Two domains of centrosomal protein CDK5RAP2, CNN1 and CNN2, link centrosomes to mitotic spindle poles. CNN1 lacking centrosomes are unable to recruit pericentriolar matrix components that mediate attachment to spindle poles.
Abstract
The centrosomal protein, CDK5RAP2, is mutated in primary microcephaly, a neurodevelopmental disorder characterized by reduced brain size. The Drosophila melanogaster homologue of CDK5RAP2, centrosomin (Cnn), maintains the pericentriolar matrix (PCM) around centrioles during mitosis. In this study, we demonstrate a similar role for CDK5RAP2 in vertebrate cells. By disrupting two evolutionarily conserved domains of CDK5RAP2, CNN1 and CNN2, in the avian B cell line DT40, we find that both domains are essential for linking centrosomes to mitotic spindle poles. Although structurally intact, centrosomes lacking the CNN1 domain fail to recruit specific PCM components that mediate attachment to spindle poles. Furthermore, we show that the CNN1 domain enforces cohesion between parental centrioles during interphase and promotes efficient DNA damage–induced G2 cell cycle arrest. Because mitotic spindle positioning, asymmetric centrosome inheritance, and DNA damage signaling have all been implicated in cell fate determination during neurogenesis, our findings provide novel insight into how impaired CDK5RAP2 function could cause premature depletion of neural stem cells and thereby microcephaly.
Introduction
The centrosome consists of a pair of centrioles surrounded by the pericentriolar matrix (PCM). In most animal cells, it is the main microtubule-organizing center. Before mitosis, the centrosome duplicates and matures to facilitate assembly of a bipolar mitotic spindle (Stearns, 2009). Bipolar spindles can form in the absence of centrosomes through a chromatin-dependent spindle assembly pathway (Heald et al., 1996; Carazo-Salas et al., 1999, 2001; Khodjakov et al., 2000; Mahoney et al., 2006; O’Connell and Khodjakov, 2007). However, when present, centrosomes are associated with the spindle poles and act as the dominant site of microtubule nucleation in mitosis (Heald et al., 1997). The positioning of centrosomes within spindle poles and the focusing of microtubule minus ends are under the control of a noncentrosomal large multiprotein complex that comprises the spindle pole–organizing protein NuMA (nuclear mitotic apparatus protein), the minus end–directed microtubule motor cytoplasmic dynein, and its activator complex, dynactin (Gaglio et al., 1995; Merdes et al., 1996, 2000; Gordon et al., 2001; Quintyne and Schroer, 2002; Silk et al., 2009). The centrosomal localization of dynein is facilitated by dynactin (Quintyne and Schroer, 2002), whereas NuMA is transported to the spindle poles in a dynein-dependent manner (Merdes et al., 2000).
CDK5RAP2 is a centrosomal gene mutated in primary microcephaly (Woods et al., 2005), a neurodevelopmental disorder characterized by a decrease in brain size (Bond et al., 2005). CNN1 (CNN motif 1) at the N terminus of CDK5RAP2 is highly conserved. Although fission yeast and Drosophila melanogaster have only one CNN1-containing protein, Mod20 and centrosomin (Cnn), respectively, vertebrates have two: CDK5RAP2 and myomegalin. All of these proteins localize to centrosomes or spindle pole bodies (Heuer et al., 1995; Li and Kaufman, 1996; Verde et al., 2001; Andersen et al., 2003; Sawin et al., 2004). Cnn and its higher eukaryotic homologues also share a C-terminal homology domain, CNN2 (Kao and Megraw, 2009). In flies, Cnn maintains the PCM and astral microtubules around the centrioles during mitosis (Lucas and Raff, 2007). In mammalian cells, CDK5RAP2 is required for cohesion between parental centrioles until centrosome separation at the G2/M transition (Meraldi and Nigg, 2001; Graser et al., 2007).
Microtubule assembly is nucleated by γ-tubulin complexes that reside within the PCM. Mod20, Cnn, and CDK5RAP2 have been shown to recruit γ-tubulin complexes to microtubule-organizing centers, a function dependent on the CNN1 domain (Megraw et al., 1999; Sawin et al., 2004; Fong et al., 2008). In addition, the anchoring of γ-tubulin complexes within the centrosome requires several PCM components, including the two pericentrin-AKAP450 centrosomal targeting (PACT) domain–containing coiled-coil proteins, pericentrin/kendrin and CG-NAP/AKAP450, as well as ninein and NEDD1 (Takahashi et al., 2002; Zimmerman et al., 2004; Delgehyr et al., 2005; Lüders et al., 2006; Haren et al., 2009).
In this study, we address the function of CDK5RAP2 in the vertebrate centrosome. Using reverse genetics in the avian B cell line DT40, we disrupted the CNN1 and CNN2 domains of CDK5RAP2. We demonstrate that both these domains of CDK5RAP2 are essential to maintain a link between the centrosome and the mitotic spindle poles. Our results indicate that the CNN1 domain is critical for the centrosomal recruitment of both AKAP450 and the dynactin subunit p150glued, thereby providing centrosomal anchorage of mitotic spindle pole proteins. Finally, we establish a novel role for CDK5RAP2 in a DNA damage–induced G2 cell cycle checkpoint.
Results
Disruption of the cdk5rap2 locus in DT40 cells
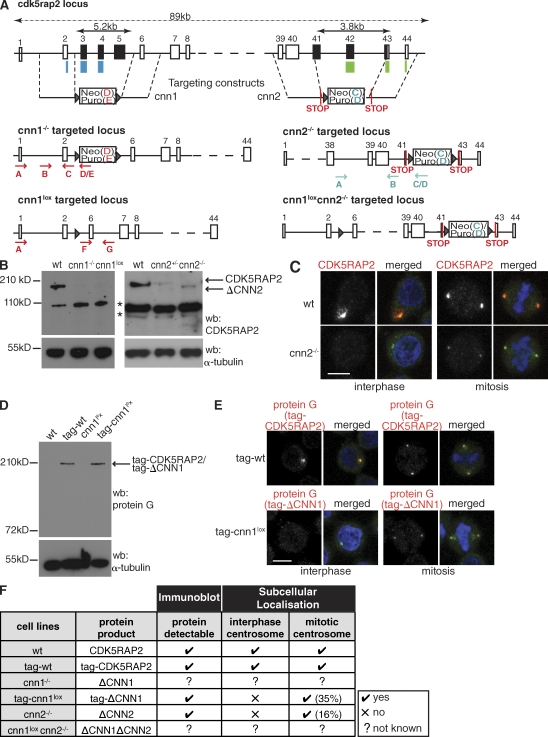
To investigate CDK5RAP2 in a clean genetic background, we assayed CDK5RAP2 function using targeted gene disruption in the chicken B cell line DT40. DT40 cells exhibit a high ratio of homologous versus nonhomologous targeting events, thus making it an ideal system to carry out precise genetic knockouts. cdk5rap2 is a large, 44-exon gene in chicken that spans 89 kb (Fig. 1 A). Gallus gallus and human CDK5RAP2 proteins share significant sequence homology (41% overall identity) and domain organization (Fig. S1 and Fig. S2 A). We hypothesized that the function of CDK5RAP2 could require its evolutionarily conserved CNN1 and CNN2 domains (Li and Kaufman, 1996; Verde et al., 2001; Sawin et al., 2004; Zhang and Megraw, 2007). We disrupted these two domains using the gene-targeting strategies shown in Fig. 1 A. In the case of CNN1 disruption, two homozygous-targeted clones were obtained: the first clone in DT40 cells (cnn1−/−; Fig. 1 A and Fig. S2 B) and the second clone in a DT40 cell line that stably carries inducible Cre recombinase (Cre-cnn1−/−; Fig. S2 C). We transiently introduced Cre recombinase into the cnn1−/− clone to remove the antibiotic-resistance cassettes and thereby derived the cnn1lox cell line (Fig. 1 A and Fig. S2 D). In the case of CNN2 disruption, we obtained multiple homozygous-targeted clones (cnn2−/−; Fig. 1 A and Fig. S2 B). We also created a double-cnn1loxcnn2−/− knockout cell line (Fig. 1 A).
Figure 1.
Disruption of the cdk5rap2 gene in DT40 cells. (A) Schematic representation of the gene-targeting strategies. The CNN1 domain maps to exons 2–4 of cdk5rap2 (blue bars). Exons 3–5 (black) were replaced by antibiotic-resistance cassettes. Antibiotic-resistance cassettes flanked by lox sites (triangles) were removed by Cre recombinase (cnn1lox). The CNN2 domain maps to exons 42–44 of cdk5rap2 (green bars). Exons 41–43 (black) were replaced by antibiotic-resistance cassettes flanked by in-frame STOP codons. The same targeting constructs were introduced into cnn1lox cells to create the cnn1loxcnn2−/− cells. (B) Western blots (wb) of wt and mutant cells. α-Tubulin serves as the loading control. Bands marked by asterisks are nonspecific because in situ tagging of CDK5RAP2 does not give rise to bands of these sizes (see D). (C) Subcellular localization of wt CDK5RAP2 and ΔCNN2. Blue, DNA; red, CDK5RAP2; green, γ-tubulin. (D) Western blot of wt, cnn1lox, tag-wt, and tag-cnn1lox cell extracts. tag-wt and tag-cnn1lox cells contain a protein G–encoding tag in one cdk5rap2 allele. α-Tubulin serves as the loading control. (E) Subcellular localization of tag-CDK5RAP2 and tag-ΔCNN1. Blue, DNA; red, protein G; green, γ-tubulin. (F) Summary of the mutant cdk5rap2 alleles generated. Not known means that these truncated protein products cannot be tracked because of disrupted antibody recognition sites. Percentages in brackets refer to the amount of protein at the centrosome relative to wt. Bars, 5 µm.
On Western blots, CDK5RAP2 antibody recognized a protein of ∼200 kD in wild-type (wt) DT40 cells (Fig. 1 B). Immunostaining revealed that CDK5RAP2 was centrosomal throughout the cell cycle in wt cells and that we could detect a diffuse staining in the proximity of the centrosome during interphase, which is reminiscent of the Golgi apparatus (Fig. 1 C). In cnn1−/− and cnn1lox cells, no CDK5RAP2 protein was detectable by Western blotting or immunostaining (Fig. 1 B). In cnn2−/− cells, a truncated protein product (ΔCNN2) was detectable by Western blotting that was absent from interphase centrosomes. However, centrosomal ΔCNN2 signal was present on separated centrosomes before nuclear envelope breakdown (NEBD; Fig. S2 E), and the protein was detectable in mitotic centrosomes (16 ± 9% of wt levels; fluorescent intensity was measured in 18 centrosomes; Fig. 1, B and C). Because the targeted region in cnn1−/− and cnn1lox cells overlaps with the recognition site of our antibody, this antibody could fail to detect a truncated protein lacking the CNN1 domain. Thus, we introduced a protein G–encoding tag (GS-TAP; Fig. S2 F) in frame into a single allele of the cdk5rap2 gene in cnn1lox (tag-cnn1lox) and wt (tag-wt) cells (Bürckstümmer et al., 2006). The protein G moiety allowed detection by anti–protein G antibody. Western blots of tag-wt and tag-cnn1lox cell extracts revealed a prominent band of 210 kD in tag-wt cells and the presence of a protein product (tag-ΔCNN1) in tag-cnn1lox cells (Fig. 1 D). In immunostaining, anti–protein G antibodies stained both interphase and mitotic centrosomes in tag-wt cells. In the tag-cnn1lox cell line, tag-ΔCNN1 protein was present in mitotic centrosomes (Fig. 1 E) corresponding to ∼35 ± 19% of tag-CDK5RAP2 levels (fluorescent intensity was measured in 42 centrosomes). Although we could not detect tag-ΔCNN1 protein in most interphase cells, a weak centrosomal signal was visible on separated centrosomes in G2 (Fig. S2 E). In summary, both cnn1lox and cnn2−/− cells express truncated protein products (Fig. 1 F) and therefore represent powerful tools to study the respective roles of the CNN1 and CNN2 domains.
