Astrocyte differentiation and maintenance is promoted by BMP signaling, which induces REST/NRSF to repress neuronal genes.
Abstract
Once they have differentiated, cells retain their individual character and repress genes that are specifically expressed in other cell lineages, but how alternative fate choice is restricted during and/or after differentiation remains unclear. In the mammalian central nervous system, neurons, astrocytes, and oligodendrocytes are generated throughout life from common tripotent neural progenitor cells (NPCs). Bone morphogenetic proteins (BMPs) are well-known astrocyte-inducing cytokines. We show here that the expression of a transcriptional repressor, RE1 silencer of transcription (REST)/neuron-restrictive silencer factor (NRSF), is up-regulated and sustained by BMP signal activation in the course of astrocytic differentiation of NPCs, and restricts neuronal differentiation. We further show that, in differentiated astrocytes, endogenous REST/NRSF associates with various neuronal genes and that disruption of its function resulted in their derepression, thereby explaining how ectopic neuronal gene expression is prevented in cells with astrocytic traits. Collectively, our results suggest that REST/NRSF functions as a molecular regulator of the nonneuronal phenotype in astrocytes.
Introduction
The mammalian cerebral cortex originates from neural progenitor cells (NPCs), which proliferate and give rise to the three major brain cell types: neurons, astrocytes, and oligodendrocytes (Gage, 2000; Temple, 2001). The fate of NPCs in the developing brain is believed to be determined by internal cellular programs and external cues that involve various types of cytokines (Gage, 2000; Schuurmans and Guillemot, 2002; Hsieh and Gage, 2004). Among the three NPC progeny types, mechanisms of differentiation have been most extensively investigated for astrocytes. The Janus kinase (JAK) signal transducer and activator of transcription (STAT) pathway (Bonni et al., 1997; Nakashima et al., 1999a; He et al., 2005) and the signaling pathway triggered by bone morphogenetic proteins (BMPs) are deeply involved in astrocytogenesis (Nakashima et al., 1999b; See et al., 2007). Despite considerable progress in unraveling how astrocytic traits are acquired, mechanisms that suppress differentiation into other cell lineages, such as neurons, during astrocytic specification remain poorly understood.
Neurogenesis is promoted by proneural basic helix-loop-helix (bHLH) transcriptional factors including Mash1 (mammalian achaete-scute homologue), Neurogenin, and NeuroD (mammalian atonal homologue), which form heterodimers with other ubiquitously expressed bHLH proteins such as the E2A gene products E12 and E47 (Bertrand et al., 2002). Consistent with their role in inducing neuronal differentiation, proneural factors are transiently expressed in neuronal progenitor cells located in the ventricular zone when neurogenesis is occurring in the developing central nervous system (CNS; Gradwohl et al., 1996; Tokunaga et al., 2004). The function of these proneural bHLH factors is inhibited by negative HLH proteins, such as the Id (inhibitor of differentiation) and Hes (hairy enhancer of split) families, at the onset of astroglial differentiation triggered by BMP2 (Nakashima et al., 2001). These findings partially explain cell fate decisions that facilitate the differentiation of one cell lineage and suppress that of the others: once NPCs are primed to differentiate into astrocytes, for example, neuronal differentiation must be inhibited. Although Id and Hes family members act by inhibiting proneural bHLH factors, thus preventing astrocytic lineage-committed NPCs from acquiring neuronal properties, their expression in vivo is generally observed in the proliferative zone, where undifferentiated but not differentiated cells reside (Duncan et al., 1992; Ellmeier and Weith, 1995; Lyden et al., 1999). Therefore, the ability to suppress neuronal properties in differentiated astrocytes should rely on regulatory pathway(s) downstream of Id and Hes, or on an as-yet-unidentified program which is subsequently established in differentiated astrocytes.
Insights into the mechanism that represses pan-neuronal gene expression in nonneural tissues have been gained from a series of studies regarding a zinc finger repressor protein, repressor element1-silencing transcription factor (REST)/neuron-restrictive silencer factor (NRSF) (Chong et al., 1995; Schoenherr et al., 1996; Chen et al., 1998; Lunyak et al., 2002; Conaco et al., 2006). The completion of neuronal differentiation is marked by the expression of proteins involved in electrophysiological processes, such as synapsin, glutamate receptors, SCG10, and type II sodium channel. Many other genes associated with the functional properties of terminally differentiated neurons have also been identified as direct targets of REST/NRSF (Bruce et al., 2004; Johnson et al., 2006; Otto et al., 2007). REST/NRSF suppresses target gene transcription through the association of its N- and C-terminal repressor domains with the mSin3/histone deacetylase-1/2(HDAC1/2) complex and with the CoREST complex, respectively (Huang et al., 1999; Lunyak et al., 2002). Suppression of neuronal gene expression by REST/NRSF is consistently observed in various cell types including PC12 cells (Ballas et al., 2001), myoblasts (Watanabe et al., 2004), and embryonic stem (ES) cells (Singh et al., 2008; Westbrook et al., 2008). Although several studies have suggested that REST/NRSF is a binary neuron/nonneuron fate regulator, its function has been examined mainly in nonneural cells and tissues. Concerning its role in the nervous system, REST/NRSF has been implicated in the acquisition of neuronal phenotype by NPCs in analyses of neuronal differentiation of ES cells (Ballas et al., 2005) and activation of REST/NRSF target genes induced by REST/NRSF-VP16 (in which the repressor domain is replaced with the activation domain of herpes simplex virus VP16; Su et al., 2004). In adult hippocampus-derived NPCs, REST/NRSF has been reported to promote neurogenesis through activation of its target genes by means of a small RNA-mediated functional switch that converts it from a suppressor to an activator of transcription (Kuwabara et al., 2004). Although these results strongly suggest the involvement of REST/NRSF in cell fate decisions of NPCs, it remains largely elusive how REST/NRSF expression is regulated in NPCs and how REST/NRSF participates in NPC fate specification and maintenance of the differentiated state of cells in the CNS.
In this study, we demonstrate that REST/NRSF is a critical suppressor of neuronal differentiation and neuronal gene expression during and/or after astroglial differentiation. The astrogliogenic cytokine BMP2 induced transcriptional activation of REST/NRSF in NPCs via the BMP-downstream transcription factor Smad, which binds to Smad-binding elements (SBEs) in the regulatory region of the REST/NRSF gene. Gain- and loss-of-function studies using wild-type REST/NRSF and a dominant-negative form of REST/NRSF (DN-REST/NRSF) in NPCs revealed a significant role for REST/NRSF in suppression of neuronal differentiation. Even in differentiated astrocytes, REST/NRSF continued to be expressed, and it associated with various neuronal genes containing the REST/NRSF binding site (RE1/NRSE). Furthermore, eliminating the repressor function of REST/NRSF led to derepression of neuronal genes in astrocytes, indicating that REST/NRSF acts to prevent astrocytes from displaying neuronal behavior.
Results
BMP2-induced expression of REST/NRSF in NPCs
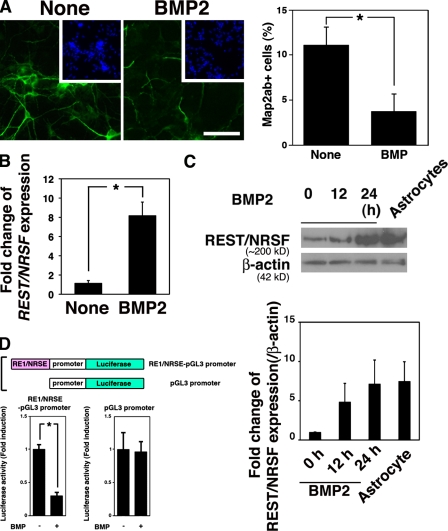
As a first step to unraveling the mechanism by which neuronal gene expression is restricted in astrocytes, we hypothesized that the repression of neuronal character in astrocytes is programmed during a fate-determination step toward astrocytes. As shown in a previous study (Nakashima et al., 2001), BMP2 inhibited neuronal differentiation of NPCs (Fig. 1 A). BMP signal activation in NPCs was confirmed by detecting phosphorylation of Smads in response to BMP stimulation (Fig. S1 A). The frequency of Map2ab-positive neurons generated from NPCs on d 4 in BMP2-treated and untreated cultures was 3.7 ± 2.0% and 11.0 ± 3.7%, respectively. We also observed a reduction in the number of NPC- and oligodendrocyte-marker–positive cells by BMP2 treatment, suggesting that BMP2 inhibits differentiation of NPCs toward nonastrocytic lineages (Fig. S1, B and C). BMP2 induced the expression of negative HLH factor genes including Id1, Id3, and Hes5 (Nakashima et al., 2001), whose products are known to inhibit neurogenic bHLH factors such as Neurogenin and Mash1. Although Ids and Hes5 thus seem to be candidate repressors of neuronal traits in differentiated astrocytes, their BMP-induced expression was transient, and they were not highly expressed in differentiated astrocytes (Nakashima et al., 2001; Fig. S1, D and E). Therefore, we set out to identify another candidate gene whose expression, in contrast to the Ids and Hes5, is induced by BMP2 and sustained. Among various transcriptional repressors, we focused on REST/NRSF because it is known as a repressor of neuronal gene expression in nonneural tissues, and found that REST/NRSF expression is indeed up-regulated by BMP2 treatment of NPCs (Fig. 1 B). Sustained REST/NRSF expression in BMP2-treated NPCs was further confirmed by Western blot (Fig. 1 C) and quantitative RT-PCR analysis (Fig. S1 E). As well as acting as a transcriptional repressor for neuronal genes, REST/NRSF is also a latent transcriptional activator in the adult CNS (Kuwabara et al., 2004). To monitor REST/NRSF function under our experimental conditions, we exploited a reporter construct carrying RE1/NRSE, a binding motif for REST/NRSF, fused upstream of an SV40 minimal promoter (Fig. 1 D, top). When this RE1/NRSE reporter was introduced into NPCs, luciferase reporter activity decreased in response to BMP2 treatment (Fig. 1 D, bottom left). On the other hand, we found no difference between control reporter activity with or without BMP2 treatment (Fig. 1 D, bottom right). Thus, these data suggest that REST/NRSF induced by BMP2 appears to function as a transcriptional repressor in embryonic NPCs.
