 0.12 | −0.35 | 0.17 | 0.32 |
| 08 Harbin | 183.49 | 947.2 | 38.35–952.85 | 391.85 | 155.89 | 0.39 | 0.59 | 0.61 | 0.35 | |
| 08 Hulan | 136.04 | 1013.96 | 23.63–1117.54 | 289.25 | 96.21 | 0.33 | −0.54 | 0.03 | 0.31 | |
| 08 Suihua | 229.18 | 1159.65 | 22.25–1189.26 | 312.00 | 89.34 | 0.28 | −0.79 | 0.59 | 0.36 | |
| TEi,f | 176.60 | 853.88 | 56.02–961.96 | 304.33 | 92.24 | 0.28 | 0.57 | 0.35 | 0.32 | |
aμg g−1
bStandard deviation
cCoefficient of variation
dBroad-sense heritability
eα-Tocopherol
fMean of all data from four different environments
gγ-Tocopherol
hδ-Tocopherol
iTotal VE
Linkage analysis
A total of 463 SSR markers were used to detect polymorphisms between the two parents. One hundred and seven of them (23.1%) were polymorphic and mapped on 20 linkage groups (LGs) according to Cregan et al. (1999) and Song et al. (2004). The map encompassed about 1,521.3 cM with a mean distance of 14.2 cM between markers (data not shown). The relative order of the SSR markers on most of LGs was similar to the public soybean genetic map (Song et al. 2004).
QTL associated with individual and total VE content
QTL associated with individual and total VE contents were identified by single-factor analysis of variance. Four QTL, QαB2_1(Sat_177, LG B2), QαC2_1(Satt376 LG C2), QαD1b_1(Satt266, LGD1b) and QαI_1(Satt440, LG I), were associated with α-Toc (Table 2). QαB2_1 explained 6.5–7.3% of the phenotypic variations at Harbin in 2 years or α-Toc means. QαC2_1 explained 5.8–16.7% of the phenotypic variations at three locations in 2 years or α-Toc overall means. QαD1b_1 explained 4.3–5.6% of the phenotypic variation at Harbin in 2 years. QαI_1 explained 5.4% of the phenotypic variation at Hulan location in 1 year.
Table 2.
QTL associated with individual tocopherols and total VE content
| Trait | QTL | LG | Marker | Environment | R 2 (%)b | P | Allelic means (μg g−1) ± SEMsa | |
|---|---|---|---|---|---|---|---|---|
| Hefeng 25 | OAC Bayfield | |||||||
| α-Tocc | QαB2_1 | B2 | Sat_177 | 07 Harbin | 6.7 | <0.0001 | 19.5 ± 1.2 | 52.4 ± 1.2 |
| 08 Harbin | 6.5 | <0.0001 | 12.5 ± 1.7 | 58.5 ± 1.7 | ||||
| α-Toc Mean | 7.3 | <0.0001 | 17.9 ± 1.4 | 49.4 ± 1.4 | ||||
| QαC2_1 | C2 | Satt376 | 07 Harbin | 7.4 | 0.0012 | 23.7 ± 1.6 | 48.8 ± 1.6 | |
| 08 Harbin | 8.6 | 0.0002 | 19.7 ± 1.6 | 49.8 ± 1.6 | ||||
| 08 Suihua | 16.7 | <0.0001 | 9.8 ± 1.6 | 35.5 ± 1.6 | ||||
| 08 Hulan | 5.8 | <0.0001 | 7.9 ± 1.5 | 37.0 ± 1.5 | ||||
