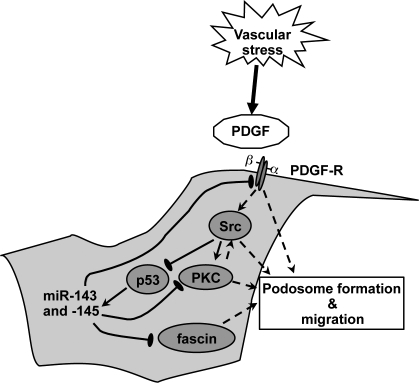
PDGF enhances podosome formation and cell migration by regulating expression of the microRNAs miR-143 and -145, which target PDGF-R, PKC-ε, and fascin.
Abstract
Smooth muscle cell (SMC) plasticity plays an important role during development and in vascular pathologies such as atherosclerosis and restenosis. It was recently shown that down-regulation of microRNA (miR)-143 and -145, which are coexpressed from a single promoter, regulates the switch from contractile to synthetic phenotype, allowing SMCs to migrate and proliferate. We show in this study that loss of miR-143/145 in vitro and in vivo results in the formation of podosomes, which are actin-rich membrane protrusions involved in the migration of several cell types, including SMCs. We further show that platelet-derived growth factor (PDGF) mediates podosome formation in SMCs through the regulation of miR-143/145 expression via a pathway involving Src and p53. Moreover, we identify key podosome regulators as targets of miR-143 (PDGF receptor α and protein kinase C ε) and miR-145 (fascin). Thus, dysregulation of the miR-143 and -145 genes is causally involved in the aberrant SMC plasticity encountered during vascular disease, in part through the up-regulation of an autoregulatory loop that promotes podosome formation.
Introduction
Vascular smooth muscle cells (SMCs [VSMCs]) can switch between differentiated (contractile) and dedifferentiated (synthetic migratory) phenotypes (Gimona et al., 1990; Sobue et al., 1999). Migration of SMCs plays a critical role in many physiological and pathological processes, including atherosclerosis, angiogenesis, smooth muscle hypertrophy, and hyperplasia. PDGF is one of the most potent stimuli for migration of mesenchymal cell types, including VSMCs. Furthermore, excessive PDGF production has been implicated in several pathological vascular disorders (Alvarez et al., 2006; Andrae et al., 2008).
An important morphological feature of VSMCs migrating in vitro is a membrane structure called a podosome (Gimona et al., 2003; Linder and Aepfelbacher, 2003). Podosomes are dynamic, short-lived, actin-rich protrusions of the plasma membrane, which are thought to mediate adhesion to and, in some cases, degradation of the surrounding extracellular matrix. Podosomes are also found in other migratory cells such as monocytes and endothelial cells (Gimona et al., 2008). Many types of human cancer cells as well as Rous sarcoma virus–transformed fibroblasts form highly related structures, termed invadopodia, whose presence is correlated with invasive and metastatic behavior (Gimona et al., 2008).
MicroRNAs (miRs) are 20–25-nt-long noncoding RNAs that negatively regulate gene expression by binding to sites in the 3′ untranslated region (UTR) of target mRNAs (Bartel, 2004). These small RNA molecules are involved in processes such as cell differentiation and proliferation (Chen et al., 2004). Recently, we and others (Boettger et al., 2009; Cheng et al., 2009; Cordes et al., 2009; Elia et al., 2009b; Xin et al., 2009) have shown that miR-143 and -145 regulate the VSMC phenotypic switch from a contractile/nonproliferative to a migrating/proliferative state (Owens, 1995). miR-143 and -145 are organized in a cluster transcribed from the same primary miR (Cordes et al., 2009; Xin et al., 2009). In this study, we have used the knockout (KO) mouse we generated, referred to as the miR-143(145) KO, in which the expression of both miRs is prevented, to investigate the molecular mechanism underlying the regulation of migration by miR-143/145.
Results and discussion
The miR-143/145 gene products inhibit podosome formation in VSMCs
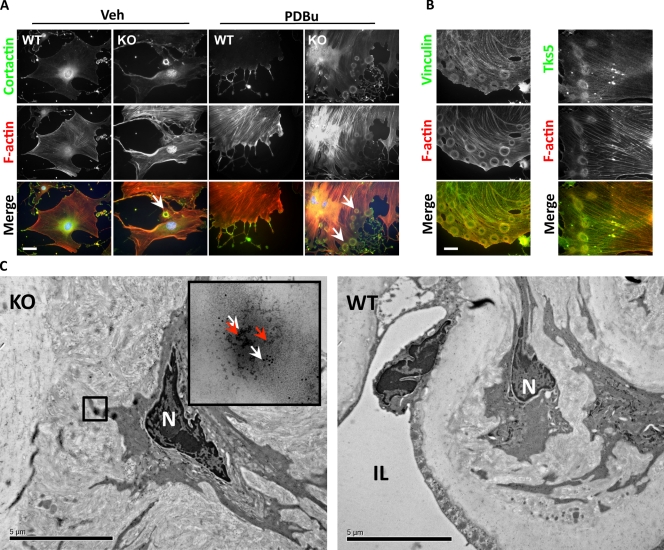
VSMCs form podosomes when they migrate and invade, so we first tested whether expression of these miRs affects podosome formation. Primary mouse aorta SMCs cultured on glass coverslips contained prominent actin stress fibers and large vinculin-containing focal adhesions. However, some (7.6 ± 2.5%) of the primary VSMCs obtained from miR-143(145) KO mouse aortas also contained podosome-like structures at the cell periphery (Fig. 1 A). Treatment with phorbol dibutyrate (PDBu), a known inducer of podosomes in SMCs (Gimona et al., 2003), greatly increased the number of KO cells presenting with these structures (91.3 ± 3.2%). These podosomes were organized into rings (known as rosettes), clusters of rosettes, and occasionally peripheral actin belts reminiscent of the structures described in primary osteoclasts (Fig. S1 A; Destaing et al., 2003). This was in stark contrast to PDBu-treated wild-type (WT) VSMCs, in which podosomes were present in <10% of the cells and found in a scattered dot-like conformation. Colocalization of proteins known to be expressed in podosomes such as cortactin, vinculin, and Tks5 (Linder and Aepfelbacher, 2003; Seals et al., 2005) confirmed that these structures were in fact podosomes (Fig. 1 B). To determine whether miR-143/145 loss facilitated podosome formation, we restored miR-143 or -145 expression in the miR-143(145) KO VSMCs with the recombinant adenoviruses Ad–miR-143 or –miR-145. Transduction with either virus completely abrogated podosome formation (Fig. S1 B), whereas a control miR-208 had no effect.