Figure 1.
BMP2 induces expression of REST/NRSF. (A) NPCs were cultured for 4 d with or without BMP2 (50 ng/ml) in the presence of bFGF (10 ng/ml), and then immunostained for Map2ab (green). Inset (blue), Hoechst nuclear staining of each field. Percentages of Map2ab-positive cells were quantified (right; mean ± SD; n = 3). Statistical significance was examined by Student’s t test (*, P < 0.05). (B) Total RNAs were extracted from NPCs treated or untreated with BMP2 (50 ng/ml) for 1 h and then subjected to quantitative RT-PCR using primer sets for REST/NRSF and G3PDH to evaluate expression of REST/NRSF mRNA. Statistical significance was examined by Student’s t test (*, P < 0.05). (C) NPCs were treated with BMP2 (50 ng/ml) for 12 and 24 h. The cells were lysed for Western blot analysis to detect expression of REST/NRSF (∼220 kD). Cell lysate from primary astrocytes from P0 mouse brain was used as a control. Fold change of REST/NRSF protein expression was analyzed by ImageJ software. (D) Schematic diagram of the construct to examine REST/NRSF activity. The RE1/NRSE element was fused upstream of the SV40 minimal promoter (top). NPCs were cotransfected with each reporter construct and pRL-CMV, and then stimulated with BMP2. After 48 h, luciferase activity was measured.
REST/NRSF is a direct target of the BMP–Smad pathway
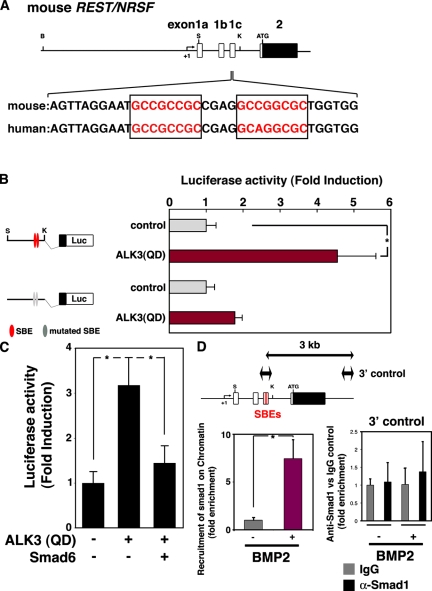
Having observed REST/NRSF up-regulation by BMP2 in NPCs, we next sought to gain mechanistic insight into BMP2-induced expression of REST/NRSF. The genomic structure of REST/NRSF has been extensively examined in previous studies (Koenigsberger et al., 2000; Kojima et al., 2001), and we identified two possible BMP-responsive SBEs (GCCGNCGC; Kusanagi et al., 2000), which are highly conserved between mouse and human, in the regulatory region of the gene (+1192 to +1199, and +1204 to +1211; Fig. 2 A). Next, we isolated a genomic fragment that contains the 5′ UTR of the mouse REST/NRSF gene, including the SBEs, to generate REST/NRSF reporter constructs. The intact reporter was activated by the expression of constitutively active BMP type I receptor (ALK3(QD); Imamura et al., 1997; Fig. 2 B), and this transactivation was severely compromised by mutating the two SBEs (Fig. 2 B). BMP2 activates Smad1, 5, and 8, and the activation is suppressed by the inhibitory Smad, Smad6 (Imamura et al., 1997). As shown in Fig. 2 C, transactivation of the REST/NRSF reporter by BMP signaling was inhibited by the expression of Smad6, and the same was true when a dominant-negative (DN) form of Smad1 (Yoshiura et al., 2007) was used instead of Smad6 (Fig. S2 A). These data, thus, suggest that activation of Smad1/5/8 is required for the induction of REST/NRSF gene expression. To determine whether Smads indeed bind to the SBE-containing endogenous REST/NRSF gene regulatory region after BMP2 stimulation, we performed a chromatin immunoprecipitation (ChIP) assay using anti-Smad1. As shown in Fig. 2 D, significant Smad binding to the SBE-containing fragment occurred in BMP2-treated cells. Collectively, these results suggest that REST/NRSF is a direct target of BMP signaling in NPCs. Although one could imagine that the sustained expression of REST/NRSF in astrocytes is established by permanent recruitment of Smads on its regulatory region, Smad1-enrichment to the region, which is induced in NPCs by BMP2 stimulation, was not observed in differentiated astrocytes, as judged by the ChIP assay (Fig. S2 B). Alternatively, we found that the regulatory region was marked by transcriptionally active histone modification, i.e., acetylated histone H3 (AcH3) and trimethylation of histone H3-lysine 4 (H3K4me3; Jenuwein and Allis, 2001; Fig. S2 C). Collectively, it is conceivable that once REST/NRSF is activated by Smads, it no longer requires Smads for sustained expression during/after astrocytic differentiation.
Figure 2.
Smad-dependent transactivation of the REST/NRSF gene. (A) Comparison of a part of REST/NRSF genomic sequences between mouse and human. Part of the mouse REST/NRSF locus is shown on the top line, and sequence data around the SBEs in mouse and human are shown below. Boxed regions are sequences that match the SBE consensus. B, BamHI; S, SacI; K, KpnI. (B) Schematic representation of the REST/NRSF reporter constructs is shown on the left. NPCs were cotransfected with the reporter construct and pRL-CMV together with a control vehicle or a construct expressing ALK3(QD). Luciferase activity was measured 24 h after transfection. Mean ± SD (n = 3). Statistical significance was examined by Student’s t test (*, P < 0.05). (C) NPCs were cotransfected with the REST/NRSF reporter construct and pRL-CMV along with the constructs indicated below the graph. Luciferase activity was measured 24 h after transfection. Mean ± SD (n = 3). Statistical significance was examined by Student’s t test (*, P < 0.05). (D) NPCs were incubated with BMP2 (50 ng/ml) for 20 min and subjected to quantitative ChIP analysis using control IgG and anti-Smad1 antibodies (bottom left graph). Mean ± SD (n = 3). Statistical significance was examined by Student’s t test (*, P < 0.05). Co-immunoprecipitated REST/NRSF gene fragments were amplified by PCR with a specific pair of primers. The amplified region is schematically indicated as the two-headed arrow spanning REST/NRSF exon 1c (top left). A primer set for amplification of a sequence 3 kb downstream of the SBEs in the REST/NRSF regulatory region (top right, two-headed arrow) was used to validate specific recruitment of Smad1 onto the SBEs (bottom right).
