Abstract
In order to obtain insights into the methicillin-resistant Staphylococcus aureus (MRSA) population structure in the Azores archipelago, 106 MRSA isolates were collected from patients attending an Azorean central hospital between January 2007 and February 2008. Antimicrobial resistance was determined for all isolates. Molecular typing was performed by pulsed-field gel electrophoresis (PFGE), spa typing, multilocus sequence typing (MLST), staphylococcal chromosome cassette mec (SCCmec) typing and the presence of Panton–Valentine leukocidin (PVL). The majority of the isolates (87%, n = 92) belonged to the EMRSA-15 clone (ST22, SCCmec-IVh), followed by the Pediatric clone (ST5-VI/IVc) (11%, n = 12). The Berlin clone (ST45-IVa) and a new clone (spa type t1839, ST1339 and SCCmec V variant) were represented by single isolates. All of the isolates carried SCCmec types IV, V or VI and a non-multiresistant antibiotic profile, resembling the currently emerging community MRSA. Moreover, PVL was described for the first time to be associated with the Pediatric clone carrying SCCmec type VI. We provided the first description of the population structure of MRSA in the Azores islands, which seems to be shaped by genetic events occurring locally, as well as by the regular population exchange between the islands, continental Portugal, the United Kingdom and the United States.
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) is a major pathogen associated with both nosocomial- and community-acquired infections (hospital-acquired [HA] MRSA and CA-MRSA, respectively) [1]. MRSA are believed to have emerged by the acquisition of the staphylococcal cassette chromosome mec (SCCmec), the mobile genetic element carrying the determinant of methicillin resistance (mecA) into methicillin-susceptible S. aureus (MSSA). Eight different SCCmec types (I to VIII) have been described so far in S. aureus, differing in their structure and size. Epidemiological studies using molecular typing methods showed that the massive geographical spread of MRSA in hospitals results from the dissemination of a few highly epidemic clones. These major epidemic clones resulted from a limited number of independent acquisitions of SCCmec into well-defined genetic backgrounds: (i) New York/Japan clone, sequence type (ST) 5, SCCmec II; (ii) Pediatric clone, ST5-IV/VI; (iii) Brazilian clone, ST239-III/IIIA; (iv) Iberian clone, ST247-IA; (v) EMRSA-15, ST22-IV; (vi) EMRSA-16, ST36-II; and (vii) Berlin clone, ST45-IV [2–4].
The MRSA prevalence in European hospitals varies from less than 3.0% in the Nordic Countries and The Netherlands to over 50% in southern European countries and the United Kingdom. Despite the significant decrease in MRSA incidence in a few countries (France, Slovenia, Belgium, Cyprus and Turkey) due to improved infection control, the incidence has increased in other countries, including Portugal, where the MRSA proportion is, nowadays, over 50%, being one of the highest in Europe [1, 5].
Surveillance studies have been conducted in Portugal since the early 1990s, showing temporal waves of MRSA clonal prevalence. In the most recent survey conducted in 2006, EMRSA-15 (ST22-IVh) was found to be the most prevalent clone in Portuguese hospitals, followed by the New York/Japan clone (ST5-II) [5, 6].
Information about the epidemiology of CA-MRSA is still scarce and non-existent in Portugal. Unlike HA-MRSA, CA-MRSA are usually only resistant to beta-lactams and one or two additional antimicrobial classes, and carry mainly SCCmec types IV and V, the smallest in size, which are believed to be more mobile. CA-MRSA frequently harbour Panton–Valentine leukocidin (PVL), but the toxin has also been detected in the hospital environment [7, 8]. Similarly to HA-MRSA, CA-MRSA epidemic clones have been described [9]. However, a much larger genetic diversity in the genetic backgrounds of CA-MRSA has been observed, suggesting a high number of SCCmec acquisitions in the community. Recent data indicates that MRSA epidemiology is suffering a new change and that the boundaries between the hospital and the community are blurring. As a result, clones traditionally linked to the hospital, like the EMRSA-15 clone, were found to be aetiologic agents of infection in the community [10] and clones specific to the community were observed as causing infections in hospitals, as is the case of the USA300 strain [11, 12]. A common trait to these clones that can survive in both environments appears to be the presence of SCCmec type IV that is, nowadays, prevalent among the current overall MRSA population [5, 6, 13–15].
Whereas the epidemiology of nosocomial MRSA has been very well characterised in the Portuguese continental territory, there is no information concerning the prevalence and clonality of MRSA in the Portuguese islands, namely in the Azores archipelago.
The aim of the present study was to identify the MRSA clonal types currently circulating in the Azores archipelago and compare them with the major MRSA clones described in continental Portugal, Europe and the United States.
Materials and methods
Hospital setting
Hospital do Divino Espírito Santo (HDES) is a 390-bed central hospital located in Ponta Delgada, São Miguel island, that services an outpatient population of 138,000 inhabitants, from the two most populous Azorean islands, S. Miguel and Santa Maria. The hospital, the largest in the archipelago, includes medicine and surgery as principal wards, and a large outpatient ambulatory unit.