Figure 1.
Podosome formation in miR-143(145) KO SMCs in vitro and in vivo. (A) Cortactin and F-actin IF to visualize podosomes in VSMCs isolated from WT and miR-143(145) KO mice treated with PDBu or with vehicle (Veh). Arrowheads indicate the rosettes of podosomes in miR-143(145) KO cells. Quantification of three separate experiments revealed podosomes in 7.6% (±2.5%) of untreated KO SMCs and in 91.3% (±3.2%) of PDBu-treated cells. (B) Colocalization of the podosome proteins vinculin (left) and Tks5 (right) with F-actin in KO VSMCs treated with PDBu. (C) Immunoelectron microscopy of miR-143(145) KO and WT aortas. White arrows indicate cortactin (10-nm gold particles), and red arrows indicate Tks5 (5-nm gold particles). The boxed area indicates what appears to be a rosette of podosomes located in an SMC of the miR-143(145) KO mouse, confirmed by the observation of the gold-labeled podosome markers Tks5 and cortactin in the higher magnification inset. N, nuclei; IL, intima layer. Bars: (A and B) 10 µm; (C) 5 µm.
Next we analyzed aortas from miR-143(145) KO and WT mice by immunoelectron microscopy to determine whether podosomes formed in vivo. KO SMCs showed the formation of circular structures remarkably similar to the ones observed in vitro. These structures costained for the podosome proteins cortactin and Tks5 and were not observed in the aortas of WT littermates (Fig. 1 C). These data suggest that podosomes form in vivo, that their morphological characteristics are retained in tissue culture, and that miR-143/145 controls podosome formation in vivo and in vitro.
Src tyrosine kinase activity promotes podosome formation (Gimona et al., 2008); the established VSMC line A7r5 forms abundant rosettes of podosomes when transformed with the constitutively active Src mutant, Src-Y527F (Furmaniak-Kazmierczak et al., 2007). We found that Src-transformed A7r5 cells had reduced expression of miR-143 and -145 compared with the parental cell line and that miR expression was partially restored by treatment with the Src inhibitor PP2 (Fig. 2 A). In addition, overexpression of either Ad–miR-143 or –miR-145 inhibited the formation of podosome rosettes, whereas overexpression of Ad–miR-208 had no effect (Fig. S2 A). Thus, Src-mediated down-regulation of the miR-143/145 gene is required for the formation of podosomes. Together, these observations strongly suggest a key role for these miRs in regulating podosome formation in VSMCs.
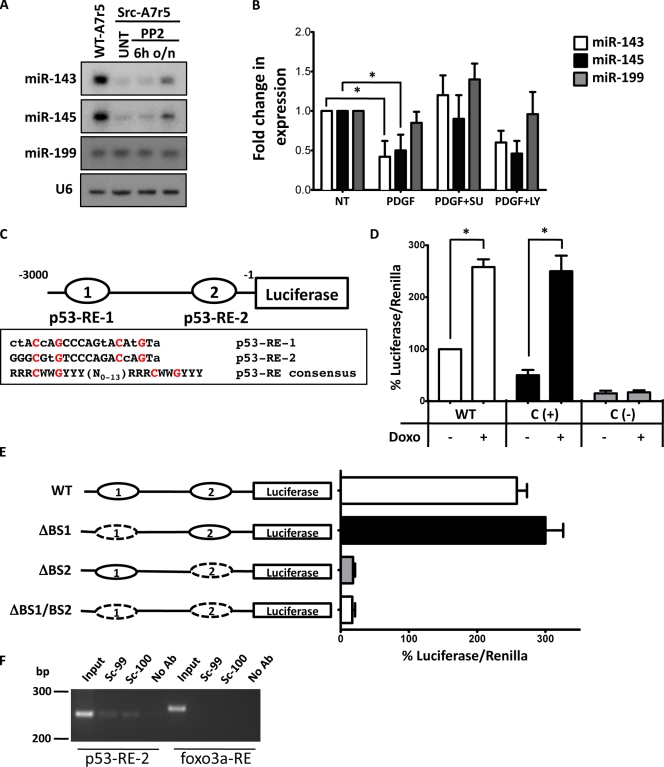
Figure 2.
miR-143 and -145 regulation by a pathway involving PDGF, Src, and p53. (A) Representative Northern blot for miR-143 and -145 in WT (WT-A7r5) and Src-Y527F–transformed (Src-A7r5) A7r5 cells treated with the Src inhibitor PP2 or with vehicle (UNT). 6h, after 6 h of treatment; o/n, after overnight treatment. (B) WT VSMCs were treated with PDGF in the presence or absence of the Src inhibitor SU6656 (SU) and the PI3-K inhibitor LY294002 (LY), and miR-143 and -145 expression was evaluated by qRT-PCR. U6 small nuclear RNA was used as internal control. All measurements were calculated as the percentage of control (NT), and error bars were calculated as propagated standard errors of the mean of triplicate measurements from each experiment. *, P < 0.05. (C) A schematic description of the putative miR-143/145 promoter with two potential p53-REs, p53-RE-1 (−3176) and p53-RE-2 (−118), as compared with the p53-RE consensus, where R: A G, W: A T, and Y: C T. Lowercase letters denote deviations from the consensus. The conserved nucleotides C and G are highlighted in red. (D) Luciferase assays with miR-143/145 promoter vector in NIH/3T3 treated or not with doxorubicin (Doxo). The promoter of the MDM2 gene was used as a positive control (C (+)), whereas the promoter of E2F was used as negative control (C (−)). (E) Deletion analysis identifies the importance of miR-143/145 p53-RE-2 in doxorubicin-mediated induction of the luciferase activity. Dashed ovals represent the deleted p53-REs. (D and E) Measurements were calculated as the percentage of control (WT promoter without doxorubicin), and error bars were calculated as propagated standard errors of the mean of triplicate measurements from each experiment on 3T3 cells. *, P < 0.05. (F) ChIP assay confirms that p53 specifically interacts with p53-RE-2. Sequences from the upstream region of the rat miR-1 gene without p53-binding sites serve as a negative control. No Ab, isotypic IgG; Sc-99, p53 antibody clone sc-99; Sc-100, p53 antibody clone sc-100.