CDK5RAP2 connects centrosomes to mitotic spindle poles
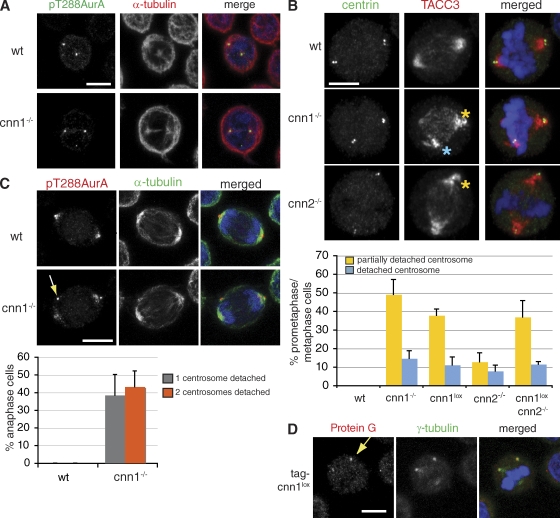
We examined the morphology of the mitotic spindles in our mutants. In prophase, centrosome-associated microtubule asters appeared indistinguishable between wt and mutant cells (Fig. 2 A). However, in prometaphase, centrioles failed to colocalize with spindle poles in cnn1−/− and cnn2−/− cells, indicating that mutant centrosomes were not tightly associated with spindle poles (Fig. 2 B). Partial and complete detachment of centrosomes (Fig. 2 B) from the mitotic spindle poles occurred in both cnn1−/− and cnn2−/− cells. cnn1loxcnn2−/− and Cre-cnn1−/− cells displayed centrosome phenotypes identical to those seen in cnn1−/− cells (Fig. S2 G). To determine whether abnormal centrosome attachment affects chromosome alignment, mitotic cells with different centrosome phenotypes were scored for the presence of the spindle assembly checkpoint component BubR1 (Taylor et al., 1998; Chan et al., 1999). Our data revealed an inverse correlation between centrosome abnormalities and chromosome congression (percentage of BubR1-positive mitotic cells with normal, partially detached, and fully detached centrosomes were 43%, 86%, and 100%, respectively; 165 mitotic cells were scored). Centrosome detachment worsened during mitosis in mutant cells, and by anaphase, almost half of cnn1−/− spindles had lost both of their centrosomes (Fig. 2 C). The presence of detached centrosomes in telophase raised the question of whether centrosomes are unevenly segregated into daughter cells. We could not find individual cnn1−/− cells that contained no centrosomes, but we noted a slight increase in both centrosome number (wt, 0.3 ± 0.4%; cnn1−/− cells with greater than two centrosomes, 2.7 ± 0.8%; n = 5; 100 cells were scored per experiment) and the frequency of multipolar spindles (wt, 4.6 ± 3%; cnn1−/−, 10 ± 3%; n = 6; 100 cells were scored per experiment). Cells with multiple centrosomes could arise from centrosome missegregation or failed mitosis and possible tetraploidization. The percentage of cells with multiple centrosomes and spindle poles remained constant over several passages, suggesting that centrosome missegregation is a rare event or that such cells are eliminated from the population.
Figure 2.
Centrosomes detach from mitotic spindle poles in cnn1−/− and cnn2−/− cells. (A) wt and cnn1−/− cells are indistinguishable in prophase. Blue, DNA; green, pT288Aur-A; red, α-tubulin. (B) In wt cells, anti–centrin-3 antibody staining colocalizes with spindle pole regions marked by anti-TACC3 antibody (top). In mutant cells, centrosomes partially (yellow asterisks) or fully (blue asterisk) detach from spindle poles. Blue, DNA; red, TACC3; green, centrin-3. The graph shows quantification of the centrosome phenotypes seen in prometaphase/metaphase cells. (C) In anaphase cnn1−/− cells, pT288Aur-A staining is located near the cortex away from the spindle poles (arrow). Blue, DNA; green, α-tubulin; red, pT288Aur-A. The graph shows quantification of centrosome phenotypes in anaphase cells. (D) Centrosomal levels of tag-ΔCNN1 protein do not correlate with centrosome detachment in tag-cnn1lox cells. Arrow marks a detached centrosome. Blue, DNA; green, γ-tubulin; red, protein G. n = 4; 150 cells per experiment. Error bars represent SD. Bars, 5 µm.
Truncated forms of CDK5RAP2, tag-ΔCNN1, and ΔCNN2 associated with detached centrosomes but not with spindle poles (Figs. 1 E and 2 D). Levels of tag-ΔCNN1 at the centrosome did not correlate with centrosome detachment, arguing that this phenotype is not caused by an overall decrease in CDK5RAP2 protein levels (Fig. 2 D). To confirm that the mitotic defects in cnn1−/− cells are linked to the disruption of the cdk5rap2 gene, we transfected cnn1lox cells with a construct encoding Flag-tagged, full-length human CDK5RAP2 (Flag-FL). Complementation of the cnn1lox cell line with a stably integrated Flag-FL rescued the detached centrosome phenotype (Fig. S3).
Centrosome detachment is a dynamic and reversible event
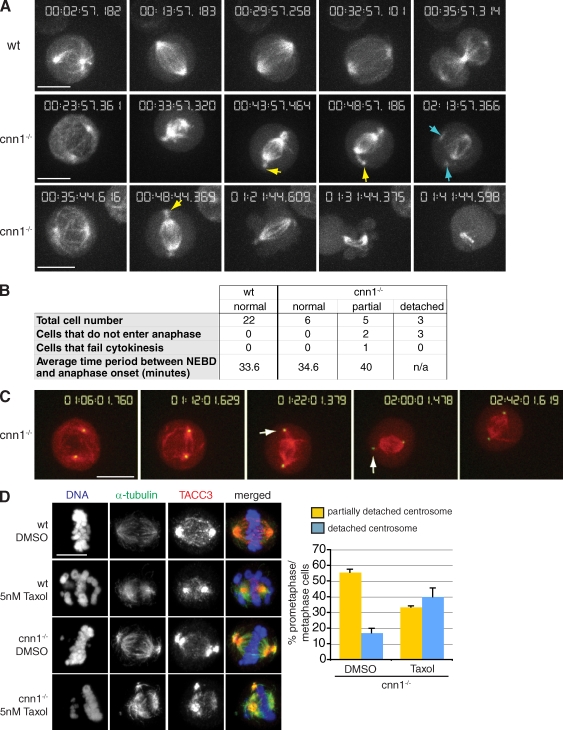
Because the centrosome detachment phenotype was more severe in cnn1−/− than in cnn2−/− cells, we focused our detailed analysis on cnn1−/− cells. To study the dynamics of centrosome detachment, time-lapse imaging was performed on cnn1−/− cells transiently expressing GFP–α-tubulin (GFP-tubulin). After NEBD, cnn1−/− cells were able to build a bipolar spindle with similar efficiency to wt (Videos 1–3). However, within a few minutes, a small aster would detach from one or both spindle poles (Fig. 3 A). Based on the striking similarities between spindle structures in live and fixed cells, we made the assumption that these small asters correspond to centrosomes. Bipolar spindle formation occurred simultaneously (within the same time point) in three out of eight cells and preceded centrosome detachment by 1–4 min in four cells and by 26 min in one cell. When fully detached from spindle poles, centrosomes moved rapidly around the cortex while still nucleating microtubules (Fig. 3 A, middle; and Video 2). Centrosomes were observed to detach and reattach to spindle poles, indicating that detachment is a dynamic and reversible event. cnn1−/− cells with normal centrosome behavior progressed through mitosis with similar timing to wt cells, whereas cnn1−/− cells with centrosome detachment phenotypes took longer to initiate anaphase (Fig. 3 B). Centrosome detachment was not caused by extended mitotic arrest in the mutants, as detachment became apparent within 9 min after NEBD in seven out of eight cells and after 28 min in one cell. Finally, to visualize microtubules and centrosomes simultaneously, we cotransfected cnn1−/− cells with mCherry–α-tubulin and a GFP fusion of PACT, the centrosomal targeting domain of AKAP450 (Gillingham and Munro, 2000). The GFP-PACT signal detached from the mitotic spindle poles as soon as a bipolar spindle became visible (Fig. 3 C and Video 4).
Figure 3.
Centrosome detachment is dynamic and reversible in cnn1−/− cells. (A) Stills from time-lapse videos of wt (Video 1) and cnn1−/− (Videos 2 and 3) cells transfected with GFP-tubulin. Note that partially detached centrosomes (yellow arrows) appear soon after NEBD and precede the detached centrosome phenotype (blue arrows). (bottom) The cell shown initiates anaphase but fails to go through cytokinesis. (B) Summary of results from A. For cnn1−/− cells, mitotic timing and outcome are shown according to centrosome phenotypes. The criteria for classification were the following: partial, cells that developed partially detached centrosomes at any point during filming; detached, cells with at least one fully detached centrosome. The three cnn1−/− cells with detached centrosomes were followed for an average of 105 min after NEBD, but they failed to initiate anaphase during filming. (C) Stills from time-lapse video of cnn1−/− cells (Video 4) transfected with mCherry-tubulin (red) and GFP-PACT (green). (D) Low dose taxol treatment of wt and cnn1−/− cells. Blue, DNA; red, TACC3; green, α-tubulin. Centrosome phenotypes were scored in fixed cells. n = 2; at least 150 cells were counted per experiment. Error bars represent SD. Bars, 5 µm.
CDK5RAP2 has been reported to regulate microtubule behavior via its interaction with the microtubule plus end–binding protein EB1 (Fong et al., 2008, 2009). Therefore, we tested whether abnormal microtubule dynamics contributed to centrosome detachment in cnn1−/− cells. Microtubule dynamic instability was suppressed with a low dose of taxol (Jordan et al., 1993; Derry et al., 1998). 2 h treatment of cnn1−/− cells with 5 nM taxol interfered with chromosome congression to the metaphase plate without inducing multipolarity (Fig. 3 D). 5 nM taxol increased the proportion of cnn1−/− cells with fully detached centrosomes (Fig. 3 D). Thus, dynamic microtubules are not essential for centrosome detachment, but instead, they may promote centrosome reattachment to spindle poles.
Mitotic spindle pole organization and centrosome structure are intact in cnn1−/− cells
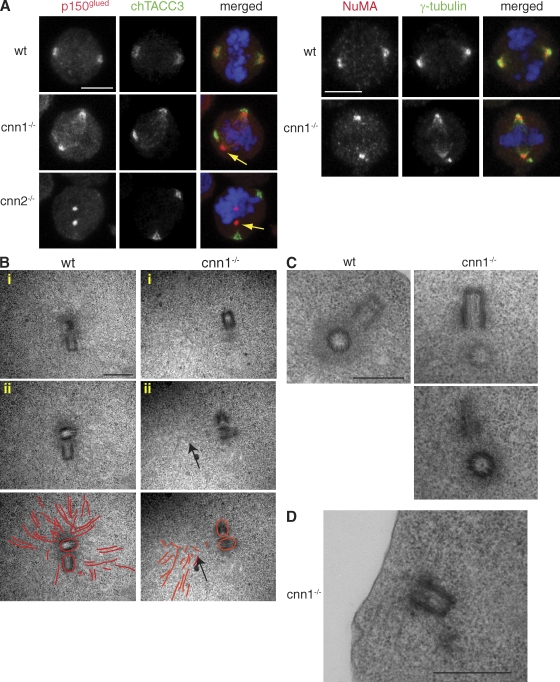
Because the connection between centrosomes and spindle poles is lost in cnn1−/− cells, we hypothesized that spindle pole organization or centrosome structure could be defective in these mutants. Spindle microtubules remain focused in cnn1−/− cells despite centrosome detachment (Fig. 3 C). To see whether spindle pole organization is intact in cnn1−/− cells, we examined the localization of dynactin and NuMA. In both wt and cnn1−/− cells, the dynactin subunit p150glued and NuMA localized in a crescent shape at the spindle poles (Fig. 4 A). A weak NuMA signal could also be detected in the detached centrosome, but we were unable to visualize the centrosomal fraction of p150glued in DT40 cells. Nonetheless, CDK5RAP2 is dispensable for the spindle pole localization and the microtubule-focusing activities of the NuMA–dynein–dynactin complex.