Bacterial isolates
Between January 2007 and February 2008, a total of 106 MRSA isolates were collected from both inpatients (n = 93, 87.7% of the isolates) and outpatients (n = 13, 12.3% of the isolates) attending HDES. The collection included single-patient isolates recovered from several sources, including sputum (n = 35, 33%), urine (n = 22, 21%), swabs from various origins (n = 19, 18%), exudates from wounds, one abscess and one ear infection (n = 16, 15%), blood (n = 10, 9.5%), catheters (n = 2, 1.9%) and auricular and peritoneal fluids (n = 1, 0.9% each). Most of the isolates (95%, n = 101) were from infection. The majority of the patients (73.5%) were inpatients hospitalised in three wards: medicine (n = 42, 39.6%), surgery (n = 24, 22.6%) and pneumology (n = 12, 11.3%). Among the 13 outpatients, three attended the haemodialysis unit.
Antimicrobial susceptibility testing and mecA detection
Susceptibility testing was performed with the semi-automatic Vitek system (bioMérieux, SA, France) according to the manufacturer’s instructions for a panel of 12 antibiotics: ciprofloxacin, clindamycin, erythromycin, gentamicin, linezolid, oxacillin, penicillin, rifampicin, trimethoprim-sulfamethoxazole, teicoplanin, tetracycline and vancomycin. Bacterial isolates were considered to belong to different antibiotypes if at least one difference was observed in the antibiotic resistance profile.
The presence of the mecA gene was confirmed in all oxacillin-resistant isolates by polymerase chain reaction (PCR) [16].
PFGE
Pulsed-field gel electrophoresis (PFGE) was performed as described by Chung et al. [17] on all 106 isolates. The resulting SmaI restriction patterns were analysed by both visual inspection using the criteria of McDougal et al. [18] and automatically with the BioNumerics software version 4.61 (Applied Maths, Sint-Martens-Latem, Belgium). Dendrograms were generated as previously described [19] using an optimisation of 0.50% and a tolerance of 1.25%. A similarity coefficient of 80% was used to define the PFGE type clusters [19].
spa typing and MLST
spa typing was carried out on at least one representative of each PFGE subtype (n = 26) and spa types were assigned through the Ridom web server (http://spaserver.ridom.de) [20]. Multilocus sequence typing (MLST) was conducted as described [21] for selected strains previously typed by PFGE and spa typing. The allelic profiles and sequence types (ST) were defined using the MLST database (http://www.mlst.net).
SCCmec and ccr typing
SCCmec was typed by the multiplex PCR strategy described by Milheiriço et al. [22]. Additionally, all type IV isolates were further subtyped as previously described [23]. Type VI isolates were confirmed by amplification of the cassette chromosome recombinase genes ccrAB4 [24]. Type V was confirmed by PCR amplification of ccrC and mec complex C, as previously described [16, 25]. Non-typeable isolates by the methods mentioned above were characterised by ccrB sequencing [26].
PVL detection
All 106 MRSA isolates were screened for the presence of PVL lukS/lukF genes by PCR, as described previously [27]. Positive results were confirmed by sequencing of the PCR amplicon.
Results
Antimicrobial susceptibility
The 106 isolates were assigned to eight different antibiotypes (Table 1) with a prevalent non-multiresistant pattern. The majority of the isolates (n = 91, 85.9%) showed antibiotype 1 (resistance to oxacillin, penicillin, ciprofloxacin and erythromycin) or 2 (resistance to oxacillin, penicillin and ciprofloxacin). The isolate with antibiotype 7 was unique in showing resistance to gentamicin. Antibiotype 8, which showed resistance to six different antimicrobials, included a single isolate.
Table 1.
Resistance profile of the 106 methicillin-resistant Staphylococcus aureus (MRSA) isolates and associated pulsed-field gel electrophoresis (PFGE) types
| Antibiotype | Resistance profilea | Total no. of isolates (%) | PFGE types (subtypes) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OXA | PEN | CIP | ERY | CLI | RIF | GEN | LZD | SXT | TEC | TET | VAN | |||
| 1 | R | R | R | R | S | S | S | S | S | S | S | S | 67 (63.2) | A (1–9); B (1, 2); D (1) |
| 2 | R | R | R | S | S | S | S | S | S | S | S | S | 24 (22.7) | A (1–6); B (1, 4); C (1) |
| 3 | R | R | S | R | S | S | S | S | S | S | S | S | 7 (6.6) | A (1, 2, 5); B (1) |
| 4 | R | R | R | R | R | S | S | S | S | S | S | S | 3 (2.8) | A (1) |
| 5 | R | R | S | S | S | S | S | S | S | S | S | S | 2 (1.9) | A (4); B (3) |
| 6 | R | R | R | S | S | R | S | S | S | S | S | S | 1 (0.9) | A (1) |
| 7 | R | R | S | S | I | R | R | S | S | S | S | S | 1 (0.9) | B (1) |
| 8 | R | R | R | R | R | R | S | S | S | S | S | S | 1 (0.9) | B (1) |
a S, susceptible; I, intermediate; R, resistant
Antibiotic abbreviations: OXA, oxacillin; PEN, penicillin; CIP, ciprofloxacin; ERY, erythromycin; CLI, clindamycin; RIF, rifampicin; GEN, gentamicin; LZD, linezolid; SXT, trimethoprim-sulfamethoxazole; TEC, teicoplanin; TET, tetracycline; VAN, vancomycin
All 106 isolates harboured the mecA gene and, therefore, were classified as MRSA.
