The microRNA miR-137 represses expression of Ezh2, a histone methyltransferase, which in turn alters the epigenetic architecture of chromatin that is important for regulation of miR-137 levels.
Abstract
Both microRNAs (miRNAs) and epigenetic regulation have important functions in stem cell biology, although the interactions between these two pathways are not well understood. Here, we show that MeCP2, a DNA methyl-CpG–binding protein, can epigenetically regulate specific miRNAs in adult neural stem cells (aNSCs). MeCP2-mediated epigenetic regulation of one such miRNA, miR-137, involves coregulation by Sox2, a core transcription factor in stem cells. miR-137 modulates the proliferation and differentiation of aNSCs in vitro and in vivo. Overexpression of miR-137 promotes the proliferation of aNSCs, whereas a reduction of miR-137 enhances aNSC differentiation. We further show that miR-137 post-transcriptionally represses the expression of Ezh2, a histone methyltransferase and Polycomb group (PcG) protein. The miR-137–mediated repression of Ezh2 feeds back to chromatin, resulting in a global decrease in histone H3 trimethyl lysine 27. Coexpression of Ezh2 can rescue phenotypes associated with miR-137 overexpression. These results demonstrate that cross talk between miRNA and epigenetic regulation contributes to the modulation of adult neurogenesis.
Introduction
Neurogenesis in adult mammalian brains occurs throughout life. This process has been observed at two locations under normal conditions: the subventricular zone of the lateral ventricles and the subgranular zone of the dentate gyrus (DG) in the hippocampus (Zhao et al., 2008). The cellular basis for adult neurogenesis is adult neural stem cells (aNSCs), which exhibit the two essential properties of stem cells: self-renewal and multipotency. Adult neurogenesis is defined as the process of generating new neurons from NSCs, which consists of the proliferation and fate determination of aNSCs, the migration and survival of young neurons, and the maturation and integration of newly produced neurons (Ming and Song, 2005; Zhao et al., 2008). Adult neurogenesis is regulated at many levels by both extrinsic factors, such as physiological and pathological conditions, and intrinsic factors, such as genetic and epigenetic programs. The maintenance and differentiation of stem cells is tightly controlled by intricate molecular networks (Li et al., 2008). Uncovering these regulatory mechanisms is crucial to understanding the functions and plasticity of adult brains.
Epigenetic regulation, including DNA methylation and histone modification, is known to play significant roles in the modulation of stem cell proliferation and differentiation, including NSCs (Abel and Zukin, 2008; Zhao et al., 2008). Recent genome-wide analyses have demonstrated a clear role for DNA methylation and chromatin remodeling, particularly by the Polycomb group (PcG) proteins, in defining the properties and regulating the functions of stem cells (Bernstein et al., 2007). The importance of epigenetic regulation in brain development and neurological disorders has been well documented (Shahbazian and Zoghbi, 2002; Abel and Zukin, 2008). For example, de novo mutations in MECP2 give rise to neurodevelopmental disorders, including Rett syndrome (Amir et al., 1999; Chahrour and Zoghbi, 2007). MeCP2 belongs to a family of DNA methyl-CpG–binding proteins (MBDs) that translate DNA methylation into gene expression regulation (Bird, 2002). Two members of the MBD family of proteins, MBD1 and MeCP2, influence either the proliferation and differentiation of aNSCs or the maturation of young neurons (Zhao et al., 2003; Kishi and Macklis, 2004; Smrt et al., 2007). Nonetheless, how these epigenetic factors regulate adult neurogenesis is unclear because of the difficulty in identifying downstream targets via classical gene expression analyses (Bienvenu and Chelly, 2006).
MicroRNAs (miRNAs) are small, noncoding RNAs that regulate gene expression and development by post-transcriptionally targeting RNA-induced silencing complex (RISC) to cognate messenger RNA (Bartel, 2004). The loss of components of the miRNA pathway, including Dicer and DGCR8, can alter the proliferation and differentiation of stem cells (Bernstein et al., 2003; Wang et al., 2007). Furthermore, specific miRNAs are known to play important roles in modulating the proliferation and differentiation of many types of stem cells (Ivey et al., 2008; Yi et al., 2008).
Here, we show that MeCP2 could epigenetically regulate specific miRNAs in mouse aNSCs. The absence of MeCP2 binding to the genomic region proximal to one such miRNA, miR-137, correlates with an altered chromatin state that is reflective of miR-137 expression. In addition, we demonstrate that the MeCP2-mediated effect on miR-137 expression could be performed through a mechanism involving Sox2, a core transcription factor regulating stem cell self-renewal (Zappone et al., 2000; Avilion et al., 2003; Ferri et al., 2004). Furthermore, we found that miR-137 influences aNSC proliferation and differentiation both in vitro and in vivo. Lastly, we identified Ezh2, a histone H3 lysine 27 methyltransferase and a member of the PcG protein family, as one of the post-transcriptionally regulated targets of miR-137; and we found that the miR-137–mediated repression of Ezh2 subsequently caused a global decrease in trimethyl H3-lysine 27 (H3-K27-Tri-Me). Functionally, coexpression of Ezh2 rescued the phenotypes associated with miR-137 overexpression. These results demonstrate that cross talk between epigenetic regulation and the miRNA pathway could play important roles in the modulation of adult neurogenesis. Furthermore, our data suggest that the loss of functional MeCP2 could alter the expression of specific miRNAs, potentially contributing to the molecular pathogenesis of Rett syndrome.
Results
Identification of miRNAs with altered expression in Mecp2-deficient aNSCs
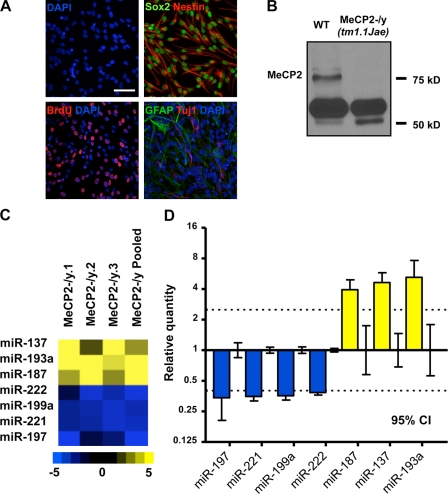
To identify miRNAs potentially regulated at the epigenetic level and determine whether Mecp2 could influence the expression of miRNAs in the context of adult neurogenesis, we profiled the expression of 218 miRNAs in primary NSCs derived from wild-type (WT) and Mecp2-deficient mice (Chen et al., 2001) using multiplex reverse transcription and miRNA-specific TaqMan assays (Fig. 1 and Table S1). Nearly all cultured NSCs were positive for the NSC markers Nestin and Sox2, which suggests a relative homogeneity in these primary aNSCs. These aNSCs incorporate the thymidine analogue BrdU under proliferating conditions and produce both β-III tubulin (TuJ1)-positive neuronal cells and glial fibrillary acidic protein (GFAP)-positive glial cells under differentiating conditions, demonstrating that they possess the same essential properties as NSCs (Fig. 1 A). We identified a subset of miRNAs that consistently displayed altered expression in the absence of Mecp2, relative to WT aNSCs (Fig. 1, C and D; and Table S1). When considering 95% confidence intervals (CIs) on mean relative quantities (RQs), we identified four miRNAs with expression decreased by ≥2.5-fold and three miRNAs with a ≥2.5-fold increase in MeCP2-/y aNSCs (Fig. 1 D). These results suggest that the loss of functional MeCP2 leads to the dysregulation of a subset of specific miRNAs in the context of neurogenesis.
Figure 1.
Identification of miRNAs with altered expression in Mecp2-deficient adult NSCs. (A) Adult NSCs cultured under proliferating conditions express Sox2 (nuclear, green) and Nestin (cytoplasmic, red), and incorporate BrdU (nuclear, red). Adult NSCs used in this study were multipotent and, when subjected to differentiation, expressed neuron-specific TuJ1 (red) and astrocyte-specific GFAP (green, DAPI is shown in blue), Bar, 50 µm. (B) Western blot showing expression of MeCP2 in WT aNSCs and the absence of MeCP2 in MeCP2-/y aNSCs (ab2828 antibody; Abcam). (C) Heat map of miRNA with a ≥2.5-fold change in expression in proliferating MeCP2-/y aNSCs. Quantities relative to WT aNSCs from each of four independent miRNA profiling experiments are shown. Relative quantity scale is shown below for reference. (D) Relative quantity of miRNA with ≥2.5-fold change in expression shown for MeCP2-/y proliferating aNSCs, calibrated to WT proliferating aNSCs. WT relative quantity = 1, mean relative quantity from three WT/MeCP2-/y pairs plus one pooled sample per genotype is plotted, with error bars representing a 95% CI. Dotted lines indicate a threshold of 2.5-fold change in expression.
To assess the potential for epigenetic regulation of these altered miRNAs, we first evaluated the genomic context of each miRNA, including CpG content and phylogenetic conservation, two hallmarks of functional nonprotein-coding regulatory elements. The region immediately upstream of one conserved single copy miRNA, miR-137, is highly conserved, contains multiple CpG-rich regions, and has a consensus binding site for Sox2, an Sry-related HMG-box transcription factor that plays important roles in stem cell function and adult neurogenesis (see Fig. 3, A–C; Jaenisch and Young, 2008; Zhao et al., 2008). Elevated expression of miR-137 in the absence of Mecp2 was verified using independent assays (Fig. 2 A). Overall, miR-137 was 6.5-fold higher in proliferating MeCP2-/y aNSCs (Fig. 2 A). Furthermore, pri–/pre–miR-137 expression was significantly increased in the absence of MeCP2, which indicates that expression of miR-137 was in fact influenced at the level of transcription (Fig. S1 C).
Figure 3.
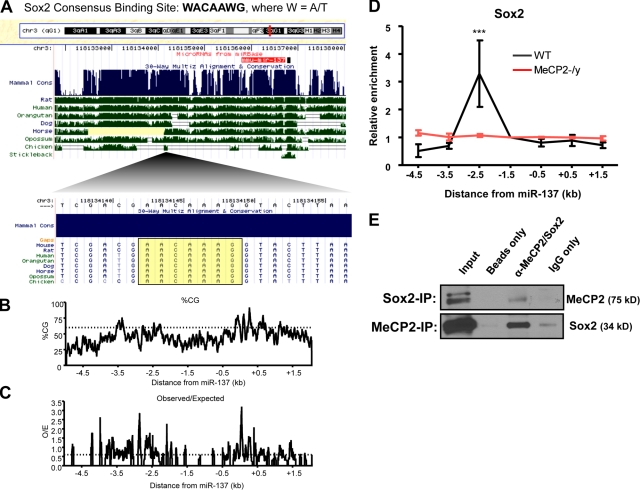
Transcriptional regulation of miR-137 involves coregulation by Sox2. (A) Schematic showing the miR-137 genomic locus and the location of a conserved Sox2 consensus-binding site within the 2.5-kb upstream region of miR-137 with which MeCP2 was also found to interact by ChIP. (B and C) Genomic structure and CpG content surrounding the miR-137 genomic locus. (B) Percentage of CG content across a 7-kb region surrounding miR-137, with a threshold indicated at 60%. (C) Ratio of observed CpG dinucleotides to the number of CpGs expected with a normal distribution across the same 7-kb region surrounding miR-137. A threshold is indicated at a ratio of 0.6 (dotted line). Data for both plots were generated using EMBOSS CpG plot with a 100-nt window size and a 1-nt window shift increment (Larsen et al., 1992). (D) Sequences 2.5 kb upstream of miR-137 enriched in a Sox2-specific ChIP relative to IgG only in WT aNSCs but not MeCP2-/y aNSCs, normalized to the directly adjacent 1.5-kb upstream region (n = 3, error bars indicate mean ± SEM, two-way ANOVA, Bonferroni post-test; ***, P < 0.001). (E) MeCP2 and Sox2 could be coimmunoprecipitated. Either MeCP2 or Sox2 was immunoprecipitated and subjected to Western blots with anti-Sox2 or anti-MeCP2 antibodies, respectively. IP beads only and IP with normal IgG were used as negative controls.
Figure 2.