| α-Toc Mean | 7.9 | 0.0009 | 12.5 ± 1.6 | 38.5 ± 1.6 | ||||
| QαD1b_1 | D1b | Satt266 | 07 Harbin | 5.6 | 0.0002 | 25.8 ± 1.4 | 53.5 ± 1.4 | |
| 08 Harbin | 4.3 | <0.0001 | 18.4 ± 1.2 | 62.1 ± 1.2 | ||||
| QαI_1 | I | Satt440 | 08 Hulan | 5.4 | <0.0001 | 4.1 ± 1.2 | 37.3 ± 1.2 | |
| α-Toc Mean | 5.0 | <0.0001 | 17.9 ± 1.2 | 64.3 ± 1.2 | ||||
| QγA2_1 | A2 | Sat_383 | 07 Harbin | 11.8 | <0.0001 | 25.4 ± 10.6 | 164.8 ± 10.7 | |
| γ-Toc Mean | 12.0 | <0.0001 | 27.2 ± 10.6 | 166.4 ± 10.6 | ||||
| γ-Tocd | QγC1_1 | C1 | Satt565 | 08 Harbin | 2.8 | <0.0001 | 248.2 ± 9.6 | 464.3 ± 9.7 |
| γ-Toc Mean | 6.7 | <0.0001 | 283.6 ± 9.7 | 537.5 ± 9.7 | ||||
| QγC2_1 | C2 | Satt286 | 08 Harbin | 7.5 | <0.0001 | 322.5 ± 8.3 | 646.1 ± 8.3 | |
| 08 Hulan | 8.6 | 0.0003 | 297.0 ± 9.4 | 438.5 ± 9.4 | ||||
| 08 Suihua | 13.0 | <0.0001 | 383.0 ± 9.6 | 725.5 ± 9.6 | ||||
| γ-Toc Mean | 6.9 | <0.0001 | 337.3 ± 11.9 | 586.2 ± 11.8 | ||||
| Qγ G_1 | G | Satt199 | 07 Harbin | 4.2 | 0.0032 | 100.7 ± 8.3 | 176.9 ± 8.3 | |
| 08 Hulan | 6.0 | <0.0001 | 247.3 ± 9.6 | 707.8 ± 9.7 | ||||
| γ-Toc Mean | 6.0 | 0.0002 | 226.9 ± 9.3 | 689.5 ± 9.3 | ||||
| QγD1b-1 | D1b | Satt266 | 07 Harbin | 3.5 | 0.0032 | 100.3 ± 10.6 | 169.9 ± 10.6 | |
| 08 Harbin | 7.4 | <0.0001 | 102.8 ± 9.4 | 437.9 ± 9.4 | ||||
| 08 Suihua | 9.0 | <0.0001 | 483.3 ± 9.7 | 829.0 ± 9.7 | ||||
| γ-Toc Mean | 5.2 | 0.0003 | 418.3 ± 9.3 | 545.6 ± 9.3 | ||||
| QγO-1 | O | Satt576 | 07 Harbin | 10.5 | 0.0032 | 97.8 ± 9.6 | 158.6 ± 9.6 | |
| 08 Harbin | 6.4 | 0.0002 | 220.4 ± 9.3 | 462.0 ± 9.3 | ||||
| 08 Suihua | 7.7 | <0.0001 | 397.2 ± 9.6 | 851.2 ± 9.6 | ||||
| 08 Hulan | 5.8 | 0.0001 | 394.6 ± 10.0 | 727.4 ± 10.0 | ||||
| γ-Toc Mean | 6.0 | 0.0001 | 227.3 ± 9.6 | 689.7 ± 9.6 | ||||
| QγE_1 | E | Satt355 | 08 Harbin | 9.2 | 0.0002 | 96.1 ± 9.4 | 427.9 ± 9.5 | |
| γ-Toc Mean | 2.4 | <0.0001 | 320.9 ± 10.0 | 537.8 ± 10.0 | ||||
| QγJ_1 | J | Satt280 | 08 Suihua | 9.0 | <0.0001 | 389.3 ± 9.2 | 840.3 ± 9.3 | |
| γ-Toc Mean | 2.5 | <0.0001 | 360.8 ± 9.2 | 578.3 ± 9.3 | ||||
| δ-Toce | QδA2_1 | A2 | Sat_383 | 07 Harbin | 4.2 | <0.0001 | 37.4 ± 5.0 | 75.4 ± 5.0 |
| 08 Suihua | 5.2 | <0.0001 | 84.3 ± 5.5 | 178.9 ± 5.5 | ||||
| δ-Toc Mean | 7.8 | <0.0001 | 123.9 ± 5.5 | 264.0 ± 5.6 | ||||
| QδD1a_1 | D1a | Satt179 | 08 Harbin | 6.5 | <0.0001 | 147.8 ± 5.0 | 428.3 ± 5.0 | |
| QδF_1 | F | Sat_262 | 08 Suihua | 7.6 | <0.0001 | 106.8 ± 5.3 | 240.5 ± 5.3 | |