REST/NRSF regulates fate specification of NPCs
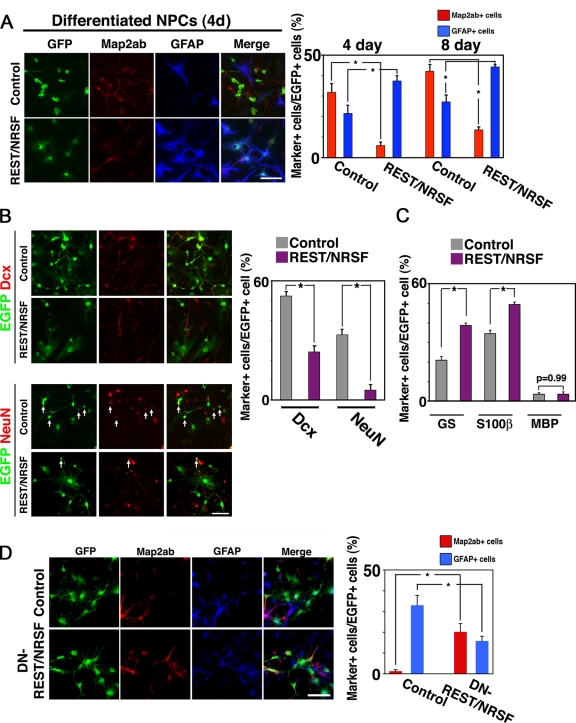
In light of the above finding that REST/NRSF expression is induced by BMP2 stimulation, we anticipated that REST/NRSF would play a critical role in suppression of neuronal differentiation by BMP2. To test this, we infected NPCs with recombinant retroviruses engineered to express GFP (control) or GFP with REST/NRSF. Because we wanted to examine first whether REST/NRSF alone inhibits neuronal differentiation of NPCs, we switched the medium on the following day to one containing 0.5% FBS without bFGF to induce spontaneous differentiation, and cultured the cells for an additional 4 d. As shown in Fig. 3 A (top left), the cells infected with control viruses effectively differentiated into Map2ab-positive neurons (4 d: 31.8 ± 4.4%; 8 d: 42.1 ± 3.3%, right graph). In contrast, REST/NRSF expression in NPCs dramatically inhibited the cells from differentiating into neurons (Fig. 3 A, bottom left; 4 d: 5.8 ± 1.6%; 8 d: 13.5 ± 1.5%, right graph). Thus, although one might anticipate that REST/NRSF would not inhibit neuronal differentiation of NPCs but simply delays it, this is not the case: inhibition of neuronal differentiation by REST/NRSF was observed in both short and prolonged culture. Doublecortin (Dcx) and NeuN are known as early and late stage markers for neuronal differentiation, respectively. We found that REST/NRSF suppressed expression of both these neuronal markers (Fig. 3 B) suggesting, therefore, that REST/NRSF inhibits pan-neuronal differentiation of NPCs. As for astroglial differentiation, there was a slight increase of GFAP-positive astrocytes in the REST/NRSF-expressing virus-infected cells (Fig. 3 A; control, 4 d: 21.5 ± 4.0%; 8 d: 27.2 ± 3.3%; REST/NRSF, 4 d: 37.4 ± 2.5%; 8 d: 44.3 ± 0.84%, right graph). The effect of REST/NRSF on astrocytic differentiation of NPCs was further confirmed using two other astrocytic markers, glutamine synthetase (GS) (Tokunaga et al., 2004) and S100β (Fig. 3 C). There seemed to be a tendency for the number of astrocytic-marker–positive cells in NPCs infected with REST/NRSF-expressing virus to increase. However, there was no significant difference in the number of GFAP-positive cells between control and REST/NRSF-expressing virus-infected cells maintained in culture medium containing bFGF (Fig. S3 A), implying that the astrocyte-inducing activity of REST/NRSF is not strong enough to overcome the inhibitory effect of bFGF in NPC differentiation. Furthermore, the proliferation and cell death rates of control and REST/NRSF-expressing virus-infected cells were similar, as judged by BrdU uptake and cleaved caspase-3 staining (Fig. S3, B and C). To minimize the potent effect of BMPs contained in the serum, we used Noggin to inhibit functional BMP signaling in this experiment, and observed significant reduction of neuronal differentiation by REST/NRSF expression (Fig. S3 D). These observations suggest that REST/NRSF is able to function as a suppressor of neuronal differentiation in NPCs.
Figure 3.
REST/NRSF is sufficient to repress neuronal differentiation and is required for BMP2-induced suppression of neuronal differentiation of NPCs. (A) NPCs were infected with recombinant retroviruses engineered to express only GFP (pMYs), or REST/NRSF together with GFP (REST/NRSF-pMYs), and cultured in medium supplemented with 0.5% FBS for 4 or 8 d to induce spontaneous differentiation. The cells were then stained with antibodies against GFP (green), Map2ab (red), and GFAP (blue). Bar, 50 µm. The percentage of marker-positive cells in each GFP-positive cell population after the 4-d culture was determined. Mean ± SD (n = 3). Statistical significance was examined by Student’s t test (*, P < 0.05). (B) Expression of the neuronal markers NeuN and Dcx was examined in cells infected with control or REST/NRSF-expressing virus. The cells were subjected to immunocytochemical analysis after culturing in 0.5% FBS for 8 d (left). Bar, 20 µm. Marker-positive cells in total EGFP-positive cells were quantified (right). Mean ± SD (n = 3). Statistical significance was examined by Student’s t test (*, P < 0.05). (C) Expression of the glial markers glutamine synthase (astrocytes), S100β (astrocytes), and MBP (oligodendrocytes) was examined in cells infected with control or REST/NRSF-expressing virus. The cells were subjected to immunocytochemical analysis after culturing with 0.5% FBS for 4 d. (D) NPCs were infected with viruses carrying control pMYs (GFP only), or DN-REST/NRSF-pMYs (DN-REST/NRSF) together with GFP, in culture medium containing bFGF (10 ng/ml), and subsequently treated with BMP2 (50 ng/ml) for 4 d. The cells were then stained with antibodies against GFP (green), Map2ab (red), and GFAP (blue). Bar, 50 µm. The percentage of marker-positive cells in each GFP-positive cell population after the 4-d culture was determined. Mean ± SD (n = 3). Statistical significance was examined by Student’s t test (*, P < 0.05).
We next examined the requirement for REST/NRSF in the BMP-induced restriction of neuronal fate by means of a dominant-negative form of REST/NRSF harboring only its DNA-binding domain (DN-REST/NRSF; Chen et al., 1998). To this end, we infected NPCs with control and DN-REST/NRSF–expressing retroviruses, and cultured them with BMP2 for 4 d. As shown in Fig. 3 D, DN-REST/NRSF expression resulted in the derepression of neuronal fate, as judged by Map2ab expression. The control retrovirus-infected cells became Map2ab-positive neurons at a frequency of 1.3 ± 0.68%, whereas 20.2 ± 4.0% of DN-REST/NRSF–infected cells were Map2ab positive (Fig. 3 D). We did not observe significant influence on cell death rates by inhibition of REST/NRSF function (Fig. S3 E). These data imply that REST/NRSF expression is necessary and sufficient for the suppression of neuronal cell fate by BMP2.
Modulating REST/NRSF function perturbs astrocytic integrity
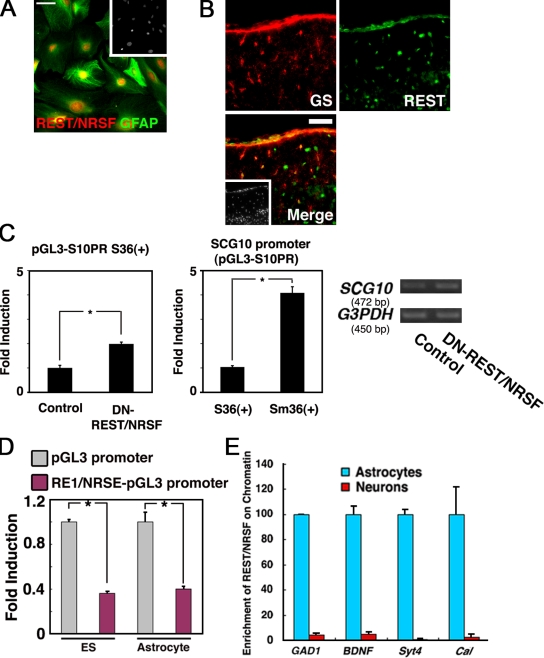
To test the idea that REST/NRSF might be a suppressor of inappropriate neuronal gene expression in astrocytes, we then examined the expression of REST/NRSF in primary cultured astrocytes derived from P0 mouse brain. We confirmed maturation of the astrocytes by determining the expression of glutamine transporter GLT-1 (Rothstein et al., 1994; Fig. S4 A). As shown in Fig. 4 A, REST/NRSF was expressed in GFAP-positive cultured astrocytes. We further examined REST/NRSF expression in vivo, and found that GS- and GFAP-positive astrocytes in postnatal day (P) 8 cortex expressed REST/NRSF (Fig. 4 B, Fig. S4 B). We further confirmed astrocytic expression of REST/NRSF compared with that in neurons by immunoblot analysis (Fig. S4 C).
Figure 4.
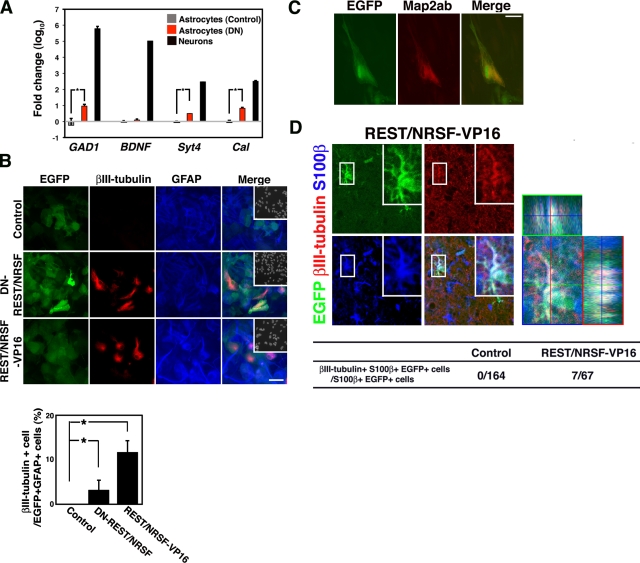
Astrocytes express functional REST/NRSF. (A) Expression of REST/NRSF in GFAP-positive astrocytes derived from P0 mouse brain. Bar, 50 µm. Inset: Hoechst nuclear staining of this field. (B) Expression of REST/NRSF in astrocytes in vivo. REST/NRSF (green) was expressed in GS (red)-positive cells in the cortical surface region of P8 mouse brain. Bar, 50 µm. Inset: Hoechst nuclear staining of the field. (C) REST/NRSF activity was examined using a reporter construct carrying the SCG10 minimal promoter (pGL3-S10PR) with intact (S36+) and mutated (Sm36+) RE1/NRSE. S36+ reporter activity was derepressed by the expression of DN-REST/NRSF in astrocytes (left). The reporter activity was enhanced by mutating the RE1/NRSE motif (middle). Mean ± SD (n = 3). Statistical significance was examined by Student’s t test (*, P < 0.05). Endogenous expression of SCG10 was also examined by RT-PCR analysis in astrocytes introduced with control or DN-REST/NRSF-expressing vector (right). (D) Transcriptional suppressor activity of REST/NRSF was observed in astrocytes and ES cells as assessed by the RE1/NRSE reporter system. Mean ± SD (n = 3). Statistical significance was examined by Student’s t test (*, P < 0.05). (E) Comparison, between neurons and astrocytes, of REST/NRSF association with RE1/NRSE-containing regions in GAD1, BDNF, Synaptotagmin4 (Syt4), and Calbindin (Cal) genes, measured by quantitative ChIP analysis. Blue columns: astrocytes. Red columns: neurons. Mean ± SD.