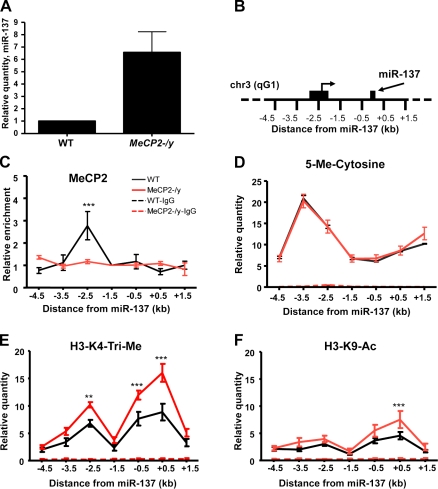
Expression of miR-137 is epigenetically regulated by MeCP2. (A) Verification of increased expression of miR-137 in MeCP2-/y proliferating aNSCs using independent real-time PCR (n = 6, mean relative quantity ± SEM, P = 0.022). (B) Schematic of the 7 kb proximal to miR-137 on chromosome 3qG1 that were assayed in ChIP experiments. The region 2.5 kb upstream is indicated, along with a previously identified transcriptional start site that lies 2.2 kb upstream of miR-137 (Shiraki et al., 2003; Carninci et al., 2005). (C) MeCP2-specific ChIP indicates the enrichment of DNA 2.5 kb upstream of the miR-137 genomic locus in WT aNSCs, but not MeCP2-/y aNSCs. Relative enrichment is calculated relative to IgG-only nonspecific control and normalized to the directly adjacent 1.5-kb upstream region (n = 3, two-way analysis of variance [ANOVA], Bonferroni post-test; ***, P < 0.001). (D) IP of 5′-Me-C (MeDIP) showing enrichment of methylated cytosines between 4.5 and 1.5 kb upstream of miR-137, with which MeCP2 may normally bind (n = 3). (E) H3-K4-Tri-Me–specific ChIP indicates increased enrichment of sequences 2.5 kb upstream of miR-137, as well as sequences in a 1-kb region directly surrounding miR-137 in MeCP2-/y aNSCs. (F) H3-K9-Ac–specific ChIP with increased enrichment of sequences in the 1 kb surrounding miR-137 in MeCP2-/y aNSCs. In all histone ChIP experiments: n = 3, two-way ANOVA, Bonferroni post-test, quantities calculated from an input DNA-generated standard curve; **, P < 0.01; ***, P < 0.001. Quantities in IgG-only nonspecific IP control experiments in E–G are near 0. Error bars indicate mean ± SEM.
The expression of miR-137 has been found to increase during neuronal differentiation, which suggests that the proper temporal expression of miR-137 may indeed influence aNSC proliferation and/or differentiation (Silber et al., 2008). Consistent with these findings, we observed a significant increase in miR-137 expression upon differentiation of both WT and MeCP2-/y aNSCs (Fig. S1 A). However, upon aNSC differentiation, we saw no significant difference in the relative increase in miR-137 expression between genotypes (Fig. S1 B). These results indicate that the absence of Mecp2 influences miR-137 expression before aNSC differentiation and that miR-137 may be prematurely expressed in MeCP2-/y aNSCs. We note that the absence of a MeCP2-mediated influence on miR-137 expression during differentiation also suggests a role for additional regulatory factors in the induction of miR-137 during this process. However, in proliferating aNSCs, precocious expression of miR-137 in the absence of MeCP2 would expose targeted mRNAs to aberrant regulation and potentially result in altered aNSC proliferation and/or differentiation. Based on these observations, we first proceeded to test the possibility of MeCP2-mediated epigenetic regulation of miR-137 in aNSCs, as well as the involvement of Sox2 in such regulation.
Expression of miR-137 is epigenetically regulated by MeCP2
To test whether MeCP2 interacts directly with genomic regions proximal to miR-137, we performed MeCP2-specific chromatin immunoprecipitation (ChIP) followed by real-time quantitative PCR across a 7-kb region surrounding miR-137, which included most of the highly conserved sequences upstream. Immunoprecipitation (IP) of chromatin chemically cross-linked to DNA in WT aNSCs with a MeCP2-specific antibody demonstrated that a region 2.5 kb upstream of miR-137 was enriched approximately threefold relative to MeCP2-/y aNSCs (Fig. 2, B and C). Between 2 kb and 4 kb upstream of miR-137, we also observed significant levels of 5′-methyl-cytosine (5′-Me-C), as detected by IP of DNA with a 5′-Me-C–specific antibody (MeDIP), which indicated the presence of methyl CpG dinucleotides to which MeCP2 could bind (Fig. 2 D; Weber et al., 2005). However, we detected no discernable difference in DNA methylation between different genotypes.
Given the role of MeCP2 in epigenetic regulation and its direct association with regions proximal to miR-137, we chose to examine the effect that loss of MeCP2 has on the chromatin state of the miR-137 locus by performing histone-specific ChIP assays in WT and MeCP2-/y aNSCs. We found that the absence of Mecp2 correlated with increased trimethyl histone H3 lysine 4 (H3-K4-Tri-Me) in the same 2.5-kb region upstream of miR-137 with which MeCP2 interacted in WT NSCs, as well as in the 1 kb directly surrounding miR-137 (Fig. 2 E). Additionally, we observed increased acetylated histone H3 lysine 9 (H3-K9-Ac) in the 1-kb region directly surrounding miR-137 (Fig. 2 F). Both H3-K4-Tri-Me and H3-K9-Ac associate with actively transcribed DNA, which correlates with the increased expression observed for miR-137 in the absence of Mecp2. In particular, the enrichment of H3K4-Tri-Me across this region strongly indicates that it is indeed a region of transcription initiation. Enrichment of H3-K4-Mono-Me and H3-K9-Tri-Me was minimal and indistinguishable between genotypes (Fig. S2, A and B). Although H3-K27-Tri-Me was enriched in the 2 kb directly surrounding miR-137, we saw no significant difference between WT and MeCP2-/y aNSCs (Fig. S2 C).
Additionally, we found that upon differentiation of WT aNSCs, there was a marked increase in H3-K4-Tri-Me and H3-K9-Ac proximal to miR-137 (Fig. S2, D and E), whereas H3-K27-Tri-Me levels decreased slightly (Fig. S2 F). Therefore, chromatin marks generally associated with active transcription appear to arise prematurely in the absence of Mecp2. Together, these data indicate the precocious establishment of a chromatin state correlating with increased miR-137 expression in the absence of MeCP2 and support a role for MeCP2 in mediating the proper epigenetic regulation of miR-137 specifically in proliferating aNSCs.
We noted that the 2.5-kb upstream region also contains a putative binding site for Sox2, a critical factor regulating stem cell self-renewal (Fig. 3 A). Because stem cell self-renewal is a process intricately linked with epigenetic regulation, we hypothesized that Sox2 may act in conjunction with MeCP2 to regulate miR-137 expression. Such regulation by Sox2 would also be supportive of a putative regulatory role for miR-137 in aNSCs. Therefore, we performed additional ChIP assays and found that the same region bound by MeCP2 was enriched ∼3.3-fold in a Sox2-specific ChIP assay (Fig. 3 D). This result correlated with previously observed Sox2 binding at the miR-137 genomic locus in embryonic stem cells (Boyer et al., 2005). Interestingly, the enrichment of Sox2 binding to this genomic region was lost in MeCP2-/y aNSCs (Fig. 3 D). These results indicate that both MeCP2 and Sox2 bind directly to the 5′ regulatory region of miR-137 in aNSCs. Consistent with such a mechanism, we also found that MeCP2 and Sox2 could be coimmunoprecipitated and that their association is DNA independent (Fig. 3 E). Thus, concurrent binding of MeCP2 and Sox2 within the 2.5-kb region upstream of miR-137 could be required for proper transcriptional regulation of miR-137 in aNSCs.
miR-137 modulates the proliferation and differentiation of aNSCs in vitro
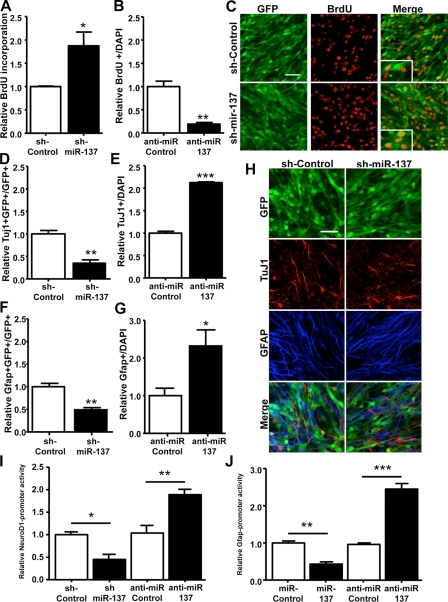
Our miRNA expression studies and ChIP assays indicated premature expression of miR-137 that was concurrent with the establishment of a chromatin state reflective of active transcription in the absence of MeCP2. An increase miR-137 expression in proliferating aNSCs would expose the distinct population of cellular mRNAs to aberrant miR-137 targeting, potentially resulting in abnormal aNSC proliferation and/or differentiation. Therefore, we sought to assess the potential regulatory effects of miR-137 by introducing or blocking miR-137 function specifically in proliferating aNSCs, and then asking what the subsequent effects on proliferation and/or differentiation were. To do this, we first created a lentiviral vector expressing a miR-137 short hairpin RNA (shRNA; sh-miR-137) under a U6 small nuclear RNA polymerase III promoter and EGFP under a cytomegalovirus (CMV) promoter (Li et al., 2008). Lentiviruses were used to infect cultured aNSCs with nearly 100% infection efficiency, as indicated by GFP expression (Figs. 4 and S3 A). We also verified overexpression of miR-137 independently of changes in several other miRNAs with altered expression in MeCP2-/y aNSCs, such that miR-137 would exert its functional effect independently of other miRNAs in aNSCs (Fig. S3 F). Quantification of BrdU-positive cells after pulse labeling indicated that miR-137–overexpressing (GFP+) aNSCs had 87.4% higher GFP+BrdU+ cells relative to sh-control lentivirus-infected aNSCs (Fig. 4, A and C). This effect did not appear to be caused by altered cell survival because we did not observe a significant difference in the apoptotic marker activated caspase-3 when comparing sh-control and sh-miR-137–treated cells (Fig. S4 A). Using a miR-137–specific inhibitor (anti-miR-137), we found that by blocking endogenous miR-137 in proliferating cells, GFP+BrdU+ cells were reduced by 81% relative to control, nonspecific anti-miR–treated cells (Fig. 4 B; Cheng et al., 2009; Zhao et al., 2009). Therefore, high levels of miR-137 led to a greater proliferative capability for aNSCs, whereas blocking endogenous miR-137 function reduced proliferation.
Figure 4.
miR-137 modulates the proliferation and differentiation of adult NSCs in vitro. (A) Cell proliferation was analyzed using BrdU pulse labeling. Quantitative analysis by stereology indicates that miR-137-overexpressing aNSCs (GFP positive) produced more BrdU+ cells, which indicates increased proliferation relative to control-treated cells (*, P = 0.0403; unpaired t test, n = 3). (B) Anti–miR-137–treated aNSCs produced fewer BrdU+ cells relative to anti–miR-control aNSCs (**, P = 0.0024; unpaired t test, n = 3). (C) Representative images of WT aNSCs infected with lentivirus expressing either sh-control or lentivirus expressing sh-miR-137, which were pulse-labeled with BrdU. Insets show 3× enlarged views of the boxed regions. Bar, 50 µm. (D) sh-miR-137–infected cells differentiated into fewer neurons compared with sh-control virus-infected cells, as determined by the percentage of TuJ1-positive cells among infected, GFP-positive cells (**, P = 0.0037; unpaired t test, n = 3). (E) Anti–miR-137 treatment reduces production of TuJ1-positive cells during differentiation (***, P < 0.0001; n = 3) (F) Among infected, GFP-positive cells, sh-miR-137–infected cells differentiated into fewer GFAP-positive cells compared with sh-control virus-infected cells (**, P = 0.0046; unpaired t test, n = 3). (G) Anti–miR-137 treatment reduces production of GFAP-positive cells during differentiation (*, P = 0.0483; n = 3). (H) Representative images of lentivirus-infected NSCs that were differentiated into Tuj1-positive neurons (red) and GFAP-positive astrocytes (blue). Bar, 50 µm. (I) Although overexpression of miR-137 leads to a decrease in NeuroD1-promoter activity relative to control treatment and as assessed by a luciferase reporter (*, P = 0.0128; unpaired t test, n ≥ 3), anti–miR-137 treatment leads to increased NeuroD1-promoter-luciferase activity (**, P = 0.0046; unpaired t test, n = 3). Control treatments are normalized to 1.0. (J) Overexpression of miR-137 leads to a decrease in GFAP promoter activity (**, P = 0.0021; unpaired t test, n = 3), whereas anti–miR-137 treatment leads to increased GFAP luciferase activity (***, P = 0.0007; unpaired t test, n = 3). Data in all panels are plotted as mean ± SEM (error bars).