| QδI_1 | I | Satt354 | 08 Hulan | 10.2 | <0.0001 | 87.4 ± 5.6 | 253.1 ± 5.6 | |
| δ-Toc Mean | 7.7 | <0.0001 | 118.6 ± 5.6 | 265.5 ± 5.6 | ||||
| Total VEf | QTVEC2_1 | C2 | Satt376 | 07 Harbin | 7.5 | <0.0001 | 166.1 ± 2.9 | 284.3 ± 3.0 |
| 08 Harbin | 9.4 | <0.0001 | 398.6 ± 8.3 | 764.3 ± 8.3 | ||||
| 08 Suihua | 8.3 | <0.0001 | 377.6 ± 9.0 | 758.6 ± 9.0 | ||||
| 08 Hulan | 7.2 | <0.0001 | 327.0 ± 9.6 | 805.4 ± 9.7 | ||||
| TVE Mean | 8.6 | 0.0001 | 402.6 ± 9.5 | 772.9 ± 10.0 | ||||
| QTVEC2_2 | C2 | Satt286 | 08 Harbin | 6.3 | <0.0001 | 394.4 ± 8.8 | 764.5 ± 8.9 | |
| 08 Hulan | 2.9 | <0.0001 | 374.5 ± 9.3 | 728.5 ± 9.3 | ||||
| 08 Suihua | 8.3 | <0.0001 | 299.8 ± 10.3 | 703.8 ± 12.0 | ||||
| TVE Mean | 6.1 | <0.0001 | 400.3 ± 8.0 | 776.8 ± 8.3 | ||||
| QTVE D1b_1 | D1b | Satt172 | 07 Harbin | 9.0 | <0.0001 | 156.7 ± 9.2 | 290.0 ± 9.3 | |
| 08 Harbin | 10.9 | <0.0001 | 287.3 ± 8.3 | 864.9 ± 8.3 | ||||
| TVE Mean | 7.3 | <0.0001 | 271.0 ± 9.0 | 789.4 ± 9.0 | ||||
| QTVEN _1 | N | Sat_125 | 08 Harbin | 6.7 | <0.0001 | 407.0 ± 9.3 | 825.5 ± 9.3 | |
| TVE Mean | 3.8 | <0.0001 | 319.1 ± 9.3 | 678.9 ± 9.3 | ||||
| QTVEO _1 | O | Satt592 | 08 Suihua | 3.5 | <0.0001 | 293.5 ± 9.3 | 647.0 ± 9.3 | |
| 08 Hulan | 5.5 | 0.0001 | 420.5 ± 9.3 | 809.4 ± 9.3 | ||||
| TVE Mean | 6.4 | <0.0001 | 357.7 ± 9.3 | 766.4 ± 9.3 | ||||
aSEM: Mean + SD√N; where N was the number of each of allele
bThe proportion of the phenotypic data explained by marker locus
cα-Tocopherol
dγ-Tocopherol
eδ-Tocopherol
fTotal VE
Eight QTL underlying γ-Toc were detected and mapped onto eight LGs and explained 2.8–13.0% of the phenotypic variation at three locations in 2 years or γ-Toc overall means (Table 2). Of them, QγC2_1 (Satt286), QγG_1 (Satt199), QγD1b-1 (Satt266) and QγO-1 (Satt576), were identified in 2–4 environments and γ-Toc means. Other QTL, including QγA2_1 (Sat_383), QγC1_1 (Satt565), QγE_1 (Satt355) and QγJ_1 (Satt280), were detected in only one environment.
Four QTL, QδA2_1 (Sat_383), QδD1a_1 (Satt179), QδF_1 (Satt262), QδI_1 (Satt354) associated with δ-Toc, were identified on LGs A2, D1a, F, and I, respectively. The phenotypic variation ranged from 4.2 to 10.2% at three locations in 2 years or δ-Toc means (Table 2). Of them, Only QδA2_1 (Sat_383) could be identified in multiple environments (two environments and δ-Toc means). The other three QTL, QδD1a_1, QδF_1, and QδI_1, were detected in only one environment.