To determine the function of REST/NRSF in astrocytes, we performed a luciferase assay using reporter constructs containing the SCG10 minimal promoter with an intact or a mutated RE1/NRSE motif (Mori et al., 1990). If REST/NRSF had repressor activity in astrocytes, the luciferase activity in cells transfected with a DN-REST/NRSF–expressing construct should be higher than that in control vehicle-transfected cells. As shown in Fig. 4 C (left), astrocytes showed increased luciferase activity after introduction of the DN-REST/NRSF–expressing construct. Furthermore, derepression of the promoter was also observed by mutating the RE1/NRSE motif (Fig. 4 C, middle), indicating that endogenous REST/NRSF functions as a transcriptional repressor in astrocytes. In support of these findings, derepression of endogenous SCG10 was detected when astrocytes were transfected with the DN-REST/NRSF–expressing construct (Fig. 4 C, right). To further confirm this repressor activity of REST/NRSF, we introduced the RE1/NRSE reporter into astrocytes and embryonic stem (ES) cells, which have been shown to express REST/NRSF (Ballas et al., 2005; Westbrook et al., 2008), and measured the reporter activity. As shown in Fig. 4 D, we found that the repressive activity of REST/NRSF was comparably strong in both ES cells and astrocytes, implying that REST/NRSF can function as a transcriptional repressor in other nonneural tissues or cell types. We also found recruitment of Smad1 to SBE-containing REST/NRSF regulatory region in ES cells (Fig. S4 D); therefore, the BMP-REST/NRSF axis apparently exists in other nonneural tissues.
The presence of repressive REST/NRSF activity in astrocytes prompted us to analyze the previously reported enrichment (Ballas et al., 2005) of endogenous REST/NRSF at its target loci. ChIP analysis revealed that REST/NRSF associated with several genes harboring RE1/NRSEs in astrocytes; in contrast, REST/NRSF binding to these target genes was negligible in neurons (Fig. 4 E).
In light of the above results, we then examined derepression of endogenous REST/NRSF target genes in astrocytes by perturbing REST/NRSF function. As shown in Fig. 5 A, transcriptional derepression of several endogenous REST/NRSF target genes was indeed observed in astrocytes transfected with the DN-REST/NRSF construct.
Figure 5.
Astrocytes are reprogrammed to express neuronal genes via perturbation of REST/NRSF function. (A) Total RNAs were extracted from astrocytes transduced with control or DN-REST/NRSF–expressing vector, and from primary cultured neurons. These RNAs were then subjected to quantitative RT-PCR with primers specific for individual REST/NRSF target genes, GAD1, BDNF, Syt4, and Cal. (B) Expression of a neuronal marker in astrocytes after perturbation of REST/NRSF activity. Introduction of a DN-REST/NRSF–expressing construct into astrocytes resulted in ectopic expression of βIII-tubulin (red) in GFAP-positive (blue) astrocytes. Insets: Hoechst nuclear staining of each field. Bar, 50 µm. The percentage of βIII-tubulin–positive cells in GFP- and GFAP-positive populations was determined (right graph); the effect of REST/NRSF-VP16 in astrocytes is also indicated. Mean ± SD (n = 3). Statistical significance was examined by Student’s t test (*, P < 0.05). (C) Expression of Map2ab in DN-REST/NRSF-expressing astrocytes. Astrocytes were cultured as in B and stained with anti-GFP and -Map2ab antibodies. (D) Ectopic expression of neuronal markers in S100β-positive astrocytes in vivo after introduction of REST/NRSF-VP16. Lentiviruses were injected stereotactically into the striatum of adult mouse brain, followed by immunohistochemical analysis 3 wk after the surgery. Insets: higher magnification image of boxed region in each field. Quantification is shown in the table (Control, 164 S100β+/EGFP+ cells from three animals; REST/NRSF-VP16, 67 S100β+/EGFP+ cells from three animals).
To determine the role played by REST/NRSF in restricting neuronal characteristics to maintain the cellular identity of astrocytes, we introduced DN-REST/NRSF into astrocytes and examined whether inappropriate expression of βIII-tubulin, a typical marker of neurons, occurred. As shown in Fig. 5 B, βIII-tubulin expression appeared in astrocytes with diminished REST/NRSF activity; we obtained a similar result using shRNA against REST/NRSF in astrocytes (Fig. S5, A–C). Another neuronal marker, Map2ab, was also expressed in DN-REST/NRSF–expressing astrocytes (Fig. 5 C).
Next, we investigated the in vivo significance of this inhibition of REST/NRSF function in astrocytes by injecting recombinant lentiviruses into the striatum of the adult mouse brain, and then examining βIII-tubulin expression in both EGFP- and S100β-positive virus-infected astrocytes 3 wk later. However, in the case of DN-REST/NRSF–expressing virus injection, we could not find any S100β-positive astrocytes that also expressed βIII-tubulin (0/107), suggesting that simple inhibition of REST/NRSF is not sufficient to induce ectopic neuronal marker expression in astrocytes in vivo. One might conclude that, although astrocytes in perinatal brain have been reported to retain broad differentiation potency (Laywell et al., 2000), astrocytes in the adult have lost the potential to express neuronal markers in response to a reduction of REST/NRSF expression. Our results, however, show that this is not the case because expression of an ectopic neuronal marker was observed even in adult astrocytes when DN-REST/NRSF was expressed (Fig. S5, D and E). Therefore, the effects observed after introducing DN-REST/NRSF into astrocytes in vivo suggest either that 3 wk was not long enough or that other stimuli were required to reprogram astrocytes to express neuronal markers in vivo. In contrast to DN-REST/NRSF, REST/NRSF-VP16 (the DNA-binding domain fused to VP16) was efficient enough to increase the frequency of astrocytes positive for the neuronal marker in vivo (Fig. 5 D). Collectively, these results suggest that REST/NRSF acts as a substantial guardian of the astrocytic phenotype.
Discussion
Our objective in the present study was to determine how, during a cell’s progression toward a particular restricted lineage (here, astrocytes), characters that are specific to an alternative lineage (neurons) are suppressed. Contribution of BMP signaling to NPC differentiation has also been examined by several genetic manipulations of BMP receptors. Although the conditional knockout of BMP type IA receptor (BMPRIA) in cortex showed minor effects on glial differentiation (Araya et al., 2008), compound disruption of BMPRIA and BMPRIB in mice resulted in a decreased number of astrocytes generated from NPCs (See et al., 2007). Moreover, several lines of evidence suggested that BMP2 is a strong astrocyte-inducing cytokine for NPCs in vitro (Nakashima et al., 1999b, 2001; Mehler et al., 2000; See et al., 2007). BMPs have also been shown to not only induce GFAP expression, but also to advance maturation of astrocytes, i.e., BMPs induce cell cycle exit and loss of NPC markers (Bonaguidi et al., 2005). Therefore, we thought that unraveling the molecular mechanisms of astrocyte differentiation and maturation induced by the activation of BMP signaling may elucidate the machinery that restricts differentiation plasticity in astrocytes.
As we reported previously, BMP2 triggers the expression of negative HLH genes (Ids and Hes5), resulting in the inhibition of such neurogenic bHLHs as Mash1 and Neurogenin. However, BMP2-induced up-regulation of these negative HLH factors in NPCs is transient (Nakashima et al., 2001; Fig. S1, D and E), and their expression is normally detected in the germinal zone of the developing brain, where undifferentiated cells are located (Duncan et al., 1992; Ellmeier and Weith, 1995; Lyden et al., 1999). Thus, we wished to identify a bona fide suppressor of neuronal phenotype that acts both during and after astrocyte differentiation. Although REST/NRSF is known as a major regulator of neuronal genes in nonneuronal tissues, and its expression is regulated during development (Chong et al., 1995), the precise regulation of REST/NRSF expression by itself has not been addressed in detail. In this study, we have now identified REST/NRSF as a BMP-regulated factor that suppresses neuronal differentiation. The transcriptional up-regulation of REST/NRSF by BMP signaling is mediated by the direct binding of Smad transcription factors to the SBEs in the REST/NRSF regulatory region (Fig. 2). Although a direct link between BMP signaling and REST/NRSF expression was not shown, it was recently reported that REST/NRSF expression is observed in the ectoderm of early Xenopus embryos where BMP signaling is known to modulate tissue patterning, and that the interference of REST/NRSF mimics several features associated with a decreased BMP function (Olguín et al., 2006). Thus, the BMP-REST/NRSF axis may exist in broader aspects of tissue organogenesis. Although we used BMP2 in the present study to examine the inhibition of neuronal differentiation of NPCs, other BMP family members, such as BMP4 and BMP7, also signal through the type IA receptor and generally exert similar effects to those of BMP2 (Yanagisawa et al., 2001). Considering this functional conservation, other type IA–using BMPs may also be able to induce REST/NRSF expression.