Figure 4.
Mitotic spindle pole organization and centrosome structure are normal in cnn1−/− cells. (A) Spindle pole–organizing proteins, p150glued (left), and NuMA (right) localize normally in cnn1−/− and cnn2−/− cells even when centrosomes fully detach (yellow arrows). (left) Blue, DNA; red, p150; green, TACC3. (right) Blue, DNA; red, NuMA; green, γ-tubulin. Bars, 5 µm. (B) Transmission electron micrographs of serially sectioned prometaphase/metaphase cells. Two sections (i and ii) are shown for a single wt (left) and cnn1−/− cell (right). (bottom) Microtubules are highlighted in red to aid visualization. Higher magnification of electron micrographs and respective images of whole cells are shown in Fig. S3. Although a large number of microtubules focus in the wt centrosome, microtubules focus outside of the cnn1−/− centrosome (arrows). Note that these are likely to be spindle microtubules, as they occupy a position between the centrosomes and the chromosomes (see whole-field view in Fig. S3 B). (C) Both wt and cnn1−/− centrioles are surrounded by an electron-dense matrix. (D) This cnn1−/− centrosome is close to the cortex and associates with microtubule bundles. Bars, 500 nm.
We investigated whether aberrant centrosome structure could underlie the centrosome detachment phenotype of cnn1−/− cells. Transmission electron microscopy on serially sectioned mitotic cnn1−/− cells revealed normal-looking, orthogonal centriole pairs (Fig. 4, B and C; and Fig. S4). An electron-dense area was visible around the centrioles in both wt and cnn1−/− centrioles, suggesting that PCM was recruited by cnn1−/− centrioles. One cnn1−/− centrosome was located near the cortex and associated with microtubules that were running along the cortex (Fig. 4 D). This pattern looked strikingly similar to the detached centrosome phenotype seen in immunofluorescence.
CDK5RAP2 is critical for the centrosomal localization of AKAP450 and p150glued/dynactin
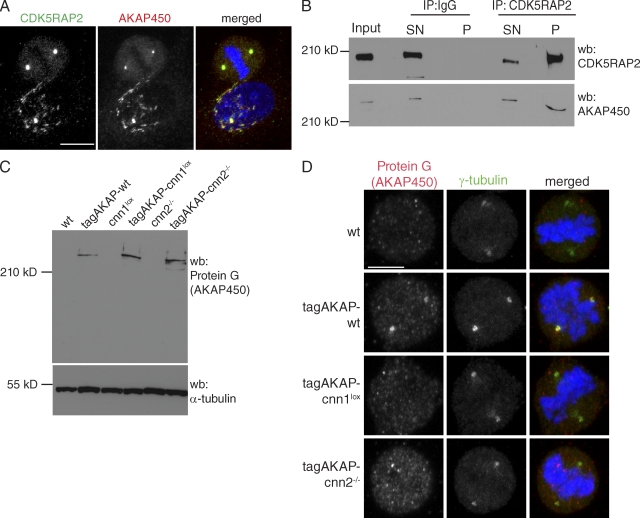
We noticed in human cells that CDK5RAP2 and the centrosomal scaffolding protein AKAP450 colocalized at the centrosome and the Golgi (Fig. 5 A; Keryer et al., 2003; Rivero et al., 2009). This reflects a molecular interaction because CDK5RAP2 antibody coimmunoprecipitated endogenous AKAP450 protein from HeLa cell extracts (Fig. 5 B). To see whether AKAP450 localization was perturbed in cnn1−/− cells, we introduced a GS-TAP tag in frame into the akap450 locus of wt, cnn1lox, and cnn2−/− cells (tagAKAP-wt, tagAKAP-cnn1lox, and tagAKAP-cnn2−/− cells; Fig. S5 A). This was necessary because mammalian AKAP450 antibodies did not cross react with the chicken protein despite extensive homology. Anti–protein G antibodies revealed a protein product (tag-AKAP450) of >210 kD in all three cell lines on Western blots (Fig. 5 C). Anti–protein G antibodies stained mitotic centrosomes in tagAKAP-wt but not in tagAKAP-cnn1lox and tagAKAP-cnn2−/− cells (Fig. 5 D and Fig. S5 B). Prominent centrosomal staining was visible in interphase tagAKAP-wt and tagAKAP-cnn2−/− cells along with a signal reminiscent of the Golgi (Fig. S5 B). However, the same antibody gave a much weaker and highly variable staining in interphase tagAKAP-cnn1lox cells (Fig. S5 B). When fused to GFP, the centrosomal targeting domain of AKAP450, PACT, was present in the centrosome of cnn1−/− cells (Fig. 3 C). Thus, the PACT domain in isolation is recruited to centrosomes independently of CDK5RAP2, but CDK5RAP2 mediates the centrosomal localization of the full-length AKAP450 protein.
Figure 5.
CDK5RAP2 interacts with and targets AKAP450 to the centrosome. (A) CDK5RAP2 colocalizes with AKAP450 in HeLa cells. Blue, DNA; red, AKAP450; green, CDK5RAP2. Bar, 10 µm. (B) CDK5RAP2 coimmunoprecipitates with AKAP450 from HeLa cell extracts. A rabbit IgG mix was used as negative control. Input lane shows cytoplasmic extract before immunoprecipitation (IP). Input and supernatant correspond to 10% of the pellet. CDK5RAP2 antibody immunoprecipitated 26% of total CDK5RAP2 and 40% of total AKAP450 protein. The extent of CDK5RAP2 immunoprecipitation may be underestimated because the CDK5RAP2 signal is saturated. SN, supernatant; P, pellet. (C) Western blots (wb) of wt, cnn1lox, cnn2−/−, tagAKAP-wt tagAKAP-cnn1lox, and tagAKAP-cnn2−/− cell extracts. α-Tubulin serves as the loading control. (D) Anti–protein G antibody stains the mitotic centrosomes of tagAKAP-wt but not of tagAKAP-cnn1lox and tagAKAP-cnn2−/− cells. Blue, DNA; red, protein G (AKAP450); green, γ-tubulin. Bar, 5 µm.
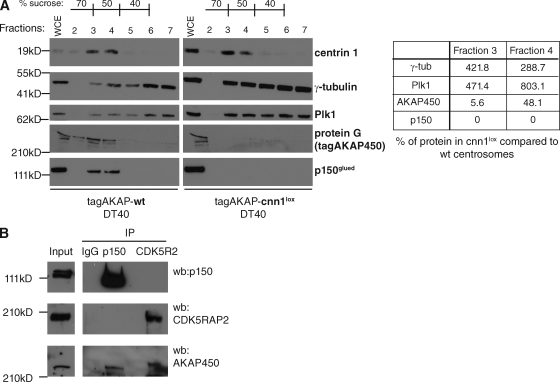
Indeed, levels of AKAP450 were substantially reduced in CNN1-deficient centrosomes purified from nocodazole-arrested cells (79.5% of wt and 80% of cnn1−/− cells were in G2/M by flow cytometry; Fig. 6 A). Because PCM components could be affected by CNN1 deficiency, centrin signal in fractions 3 and 4 was used for normalization. In addition to a decrease in AKAP450 levels, the p150glued subunit of dynactin was absent from CNN1-deficient centrosomes (Fig. 6 A). AKAP450 has been previously shown to interact with p150glued (Kim et al., 2007), and this interaction is maintained during mitosis (Fig. 6 B). Although CDK5RAP2 did not coimmunoprecipitate with p150glued (Fig. 6 B), our results raise the possibility that AKAP450 could act as a receptor for dynactin in the mitotic PCM, thereby establishing a stable link between spindle poles and the centrosome.
Figure 6.
The CNN1 domain is essential for targeting AKAP450 and p150glued to the centrosome. (A) Protein fractions of centrosomes purified from nocodazole-arrested tagAKAP-wt (left) or tagAKAP-cnn1lox (right) cells were immunoblotted with antibodies against centrin-1 and specific PCM components. Signal intensities in fractions 3 and 4 were normalized against the centrin signal in the same fraction. Table shows the percentage of each PCM component in tagAKAP-cnn1lox centrosomal fractions compared with tagAKAP-wt fractions. Whole cell extract (WCE) represents 0.15% of total cell extract; 50% of final centrosome pellet was loaded in each fraction. (B) AKAP450 coimmunoprecipitates with p150glued and CDK5RAP2 (CDK5R2) from mitotic extracts of nocodazole-arrested HeLa cells. A rabbit IgG mix was used as negative control. Input lane shows cytoplasmic extract before immunoprecipitation (IP). wb, Western blotting.
The CNN1 domain of CDK5RAP2 is dispensable for microtubule nucleation and centrosome maturation
AKAP450 and CDK5RAP2 have been implicated in anchoring γ-tubulin complexes within the centrosome (Takahashi et al., 2002; Fong et al., 2008). Because the γ-TuRC–binding site of CDK5RAP2 has been mapped to the CNN1 domain, we expected γ-tubulin recruitment to be perturbed in cnn1−/− centrosomes (Fig. S1); Fong et al., 2008). However, we could not detect any abnormalities in γ-tubulin localization in interphase cnn1−/− cells (Fig. S6 A). In G2 and mitosis, the mean fluorescent signal of γ-tubulin was similar between wt and mutant centrosomes, whereas the total volume of centrosomal γ-tubulin staining was slightly reduced in cnn1−/− centrosomes (Fig. S6 A). Because γ-tubulin is a key factor in centrosomal microtubule nucleation, we evaluated the nucleation ability of cnn1−/− centrosomes (Raynaud-Messina and Merdes, 2007). Microtubule recovery from nocodazole-induced depolymerization was indistinguishable between mitotic wt and cnn1−/− centrosomes, indicating that cnn1−/− centrosomes are not defective in microtubule nucleation (Fig. S6 B). Furthermore, γ-tubulin was highly enriched in CNN1-deficient centrosomes purified from a mitotic-enriched cell population (Fig. 6 A). Perturbations in centrosomal γ-tubulin levels are therefore unlikely to explain the centrosome detachment phenotype. Finally, we also assayed the localization of ninein, a centrosomal protein required for microtubule anchoring and nucleation during interphase (Delgehyr et al., 2005), but again, it was unperturbed in cnn1−/− centrosomes (Fig. S6 C).
In Drosophila somatic cells, Cnn is essential for centrosome maturation and is codependent for its centrosomal localization with polo kinase (Dobbelaere et al., 2008). Immunoblotting of purified centrosomes and immunostaining of mitotic centrosomes with an antibody against polo-like kinase 1, the vertebrate orthologue of polo, failed to reveal a difference between wt and cnn1−/− cells (Fig. 6 A and Fig. S6 D). The localization of aurora A, a protein kinase involved in centrosome maturation and separation (Barr and Gergely, 2007), was also unperturbed in cnn1−/− centrosomes (unpublished data). Moreover, aurora A kinase autophosphorylates its Thr 288 residue, and an antibody recognizing this phospho residue colocalized with cnn1−/− centrosomes (Fig. 2, A and C; Barr and Gergely, 2007). Our results indicate that the role of the CNN1 domain in centrosome to spindle pole attachment is independent of centrosome maturation. TACC3 staining often appeared disorganized between partially detached centrosomes and spindle poles (Fig. 2 B), a pattern likely to reflect the presence of microtubule minus ends because TACC proteins concentrate at spindle regions rich in microtubule minus end (Gergely et al., 2000; Cullen and Ohkura, 2001; Barros et al., 2005). The TACC3-binding partner, ch-Tog, is also recruited normally to cnn1−/− centrosomes (Fig. S6 E). Therefore, in contrast to Drosophila, the CNN1 domain is dispensable for the centrosomal targeting of TACC3 and ch-Tog in vertebrates (Zhang and Megraw, 2007).