QTL underlying total seed VE content, QTVEC2_1 (Satt376), QTVEC2_2 (Satt286), QTVE D1b_1 (Satt172), QTVEN _1 (Sat_125), and QTVEO _1 (Satt592) were identified in 1–4 environments and total VE overall means and mapped onto four LGs (Table 2). They explained 2.9–10.9% of the phenotypic variation.
Satt376 on LG C2 was associated both with α-Toc (QaC2-1) and total VE (QTVEC2-1) across four environments on linkage group C2 with beneficial allele from ‘OAC Bayfield’. QγC2_1 was associated with γ-Toc across three environment and γ-Toc means, and QTVEC2_2 was associated with total VE across three environments and the total VE means. Both were linked with the same SSR marker (Satt286) on linkage group C2. QαD1b_1, associated with α-Toc across two environments, QγD1b-1, associated with γ-Toc across three environments, and the γ-Toc means, were both associated with the same SSR marker (Satt266) on linkage group D1b.
Analyses of QTL by environment interaction
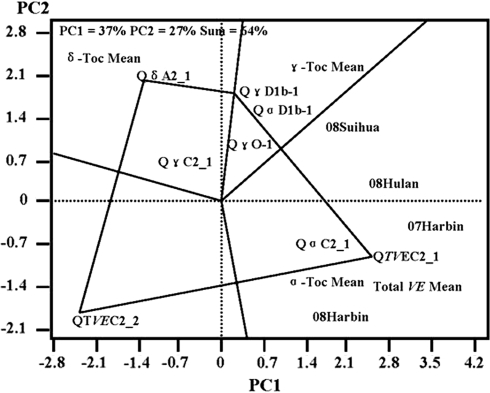
GT biplot analysis (Yan 2001) of all QTL with individual and total VE contents against four environments, and individual or total VE mean, showed that these QTL explained 64.0% of the total variation of VE. The performance of different QTL in each environment was evaluated. When QTL QδA2_1, QγD1b_1, QTVEC2_1 and QTVEC2_2 were set as the corner QTL, four different environments and individual or total VE means fell in the sector in which QTVEC2_1 were the best QTL for four environments (at Harbin in 2007 and 2008, at Hulan in 2008, at Suihua in 2008) for α-Toc and total VE means. QδA2_1 and QγD1b_1 were the best QTL for δ-Toc and γ-Toc means, respectively. No tested location fell into sectors with QTVEC2_2 as vectors, indicating it was the poorest QTL for any of four different locations tested (Fig. 1).
Fig. 1.
GT biplot analysis for the relatedness of QTL and environment or individual and total VE mean. PC1 was the first principle component; and PC2 the second principle component. Different environments were represented by year and location, for example ‘at Harbin location in 2007’ was represented by 07 Harbin
Discussion
‘OAC Bayfield’ had high VE and α-Toc contents in Canada over many years. Therefore, there is great interest in transferring high VE and α-Toc contents into the cultivars in Northeastern China to increase VE activity, and understanding the breeding potential among the progeny. Because VE content in soybean is difficult to evaluate by phenotype, increasing the genotype selection intensity by MAS should improve the selection efficiency.
Phenotypic variability of individual and total VE content in soybean seed was high, whether in cultivars (Dolde et al. 1999; Ujiie et al. 2005; Concordia et al. 2007) or a cross derived population (Dwiyanti et al. 2007; Table 1). Many studies have demonstrated that the interactions between environmental factors, such as year by location and genotype by environment, were the primary sources of variation of VE contents in soybean seeds (Almonor et al. 1998; Dolde et al. 1999). However, main genotypic effects of total and individual VE were large enough for effective cultivar improvement (Rani et al. 2007).
Four QTL associated with α-Toc, eight QTL associated with γ-Toc, four QTL associated with δ-Toc and five QTL associated with total VE, were mapped onto four, eight, four and four LGs, respectively. These QTL explained 2.8–16.7% of phenotypic variation for individual and total VE in different environments, and most of the variation was less than 10.0%. The low level of phenotypic variation evaluated by these QTL indicated the quantitative nature of individual and total VE inheritance in soybean seeds. These results seemed to verify the difficulty of genetic improvement by phenotype selection.