Unlike that of Ids and Hes5, expression of REST/NRSF is maintained even in differentiated astrocytes (Fig. 1 C and Fig. 4 B), yet how this difference occurs is currently unknown. REST/NRSF is known to be regulated by canonical Wnt signaling (Nishihara et al., 2003), and certain types of astrocytes secrete Wnt3 to provide a niche for adult NPCs (Lie et al., 2005). It will therefore be of interest to test whether activation of Wnt pathways plays crucial roles, whether autocrine or paracrine, to maintain an astrocytic niche. An alternative possibility is that REST/NRSF expression is maintained by mammalian SWI/SNF chromatin remodeling complexes (Martens and Winston, 2003) after a transcriptional priming effect supplied by the BMP–Smad pathway. Brg1, one of the two catalytic ATPase subunits of the SWI/SNF complex, has been shown using CNS-specific Brg1 knockout animals to be required for gliogenesis (Matsumoto et al., 2006). Nevertheless, the contribution of SWI/SNF complex to REST/NRSF expression and repression of neuronal genes in differentiated astrocytes must await future study in an animal model of glia-specific depletion of Brg1.
Similar to Ids, as reported in our previous study (Nakashima et al., 2001), the expression of REST/NRSF in NPCs does not seem to actively induce astrogliogenesis because forced expression of REST/NRSF alone was unable to induce GFAP-positive astrocytic differentiation (Fig. S3 A). The contribution of REST/NRSF to an early step of astrocytic differentiation may be exerted by repression either of neurogenic bHLH factors such as NeuroD (Lunyak et al., 2002; Otto et al., 2007), or of an orphan nuclear receptor like Tlx (Otto et al., 2007); the transcriptional repressor Tlx represses astrocyte differentiation (Miyawaki et al., 2004; Shi et al., 2004; Uemura et al., 2006), in part by binding directly to the promoter region of the astrocytic gene gfap (Shi et al., 2004). Because we did not observe active promotion of astroglial differentiation of NPCs by REST/NRSF overexpression (Fig. S3 A), we cannot rule out the possibility that REST/NRSF-induced suppression of genes which repress astrocyte differentiation is involved in BMP2-induced astrogliogenesis. Considering that RE1/NRSE motifs have been identified in hundreds of neuronal genes, including ion channels, neurotransmitter receptors, neurotrophins, synaptic vesicle proteins, and cytoskeletal and adhesion molecules (Bruce et al., 2004), REST/NRSF potentially could suppress a diverse array of neuronal genes simultaneously. Therefore, although REST/NRSF induction is probably sufficient for the inhibition of neuronal differentiation, it is conceivable that BMP signaling needs to collaborate with other signaling pathways such as the JAK/STAT pathway (Nakashima et al., 1999a; He et al., 2005) for effective induction of astrocyte differentiation.
Once cells acquire the properties of a particular lineage, they specifically repress genes that are expressed in alternative cell lineages. This restriction of cellular plasticity must be tightly regulated to ensure cellular or tissue homeostasis. In the case of neurons, astrocytic gene expression is, in part, repressed by epigenetic modification through DNA methylation (Takizawa et al., 2001; Setoguchi et al., 2006; Kohyama et al., 2008; Namihira et al., 2009). As shown in Figs. 1, 4, and S1 E, expression of REST/NRSF is up-regulated and maintained during astrocyte differentiation, and REST/NRSF is bound to various kinds of neuronal genes in differentiated astrocytes (Fig. 4 E). We have here provided evidence that perturbing the function of REST/NRSF by both DN-REST/NRSF and REST/NRSF-VP16 resulted in ectopic neuronal marker expression in astrocytes in vitro (Fig. 5 A). However, we could not observe βIII-tubulin expression in astrocytes in vivo by simple inhibition of REST/NRSF function with DN-REST/NRSF, whereas REST/NRSF-VP16 was able to induce the neuronal marker ectopically. This difference between the two REST/NRSF mutants may be caused by as-yet-unknown mechanisms that are functioning to further repress neuronal genes in astrocytes in vivo. Although we must await further investigation to identify these mechanisms, it is conceivable that REST/NRSF plays at least some part in restricting the differentiation plasticity of astrocytes by suppressing neuronal gene expression. However, we have not yet been able to rule out the possibility that off-target variable effects of REST/NRSF-VP16 contribute to ectopic neuronal gene expression in astrocytes in vivo.
Materials and methods
Cell culture
NPCs were prepared from embryonic day (E) 14 mouse telencephalons as described previously (Nakashima et al., 1999b). Freshly isolated cells were plated onto dishes precoated with poly-l-ornithine (Sigma-Aldrich) and fibronectin (Sigma-Aldrich) and cultured in N2-supplemented DME/F-12 medium (N2/DME/F12) with 10 ng/ml basic fibroblast growth factor (bFGF; PeproTech) for 4 d. bFGF was included unless otherwise indicated. Cells were then detached and replated onto 8-well chamber slides (Thermo Fisher Scientific) precoated with poly-l-ornithine and fibronectin (6 × 104 cells per well) and 12-well plates (4 × 105 cells per well; Thermo Fisher Scientific) for immunocytochemistry and luciferase assay, respectively. Primary astrocytes from cortex of postnatal day (P) 0 and adult mice were prepared essentially as described previously (Song et al., 2002). In brief, confluent astrocytes in medium were treated with 20 µM cytosine arabinoside for 72 h to eliminate proliferating cells, followed by recovery in fresh medium for 24 h. Under these conditions, most of the cells were GFAP-positive astrocytes with little contamination by other cell types. Maturation of astrocytes was assessed by examining the expression of glutamine transporter GLT-1 (Rothstein et al., 1994). Primary neurons from E14 cortex were prepared as described previously (Martinowich et al., 2003).
Immunocytochemistry
Cells cultured on coated chamber slides were washed with PBS, fixed in 4% paraformaldehyde (PFA) in PBS, and stained with one of the following primary antibodies: rabbit anti-GFAP (1:1,500; Dako), rabbit anti-REST/NRSF (1:500; Millipore), rabbit anti-GFP (1:1,000; MBL), rat anti-GFP (1:200; Nacalai Tesque), mouse anti–microtubule-associated proteins 2a and 2b (MAP2ab) (1:1,000; Sigma-Aldrich), mouse anti-Nestin (1:1,000; Millipore), mouse anti-S100β (1:1,000; Sigma-Aldrich), guinea pig anti-GFAP (1:2,000; Advanced Immunochemical), guinea pig anti-GLT1 (Millipore), mouse anti-GS (BD), rabbit anti-Id1 (1:100; Santa Cruz Biotechnology, Inc.), and rabbit anti-cleaved caspase-3 (Asp175; 1:500; Cell Signaling Technology). The following secondary antibodies were used: Alexa 488–conjugated goat anti–mouse IgG (1:500; Invitrogen), Cy3-conjugated goat anti–mouse IgG (1:500; Jackson ImmunoResearch Laboratories, Inc.), Alexa 488–conjugated goat anti–rabbit (1:500; Invitrogen), FITC-conjugated donkey anti–rat (1:500; Jackson ImmunoResearch Laboratories, Inc.), FITC-conjugated donkey anti–guinea pig (1:500; Jackson ImmunoResearch Laboratories, Inc.), Cy3-conjugated donkey anti–rabbit (1:250; Jackson ImmunoResearch Laboratories, Inc.), and Cy5-conjugated donkey anti–guinea pig (1:250; Jackson ImmunoResearch Laboratories, Inc.). Nuclei were stained using bisbenzimide H33258 fluorochrome trihydrochloride (Nacalai Tesque). The fluorescence images were acquired using a fluorescence microscope (Axiovert 200M; Carl Zeiss, Inc.), 20x and 40x objectives, and LSM Image Browser software. Images were combined for figures using Adobe Photoshop CS. All experiments were independently replicated at least three times.
Immunohistochemistry
P8 pups were perfused through the left ventricle with 4% PFA in PBS (pH 7.4). Brains were dissected out and postfixed overnight at 4°C, cryoprotected in 30% sucrose in PBS overnight at 4°C, and then embedded in OCT compound (Tissue Tek). Cryostat sections (12 µm) were cut and affixed to APS-coated glass slides (Matsunami Glass). The sections were then permeabilized in TBS-T (TBS containing 0.05% Tween 20) for 10 min and blocked with 10% normal donkey serum (Millipore) in PBS for 1 h at room temperature. The sections were reacted overnight at 4°C with primary antibodies in blocking solution. After three washes with PBS, the sections were incubated in PBS containing the secondary antibodies for 1 h. Optical sections were viewed using a scanning-laser confocal imaging system (LSM510; Carl Zeiss, Inc.) with 40x and 63x objectives. Image acquisition was performed with LSM Image Browser software (Carl Zeiss, Inc.). Images were combined for figures using Adobe Photoshop CS.
