Abstract
It is a long-standing view that the circadian clock functions to proactively align internal physiology with the 24-h rotation of the earth. Recent studies, including one by Schmutz and colleagues (pp. 345–357) in the February 15, 2010, issue of Genes & Development, delineate strikingly complex connections between molecular clocks and nuclear receptor signaling pathways, implying the existence of a large-scale circadian regulatory network coordinating a diverse array of physiological processes to maintain dynamic homeostasis.
Keywords: Circadian oscillator, transcription, protein–protein interaction
Light from the sun sustains life on earth. The 24-h rotation of the earth exposes a vast number of plants and animals to the light/dark cycle. Consequently, the behavior and physiology of numerous living organisms exhibit circadian rhythms. The word “circadian” is derived from Latin circa diem, which means “about a day.” Behavioral rhythms such as sleeping, food seeking, and predator avoidance are thought to help animals survive. Physiological rhythms such as body temperature, blood pressure, and metabolism also anticipate and adapt to predictable changes in the environment to maintain the overall well-being of animals (Young 2000).
Circadian rhythms are controlled by evolutionarily conserved internal clocks residing in most tissues of the body. The central clock is located in the suprachiasmatic nucleus (SCN) of the hypothalamus and is entrained directly by light (Hatings et al. 2008). This master pacemaker can synchronize circadian oscillators in peripheral tissues, yet underlying neural and humoral mechanisms remain obscure. Besides light, other external cues such as feeding and ambient temperature are also powerful Zeitgebers (from German for time givers) for peripheral clocks (Damiola et al. 2000; Brown et al. 2002). How these time cues act in concert to entrain tissue-specific oscillators and evoke diverse physiological responses is poorly understood. Nevertheless, these processes clearly involve the endocrine system.
The rhythmic production and circulation of many hormones and metabolites within the endocrine system is instrumental in regulating regular physiological processes such as reproduction, blood pressure, and metabolism. Levels of circulating estrogen and progesterone fluctuate with the menstrual cycle, which in turn affect circadian rhythms in women (Shechter and Boivin 2010). In parallel with a diurnal rhythm in circulating adrenocorticotropic hormone, secretion of glucocorticoids and aldosterone from the adrenal gland rises before awakening (Weitzman 1976). Glucocorticoids boost energy production, and aldosterone increases blood pressure, together gearing up the body for the activity phase. Similarly, plasma levels of thyroid-stimulating hormone and triiodothyronine have a synchronous diurnal rhythm (Russell et al. 2008). A broad range of metabolites—such as glucose, free fatty acids, cholesterol, and bile acids—also exhibit diurnal fluctuation. A number of these hormones and metabolites serve as ligands for nuclear receptors that direct a large array of transcriptional programs involved in lipid and carbohydrate metabolism (Chawla et al. 2001). A survey of the diurnal expression profile of the mRNAs encoding all 49 mouse nuclear receptors revealed that more than half of receptors follow rhythmic cycles in key metabolic tissues (Yang et al. 2006). Together, these observations suggest a complex interaction between the circadian clock and nuclear receptor signaling (Yang et al. 2007; Teboul et al. 2008). Several recent studies lend further insight into an elaborate “wheel of time” composed of molecular clocks and nuclear receptors, which together help shape an emerging perspective on “design principles” and biological implications of the clock–receptor signaling network.
Hub of time: nuclear receptors at the core of molecular clocks
Circadian clocks are self-sustained, robust, and tunable molecular oscillators. At the core of both the central and peripheral clocks are two interlocked transcriptional/post-translational feedback loops (Ko and Takahashi 2006). In the negative feedback loop, BMAL1/CLOCK heterodimers activate the transcription of Period genes (Per1, Per2, and Per3) and Cryptochrome genes (Cry1 and Cry2) by recognizing E-box cis elements in their promoters. The PER/CRY complex in turn inhibits the transcription of their own genes by blocking BMAL1–CLOCK activity. The second feedback loop involves the ROR (α, β, and γ) and REV-ERB (α and β) proteins, members of a subfamily of nuclear receptors that recognize similar cis-regulatory elements (ROREs) on target genes. RORs act as transcriptional activators, and REV-ERBs are repressors. BMAL1/CLOCK binds to E-box elements present in Ror and Rev-erb genes and activate their transcription. RORs and REV-ERBs in turn drive rhythmic transcription of the Bmal1 gene by alternately binding to ROREs in its promoter (Preitner et al. 2002; Liu et al. 2008).
Theoretically, a negative feedback loop with time delay is sufficient to generate an oscillator. Why does the molecular clock adopt the “two-loop” design principle? Recent studies with synthetic oscillators in bacteria and mammalian cells as well as computational modeling demonstrate that a second feedback loop would make an oscillator robust and tunable (Stricker et al. 2008; Tigges et al. 2009). In the clockwork, these features are likely to be accomplished by the ROR/REV-ERB/BMAL1 loop.