QTL specific to one environment were also found among other nutrient components of soybean (Zeng et al. 2009). Inconsistent QTL detection across multi-environments could be due to non- or weak-expression of the QTL, QTL × environment interaction in the opposite direction to the main QTL effects, and/or epistasis. The results here demonstrated that QTL × environment interaction was an important property of many QTL of individual and total VE. The interactions between QTL and environment indicated that Satt376, associated with QTEVC2_1 for total VE and with QaC2_1 for α-Toc was the best molecular marker for MAS for α-Toc and total VE across environments (at Harbin in 2007 and 2008, at Hulan in 2008, and at Suihua in 2008). Therefore, QTL × environment interactions should be considered during marker-assisted selection of VE contents.
In this study, Q alpha C2_1, which was associated with α-Toc across four environments and α-Toc means, and QTVEC2_1, which was associated with total VE across four environments and total VE means, were both linked with the same SSR marker (Satt376 in LG C2; Table 2). Qgamma C2_1, associated with γ-Toc across three environments and γ-Toc means, and QTVEC2_2 associated with total VE across three environments and total VE means, also were both associated with the same SSR marker (Satt286 in LG C2). QalphaD1b_1, associated with α-Toc across two environments, and QγD1b-1, associated with γ-Toc across three environments and γ-Toc means, also were both associated with the same SSR marker (Satt266 in LG D1b). The SSR markers Satt376, Satt286 and Satt266 were associated with many tocopherols and total VE content in seed across multi-environment conditions. Therefore, these three markers might have good potential for application in MAS for high VE variety development.
The availability of QTL associated with individual and total VE in soybean seed could facilitate MAS in breeding programs aiming to transfer high VE and α-Toc content from soybean cultivar ‘OAC Bayfield’ to other elite breeding lines in Northeastern China. Knowledge of QTL controlling individual and total VE in soybean will allow the design and implementation of more efficient selection schemes to develop high-VE or α-Toc soybean cultivars. So far, progress in breeding high-VE and α-Toc cultivars is slow due to large environmental effects on this trait. However, MAS will be an efficient and cost effective breeding strategy for developing high VE and α-Toc soybean cultivars.
Acknowledgments
We thank Dr. Istvan Rajcan (University of Guelph, Canada) for the supply of cultivar ‘OAC Bayfield’. This study was conducted in the Key Laboratory of Soybean Biology of Chinese Education Ministry and financially supported by National High Technology Project (Contract No. 2006AA10Z1F1 and 2006AA100104-4), 948 project of Agricultural ministry of China (Contract No. 2006-G5), National 973 Project for molecular breeding of soybean and corn (Contract No. 2009CB118405).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
H. Li, H. Liu, Y. Han and X. Wu contributed equally to this work.
References
- Adachi N, Migita M, Ohta T, Higashi A, Matsuda I. Depressed natural killer cell activity due to decreased natural killer cell population in a vitamin E-deficient patient with Shwachman syndrome: reversible natural killer cell abnormality by α-tocopherol supplementation. Eur J Pediatr. 1997;156:444–448. doi: 10.1007/s004310050634. [DOI] [PubMed] [Google Scholar]
- Allen FL. Usefulness of plant genome mapping to plant breeding. In: Gresshoff P, editor. Plant genome analysis. Boca Raton: CRC Press; 1994. pp. 11–18. [Google Scholar]