To assess the effect of precocious miR-137 expression on subsequent aNSC differentiation, sh-miR-137 or sh-control lentivirus-infected aNSCs were subjected to a differentiation protocol. Neuronal differentiation was assessed by both TuJ1 immunostaining and the promoter activity of a pan-neuronal transcription factor, NeuroD1. Astrocyte differentiation was determined by GFAP immunostaining as well as GFAP promoter activity (Zhao et al., 2003; Barkho et al., 2006; Smrt et al., 2007). Sh-miR-137 lentivirus-infected aNSCs exhibited a 65.0% decrease in neuronal differentiation (Fig. 4, D and H) and a 51.0% decrease in astrocyte differentiation (Fig. 4, F and H) relative to sh-control lentivirus-infected aNSCs. Consistent with these observations, transfection of sh-miR-137 or synthetic duplex miR-137 reduced the activities of both GFAP and NeuroD1 promoters (Fig. 4, I and J). Furthermore, anti–miR-137 treatment had opposite effects compared with miR-137 overexpression on neuronal and astrocyte differentiation. The number of TuJ1-positive cells increased by 2.13-fold, whereas GFAP-positive cells were 2.32-fold more common relative to nonspecific anti–miR-control treatments (Fig. 4, E and G). Similarly, blocking the function of endogenous miR-137 led to enhancement of cotransfected NeuroD1- and GFAP-promoter-luciferase activity assayed at 24–48 h of differentiation, whereas a nonspecific anti-miR had no effect (Fig. 4, I and J). These results indicate that the dosage of miR-137 in aNSCs is critical for modulating the proliferation and differentiation of aNSCs. Overexpression of miR-137 promoted the proliferation of aNSCs at the expense of aNSC differentiation, whereas antagonizing miR-137 enhanced aNSC differentiation and reduced proliferation.
miR-137 modulates the proliferation and differentiation of aNSCs in vivo
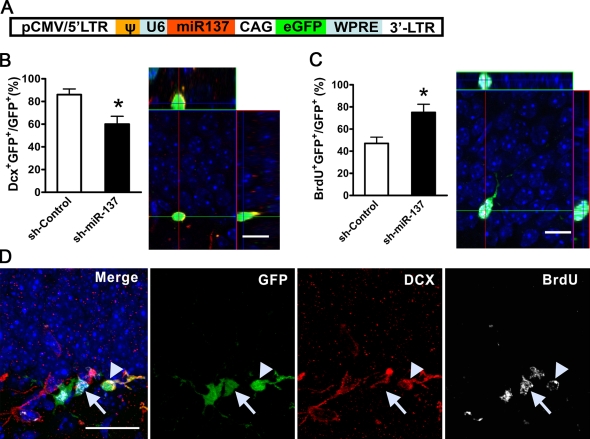
To assess the function of miR-137 in vivo, we took advantage of the persistent neurogenesis in the DG of the postnatal hippocampus, which recapitulates the neurogenic process during development (van Praag et al., 2002; Ge et al., 2006; Smrt et al., 2007). Recombinant retroviruses capable of infecting dividing cells have previously been used to label and follow the differentiation of NSCs in postnatal DG (van Praag et al., 2002; Ge et al., 2006; Smrt et al., 2007). We therefore engineered a retroviral vector that expresses both sh-miR-137 under an U6 promoter and EGFP under a chicken actin promoter (Fig. 5 A). The retrovirus expressing sh-miR-137 was stereotaxically grafted into the right side of the DG, and retrovirus-expressing sh-control was grafted into the left side of the DG of the same animal. To assess proliferation of retrovirus-labeled progenitors, mice also received BrdU injections immediately after the surgery. At 1 wk after viral grafting, a time when some of the retrovirus-labeled NSCs were expected to differentiate, we analyzed BrdU incorporation and expression of the early neuronal marker doublecortin (DCX) using triple fluorescence immunohistology and confocal microscopy (van Praag et al., 2002; Ge et al., 2006; Smrt et al., 2007). Many retrovirus-labeled aNSCs (GFP+) were also positively labeled with BrdU (Fig. 5 C), which indicates that these infected cells were in fact dividing after surgery. Some of these retrovirus-labeled cells had initiated neuronal differentiation and were, therefore, positive for DCX expression (Fig. 5 C). Using z-stack images of confocal microscopy at 1-µm resolution, we quantified the percentage of retrovirus-labeled GFP+ cells that expressed either DCX or incorporated BrdU. Compared with sh-control retrovirus-infected cells, a lower percentage of sh-miR-137 retrovirus-infected cells expressed DCX (Fig. 5 B, DCX+ GFP+/GFP+) and a higher percentage of sh-miR-137 retrovirus-infected cells incorporated BrdU (Fig. 5 B, BrdU+GFP+/GFP+), which suggests that miR-137–overexpressing aNSCs proliferated more and exhibited reduced neuronal differentiation capacity in vivo. Therefore, both our in vitro cell culture assay and in vivo single-cell genetic analyses indicate that a high level of miR-137 promoted aNSC proliferation, but repressed neuronal differentiation.
Figure 5.
miR-137 modulates the proliferation and differentiation of adult NSCs in vivo. (A) A schematic diagram of the retroviral vector used for in vivo miRNA expression. (B, left) sh-miR-137–infected cells differentiated into fewer DCX+ early neurons compared with sh-control virus-infected cells (n ≥ 3; *, P < 0.05; error bars indicate mean ± SEM). (B, right) A representative confocal 3D z-stack image used for quantification of DCX+GFP+ cells. (C, left) More sh-miR-137–infected cells proliferated than control virus-infected cells (n = 3; *, P < 0.05; error bars indicate mean ± SEM). (C, right) A representative 3D z-stack image used for quantification of BrdU+GFP+ cells (Bar, 5 µm). (D) A representative image of immunohistological analysis of virus-infected endogenous aNSCs in the subgranular zone of the DG. Arrowheads indicate a cell that is positive for GFP (green), BrdU (white), and DCX (red), which suggests that this cell was proliferating during the viral grafting period and has differentiated into early neurons within 1 wk of grafting. Arrows indicate a cell that is positive for GFP and BrdU, but not for DCX, which indicates that this cell did not differentiate. Bars: (B and C) 5 µm; (D) 20 µm.
Ezh2, a histone methyltransferase, is a functional target of miR-137 in aNSCs
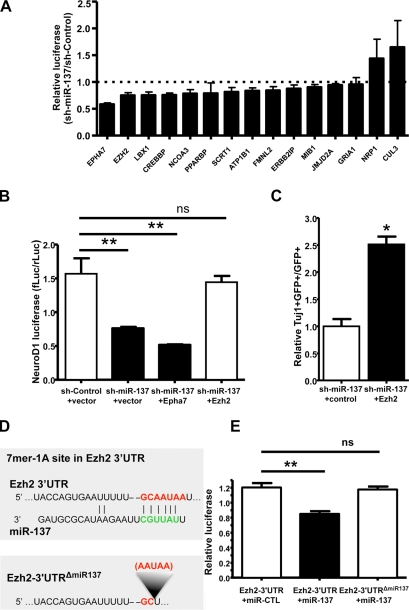
To understand the mechanisms by which miR-137 modulates adult neurogenesis, we cross-referenced TargetScan 4.1, PicTar, and miRanda to identify potential miR-137 targets (Lewis et al., 2003; John et al., 2004; Krek et al., 2005). Given the effects of miR-137 on aNSC proliferation and differentiation, 20 candidate miR-137 targets were selected for further analyses on the basis of three criteria: conservation, context score of target “seed sequences,” and known ontology relevant to neurodevelopment. To test whether miR-137 could indeed target to any of these candidates, we cloned 3′ untranslated regions (UTRs) directly from aNSC cDNA, ensuring expression of the putative target within aNSCs. We successfully cloned 15 3′ UTRs into a dual luciferase 3′ UTR reporter construct, allowing for the assessment of luciferase expression as dependent on a given 3′ UTR in response to miR-137 (Table S2). With these constructs, we performed a series of reporter assays in cell culture and found that miR-137 overexpression in HEK293T cells could suppress the activity of multiple 3′ UTR luciferase reporter genes (Fig. 6 A). To determine which genes could be involved in miR-137–mediated modulation of adult neurogenesis, we then performed functional rescue experiments. Based on the observation that overexpression of miR-137 could suppress the activity of a transfected NeuroD1-promoter-luciferase reporter at 24 h of differentiation in WT aNSCs, we asked whether coexpression of any of the genes suppressed by miR-137 in our reporter assays could alleviate the reduction in NeuroD1-luciferase reporter activity. We found that coexpression of Ezh2, but not EphA7, could indeed rescue the decreased NeuroD1-luciferase activity caused by the overexpression of miR-137. Additionally, direct knockdown of Ezh2 using a specific short hairpin directed against the endogenous Ezh2 mRNA significantly reduced NeuroD1 promoter luciferase activity. This effect was similar to that seen by miR-137 overexpression, which suggested that rescue was not simply an artifact of overexpression (Fig. S4, C and D). Ezh2 overexpression also had no effect miR-137 levels (Fig. 6 B and not depicted). To subsequently verify the ability of Ezh2 to rescue the miR-137–mediated deficit in neuronal differentiation, we also assayed the effect of Ezh2 coexpression on the production of TuJ1-positive cells by immunostaining. Indeed, Ezh2 coexpression significantly increased the number of TuJ1-positive cells relative to control treatment (Fig. 6 C). Ezh2 expression was further capable of significantly reducing proliferation as assayed by BrdU incorporation, an effect opposite to the one seen with miR-137 overexpression (Fig. S4 B). This suggests that Ezh2 coexpression could also rescue the miR-137–mediated increase in aNSC proliferation. Furthermore, targeting of Ezh2 by miR-137 was specific because mutating the seed sequence targeted by miR-137 within the luciferase-Ezh2–3′ UTR reporter alleviated miR-137–mediated suppression (Fig. 6, D and E).
Figure 6.
Ezh2 is a functional target of miR-137. (A) Primary screen of predicted miR-137 targets in HEK293FT cells. 3′ UTR–dependent luciferase assays were performed using both sh-control and sh-miR-137 for each of 15 predicted miR-137 targets. For each 3′ UTR, luciferase expression was normalized (hRluc/hluc) to 1 for the sh-control control treatment, as indicated by the dotted line. The effect of sh-miR-137 was then calculated relative to sh-control (sh-miR-137/sh-control). For all experiments, n ≥ 3, error bars indicate mean ± SEM. (B) Coexpression of Ezh2 rescued the decreased NeuroD1-luciferase expression caused by the overexpression of miR-137 in aNSCs. NeuroD1 promoter luciferase activity was normalized to coelectroporated E1a-Renilla luciferase activity (n ≥ 3 for all experiments, one-way ANOVA with Bonferroni post-test; **, P < 0.01; ns, P > 0.05). (C) Coexpression of Ezh2 rescued the decreased neuronal differentiation caused by the overexpression of miR-137 in aNSCs, as determined by the percentage of TuJ1-positive cells among infected, GFP-positive cells (unpaired t test, n = 3; *, P = 0.0016). (D) The miR-137 7mer-1A target site in the Ezh2 3′ UTR as predicted by TargetScan. (E) Ezh2-3′ UTR–dependent expression of a luciferase reporter gene was suppressed by miR-137 in HEK293FT cells. MiR-137–mediated suppression of luciferase was specific, as deletion of the miR-137 target site in the Ezh2 3′ UTR abolished repression by miR-137. Renilla luciferase-Ezh2–3′ UTR expression was normalized to firefly luciferase (n = 6 for HEK293FT cells, unpaired t test; **, P < 0.05).