RNA isolation and reverse-transcription PCR (RT-PCR)
Total RNAs were isolated using Sepasol RNAI (Nacalai Tesque) and treated with DNase I (Promega). First-strand cDNAs were synthesized from 1 µg total RNA with Superscript II (Invitrogen). The RT products (1 µl) were used as templates for PCR amplification (AmpliTaq Gold; Applied Biosystems) in 25 µl reaction solution containing 2 µM gene-specific primers. Quantitative real-time PCR was performed in MX3000p (Agilent Technologies) using SYBR-Green PCR Master Mix (Takara Bio Inc.). The gene-specific primers were as follows: mouse REST/NRSF: REST-S, 5′-gaactcacacaggagaacgcc-3′; REST-AS, 5′-gattacacttcttagaagccg-3′; mouse G3PDH: G3PDH-S, 5′-accacagtccatgccatcac-3′; G3PDH-AS, 5′-tccaccaccctgttgctgta-3′; mouse SCG10: SCG10-S, 5′-gagctgtctatgctgtcactg-3′; SCG10-AS, 5′-tgttcctgcgaacctctgca-3′; mouse Hes5: Hes5-S, 5′-aagagcctgcaccaggacta-3′; Hes5-AS, 5′-cgctggaagtggtaaagca-3′; mouse Id1: Id1-S, 5′-ggtggatccaccatgaaggtcgccagtg-3′; Id1-AS, 5′-ggtggatccgtccatctggtccctcagtgc-3′. For analysis of the REST/NRSF target genes GAD1, BDNF, Synaptotagmin4, and Calbindin, primer sets were designed according to a previous report (Ballas et al., 2005).
Western blot analysis
Western blot analysis was performed by an established method. 20-μg protein samples of each total cell extract were separated by 5–20% gradient SDS-PAGE, transferred to a nitrocellulose membrane, and probed with anti-REST (rabbit IgG, Millipore), anti–β-actin (mouse IgG; Sigma-Aldrich), anti-Smad1 (mouse IgG; Santa Cruz Biotechnology, Inc.), and anti-phosphorylated Smad1/5/8 (Cell Signaling Technology) antibodies. Signals were detected with HRP-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, Inc.) using an ECL kit (GE Healthcare). The amounts of proteins loaded in each slot were normalized to those of β-actin.
Luciferase assay
NPCs, primary astrocytes, and ES cells were transfected with luciferase reporter plasmids, pGL3 promoter (Promega) with or without RE1/NRSE (5′-TTCAGCACCACGGACAGCGCC-3′), using Trans-IT LT-1 (Mirus) or Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. Luciferase reporter constructs with a REST/NRSF gene regulatory fragment (+65 bp to +1607 bp) or harboring Smad binding element mutations were used. To manipulate BMP signaling, three expression constructs were also used: smad6 (inhibitory smad) and ALK3QD (constitutively activated BMP type I receptor) cDNA inserted into pMY vector (Morita et al., 2000), DN-Smad1 in pCI vector (Promega). As an internal control, a plasmid containing a sea pansy luciferase expression construct (pRL-CMV; Promega) was cotransfected with the reporter constructs described above. To evaluate repressor activity of REST/NRSF on SCG10, pGL3-basic vectors carrying a minimum promoter region of SCG10 with intact (pGL3-S10PR S36(+); 5′-TTCAGCACCACGGAGAGTGCC-3′) and mutated (pGL3-S10PR Sm36(+); 5′-TTCAGCACCACTTAGAGTGCC-3′) RE1/NRSE were also used (underlined GG dinucleotides were substituted by TT dinucleotides). On the following day, cells were solubilized and luciferase activity was measured according to the procedures recommended for the dual luciferase reporter assay system (Promega). ARVO (PerkinElmer) was used for quantification.
Recombinant retrovirus construction and infection
cDNAs encoding full-length mouse REST/NRSF and its DNA-binding domain were cloned into expression vector pMY containing an internal ribosome entry site followed by the region upstream of the GFP gene (Kitamura et al., 2003). The Plat-E packaging cell line was transiently transfected with this construct by Trans-IT 293 (Mirus). On the following day, the medium was replaced with N2/DMEM/F12 and the cells were cultured in this medium for 1 d before virus was collected.
Lentivirus construction and infection
The DNA-binding domain of REST/NRSF and the DNA-binding domain of REST/NRSF fused with VP16 were subcloned into CSC PW lentiviral vector (Kuwabara et al., 2004). The production and infection of lentivirus followed previous reports (Miyoshi et al., 1999; Kohyama et al., 2005). The shRNA sequence targeted for REST/NRSF was 5′-gaggcagtctcttcaacaa-3′. This shRNA cassette was subcloned into CSC PW lentiviral vector containing both CMV-EGFP and murine U6 promoter cassettes derived from the original CSC PW plasmid (Kuwabara et al., 2004). Lentiviruses (200 nl) were stereotactically injected into the striatum of the adult mouse brain (0 mm anterior, ± 2 mm lateral to bregma, and 2.9 mm below the brain surface).
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) was performed according to a protocol published by Upstate Biotechnology. Undifferentiated NPCs and differentiated cells were exposed to formaldehyde, at a final concentration of 1%, added directly to the tissue culture medium. Co-immunoprecipitated DNA was used as a template for PCR of the genomic region containing two smad-binding elements in the REST/NRSF gene with the following sets of primers: REST/NRSF chipS (5′-GCCCAAGTTTGCAAAGAGCTG-3′) and REST/NRSF chipAS (5′-CAACAAAAAGTTGAGCCCGAATG-3′). Quantitative real-time PCR was performed in the Mx3000p system (Agilent Technologies) using SYBR-Green PCR Master Mix (Takara Bio Inc.). For analysis of REST/NRSF target genes including GAD1, BDNF, Synaptotagmin4, and Calbindin, primer sets were designed according to a previous report (Ballas et al., 2005). Antibodies used for the ChIP assay were rabbit anti-REST/NRSF (Millipore), mouse anti-Smad1 (Santa Cruz Biotechnology, Inc.), rabbit anti-acetylated histone H3 (Millipore), rabbit anti-trimethylated histone H3-lysine 4 (Abcam), and mouse/rabbit control IgGs (Santa Cruz Biotechnology, Inc.).
Online supplemental material
Fig. S1 shows that phosphorylation of Smad1/5/8 induced by BMP2 exposure in NPCs and the effect of BMP2 on oligodendrocyte differentiation of NPCs. Expression kinetics of BMP signaling-target genes are also shown. Fig. S2 shows reporter assays using a dominant-negative form of Smad1 and ChIP assays using antibodies against Smad1, H3K4me3, and AcH3 in NPCs, astrocytes, and neurons. Fig. S3 shows the effect of REST/NRSF or DN-REST/NRSF expression on cell proliferation and cell death in NPCs. Fig. S4 shows expression of GLT-1 in cultured astrocytes and REST/NRSF expression in the different regions of mouse brain. Smad1 occupancy in REST/NRSF regulatory region in ES cells is shown. Fig. S5 A shows a knockdown study using an RNAi approach in cultured astrocytes. We also show the effect of DN-REST/NRSF in cultured astrocytes derived from adult mice. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200908048/DC1.
Acknowledgments
We thank Dr. T. Kitamura (Tokyo University) for pMY vector and Plat-E cells; Dr. A. Smith (Cambridge University) for ES cells; Dr. N. Mori (Nagasaki University) for REST/NRSF promoter, SCG10 promoter, and REST/NRSF expression vector; Dr. R. Kageyama for DN-Smad1 construct; and Astellas Pharma for BMP2. We appreciate Drs. T. Sunabori, M. Namihira, Y. Bessho, and T. Matsui for valuable discussions. We also thank Dr. I. Smith for helpful comments and critical reading of the manuscript. We are very grateful to N. Ueda and M. Tano for excellent secretarial assistance. Many thanks to S. Urayama and N. Namihira for technical help.