AKAP450 maintains centrosome cohesion during interphase
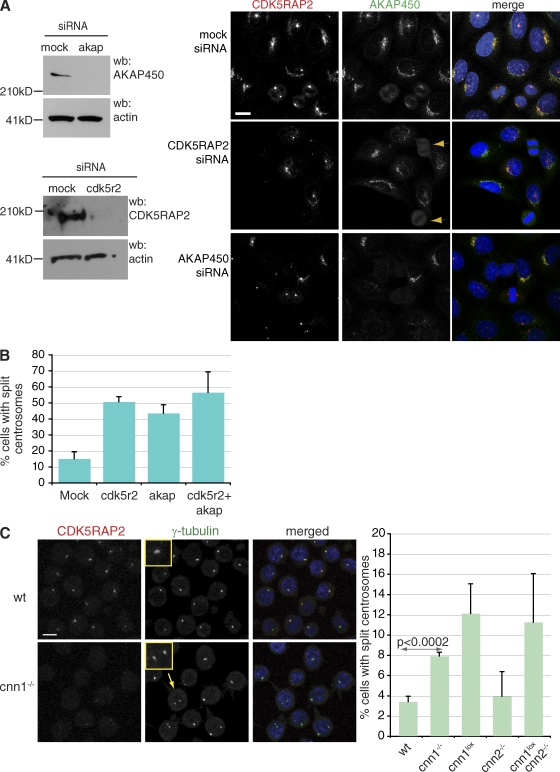
To determine whether CDK5RAP2 and AKAP450 are codependent for their localization in mammalian cells, we depleted the two proteins in HeLa cells by siRNA (Fig. 7 A). Depleting AKAP450 did not perturb the localization of CDK5RAP2 in HeLa cells. In contrast, when CDK5RAP2 levels were depleted, AKAP450 localized normally in interphase but failed to concentrate on mitotic centrosomes (Fig. 7 A). Thus, CDK5RAP2 is required for the mitotic localization of AKAP450 in both DT40 and HeLa cells, but the interphase targeting of AKAP450 seems independent of CDK5RAP2 in mammalian cells. One possibility is that depletion of CDK5RAP2 is incomplete in HeLa cells, and a small amount of CDK5RAP2 can maintain AKAP450 localization. Indeed, in most depleted cells, residual CDK5RAP2 staining is visible in the centrosome (metaphase and anaphase cells; Fig. 7 A), which may also explain the lack of centrosome detachment in these cells. Another possibility is that the second vertebrate Cnn homologue, myomegalin, functions redundantly with CDK5RAP2 in HeLa cells. Although myomegalin is expressed in DT40 cells (unpublished data), we have so far been unable to identify a CNN1 domain in the G. gallus myomegalin gene using various expressed sequence tags and genomic databases. Thus, in DT40 cells, CDK5RAP2 may be the only CNN1 domain–containing protein.
Figure 7.
AKAP450 is required for centrosome cohesion. (A) Western blots (wb) showing siRNA-mediated depletion of CDK5RAP2 (CDK5R2) and AKAP450 (akap) in HeLa cells. Depletion of CDK5RAP2 does not interfere with the localization of AKAP450 to the Golgi and the centrosome during interphase, but it prevents its accumulation in the mitotic centrosome (arrowheads). Depletion of AKAP450 does not prevent the localization of CDK5RAP2 to the centrosome. Note that the Golgi disperses in AKAP450-depleted cells (Larocca et al., 2004), so CDK5RAP2 cannot accumulate in the Golgi. Blue, DNA; red, CDK5RAP2; green, AKAP450. Bar, 10 µm. (B) Graph shows quantification of centrosome splitting in mock and specific siRNA-treated cells. n = 3; at least 150 cells per experiment. (C) Centrosomes split in cnn1−/− DT40 cells. Anti-CDK5RAP2 antibody does not stain the centrosome in interphase cnn1−/− cells, but centrosomal γ-tubulin signal is comparable between wt and cnn1−/− centrosomes. Insets show close ups of normal (wt) and split (cnn1−/−) centrosomes. Blue, DNA; red, CDK5RAP2; green, γ-tubulin. Arrow marks the identity of the cell in inset. Bar, 5 µm. Graph shows quantification of centrosome splitting in wt and mutant cells (n = 4; 150 cells per experiment). P-values were calculated by two-tailed unpaired Student’s t test. Error bars represent SD.
We noted that AKAP450-depleted cells displayed loss of cohesion between parental centrioles in interphase (termed centrosome splitting hereafter; Fig. 7 B). The extent of this effect was very similar to that reported in CDK5RAP2-depleted human cells (Graser et al., 2007). Thus, although AKAP450 and CDK5RAP2 may be targeted independently to the interphase centrosome, both proteins must be present for cohesion between parental centrioles (termed centrosome cohesion hereafter). Split centrosomes were also more frequent in cnn1−/− than in wt cells (Fig. 7 C). As this could be caused by either a delay in G2 or loss of cohesion between disengaged centrioles, we costained cells with anti–γ-tubulin and anti-TACC3 antibodies. Like its human counterpart, chicken TACC3 antibody associates with G2 centrosomes (Gergely et al., 2000, 2003). We found no evidence for a G2 delay in cnn1−/− cells, but when scoring TACC3-negative cells, we observed an increase in split centrosomes. Loss of cohesion between centrosomes is specific to the disruption of the CNN1 domain because cnn2−/− cells behaved as wt. How the truncated ΔCNN2 product maintains centrosome cohesion and recruits AKAP450 to interphase centrosomes is an interesting question, as ΔCNN2 product seems absent from interphase centrosomes (Fig. 1 C and Fig. S5 B). Thus, these functions may require only small amounts (below detection level) of ΔCNN2. Alternatively, it may be during mitosis when the CNN1 domain of CDK5RAP2 establishes centrosome cohesion and primes the centrosome for subsequent AKAP450 recruitment. The extent of centrosome splitting was less prominent in DT40 than in HeLa cells, representing a 2.5-fold instead of a 3.5-fold increase over background, but the small cytoplasmic volume of B cells may limit the distances split centrosomes can travel.
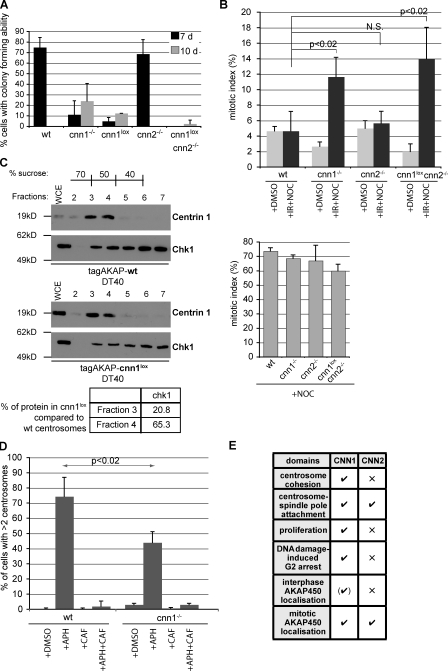
The CNN1 domain is essential for normal proliferation and clonogenic potential
We tested the capacity of individual cells to give rise to clones (i.e., clonogenic potential) by seeding single cells into 96-well plates and recording the percentage of cells that produced visible clones after 7 or 10 d (Fig. 8 A). cnn1−/−, cnn1lox, and cnn1loxcnn2−/− cell lines displayed reduced clonogenic potential. This phenotype was not caused by impairment in cell-to-cell attachment because we did not detect floating cells in the wells. In contrast, wt and cnn2−/− cells had similar clonogenic potential. Time-lapse differential interference contrast microscopy confirmed a proliferation defect in cnn1−/− cells. When followed over a 26-h period, we observed fewer cell divisions in cnn1−/− cells (42% of wt and 17% of cnn1−/− cells divided twice in 26 h). Moreover, cnn1−/− cells were also more prone to cell death; within 26 h, 11% of cnn1−/− cells died compared with 1% in wt cells (9 out of 77 cnn1−/− cells and 1 out of 92 wt cells). Although we observed mitotic death in some cnn1−/− cells (two out of nine cells), the majority of cells died in interphase (seven out of nine cells), indicating that an increased rate of spontaneous cell death is likely to account for the lower proliferation potential.
Figure 8.
CDK5RAP2 promotes G2 arrest in the presence of DNA damage. (A) Colony-forming ability of cells assayed 7 or 10 d after plating. n = 4; 40 cells per experiment. (B) Mitotic index was determined for wt and gene-disrupted DT40 cells that were incubated in DMSO for 10 h (+DMSO) or first irradiated with 20 Gy and then incubated in nocodazole for 10 h (+IR+NOC) or incubated in nocodazole for 10 h without irradiation (+NOC; bottom graph). n = 3; minimum of 3,000 cells was scored per condition per experiment for each genotype. P-values were calculated by two-tailed unpaired Student’s t test. (C) Levels of Chk1 protein are reduced in CNN1-deficient centrosomes. Protein fractions containing centrosomes purified from tagAKAP-wt or tagAKAP-cnn1lox cells (same purification as in Fig. 6 A) were immunoblotted with antibodies against centrin-1 and Chk1 kinase. Signal intensities of Chk1 in fractions 3 and 4 were normalized against the centrin signal in the same fraction. Table shows the percentage of Chk1 in tagAKAP-cnn1lox compared with tagAKAP-wt centrosomal fractions. WCE, whole cell extract. (D) 16 h treatment with aphidicolin (+APH) induces centrosome overduplication in DT40 cells. Caffeine alone (+CAF) or with aphidicolin (+APH+CAF) does not cause centrosome overduplication. Centrosome number was determined using γ-tubulin staining. Note that cnn1−/− cells contain slightly elevated centrosome numbers under all conditions. n = 3; at least 150 cells were scored per condition per experiment for each genotype. P-value was calculated by two-tailed unpaired Student’s t test. (E) Summary of the respective roles of the CNN1 and CNN2 domains in vertebrate cells. √, essential; ×, dispensable. Error bars represent SD.
The CNN1 domain is required for effective DNA damage–induced G2 arrest
Despite both cnn1−/− and cnn2−/− cells displaying abnormal centrosome to spindle pole attachments, only cnn1−/− cells are growth impaired. Thus, the mitotic phenotype cannot account solely for this impairment. A decrease in proliferation rate has been observed in knockouts of cell cycle checkpoint genes such as chk1 in DT40 cells (Zachos et al., 2003). Several centrosomal components have been reported to participate in maintaining G2 arrest in response to DNA-damaging agents (Fletcher and Muschel, 2006). To address whether CDK5RAP2 plays a role in this process, we irradiated asynchronous DT40 cells. The irradiation dose of 20 Gy causes extensive DNA damage in DT40 cells, but as these cells are deficient for p53, they cycle through S phase and accumulate in G2 phase in a Chk1 kinase–dependent manner (Takao et al., 1999; Zachos et al., 2003). After irradiation, cells were incubated in nocodazole for 10 h to trap cells that entered mitosis after DNA damage. The mitotic index of these cells is therefore in inverse relation with their ability to arrest in G2. Consistent with a tight cell cycle arrest, mitotic indices of wt and cnn2−/− cells did not increase after irradiation (Fig. 8 B). In marked contrast, such an increase was detected in cnn1−/− and cnn1loxcnn2−/− cells, which is indicative of an impaired G2 checkpoint (Fig. 8 B).