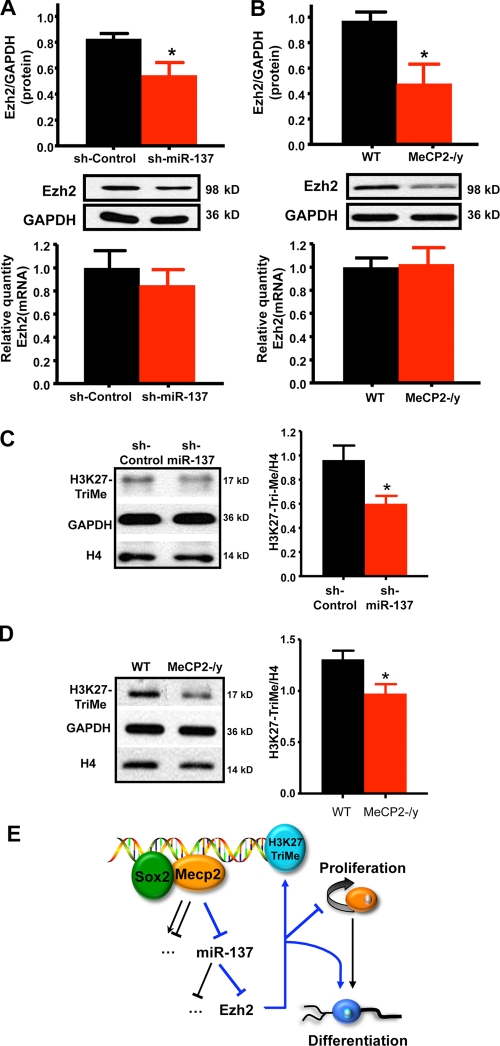
We went on to examine the effect of miR-137 on endogenous Ezh2 expression and saw a ∼35% reduction of endogenous Ezh2 protein in WT aNSCs infected by sh-miR-137–expressing lentivirus (Fig. 7 A). However, the reduction in protein did not correlate with a proportional reduction in steady-state mRNA expression, which indicates post-transcriptional regulation of Ezh2 mRNA by miR-137 (Fig. 7 A). Importantly, Ezh2 protein expression was also reduced in MeCP2-/y aNSCs, where expression of miR-137 is increased similarly (Fig. 7 B). To further verify the miR-137–mediated repression of Ezh2, we transfected anti–miR-137 into MeCP2-/y cells and asked whether blocking miR-137 expression in MeCP2-/y aNSCs could restore Ezh2 expression to levels similar to those seen in WT aNSCs. Indeed, Ezh2 protein expression recovered to levels comparable with those seen in WT aNSCs (Fig. S4 E). Together, these data demonstrate that the effect of miR-137 on Ezh2 protein expression is repressive, post-transcriptional, and specific. Importantly, they also suggest that Ezh2 is a functional target of miR-137 in the context of adult neurogenesis.
Figure 7.
Overexpression of miR-137 suppresses the expression of Ezh2 post-transcriptionally and results in an overall reduction in H3K27-TriMe. (A) Overexpression of miR-137 in WT aNSCs led to the reduction of endogenous Ezh2 protein expression (top, n = 4; *, P = 0.0366) without a proportional reduction in Ezh2 mRNA (bottom, n = 3, error bars indicate the mean with a 95% CI). (B) Loss of MeCP2 in aNSCs led to a similar reduction in endogenous Ezh2 protein expression (top, n = 4; *, P = 0.0417) without a proportional reduction in Ezh2 mRNA (bottom, n = 3, error bars indicate the mean with a 95% CI). (C) Overexpression miR-137 resulted in a reduction in H3K27-TriMe relative to histone H4 (*, P = 0.0379; n ≥ 3, unpaired t test). (D) H3K27-TriMe is also reduced relative to histone H4 in MeCP2-/y aNSCs (*, P = 0.0367; n ≥ 3, unpaired t test). (E) Model for the cross talk between MeCP2, miR-137, and Ezh2 in modulating adult neurogenesis. MeCP2 along with Sox2 mediates the epigenetic regulation of miR-137 in aNSCs, where increased expression of miR-137 promotes aNSC proliferation and inhibits aNSC differentiation, whereas decreased expression of miR-137 promotes differentiation of aNSCs. One target gene involved in this process is Ezh2. The miR-137–mediated suppression of Ezh2 feeds back to chromatin by decreasing global H3K27-TriMe. Error bars indicate mean ± SEM.
We next asked whether the functional targeting of Ezh2 by miR-137 correlated with an altered epigenetic state, in terms of chromatin, by assaying the effect of miR-137 overexpression on global H3-K27-Tri-Me. In both miR-137–overexpressing cells and MeCP2-/y cells, the miR-137–mediated decrease in Ezh2 protein correlated with an overall reduction in H3-K27-Tri-Me (Fig. 7, C and D). This indicates the integration of epigenetic regulation in aNSCs through MeCP2-mediated control of miR-137, the subsequent repression of Ezh2, and the feedback to chromatin in the form of decreased H3-K27-Tri-Me.
Discussion
Epigenetic regulation is proposed to play important roles in neurogenesis. Emerging evidence implicates both chromatin remodeling and epigenetic modifications as critical to the regulation of various aspects of adult neurogenesis (Zhao et al., 2003; Lim et al., 2009; Ma et al., 2009); however, the identification of downstream targets has proved elusive (Hsieh and Gage, 2004; Cheng et al., 2005; Abel and Zukin, 2008). Our results suggest that one such class of targets is miRNAs, including miR-137. We show that miR-137 is an intrinsic modulator of adult neurogenesis. The finding that overexpression and inhibition of miR-137 have distinctly opposite effects on aNSCs suggests that the dosage of miR-137 is critical to the modulation of adult neurogenesis. Previous studies have found that miR-137 could inhibit proliferation and induce differentiation of tumor cells (Kozaki et al., 2008; Silber et al., 2008); CDK6 was identified as the miR-137 target relevant to the proliferation phenotype that was observed (Kozaki et al., 2008; Silber et al., 2008). We have directly assayed CDK6 expression in aNSCs overexpressing miR-137 (both MeCP2-/y aNSCs and sh-miR-137–treated aNSCs) and observed an increase in CDK6 protein, an effect opposite to previous results (Fig. S5). This difference between primary cells and transformed tumor cells is not surprising because expression profiles of miRNAs have been shown to distinguish human tumor cells of different origins from their normal tissue counterparts more effectively than the expression profiles of mRNAs (Jay et al., 2007; Barbarotto et al., 2008). Our data further support the notion that miRNAs may play distinct roles in normal and tumor stem cells by post-transcriptionally regulating different target mRNAs.
We show that the expression of miR-137 is subject to epigenetic regulation mediated by MeCP2. Furthermore, we observe a novel interaction between MeCP2 and Sox2, a core transcriptional regulator in stem cells, in the genomic region proximal to miR-137. In MeCP2-/y aNSCs, we find a chromatin state reflective of premature miR-137 expression that also correlated with a loss of Sox2 binding upstream of miR-137. These results indicate a role for MeCP2 in establishing or maintaining an epigenetic state in the chromatin surrounding miR-137. Such chromatin states may allow for proper transcriptional coordination by Sox2, which has been shown to play important roles in the maintenance of stem cell properties and the regulation of adult neurogenesis (Bylund et al., 2003; Graham et al., 2003; Ellis et al., 2004; Ferri et al., 2004; Taranova et al., 2006; Suh et al., 2007). Together, these observations reinforce the importance of controlling miRNA expression in stem cells and reveal epigenetic regulation as one such mechanism by which this could be accomplished.
Our data has clearly demonstrated that the dosage of miR-137 in aNSCs is important for aNSC function and that the transcriptional control by MeCP2 helps to retain the levels of miR-137 at its proper level. Overexpression of miR-137 in the absence of MeCP2 results in an influence of miR-137 on subsequent aNSC proliferation/differentiation that was not expected based on the normal expression profile of miR-137 during aNSC differentiation. This indicates that miR-137 overexpression specifically in proliferating aNSCs has a regulatory effect distinct from that which it may have during differentiation. This may not be surprising in light of our findings that miR-137 dosage is critical toward its regulatory role in aNSCs and the fact that proliferating and differentiated aNSCs are likely to express quantitatively distinct populations of mRNAs that would be subjected to differential miR-137 targeting. As a result, it will be interesting and important to further identify miR-137 targets specific to differentiated aNSCs, or even mature neurons, as compared with those targets regulated in proliferating aNSCs.
We identified Ezh2 as a functional miR-137 target with relevance to adult neurogenesis. Ezh2 is a H3-K27 methyltransferase and component of the PcG of protein complexes known to perform important functions in stem cells (O’Carroll et al., 2001; Cao and Zhang, 2004; De Haan and Gerrits, 2007; Lee et al., 2007). PcG proteins function by forming and maintaining the bivalent chromatin state of stem cells (Boyer et al., 2006; Lee et al., 2006). This so-called “bivalent chromatin state” allows cell/tissue-specific genes to be “primed” for expression but “held in check” by opposing chromatin modifications (Boyer et al., 2006; Lee et al., 2006). Therefore, a bivalent chromatin state is likely a common mechanism in many types of stem and progenitor cells for maintaining their differentiation potential, with PcG proteins being critical factors in this regulatory mechanism. Not surprisingly, Ezh2 is found to be a critical regulator of neuroprogenitor cell maintenance and differentiation (Shen et al., 2008; Sher et al., 2008; Ezhkova et al., 2009). Our finding that miR-137 regulates the expression of Ezh2 in aNSCs reveals a novel interaction between PcG proteins and miRNAs; hence, distinct miRNAs may be involved in establishing and maintaining the bivalent chromatin state of stem cells. This further suggests a potential epigenetic circuitry in the modulation of aNSCs, with a feedback regulatory mechanism mediated by miRNAs (Fig. 7 E). In this particular example, precocious overexpression of miR-137 results in a reduction of Ezh2 and an altered epigenetic state of aNSCs in the form of a global decrease in H3-K27-Tri-Me. It is likely that altered H3-K27-Tri-Me at specific key developmental genes contributes to the phenotypes observed with miR-137 overexpression, and identification of these loci will be an important question moving forward. These results provide direct evidence for the hypothesis that cross talk between epigenetic regulation and the miRNA pathway plays an important role in modulating adult neurogenesis, which reflects the complexity of the network regulating proliferation and differentiation of stem cells in general.
The postnatal neurodevelopmental disorder known as Rett syndrome is largely caused by mutations in MECP2 (Amir et al., 1999). Recent studies have also shown that MeCP2 aberrations result in a constellation of neuropsychiatric abnormalities, wherein both loss of function and a gain in MeCP2 dosage lead to similar neurological phenotypes (Chahrour and Zoghbi, 2007). One challenge in understanding the etiology of MeCP2-related disorders is that MeCP2 processes multiple function domains, post-translational modifications, and cofactors, which allows the protein to be involved in numerous cellular pathways (Zhou et al., 2006; Chahrour et al., 2008; Ballas et al., 2009; Tao et al., 2009). The identification of key downstream targets of MeCP2 is critical for understanding the molecular basis of MeCP2-related neurological disorders. Earlier studies have focused mainly on protein-coding RNAs (Chahrour and Zoghbi, 2007). Here, we demonstrate that MeCP2 can also regulate the expression of small noncoding RNAs, particularly a subset of miRNAs, which reveals another layer of gene regulation mediated by MeCP2. miRNAs are particularly abundant in the brain and play key roles in neuronal function and plasticity (Cheng et al., 2005). Although our present study focuses on miR-137, our results indicate that there are additional miRNAs that could be regulated by MeCP2. In consideration of previous studies suggesting a role for MeCP2 in regulating neural precursor maturation (Kishi and Macklis, 2004; Smrt et al., 2007), it will be important to further evaluate how the miRNAs regulated by MeCP2 could interact with each other in neurodevelopmental contexts, thereby contributing to the molecular pathogenesis of Rett syndrome and other MeCP2-related disorders.
Materials and methods
Isolation and culture of adult NSCs
All animal procedures were performed according to protocols approved by the University of New Mexico Animal Care and Use Committee. The Mecp2 mutant mice (Mecp2tm1.1Jae) used in this study were created by deleting exon 3 containing the MBD domain of Mecp2 (Chen et al., 2001). These mice have been bred over 40 generations on an ICR background. They begin to show neurological symptoms between 5 and 8 wk of age and die at ∼10–11 wk. Mice 7 wk of age and without severe physical symptoms were used for cell isolation, as were WT littermates. The isolation of adult mouse brain-derived NSCs was performed according to an established protocol with modifications (Zhao et al., 2003). In brief, the forebrain without the olfactory bulb was dissociated mechanically followed by enzymatic digestion based on the MACS Neural Tissue Dissociation kit (Miltenyi Biotec). After enzymatic digestion was stopped using DME/F-12 containing 10% FBS (Sigma-Aldrich), 2 mM l-glutamine (Invitrogen), and 1% antibiotic-antimycotic (Invitrogen), the cell suspension was filtered through a 70-µm cell strainer (BD), and the single-cell suspension was loaded onto 50% Percoll (GE Healthcare). The NSCs were separated from other cells by ultracentrifugation at 127,000 rpm for 30 min at 20°C using a SW41 Rotor (Beckman Coulter). The fraction containing NSCs was collected and cultured in DME/F-12 medium containing 20 ng/ml basic FGF (FGF-2; PeproTech), 20 ng/ml EGF (PeproTech), 1% N2 supplement (Invitrogen), 1% antibiotic-antimycotic, and 2 mM l-glutamine in a 5% CO2 incubator at 37°C. We replaced half the medium every 2–3 d.