The clock control of nuclear receptor pathways
One of the major efforts in circadian biology is to define molecular pathways by which the core clockwork regulates physiology and behavior. It has been demonstrated that many nuclear receptors exhibit circadian-like patterns of expression (Yang et al. 2006). Recent studies reveal a few of the nuclear receptor genes as being direct targets of the circadian clock. Peroxisome proliferator-activated receptor-α (PPARα) and PPARγ regulate lipid metabolism and energy homeostasis by coordinated actions in a variety of tissues (Evans et al. 2004). Pparα transcription is induced by CLOCK and BMAL1 via an intronic E-box-rich region (Oishi et al. 2005; Canaple et al. 2006). In contrast, PPARγ expression is regulated by two clock-controlled genes, albumin gene D-site-binding protein (Dbp) and E4 promoter A-binding protein 4 (E4bp4). DBP is an activator, whereas E4BP4 is a repressor. Together, they induce oscillation of PPARγ expression by binding reciprocally to the D-boxes located in the first exon (Takahashi et al. 2010). Constitutive androstane receptor (CAR) is a potent regulator of xenobiotic metabolism (Qatanani and Moore 2005). Gachon et al. (2006) showed that CAR cycling is regulated by the clock-controlled PARbZIP family of transcription factors, including DBP, HLF, and TEF. Mice devoid of all three of these proteins showed the loss of rhythmic expression of CAR and its target genes and defects in xenobiotic detoxification, providing a compelling example of nuclear receptor signaling linking the clock to xenobiotic metabolism (Gachon et al. 2006).
Nuclear receptor signaling is dependent on levels of receptors as well as ligands. It has long been known that plasma levels of hormonal ligands such as glucocorticoids and aldosterone have daily rhythms (Williams et al. 1972). Very recently, several studies provided direct evidence that endogenous clocks can regulate ligand production in the endocrine glands. A survey of the adrenal transcriptome revealed that a number of genes involved in steroid biosynthesis are expressed in a rhythmic fashion (Oster et al. 2006). Disruption of the adrenal clock by tissue-specific knockdown of BMAL1 abolishes circadian rhythms in steroidogenic gene expression and glucocorticoid production (Son et al. 2008). Cry-null mice showed a dramatic increase in aldosterone production and resultant hypertension, which is due to the loss of repression of Hsd3b6, a gene involved specifically in steroidogenesis in the adrenal cortex (Doi et al. 2010).
A range of ligands can be produced in local tissues. Heme is a porphyrin that functions as a prosthetic group for enzymes involved in oxygen and electron transport. Recently, two independent studies identified heme as an endogenous ligand for REV-ERBα (Raghuram et al. 2007; Yin et al. 2007). Interestingly, the circadian clock and heme biosynthesis are reciprocally regulated (Kaasik and Lee 2004). Expression of ALAS1, the rate-limiting enzyme in heme biosynthesis, is under clock control, resulting in oscillation in the intracellular level of heme and its association with REV-ERBα. Another example is that the circadian clock regulates cyclic expression of HMG-CoA reductase and cholesterol 7α-hydroxylase, the rate-limiting enzymes in cholesterol and bile acid biosynthesis, respectively (Le Martelot et al. 2009). This is believed to lead to cyclic production of oxysterols and bile acids, and activation of their corresponding receptors, LXR and FXR. Therefore, cyclic production of ligands for nuclear receptors provides a potential means of circadian regulation of metabolism.
The clock entrainment by nuclear receptors
Peripheral clocks appear to act as the integrators of signals from the light-sensing central clock and other physiological cues. The nature of the signals that entrain peripheral clocks in individual tissues remains obscure. Serving as endocrine and metabolic sensors, a number of nuclear receptors have been implicated in clock entrainment (Yang et al. 2007; Kovac et al. 2009).
Early studies showed that glucocorticoids are involved in the phase resetting of peripheral clocks in response to restricted feeding (Le Minh et al. 2001). Recent analysis revealed that glucocorticoid response elements (GREs) are present at multiple clock genes, including Per1 and Per2 (So et al. 2009). Through a distal GRE in the 5′-flanking region, Per1 is induced in peripheral tissues by acute physical stress, supporting the notion that PER1 is likely to mediate early response to time cues (Yamamoto et al. 2005). A recent report showed that chronic administration of glucocorticoids impairs glucose metabolism in a PER2-dependent manner (So et al. 2009). These findings reinforce the idea that glucocorticoid signaling may be partially involved in clock entrainment by metabolic cues.
It was mentioned earlier that the molecular clock modulates the expression of PPARα and PPARγ. Conversely, PPARα and PPARγ directly regulate peripheral clocks. PPARα affects the amplitude of Bmal1 expression in the liver. This likely occurs through PPRE cis elements at the promoters of Bmal1 and Rev-erbα genes (Canaple et al. 2006). Ablation of PPARγ in vascular cells reduces diurnal variations in blood pressure and heart rate, which is associated with impaired rhythmicity of the core clock genes in the aorta (Wang et al. 2008). Wang et al. (2008) further demonstrated that Bmal1 is a direct target of vascular PPARγ. These studies suggest that different nuclear receptors may mediate clock entrainment in different tissues and, furthermore, that the molecular clock and nuclear receptors are intertwined through multiple feedback loops.
