Abstract
The developmental switch from human fetal (γ) to adult (β) hemoglobin represents a clinically important example of developmental gene regulation. The transcription factor BCL11A is a central mediator of γ-globin silencing and hemoglobin switching. Here we determine chromatin occupancy of BCL11A at the human β-globin locus and other genomic regions in vivo by high-resolution chromatin immunoprecipitation (ChIP)–chip analysis. BCL11A binds the upstream locus control region (LCR), ɛ-globin, and the intergenic regions between γ-globin and δ-globin genes. A chromosome conformation capture (3C) assay shows that BCL11A reconfigures the β-globin cluster by modulating chromosomal loop formation. We also show that BCL11A and the HMG-box-containing transcription factor SOX6 interact physically and functionally during erythroid maturation. BCL11A and SOX6 co-occupy the human β-globin cluster along with GATA1, and cooperate in silencing γ-globin transcription in adult human erythroid progenitors. These findings collectively demonstrate that transcriptional silencing of γ-globin genes by BCL11A involves long-range interactions and cooperation with SOX6. Our findings provide insight into the mechanism of BCL11A action and new clues for the developmental gene regulatory programs that function at the β-globin locus.
Keywords: Hemoglobin switching, fetal hemoglobin, BCL11A, transcription regulation
The human β-globin locus on chromosome 11 contains the linked embryonic (ɛ), fetal (Gγ and Aγ), and adult (δ and β) globin genes that are expressed sequentially during development. The β-globin locus provides a paradigm for tissue-specific and developmentally regulated transcription. Upstream of the locus lies a cluster of erythroid-specific cis-regulatory elements demarcated by DNase I-hypersensitive sites (HSs), known as the locus control region (LCR), which ensure high-level expression of the linked globin genes (Tuan et al. 1985; Forrester et al. 1986; Grosveld et al. 1987; Bender et al. 2000). Earlier studies using transgenic and knockout β-globin mouse models led to fundamental insight into the developmental regulation of gene expression and gene competition, and the function of distal transcriptional regulatory elements (Palstra et al. 2008). Recently, the β-globin locus has provided the basis for studying long-range interactions mediated by inter- and intrachromosomal looping. For example, the LCR of the β-globin locus has been shown to be in close proximity to the active promoters, and also contacts other chromosomal regions in an erythroid cell and developmental stage-specific manner (Tolhuis et al. 2002; Palstra et al. 2003; Simonis et al. 2006; Schoenfelder et al. 2010).
The switch from human fetal (γ) to adult (β) expression is of particular interest due to its clinical importance. Delay of the switch or reactivation of fetal hemoglobin (HbF) in the adult stage greatly ameliorates clinical severity in the principal β-hemoglobin disorders sickle cell anemia and β-thalassemias. Over the last three decades, numerous approaches have been taken to elucidate the molecular events regulating normal human hemoglobin switching and the potential reactivation of HbF in adult hematopoietic cells (Stamatoyannopoulos 2005). Various models—including gene silencing, gene competition, chromosomal looping, and tracking—have been presented for the fetal-to-adult globin switch (Orkin 1990; Stamatoyannopoulos 2005; Bank 2006). However, despite intensive molecular studies, the proteins regulating the switch and the mechanisms controlling the process have been largely elusive.
Recent human genetic studies focused on natural variation in the level of HbF expression in human populations shed new light on this complex regulatory process (Thein and Menzel 2009; Thein et al. 2009). More specifically, genome-wide association studies (GWAS) led to the identification of a new HbF-associated locus on chromosome 2, located within the gene BCL11A (Menzel et al. 2007; Thein et al. 2007; Lettre et al. 2008; Sedgewick et al. 2008; So et al. 2008; Uda et al. 2008; Thein and Menzel 2009). Subsequently, the gene BCL11A (also known as Evi9, Ctip1), encoding a zinc finger transcription factor, was shown to function as a regulator of HbF expression (Sankaran et al. 2008). Abundant expression of full-length forms of BCL11A is developmentally restricted to adult erythroid cells (Sankaran et al. 2008, 2009). Down-regulation of BCL11A expression in adult human erythroid precursors leads to robust induction of HbF (Sankaran et al. 2008). Knockout of BCL11A in transgenic mice harboring the human β-globin locus prevents proper silencing of endogenous mouse β-like embryonic genes and human γ-globin genes in adult erythroid cells of the fetal liver (Sankaran et al. 2009). Additionally, BCL11A interacts with the Mi-2/NuRD chromatin remodeling complexes, as well as the erythroid transcription factors GATA1 and FOG1, in erythroid progenitors (Sankaran et al. 2008). These findings established BCL11A as the first genetically validated regulator of both developmental control of hemoglobin switching and silencing of γ-globin expression in adults.