Relative quantification of mature miRNAs by real-time PCR
Profiling of mature miRNA expression was performed using TaqMan miRNA assays (Applied Biosystems) with 48-plex reverse transcription and individual TaqMan miRNA real-time PCR assays according to the manufacturer’s instructions (Lao et al., 2006). In brief, 8 pools of 48 reverse transcription primers each were used in 20-µl reactions consisting of: 20 ng total RNA, 1× TaqMan miRNA reverse transcription primer pool, 0.5 mM of each deoxyribonucleotide triphosphate (dNTP), 10.0 U/µl MultiScribe (Applied Biosystems) reverse transcription, 1× reverse transcription buffer, 0.25 U/µl RNase inhibitor, and nuclease-free water. The reactions were incubated at 16°C for 30 min, 42°C for 30 min, and 85°C for 5 min. Reactions were diluted 1:10 with nuclease-free water for use in the TaqMan real-time PCR reactions. Individual TaqMan miRNA real-time PCR reactions for profiling experiments were performed on a PCR system (7900HT SDS; Applied Biosystems) in a 384-well format running WT and MeCP2-/y pairs in parallel for each cDNA pool generated in the reverse transcription step. PCR reactions were performed in triplicate for each sample and each miRNA. The 10-µl reactions consisted of 1× TaqMan Universal Master Mix, No AmpErase UNG, 1× TaqMan miRNA assay mix, 0.8 µl of 1:10 diluted cDNA, and nuclease-free water. All TaqMan PCR reactions were prepared and aliquoted using a custom method on an automated pipette (Biomek FX; Beckman Coulter). PCR reaction conditions were run according to the standard protocol without the 50°C incubation using version 2.3 of the SDS software, with reactions incubated at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, and 60°C for 1 min. RQ of miRNA were determined using the ΔΔCt method (Livak and Schmittgen, 2001). RQ values and the associated error were determined using SDS v1.2 RQ manager software (Applied Biosystems) to calculate mean RQ and RQ min/max values based on a 95% CI. All paired samples were incorporated into a single analysis within the SDS version 1.2 RQ manager to obtain the reported values. Reverse transcription primer pool-specific endogenous controls were chosen based on miRNA with the least variable expression among all samples tested. All relative quantity calculations were calibrated to WT samples.
Individual reverse transcription and TaqMan miRNA assays were performed on a PCR system (7500 Fast Instrument; Applied Biosystems). The 15-µl reverse transcription reactions consisted of 10 ng of total RNA isolated with TRIZOL (Invitrogen), 5 U MultiScribe Reverse transcription, 0.5 mM of each dNTP, 1× reverse transcription buffer, 4 U RNase inhibitor, and nuclease-free water. Reverse transcription reactions were incubated at 16°C for 30 min, 42°C for 30 min, and 85°C for 5 min, and then stored at 4°C until use in TaqMan assays. The 10-µl TaqMan real-time PCR reactions consisted of 1× TaqMan Universal PCR Master Mix, No AmpErase UNG, 1× TaqMan miRNA assay, 1.33 µl of undiluted cDNA, and nuclease-free water. Each TaqMan assay was performed in either triplicate or quadruplicate for each sample tested. RQs were calculated using the ΔΔCt method with RNU6B TaqMan miRNA control assay as the endogenous control and calibrated to the WT samples (Livak and Schmittgen, 2001). Reactions were run with the standard 7500 default cycling protocol without the 50°C incubation stage using the SDS 7500 Fast Real-Time PCR System Software (version 1.3.1), with reactions incubated at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, and 60°C for 1 min. Fluorescence readings were taken during the 60°C step.
Primary/precursor miR-137 was detected by polyadenylating total RNA using SuperScript III oligo-dT reverse transcription (Invitrogen) to generate first-strand cDNA and real-time PCR targeting the pri–/pre–miR-137. Polyadenylation and reverse transcription were performed according to the manufacturer’s instructions using the Ncode miRNA First-Strand cDNA Synthesis kit (Invitrogen), with 1 µg of total RNA isolated with TRIZOL as input. Relative quantification was performed by real-time PCR using 1:10 diluted cDNA, 1× Power SYBR Green Master Mix (Applied Biosystems), 0.5 µM forward (5′-GTGACGGGTATTCTTGGGT-3′) and reverse primers (Universal qPCR primer provided with kit), and nuclease-free water. 18S rRNA was used as an endogenous control for all samples. Reactions were run in triplicate on four samples per genotype using an SDS 7500 Fast Instrument Standard 7500 default cycling protocol and SDS 7500 Fast System Software version 1.3.1 without the 50°C incubation.
ChIP
ChIP was performed as described previously (Coffee et al., 1999). In brief, cells grown on 3–6 confluent 10-cm cell culture plates were fixed by 1% formaldehyde (Sigma-Aldrich) to culture medium for 10 min at room temperature. After washing with cold PBS, cells were collected with cold PBS, washed, and suspended in 1 ml of cold cell lysis buffer (5 mM Pipes, pH 8.0, 85 mM KCl, 0.5% NP-40, and 1× complete proteinase inhibitor [Roche]), then incubated on ice for 5 min. Cell lysates were pelleted by centrifugation at 3,000 rpm for 5 min, resuspended again in 1 ml of cold cell lysis buffer for 5 min on ice, and then repelleted to collect nuclei. Nuclei were lysed at room temperature with 500 µl of nuclei lysis buffer (50 mM Tris, pH 8.1, 10 mM EDTA, 1% SDS, and 1× complete protease inhibitor). Nuclear lysates were sonicated using a sonicator (Sonicator 3000; Misonix). The size of the sonicated chromatin (mean size of ∼500–600 bp) was verified by treating 5-µl aliquots with 1 µl of 20 mg/ml proteinase A for 20 min at 50°C and running on a 1.5% agarose gel stained with ethidium bromide. For IP reactions, we used 50 µl of sonicated chromatin precleared with salmon sperm/tRNA-blocked protein A agarose for 60 min at 4°C in 950 µl of IP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 20 mM Tris, pH 8.1, and 500 mM NaCl). Precleared chromatin was rotated at 4°C overnight with 10 µg of the appropriate antibody.
Antibodies used were: normal rabbit IgG (Millipore), rabbit polyclonal to MeCP2 (ChIP grade; ab2828; Abcam), rabbit anti-Sox2 (AB5603; Millipore), rabbit polyclonal to H3 (tri-methyl K4, ChIP grade; ab8580; Abcam), anti–acetyl-histone-H3 (Lys9; Millipore), anti–monomethyl-histone H3 (Lys4; Millipore), and rabbit polyclonal to H3 (tri-methyl K9, ChIP grade; ab8898; Abcam).
Antibodies were pulled down with 60-µl blocked protein A–agarose beads for 1 h at 4°C with rotation. The beads were washed sequentially two times each in IP dilution buffer, TSE-500 solution (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris, pH 8.1, and 500 mM NaCl), freshly prepared Li/Cl wash solution (100 mM Tris, pH 8.1, 300 mM LiCl, 1% NP-40, and 1% deoxycholic acid), and 1× Tris-EDTA buffer (TE) for 10 min at 4°C. Protein–DNA complexes were eluted from the protein A–agarose beads twice with 250 µl of IP elution buffer (50 mM NaHCO3 and 1% SDS) for 15 min at 37°C with rotation. Formaldehyde-induced protein–DNA cross-linking was heat reversed by incubating protein–DNA complexes at 65°C overnight. DNA was purified using phenol/chloroform/isoamyl alcohol (25:25:1) isolations and precipitated with two volumes of 100% ethanol and 10 µg linear acrylamide at −35°C overnight. Immunoprecipitated and purified DNA fragments were resuspended in nuclease-free water, concentrations were determined by NanoDrop (Thermo Fisher Scientific), and each sample was diluted to 1 ng/µl. We used 8 ng of DNA in 20-µl SYBR Green real-time PCR reactions consisting of 1× Power SYBR Green Master Mix and 0.5 µM forward and reverse primers. Reactions were run on an SDS 7500 Fast Instrument using the standard 7500 default cycling protocol and SDS 7500 Fast System Software version 1.3.1 without the 50°C incubation. Primer sequences spaced at 1-kb intervals spanning from 4.5 kb upstream to 1.5 kb downstream of mmu-miR-137 were designed using Primer Express 3.0 software (Applied Biosystems) and were as follows. 4.5 kb upstream: forward, 5′-ACATTGCCATATCACTCCTATCAAAT-3′; and reverse, 5′-CCCTCCTCCCACCCATACA-3′. 3.5 kb upstream: forward, 5′-TCCCTTCCCAGGGCTTGT-3′; and reverse, 5′-GGAGCCGCTGCTCTCTGA-3′. 2.5 kb upstream: forward, 5′-AGCTTAAGGAGGTTTGAATTGAATATG-3′; and reverse, 5′-CTTACGAGAACACCATTTCCTACGA-3′. 1.5 kb upstream: forward, 5′-GGAATTCATGTTGGTTTTTCTACTTG-3′; and reverse, 5′-CACTTCTCCAGGTAGACCAACTCA-3′. 0.5 kb upstream: forward, 5′-AAAGCACTTGTCTGTTGTGTGTTAAGT-3′; and reverse, 5′-TGGCTGTTCTATTTCCAATTCTGA-3′. 0.5 kb downstream: forward, 5′-GCCGAGCTGCTCAGCAA-3′; and reverse, 5′-CCCCGCCCCTTCCTTAG-3′. 1.5 kb downstream: forward, 5′-TGCCTTGAAGAGCAAGCTGAA-3′; and reverse, 5′-AAGGCTGTTTTTCCAGGGTTCT-3′.
DNA relative enrichment was determined by taking the absolute quantity ratios of specific IPs to nonspecific IPs (normal rabbit IgG only), IP/IgG, and normalizing to a control genomic region that was not enriched in specific IPs relative to nonspecific IPs. Absolute quantification was based upon standard curves generated from four 10-fold dilutions ranging from 0.08–80 ng of input DNA treated in parallel with immunoprecipitated DNA during reverse cross-linking and purification steps. For histone ChIP experiments, quantity was determined based upon input DNA-generated standard curves and reported directly for both specific and IgG nonspecific IPs. All ChIP experiments were from obtained independent chromatin preparations, and all real-time PCR reactions were performed in triplicate for each sample on each amplicon. All primer sets were subjected to a dissociation curve analysis and produced single peaks on a derivative plot of raw fluorescence.
5′-Me-C IP (MeDIP)
MeDIP was performed as described previously (Weber et al., 2005). 4 µg of sheared input DNA isolated during histone-specific ChIP experiments was diluted into 450 µl of 1× TE. DNA was denatured for 10 min at 100°C in a dry heat block, then immediately placed on ice for 5–10 min in 51 µl of 10× IP buffer (100 mM sodium-phosphate, pH 7.0, made from a 1 M stock solution [2 M monobasic sodium phosphate, 2 M dibasic sodium phosphate at a 1:1.564 ratio, and equal volume H2O], 1.4 M NaCl, and 0.5% Triton X-100) was added along with 10 µg 5-methylcytidine antibody (BI-MECY-0500; Eurogentec) or 10 µg of normal mouse IgG (Millipore). IP was performed at 4°C with rotation for 2 h. Antibody–DNA complexes were pulled down by adding 40 µl of Dynabeads (M-280) sheep anti–mouse IgG (Invitrogen) directly to the IP reaction at 4°C for 2 h with rotation. Beads were collected with a 1.5-ml microcentrifuge tube holder magnet and washed three times in 1× IP buffer at room temperature, 10 min per wash with rotation. Washed beads were collected with a magnet and resuspended in 250 µl of proteinase K digestion buffer (50 mM Tris, pH 8.0, 10 mM EDTA, and 0.5% SDS). 3.5 µl of 20 mg/ml proteinase K was added, and digestion was performed at 50°C for 3 h in a thermomixer (Eppendorf) set at 800 rpm. Beads were collected with a magnet, and DNA was extracted from the supernatant once with phenol and once with chloroform. DNA was precipitated with 400 mM NaCl, 15 µg linear acrylamide, and two volumes of 100% ethanol at −35°C overnight. Precipitated DNA was resuspended in 11 µl of nuclease-free H2O, and concentrations were determined by NanoDrop. Samples were diluted to 1 ng/µl, and 8 ng was used in a SYBR Green real-time PCR reaction identical to the one used for ChIP experiments, including primer pairs. In the case of the normal mouse IgG nonspecific IPs, not enough DNA was immunoprecipitated from each of the three individual MeDIP experiments to assay all seven genomic regions, and so DNA from all three experiments from both genotypes was pooled and diluted to 1 ng/µl. Quantities were determined against a non–immuno-enriched input DNA-generated standard curve and reported directly for both specific and IgG nonspecific IPs.