Previously, we demonstrated the role of the miR-143/145 cluster in maintaining the differentiation status of VSMCs and proposed that their dysregulation underlies the histological changes observed in vessels in pathologies such as atherosclerosis and restenosis (Elia et al., 2009b). In this study, we show the ability of these cells to form podosomes, which are structures involved in cell migration and invasiveness, in vitro and in vivo. This is the first direct correlation between podosome formation and vascular pathologies such as atherosclerosis and restenosis. The miR-143/145 cluster has also been documented to play an important role in cancer. Dysregulation of miR-143 and -145 as the result of chromosomal deletion at 5q32 occurs in colon (Michael et al., 2003) and breast (Iorio et al., 2005) adenocarcinomas. Most B cell malignancies (Akao et al., 2007) and cancer cell lines (Michael et al., 2003) have down-regulation of both miRs. The presence of invadopodia, which are structurally related to podosomes, has been correlated with invasiveness of cancer cells (Weaver, 2006). In the future, it will be important to determine whether miR-143 and -145, or indeed other miRs, regulate invadopodia formation and invasiveness in human cancer cells.
PDGF regulates miR-143 and -145 expression through Src and p53 activity
PDGF is a known regulator of SMC differentiation and migration (Owens et al., 2004), and Src is known to be a key mediator of the PDGF signal transduction pathway (Bromann et al., 2004). Previous studies have shown that Src family kinase signaling is required to elicit both mitogenicity and motogenicity in response to PDGF (Kypta et al., 1990; Twamley et al., 1992; Klinghoffer et al., 1999). It has also been shown that PDGF can reduce miR-145 expression (Cheng et al., 2009). In this study, we found that PDGF had a similar effect on miR-143 (Fig. 2 B). Because we demonstrated that activated Src is able to down-regulate miR-143/145, we next tested whether Src activation is the mechanism by which PDGF regulates these genes. Treatment with the Src family kinase inhibitor SU6656 (Blake et al., 2000) restored miR-143 and -145 levels in PDGF-treated cells (Fig. 2 B). In contrast, the PI3-K (phosphatidylinositol 3-kinase) inhibitor LY294002 had no effect. Next, we asked how Src regulates these two miRs. It has been shown that, once activated by PDGF, Src activity overcomes a p53-mediated inhibition of cell cycle progression (Broome and Courtneidge, 2000) and that inhibition of p53 expression is a key phenomenon for Src-mediated podosome formation (Mukhopadhyay et al., 2009). Moreover, p53 activity increases miR-143 and -145 levels in cancer cells in a manner that does not involve transcription (Suzuki et al., 2009). We hypothesized that in response to PDGF, Src down-regulates miR expression through p53 inhibition. In keeping with this, doxorubicin, a known p53 activator, restored miR-143 and -145 levels in PDGF-stimulated cells (Fig. S2 B).
We analyzed the promoter region of the primary miR gene encoding miR-143/145 and identified two potential binding sites for p53 (Fig. 2 C). To define the role of these potential p53 response elements (p53-REs) in miR-143/145 induction, we cloned a fragment from rat chromosome 18 containing the two potential p53-binding sites into a luciferase vector and evaluated promoter activity by luciferase assay. As shown in Fig. 2 D, p53 activation by doxorubicin treatment induced a marked elevation of miR-143/145 promoter activity. Deletion of p53-RE-2 resulted in total inhibition of promoter activity, whereas deletion of p53-RE-1 had no effect (Fig. 2 E). To test whether p53 can bind the p53-RE-2 sequence in the endogenous promoter sequence of miR-143/145, we performed chromatin immunoprecipitation (ChIP) assays. We detected one specific PCR product derived from p53-RE-2 using two different antibodies against p53 (Fig. 2 F). Collectively, these data suggest that p53 can transcriptionally regulate miR-143/145 expression through p53-RE-2.
Excessive activity of PDGF has been associated with several human disorders, including atherosclerosis and restenosis (Alvarez et al., 2006; Andrae et al., 2008), at least in part through the regulation of SMC plasticity. The mechanism underling this phenomenon is not completely understood, but it was recently shown that miR-143 and -145 regulate the SMC switch from contractile to synthetic phenotype (Boettger et al., 2009; Cheng et al., 2009; Cordes et al., 2009; Elia et al., 2009b; Xin et al., 2009). In this study, we show for the first time that PDGF is able to regulate miR-143/145 expression via a pathway involving Src and p53. The mechanism by which Src regulates p53 level and/or function is the subject of active investigation in our laboratory. Nevertheless, these results open up new potential avenues to use Src and/or p53 inhibitors to treat vascular pathologies (Elia et al., 2009b).
PDGF induces podosome formation in SMCs through miR-143 and -145 down-regulation
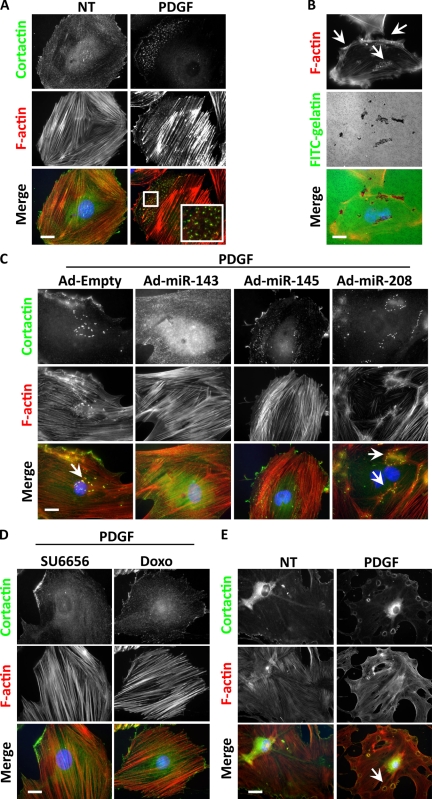
PDGF is a known inducer of migration in SMCs, yet stimulation of podosome formation by PDGF has not previously been described. We pretreated A7r5 cells with PDGF for 24 h (allowing the complete down-regulation of the two miRs) and then treated the cells again with PDGF for 3 h more. After fixation and F-actin staining, we observed punctate structures in the cells (57.3 ± 2.5%; Fig. 3 A). In cells that were not pretreated with PDGF, far fewer cells (9.0 ± 3.6%) formed the same structures. When plated on gelatin-coated glass coverslips, these punctate structures, which costained for cortactin and Tks5, were able to digest the substrate (Fig. 3 B and Fig. S1 C). Confocal microscopy demonstrated that the actin puncta colocalized in the same plane with the degradation spots (Fig. S2 C), confirming that they were indeed podosomes. Overexpression of miR-143 or -145 during the PDGF treatment inhibited podosomes, demonstrating that the down-regulation of miR-143 and -145 is a key step in PDGF-stimulated podosome formation (Fig. 3 C). When the PDGF pretreatment was conducted in the presence of the Src inhibitor SU6656 or the p53 inducer doxorubicin, podosomes were not observed (Fig. 3 D). Finally, in keeping with the notion that the 24 h of pretreatment with PDGF is required to down-regulate the miRs, 80% of SMCs from miR-143(145) KO mouse aortas formed podosomes after only 3 h of PDGF treatment (75.3 ± 5%; Fig. 3 E). Before this study, only nonphysiological treatments such as phorbol esters had been shown to elicit podosome formation in SMCs. We speculate that the lengthy pretreatment with PDGF and subsequent down-regulation of the miRs might represent a physiological control mechanism that provides a threshold to be overcome to elicit the synthetic/motile response.