Early mitotic events rely on the cdc25 phosphatase–dependent activation of cyclin B–Cdk1 at the centrosome (De Souza et al., 2000; Jackman et al., 2003). Chk1 kinase at the centrosome mediates the inhibition of cdc25 phosphatases to prevent premature mitotic entry during unperturbed cell cycles as well as in the presence of DNA damage (Krämer et al., 2004; Tibelius et al., 2009). Therefore, we asked whether levels of centrosomal Chk1 were affected in cnn1−/− DT40 cells. Anti-Chk1 antibody did not recognize mitotic centrosomes and gave a very weak and variable staining in interphase DT40 cells, which we were unable to use for quantification (unpublished data). Centrosomes purified from mutant cells contained less Chk1 than those from wt cells (Fig. 8 C). These centrosomes were isolated from a nocodazole-arrested population (same experiment as in Fig. 6 A) in which 79.5% of wt and 80% of cnn1−/− cells were in G2/M based on flow cytometry. Thus, at least 20% of purified centrosomes originate from cells in G1 or S phase. Because we cannot detect Chk1 in mitotic centrosomes, it is likely that the signal in purified centrosomes in Fig. 8 C is mostly contributed by interphase centrosomes. Centrosome overduplication requires the accumulation of Chk1 kinase in the centrosome and intact ataxia telangiectasia mutated (ATM)/ATM- and Rad3-related (ATR) kinase activities (Dodson et al., 2004; Bourke et al., 2007; Löffler et al., 2007). DT40 cells are known to overduplicate their centrosomes in the presence of DNA replication fork–stalling agents such as aphidicolin (Dodson et al., 2004). After a 16-h aphidicolin treatment, 74% of wt DT40 cells contained two or more centrosomes, but, consistent with lower levels of centrosomal Chk1, this value was only 44% in cnn1−/− cells (Fig. 8 D). ATM/ATR kinases mediated overduplication in both wt and cnn1−/− cells because cotreating cells with aphidicolin and caffeine, an inhibitor of ATM and ATR kinase activities, abolished centrosome overduplication. In summary, we show that although the CNN1 domain is required for efficient DNA damage–induced G2 arrest, the CNN2 domain is not, thereby revealing another example of nonoverlapping functions of these two domains (Fig. 8 E).
Discussion
CDK5RAP2 is a critical regulator of the AKAP450–dynactin complex in the centrosome
Our results suggest that CDK5RAP2 enables the centrosome to remain connected to spindle poles in the presence of microtubule-based forces such as the tension generated by kinetochore capture and biorientation. We believe that the molecular interplay between AKAP450 and CDK5RAP2 explains many of the phenotypes of cnn1−/− cells. Loss of AKAP450 from mitotic centrosomes correlates with centrosome detachment defects in cnn1lox and cnn2−/− cells. Because AKAP450 interacts with p150glued (Kim et al., 2007) and the NuMA–dynein–dynactin protein complex is required for centrosome to spindle pole attachment (Merdes et al., 1996; Silk et al., 2009), AKAP450 could connect centrosomes with spindle poles by acting as a PCM-resident receptor for spindle pole–associated dynactin molecules. The absence of p150glued from cnn1lox centrosomes clearly favors this possibility (Fig. 6 A). Similar to their role on the Golgi (Kim et al., 2007; Rivero et al., 2009), dynactin–AKAP450 complexes could also contribute to microtubule anchoring in the mitotic PCM. Although both the CNN1 and CNN2 domains are required for the mitotic recruitment of AKAP450, the weaker centrosome detachment phenotype of cnn2−/− cells indicates that an intact CNN1 domain can partially aid centrosome attachment even when AKAP450 is absent. Intriguingly, the depletion of the centrosomal protein Cep72 has been shown to delocalize centrosomal AKAP450 during mitosis, leading to impaired spindle pole focusing and centrosome detachment from spindle poles (Oshimori et al., 2009). AKAP450 has been shown to be important for centriole duplication in mammalian cells (Keryer et al., 2003), but our data from DT40 cells suggest that this function is likely to be independent of CDK5RAP2 or at least of the CNN1 and CNN2 domains. However, we show that AKAP450, like CDK5RAP2 (Graser et al., 2007), is required for interphase centrosome cohesion in human cells. Our results also reveal an inverse correlation between interphase centrosomal levels of AKAP450 and centrosome cohesion in cnn1−/− and cnn2−/− cells (Fig. 8 E). CDK5RAP2 and AKAP450 could be part of an intercentriolar linker (Graser et al., 2007) or mediate centrosome cohesion by other means, for instance by regulating microtubule anchoring or dynamics (Meraldi and Nigg, 2001; Fong et al., 2009). Interestingly, inhibiting dynactin function has also been found to impair centrosome cohesion (Quintyne and Schroer, 2002). AKAP450, CDK5RAP2, and dynactin could therefore be essential components of a molecular pathway that mediates centrosome cohesion during interphase and centrosome to spindle pole attachment during mitosis.
The role of CDK5RAP2 in DNA damage–induced G2 arrest
In response to DNA damage or replication-blocking agents, eukaryotic cells trigger checkpoint responses that facilitate DNA repair and delay cell cycle progression. The phenotypes observed in cnn1−/− cells, in particular, the inefficient G2 arrest after irradiation, are reminiscent of findings reported in Chk1 knockout DT40 cells (Zachos et al., 2003). However, unlike Chk1 knockout cells, the majority of cnn1−/− cells can still arrest in G2. This is to be expected because cnn1−/− cells exhibit only a reduction instead of a complete loss of centrosomal Chk1 (Fig. 8 C). Nonetheless, CDK5RAP2 facilitates the G2 checkpoint, and its role in this process is likely to involve the recruitment or maintenance of Chk1 at the centrosome. This is a particularly exciting possibility because pericentrin and microcephalin, two centrosomal proteins implicated in primary microcephaly (Jackson et al., 2002; Rauch et al., 2008), have also been found to mediate Chk1 localization to the centrosome (Tibelius et al., 2009). The role of the centrosome in DNA damage signaling could therefore be critical during the rapid proliferation of early neural progenitors, a developmental process profoundly affected by deficiencies in DNA repair (McKinnon, 2009).
Linking the mitotic function of CDK5RAP2 to microcephaly
Why do mutations in cdk5rap2 perturb cell division specifically in the neuroepithelium when CDK5RAP2 is a ubiquitously expressed protein? We cannot exclude that in humans, a CNN1-independent pathway connects spindle poles with centrosomes in all cell types but neural progenitors. However, we favor the explanation that CDK5RAP2 functions redundantly with myomegalin. The CNN1 domain is absent from at least two splicing variants of human myomegalin. Thus, if these variants are expressed in the developing neuroepithelium (or if myomegalin is not expressed at all), CDK5RAP2 may be the only CNN1-containing protein available in the progenitors. In mouse brain, the orientation of the mitotic division plane determines whether apical progenitors undergo self-renewing or neurogenic cell divisions in the neuroepithelium (Farkas and Huttner, 2008). Moreover, asymmetric centrosome inheritance (i.e., old vs. new centrosome) has been found to be decisive in cell fate determination in mouse brain (Wang et al., 2009). Disrupting the link between centrosomes and mitotic spindle poles would not only prevent centrosomal microtubules from positioning the mitotic spindle but would also randomize the inheritance of the old and new centrosome by daughter cells. Therefore, the combined roles of the centrosome in cell fate determination and DNA damage signaling could underlie the pathogenesis of inherited microcephalic syndromes.
Materials and methods
Cell culture and drug treatments
DT40 cells were maintained in suspension in RPMI (Invitrogen) supplemented with 10% FBS, 1% chicken serum, penicillin, streptomycin, and 10−5 M β-mercaptoethanol at 40°C with 5% CO2. HeLa cells were maintained in DME (Invitrogen) supplemented with 10% FBS, penicillin, and streptomycin. Nocodazole (Sigma-Aldrich) was used at 0.5 µg/ml unless otherwise stated. Taxol (paclitaxel; Sigma-Aldrich) was used at 5 nM, whereas RO3306 (Enzo Life Sciences, Inc.) was used at 6 µM.
Generating cnn1−/− DT40 cells
To make the targeting construct, left and right arm homology regions were PCR amplified from DT40 genomic DNA using polymerase (LA; Takara Bio Inc.). The primers used to amplify the left arm were 5′-GATATCACCATGAAGGACTAAGAAAATGTAAGC-3′ and 5′-GGATCCTGAGCCAACTACGTCATGAGTTTC-3′. The primers used to amplify the right arm were 5′-ACTAGTGTGAGTGCCTAAAATGGAATGAAT-3′ and 5′-GCGGCCGCAGAGCTCCTTACTCCACACAGCCT-3′. Homology arms were cloned into pBluescript II SK−, with the left arm between EcoRV and BamHI sites and the right arm between SpeI and NotI sites. Selection markers (neomycin, puromycin, or blasticidin resistance cassettes) were cloned into these vectors between BamHI sites. Two rounds of targeting were performed to disrupt the two alleles. Gene targeting was performed by electroporation using a gene pulsar (Bio-Rad Laboratories). In brief, 60 µg linearized DNA was mixed with 2 × 107 cells in chilled PBS in a 4-mm cuvette and electroporated at 550 V and 25 µF. DT40 or Cre recombinase–expressing DT40 cells were plated into six 96-well plates. After 24 h, antibiotics were added for 7–10 d at the following concentrations: 1.5 mg/ml neomycin (Invitrogen), 0.5 µg/ml puromycin (Acros Organics), and 50 µg/ml blasticidin (Invitrogen). Antibiotic-resistant clones were expanded, and genomic DNA was extracted. Targeted integrations were screened by PCR amplification using the following primers (letters in parentheses correspond to the abbreviations used in Fig. 1 A): neomycin (D), 5′-CTGAATGAACTGCAGGACGAG-3′; puromycin (E), 5′-ACGACCCCATGGCTCCGACCGAAG-3′; and CDK5RAP2CHK (A), 5′-CTGCTGGTATTCCCTGGGATG-3′.
cnn1−/− knockout was generated with neomycin and puromycin cassettes, whereas Cre-cnn1−/− clone was generated with blasticidin and puromycin cassettes. cnn1lox cells were generated by removal of antibiotic-resistance cassettes from cnn1−/− cells. 30 µg pCre recombinase (provided by J.-M. Buerstedde, Institute for Molecular Radiobiology, Neuherberg, Germany) was mixed with 3 × 106 cells in nucleofection solution T (Lonza). Cells were transfected using the nucleofector and recovered in medium. 24 h after transfection, cells were counted and plated at a density of one cell per well of a 96-well plate. Cells were incubated for 1 wk after which colony-containing wells were expanded and analyzed for loss of drug resistance. Those colonies that lost resistance to both puromycin and neomycin were further analyzed by PCR for loss of drug resistance cassettes (Fig. S2 D).
Generating cnn2−/− and cnn1loxcnn2−/− DT40 cells
Primers used to amplify the left homology arm were 5′-ATCGATTGCCCTGCTGCCAGCAG-3′ and 5′-GAATTCTTGCAGCGATCACTGGTCTGGTAC-3′. Primers used to amplify the right homology arm were 5′-ACTAGTAAGGCTAGATGAAACCTGGAAG-3′ and 5′-GCGGCCGCGATAGAGCTGTGAGCAG-3′. Homology arms were cloned into pBluescript II SK−, with the left arm between ClaI and EcoRI and the right arm between SpeI and NotI sites. Selection markers (neomycin or puromycin resistance cassettes) were cloned into these vectors between BamHI sites. The cnn2 knockout constructs were introduced sequentially into wt DT40 cells to generate cnn2−/− or into cnn1lox DT40 cells to generate cnn1loxcnn2−/− cells. Targeting was performed as described in Generating cnn1−/− DT40 cells. Targeted integrations were screened by PCR amplification using the following primers: neomycin (C), 5′-CTGAATGAACTGCAGGACGAG-3′; puromycin (D), 5′-ACGACCCCATGGCTCCGACCGAAG-3′; and Rev_CNN2 (A), 5′-TTGCTTCGGAGCATCTCG-3′.