IP
Whole cell lysates were prepared using the Nuclear Complex Co-IP kit (Active Motif) according to the manufacturer’s instructions. 1 mg of nuclear lysate was precleared with 60 µl of anti–rabbit Ig IP beads (eBioscience). 5% of precleared nuclear lysate was used as an input in Western blot analysis. The remaining nuclear lysate was immunoprecipitated with no antibody (beads alone), 2.5 µg of specific antibody, or 2.5 µg normal rabbit IgG. IP antibodies used were identical to those used in ChIP experiments, including normal rabbit IgG. 80 µl of anti–rabbit Ig IP beads were used to bind IP antibodies. IP incubations and IP washes were done using 1× IP low buffer. 50% of immunoprecipitated protein was used for Western blot analysis. MeCP2 was detected by Western blotting using Anti-MeCP2 (07–013; Millipore) at a dilution of 1:1,000. Sox2 was detected by Western blotting using anti-Sox2 (AB5603; Millipore) at a dilution of 1:1,000. Rabbit TrueBlot: HRP anti-rabbit IgG (eBioscience) was used as the secondary antibody at a dilution of 1:1,000. Detection of HRP was performed using ECL Western Blotting Detection reagents (GE Healthcare).
shRNA expression constructs
PCR-based generation of the miR-137 shRNA driven by a U6 Pol III promoter was done using a PCR-shagging approach as described previously (Paddison et al., 2002) with the following PAGE-purified long oligos.
shRNA miR-137 (sh-miR-137): 5′-TATCGATAAAAAAATTATTGCTTAAGAATACGCGTAGTCTCTTGAACTACGCGTATTCTTAAGCAATAAAAACAAGGCTTTTCTCCAAGGGA-3′; and shRNA control (sh-control): 5′-TATCGATAAAAAAAAATTCTCCGAACGTGTCACGTTCTCTTGAAACGTGACACGTTCGGAGAATTAAACAAGGCTTTTCTCCAAGGGA-3′.
Long oligos were used as reverse primers in combination with a common forward primer complementary to the 5′ end of the U6 promoter (5′-AAAGTTAACTAGTGGATCCGACGCCGCCATCTC-3′) to amplify the entire U6 promoter and shRNA in a single PCR product. Amplification was done using 20 ng of a previously generated U6-shRNA lentiviral construct (a gift from F.H. Gage, Salk Institute, La Jolla, CA) with AmpliTaq Gold PCR (Applied Biosystems; 1× PCR buffer, 2.2 mM MgCl2, 0.2 mM dNTP mix, 0.2 mM forward primer, 0.2 mM reverse primer, and 1.75 U AmpliTaq Gold) at 95°C for 9 min, 40 cycles of 94°C for 1 min, and 60°C for 1 min, followed by 60°C for 10 min, then stored at 4°C. We used 2 µl of PCR product in a TOPO TA cloning reaction with pCR2.1 vector and chemical transformation of TOP-10–competent cells (Invitrogen). U6-shRNA expression cassettes were removed from the TOPO vector and transferred to lentiviral and retroviral vectors by HpaI and ClaI restriction digestion followed by ligation. The lentiviral vectors expressing miR-137 or control shRNA were then verified by sequencing.
Production of recombinant lentivirus, differentiation and proliferation of lentivirus-infected NSCs
We produced lentivirus as described previously (Barkho et al., 2006). In brief, lentiviral transfer vector DNA and packaging plasmid DNA were transfected into cultured 293T cells using calcium phosphate methods. The medium containing lentivirus was collected at 40, 64, and 88 h after transfection, then pooled, filtered through a 0.2-µm filter, and concentrated using an ultracentrifuge at 19,400 rpm for 2 h at 20°C using a rotor (SW27; Beckman Coulter). The virus was washed once and then resuspended in 500 µl PBS. We routinely obtained 0.5–1 × 109 infectious viral particles/ml. To study the effects of miR-137 on the proliferation and differentiation of NSCs, ∼60 µl of lentivirus was added to the WT aNSCs cultured under proliferating conditions on a 10-cm tissue culture plate. After a 3-d incubation, infected aNSCs were either collected for RNA analysis or trypsinized and plated into chamber slides (Thermo Fisher Scientific) at a density of 5–7 × 104 cells per well for differentiation or proliferation analyses. To assess the effects of miR-137 on cell survival, an anti-caspase 3 (active) antibody (AB3623; Millipore) was used for immunostaining.
In vitro NSC proliferation and differentiation assays were performed as described previously (Zhao et al., 2003; Barkho et al., 2006). To assay proliferation, at 6–8 h after plating, 5 µM BrdU (Sigma-Aldrich) was added to the cells, incubated for 16 h, fixed with 4% paraformaldehyde for 30 min, and followed by immunohistochemical analysis. To detect BrdU incorporation, cells were pretreated with 1 M HCl for 30 min at 37°C followed by washing with borate buffer, pH 8.5, for 30 min, before being subjected to a standard immunohistochemistry protocol. For the differentiation assay, at 24 h after plating, cells were transferred into differentiation medium, DME/F12 (1:1) containing 5 µM forskolin (Sigma-Aldrich) and 1 µM retinoic acid (Sigma-Aldrich) and incubated for 4 d, followed by fixation with 4% paraformaldehyde for 30 min and washing with Dulbecco’s phosphorylated buffered saline (DPBS), pH 7.4, for 30 min. Immunocytochemistry staining was performed as described previously (Zhao et al., 2003). In brief, cells were first pre-blocked using DPBS containing 5% normal goat serum (Vector Laboratories) and 0.1% Triton X-100 for 30 min, followed by overnight incubation with primary antibodies: mouse neuron-specific type III tubulin (Tuj1, 1:4,000; G712A; Promega), rabbit GFAP (1:1,000; Z-0334; Dako), rat anti-BrdU (1:3,000; ab-6326; Abcam), chicken anti-GFP (1:500; A10262; Invitrogen), or rabbit anti-GFP (1:1,000; A11122; Invitrogen). After washing with DPBS, cells were incubated with secondary antibodies that included goat anti–mouse Alexa Fluor 568 (1:500; A11031; Invitrogen), goat anti–rabbit Alexa Fluor 647 (1:500; A21245; Invitrogen), goat anti–rat Alexa Fluor 568 (1:500; A11077; Invitrogen), goat anti–rabbit Alexa Fluor 488 (1:500; A11008; Invitrogen), and goat anti–chicken Alexa Fluor 488 (1:500; A11039; Invitrogen), followed by counterstaining with the fluorescent nuclear dye DAPI (B2261; Sigma-Aldrich). After the cells were mounted with Vectashield (Vector Laboratories), the numbers of double positive cells (GFP+Tuj1+, GFP+GFAP+, and GFP+BrdU+) were quantified using a microscope (BX51; Olympus) equipped with a MicroFire digital camera (Optronics, Inc.) and a motorized stage. The quantification was performed using unbiased stereology with the aid of StereoInvestigator software (MicroBrightField, Inc.). The data were analyzed using a two-tailed unpaired t test.
To block the activity of endogenous miR-137, WT aNSCs were plated onto coated coverslips in a 24-well plate. Anti–miR-control or anti–miR-137 were transfected at a final concentration of 0.5 µM with FuGENE HD transfection reagent (Roche). 24 h after transfection, cells were treated with 5 µM of BrdU for 3 h or cultured with differentiation medium for 3 d. MeCP2-/y NSCs were plated into 6-well plates and transfected with anti–miR-control or anti–miR-137 at a final concentration of 0.5 µM, and collected 24 h later for analysis of Ezh2 expression by Western blotting.
For Ezh2 rescue studies, WT aNSCs were infected with lentivirus expressing sh-control + lentivirus expressing GFP as the control treatment or lentivirus expressing sh-miR-137 + lentivirus expressing Ezh2. The infected cells were collected and plated onto coated coverslips for proliferation or differentiation assays.
Electroporation and luciferase assay
miR-137, anti–miR-137, and controls were obtained from Applied Biosystems. NeuroD1-luciferase DNA, GFAP-luciferase, and internal control E1α-Rluc DNA plasmids have been described previously (Barkho et al., 2006). Ezh2 and EphA7 expression plasmids were obtained from Thermo Fisher Scientific. Electroporation of these RNA and DNA into aNSCs was performed using a nucleofector electroporator based on the manufacturer’s instructions (Lonza). In brief, 2 × 106 cells were trypsinized, resuspended in Nucleofector solution, mixed with DNA and miRNAs, and electroporated using a preset program for mouse aNSCs (A033). The cells were then plated onto polyornithin/laminin-coated 24-well plates in aNSC proliferation medium (see “Isolation and culture of adult NSCs”). 24 h after plating, cells were transferred into differentiation medium (see above) for 24 h. The cells were then collected using cell lysis buffer from a Dual-Luciferase Reporter Assay System kit (Promega). Luciferase activity was measured using a microplate luminometer (Veritas; Promega) as described previously (Barkho et al., 2006). The luciferase counts were then normalized to R-Luc counts to obtain final NeuroD1 or GFAP promoter activities.
Construction of retroviral vector expressing sh-miR-137 and in vivo retroviral grafting
A retroviral vector expressing both miR-137 and EGFP was engineered by deleting the original HpaI and ClaI sites in the CAG-EGFP vector (Zhao et al., 2006; Smrt et al., 2007) and inserting new HpaI and ClaI sites 5′ upstream from the CAG promoter. The U6-shRNA cassettes were then inserted between the HapI and ClaI sites.
Retrovirus production was performed as described previously (Zhao et al., 2006; Smrt et al., 2007). In brief, retroviral transfer vector DNA and packaging plasmid DNA were cotransfected with the packaging plasmids pCMV-gag-pol and pCMV-Vsvg into HEK293T cells using the calcium phosphate method. The medium containing retrovirus was collected at 40, 64, and 88 h after transfection, then pooled, filtered through a 0.2-µm filter, and concentrated via ultracentrifugation at 19,400 rpm for 2 h at 20°C (SW27 Rotor). The virus was washed once with PBS and then resuspended in 150 µl PBS. We routinely obtain >0.5–1 × 109 infectious viral particles/ml.
In vivo retroviral grating was performed as described previously (Zhao et al., 2006; Smrt et al., 2007). In brief, 7- to 8-wk-old C57B/L6 male mice were anesthetized with isoflurane, and virus (1.5 µl with titer >5 × 105/µl) was injected stereotaxically into the DG using the following coordinates relative to bregma: anteroposterior, −(1/2) × d mm; lateral, ±1.8 mm (if d > 1.6), or otherwise ±1.7 mm; and ventral, −1.9 mm (from dura). For each mouse, the sh-control virus was injected into the left DG, and the miR-137 virus was injected into the right DG. Mice received two BrdU injections per day (50 mg/kg, i.p.) for a total of seven injections, immediately after viral grafting. 1 wk after viral grafting, mice were deeply anesthetized with pentobarbital and perfused with saline followed by 4% PFA. Brains were dissected out, postfixed overnight in 4% PFA, and then equilibrated in 30% sucrose. 40-µm brain sections were generated using a sliding microtome and were stored in a −20°C freezer as floating sections in 96-well plates filled with cryoprotectant solution (glycerol, ethylene glycol, and 0.2 M phosphate buffer, pH 7.4, 1:1:2 by volume).