Predominance of EMRSA-15 clone
The characterisation of the 106 MRSA strains by PFGE, spa typing, MLST and SCCmec typing clustered the isolates into four clonal types (Fig. 1). The majority of the isolates (n = 92, 87%) belonged to PFGE type A. PFGE type B included 11% (n = 12) of the isolates and types C and D were represented by single isolates only (1% each).
Fig. 1.
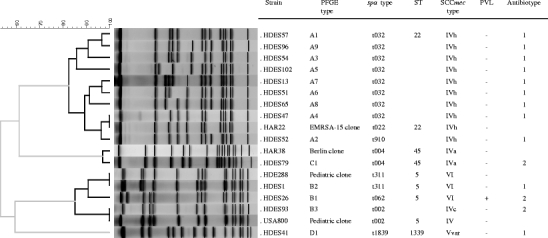
Molecular characterisation of representative methicillin-resistant Staphylococcus aureus (MRSA) isolates of each pulsed-field gel electrophoresis (PFGE) subtype and comparison with MRSA pandemic clones
PFGE type A was subdivided into nine subtypes; subtypes A1 (n = 40) and A2 (n = 22) represented 67% of all type A isolates. The representative isolate of PFGE type A (subtype A1) showed spa type t032, ST22 and SCCmec type IVh, which are characteristics of the internationally disseminated EMRSA-15 clone [28]. Interestingly, subtype A4 showed the same restriction profile as the EMRSA-15 clone reference strain, HAR22, but a different spa type (t032), including one additional repeat sequence (Fig. 1). All representative isolates of type A shared the same spa type t032, except one isolate (t910), which differs from t032 by the deletion of five repeats. HDES85 was the unique isolate of PFGE type A1 defined as non-typeable by spa typing, since no PCR product was obtained. Another single type A1 isolate, HDES73, showed a SCCmec cassette type IV non-subtypeable. PFGE type A included isolates from six out of the eight antibiotypes described, but 95% (n = 53) of the isolates belonged to antibiotypes 1 and 2 (Table 1).
Evidence of horizontal gene transfer among strains of the Pediatric clone
PFGE type B was the second most common PFGE type and included three subtypes, of which B1 represented 75% (n = 9) of type B isolates (Fig. 1). Subtypes B1 and B2 showed ST5, SCCmec VI and spa types t062, t2049, t2724 (B1)/t311 (B2), which are typical of the Pediatric clone. SCCmec type VI was confirmed in five isolates by amplification of the characteristic ccrAB4 genes. The remaining type VI isolates did not amplify the ccrAB4 genes by uniplex PCR but showed a ccrB sequence with 100% homology with the Pediatric clone reference strain HDE288 [29]. Interestingly, subtype B3 carried SCCmec type IVc instead of the SCCmec type VI, which may indicate the occurrence of independent acquisition of SCCmec in the Pediatric clone background.
Moreover, we found that three isolates belonging to PFGE subtype B1 (ST5-VI) carried the PVL genes.
The remaining two isolates belonged to PFGE types C and D and were, respectively, recovered from an outpatient and an inpatient. PFGE C isolate, spa type t004 and ST45-IVa showed a single band difference relative to HAR38, the prototype isolate of the Berlin clone (spa type t004, ST45-IVa) [30, 31]. PFGE type D isolate showed spa type t1839, a novel sequence type (ST1339) not related to any major clonal complex (CC) and a variant of SCCmec type V.
High genetic diversity and high frequency of SCCmec types IV, V and VI
SCCmec type IV was found to be the prevalent type (n = 13) among the 24 isolates tested. Three different SCCmec IV subtypes were found in this collection, associated to three unrelated genetic backgrounds (ST5: SCCmec IVc, ST22: SCCmec IVh/IVnon-subtypeable, and ST45: SCCmec IVa). SCCmec type VI was the second most prevalent cassette type, which appeared to be associated to isolates with PFGE types B1 and B2. A variant of SCCmec type V was shown on a single isolate that harboured the mec complex C and ccrC genes, but missed the specific type V J1 region. No isolates carrying SCCmec types I, II or III were identified in this collection.
Discussion
In order to describe the MRSA population structure in the Atlantic Azores islands, a collection of 106 isolates recovered during a 13-month period was characterised by a combination of up-to-date molecular typing methods. The analysis showed the predominance of two major international MRSA clones: the EMRSA-15 clone and the Pediatric clone.