This notion is also supported by studies on gonadal hormones. It has long been known that the SCN controls daily rhythms in gonadal hormone secretion (Turek et al. 1984). Interestingly, the SCN itself expresses the androgen receptor, through which androgen is likely to modulate the responses of the central clock to light (Karatsoreos et al. 2007). Estrogen has a potent effect on the circadian period of locomotor activity (Morin et al. 1977). The estrogen receptors are absent in the SCN but enriched in several brain areas, such as the medial preoptic area and the dorsal raphe. The medial preoptic area is the primary brain site responsible for the behavioral effect of estrogen (Ogawa et al. 2003). It is tempting to speculate that estrogen directly affects the local clock in this area. In the dorsal raphe, estradiol treatment modulates the light-induced response of serotonergic neurons, which may in turn affect the SCN through a median raphe projection (Abizaid et al. 2005). In addition to the brain, circadian rhythms in other tissues are also modulated by estrogen. Estradiol treatment alters the phase and amplitude of Per1 expression in the liver and kidney, but induces biphasic rhythms in Per1 and Per2 transcripts in the uterus (Nakamura et al. 2005). These findings stem from pharmacological intervention of the circadian timing system. Physiological roles for gonadal hormones in clock entrainment will need to be examined.
Additional level of complexity within the nuclear receptor–clock network
The above sections outlined the bidirectional connections between the circadian clock and nuclear receptor pathways. The circadian oscillators directly govern cyclic expression of nuclear receptors or regulate cyclic production of their ligands. Nuclear receptors in turn regulate the circadian oscillators through their cis-regulatory elements on specific clock genes. In this way, nuclear receptors and the molecular clock may be organized into tissue-specific transcriptional circuits that relay temporal and nutritional cues to control metabolic physiology. Nonetheless, this scheme seems to be oversimplified. Direct protein–protein interactions between clock components and nuclear receptors are emerging as a crucial mechanism for the working of the circadian clock.
Physical interactions between clock proteins and nuclear receptors were first demonstrated in the vasculature. McNamara et al. (2001) found that the retinoic acid receptors RARα/RXRα associate with CLOCK in a ligand-dependent fashion. This association blocks the binding of the CLOCK/BMAL1 heterodimer to E-box elements and activation of target genes. Through this mechanism, retinoic acid can reset the vascular clock in mice (McNamara et al. 2001).
The above study clearly indicates that nuclear receptors can function as cofactors for clock components. In fact, the converse also holds true. In the February 15, 2010, issue of Genes & Development, Schmutz et al. (2010) report that PER2 interacts physically with a handful of nuclear receptors, including REV-ERBα, PPARα, HNF4α, and TRα. In a meticulous effort to explore functional roles of these interactions, Schmutz et al. (2010) found that REV-ERBα recruits PER2 to the RORE region of the Bmal1 promoter, where PER2 contributes to REV-ERBα-mediated repression. In a different phase, PER2 is brought to the upstream PPRE region and acts as a coactivator for PPARα to induce Bmal1 transcription. PER2 is also rhythmically bound to other nuclear receptor target genes in liver, such as HNf1α and Glucose-6-phosphatase, suggesting a novel role for PER2 as a nuclear receptor cofactor to transform clock information into metabolic responses (Schmutz et al. 2010).
A similar scenario occurs with Dec1 and Dec2, which are clock-controlled genes that encode E-box-binding transcriptional repressors. They are involved in circadian regulation by competing with BMAL1/CLOCK for E-box association. A recent study showed that DEC1 and DEC2 can bind to RXRα and repress the transactivation potency of a number of RXRα heterodimers on the nuclear receptor elements (Cho et al. 2009), further supporting the concept that there is pervasive cross-talk between clock proteins and nuclear receptors via their cis-regulatory elements. Given that both clock protein-binding sites and nuclear receptor-binding sites are highly abundant in the genome, this mode of cross-talk may expand the regulatory capacity of the clock–receptor network in the order of magnitude.
Connections between the clock and metabolism via nuclear receptors?
From the evidence presented above, it seems clear that the circadian clock and nuclear receptor pathways are interlocked at the molecular level. However, whether nuclear receptors serve as a direct link between metabolic cycles and the core clock circuitry remains largely elusive. This is due partly to the lack of the clock/receptor double-mutant mouse models for metabolic phenotype analysis. It is further hindered by the fact that metabolic regulators often function in both nuclear receptor-dependent and receptor-independent manners. One such example is the NAD+-dependent deacetylase SIRT1.