Generating tag-wt and tag-cnn1lox DT40 cells
Primers used to amplify the left homology arm were 5′-GAATTCGTCAGTTTCTAGTTCCAG-3′ and 5′-TCTAGAGAGAATCCTTTTTTTGCT-3′. Primers used to amplify the right arm were 5′-ACTAGTTGAAGGAGAAAGGTCCAC-3′ and 5′- GCGGCCGCCACTCTGCACATGGAATG-3′.
Homology arms were cloned into a vector carrying the GS-TAP cassette (provided by K.J. Patel, Medical Research Council Laboratory of Molecular Biology, Cambridge, England, UK), with the left arm between EcoRI and XbaI and the right arm between SpeI and NotI sites. Selection markers (neomycin or puromycin resistance cassettes) were cloned into these vectors between BamHI sites. Constructs were transfected into cells as described in Generating cnn1−/− DT40 cells.
Generating tag-AKAP DT40 cells
Primers used to amplify the left homology arm were 5′-GAATTCCCAGCAGCAAACTGTTGC-3′ and 5′-TCTAGATCTTCTCATAGCAGAGTG-3′. Primers used to amplify the right arm were 5′-ACTAGTTCCATCACTGGCAGCGAT-3′ and 5′-GCGGCCGCTTATATTGGAGTTTATTC-3′.
Homology arms were cloned into a vector carrying the GS-TAP cassette (provided by K.J. Patel), with the left arm between EcoRI and XbaI and the right arm between SpeI and NotI sites. Selection markers (neomycin, blasticidin, and puromycin resistance cassettes) were cloned into these vectors between BamHI sites. Constructs were transfected into wt, cnn1lox−, and cnn2−/− cells as described in Generating cnn1−/− DT40 cells.
Plasmid transfection into DT40 cells
For transient transfections of GFP-tubulin (provided by P. Coopman, Centre National de la Recherche Scientifique, Montpellier, France), the nucleofector was used as described in Generating cnn1−/− DT40 cells. Cells were imaged 16–24 h after transfection. For random integration of Flag-tagged Homo sapiens CDK5RAP2 into cnn1lox, 10 µg plasmid was mixed with 107 cells in PBS in a 4-mm cuvette. Cells were electroporated using a gene pulser (Bio-Rad Laboratories) at 250 V and 950 µF. Cells were plated out into three 96-well plates. After 24 h, neomycin selection was added to a final concentration of 1.5 mg/ml. After 7–10 d, drug-resistant colonies were selected and screened for the presence of insertion.
RNA interference and transfection in mammalian cells
Cells were seeded the day before transfection such that they would be 40% confluent on the day of transfection. Cells were washed three times in PBS and put into fresh OPTIMEM (Invitrogen). 15 pmol siRNA was transfected per 24-well using Oligofectamine (Invitrogen) with the volumes and concentrations recommended by the manufacturer. 4 h after transfection, the medium was changed. Cells were processed 72 h after transfection. siRNAs used in this study were CDK5RAP2 (#132391; Applied Biosystems) and AKAP450 (5′-AUCCCUUGCCAGCACAUGATT-3′; based on Larocca et al., 2004).
Antibody generation and primary antibodies
Antibodies were raised in rabbits against bacterially expressed and purified glutathione S-transferase or maltose-binding protein (NEB) fusion proteins that contained amino acids 40–375 of the H. sapiens CDK5RAP2 protein and amino acids 126–442 of G. gallus TACC3. Antibodies were produced by Eurogentec and were subsequently affinity purified against fusion proteins. Additional primary antibodies used in this study were AKAP450 (BD), pT288 aurora A (BD), aurora A (Abcam), BubR1 (provided by W. Earnshaw, Wellcome Trust Centre for Cell Biology, University of Edinburgh, Edinburgh, Scotland, UK), CDK5RAP2 (Bethyl Laboratories, Inc.; used for immunoprecipitations in Figs. 5 and 6), centrin-1 (Sigma-Aldrich), centrin-2 and centrin-3 (Abnova), Chk1 (Sigma-Aldrich), ch-Tog (provided by L. Cassimeris, Lehigh University, Bethlehem, PA; Cassimeris and Morabito, 2004), Flag-M2 (Sigma-Aldrich), GM130 (BD), NuMA (BD and Abcam), p150glued (BD), protein G and protein G–HRP (Abcam), ninein and phospho–histone H3 and Plk1 (Abcam), TGN46 (Abnova), α-tubulin (Dm1α; Sigma-Aldrich), and γ-tubulin (GTU88; Sigma-Aldrich). DNA was detected with Hoechst (Sigma-Aldrich). All antibodies were stored as a 50% glycerol stock.
Immunofluorescence
DT40 cells were spotted onto poly-L-lysine–coated coverslips and left to settle for 10–15 min at 40°C. For visualization of mitotic spindles and centrosomal proteins, cells were fixed in 100% methanol for 5 min at −20°C. For anti-Plk1, -Chk1, and -ninein antibody stainings, DT40 cells were extracted in PBS/1% Triton X-100/0.5% NP-40 for 5 min after methanol fixation. To visualize p150glued in cells (Fig. 4 A), DT40 cells were fixed in 3% paraformaldehyde in PHEM buffer (60 mM Pipes, 25 mM Hepes, 2 mM MgCl2, and 1 mM EGTA, pH 6.9) containing 0.2% Triton X-100 at RT for 20 min followed by further permeabilization with PBS/0.2% Triton X-100. All primary antibodies were diluted 1:1,000 in blocking buffer and incubated with coverslips for 2 h at 37°C or overnight at 4°C. Secondary antibodies conjugated to Alexa Fluor 488, 555, and 637 were diluted 1:1,000 in blocking buffer and incubated with coverslips for 1 h at 37°C. Coverslips were mounted using antifade medium (ProLong Gold; Invitrogen) containing 1.5 µg/ml Hoechst. Cells in Fig. S6 B were first stained with antibodies against phospho–histone H3 and centrin-3 and washed in PBS/0.1% Tween eight times for 30 min per wash. Cells were stained with Dm1α and secondary antibodies and processed as before. For immunofluorescence of HeLa cells, cells were grown on coverslips and were fixed and treated as in Barr and Gergely (2008).
Image acquisition and analysis
For fixed cell analysis, images were acquired as z stacks (0.4-µm steps) using a microscope (Eclipse 90i; Nikon) fitted with a camera (Eclipse C1Si; Nikon). Images were acquired using EZ-C1 software (Nikon). HeLa and DT40 cells were imaged using a 60× oil 1.40 NA (Nikon) and a 100× oil 1.40 NA (Nikon) objective, respectively. Images were imported into Volocity (version 4.0; PerkinElmer) and exported as 2D volume-rendered images into Photoshop (CS3; Adobe). Images were adjusted to use the full range of pixel intensities in Photoshop. All images from a single experiment were treated in exactly the same way. The fluorochromes used in this study were Alexa Fluor 488, 555, and 637 (Invitrogen). DNA was visualized by Hoechst staining.
For time-lapse imaging, DT40 cells were settled onto 60-µm dishes (Ibidi). Cells were imaged in Leibovitz medium (Invitrogen) containing 10% FBS. Imaging was performed with a 100× oil 1.40 NA objective (Nikon). Cells were kept at 40°C in a hit incubation chamber (Tokai). mCherry-tubulin was provided by V. Draviam (University of Cambridge, Cambridge, England, UK) and GFP-PACT was provided by S. Munro (Medical Research Council Laboratory of Molecular Biology). For imaging GFP-tubulin or mCherry-tubulin/GFP-PACT–expressing DT40 cells, we used a spinning-disc confocal system (PerkinElmer) mounted on an inverted microscope (Eclipse TE2000-S; Nikon) equipped with an electron microscopy charge-coupled device digital camera (C9100-13; Hamamatsu Photonics). Images were acquired at 1-min (Fig. 3 A and Videos 1–3) or 2-min (Fig. 3 C and Video 4) intervals as z stacks (1.5-µm steps) using Volocity. 2D volume-rendered image sequences were exported as QuickTime files. For video stills, snapshot images were taken in Volocity and processed as described in the previous paragraph. For differential interference contrast microscopy of DT40 cells, images were acquired on a microscope (Eclipse TE2000-E; Nikon) fitted with a digital sight camera (DS-2MBW; Nikon) using a 60× oil 1.40 NA objective (Nikon) with advanced research software (NIS-Elements; Nikon). Images were acquired every 15 min at 40°C and 5% CO2.
For all quantifications, z-stack images were acquired on a confocal microscope (Nikon) and imported into Volocity. To quantify CDK5RAP2 and protein G levels in the centrosomes of wt, tag-cnn1lox, and cnn2−/− cells (Fig. 1, C, E, and F), γ-tubulin–positive 3D volumes (cut-off value of 1,300; range of 0–4,095) were selected, and the mean signal intensities of CDK5RAP2 (Fig. 1 C) or protein G (Fig. 1 E) were determined across the volume. To quantify α- and γ-tubulin signals in Fig. S6 (A and B), we selected volumes that contained α-tubulin staining intensity over the cut-off value of 1,500 or γ-tubulin staining intensity over the cut-off value of 900, thus defining α- and γ-tubulin–positive volumes. Volumes were converted to micrometers cubed. In the case of α-tubulin, the cut-off value was determined to allow separation of centrosomal asters from chromatin-associated microtubule asters. In the case of γ-tubulin, the cut-off value allowed us to exclude contribution from spindle-bound protein.
Electron microscopy
DT40 cell pellets were fixed in prewarmed 1% glutaraldehyde in PHEM for 1 h at 37°C. Cell pellets were washed in 0.1 M Na cacodylate buffer, pH 7.2, at RT for 5 min. Pellets were postfixed in 1% osmium tetroxide in Na cacodylate buffer for 1 h at RT. Pellets were washed in Na cacodylate buffer two times for 30 min. Pellets were dehydrated in a graded ethanol series from 50, 70, 90, and 3× 100% ethanol. Dehydrated cell pellets were embedded using the Epoxy Embedding kit (Fluka) according to the manufacturer’s instructions.
Microtubule regrowth in DT40 cells
Microtubules were depolymerized by addition of 2 µg/ml nocodazole for 2 h at 40°C. Cells were washed three times in ice-cold PBS (supplemented with 1:1,000 vol DMSO) by centrifuging at 1,100 g for 2 min. Cells were resuspended in 100 µl prewarmed culture medium and spotted onto poly-L-lysine–coated coverslips. Coverslips were incubated at 40°C for 5 min before methanol fixation. Fixed cells were immunostained for centrin-3 and phospho–histone H3 and poststained with Dm1α antibodies.
Taxol treatment of DT40 cells
DT40 cells were treated with 6 µM of the Cdk1 inhibitor RO3306 for 3 h to ensure that cells to be scored for spindle morphology entered mitosis in the presence of taxol. Cells were washed three times in prewarmed culture medium to remove RO3306 and resuspended in fresh medium containing either 5 nM taxol or DMSO. Cells were incubated in taxol or DMSO for 2 h before being fixed and processed for immunofluorescence.