Massive dominance of the EMRSA-15 clone
In the present study, 87% of the isolates showed molecular characteristics of the highly internationally disseminated EMRSA-15 clone (ST22-IVh) [28], including a common spa type t032. In addition, a single PFGE type A2 isolate showed t910, a spa type already reported in a CC22 isolate from Germany [32]. This fact is not surprising, since the spa gene is a variable repeat region prone to the introduction or deletion of repeats. Regional spa clusters frequently include highly related spa types characterising clonal related isolates, which translates a local evolution of the spa locus [8, 33]. Unusually, a non-typeable isolate was found in the collection, which should be due to a mutation or insertion/deletion in spa rather than non-existence of the gene [34]. Interestingly, the majority of the EMRSA-15 isolates described in this study were recovered from sources other than blood (sputum, swabs, exudates and urine), corresponding to the major pathologies presented by the patients attending HDES (respiratory tract and skin related). The non-multiresistant EMRSA-15 clone emerged in the United Kingdom in 1991 and rapidly became a dominant clone worldwide, particularly in Europe [8, 35]. Recently, an EMRSA-15 variant was reported in central United States [36]. In the Azores islands, the clear dominance of the EMRSA-15 clone is, in part, in agreement with the situation in continental Portugal [5, 6]. In fact, in a study involving isolates recovered in the same period from 11 hospitals scattered over continental Portugal, EMRSA-15 was found to be the most prevalent clone. However, and in contrast with the present study, the New York/Japan clone (ST5-II) was shown to represent a new wave of MRSA in the country, whereas the Pediatric clone (ST5-VI/IV) was exceedingly rare [5]. In the Portuguese neighbouring country Spain, EMRSA-15 was found in continental territory in co-existence with EMRSA-16 and two CC5 clones (ST125-IV and ST146-IV), but not as a major clone as in the Spanish islands (Majorca and Canarias) and as in our study [14, 37, 38]. The frequent touristic flux from the United Kingdom, hypothesised as the cause of the importation of EMRSA-15 to the Mediterranean island of Malta [39], could also support, in part, the emergence of this clone in the most touristic Azores islands. In parallel, the regular exchange of inhabitants between the islands and the continent seem to be an important vehicle of clonal importation.
Emergence of PVL and SCCmec variability: evidence for local evolution
Recent studies revealed that the Pediatric MRSA clone, isolated for the first time in 1992 in a Portuguese hospital [29], was no longer present in that hospital in 2006 [5]. However, it appeared in the present collection as the second major clone.
Interestingly, in this study, we observed that strains of the Pediatric clone differing in three to five bands in PFGE pattern, isolated in the same hospital, carried either the typical SCCmec types VI or IVc. ST5-VI MRSA isolates, which had been restricted to isolates from continental Portugal, were recently found in a collection of nosocomial isolates recovered from 23 hospitals located throughout France [24, 40]. The hypothesis of a recent introduction of this clone from continental Portugal seems to be quite remote, since, in a recent national study, this clone was isolated from a single patient only in a hospital in the South of Portugal [5]. On the other hand, the Azores islands have a high emigrant community in the United States, who frequently travel to the national territory, constituting also possible vehicles of dissemination of the Pediatric clone strains already reported in this country.
ST5 lineage is known to frequently acquire mobile genetic elements, such as different SCCmec, antibiotic resistance and toxin genes [4, 41]. In a recent study, Nübel et al. showed that at least 23 SCCmec acquisitions events have occurred within ST5 lineage, on multiple and independent occasions, showing evidence that the MRSA emerged in numerous circumstances and in distinct locations rather than by the geographic dispersal of isolates [4]. Moreover, besides being originally identified in a community-associated USA400 strain, the first introduction of SCCmecIV in S. aureus might have occurred in the ST5 Pediatric clone background that was circulating in hospitals in the 1990s [9]. Similarly, our results suggest that SCCmec cassettes VI and IVc might have been introduced de novo and recently in these ST5 strains and that the Pediatric clone may be evolving locally. Another observation that supports the hypothesis of recent local evolution of the Pediatric clone is the presence of PVL-positive and PVL-negative strains with closely related PFGE profiles. Noteworthy, this is the first report of PVL among isolates belonging to the SCCmec type VI Pediatric clone. The introduction of the PVL genes in a genetic background with such a high epidemic potential is of particular concern. However, we cannot exclude the reverse hypothesis of a local evolution through the acquisition of SCCmec type VI by a PVL-positive MSSA. Only the analysis of the genetic background of susceptible S. aureus in the Azores islands would clarify the chronological acquisition of PVL and SCCmec in these Pediatric clone strains.
The Berlin MRSA clone (ST45-IVa), first observed in Berlin hospitals in 1993, is currently spread in some European countries [8, 42], showing a high epidemic potential not only in the hospital environment but also in the community [43, 44]. This clone was found in a single MRSA isolate in the Azores island collection. The same genetic background (ST45) had been previously described in Portuguese hospitals, but exclusively among MSSA isolates [45]. The collection of a single MRSA isolate carrying SCCmec IVa suggests another de novo SCCmec acquisition, probably in an already established MSSA clone.