Figure 3.
Podosome formation induced by PDGF in WT and KO SMCs. (A) Cortactin and F-actin IF to visualize podosomes in A7r5 treated with 10 ng/ml PDGF for 24 + 3 h or only for 3 h. Quantification of three independent experiments showed no podosome formation under normal conditions, whereas 57.3% (±2.5%) of cells had podosomes after PDGF treatment. The inset indicates an area of podosome formation (boxed area). (B) FITC-gelatin degradation assay on A7r5 cells treated with PDGF. (C) Cortactin and F-actin IF to visualize podosomes in A7r5 cells induced by PDGF and treated with different adenoviruses (Ad). (D) Cortactin and F-actin IF to visualize podosomes in A7r5 cells induced by PDGF and treated with different compounds (SU6656 and doxorubicin [Doxo]). (E) Cortactin and F-actin IF to visualize podosomes in VSMCs isolated from miR-143(145) KO mice treated with 10 ng/ml PDGF or not treated (NT). In three separate experiments, 75.3% (±5%) of KO VSMC formed rosettes of podosomes after PDGF treatment. (B, C, and E) Arrows indicate areas of podosome formation. Bars, 10 µm.
Identification of targets of miR-143 and -145
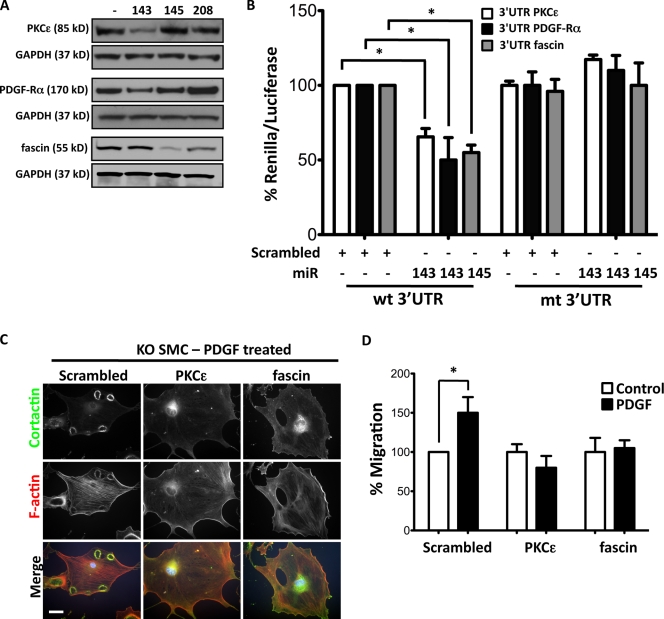
Using bioinformatics to search for miR-143 and -145 targets, we identified likely candidates to be PKC-ε and PDGF receptor α (PDGF-Rα) for miR-143 and fascin for miR-145. These genes have highly conserved seed sequences for miR-143 or -145 in a variety of species (Fig. S3 A). Transduction of miR-143(145) KO VSMCs with Ad–miR-143 decreased the abundance of PKC-ε and PDGF-Rα, whereas transduction with Ad–miR-145 decreased fascin without effecting a change in mRNA levels (Fig. 4 A and Fig. S3 B). Luciferase assays with WT and mutated 3′ UTR–binding sites confirmed that these mRNAs are targets for miR-143 and -145 (Fig. 4 B). All three proteins were up-regulated in KO VSMCs (Fig. S3 C). Both PKC-ε and fascin were localized to podosomes of miR-143(145) KO VSMCs (Fig. S3 D), and RNA interference of either gene reduced podosome formation and migration in miR-143(145) KO VSMCs and Src-3T3 cells (Fig. 4, C and D; and Fig. S3, E and F).
Figure 4.
miR-143 and -145 targets. (A) Representative immunoblot of primary VSMCs isolated from miR-143(145) KO mice transduced with adenovirus (Ad-miR) with an empty expression cassette (−) or expressing miR-143, -145, or -208. (B) Luciferase reporter assay on 3T3 cells performed by cotransfection of 20 nM miR-143, miR-145, or scrambled oligonucleotide (+) with a renilla reporter gene linked to 10 ng WT (wt) or mutated (mt) 3′ UTR of PKC-ε, PDGF-Rα, or fascin. All measurements were calculated as the percentage of control (WT 3′ UTR), and error bars were calculated as propagated standard errors of the mean of triplicate measurements from each experiment. *, P < 0.03 versus control scrambled miR. (C) Morphology of KO VSMCs with a knockdown of PKC-ε and fascin by lentiviral shRNA interference. (D) Migration of KO VSMCs with a knockdown of PKC-ε and fascin by lentiviral shRNA interference. All measurements were calculated as the percentage of scrambled control, and error bars were calculated as propagated standard errors of the mean of triplicate measurements from each experiment. *, P < 0.05. Bar, 10 µm.
The serine/threonine kinase PKC-ε is involved in cell migration and proliferation. It is up-regulated in skin cancer, where it is responsible for increased proliferation (Breitkreutz et al., 2007). Although PKC activity is known to be required for podosome formation (Hai et al., 2002), there are no prior studies on the involvement of this isoform. PDGF-Rα is a receptor tyrosine kinase that stimulates both migration and proliferation in response to ligand binding (Heldin and Westermark, 1999). In SMCs, the likely ligand is PDGF-AA (Barrett and Benditt, 1988). Fascin is an actin-bundling protein with roles in diverse forms of cell protrusions and in cytoplasmic actin bundles (Adams, 2004). It has been shown to localize to the leading edge of cancer cells and play a role in their migration (Vignjevic et al., 2007), but it has not previously been described to be involved in podosome formation. We show in this study that each of these targets is required for podosome formation in VSMCs, and their inhibition, at least in part, explains the role of the miR-143/145 gene as an inhibitor of cell migration.