- Almonor GO, Fenner GP, Wilson RF. Temperature effects on tocopherols composition in soybeans with genetically improved oil quality. J Am Oil Chem Soc. 1998;75:591–596. doi: 10.1007/s11746-998-0070-3. [DOI] [Google Scholar]
- Bramley PM, Elmadfa I, Kafatos A, Kelly FJ, Manios Y, Roxborough HE, Schuch W, Sheehy PJA, Wagner KH. Vitamin E. Sci Food Agric. 2000;80:913–938. doi: 10.1002/(SICI)1097-0010(20000515)80:7<913::AID-JSFA600>3.0.CO;2-3. [DOI] [Google Scholar]
- Britz ST, Kremer F. Warm temperature or drought during seed maturation increase free a-tocopherol in seeds of soybean (Glycine max [L.] Merr.) J Agric Food Chem. 2002;50:6058–6063. doi: 10.1021/jf0200016. [DOI] [PubMed] [Google Scholar]
- Cahoon EB, Hall SE, Ripp KG, Ganzke TS, Hitz WD, Coughlan SJ. Metabolic redesign of vitamin E biosynthesis in plants for tocotrienol production and increased antioxidant content. Nat Biotechnol. 2003;21:1082–1087. doi: 10.1038/nbt853. [DOI] [PubMed] [Google Scholar]
- Cho EA, Lee CA, Kim YS, Baek SH, Reyes BG, Yun SJ. Expression of γ-Tocopherol methyltransferase transgene improves tocopherol composition in Lettuce (Latuca sativa L.) Mol Cells. 2005;19(1):16–22. [PubMed] [Google Scholar]
- Conco’rdia M, Panizzi C, Erhan S. Environmental and genetic variation of soybean tocopherol content under Brazilian growing conditions. J Am Oil Chem Soc. 2007;84:921–928. doi: 10.1007/s11746-007-1128-3. [DOI] [Google Scholar]
- Cregan PB, Jarvik T, Bush AL, Shoemaker RC, Lark KG, Kahler AL, VanToai TT, Lohnes DG, Chung J, Specht JE. An integrated genetic linkage map of the soybean genome. Crop Sci. 1999;39:1464–1490. [Google Scholar]
- Dolde D, Vlahakis C, Hazebroek J. Tocopherol in breeding lines and effects of planting location, fatty acid composition, and temperature during development. J Am Oil Chem Soc. 1999;76:349–355. doi: 10.1007/s11746-999-0242-9. [DOI] [Google Scholar]
- Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–15. [Google Scholar]
- Dwiyanti MS, Ujiie A, Thuy LTB, Yamda T, Kitamura K. Genetic analysis of high α-tocopherol content in soybean seeds. Breed Sci. 2007;57:23–28. doi: 10.1270/jsbbs.57.23. [DOI] [Google Scholar]
- Eitenmiller R. Vitamin E Content of fats and oils-nutritional implications: tocopherols and tocotrienols play important roles in health and disease. Food Technol. 1997;51:78–81. [Google Scholar]
- Kamal-Eldin A, Appelqvist LA. The chemistry and antioxidant properties of tocopherols and tocotrienols. Lipids. 1996;31:671–701. doi: 10.1007/BF02522884. [DOI] [PubMed] [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow A, Daly M, Lincoln S, Newburg L. Mapmaker: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- Munne-Bosch S, Alegre L. The function of tocopherols and tocotrienols in plants. Crit Rev Plant Sci. 2002;21:31–57. doi: 10.1016/S0735-2689(02)80037-5. [DOI] [Google Scholar]
- Prasad KN, Kumar A, Kochupillai V, Cole WC. High doses of multiple antioxidant vitamins: essential ingredients in improving the efficacy of standard cancer therapy. J Am Coll Nutr. 1999;18:13–25. doi: 10.1080/07315724.1999.10718822. [DOI] [PubMed] [Google Scholar]
- Primomo VS, Poysa V, Ablett GR, Jackson CJ, Gijzen M, Rajcan I. Mapping QTL for individual and total isoflavone content in soybean seeds. Crop Sci. 2005;45:2454–2464. doi: 10.2135/cropsci2004.0672. [DOI] [Google Scholar]
- Pryor WA. Vitamin E and heart disease: basic science to clinical intervention trials. Free Radic Biol Med. 2000;28:141–164. doi: 10.1016/S0891-5849(99)00224-5. [DOI] [PubMed] [Google Scholar]
- Rani A, Kumar V, Verma SK, Shakya AK, Chauhan GS. Tocopherol content and profile of soybean: genotypic variability and correlation studies. J Am Oil Chem Soc. 2007;84:377–383. doi: 10.1007/s11746-007-1040-x. [DOI] [Google Scholar]