Likewise, the introduction of a variant of SCCmec type V seems to have occurred in an isolate with spa type t1839 and a novel sequence type (ST1339) that is a singleton. This strain is not related to the single SCCmec type V variant isolate (ST45) previously reported in Portugal [5]. Interestingly, this isolate was recovered from a patient within the first 48 h of hospitalisation, from a skin and soft tissue infection (erysipelas), the main source of CA-MRSA isolates. However, the isolate could not be considered as having a community origin, since no additional information regarding patient risk factors was available.
Emergence of HA-MRSA isolates with community traits
SCCmec type IV is one of the most frequent SCCmec types found in the community and is, nowadays, becoming the predominant type also in the nosocomial setting [5, 6, 9, 13–15, 46]. Moreover, this SCCmec type has been usually associated to a pattern of susceptibility to almost all antimicrobial classes. These same characteristics were observed in isolates from our study: SCCmec type IV was found to be the most frequent SCCmec type and all isolates were highly susceptible. At least three different SCCmec subtypes were identified (IVh: EMRSA-15, IVc: Pediatric clone, IVa: Berlin clone), showing evidence of a high genetic diversity among the pool of SCCmec type IV in the Azores islands. Interestingly, the detection of one isolate belonging to the EMRSA-15 clone, SCCmec type IV non-subtypeable (only positive for the ccrB2 allele in the subtyping multiplex), suggests the presence of a new uncharacterised SCCmec type IV subtype in addition to the ones already described [23].
Besides SCCmec type IV, the only SCCmec types found among the whole collection were the small SCCmec V and VI, reflecting the actual tendency for the dominance of low resistant and fitness cost clones within the hospital setting, which is parallel with what is observed in the community environment [9]. The introduction of well-established community isolates into the hospital setting is not new, as observed in Taiwan, where the ST59 clonal type, highly prevalent in the community, emerged in the hospital setting as the second most prevalent clone [38, 47, 48]. Although ST22-IV EMRSA-15-related isolates as well as ST5-IV were already reported in the community [10, 46], no conclusions can be drawn for the flow direction in the present study. Even if a high percentage (30%) of the isolates studied was recovered within the first 48 h of admission or in an outpatient unit, no data of previous hospital contact was available for the patients.
In the present study, we provided the first description of the population structure of MRSA in the Azores islands and showed a prevalence of the epidemic EMRSA-15 clone in co-existence with the Pediatric clone. Our findings were in line with recent reports from continental Portuguese and European hospitals. The horizontal gene transfer and the de novo acquisition of mobile genetic elements seem to have contributed to the local genetic diversity and clonal evolution. Moreover, the population exchange between the islands and continental Portugal or the United States, in addition to the high flux of tourists, particularly from the United Kingdom, may explain the dominance and spread of the EMRSA-15 and Pediatric clones. Further surveillance studies concerning both HA-MRSA and MSSA populations and CA-MRSA in the geographically restricted Azores islands will be fundamental to support the S. aureus localised evolution, as well as to understand the links between the hospital and the community.
Acknowledgements
This work was supported by project TROCAR (contract HEALTH-F3-2008-223031) and CONCORD (contract HEALTHF3-2008-222718) from the European Commission. T. Conceição and A. Tavares were supported by grants SFRH/BD/21424/2005 and SFRH/BD/44220/2008, respectively, from Fundação para a Ciência e a Tecnologia, Lisbon, Portugal.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.European Antimicrobial Resistance Surveillance System (2009) EARSS annual report 2008. EARSS, Bilthoven, The Netherlands. Available online at: http://www.rivm.nl/earss/Images/EARSS%202008_final_tcm61-65020.pdf, pp 55–58
- 2.Oliveira DC, Tomasz A, de Lencastre H. Secrets of success of a human pathogen: molecular evolution of pandemic clones of meticillin-resistant Staphylococcus aureus. Lancet Infect Dis. 2002;2(3):180–189. doi: 10.1016/S1473-3099(02)00227-X. [DOI] [PubMed] [Google Scholar]
- 3.Enright MC. The evolution of a resistant pathogen—the case of MRSA. Curr Opin Pharmacol. 2003;3(5):474–479. doi: 10.1016/S1471-4892(03)00109-7. [DOI] [PubMed] [Google Scholar]