Immunohistochemistry and confocal imaging analysis were performed as described previously (Smrt et al., 2007). Floating brain sections containing EGFP+ cells were selected for staining and matched by DG region. Sections were pretreated with 1 M HCl, as described previously (Tang et al., 2007). The primary antibodies used were chicken anti-GFP (A10262; Invitrogen), rat anti-BrdU (ab-6326; Abcam), and rabbit anti-DCX (4604; Cell Signaling Technology). The secondary antibodies used were anti–chicken Alexa Fluor 488 (A11039; Invitrogen), goat anti–rat Alexa Fluor 647 (A21242; Invitrogen), and goat anti–rabbit Alexa Fluor 568 (A11036; Invitrogen). The z-stack images of GFP-BrdU-DCX staining were taken at 1-µm resolution using a TE2000 (Nikon) equipped with a spin disc confocal microscope with an oil-immersion objective lens (40×, NA = 1.3; Carl Zeiss, Inc.) and MetaMorph quantification software (MDS Analytical Technologies); we then counted the proportion of GFP+DCX+ or GFP+BrdU+ out of total GFP+ cells. For colocalization analysis, roughly 70 GFP+ cells per animal were imaged. The data were analyzed using the Student’s t test.
3′ UTR dual luciferase assays of candidate miR-137 target mRNA
3′ UTR sequences of candidate mRNAs were PCR amplified directly from proliferating aNSC first-strand cDNA generated from 5 µg of TRIZOL-isolated total RNA using oligo-dT SuperScript III reverse transcription, according to the manufacturer’s instructions (Invitrogen). All primers were designed incorporating XhoI and NotI restriction sites and 4 bp of extra random sequence to aid in restricting digestion. XhoI- and NotI-digested PCR products were cloned into XhoI- and NotI-digested psiCHECK-2 dual luciferase vector (Promega). As a primary screen of candidate miR-137 targets, 293FT cells (3 × 103 cells per well, 96-well plate, grown overnight before transfection) were transfected with sh-miR-137 cloned into a pCR2.1 TOPO vector (sh-miR-137 TOPO) and psiCHECK-2–3′ UTR using TransFast Transfection reagent (Promega) according to the manufacturer’s instructions. As a control, psiCHECK-2 plasmid with no 3′ UTR and U6-neg-shRNA were cotransfected with U6-mir-137-shRNA TOPO or psiCHECK-2–3′ UTR, respectively. All transfections used a total of 1 µg of plasmid DNA. The ratio of luciferase–3′ UTR/shRNA plasmid was 1:2 for all experiments. Luciferase expression was detected using the Dual-Luciferase Reporter 1000 System (Promega) according to the manufacturer’s instructions. 48 h after transfection, hRLuc activity was normalized to hLuc+ activity to account for variation in transfection efficiencies, and miR-137–mediated knockdown of hRLuc activity was calculated as the ratio of normalized hRLuc activity in the U6-miR-137-shRNA treatments to normalized hRLuc activity in the U6-neg-shRNA treatments. All luciferase readings were taken from either three or four individual wells for each psiCHECK-2–3′ UTR construct and control construct tested. Each transfection experiment was repeated at least three times.
The miR-137 target site in the Ezh2 3′ UTR was deleted using the QuikChange Site-Directed Mutagenesis kit (Agilent Technologies) to delete 5 bases (AAUAA) from the 7mer-1a miR-137 seed site in the Ezh2 3′ UTR luciferase reporter. Target site deletion was verified by Sanger sequencing.
To confirm the specificity of miR-137 targeting the Ezh2 3′ UTR, the Ezh2 3′ UTR and Ezh2 3′ UTRΔmiR-137 were transferred by XhoI–NotI double digestion and T4 DNA ligation from psiCHECK2 into a pIS2 renilla luciferase vector modified with the addition of an XhoI restriction site and deletion of the SpeI restriction site. MiR-137–dependent Ezh2 3′ UTR luciferase assays were performed as described previously using 10 pmol of miR-137 duplex RNA or control miRNA duplex, and Promega Dual Luciferase Reporter System and pIS0 firefly luciferase as a control (Yekta et al., 2004). To test whether Ezh2 knockdown modulates NSC differentiation, Ezh2 shRNA plasmid was electroporated into WT NSCs along with NeuroD1-luciferase DNA and internal control E1α-Rluc DNA plasmids. Both Ezh2 mRNA and NeuroD1-luciferase activity were determined after 24 h of differentiation.
Western blot analyses
Protein samples were separated on SDS-PAGE gels and then transferred to polyvinylidene fluoride membranes (Millipore). Membranes were processed according to the ECL Western Blotting Protocol (GE Healthcare). anti-MeCP2 (ab2828; Abcam), anti-Ezh2 (4905; Cell Signaling Technology), anti–tri-methyl-histone H3 (Lys27, C36B11; 9733; Cell Signaling Technology), and anti-histone H4 (ab10158; Abcam) were used as primary antibodies at a 1:1,000 dilution. HRP-labeled secondary antibodies were obtained from Sigma-Aldrich (A0545) and were used at a dilution of 1:5,000. For loading controls, membranes were stripped and re-probed with the antibody against glyceraldehyde 3-phosphate dehydrogenase (GAPDH; Applied Biosystems AM4300). All Western blot quantifications were performed using ImageJ software from the National Institutes of Health.
Real-time PCR relative quantification of Ezh2 mRNA
500 ng of total RNA isolated by TRIZOL from lentiviral sh-miR-137 and lentiviral sh-control–infected aNSCs was reverse transcribed using random hexamers to generate first-strand cDNA with SuperScript III according to the manufacturer’s instructions. 1 µl of cDNA was used directly in 20 µl SYBR Green real-time PCR reactions that consisted of 1× Power SYBR Green Master Mix, 0.5 µM forward and reverse primers, and nuclease-free water. 18S rRNA was used as an endogenous control for all samples, with 1 µl of cDNA diluted 1:10 in the nuclease-free water used. Reactions were run on an Applied Biosystems SDS 7500 Fast Instrument using the Standard 7500 default cycling protocol and SDS 7500 Fast System Software version 1.3.1 without the 50°C incubation. Primers for Ezh2 mRNA and 18S rRNA were designed using Primer Express 3.0 software (Applied Biosystems) and were as follows. Ezh2: forward, 5′-GGTGAAGAGTTGTTTTTTGATTACAGA-3′; and reverse, 5′-TCTCGTTCGATGCCCACATA-3′. 18S: forward, 5′-CGGCTACCACATCCAAGGAA-3′; and reverse, 5′-CCTGTATTGTTATTTTTCGTCACTACCT-3′. All real-time PCR reactions were performed in triplicate, and RQs were calculated using the ΔΔCt method (95% CI) with calibration to sh-control–treated samples. All primer sets were subjected to a dissociation curve analysis and produced single peaks on a derivative plot of raw fluorescence.
Online supplemental materials
Fig. S1 shows miR-137 and primary/precursor miR-137 expression in aNSCs. Fig. S2 shows determination of the epigenetic state of the miR-137 genomic locus using additional histone ChIP assays. Fig. S3 shows efficient delivery of control and miR-137 shRNA constructs by lentivirus. Fig. S4 shows verification of a functional interaction between miR-137 and Ezh2. Fig. S5 shows that miR-137 has distinct effects on the expression of CDK6 in different cell types. Table S1 shows miRNA profiling of aNSCs. Table S2 lists candidate miR-137 targets. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200908151/DC1.
Acknowledgments
We would like to thank S. Warren, C. Strauss, and S. Chang for their helpful discussions and critical reading of the manuscript, as well as Erin Finn-Flesher for technical assistance. We thank F. H. Gage for providing us the lentiviral vector used to engineer the expression vector for miR-137.
P. Jin is supported by grants from the International Rett Syndrome Foundation and the National Institutes of Health (NIH; NS051630 and MH076090). P. Jin is the recipient of a Beckman Young Investigator Award and the Basil O’Connor Scholar Research Award, and is an Alfred P. Sloan Research Fellow in Neuroscience. X. Zhao is supported by grants from the International Rett Syndrome Foundation and the NIH (MH080434 and MH078972). X. Li is a recipient of the Autism Speaks Postdoctoral Fellowship. R.D. Smrt is supported by a Minority Supplement to NIH grant (MH080434). N.J. Santistevan is supported by the NIH/Minority Access to Research Career (MARC) program.
Footnotes
Keith E. Szulwach and Xuekun Li contributed equally to this paper.
Abbreviations used in this paper:
- 5′-Me-C
- 5′-methyl-cytosine
- ANOVA
- analysis of variance
- aNSC
- adult neural stem cell
- ChIP
- chromatin immunoprecipitation
- CI
- confidence interval
- CMV
- cytomegalovirus
- DG
- dentate gyrus
- dNTP
- deoxyribonucleotide triphosphate
- DPBS
- Dulbecco’s phosphorylated buffered saline
- GFAP
- glial fibrillary acidic protein
- IP
- immunoprecipitation
- miRNA
- microRNA
- PcG
- Polycomb group
- RQ
- relative quantities
- shRNA
- short hairpin RNA
- UTR
- untranslated region
References
- Abel T., Zukin R.S. 2008. Epigenetic targets of HDAC inhibition in neurodegenerative and psychiatric disorders. Curr. Opin. Pharmacol. 8:57–64 10.1016/j.coph.2007.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir R.E., Van den Veyver I.B., Wan M., Tran C.Q., Francke U., Zoghbi H.Y. 1999. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 23:185–188 10.1038/13810 [DOI] [PubMed] [Google Scholar]
- Avilion A.A., Nicolis S.K., Pevny L.H., Perez L., Vivian N., Lovell-Badge R. 2003. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 17:126–140 10.1101/gad.224503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballas N., Lioy D.T., Grunseich C., Mandel G. 2009. Non-cell autonomous influence of MeCP2-deficient glia on neuronal dendritic morphology. Nat. Neurosci. 12:311–317 10.1038/nn.2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbarotto E., Schmittgen T.D., Calin G.A. 2008. MicroRNAs and cancer: profile, profile, profile. Int. J. Cancer. 122:969–977 10.1002/ijc.23343 [DOI] [PubMed] [Google Scholar]
- Barkho B.Z., Song H., Aimone J.B., Smrt R.D., Kuwabara T., Nakashima K., Gage F.H., Zhao X. 2006. Identification of astrocyte-expressed factors that modulate neural stem/progenitor cell differentiation. Stem Cells Dev. 15:407–421 10.1089/scd.2006.15.407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 116:281–297 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- Bernstein E., Kim S.Y., Carmell M.A., Murchison E.P., Alcorn H., Li M.Z., Mills A.A., Elledge S.J., Anderson K.V., Hannon G.J. 2003. Dicer is essential for mouse development. Nat. Genet. 35:215–217 10.1038/ng1253 [DOI] [PubMed] [Google Scholar]
- Bernstein B.E., Meissner A., Lander E.S. 2007. The mammalian epigenome. Cell. 128:669–681 10.1016/j.cell.2007.01.033 [DOI] [PubMed] [Google Scholar]
- Bienvenu T., Chelly J. 2006. Molecular genetics of Rett syndrome: when DNA methylation goes unrecognized. Nat. Rev. Genet. 7:415–426 10.1038/nrg1878 [DOI] [PubMed] [Google Scholar]
- Bird A. 2002. DNA methylation patterns and epigenetic memory. Genes Dev. 16:6–21 10.1101/gad.947102 [DOI] [PubMed] [Google Scholar]
- Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., et al. 2005. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 122:947–956 10.1016/j.cell.2005.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer L.A., Plath K., Zeitlinger J., Brambrink T., Medeiros L.A., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K., et al. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 441:349–353 10.1038/nature04733 [DOI] [PubMed] [Google Scholar]
- Bylund M., Andersson E., Novitch B.G., Muhr J. 2003. Vertebrate neurogenesis is counteracted by Sox1-3 activity. Nat. Neurosci. 6:1162–1168 10.1038/nn1131 [DOI] [PubMed] [Google Scholar]
- Cao R., Zhang Y. 2004. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr. Opin. Genet. Dev. 14:155–164 10.1016/j.gde.2004.02.001 [DOI] [PubMed] [Google Scholar]
- Carninci P., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., et al. 2005. The transcriptional landscape of the mammalian genome. Science. 309:1559–1563 10.1126/science.1112014 [DOI] [PubMed] [Google Scholar]
- Chahrour M., Zoghbi H.Y. 2007. The story of Rett syndrome: from clinic to neurobiology. Neuron. 56:422–437 10.1016/j.neuron.2007.10.001 [DOI] [PubMed] [Google Scholar]