- Savidge B, Weiss JD, Wong JH, Lassner MW, Mitsky TA, Shewmaker CK, Post-Beittenmiller D, Valentin HE. Isolation and characterization of homogentisate phytyltransferase genes from Synechocystis sp. PCC6803 and Arabidopsis. Plant Physiol. 2002;129:321–332. doi: 10.1104/pp.010747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard AJ, Pennington JA, Weihrauch JL. Analysis and distribution of vitamin E in vegetable oils and foods. In: Packer L, Fuchs J, editors. Vitamin E in health and disease. New York: Marcel Dekker; 1993. pp. 9–31. [Google Scholar]
- Shintani D, DellaPenna D. Elevating the vitamin E content of plants through metabolic engineering. Science. 1998;282:2098–2100. doi: 10.1126/science.282.5396.2098. [DOI] [PubMed] [Google Scholar]
- Soll J, Schultz G. 2-Methyl-6-phytylquinol and 2, 3-dimethyl-5-phytylquinol as precursors of tocopherol synthesis in spinach chloroplasts. Phytochemistry. 1980;19:215–218. doi: 10.1016/S0031-9422(00)81963-9. [DOI] [Google Scholar]
- Soll J, Schultz G, Joyard J, Douce R, Block MA. Localization and synthesis of prenylquinones in isolated outer and inner envelope membranes from spinach chloroplasts. Arch Biochem Biophys. 1985;238:290–299. doi: 10.1016/0003-9861(85)90167-5. [DOI] [PubMed] [Google Scholar]
- Song QJ, Marek LF, Shoemaker RC, Lark KG, Concibido VC, Delannay X, Specht JE, Cregan PB. A new integrated genetic linkage map of the soybean. Theor Appl Genet. 2004;109:122–128. doi: 10.1007/s00122-004-1602-3. [DOI] [PubMed] [Google Scholar]
- Stuber CW, Lincoln SE, Wolff DW, Helentjaris T, Lander ES. Identification of genetic factors contributing to heterosis in a hybrid from two elite maize inbred lines using molecular markers. Genetics. 1992;132:832–839. doi: 10.1093/genetics/132.3.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner JW, Luzzi BM, Gostovic P, Montminy W, Hume DJ. OAC Bayfield soybean. Can J Plant Sci. 1998;78:625–626. [Google Scholar]
- Tavva VS, Kim YH, Kagan IA, Dinkins RD, Kim KH, Collins GB. Increased α-tocopherol content in soybean seed overexpressing the Perilla frutescens γ-tocopherol methyltransferase gene. Plant Cell Rep. 2007;26:61–70. doi: 10.1007/s00299-006-0218-2. [DOI] [PubMed] [Google Scholar]
- Trigizano RN, Caetano-Anolles G. Laboratory exercises on DNA amplification fingerprinting for evaluating the molecular diversity of horticultural species. Hortic Technol. 1998;8:413–423. [Google Scholar]
- Ujiie A, Yamada T, Fujimoto K, Endo Y, Kitamura K. Identification of soybean varieties with high a-tocopherol content. Breed Sci. 2005;55:123–125. doi: 10.1270/jsbbs.55.123. [DOI] [Google Scholar]
- Van Eenennaam AL, Kim Lincoln, Durrett TP, Valentin HE, Shewmaker CK, Thorne GM, Jiang J, Baszis SR, Levering CK, Aasen ED, Hao M, Stein JC, Norris SR, Last RL. Engineering vitamin E content: from Arabidopsis mutant to soy oil. Plant Cell. 2003;15:3007–3019. doi: 10.1105/tpc.015875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W. GGE biplot-a windows application for grahical analysis of multienvironment trial data and other types of two way data. Agron J. 2001;93:1111–1117. [Google Scholar]
- Zeng GL, Li DM, Han YP, Teng WL, Wang JA, Qiu LJ, Li WB. Identification of QTL underlying isoflavone contents in soybean seeds among multiple environments. Theor Appl Genet. 2009;118:1455–1463. doi: 10.1007/s00122-009-0994-5. [DOI] [PubMed] [Google Scholar]
- Zou J, Lee J, Singh R, Xu SS, Cregan PB, Hymowitz T. Assignment of molecular linkage groups to the soybean chromosomes by primary trisomics. Theor Appl Genet. 2003;107:745–750. doi: 10.1007/s00122-003-1304-2. [DOI] [PubMed] [Google Scholar]