- 4.Nübel U, Roumagnac P, Feldkamp M, Song JH, Ko KS, Huang YC, Coombs G, Ip M, Westh H, Skov R, Struelens MJ, Goering RV, Strommenger B, Weller A, Witte W, Achtman M. Frequent emergence and limited geographic dispersal of methicillin-resistant Staphylococcus aureus. Proc Natl Acad Sci U S A. 2008;105(37):14130–14135. doi: 10.1073/pnas.0804178105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aires-de-Sousa M, Correia B, de Lencastre H. Changing patterns in frequency of recovery of five methicillin-resistant Staphylococcus aureus clones in Portuguese hospitals: surveillance over a 16-year period. J Clin Microbiol. 2008;46(9):2912–2917. doi: 10.1128/JCM.00692-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amorim ML, Faria NA, Oliveira DC, Vasconcelos C, Cabeda JC, Mendes AC, Calado E, Castro AP, Ramos MH, Amorim JM, de Lencastre H. Changes in the clonal nature and antibiotic resistance profiles of methicillin-resistant Staphylococcus aureus isolates associated with spread of the EMRSA-15 clone in a tertiary care Portuguese hospital. J Clin Microbiol. 2007;45(9):2881–2888. doi: 10.1128/JCM.00603-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tristan A, Ferry T, Durand G, Dauwalder O, Bes M, Lina G, Vandenesch F, Etienne J. Virulence determinants in community and hospital meticillin-resistant Staphylococcus aureus. J Hosp Infect. 2007;65(Suppl 2):105–109. doi: 10.1016/S0195-6701(07)60025-5. [DOI] [PubMed] [Google Scholar]
- 8.Grundmann H, Aanensen DM, van den Wijngaard CC, Spratt BG, Harmsen D, Friedrich AW. Geographic distribution of Staphylococcus aureus causing invasive infections in Europe: a molecular-epidemiological analysis. PLoS Med. 2010;7(1):e1000215. doi: 10.1371/journal.pmed.1000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chambers HF, Deleo FR. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol. 2009;7(9):629–641. doi: 10.1038/nrmicro2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O’Brien FG, Lim TT, Chong FN, Coombs GW, Enright MC, Robinson DA, Monk A, Saïd-Salim B, Kreiswirth BN, Grubb WB. Diversity among community isolates of methicillin-resistant Staphylococcus aureus in Australia. J Clin Microbiol. 2004;42(7):3185–3190. doi: 10.1128/JCM.42.7.3185-3190.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saiman L, O’Keefe M, Graham PL, 3rd, Wu F, Saïd-Salim B, Kreiswirth B, LaSala A, Schlievert PM, Della-Latta P. Hospital transmission of community-acquired methicillin-resistant Staphylococcus aureus among postpartum women. Clin Infect Dis. 2003;37(10):1313–1319. doi: 10.1086/379022. [DOI] [PubMed] [Google Scholar]
- 12.Seybold U, Kourbatova EV, Johnson JG, Halvosa SJ, Wang YF, King MD, Ray SM, Blumberg HM. Emergence of community-associated methicillin-resistant Staphylococcus aureus USA300 genotype as a major cause of health care-associated blood stream infections. Clin Infect Dis. 2006;42(5):647–656. doi: 10.1086/499815. [DOI] [PubMed] [Google Scholar]
- 13.International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (IWG-SCC) Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob Agents Chemother. 2009;53(12):4961–4967. doi: 10.1128/AAC.00579-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alcoceba E, Mena A, Cruz Pérez M, Ruiz de Gopegui E, Padilla E, Gil J, Ramírez A, Gallegos C, Serra A, Pérez JL, Oliver A. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in Majorcan hospitals: high prevalence of the epidemic clone EMRSA-15. Clin Microbiol Infect. 2007;13(6):599–605. doi: 10.1111/j.1469-0691.2007.01703.x. [DOI] [PubMed] [Google Scholar]
- 15.Vindel A, Cuevas O, Cercenado E, Marcos C, Bautista V, Castellares C, Trincado P, Boquete T, Pérez-Vázquez M, Marín M, Bouza E. Methicillin-resistant Staphylococcus aureus in Spain: molecular epidemiology and utility of different typing methods. J Clin Microbiol. 2009;47(6):1620–1627. doi: 10.1128/JCM.01579-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okuma K, Iwakawa K, Turnidge JD, Grubb WB, Bell JM, O’Brien FG, Coombs GW, Pearman JW, Tenover FC, Kapi M, Tiensasitorn C, Ito T, Hiramatsu K. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J Clin Microbiol. 2002;40(11):4289–4294. doi: 10.1128/JCM.40.11.4289-4294.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chung M, de Lencastre H, Matthews P, Tomasz A, Adamsson I, Aires de Sousa M, Camou T, Cocuzza C, Corso A, Couto I, Dominguez A, Gniadkowski M, Goering R, Gomes A, Kikuchi K, Marchese A, Mato R, Melter O, Oliveira D, Palacio R, Sá-Leão R, Santos Sanches I, Song JH, Tassios PT, Villari P. Molecular typing of methicillin-resistant Staphylococcus aureus by pulsed-field gel electrophoresis: comparison of results obtained in a multilaboratory effort using identical protocols and MRSA strains. Microb Drug Resist. 2000;6(3):189–198. doi: 10.1089/mdr.2000.6.189. [DOI] [PubMed] [Google Scholar]