We have demonstrated that Src activity can inhibit miR-143/145 gene expression, likely through the inhibition of p53 (Mukhopadhyay et al., 2009), which is a direct transcription factor for this gene cluster. Src is an activator of podosome formation in VSMCs (Gimona et al., 2008; Mukhopadhyay et al., 2009), and it has been shown that the PKC induction of podosomes is a result of Src activation (Gatesman et al., 2004) and that Src and PKC-ε can associate with each other (Vondriska et al., 2001). Furthermore, Src family kinases are activated by PDGF-Rs (Kypta et al., 1990), as is PKC-ε (Ha and Exton, 1993). We hypothesize the presence of an autoregulatory loop, initiated by PDGF production in response to vessel injury. Stimulation of the PDGF-R activates PKC and Src, which in turn promotes migration and podosome formation, in part through Src inhibition of p53 and thus miR-143 and -145 expression. This allows the subsequent up-regulation and Src phosphorylation of key podosome proteins as well as increased expression of the PDGF-R, which further boosts signaling (Fig. 5).
Figure 5.
Model for the role of miR-143 and -145 in VSMC migration and podosome formation. Vascular stress triggers the PDGF response, which activates Src, which in turn inhibits p53 and thus represses miR-143 and -145 expression. This relieves miR-143 repression of the expression of PKC-ε and PDGF-Rα and miR-145 repression of fascin, allowing the formation of podosomes and an increased migratory capacity. Thus, loss of both miRs increases the activity of pathways involved in cell migration.
Materials and methods
Materials
The following antibodies were used: cortactin, Tks5, and fascin (Millipore); vinculin, smooth muscle α-actin, and CD31 (Sigma-Aldrich); PKC-ε, PDGF-Rα, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH; Cell Signaling Technology); and p53 clones sc-99 and sc-100 and isotypic IgG clone sc-2025 (Santa Cruz Biotechnology, Inc.). Secondary antibodies conjugated to Alexa Fluor 488 and 568 and Alexa Fluor 594–conjugated phalloidin were obtained from Invitrogen. PDBu, PDGF-BB, PP2, SU6656, doxorubicin, LY294002, and UO126 were purchased from Sigma-Aldrich and used at the following concentrations: 10 µM PDBu, 10 and 20 ng/ml PDGF-BB, 10 µM PP2, 5 µM SU6656, 1 µM doxorubicin, 5 µM LY294002, and 10 µM UO126.
Cell lines
A7r5 rat SMCs and 3T3 cells were maintained in DME (Invitrogen) supplemented with 1.5 g/l-glucose, 4 mM l-glutamine, 10% fetal bovine serum, and penicillin/streptomycin at 37°C in a 5% CO2 atmosphere. Primary VSMCs were obtained from WT and KO mouse aortas, and purity was assessed by staining with antibodies against smooth muscle α-actin and CD31 as previously described (Ray et al., 2001). 99% of the cells were positive for smooth muscle α-actin.
Animals
Animals were housed in accordance with the guidelines of the American Association for Laboratory Animal Care. The miR-143(145) KO mice were previously described (Elia et al., 2009b).
Electron microscopy
Electronic microscopy was performed at the Veterans Medical Research Foundation, San Diego, CA. Adult aortas from WT and KO mice were dissected and fixed in 2% paraformaldehyde, 0.1% glutaraldehyde, and 0.1 M sodium phosphate in PBS, pH 7.2, for 24 h. After fixation, aortas were treated with 1% osmium tetrachloride and dehydrated using graded ethanol. The samples were then infiltrated with and embedded in LX112 epoxy resin (Ted Pella, Inc.). Ultrathin sections (83 nm) were made using an Ultracut II (Leica) and placed on 150-mesh nickel grids. Sections were then steamed for 20 min in a citrate-based antigen retrieval solution. When cool, the sections were then blocked using serum-free protein block. Each section was then probed for the primary antibodies for 2 h, followed by gold secondary antibodies (5- and 10-nm gold particle from Ted Pella, Inc.) at 1:20 dilution in TBS, pH 7.6, for 1 h. Sections were viewed, and images were obtained using an electron microscope (EM10C; Carl Zeiss, Inc.).
Immunofluorescence (IF)
IF was performed on paraformaldehyde-fixed cells. Cells were permeabilized with 0.1% Triton X-100 in PBS and then blocked with 0.1% Triton X-100 and 5% BSA in PBS for 1 h, washed, and incubated overnight with the related antibody at 4°C. Antibodies conjugated to Alexa Fluor 488 and 568 were used as secondary antibodies. Actin was visualized with Alexa Fluor 594–conjugated phalloidin, and nuclei were stained with DAPI. Coverslips were mounted with Mounting Medium for Fluorescence (Vectashield; Vector Laboratories). Fluorescence microscopy images were obtained with a microscope (Axioplan2; Carl Zeiss, Inc.) equipped with a charge-coupled device camera (Axiocam HRm; Carl Zeiss, Inc.) using Axiovision software (Carl Zeiss, Inc.) at room temperature. All images of primary cells were taken with a 40× NA 0.75 objective. The A7r5 cells were imaged with a 63× NA 1.40 oil immersion objective. Confocal images were obtained with a microscope (Radiance 2100 MP; Bio-Rad Laboratories) using Laser Sharp 2000 software (Bio-Rad Laboratories) at room temperature (60× NA 1.4, zoom 7, Pix0.027 oil immersion objective). Postacquisition analyses were performed using Volocity software (PerkinElmer).
Podosome function and migration assays
Functional assays were performed essentially as described previously (Berdeaux et al., 2004). Quantification of podosomes was performed on merged phalloidin-cortactin–stained samples on at least 50 randomly chosen fields representing ∼100 total cells per experimental point. Primary WT and KO cells containing at least one complete rosette of podosomes were scored as positive. A7r5 cells containing at least two punctate F-actin–rich podosomes were scored as positive. Total cell numbers were calculated by scoring the number of nuclei on the same field. Motility and invasion assays using Transwell chambers were conducted as described previously (Seals et al., 2005), except that 10,000 cells were used in each assay and it was performed for 6 h. Adenovirus infection was performed 24 h before the assay, whereas PDBu was added directly to the culture medium during the seeding of the Boyden chamber (BD). PDGF was used as a chemoattractant.
Generation of adenoviral vectors harboring miRs
Adenovirus vectors were prepared as previously described (Carè et al., 2007; Elia et al., 2009a). Cells were transduced at an MOI of 50 and checked for adequate transduction by quantitative RT-PCR (qRT-PCR).
Bioinformatics
Potential miR-143 and -145 targets were identified using the algorithms miRanda (http://www.microrna.org), TargetScan (http://www.targetscan.org), and PicTar (http://pictar.mdc-berlin.de/) as previously described (Carè et al., 2007; Elia et al., 2009a).