Western blotting and immunoprecipitation
DT40 cell pellets were lysed in hypotonic buffer (10 mM Tris-HCl, pH 7.4, 10 mM KCl, 1.5 mM MgCl2, 10 µM β-mercaptoethanol, and protease inhibitor cocktail [Sigma-Aldrich]) by passing cells through a 19-G needle. Cytoplasmic extracts were obtained by pelleting (16,000 g for 15 min at 4°C) and discarding chromatin. Extracts were separated by SDS-PAGE using 3–8% Tris-acetate NuPAGE gels (Invitrogen) and blotted to nitrocellulose as described previously (Towbin et al., 1979). Blots were incubated with primary antibodies at 0.5 µg/ml final concentration. Secondary antibodies conjugated to HRP (Dako) were diluted 1:2,000. Antibody binding was detected using an ECL system (Lumilight; Roche) according to the manufacturer’s instructions. For immunoprecipitation, HeLa cells were lysed at 4°C in lysis buffer (25 mM Tris-HCl, pH 7.4, 0.5% NP-40, 100 mM NaCl, 5 mM MgCl2, 5 mM NaF, 1 mM DTT, and protease inhibitor cocktail). Chromatin was pelleted and discarded. Antibodies were cross-linked with protein G Dynabeads according to the manufacturer’s instructions (Invitrogen) and were incubated with cell lysates for 2 h at 4°C. Bound protein complexes were eluted using 0.1 M glycine, pH 2.2, and neutralized with Tris-HCl, pH 8.0. Extracts were separated by SDS-PAGE. Quantification of chemiluminescence signal was performed using ImageJ software (National Institutes of Health).
Centrosome purification
Centrosome purification was performed as described previously (Bornens and Moudjou, 1999) with the following modifications. In brief, 8 × 107 tagAKAP-wt and tagAKAP-cnn1lox DT40 cells were arrested in mitosis by incubation with 500 ng/ml nocodazole for 12 h. Cell lysates were spun onto 2 ml 60% sucrose cushion in a rotor (SW40; Beckman Coulter) at 10,000 g for 30 min. The discontinuous sucrose gradient consisted of 1 ml 70% sucrose, 600 µl 50% sucrose, and 600 µl 40% sucrose. After addition of the centrosome-containing supernatant onto this gradient, samples were centrifuged for 2 h at 120,000 g in a rotor (SW55 Ti; Beckman Coulter). 400 µl fractions were collected by punching a small hole in the bottom of the centrifuge tube and collecting droplets. After addition of 10 mM Pipes-KOH, pH 7.2, centrosomes in each fraction were pelleted at 115,000 g in a rotor (MLA 55; Beckman Coulter) in an ultracentrifuge (Beckman Coulter). The supernatant was discarded, and centrosome pellets were resuspended in 40 µl boiling SDS Laemmli buffer.
Cell cycle analysis, colony formation, and ionizing radiation assays
For analysis of cell cycle profiles, samples were centrifuged at 800 g for 3 min, and cell pellets were resuspended in 0.75 ml 70% ethanol and fixed at −20°C overnight. Tubes were topped up with 1× PBS and spun at 800 g for 5 min. The supernatant was removed, and cell pellets were resuspended in 0.5 ml of PBS/0.1% Triton X-100 containing 20 µg/ml propidium iodide (Sigma-Aldrich) and 0.2 mg/ml RNase A (Sigma-Aldrich). Cells were filtered into 5-ml falcon tubes (BD) through the filter cap to remove cell clumps. Samples were analyzed using FACS (Calibur and FlowJo; BD). For colony-formation assays, DT40 cells were diluted and seeded at a density of 40 cells per 96-wells. Cells were incubated at 40°C for 7 or 10 d, and the number of colonies per plate was scored. To assess the efficiency of G2 arrest after DNA damage, 106 cells were either irradiated with 20 Gy using a caesium-137 γ-irradiator and treated with 1 µg/ml nocodazole for 10 h or treated with nocodazole only or DMSO only for 10 h. Cells were fixed in methanol, processed for immunofluorescence, and stained with anti-phospho–histone H3 antibodies and Hoechst. Tiled images of slides were acquired using the iCys system (CompuCyte) coupled to a microscope (IX71; Olympus) fitted with a charge-coupled device camera (Sony). Images were acquired using a 40× air 0.75 NA objective (Olympus). Analysis of phospho–histone H3-positive cells was performed in an automated fashion using the iCys software.
Online supplemental material
Fig. S1 shows alignment of the human and chicken CDK5RAP2 proteins. Fig. S2 shows key controls to prove gene-targeting events. Fig. S3 demonstrates that centrosome detachment in cnn1−/− cells is rescued by complementation with human CDK5RAP2 protein. Fig. S4 shows high magnifications of electron micrographs from Fig. 4 B. Fig. S5 illustrates the targeting strategy for the generation of tag-AKAP cell lines and provides additional localization data. Fig. S6 demonstrates that microtubule nucleation and maturation are not impaired in cnn1−/− cells. Videos 1–3 correspond to Fig. 3 A, and Video 4 corresponds to Fig. 3 C. Video 1 shows mitosis in a wt DT40 cell transfected with GFP–α-tubulin. Videos 2 and 3 show mitosis in a cnn1−/− DT40 cell transfected with GFP–α-tubulin. Video 4 shows mitosis in a cnn1−/− DT40 cell transfected with mCherry-tubulin and GFP-PACT. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200912163/DC1.
Acknowledgments
We thank L. Cassimeris and W. Earnshaw for antibodies and P. Coopman, V. Draviam, S. Munro, and K.J. Patel for plasmids. We thank the Cambridge Research Institute core facilities and especially H. Zecchini for her help. We thank members of our laboratory for helpful suggestions.
F. Gergely is the recipient of a Royal Society University Research Fellowship. A.R. Barr is supported by a Cancer Research UK Ph.D studentship. Research in F. Gergely’s laboratory is funded by Cancer Research UK.
Footnotes
Abbreviations used in this paper:
- ATM
- ataxia telangiectasia mutated
- ATR
- ATM and Rad3 related
- Cnn
- centrosomin
- NEBD
- nuclear envelope breakdown
- NuMA
- nuclear mitotic apparatus protein
- PACT
- pericentrin-AKAP450 centrosomal targeting
- PCM
- pericentriolar matrix
- wt
- wild type
References
- Andersen J.S., Wilkinson C.J., Mayor T., Mortensen P., Nigg E.A., Mann M. 2003. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 426:570–574 10.1038/nature02166 [DOI] [PubMed] [Google Scholar]
- Barr A.R., Gergely F. 2007. Aurora-A: the maker and breaker of spindle poles. J. Cell Sci. 120:2987–2996 10.1242/jcs.013136 [DOI] [PubMed] [Google Scholar]
- Barr A.R., Gergely F. 2008. MCAK-independent functions of ch-Tog/XMAP215 in microtubule plus-end dynamics. Mol. Cell. Biol. 28:7199–7211 10.1128/MCB.01040-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros T.P., Kinoshita K., Hyman A.A., Raff J.W. 2005. Aurora A activates D–TACC–Msps complexes exclusively at centrosomes to stabilize centrosomal microtubules. J. Cell Biol. 170:1039–1046 10.1083/jcb.200504097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond J., Roberts E., Springell K., Lizarraga S.B., Lizarraga S., Scott S., Higgins J., Hampshire D.J., Morrison E.E., Leal G.F., et al. 2005. A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat. Genet. 37:353–355 10.1038/ng1539 [DOI] [PubMed] [Google Scholar]
- Bornens M., Moudjou M. 1999. Studying the composition and function of centrosomes in vertebrates. Methods Cell Biol. 61:13–34 10.1016/S0091-679X(08)61973-1 [DOI] [PubMed] [Google Scholar]
- Bourke E., Dodson H., Merdes A., Cuffe L., Zachos G., Walker M., Gillespie D., Morrison C.G. 2007. DNA damage induces Chk1-dependent centrosome amplification. EMBO Rep. 8:603–609 10.1038/sj.embor.7400962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürckstümmer T., Bennett K.L., Preradovic A., Schütze G., Hantschel O., Superti-Furga G., Bauch A. 2006. An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat. Methods. 3:1013–1019 10.1038/nmeth968 [DOI] [PubMed] [Google Scholar]
- Carazo-Salas R.E., Guarguaglini G., Gruss O.J., Segref A., Karsenti E., Mattaj I.W. 1999. Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature. 400:178–181 10.1038/22133 [DOI] [PubMed] [Google Scholar]
- Carazo-Salas R.E., Gruss O.J., Mattaj I.W., Karsenti E. 2001. Ran-GTP coordinates regulation of microtubule nucleation and dynamics during mitotic-spindle assembly. Nat. Cell Biol. 3:228–234 10.1038/35060009 [DOI] [PubMed] [Google Scholar]
- Cassimeris L., Morabito J. 2004. TOGp, the human homolog of XMAP215/Dis1, is required for centrosome integrity, spindle pole organization, and bipolar spindle assembly. Mol. Biol. Cell. 15:1580–1590 10.1091/mbc.E03-07-0544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan G.K., Jablonski S.A., Sudakin V., Hittle J.C., Yen T.J. 1999. Human Bubr1 is a mitotic checkpoint kinase that monitors CENP-E functions at kinetochores and binds the cyclosome/APC. J. Cell Biol. 146:941–954 10.1083/jcb.146.5.941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen C.F., Ohkura H. 2001. Msps protein is localized to acentrosomal poles to ensure bipolarity of Drosophila meiotic spindles. Nat. Cell Biol. 3:637–642 10.1038/35083025 [DOI] [PubMed] [Google Scholar]
- De Souza C.P., Ellem K.A., Gabrielli B.G. 2000. Centrosomal and cytoplasmic Cdc2/cyclin B1 activation precedes nuclear mitotic events. Exp. Cell Res. 257:11–21 10.1006/excr.2000.4872 [DOI] [PubMed] [Google Scholar]
- Delgehyr N., Sillibourne J., Bornens M. 2005. Microtubule nucleation and anchoring at the centrosome are independent processes linked by ninein function. J. Cell Sci. 118:1565–1575 10.1242/jcs.02302 [DOI] [PubMed] [Google Scholar]
- Derry W.B., Wilson L., Jordan M.A. 1998. Low potency of taxol at microtubule minus ends: implications for its antimitotic and therapeutic mechanism. Cancer Res. 58:1177–1184 [PubMed] [Google Scholar]
- Dobbelaere J., Josué F., Suijkerbuijk S., Baum B., Tapon N., Raff J. 2008. A genome-wide RNAi screen to dissect centriole duplication and centrosome maturation in Drosophila. PLoS Biol. 6:e224 10.1371/journal.pbio.0060224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson H., Bourke E., Jeffers L.J., Vagnarelli P., Sonoda E., Takeda S., Earnshaw W.C., Merdes A., Morrison C. 2004. Centrosome amplification induced by DNA damage occurs during a prolonged G2 phase and involves ATM. EMBO J. 23:3864–3873 10.1038/sj.emboj.7600393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas L.M., Huttner W.B. 2008. The cell biology of neural stem and progenitor cells and its significance for their proliferation versus differentiation during mammalian brain development. Curr. Opin. Cell Biol. 20:707–715 10.1016/j.ceb.2008.09.008 [DOI] [PubMed] [Google Scholar]
- Fletcher L., Muschel R.J. 2006. The centrosome and the DNA damage induced checkpoint. Cancer Lett. 243:1–8 10.1016/j.canlet.2006.01.006 [DOI] [PubMed] [Google Scholar]