- 18.McDougal LK, Steward CD, Killgore GE, Chaitram JM, McAllister SK, Tenover FC. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J Clin Microbiol. 2003;41(11):5113–5120. doi: 10.1128/JCM.41.11.5113-5120.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Faria NA, Carrico JA, Oliveira DC, Ramírez M, de Lencastre H. Analysis of typing methods for epidemiological surveillance of both methicillin-resistant and methicillin-susceptible Staphylococcus aureus strains. J Clin Microbiol. 2008;46(1):136–144. doi: 10.1128/JCM.01684-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aires-de-Sousa M, Boye K, de Lencastre H, Deplano A, Enright MC, Etienne J, Friedrich A, Harmsen D, Holmes A, Huijsdens XW, Kearns AM, Mellmann A, Meugnier H, Rasheed JK, Spalburg E, Strommenger B, Struelens MJ, Tenover FC, Thomas J, Vogel U, Westh H, Xu J, Witte W. High interlaboratory reproducibility of DNA sequence-based typing of bacteria in a multicenter study. J Clin Microbiol. 2006;44(2):619–621. doi: 10.1128/JCM.44.2.619-621.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crisóstomo MI, Westh H, Tomasz A, Chung M, Oliveira DC, de Lencastre H. The evolution of methicillin resistance in Staphylococcus aureus: similarity of genetic backgrounds in historically early methicillin-susceptible and -resistant isolates and contemporary epidemic clones. Proc Natl Acad Sci U S A. 2001;98(17):9865–9870. doi: 10.1073/pnas.161272898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Milheiriço C, Oliveira DC, de Lencastre H. Update to the multiplex PCR strategy for assignment of mec element types in Staphylococcus aureus. Antimicrob Agents Chemother. 2007;51(9):3374–3377. doi: 10.1128/AAC.00275-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Milheiriço C, Oliveira DC, de Lencastre H. Multiplex PCR strategy for subtyping the staphylococcal cassette chromosome mec type IV in methicillin-resistant Staphylococcus aureus: ‘SCCmec IV multiplex’. J Antimicrob Chemother. 2007;60(1):42–48. doi: 10.1093/jac/dkm112. [DOI] [PubMed] [Google Scholar]
- 24.Oliveira DC, Milheiriço C, de Lencastre H. Redefining a structural variant of staphylococcal cassette chromosome mec, SCCmec type VI. Antimicrob Agents Chemother. 2006;50(10):3457–3459. doi: 10.1128/AAC.00629-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ito T, Ma XX, Takeuchi F, Okuma K, Yuzawa H, Hiramatsu K. Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC. Antimicrob Agents Chemother. 2004;48(7):2637–2651. doi: 10.1128/AAC.48.7.2637-2651.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oliveira DC, Milheiriço C, Vinga S, de Lencastre H. Assessment of allelic variation in the ccrAB locus in methicillin-resistant Staphylococcus aureus clones. J Antimicrob Chemother. 2006;58(1):23–30. doi: 10.1093/jac/dkl208. [DOI] [PubMed] [Google Scholar]
- 27.Lina G, Piémont Y, Godail-Gamot F, Bes M, Peter MO, Gauduchon V, Vandenesch F, Etienne J. Involvement of Panton–Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis. 1999;29(5):1128–1132. doi: 10.1086/313461. [DOI] [PubMed] [Google Scholar]
- 28.Moore PC, Lindsay JA. Molecular characterisation of the dominant UK methicillin-resistant Staphylococcus aureus strains, EMRSA-15 and EMRSA-16. J Med Microbiol. 2002;51(6):516–521. doi: 10.1099/0022-1317-51-6-516. [DOI] [PubMed] [Google Scholar]
- 29.Sá-Leão R, Santos Sanches I, Dias D, Peres I, Barros RM, de Lencastre H. Detection of an archaic clone of Staphylococcus aureus with low-level resistance to methicillin in a pediatric hospital in Portugal and in international samples: relics of a formerly widely disseminated strain? J Clin Microbiol. 1999;37(6):1913–1920. doi: 10.1128/jcm.37.6.1913-1920.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murchan S, Kaufmann ME, Deplano A, de Ryck R, Struelens M, Zinn CE, Fussing V, Salmenlinna S, Vuopio-Varkila J, El Solh N, Cuny C, Witte W, Tassios PT, Legakis N, van Leeuwen W, van Belkum A, Vindel A, Laconcha I, Garaizar J, Haeggman S, Olsson-Liljequist B, Ransjo U, Coombes G, Cookson B. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J Clin Microbiol. 2003;41(4):1574–1585. doi: 10.1128/JCM.41.4.1574-1585.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cookson BD, Robinson DA, Monk AB, Murchan S, Deplano A, de Ryck R, Struelens MJ, Scheel C, Fussing V, Salmenlinna S, Vuopio-Varkila J, Cuny C, Witte W, Tassios PT, Legakis NJ, van Leeuwen W, van Belkum A, Vindel A, Garaizar J, Haeggman S, Olsson-Liljequist B, Ransjo U, Muller-Premru M, Hryniewicz W, Rossney A, O’Connell B, Short BD, Thomas J, O’Hanlon S, Enright MC. Evaluation of molecular typing methods in characterizing a European collection of epidemic methicillin-resistant Staphylococcus aureus strains: the HARMONY collection. J Clin Microbiol. 2007;45(6):1830–1837. doi: 10.1128/JCM.02402-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strommenger B, Braulke C, Heuck D, Schmidt C, Pasemann B, Nübel U, Witte W. spa typing of Staphylococcus aureus as a frontline tool in epidemiological typing. J Clin Microbiol. 