SIRT1 is an energy sensor, as its activity depends on intracellular NAD+/NADH ratio. Two groups reported recently that levels of NAD+ display circadian oscillation in mouse embryonic fibroblasts and in liver (Nakahata et al. 2009; Ramsey et al. 2009). This oscillation is attributed to a negative feedback loop between the clock and the salvage pathway of NAD+ biosynthesis. The gene encoding nicotinamide phosphoribosyltransferase (Nampt), the rate-limiting enzyme for NAD+ biosynthesis, is a direct target of BMAL1/CLOCK. Oscillation in the levels of NAMPT and NAD+ leads to cyclic activation of SIRT1, which feeds back on the core clock by deacetylating and destabilizing PER2 and probably BMAL1 (Asher et al. 2008; Nakahata et al. 2008). Although this feedback loop does not directly involve nuclear receptors, SIRT1 has been shown to deacetylate and destabilize LXR and FXR (Li et al. 2007; Kemper et al. 2009). Previous work revealed that levels of LXR and FXR transcripts are constant in liver (Yang et al. 2006). These findings raise an intriguing possibility that LXR and FXR may be subject to circadian regulation by SIRT1 through a post-translational mechanism.
The nuclear receptor coactivator PGC-1α is a key regulator of mitochondrial biogenesis, hepatic gluconeogenesis, fatty acid β oxidation, and heme biosynthesis. Expression of PGC-1α in metabolic tissues shows robust circadian rhythms (Yang et al. 2006; Liu et al. 2007). This is at least in part due to direct transcriptional repression by REV-ERBα (Wu et al. 2009). Conversely, PGC-1α stimulates expression of clock genes, including Bmal1 and Rev-erbα, through coactivation of RORα and RORγ (Liu et al. 2007). This regulatory circuit further integrates the circadian clock and energy metabolism through nuclear receptors (such as PPARα, ERRα, HNF4α, and GR) as well as other transcription factors (such as FOXO1 and NRF-1) (Li and Lin 2009). Furthermore, it has been shown recently that feedback regulation between REV-ERBα and PGC-1α dictates circadian oscillation in the levels of heme and an endocrine hormone referred to as FGF21 (Estall et al. 2009; Wu et al. 2009). FGF21 in turn regulates carbohydrate and lipid metabolism, partially through induction of PGC-1α expression (Potthoff et al. 2009). Taken together, these studies underscore the evolving complexity of connections between the circadian clock and metabolism, and pose a daunting challenge to understanding the circadian metabolic network by the reductionist approach.
On the rim of time: more than metabolic rhythms
Virtually all physiological processes—such as growth and differentiation, immune responses, and reproduction—have intrinsic rhythms. The links between the circadian clock and rhythmic cellular and physiological processes are just beginning to be unveiled.
A remarkable example is the discovery of the connection between the circadian clock and the cell cycle (Hunt and Sassone-Corsi 2007). Expression of several cell cycle genes, including Wee1, c-Myc, cyclin D1, Gadd45, and Mdm2, oscillate in a circadian manner (Fu et al. 2002; Matsuo et al. 2003). Wee1 and c-Myc are directly regulated by the molecular clock via the E-box elements at their promoters and, as a result, their expression is severely disturbed in mice deficient in Clock, Cry, or Per2. These are associated with impaired liver regeneration in Cry-deficient mice and increased tumor incidence in Per2 mutant mice, suggesting that the circadian clock may protect against aberrant cell proliferation under adverse conditions. It should be noted that several mouse strains deficient in core clock genes are normal in growth and development. Whether the circadian clock influences the cell cycle in a normal physiological setting is yet to be addressed. The circadian clock is likely to serve as a fine-tuning mechanism rather than a determinant of cell growth and differentiation.
The immune system exhibits distinct diurnal features. It has been known for 50 years that mice have diurnal variation in survival rate after endotoxin shock (Halberg et al. 1960). Circulating immune cell numbers, serum cytokine levels, and amounts of cell surface cytokine receptors vary in a diurnal manner (Coogan and Wyse 2008). Recent studies provide compelling evidence that these diurnal variations are ascribed to intrinsic clockworks in immune cells (Arjona and Sarkar 2006; Keller et al. 2009). This is corroborated by genetic evidence showing a phase-delayed circadian pattern in lymphocyte numbers in Clock mutant mice and a blunted rhythm in interferon-γ expression in Per2 mutant mice (Coogan and Wyse 2008). Conversely, exogenous cytokines can modulate clock gene expression in the liver and cultured fibroblasts, although the physiological relevance of this observation is not clear (Cavadini et al. 2007). The regulation of the circadian system by the immune system has been better revealed in the brain. Endotoxin can induce a phase shift in mouse locomotor activity and the expression of c-Fos in the SCN, an immediate early response gene involved in the central clock entrainment (Marpegan et al. 2005). Interferon-α and tumor necrosis factor-α can also modulate locomotor rhythms and the expression of the clock genes in the SCN (Ohdo et al. 2001; Cavadini et al. 2007). These observations suggest that cytokines may play a role in the central clock function. However, whether direct or indirect mechanisms are involved is debatable, because cytokines may act at other brain sites that control sleep, feeding, or activity. In aggregate, a bidirectional relationship between the circadian and immune systems is becoming evident.