- Fong K.-W., Choi Y.-K., Rattner J.B., Qi R.Z. 2008. CDK5RAP2 is a pericentriolar protein that functions in centrosomal attachment of the gamma-tubulin ring complex. Mol. Biol. Cell. 19:115–125 10.1091/mbc.E07-04-0371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong K.W., Hau S.Y., Kho Y.S., Jia Y., He L., Qi R.Z. 2009. Interaction of CDK5RAP2 with EB1 to track growing microtubule tips and to regulate microtubule dynamics. Mol. Biol. Cell. 20:3660–3670 10.1091/mbc.E09-01-0009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglio T., Saredi A., Compton D.A. 1995. NuMA is required for the organization of microtubules into aster-like mitotic arrays. J. Cell Biol. 131:693–708 10.1083/jcb.131.3.693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gergely F., Karlsson C., Still I., Cowell J., Kilmartin J., Raff J.W. 2000. The TACC domain identifies a family of centrosomal proteins that can interact with microtubules. Proc. Natl. Acad. Sci. USA. 97:14352–14357 10.1073/pnas.97.26.14352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gergely F., Draviam V.M., Raff J.W. 2003. The ch-TOG/XMAP215 protein is essential for spindle pole organization in human somatic cells. Genes Dev. 17:336–341 10.1101/gad.245603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillingham A.K., Munro S. 2000. The PACT domain, a conserved centrosomal targeting motif in the coiled-coil proteins AKAP450 and pericentrin. EMBO Rep. 1:524–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon M.B., Howard L., Compton D.A. 2001. Chromosome movement in mitosis requires microtubule anchorage at spindle poles. J. Cell Biol. 152:425–434 10.1083/jcb.152.3.425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graser S., Stierhof Y.-D., Nigg E.A. 2007. Cep68 and Cep215 (Cdk5rap2) are required for centrosome cohesion. J. Cell Sci. 120:4321–4331 10.1242/jcs.020248 [DOI] [PubMed] [Google Scholar]
- Haren L., Stearns T., Lüders J. 2009. Plk1-dependent recruitment of gamma-tubulin complexes to mitotic centrosomes involves multiple PCM components. PLoS One. 4:e5976 10.1371/journal.pone.0005976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald R., Tournebize R., Blank T., Sandaltzopoulos R., Becker P., Hyman A., Karsenti E. 1996. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature. 382:420–425 10.1038/382420a0 [DOI] [PubMed] [Google Scholar]
- Heald R., Tournebize R., Habermann A., Karsenti E., Hyman A. 1997. Spindle assembly in Xenopus egg extracts: respective roles of centrosomes and microtubule self-organization. J. Cell Biol. 138:615–628 10.1083/jcb.138.3.615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuer J.G., Li K., Kaufman T.C. 1995. The Drosophila homeotic target gene centrosomin (cnn) encodes a novel centrosomal protein with leucine zippers and maps to a genomic region required for midgut morphogenesis. Development. 121:3861–3876 [DOI] [PubMed] [Google Scholar]
- Jackman M., Lindon C., Nigg E.A., Pines J. 2003. Active cyclin B1-Cdk1 first appears on centrosomes in prophase. Nat. Cell Biol. 5:143–148 10.1038/ncb918 [DOI] [PubMed] [Google Scholar]
- Jackson A.P., Eastwood H., Bell S.M., Adu J., Toomes C., Carr I.M., Roberts E., Hampshire D.J., Crow Y.J., Mighell A.J., et al. 2002. Identification of microcephalin, a protein implicated in determining the size of the human brain. Am. J. Hum. Genet. 71:136–142 10.1086/341283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan M.A., Toso R.J., Thrower D., Wilson L. 1993. Mechanism of mitotic block and inhibition of cell proliferation by taxol at low concentrations. Proc. Natl. Acad. Sci. USA. 90:9552–9556 10.1073/pnas.90.20.9552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao L.R., Megraw T.L. 2009. Centrocortin cooperates with centrosomin to organize Drosophila embryonic cleavage furrows. Curr. Biol. 19:937–942 10.1016/j.cub.2009.04.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keryer G., Witczak O., Delouvée A., Kemmner W.A., Rouillard D., Tasken K., Bornens M. 2003. Dissociating the centrosomal matrix protein AKAP450 from centrioles impairs centriole duplication and cell cycle progression. Mol. Biol. Cell. 14:2436–2446 10.1091/mbc.E02-09-0614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A., Cole R.W., Oakley B.R., Rieder C.L. 2000. Centrosome-independent mitotic spindle formation in vertebrates. Curr. Biol. 10:59–67 10.1016/S0960-9822(99)00276-6 [DOI] [PubMed] [Google Scholar]
- Kim H.S., Takahashi M., Matsuo K., Ono Y. 2007. Recruitment of CG-NAP to the Golgi apparatus through interaction with dynein-dynactin complex. Genes Cells. 12:421–434 10.1111/j.1365-2443.2007.01055.x [DOI] [PubMed] [Google Scholar]
- Krämer A., Mailand N., Lukas C., Syljuåsen R.G., Wilkinson C.J., Nigg E.A., Bartek J., Lukas J. 2004. Centrosome-associated Chk1 prevents premature activation of cyclin-B-Cdk1 kinase. Nat. Cell Biol. 6:884–891 10.1038/ncb1165 [DOI] [PubMed] [Google Scholar]
- Larocca M.C., Shanks R.A., Tian L., Nelson D.L., Stewart D.M., Goldenring J.R. 2004. AKAP350 interaction with cdc42 interacting protein 4 at the Golgi apparatus. Mol. Biol. Cell. 15:2771–2781 10.1091/mbc.E03-10-0757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Kaufman T.C. 1996. The homeotic target gene centrosomin encodes an essential centrosomal component. Cell. 85:585–596 10.1016/S0092-8674(00)81258-1 [DOI] [PubMed] [Google Scholar]
- Löffler H., Bochtler T., Fritz B., Tews B., Ho A.D., Lukas J., Bartek J., Krämer A. 2007. DNA damage-induced accumulation of centrosomal Chk1 contributes to its checkpoint function. Cell Cycle. 6:2541–2548 [DOI] [PubMed] [Google Scholar]
- Lucas E.P., Raff J.W. 2007. Maintaining the proper connection between the centrioles and the pericentriolar matrix requires Drosophila centrosomin. J. Cell Biol. 178:725–732 10.1083/jcb.200704081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüders J., Patel U.K., Stearns T. 2006. GCP-WD is a gamma-tubulin targeting factor required for centrosomal and chromatin-mediated microtubule nucleation. Nat. Cell Biol. 8:137–147 10.1038/ncb1349 [DOI] [PubMed] [Google Scholar]
- Mahoney N.M., Goshima G., Douglass A.D., Vale R.D. 2006. Making microtubules and mitotic spindles in cells without functional centrosomes. Curr. Biol. 16:564–569 10.1016/j.cub.2006.01.053 [DOI] [PubMed] [Google Scholar]
- McKinnon P.J. 2009. DNA repair deficiency and neurological disease. Nat. Rev. Neurosci. 10:100–112 10.1038/nrn2559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw T.L., Li K., Kao L.R., Kaufman T.C. 1999. The centrosomin protein is required for centrosome assembly and function during cleavage in Drosophila. Development. 126:2829–2839 [DOI] [PubMed] [Google Scholar]
- Meraldi P., Nigg E.A. 2001. Centrosome cohesion is regulated by a balance of kinase and phosphatase activities. J. Cell Sci. 114:3749–3757 [DOI] [PubMed] [Google Scholar]
- Merdes A., Ramyar K., Vechio J.D., Cleveland D.W. 1996. A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell. 87:447–458 10.1016/S0092-8674(00)81365-3 [DOI] [PubMed] [Google Scholar]
- Merdes A., Heald R., Samejima K., Earnshaw W.C., Cleveland D.W. 2000. Formation of spindle poles by dynein/dynactin-dependent transport of NuMA. J. Cell Biol. 149:851–862 10.1083/jcb.149.4.851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell C.B., Khodjakov A.L. 2007. Cooperative mechanisms of mitotic spindle formation. J. Cell Sci. 120:1717–1722 10.1242/jcs.03442 [DOI] [PubMed] [Google Scholar]
- Oshimori N., Li X., Ohsugi M., Yamamoto T. 2009. Cep72 regulates the localization of key centrosomal proteins and proper bipolar spindle formation. EMBO J. 28:2066–2076 10.1038/emboj.2009.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintyne N.J., Schroer T.A. 2002. Distinct cell cycle–dependent roles for dynactin and dynein at centrosomes. J. Cell Biol. 159:245–254 10.1083/jcb.200203089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch A., Thiel C.T., Schindler D., Wick U., Crow Y.J., Ekici A.B., van Essen A.J., Goecke T.O., Al-Gazali L., Chrzanowska K.H., et al. 2008. Mutations in the pericentrin (PCNT) gene cause primordial dwarfism. Science. 319:816–819 10.1126/science.1151174 [DOI] [PubMed] [Google Scholar]
- Raynaud-Messina B., Merdes A. 2007. Gamma-tubulin complexes and microtubule organization. Curr. Opin. Cell Biol. 19:24–30 10.1016/j.ceb.2006.12.008 [DOI] [PubMed] [Google Scholar]
- Rivero S., Cardenas J., Bornens M., Rios R.M. 2009. Microtubule nucleation at the cis-side of the Golgi apparatus requires AKAP450 and GM130. EMBO J. 28:1016–1028 10.1038/emboj.2009.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin K.E., Lourenco P.C.C., Snaith H.A. 2004. Microtubule nucleation at non-spindle pole body microtubule-organizing centers requires fission yeast centrosomin-related protein mod20p. Curr. Biol. 14:763–775 10.1016/j.cub.2004.03.042 [DOI] [PubMed] [Google Scholar]
- Silk A.D., Holland A.J., Cleveland D.W. 2009. Requirements for NuMA in maintenance and establishment of mammalian spindle poles. J. Cell Biol. 184:677–690 10.1083/jcb.200810091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns T. 2009. Centrosome duplication and maturation. Annu. Rev. Cell Dev. Biol. 10.1146/annurev.cellbio.042308.113252 [DOI] [Google Scholar]
- Takahashi M., Yamagiwa A., Nishimura T., Mukai H., Ono Y. 2002. Centrosomal proteins CG-NAP and kendrin provide microtubule nucleation sites by anchoring gamma-tubulin ring complex. Mol. Biol. Cell. 13:3235–3245 10.1091/mbc.E02-02-0112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takao N., Kato H., Mori R., Morrison C., Sonada E., Sun X., Shimizu H., Yoshioka K., Takeda S., Yamamoto K. 1999. Disruption of ATM in p53-null cells causes multiple functional abnormalities in cellular response to ionizing radiation. Oncogene. 18:7002–7009 10.1038/sj.onc.1203172 [DOI] [PubMed] [Google Scholar]
- Taylor S.S., Ha E., McKeon F. 1998. The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1-related protein kinase. J. Cell Biol. 142:1–11 10.1083/jcb.142.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibelius A., Marhold J., Zentgraf H., Heilig C.E., Neitzel H., Ducommun B., Rauch A., Ho A.D., Bartek J., Krämer A. 2009. Microcephalin and pericentrin regulate mitotic entry via centrosome-associated Chk1. J. Cell Biol. 185:1149–1157 10.1083/jcb.200810159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA. 76:4350–4354 10.1073/pnas.76.9.4350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde I., Pahlke G., Salanova M., Zhang G., Wang S., Coletti D., Onuffer J., Jin S.L.C., Conti M. 2001. Myomegalin is a novel protein of the golgi/centrosome that interacts with a cyclic nucleotide phosphodiesterase. J. Biol. Chem. 276:11189–11198 10.1074/jbc.M006546200 [DOI] [PubMed] [Google Scholar]
- Wang X., Tsai J.W., Imai J.H., Lian W.N., Vallee R.B., Shi S.H. 2009. Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature. 461:947–955 10.1038/nature08435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods C.G., Bond J., Enard W. 2005. Autosomal recessive primary microcephaly (MCPH): a review of clinical, molecular, and evolutionary findings. Am. J. Hum. Genet. 76:717–728 10.1086/429930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachos G., Rainey M.D., Gillespie D.A. 2003. Chk1-deficient tumour cells are viable but exhibit multiple checkpoint and survival defects. EMBO J. 22:713–723 10.1093/emboj/cdg060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Megraw T.L. 2007. Proper recruitment of gamma-tubulin and D-TACC/Msps to embryonic Drosophila centrosomes requires centrosomin motif 1. Mol. Biol. Cell. 18:4037–4049 10.1091/mbc.E07-05-0474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman W.C., Sillibourne J., Rosa J., Doxsey S.J. 2004. Mitosis-specific anchoring of gamma tubulin complexes by pericentrin controls spindle organization and mitotic entry. Mol. Biol. Cell. 15:3642–3657 10.1091/mbc.E03-11-0796 [DOI] [PMC free article] [PubMed] [Google Scholar]