2008;46(2):574–581. doi: 10.1128/JCM.01599-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harmsen D, Claus H, Witte W, Rothgänger J, Claus H, Turnwald D, Vogel U. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol. 2003;41(12):5442–5448. doi: 10.1128/JCM.41.12.5442-5448.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baum C, Haslinger-Löffler B, Westh H, Boye K, Peters G, Neumann C, Kahl BC. Non-spa-typeable clinical Staphylococcus aureus strains are naturally occurring protein A mutants. J Clin Microbiol. 2009;47(11):3624–3629. doi: 10.1128/JCM.00941-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson AP, Pearson A, Duckworth G. Surveillance and epidemiology of MRSA bacteraemia in the UK. J Antimicrob Chemother. 2005;56(3):455–462. doi: 10.1093/jac/dki266. [DOI] [PubMed] [Google Scholar]
- 36.Wolter DJ, Chatterjee A, Varman M, Goering RV. Isolation and characterization of an epidemic methicillin-resistant Staphylococcus aureus 15 variant in the central United States. J Clin Microbiol. 2008;46(10):3548–3549. doi: 10.1128/JCM.00985-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vindel A, Trincado P, Gómez E, Cabrera R, Boquete T, Solá C, Valdezate S, Saez-Nieto JA. Prevalence and evolution of methicillin-resistant Staphylococcus aureus in Spanish hospitals between 1996 and 2002. J Clin Microbiol. 2006;44(1):266–270. doi: 10.1128/JCM.44.1.266-270.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pérez-Roth E, Lorenzo-Díaz F, Batista N, Moreno A, Méndez-Alvarez S. Tracking methicillin-resistant Staphylococcus aureus clones during a 5-year period (1998 to 2002) in a Spanish hospital. J Clin Microbiol. 2004;42(10):4649–4656. doi: 10.1128/JCM.42.10.4649-4656.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scicluna EA, Shore AC, Thürmer A, Ehricht R, Slickers P, Borg MA, Coleman DC, Monecke S. Characterisation of MRSA from Malta and the description of a Maltese epidemic MRSA strain. Eur J Clin Microbiol Infect Dis. 2010;29:163–170. doi: 10.1007/s10096-009-0834-1. [DOI] [PubMed] [Google Scholar]
- 40.Dauwalder O, Lina G, Durand G, Bes M, Meugnier H, Jarlier V, Coignard B, Vandenesch F, Etienne J, Laurent F. Epidemiology of invasive methicillin-resistant Staphylococcus aureus clones collected in France in 2006 and 2007. J Clin Microbiol. 2008;46(10):3454–3458. doi: 10.1128/JCM.01050-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Enright MC, Robinson DA, Randle G, Feil EJ, Grundmann H, Spratt BG. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA) Proc Natl Acad Sci U S A. 2002;99(11):7687–7692. doi: 10.1073/pnas.122108599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wannet WJ, Spalburg E, Heck ME, Pluister GN, Willems RJ, De Neeling AJ. Widespread dissemination in The Netherlands of the epidemic Berlin methicillin-resistant Staphylococcus aureus clone with low-level resistance to oxacillin. J Clin Microbiol. 2004;42(7):3077–3082. doi: 10.1128/JCM.42.7.3077-3082.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Witte W, Braulke C, Cuny C, Heuck D, Kresken M. Changing pattern of antibiotic resistance in methicillin-resistant Staphylococcus aureus from German hospitals. Infect Control Hosp Epidemiol. 2001;22(11):683–686. doi: 10.1086/501845. [DOI] [PubMed] [Google Scholar]
- 44.Regev-Yochay G, Carmeli Y, Raz M, Pinco E, Etienne J, Leavitt A, Rubinstein E, Navon-Venezia S. Prevalence and genetic relatedness of community-acquired methicillin-resistant Staphylococcus aureus in Israel. Eur J Clin Microbiol Infect Dis. 2006;25(11):719–722. doi: 10.1007/s10096-006-0210-3. [DOI] [PubMed] [Google Scholar]
- 45.Aires de Sousa M, Conceição T, Simas C, de Lencastre H. Comparison of genetic backgrounds of methicillin-resistant and -susceptible Staphylococcus aureus isolates from Portuguese hospitals and the community. J Clin Microbiol. 2005;43(10):5150–5157. doi: 10.1128/JCM.43.10.5150-5157.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Witte W. Community-acquired methicillin-resistant Staphylococcus aureus: what do we need to know? Clin Microbiol Infect. 2009;15(Suppl 7):17–25. doi: 10.1111/j.1469-0691.2009.03097.x. [DOI] [PubMed] [Google Scholar]
- 47.Montesinos I, Castro B, Barroso I, Delgado T, Ramos MJ, Lecuona M, Sierra A. Evolution of the MRSA clones at a university hospital in the Canary islands. Clin Microbiol Infect. 2009;15(Suppl 4):S447. [Google Scholar]
- 48.Huang YC, Su LH, Wu TL, Lin TY. Changing molecular epidemiology of methicillin-resistant Staphylococcus aureus bloodstream isolates from a teaching hospital in Northern Taiwan. J Clin Microbiol. 2006;44(6):2268–2270. doi: 10.1128/JCM.00776-06. [DOI] [PMC free article] [PubMed] [Google Scholar]