This work has been supported by a Grant-in-Aid for Young Scientists (Start-up) and a Grant-in-Aid for Scientific Research on Priority Areas-Molecular Brain Science, and the NAIST Global COE Program (Frontier Biosciences: Strategies for survival and adaptation in a changing global environment) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Footnotes
Abbreviations used in this paper:
- bHLH
- basic helix-loop-helix
- BMP
- bone morphogenetic protein
- ChIP
- chromatin immunoprecipitation
- CNS
- central nervous system
- DN
- dominant negative
- ES
- embryonic stem
- Hes
- hairy enhancer of split
- Id
- inhibitor of differentiation
- NPC
- neural progenitor cell
- NRSF
- neuron-restrictive silencer factor
- REST
- RE1 silencer of transcription
- SBE
- Smad-binding element
References
- Araya R., Kudo M., Kawano M., Ishii K., Hashikawa T., Iwasato T., Itohara S., Terasaki T., Oohira A., Mishina Y., Yamada M. 2008. BMP signaling through BMPRIA in astrocytes is essential for proper cerebral angiogenesis and formation of the blood-brain-barrier. Mol. Cell. Neurosci. 38:417–430 10.1016/j.mcn.2008.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballas N., Battaglioli E., Atouf F., Andres M.E., Chenoweth J., Anderson M.E., Burger C., Moniwa M., Davie J.R., Bowers W.J., et al. 2001. Regulation of neuronal traits by a novel transcriptional complex. Neuron. 31:353–365 10.1016/S0896-6273(01)00371-3 [DOI] [PubMed] [Google Scholar]
- Ballas N., Grunseich C., Lu D.D., Speh J.C., Mandel G. 2005. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 121:645–657 10.1016/j.cell.2005.03.013 [DOI] [PubMed] [Google Scholar]
- Bertrand N., Castro D.S., Guillemot F. 2002. Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci. 3:517–530 10.1038/nrn874 [DOI] [PubMed] [Google Scholar]
- Bonaguidi M.A., McGuire T., Hu M., Kan L., Samanta J., Kessler J.A. 2005. LIF and BMP signaling generate separate and discrete types of GFAP-expressing cells. Development. 132:5503–5514 10.1242/dev.02166 [DOI] [PubMed] [Google Scholar]
- Bonni A., Sun Y., Nadal-Vicens M., Bhatt A., Frank D.A., Rozovsky I., Stahl N., Yancopoulos G.D., Greenberg M.E. 1997. Regulation of gliogenesis in the central nervous system by the JAK-STAT signaling pathway. Science. 278:477–483 10.1126/science.278.5337.477 [DOI] [PubMed] [Google Scholar]
- Bruce A.W., Donaldson I.J., Wood I.C., Yerbury S.A., Sadowski M.I., Chapman M., Göttgens B., Buckley N.J. 2004. Genome-wide analysis of repressor element 1 silencing transcription factor/neuron-restrictive silencing factor (REST/NRSF) target genes. Proc. Natl. Acad. Sci. USA. 101:10458–10463 10.1073/pnas.0401827101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.F., Paquette A.J., Anderson D.J. 1998. NRSF/REST is required in vivo for repression of multiple neuronal target genes during embryogenesis. Nat. Genet. 20:136–142 10.1038/2431 [DOI] [PubMed] [Google Scholar]
- Chong J.A., Tapia-Ramírez J., Kim S., Toledo-Aral J.J., Zheng Y., Boutros M.C., Altshuller Y.M., Frohman M.A., Kraner S.D., Mandel G. 1995. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell. 80:949–957 10.1016/0092-8674(95)90298-8 [DOI] [PubMed] [Google Scholar]
- Conaco C., Otto S., Han J.J., Mandel G. 2006. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc. Natl. Acad. Sci. USA. 103:2422–2427 10.1073/pnas.0511041103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan M., DiCicco-Bloom E.M., Xiang X., Benezra R., Chada K. 1992. The gene for the helix-loop-helix protein, Id, is specifically expressed in neural precursors. Dev. Biol. 154:1–10 10.1016/0012-1606(92)90042-F [DOI] [PubMed] [Google Scholar]
- Ellmeier W., Weith A. 1995. Expression of the helix-loop-helix gene Id3 during murine embryonic development. Dev. Dyn. 203:163–173 [DOI] [PubMed] [Google Scholar]
- Gage F.H. 2000. Mammalian neural stem cells. Science. 287:1433–1438 10.1126/science.287.5457.1433 [DOI] [PubMed] [Google Scholar]
- Gradwohl G., Fode C., Guillemot F. 1996. Restricted expression of a novel murine atonal-related bHLH protein in undifferentiated neural precursors. Dev. Biol. 180:227–241 10.1006/dbio.1996.0297 [DOI] [PubMed] [Google Scholar]
- He F., Ge W., Martinowich K., Becker-Catania S., Coskun V., Zhu W., Wu H., Castro D., Guillemot F., Fan G., et al. 2005. A positive autoregulatory loop of Jak-STAT signaling controls the onset of astrogliogenesis. Nat. Neurosci. 8:616–625 10.1038/nn1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh J., Gage F.H. 2004. Epigenetic control of neural stem cell fate. Curr. Opin. Genet. Dev. 14:461–469 10.1016/j.gde.2004.07.006 [DOI] [PubMed] [Google Scholar]
- Huang Y., Myers S.J., Dingledine R. 1999. Transcriptional repression by REST: recruitment of Sin3A and histone deacetylase to neuronal genes. Nat. Neurosci. 2:867–872 10.1038/13165 [DOI] [PubMed] [Google Scholar]
- Imamura T., Takase M., Nishihara A., Oeda E., Hanai J., Kawabata M., Miyazono K. 1997. Smad6 inhibits signalling by the TGF-beta superfamily. Nature. 389:622–626 10.1038/39355 [DOI] [PubMed] [Google Scholar]
- Jenuwein T., Allis C.D. 2001. Translating the histone code. Science. 293:1074–1080 10.1126/science.1063127 [DOI] [PubMed] [Google Scholar]
- Johnson R., Gamblin R.J., Ooi L., Bruce A.W., Donaldson I.J., Westhead D.R., Wood I.C., Jackson R.M., Buckley N.J. 2006. Identification of the REST regulon reveals extensive transposable element-mediated binding site duplication. Nucleic Acids Res. 34:3862–3877 10.1093/nar/gkl525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura T., Koshino Y., Shibata F., Oki T., Nakajima H., Nosaka T., Kumagai H. 2003. Retrovirus-mediated gene transfer and expression cloning: powerful tools in functional genomics. Exp. Hematol. 31:1007–1014 [PubMed] [Google Scholar]
- Koenigsberger C., Chicca J.J., II, Amoureux M.C., Edelman G.M., Jones F.S. 2000. Differential regulation by multiple promoters of the gene encoding the neuron-restrictive silencer factor. Proc. Natl. Acad. Sci. USA. 97:2291–2296 10.1073/pnas.050578797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohyama J., Tokunaga A., Fujita Y., Miyoshi H., Nagai T., Miyawaki A., Nakao K., Matsuzaki Y., Okano H. 2005. Visualization of spatiotemporal activation of Notch signaling: live monitoring and significance in neural development. Dev. Biol. 286:311–325 10.1016/j.ydbio.2005.08.003 [DOI] [PubMed] [Google Scholar]
- Kohyama J., Kojima T., Takatsuka E., Yamashita T., Namiki J., Hsieh J., Gage F.H., Namihira M., Okano H., Sawamoto K., Nakashima K. 2008. Epigenetic regulation of neural cell differentiation plasticity in the adult mammalian brain. Proc. Natl. Acad. Sci. USA. 105:18012–18017 10.1073/pnas.0808417105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima T., Murai K., Naruse Y., Takahashi N., Mori N. 2001. Cell-type non-selective transcription of mouse and human genes encoding neural-restrictive silencer factor. Brain Res. Mol. Brain Res. 90:174–186 10.1016/S0169-328X(01)00107-3 [DOI] [PubMed] [Google Scholar]
- Kusanagi K., Inoue H., Ishidou Y., Mishima H.K., Kawabata M., Miyazono K. 2000. Characterization of a bone morphogenetic protein-responsive Smad-binding element. Mol. Biol. Cell. 11:555–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara T., Hsieh J., Nakashima K., Taira K., Gage F.H. 2004. A small modulatory dsRNA specifies the fate of adult neural stem cells. Cell. 116:779–793 10.1016/S0092-8674(04)00248-X [DOI] [PubMed] [Google Scholar]
- Laywell E.D., Rakic P., Kukekov V.G., Holland E.C., Steindler D.A. 2000. Identification of a multipotent astrocytic stem cell in the immature and adult mouse brain. Proc. Natl. Acad. Sci. USA. 97:13883–13888 10.1073/pnas.250471697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lie D.C., Colamarino S.A., Song H.J., Désiré L., Mira H., Consiglio A., Lein E.S., Jessberger S., Lansford H., Dearie A.R., Gage F.H. 2005. Wnt signalling regulates adult hippocampal neurogenesis. Nature. 437:1370–1375 10.1038/nature04108 [DOI] [PubMed] [Google Scholar]
- Lunyak V.V., Burgess R., Prefontaine G.G., Nelson C., Sze S.H., Chenoweth J., Schwartz P., Pevzner P.A., Glass C., Mandel G., Rosenfeld M.G. 2002. Corepressor-dependent silencing of chromosomal regions encoding neuronal genes. Science. 298:1747–1752 10.1126/science.1076469 [DOI] [PubMed] [Google Scholar]