- Chahrour M., Jung S.Y., Shaw C., Zhou X., Wong S.T., Qin J., Zoghbi H.Y. 2008. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 320:1224–1229 10.1126/science.1153252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.Z., Akbarian S., Tudor M., Jaenisch R. 2001. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat. Genet. 27:327–331 10.1038/85906 [DOI] [PubMed] [Google Scholar]
- Cheng L.C., Tavazoie M., Doetsch F. 2005. Stem cells: from epigenetics to microRNAs. Neuron. 46:363–367 10.1016/j.neuron.2005.04.027 [DOI] [PubMed] [Google Scholar]
- Cheng L.C., Pastrana E., Tavazoie M., Doetsch F. 2009. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat. Neurosci. 12:399–408 10.1038/nn.2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffee B., Zhang F., Warren S.T., Reines D. 1999. Acetylated histones are associated with FMR1 in normal but not fragile X-syndrome cells. Nat. Genet. 22:98–101 10.1038/8807 [DOI] [PubMed] [Google Scholar]
- De Haan G., Gerrits A. 2007. Epigenetic control of hematopoietic stem cell aging the case of Ezh2. Ann. N. Y. Acad. Sci. 1106:233–239 10.1196/annals.1392.008 [DOI] [PubMed] [Google Scholar]
- Ellis P., Fagan B.M., Magness S.T., Hutton S., Taranova O., Hayashi S., McMahon A., Rao M., Pevny L. 2004. SOX2, a persistent marker for multipotential neural stem cells derived from embryonic stem cells, the embryo or the adult. Dev. Neurosci. 26:148–165 10.1159/000082134 [DOI] [PubMed] [Google Scholar]
- Ezhkova E., Pasolli H.A., Parker J.S., Stokes N., Su I.H., Hannon G., Tarakhovsky A., Fuchs E. 2009. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 136:1122–1135 10.1016/j.cell.2008.12.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferri A.L., Cavallaro M., Braida D., Di Cristofano A., Canta A., Vezzani A., Ottolenghi S., Pandolfi P.P., Sala M., DeBiasi S., Nicolis S.K. 2004. Sox2 deficiency causes neurodegeneration and impaired neurogenesis in the adult mouse brain. Development. 131:3805–3819 10.1242/dev.01204 [DOI] [PubMed] [Google Scholar]
- Ge S., Goh E.L., Sailor K.A., Kitabatake Y., Ming G.L., Song H. 2006. GABA regulates synaptic integration of newly generated neurons in the adult brain. Nature. 439:589–593 10.1038/nature04404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham V., Khudyakov J., Ellis P., Pevny L. 2003. SOX2 functions to maintain neural progenitor identity. Neuron. 39:749–765 10.1016/S0896-6273(03)00497-5 [DOI] [PubMed] [Google Scholar]
- Hsieh J., Gage F.H. 2004. Epigenetic control of neural stem cell fate. Curr. Opin. Genet. Dev. 14:461–469 10.1016/j.gde.2004.07.006 [DOI] [PubMed] [Google Scholar]
- Ivey K.N., Muth A., Arnold J., King F.W., Yeh R.F., Fish J.E., Hsiao E.C., Schwartz R.J., Conklin B.R., Bernstein H.S., Srivastava D. 2008. MicroRNA regulation of cell lineages in mouse and human embryonic stem cells. Cell Stem Cell. 2:219–229 10.1016/j.stem.2008.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R., Young R. 2008. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 132:567–582 10.1016/j.cell.2008.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay C., Nemunaitis J., Chen P., Fulgham P., Tong A.W. 2007. miRNA profiling for diagnosis and prognosis of human cancer. DNA Cell Biol. 26:293–300 10.1089/dna.2006.0554 [DOI] [PubMed] [Google Scholar]
- John B., Enright A.J., Aravin A., Tuschl T., Sander C., Marks D.S. 2004. Human MicroRNA targets. PLoS Biol. 2:e363 10.1371/journal.pbio.0020363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi N., Macklis J.D. 2004. MECP2 is progressively expressed in post-migratory neurons and is involved in neuronal maturation rather than cell fate decisions. Mol. Cell. Neurosci. 27:306–321 [DOI] [PubMed] [Google Scholar]
- Kozaki K., Imoto I., Mogi S., Omura K., Inazawa J. 2008. Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Res. 68:2094–2105 10.1158/0008-5472.CAN-07-5194 [DOI] [PubMed] [Google Scholar]
- Krek A., Grün D., Poy M.N., Wolf R., Rosenberg L., Epstein E.J., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., Rajewsky N. 2005. Combinatorial microRNA target predictions. Nat. Genet. 37:495–500 10.1038/ng1536 [DOI] [PubMed] [Google Scholar]
- Lao K., Xu N.L., Yeung V., Chen C., Livak K.J., Straus N.A. 2006. Multiplexing RT-PCR for the detection of multiple miRNA species in small samples. Biochem. Biophys. Res. Commun. 343:85–89 10.1016/j.bbrc.2006.02.106 [DOI] [PubMed] [Google Scholar]
- Larsen F., Gundersen G., Lopez R., Prydz H. 1992. CpG islands as gene markers in the human genome. Genomics. 13:1095–1107 10.1016/0888-7543(92)90024-M [DOI] [PubMed] [Google Scholar]
- Lee T.I., Jenner R.G., Boyer L.A., Guenther M.G., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K., et al. 2006. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 125:301–313 10.1016/j.cell.2006.02.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E.R., Murdoch F.E., Fritsch M.K. 2007. High histone acetylation and decreased polycomb repressive complex 2 member levels regulate gene specific transcriptional changes during early embryonic stem cell differentiation induced by retinoic acid. Stem Cells. 25:2191–2199 10.1634/stemcells.2007-0203 [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B. 2003. Prediction of mammalian microRNA targets. Cell. 115:787–798 10.1016/S0092-8674(03)01018-3 [DOI] [PubMed] [Google Scholar]
- Li X., Barkho B.Z., Luo Y., Smrt R.D., Santistevan N.J., Liu C., Kuwabara T., Gage F.H., Zhao X. 2008. Epigenetic regulation of the stem cell mitogen Fgf-2 by Mbd1 in adult neural stem/progenitor cells. J. Biol. Chem. 283:27644–27652 10.1074/jbc.M804899200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim D.A., Huang Y.C., Swigut T., Mirick A.L., Garcia-Verdugo J.M., Wysocka J., Ernst P., Alvarez-Buylla A. 2009. Chromatin remodelling factor Mll1 is essential for neurogenesis from postnatal neural stem cells. Nature. 458:529–533 10.1038/nature07726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 25:402–408 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Ma D.K., Jang M.H., Guo J.U., Kitabatake Y., Chang M.L., Pow-Anpongkul N., Flavell R.A., Lu B., Ming G.L., Song H. 2009. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 323:1074–1077 10.1126/science.1166859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming G.L., Song H. 2005. Adult neurogenesis in the mammalian central nervous system. Annu. Rev. Neurosci. 28:223–250 10.1146/annurev.neuro.28.051804.101459 [DOI] [PubMed] [Google Scholar]
- O’Carroll D., Erhardt S., Pagani M., Barton S.C., Surani M.A., Jenuwein T. 2001. The polycomb-group gene Ezh2 is required for early mouse development. Mol. Cell. Biol. 21:4330–4336 10.1128/MCB.21.13.4330-4336.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddison P.J., Caudy A.A., Bernstein E., Hannon G.J., Conklin D.S. 2002. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 16:948–958 10.1101/gad.981002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazian M.D., Zoghbi H.Y. 2002. Rett syndrome and MeCP2: linking epigenetics and neuronal function. Am. J. Hum. Genet. 71:1259–1272 10.1086/345360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., Liu Y., Hsu Y.J., Fujiwara Y., Kim J., Mao X., Yuan G.C., Orkin S.H. 2008. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol. Cell. 32:491–502 10.1016/j.molcel.2008.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sher F., Rössler R., Brouwer N., Balasubramaniyan V., Boddeke E., Copray S. 2008. Differentiation of neural stem cells into oligodendrocytes: involvement of the polycomb group protein Ezh2. Stem Cells. 26:2875–2883 10.1634/stemcells.2008-0121 [DOI] [PubMed] [Google Scholar]
- Shiraki T., Kondo S., Katayama S., Waki K., Kasukawa T., Kawaji H., Kodzius R., Watahiki A., Nakamura M., Arakawa T., et al. 2003. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc. Natl. Acad. Sci. USA. 100:15776–15781 10.1073/pnas.2136655100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silber J., Lim D.A., Petritsch C., Persson A.I., Maunakea A.K., Yu M., Vandenberg S.R., Ginzinger D.G., James C.D., Costello J.F., et al. 2008. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 6:14 10.1186/1741-7015-6-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smrt R.D., Eaves-Egenes J., Barkho B.Z., Santistevan N.J., Zhao C., Aimone J.B., Gage F.H., Zhao X. 2007. Mecp2 deficiency leads to delayed maturation and altered gene expression in hippocampal neurons. Neurobiol. Dis. 27:77–89 10.1016/j.nbd.2007.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh H., Consiglio A., Ray J., Sawai T., D’Amour K.A., Gage F.H. 2007. In vivo fate analysis reveals the multipotent and self-renewal capacities of Sox2+ neural stem cells in the adult hippocampus. Cell Stem Cell. 1:515–528 10.1016/j.stem.2007.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Falls D.L., Li X., Lane T., Luskin M.B. 2007. Antigen-retrieval procedure for bromodeoxyuridine immunolabeling with concurrent labeling of nuclear DNA and antigens damaged by HCl pretreatment. J. Neurosci. 27:5837–5844 10.1523/JNEUROSCI.5048-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao J., Hu K., Chang Q., Wu H., Sherman N.E., Martinowich K., Klose R.J., Schanen C., Jaenisch R., Wang W., Sun Y.E. 2009. Phosphorylation of MeCP2 at Serine 80 regulates its chromatin association and neurological function. Proc. Natl. Acad. Sci. USA. 106:4882–4887 10.1073/pnas.0811648106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taranova O.V., Magness S.T., Fagan B.M., Wu Y., Surzenko N., Hutton S.R., Pevny L.H. 2006. SOX2 is a dose-dependent regulator of retinal neural progenitor competence. Genes Dev. 20:1187–1202 10.1101/gad.1407906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Praag H., Schinder A.F., Christie B.R., Toni N., Palmer T.D., Gage F.H. 2002. Functional neurogenesis in the adult hippocampus. Nature. 415:1030–1034 10.1038/4151030a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Medvid R., Melton C., Jaenisch R., Blelloch R. 2007. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat. Genet. 39:380–385 10.1038/ng1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M., Davies J.J., Wittig D., Oakeley E.J., Haase M., Lam W.L., Schübeler D. 2005. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 37:853–862 10.1038/ng1598 [DOI] [PubMed] [Google Scholar]
- Yekta S., Shih I.H., Bartel D.P. 2004. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 304:594–596 10.1126/science.1097434 [DOI] [PubMed] [Google Scholar]
- Yi R., Poy M.N., Stoffel M., Fuchs E. 2008. A skin microRNA promotes differentiation by repressing ‘stemness’. Nature. 452:225–229 10.1038/nature06642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappone M.V., Galli R., Catena R., Meani N., De Biasi S., Mattei E., Tiveron C., Vescovi A.L., Lovell-Badge R., Ottolenghi S., Nicolis S.K. 2000. Sox2 regulatory sequences direct expression of a (beta)-geo transgene to telencephalic neural stem cells and precursors of the mouse embryo, revealing regionalization of gene expression in CNS stem cells. Development. 127:2367–2382 [DOI] [PubMed] [Google Scholar]
- Zhao X., Ueba T., Christie B.R., Barkho B., McConnell M.J., Nakashima K., Lein E.S., Eadie B.D., Willhoite A.R., Muotri A.R., et al. 2003. Mice lacking methyl-CpG binding protein 1 have deficits in adult neurogenesis and hippocampal function. Proc. Natl. Acad. Sci. USA. 100:6777–6782 10.1073/pnas.1131928100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Teng E.M., Summers R.G., Jr., Ming G.L., Gage F.H. 2006. Distinct morphological stages of dentate granule neuron maturation in the adult mouse hippocampus. J. Neurosci. 26:3–11 10.1523/JNEUROSCI.3648-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Deng W., Gage F.H. 2008. Mechanisms and functional implications of adult neurogenesis. Cell. 132:645–660 10.1016/j.cell.2008.01.033 [DOI] [PubMed] [Google Scholar]
- Zhao C., Sun G., Li S., Shi Y. 2009. A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat. Struct. Mol. Biol. 16:365–371 10.1038/nsmb.1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Hong E.J., Cohen S., Zhao W.N., Ho H.Y., Schmidt L., Chen W.G., Lin Y., Savner E., Griffith E.C., et al. 2006. Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth, and spine maturation. Neuron. 52:255–269 10.1016/j.neuron.2006.09.037 [DOI] [PMC free article] [PubMed] [Google Scholar]