Formaldehyde cross-linking and ChIP assay
ChIP assays were performed as previously described (Fazi et al., 2005). In brief, cross-linking of proteins to DNA was obtained by the direct addition of formaldehyde to cultured cells (2 × 106) to a final concentration of 1%. After incubation for 10 min at 37°C, the mixture was sonicated, and chromatin was immunoprecipitated overnight with 5 µl of p53 antibodies. miR-143/145 promoter analysis was performed using MatInspector (http://www.genomatix.de/products/MatInspector). Binding sites were amplified using the primers p53-RE-2–5′ (5′-TAAGCCCCTCTCATCTCCCAACAG-3′), p53-RE-2–3′ (5′-GGAACAATGGGATGGGGAGGTATG-3′), foxo3a-RE–5′ (5′-GACTCTGTATTAATCTGCTTTAC-3′), and foxo3a-RE–3′ (5′-GCAGTGCTCAAATGGTACCAG-3′).
Reporter assays
The miR-143/145 promoter, which contains two potential binding sites for p53, was amplified from rat chromosome 18 using KOD Taq polymerase (EMD) and the primers p53-5′ (5′-TAAGCCCCTCTCATCTCCCA-3′) and p53-3′ (5′-GACTCCCAACTGACCAGAGA-3′). The promoter was cloned into the reporter plasmid pGL3-basic (Promega). For the promoter experiment, 3T3 cells were seeded in 24-well plates (Nunc) and transfected with 100 ng of the miR-143/145 promoter-luciferase reporter construct and 10 ng of the renilla plasmid using Lipofectamine 2000 (Invitrogen) and then treated with 1 µM doxorubicin. The deleted promoter sequences were obtained using the primers ΔBS1-5′ (5′-CCTCCCCATCCCATTGTTCC-3′), ΔBS1-3′ (5′-GACTCCCAACTGACCAGAGA-3′), ΔBS2-5′ (5′-TAAGCCCCTCTCATCTCCCA-3′), and ΔBS2-3′ (5′-ACGTCCATCTTCGTGTCCTTTG-3′). For the 3′ UTR reporter assay, experiments were performed in 3T3 cells. 3′ UTR segments were subcloned by standard procedures into the psiCHECK-2 (Promega) immediately downstream of the stop codon of the renilla gene using the primers PKC-ε–5′ (5′-GAGGCTGCTTCGGATGGAGGGA-3′), PKC-ε–3′ (5′-TAAGATTTAACATCATCATATT-3′), fascin-5′ (5′-GGCCACCTGCCCTCTGCAGG-3′), fascin-3′ (5′-GGCCGCAGACTTGATTTTAC-3′), PDGF-Rα–5′ (′5-CTGACACGCTCCGGGTATCA-3′), and PDGF-Rα–3′ (5′-AACGTTAAGTCATATATAAT-3′). Seed sequence mutagenesis was performed as described by the manufacturer (Agilent Technologies). 3T3 cells were transfected with 10 ng of the reporter plasmid and 20 nM of the miR mimics or scrambled sequence (Thermo Fisher Scientific) using Lipofectamine 2000 according to the manufacturer’s protocol (Invitrogen). Cells were lysed and assayed for luciferase activity at 48 h after transfection, and the luciferase gene coexpressed in the same plasmid was used as internal control. All luciferase assays were performed with Dual Luciferase kit (Promega) as previously described (Carè et al., 2007; Elia et al., 2009a).
RNA interference
Smart pool siRNAs for scrambled, PKC-ε, and fascin were purchased from Thermo Fisher Scientific and transfected into Src-3T3 using Lipofectamine 2000. Lentiviruses with scrambled, PKC-ε, and fascin short hairpin RNA (shRNA) were purchased from Santa Cruz Biotechnology, Inc., and KO SMCs were infected at MOI 5 using 5 µg/ml polybrene. Cells were selected for puromycin resistance according to the manufacturer’s protocol. RNA silencing was evaluated by qRT-PCR, and the residual levels of RNA for each target were as follows: PKC-ε, 15 ± 0.9 in Src-3T3 and 10 ± 1.3 in SMCs; and fascin, 34 ± 4 in Src-3T3 and 30 ± 1.4 in SMCs.
RNA quantification
For qRT-PCR analysis, total RNA was extracted using the TRIZOL reagent (Invitrogen) according to the manufacturer’s protocol. cDNA was prepared using SuperScript Reverse transcription cDNA kit (Invitrogen). SYBR green qRT-PCR was performed using the primers PKC-ε–5′ (5′-CTGGACGTGGACTCCTGATT-3′), PKC-ε–3′ (5′-ACTCAGGCTCCTTCCATCA-3′), fascin-5′ (5′-AACCCCTTGCCTTTCAAACT-3′), fascin-3′ (5′-CATGGAAAGAAGGGGACAGA-3′), PDGF-Rα–5′ (5′-AGGACTTGGGTGATGTGGAG-3′), PDGF-Rα–3′ (5′-TGTGGCCAGTCACTCTCTTG-3′), GAPDH-5′ (5′-GACGGCCGCATCTTCTTGT-3′), and GAPDH-3′ (5′-CACACCGACCTTCACCATTTT-3′). For miR qRT-PCR, total RNA was extracted using TRIZOL. Primers and probes specific for mouse/rat miR-143, -145, and -199 and internal control, Sno202 RNA, and U6 small nuclear RNA were purchased from Applied Biosystems. Amplification and detection was performed with a sequence detection system (model 7300; ABI) using 40 cycles of denaturation at 95°C (15 s) and annealing/extension at 60°C (60 s). This was preceded by reverse transcription at 42°C for 30 min and denaturation at 85°C for 5 min. Northern blotting was performed to confirm the expression levels of miR-143 and -145. Probes, antisense oligonucleotides against mature miR-143, -145, and -199, and U6 were LNA (locked nucleic acid) based (Exiqon).
Statistical analysis
Luciferase, RNA, protein, and migration values were compared using the two-tailed analysis of variance (ANOVA) test. A value of P < 0.05 or less was considered to be statistically significant.
Online supplemental material
Fig. S1 shows clustering of KO SMC podosomes and inhibition by miR-143 and -145 expression. Fig. S2 shows the effect of miR-143 and -145 on podosome formation and the effect of doxorubicin on miR-143 and -145 levels. Fig. S3 shows the cellular localization and effect of knockdown of miR-143 and -145 targets in VSMCs. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200912096/DC1.
Acknowledgments
We are grateful to Paula Sicurello (Veterans Medical Research Foundation, San Diego, CA) for technical support with the electron microscopy, Ju Chen for miR-143(145) KO mice, and Joseph Russo for assistance with the confocal images.