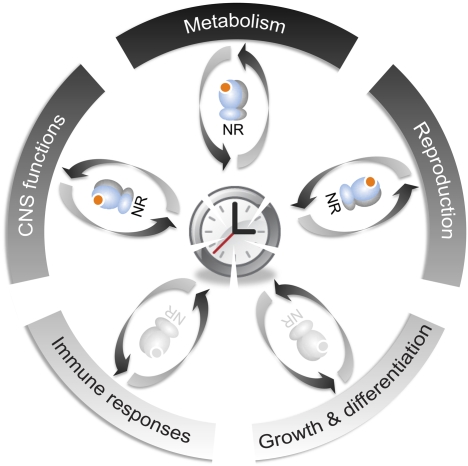
The question arises as to whether nuclear receptors play any roles. By surveying the expression of all 49 mouse nuclear receptors on a system-wide scale, Bookout et al. (2006) reveal the existence of a high-order regulatory network tying nuclear receptor function to reproduction, development, the CNS, and immunity, as well as nutrient metabolism and energy homeostasis. This opens the door to further investigation into the integration and orchestration of distinct physiological processes through the circadian clock–nuclear receptor network on an organismal scale (Fig. 1).
Figure 1.
A hypothetical schematic of wheel-like connections between the circadian clock, nuclear receptors, and various physiological processes. The circadian clock has a bidirectional relationship with diverse physiological processes, including metabolism, reproduction, CNS functions, growth and differentiation, and immune responses. In a specific tissue, cell-autonomous circadian clocks may integrate both environmental cues and internal physiological cues derived from other tissues, leading to concerted actions of distinct parts of the body. A large family of nuclear receptors (NRs) function to mediate two-way communications between molecular clocks and various physiological processes through a myriad of transcriptional circuits. These circuits may be organized across tissues into a large-scale network to maintain dynamic homeostasis of the whole body.
The dissolving boundaries
To date, the prevailing view of the circadian system is a hierarchical structure in which the light-sensing master pacemaker and other environmental cues synchronize numerous peripheral oscillators via the “input” pathways and, subsequently, drive rhythmic physiologic “outputs.” Much effort is focused on the identification of molecular components of the input and output pathways. However, as exemplified by the interactions between the circadian clock and nuclear receptors, feedback loops are pervasively present at the molecular, cellular, tissue, and systems levels. The boundary between the input and output pathways is dissolving. Thus, it is probably time to revisit the role of the circadian system in whole-body physiology. In addition to keeping internal physiology synchronized with the environment—predominantly the light/dark cycle—circadian clocks may serve at least two other ancient purposes: (1) to temporally separate chemically incompatible metabolic processes, such as anabolism and catabolism; and (2) to coordinate distinct physiological processes to maintain dynamic homeostasis. Evidence for these scenarios is emerging. The Clock mutant mice display a vast array of physiological defects, including metabolic syndrome, mania-like behavior, disrupted reproductive function, and reduced embryonic fibroblast proliferation (Turek et al. 2005; Miller et al. 2007; Roybal et al. 2007). Similarly, Bmal1-deficient mice have altered glucose metabolism, arrhythmic blood pressure, infertility, impaired adipocyte differentiation, and early aging (Shimba et al. 2005; Kondratov et al. 2006; Curtis et al. 2007; Alvarez et al. 2008; Lamia et al. 2008). These findings highlight the broad scope of physiology controlled by molecular clocks. As illustrated in Figure 1, it seems that connections between the circadian clock and most (if not all) physiological processes are bidirectional. Therefore, the circadian system might provide a potential means of communications between different physiological domains. In view of the dissolving boundary between different physiological processes, the circadian clock is probably not merely a timekeeper, but also a guardian of physiological homeostasis.
Acknowledgments
I thank Hai-Bin Ruan for preparation of the artwork, and Mindian Li for inspiring discussions. Research in my laboratory is supported by the Yale start-up package and American Diabetes Association Junior Faculty Award (1-10-JF-56).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1920710.