- Lyden D., Young A.Z., Zagzag D., Yan W., Gerald W., O’Reilly R., Bader B.L., Hynes R.O., Zhuang Y., Manova K., Benezra R. 1999. Id1 and Id3 are required for neurogenesis, angiogenesis and vascularization of tumour xenografts. Nature. 401:670–677 10.1038/44334 [DOI] [PubMed] [Google Scholar]
- Martens J.A., Winston F. 2003. Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr. Opin. Genet. Dev. 13:136–142 10.1016/S0959-437X(03)00022-4 [DOI] [PubMed] [Google Scholar]
- Martinowich K., Hattori D., Wu H., Fouse S., He F., Hu Y., Fan G., Sun Y.E. 2003. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 302:890–893 10.1126/science.1090842 [DOI] [PubMed] [Google Scholar]
- Matsumoto S., Banine F., Struve J., Xing R., Adams C., Liu Y., Metzger D., Chambon P., Rao M.S., Sherman L.S. 2006. Brg1 is required for murine neural stem cell maintenance and gliogenesis. Dev. Biol. 289:372–383 10.1016/j.ydbio.2005.10.044 [DOI] [PubMed] [Google Scholar]
- Mehler M.F., Mabie P.C., Zhu G., Gokhan S., Kessler J.A. 2000. Developmental changes in progenitor cell responsiveness to bone morphogenetic proteins differentially modulate progressive CNS lineage fate. Dev. Neurosci. 22:74–85 10.1159/000017429 [DOI] [PubMed] [Google Scholar]
- Miyawaki T., Uemura A., Dezawa M., Yu R.T., Ide C., Nishikawa S., Honda Y., Tanabe Y., Tanabe T. 2004. Tlx, an orphan nuclear receptor, regulates cell numbers and astrocyte development in the developing retina. J. Neurosci. 24:8124–8134 10.1523/JNEUROSCI.2235-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi H., Smith K.A., Mosier D.E., Verma I.M., Torbett B.E. 1999. Transduction of human CD34+ cells that mediate long-term engraftment of NOD/SCID mice by HIV vectors. Science. 283:682–686 10.1126/science.283.5402.682 [DOI] [PubMed] [Google Scholar]
- Mori N., Stein R., Sigmund O., Anderson D.J. 1990. A cell type-preferred silencer element that controls the neural-specific expression of the SCG10 gene. Neuron. 4:583–594 10.1016/0896-6273(90)90116-W [DOI] [PubMed] [Google Scholar]
- Morita S., Kojima T., Kitamura T. 2000. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 7:1063–1066 10.1038/sj.gt.3301206 [DOI] [PubMed] [Google Scholar]
- Nakashima K., Wiese S., Yanagisawa M., Arakawa H., Kimura N., Hisatsune T., Yoshida K., Kishimoto T., Sendtner M., Taga T. 1999a. Developmental requirement of gp130 signaling in neuronal survival and astrocyte differentiation. J. Neurosci. 19:5429–5434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K., Yanagisawa M., Arakawa H., Kimura N., Hisatsune T., Kawabata M., Miyazono K., Taga T. 1999b. Synergistic signaling in fetal brain by STAT3-Smad1 complex bridged by p300. Science. 284:479–482 10.1126/science.284.5413.479 [DOI] [PubMed] [Google Scholar]
- Nakashima K., Takizawa T., Ochiai W., Yanagisawa M., Hisatsune T., Nakafuku M., Miyazono K., Kishimoto T., Kageyama R., Taga T. 2001. BMP2-mediated alteration in the developmental pathway of fetal mouse brain cells from neurogenesis to astrocytogenesis. Proc. Natl. Acad. Sci. USA. 98:5868–5873 10.1073/pnas.101109698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namihira M., Kohyama J., Semi K., Sanosaka T., Deneen B., Taga T., Nakashima K. 2009. Committed neuronal precursors confer astrocytic potential on residual neural precursor cells. Dev. Cell. 16:245–255 10.1016/j.devcel.2008.12.014 [DOI] [PubMed] [Google Scholar]
- Nishihara S., Tsuda L., Ogura T. 2003. The canonical Wnt pathway directly regulates NRSF/REST expression in chick spinal cord. Biochem. Biophys. Res. Commun. 311:55–63 10.1016/j.bbrc.2003.09.158 [DOI] [PubMed] [Google Scholar]
- Olguín P., Oteíza P., Gamboa E., Gómez-Skármeta J.L., Kukuljan M. 2006. RE-1 silencer of transcription/neural restrictive silencer factor modulates ectodermal patterning during Xenopus development. J. Neurosci. 26:2820–2829 10.1523/JNEUROSCI.5037-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto S.J., McCorkle S.R., Hover J., Conaco C., Han J.J., Impey S., Yochum G.S., Dunn J.J., Goodman R.H., Mandel G. 2007. A new binding motif for the transcriptional repressor REST uncovers large gene networks devoted to neuronal functions. J. Neurosci. 27:6729–6739 10.1523/JNEUROSCI.0091-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein J.D., Martin L., Levey A.I., Dykes-Hoberg M., Jin L., Wu D., Nash N., Kuncl R.W. 1994. Localization of neuronal and glial glutamate transporters. Neuron. 13:713–725 10.1016/0896-6273(94)90038-8 [DOI] [PubMed] [Google Scholar]
- Schoenherr C.J., Paquette A.J., Anderson D.J. 1996. Identification of potential target genes for the neuron-restrictive silencer factor. Proc. Natl. Acad. Sci. USA. 93:9881–9886 10.1073/pnas.93.18.9881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuurmans C., Guillemot F. 2002. Molecular mechanisms underlying cell fate specification in the developing telencephalon. Curr. Opin. Neurobiol. 12:26–34 10.1016/S0959-4388(02)00286-6 [DOI] [PubMed] [Google Scholar]
- See J., Mamontov P., Ahn K., Wine-Lee L., Crenshaw E.B., III, Grinspan J.B. 2007. BMP signaling mutant mice exhibit glial cell maturation defects. Mol. Cell. Neurosci. 35:171–182 10.1016/j.mcn.2007.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setoguchi H., Namihira M., Kohyama J., Asano H., Sanosaka T., Nakashima K. 2006. Methyl-CpG binding proteins are involved in restricting differentiation plasticity in neurons. J. Neurosci. Res. 84:969–979 10.1002/jnr.21001 [DOI] [PubMed] [Google Scholar]
- Shi Y., Chichung Lie D., Taupin P., Nakashima K., Ray J., Yu R.T., Gage F.H., Evans R.M. 2004. Expression and function of orphan nuclear receptor TLX in adult neural stem cells. Nature. 427:78–83 10.1038/nature02211 [DOI] [PubMed] [Google Scholar]
- Singh S.K., Kagalwala M.N., Parker-Thornburg J., Adams H., Majumder S. 2008. REST maintains self-renewal and pluripotency of embryonic stem cells. Nature. 453:223–227 10.1038/nature06863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H., Stevens C.F., Gage F.H. 2002. Astroglia induce neurogenesis from adult neural stem cells. Nature. 417:39–44 10.1038/417039a [DOI] [PubMed] [Google Scholar]
- Su X., Kameoka S., Lentz S., Majumder S. 2004. Activation of REST/NRSF target genes in neural stem cells is sufficient to cause neuronal differentiation. Mol. Cell. Biol. 24:8018–8025 10.1128/MCB.24.18.8018-8025.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa T., Nakashima K., Namihira M., Ochiai W., Uemura A., Yanagisawa M., Fujita N., Nakao M., Taga T. 2001. DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Dev. Cell. 1:749–758 10.1016/S1534-5807(01)00101-0 [DOI] [PubMed] [Google Scholar]
- Temple S. 2001. The development of neural stem cells. Nature. 414:112–117 10.1038/35102174 [DOI] [PubMed] [Google Scholar]
- Tokunaga A., Kohyama J., Yoshida T., Nakao K., Sawamoto K., Okano H. 2004. Mapping spatio-temporal activation of Notch signaling during neurogenesis and gliogenesis in the developing mouse brain. J. Neurochem. 90:142–154 10.1111/j.1471-4159.2004.02470.x [DOI] [PubMed] [Google Scholar]
- Uemura A., Kusuhara S., Wiegand S.J., Yu R.T., Nishikawa S. 2006. Tlx acts as a proangiogenic switch by regulating extracellular assembly of fibronectin matrices in retinal astrocytes. J. Clin. Invest. 116:369–377 10.1172/JCI25964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y., Kameoka S., Gopalakrishnan V., Aldape K.D., Pan Z.Z., Lang F.F., Majumder S. 2004. Conversion of myoblasts to physiologically active neuronal phenotype. Genes Dev. 18:889–900 10.1101/gad.1179004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westbrook T.F., Hu G., Ang X.L., Mulligan P., Pavlova N.N., Liang A., Leng Y., Maehr R., Shi Y., Harper J.W., Elledge S.J. 2008. SCFbeta-TRCP controls oncogenic transformation and neural differentiation through REST degradation. Nature. 452:370–374 10.1038/nature06780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa M., Nakashima K., Takizawa T., Ochiai W., Arakawa H., Taga T. 2001. Signaling crosstalk underlying synergistic induction of astrocyte differentiation by BMPs and IL-6 family of cytokines. FEBS Lett. 489:139–143 10.1016/S0014-5793(01)02095-6 [DOI] [PubMed] [Google Scholar]
- Yoshiura S., Ohtsuka T., Takenaka Y., Nagahara H., Yoshikawa K., Kageyama R. 2007. Ultradian oscillations of Stat, Smad, and Hes1 expression in response to serum. Proc. Natl. Acad. Sci. USA. 104:11292–11297 10.1073/pnas.0701837104 [DOI] [PMC free article] [PubMed] [Google Scholar]