This work was supported by the Ministero dell’Istruzione dell’Università e della Ricerca, CARIPLO, and a Fondation Leducq grant to G. Condorelli and National Institutes of Health/National Cancer Institute grants CA098383 and CA129686 to S.A. Courtneidge.
Footnotes
Abbreviations used in this paper:
- ChIP
- chromatin immunoprecipitation
- GAPDH
- glyceraldehyde 3-phosphate dehydrogenase
- IF
- immunofluorescence
- KO
- knockout
- miR
- microRNA
- PDBu
- phorbol dibutyrate
- PDGF-R
- PDGF receptor
- qRT-PCR
- quantitative RT-PCR
- shRNA
- short hairpin RNA
- SMC
- smooth muscle cell
- UTR
- untranslated region
- VSMC
- vascular SMC
- WT
- wild type
References
- Adams J.C. 2004. Roles of fascin in cell adhesion and motility. Curr. Opin. Cell Biol. 16:590–596 10.1016/j.ceb.2004.07.009 [DOI] [PubMed] [Google Scholar]
- Akao Y., Nakagawa Y., Kitade Y., Kinoshita T., Naoe T. 2007. Downregulation of microRNAs-143 and -145 in B-cell malignancies. Cancer Sci. 98:1914–1920 10.1111/j.1349-7006.2007.00618.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez R.H., Kantarjian H.M., Cortes J.E. 2006. Biology of platelet-derived growth factor and its involvement in disease. Mayo Clin. Proc. 81:1241–1257 10.4065/81.9.1241 [DOI] [PubMed] [Google Scholar]
- Andrae J., Gallini R., Betsholtz C. 2008. Role of platelet-derived growth factors in physiology and medicine. Genes Dev. 22:1276–1312 10.1101/gad.1653708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T.B., Benditt E.P. 1988. Platelet-derived growth factor gene expression in human atherosclerotic plaques and normal artery wall. Proc. Natl. Acad. Sci. USA. 85:2810–2814 10.1073/pnas.85.8.2810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 116:281–297 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- Berdeaux R.L., Díaz B., Kim L., Martin G.S. 2004. Active Rho is localized to podosomes induced by oncogenic Src and is required for their assembly and function. J. Cell Biol. 166:317–323 10.1083/jcb.200312168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake R.A., Broome M.A., Liu X., Wu J., Gishizky M., Sun L., Courtneidge S.A. 2000. SU6656, a selective src family kinase inhibitor, used to probe growth factor signaling. Mol. Cell. Biol. 20:9018–9027 10.1128/MCB.20.23.9018-9027.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettger T., Beetz N., Kostin S., Schneider J., Krüger M., Hein L., Braun T. 2009. Acquisition of the contractile phenotype by murine arterial smooth muscle cells depends on the Mir143/145 gene cluster. J. Clin. Invest. 119:2634–2647 10.1172/JCI38864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitkreutz D., Braiman-Wiksman L., Daum N., Denning M.F., Tennenbaum T. 2007. Protein kinase C family: on the crossroads of cell signaling in skin and tumor epithelium. J. Cancer Res. Clin. Oncol. 133:793–808 10.1007/s00432-007-0280-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromann P.A., Korkaya H., Courtneidge S.A. 2004. The interplay between Src family kinases and receptor tyrosine kinases. Oncogene. 23:7957–7968 10.1038/sj.onc.1208079 [DOI] [PubMed] [Google Scholar]
- Broome M.A., Courtneidge S.A. 2000. No requirement for src family kinases for PDGF signaling in fibroblasts expressing SV40 large T antigen. Oncogene. 19:2867–2869 10.1038/sj.onc.1203608 [DOI] [PubMed] [Google Scholar]
- Carè A., Catalucci D., Felicetti F., Bonci D., Addario A., Gallo P., Bang M.L., Segnalini P., Gu Y., Dalton N.D., et al. 2007. MicroRNA-133 controls cardiac hypertrophy. Nat. Med. 13:613–618 10.1038/nm1582 [DOI] [PubMed] [Google Scholar]
- Chen C.Z., Li L., Lodish H.F., Bartel D.P. 2004. MicroRNAs modulate hematopoietic lineage differentiation. Science. 303:83–86 10.1126/science.1091903 [DOI] [PubMed] [Google Scholar]
- Cheng Y., Liu X., Yang J., Lin Y., Xu D.Z., Lu Q., Deitch E.A., Huo Y., Delphin E.S., Zhang C. 2009. MicroRNA-145, a novel smooth muscle cell phenotypic marker and modulator, controls vascular neointimal lesion formation. Circ. Res. 105:158–166 10.1161/CIRCRESAHA.109.197517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes K.R., Sheehy N.T., White M.P., Berry E.C., Morton S.U., Muth A.N., Lee T.H., Miano J.M., Ivey K.N., Srivastava D. 2009. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 460:705–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Destaing O., Saltel F., Géminard J.C., Jurdic P., Bard F. 2003. Podosomes display actin turnover and dynamic self-organization in osteoclasts expressing actin-green fluorescent protein. Mol. Biol. Cell. 14:407–416 10.1091/mbc.E02-07-0389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia L., Contu R., Quintavalle M., Varrone F., Chimenti C., Russo M.A., Cimino V., De Marinis L., Frustaci A., Catalucci D., Condorelli G. 2009a. Reciprocal regulation of microRNA-1 and insulin-like growth factor-1 signal transduction cascade in cardiac and skeletal muscle in physiological and pathological conditions. Circulation. 120:2377–2385 10.1161/CIRCULATIONAHA.109.879429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia L., Quintavalle M., Zhang J., Contu R., Cossu L., Latronico M.V., Peterson K.L., Indolfi C., Catalucci D., Chen J., et al. 2009b. The knockout of miR-143 and -145 alters smooth muscle cell maintenance and vascular homeostasis in mice: correlates with human disease. Cell Death Differ. 16:1590–1598 10.1038/cdd.2009.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazi F., Rosa A., Fatica A., Gelmetti V., De Marchis M.L., Nervi C., Bozzoni I. 2005. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 123:819–831 10.1016/j.cell.2005.09.023 [DOI] [PubMed] [Google Scholar]