References
- Abizaid A, Mezei G, Thanarajasingam G, Horvath TL 2005. Estrogen enhances light-induced activation of dorsal raphe serotonergic neurons. Eur J Neurosci 21: 1536–1546 [DOI] [PubMed] [Google Scholar]
- Alvarez JD, Hansen A, Ord T, Bebas P, Chappell PE, Giebultowicz JM, Williams C, Moss S, Sehgal A 2008. The circadian clock protein BMAL1 is necessary for fertility and proper testosterone production in mice. J Biol Rhythms 23: 26–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arjona A, Sarkar DK 2006. Evidence supporting a circadian control of natural killer cell function. Brain Behav Immun 20: 469–476 [DOI] [PubMed] [Google Scholar]
- Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U 2008. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 134: 317–328 [DOI] [PubMed] [Google Scholar]
- Bookout AL, Jeong Y, Downes M, Yu RT, Evans RM, Mangelsdorf DJ 2006. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell 126: 789–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SA, Zumbrunn G, Fleury-Olela F, Preitner N, Schibler U 2002. Rhythms of mammalian body temperature can sustain peripheral circadian clocks. Curr Biol 12: 1574–1583 [DOI] [PubMed] [Google Scholar]
- Canaple L, Rambaud J, Dkhissi-Benyahya O, Rayet B, Tan NS, Michalik L, Delaunay F, Wahli W, Laudet V 2006. Reciprocal regulation of brain and muscle Arnt-like protein 1 and peroxisome proliferator-activated receptor α defines a novel positive feedback loop in the rodent liver circadian clock. Mol Endocrinol 20: 1715–1727 [DOI] [PubMed] [Google Scholar]
- Cavadini G, Petrzilka S, Kohler P, Jud C, Tobler I, Birchler T, Fontana A 2007. TNF-α suppresses the expression of clock genes by interfering with E-box-mediated transcription. Proc Natl Acad Sci 104: 12843–12848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ 2001. Nuclear receptors and lipid physiology: Opening the X-files. Science 294: 1866–1870 [DOI] [PubMed] [Google Scholar]
- Cho Y, Noshiro M, Choi M, Morita K, Kawamoto T, Fujimoto K, Kato Y, Makishima M 2009. The basic helix–loop–helix proteins differentiated embryo chondrocyte (DEC) 1 and DEC2 function as corepressors of retinoid X receptors. Mol Pharmacol 76: 1360–1369 [DOI] [PubMed] [Google Scholar]
- Coogan AN, Wyse CA 2008. Neuroimmunology of the circadian clock. Brain Res 1232: 104–112 [DOI] [PubMed] [Google Scholar]
- Curtis AM, Cheng Y, Kapoor S, Reilly D, Price TS, Fitzgerald GA 2007. Circadian variation of blood pressure and the vascular response to asynchronous stress. Proc Natl Acad Sci 104: 3450–3455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U 2000. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes & Dev 14: 2950–2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi M, Takahashi Y, Komatsu R, Yamazaki F, Yamada H, Haraguchi S, Emoto N, Okuno Y, Tsujimoto G, Kanematsu A, et al. 2010. Salt-sensitive hypertension in circadian clock-deficient Cry-null mice involves dysregulated adrenal Hsd3b6. Nat Med 16: 67–74 [DOI] [PubMed] [Google Scholar]
- Estall JL, Ruas JL, Choi CS, Laznik D, Badman M, Maratos-Flier E, Shulman GI, Spiegelman BM 2009. PGC-1α negatively regulates hepatic FGF21 expression by modulating the heme/Rev-Erbα axis. Proc Natl Acad Sci 106: 22510–22515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans RM, Barish GD, Wang YX 2004. PPARs and the complex journey to obesity. Nat Med 10: 355–361 [DOI] [PubMed] [Google Scholar]
- Fu L, Pelicano H, Liu J, Huang P, Lee C 2002. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell 111: 41–50 [DOI] [PubMed] [Google Scholar]
- Gachon F, Olela FF, Schaad O, Descombes P, Schibler U 2006. The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metab 4: 25–36 [DOI] [PubMed] [Google Scholar]
- Halberg F, Johnson EA, Brown BW, Bittner JJ 1960. Susceptibility rhythm to E. coli endotoxin and bioassay. Proc Soc Exp Biol Med 103: 142–144 [DOI] [PubMed] [Google Scholar]
- Hastings MH, Maywood ES, Reddy AB 2008. Two decades of circadian time. J Neuroendocrinol 20: 812–819 [DOI] [PubMed] [Google Scholar]
- Hunt T, Sassone-Corsi P 2007. Riding tandem: Circadian clocks and the cell cycle. Cell 129: 461–464 [DOI] [PubMed] [Google Scholar]
- Kaasik K, Lee CC 2004. Reciprocal regulation of haem biosynthesis and the circadian clock in mammals. Nature 430: 467–471 [DOI] [PubMed] [Google Scholar]
- Karatsoreos IN, Wang A, Sasanian J, Silver R 2007. A role for androgens in regulating circadian behavior and the suprachiasmatic nucleus. Endocrinology 148: 5487–5495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller M, Mazuch J, Abraham U, Eom GD, Herzog ED, Volk HD, Kramer A, Maier B 2009. A circadian clock in macrophages controls inflammatory immune responses. Proc Natl Acad Sci 106: 21407–21412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper JK, Xiao Z, Ponugoti B, Miao J, Fang S, Kanamaluru D, Tsang S, Wu SY, Chiang CM, Veenstra TD 2009. FXR acetylation is normally dynamically regulated by p300 and SIRT1 but constitutively elevated in metabolic disease states. Cell Metab 10: 392–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko CH, Takahashi JS 2006. Molecular components of the mammalian circadian clock. Hum Mol Genet 15: R271–R277 doi: 10.1093/hmg/ddl207 [DOI] [PubMed] [Google Scholar]
- Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP 2006. Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock. Genes & Dev 20: 1868–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovac J, Husse J, Oster H 2009. A time to fast, a time to feast: The crosstalk between metabolism and the circadian clock. Mol Cells 28: 75–80 [DOI] [PubMed] [Google Scholar]
- Lamia KA, Storch KF, Weitz CJ 2008. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci 105: 15172–15177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Martelot G, Claudel T, Gatfield D, Schaad O, Kornmann B, Sasso GL, Moschetta A, Schibler U 2009. REV-ERBα participates in circadian SREBP signaling and bile acid homeostasis. PLoS Biol 7: e1000181 doi: 10.1371/journal.pbio.1000181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Minh N, Damiola F, Tronche F, Schutz G, Schibler U 2001. Glucocorticoid hormones inhibit food-induced phase-shifting of peripheral circadian oscillators. EMBO J 20: 7128–7136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Lin JD 2009. Molecular control of circadian metabolic rhythms. J Appl Physiol 107: 1959–1964 [DOI] [PubMed] [Google Scholar]
- Li X, Zhang S, Blander G, Tse JG, Krieger M, Guarente L 2007. SIRT1 deacetylates and positively regulates the nuclear receptor LXR. Mol Cell 28: 91–106 [DOI] [PubMed] [Google Scholar]
- Liu C, Li S, Liu T, Borjigin J, Lin JD 2007. Transcriptional coactivator PGC-1α integrates the mammalian clock and energy metabolism. Nature 447: 477–481 [DOI] [PubMed] [Google Scholar]
- Liu AC, Tran HG, Zhang EE, Priest AA, Welsh DK, Kay SA 2008. Redundant function of REV-ERBα and β and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet 4: e1000023 doi: 10.1371/journal.pgen.1000023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marpegan L, Bekinschtein TA, Costas MA, Golombek DA 2005. Circadian responses to endotoxin treatment in mice. J Neuroimmunol 160: 102–109 [DOI] [PubMed] [Google Scholar]
- Matsuo T, Yamaguchi S, Mitsui S, Emi A, Shimoda F, Okamura H 2003. Control mechanism of the circadian clock for timing of cell division in vivo. Science 302: 255–259 [DOI] [PubMed] [Google Scholar]
- McNamara P, Seo SP, Rudic RD, Sehgal A, Chakravarti D, FitzGerald GA 2001. Regulation of CLOCK and MOP4 by nuclear hormone receptors in the vasculature: A humoral mechanism to reset a peripheral clock. Cell 105: 877–889 [DOI] [PubMed] [Google Scholar]
- Miller BH, McDearmon EL, Panda S, Hayes KR, Zhang J, Andrews JL, Antoch MP, Walker JR, Esser KA, Hogenesch JB, et al. 2007. Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proc Natl Acad Sci 104: 3342–3347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin LP, Fitzgerald KM, Zucker I 1977. Estradiol shortens the period of hamster circadian rhythms. Science 196: 305–307 [DOI] [PubMed] [Google Scholar]
- Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P 2008. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 134: 329–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P 2009. Circadian control of the NAD+ salvage pathway by CLOCK–SIRT1. Science 324: 654–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura TJ, Moriya T, Inoue S, Shimazoe T, Watanabe S, Ebihara S, Shinohara K 2005. Estrogen differentially regulates expression of Per1 and Per2 genes between central and peripheral clocks and between reproductive and nonreproductive tissues in female rats. J Neurosci Res 82: 622–630 [DOI] [PubMed] [Google Scholar]
- Ogawa S, Chan J, Gustafsson JA, Korach KS, Pfaff DW 2003. Estrogen increases locomotor activity in mice through estrogen receptor α: Specificity for the type of activity. Endocrinology 144: 230–239 [DOI] [PubMed] [Google Scholar]
- Ohdo S, Koyanagi S, Suyama H, Higuchi S, Aramaki H 2001. Changing the dosing schedule minimizes the disruptive effects of interferon on clock function. Nat Med 7: 356–360 [DOI] [PubMed] [Google Scholar]
- Oishi K, Shirai H, Ishida N 2005. CLOCK is involved in the circadian transactivation of peroxisome-proliferator-activated receptor α (PPARα) in mice. Biochem J 386: 575–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oster H, Damerow S, Hut RA, Eichele G 2006. Transcriptional profiling in the adrenal gland reveals circadian regulation of hormone biosynthesis genes and nucleosome assembly genes. J Biol Rhythms 21: 350–361 [DOI] [PubMed] [Google Scholar]
- Potthoff MJ, Inagaki T, Satapati S, Ding X, He T, Goetz R, Mohammadi M, Finck BN, Mangelsdorf DJ, Kliewer SA, et al. 2009. FGF21 induces PGC-1α and regulates carbohydrate and fatty acid metabolism during the adaptive starvation response. Proc Natl Acad Sci 106: 10853–10858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U 2002. The orphan nuclear receptor REV-ERBα controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 110: 251–260 [DOI] [PubMed] [Google Scholar]
- Qatanani M, Moore DD 2005. CAR, the continuously advancing receptor, in drug metabolism and disease. Curr Drug Metab 6: 329–339 [DOI] [PubMed] [Google Scholar]