- Furmaniak-Kazmierczak E., Crawley S.W., Carter R.L., Maurice D.H., Côté G.P. 2007. Formation of extracellular matrix-digesting invadopodia by primary aortic smooth muscle cells. Circ. Res. 100:1328–1336 10.1161/CIRCRESAHA.106.147744 [DOI] [PubMed] [Google Scholar]
- Gatesman A., Walker V.G., Baisden J.M., Weed S.A., Flynn D.C. 2004. Protein kinase Calpha activates c-Src and induces podosome formation via AFAP-110. Mol. Cell. Biol. 24:7578–7597 10.1128/MCB.24.17.7578-7597.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimona M., Herzog M., Vandekerckhove J., Small J.V. 1990. Smooth muscle specific expression of calponin. FEBS Lett. 274:159–162 10.1016/0014-5793(90)81353-P [DOI] [PubMed] [Google Scholar]
- Gimona M., Kaverina I., Resch G.P., Vignal E., Burgstaller G. 2003. Calponin repeats regulate actin filament stability and formation of podosomes in smooth muscle cells. Mol. Biol. Cell. 14:2482–2491 10.1091/mbc.E02-11-0743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimona M., Buccione R., Courtneidge S.A., Linder S. 2008. Assembly and biological role of podosomes and invadopodia. Curr. Opin. Cell Biol. 20:235–241 10.1016/j.ceb.2008.01.005 [DOI] [PubMed] [Google Scholar]
- Ha K.S., Exton J.H. 1993. Differential translocation of protein kinase C isozymes by thrombin and platelet-derived growth factor. A possible function for phosphatidylcholine-derived diacylglycerol. J. Biol. Chem. 268:10534–10539 [PubMed] [Google Scholar]
- Hai C.M., Hahne P., Harrington E.O., Gimona M. 2002. Conventional protein kinase C mediates phorbol-dibutyrate-induced cytoskeletal remodeling in a7r5 smooth muscle cells. Exp. Cell Res. 280:64–74 10.1006/excr.2002.5592 [DOI] [PubMed] [Google Scholar]
- Heldin C.H., Westermark B. 1999. Mechanism of action and in vivo role of platelet-derived growth factor. Physiol. Rev. 79:1283–1316 [DOI] [PubMed] [Google Scholar]
- Iorio M.V., Ferracin M., Liu C.G., Veronese A., Spizzo R., Sabbioni S., Magri E., Pedriali M., Fabbri M., Campiglio M., et al. 2005. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 65:7065–7070 10.1158/0008-5472.CAN-05-1783 [DOI] [PubMed] [Google Scholar]
- Klinghoffer R.A., Sachsenmaier C., Cooper J.A., Soriano P. 1999. Src family kinases are required for integrin but not PDGFR signal transduction. EMBO J. 18:2459–2471 10.1093/emboj/18.9.2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kypta R.M., Goldberg Y., Ulug E.T., Courtneidge S.A. 1990. Association between the PDGF receptor and members of the src family of tyrosine kinases. Cell. 62:481–492 10.1016/0092-8674(90)90013-5 [DOI] [PubMed] [Google Scholar]
- Linder S., Aepfelbacher M. 2003. Podosomes: adhesion hot-spots of invasive cells. Trends Cell Biol. 13:376–385 10.1016/S0962-8924(03)00128-4 [DOI] [PubMed] [Google Scholar]
- Michael M.Z., O’ Connor S.M., van Holst Pellekaan N.G., Young G.P., James R.J. 2003. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol. Cancer Res. 1:882–891 [PubMed] [Google Scholar]
- Mukhopadhyay U.K., Eves R., Jia L., Mooney P., Mak A.S. 2009. p53 suppresses Src-induced podosome and rosette formation and cellular invasiveness through the upregulation of caldesmon. Mol. Cell. Biol. 29:3088–3098 10.1128/MCB.01816-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens G.K. 1995. Regulation of differentiation of vascular smooth muscle cells. Physiol. Rev. 75:487–517 [DOI] [PubMed] [Google Scholar]
- Owens G.K., Kumar M.S., Wamhoff B.R. 2004. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol. Rev. 84:767–801 10.1152/physrev.00041.2003 [DOI] [PubMed] [Google Scholar]
- Ray J.L., Leach R., Herbert J.M., Benson M. 2001. Isolation of vascular smooth muscle cells from a single murine aorta. Methods Cell Sci. 23:185–188 10.1023/A:1016357510143 [DOI] [PubMed] [Google Scholar]
- Seals D.F., Azucena E.F., Jr., Pass I., Tesfay L., Gordon R., Woodrow M., Resau J.H., Courtneidge S.A. 2005. The adaptor protein Tks5/Fish is required for podosome formation and function, and for the protease-driven invasion of cancer cells. Cancer Cell. 7:155–165 10.1016/j.ccr.2005.01.006 [DOI] [PubMed] [Google Scholar]
- Sobue K., Hayashi K., Nishida W. 1999. Expressional regulation of smooth muscle cell-specific genes in association with phenotypic modulation. Mol. Cell. Biochem. 190:105–118 10.1023/A:1006941621170 [DOI] [PubMed] [Google Scholar]
- Suzuki H.I., Yamagata K., Sugimoto K., Iwamoto T., Kato S., Miyazono K. 2009. Modulation of microRNA processing by p53. Nature. 460:529–533 10.1038/nature08199 [DOI] [PubMed] [Google Scholar]
- Twamley G.M., Kypta R.M., Hall B., Courtneidge S.A. 1992. Association of Fyn with the activated platelet-derived growth factor receptor: requirements for binding and phosphorylation. Oncogene. 7:1893–1901 [PubMed] [Google Scholar]
- Vignjevic D., Schoumacher M., Gavert N., Janssen K.P., Jih G., Laé M., Louvard D., Ben-Ze’ev A., Robine S. 2007. Fascin, a novel target of beta-catenin-TCF signaling, is expressed at the invasive front of human colon cancer. Cancer Res. 67:6844–6853 10.1158/0008-5472.CAN-07-0929 [DOI] [PubMed] [Google Scholar]
- Vondriska T.M., Zhang J., Song C., Tang X.L., Cao X., Baines C.P., Pass J.M., Wang S., Bolli R., Ping P. 2001. Protein kinase C epsilon-Src modules direct signal transduction in nitric oxide-induced cardioprotection: complex formation as a means for cardioprotective signaling. Circ. Res. 88:1306–1313 10.1161/hh1201.092994 [DOI] [PubMed] [Google Scholar]
- Weaver A.M. 2006. Invadopodia: specialized cell structures for cancer invasion. Clin. Exp. Metastasis. 23:97–105 10.1007/s10585-006-9014-1 [DOI] [PubMed] [Google Scholar]
- Xin M., Small E.M., Sutherland L.B., Qi X., McAnally J., Plato C.F., Richardson J.A., Bassel-Duby R., Olson E.N. 2009. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev. 23:2166–2178 10.1101/gad.1842409 [DOI] [PMC free article] [PubMed] [Google Scholar]