- Raghuram S, Stayrook KR, Huang P, Rogers PM, Nosie AK, McClure DB, Burris LL, Khorasanizadeh S, Burris TP, Rastinejad F 2007. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBα and REV-ERBβ. Nat Struct Mol Biol 14: 1207–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong HK, Chong JL, Buhr ED, Lee C, et al. 2009. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science 324: 651–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roybal K, Theobold D, Graham A, DiNieri JA, Russo SJ, Krishnan V, Chakravarty S, Peevey J, Oehrlein N, Birnbaum S, et al. 2007. Mania-like behavior induced by disruption of CLOCK. Proc Natl Acad Sci 104: 6406–6411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell W, Harrison RF, Smith N, Darzy K, Shalet S, Weetman AP, Ross RJ 2008. Free triiodothyronine has a distinct circadian rhythm that is delayed but parallels thyrotropin levels. J Clin Endocrinol Metab 93: 2300–2306 [DOI] [PubMed] [Google Scholar]
- Schmutz I, Ripperger JA, Baeriswyl-Aebischer S, Albrecht U 2010. The mammalian clock component PERIOD2 coordinates circadian output by interaction with nuclear receptors. Genes & Dev 24: 345–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shechter A, Boivin DB 2010. Sleep, hormones, and circadian rhythms throughout the menstrual cycle in healthy women and women with premenstrual dysphoric disorder. Int J Endocrinol 2010: Article ID 259345. doi: 10.1155/2010/259345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimba S, Ishii N, Ohta Y, Ohno T, Watabe Y, Hayashi M, Wada T, Aoyagi T, Tezuka M 2005. Brain and muscle Arnt-like protein-1 (BMAL1), a component of the molecular clock, regulates adipogenesis. Proc Natl Acad Sci 102: 12071–12076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- So AY, Bernal TU, Pillsbury ML, Yamamoto KR, Feldman BJ 2009. Glucocorticoid regulation of the circadian clock modulates glucose homeostasis. Proc Natl Acad Sci 106: 17582–17587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son GH, Chung S, Choe HK, Kim HD, Baik SM, Lee H, Lee HW, Choi S, Sun W, Kim H, et al. 2008. Adrenal peripheral clock controls the autonomous circadian rhythm of glucocorticoid by causing rhythmic steroid production. Proc Natl Acad Sci 105: 20970–20975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stricker J, Cookson S, Bennett MR, Mather WH, Tsimring LS, Hasty J 2008. A fast, robust and tunable synthetic gene oscillator. Nature 456: 516–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Inoue I, Nakajima Y, Seo M, Nakano T, Yang F, Kumagai M, Komoda T, Awata T, Ikeda M, et al. 2010. A promoter in the novel exon of hPPARγ directs the circadian expression of PPARγ. J Atheroscler Thromb 17: 73–83 [DOI] [PubMed] [Google Scholar]
- Teboul M, Guillaumond F, Grechez-Cassiau A, Delaunay F 2008. The nuclear hormone receptor family round the clock. Mol Endocrinol 22: 2573–2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tigges M, Marquez-Lago TT, Stelling J, Fussenegger M 2009. A tunable synthetic mammalian oscillator. Nature 457: 309–312 [DOI] [PubMed] [Google Scholar]
- Turek FW, Swann J, Earnest DJ 1984. Role of the circadian system in reproductive phenomena. Recent Prog Horm Res 40: 143–183 [DOI] [PubMed] [Google Scholar]
- Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, et al. 2005. Obesity and metabolic syndrome in circadian Clock mutant mice. Science 308: 1043–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Yang G, Jia Z, Zhang H, Aoyagi T, Soodvilai S, Symons JD, Schnermann JB, Gonzalez FJ, Litwin SE, et al. 2008. Vascular PPARγ controls circadian variation in blood pressure and heart rate through Bmal1. Cell Metab 8: 482–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzman ED 1976. Circadian rhythms and episodic hormone secretion in man. Annu Rev Med 27: 225–243 [DOI] [PubMed] [Google Scholar]
- Williams GH, Cain JP, Dluhy RG, Underwood RH 1972. Studies of the control of plasma aldosterone concentration in normal man. I. Response to posture, acute and chronic volume depletion, and sodium loading. J Clin Invest 51: 1731–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N, Yin L, Hanniman EA, Joshi S, Lazar MA 2009. Negative feedback maintenance of heme homeostasis by its receptor, Rev-erbα. Genes & Dev 23: 2201–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Nakahata Y, Tanaka M, Yoshida M, Soma H, Shinohara K, Yasuda A, Mamine T, Takumi T 2005. Acute physical stress elevates mouse period1 mRNA expression in mouse peripheral tissues via a glucocorticoid-responsive element. J Biol Chem 280: 42036–42043 [DOI] [PubMed] [Google Scholar]
- Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, Mangelsdorf DJ, Evans RM 2006. Nuclear receptor expression links the circadian clock to metabolism. Cell 126: 801–810 [DOI] [PubMed] [Google Scholar]
- Yang X, Lamia KA, Evans RM 2007. Nuclear receptors, metabolism, and the circadian clock. Cold Spring Harb Symp Quant Biol 72: 387–394 [DOI] [PubMed] [Google Scholar]
- Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, Reid RA, Waitt GM, Parks DJ, Pearce KH, Wisely GB, et al. 2007. Rev-erbα, a heme sensor that coordinates metabolic and circadian pathways. Science 318: 1786–1789 [DOI] [PubMed] [Google Scholar]
- Young MW 2000. The tick-tock of the biological clock. Sci Am 282: 64–71 [DOI] [PubMed] [Google Scholar]