SOX6 is a member of the Sry-related high-mobility group (HMG) box transcription factors, many of which serve as determinants of cell fate and differentiation in various lineages (Wegner 1999; Schepers et al. 2002). A potential role for SOX6 in globin gene regulation was first recognized by analysis of the Sox6-deficient mouse. At the fetal liver stage, expression of mouse embryonic β-like globins (ɛy and βh1) was dramatically elevated in the Sox6-deficient p100H mouse (Yi et al. 2006). While βh1 was down-regulated rapidly in late fetal livers, the ɛy-globin gene was expressed persistently until birth (Yi et al. 2006). SOX6 also represses ɛy-globin and, to a lesser extent, βh1-globin expression in definitive erythropoiesis of adult mice, as revealed by transplantation of fetal liver cells from Sox6-deficient mice into wild-type adult mice (Cohen-Barak et al. 2007). SOX6 acts as a repressor of ɛy-globin expression by direct binding to the ɛy promoter (Yi et al. 2006). SOX6 has also been suggested to enhance definitive erythropoiesis in mice by stimulating erythroid cell survival, proliferation, and terminal maturation (Dumitriu et al. 2006). A role for SOX6 in human globin gene regulation has not yet been examined directly, although recent work in human erythroid progenitors suggests that variation in the levels of SOX6 correlates with γ-globin gene expression (Sripichai et al. 2009).
While a precise role for SOX6 in hemoglobin switching remains to be determined, SOX6 appears to act as either an activator or a repressor, depending on its interacting proteins and promoter context (Lefebvre et al. 1998; Murakami et al. 2001). In addition, Sox transcription factors bind to the minor groove of DNA and cause a drastic bend of the DNA that leads to local conformational changes (Ferrari et al. 1992; Connor et al. 1994). Therefore, it has been suggested that SOX6 may perform part of its function as an architectural factor by organizing local chromatin structure and assembling other DNA-bound transcriptional factors into biologically active, sterically defined multiprotein transcriptional complexes (Yi et al. 2006).
Here we show that full-length BCL11A protein occupies the β-globin locus in vivo in primary adult human erythroid cells. BCL11A associates with distal regulatory elements and reconfigures the β-globin cluster. We show that SOX6 is also required to maintain silencing of human γ-globin genes in definitive erythropoiesis. BCL11A and SOX6 are coexpressed, interact physically, co-occupy the human β-globin cluster, and act collaboratively in silencing γ-globin transcription in adult human erythroid cells. Our findings provide important insights into the developmental gene regulatory programs operating at the human β-globin locus, and new clues for manipulating globin gene expression as an approach to treat human blood diseases.
Results
BCL11A occupies the human β-globin cluster in vivo
The identification of BCL11A as a critical switching factor allows focused mechanistic studies to dissect the globin switching process. In principle, BCL11A might silence γ-globin expression either directly by interacting with cis-regulatory elements within the β-globin cluster, or indirectly by affecting other pathways that ultimately impact γ-globin transcription. To investigate whether BCL11A silences γ-globin expression by direct association with the human β-globin locus, we previously used chromatin immunoprecipitation (ChIP) followed by quantitative PCR to examine in vivo binding of BCL11A protein to the β-globin locus in adult human erythroid progenitors (Sankaran et al. 2008). The results indicated that BCL11A occupies several discrete regions within the human β-globin cluster. These include the third HS (HS3) of the LCR and an intergenic region between Aγ-globin and δ-globin genes (Sankaran et al. 2008). Such results argue that BCL11A acts within the β-globin locus. A fuller accounting of the mechanism necessitates comprehensive analysis of chromatin occupancy of BCL11A and its partner proteins in the β-globin cluster.
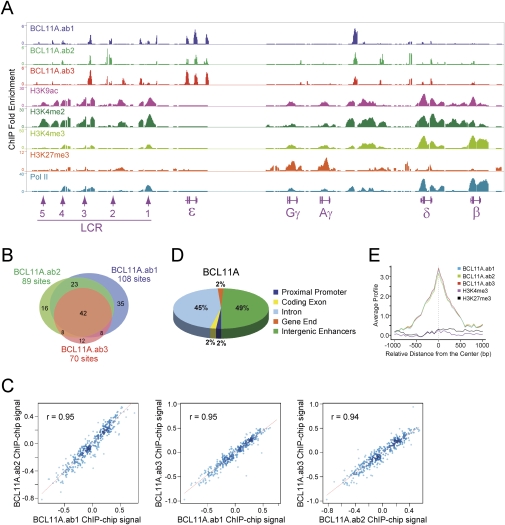
To obtain high-resolution chromatin occupancy of BCL11A in the β-globin locus and other genomic regions, we performed ChIP–chip analysis in primary adult human erythroid progenitors. We used the human ENCODE 2.0R array, which is comprised of 30 Mb of DNA or ∼1% of the human genome. The ENCODE array encompasses the entire human β-globin cluster and offers exceptionally high resolution (7-base-pair [bp] tiling) in the most detailed genomic view (The ENCODE Project Consortium 2004). BCL11A has at least four alternatively spliced isoforms (Supplemental Fig. 1A). We previously described that BCL11A is expressed as short variant proteins at stages that robustly expressed the γ-globin genes (primitive and fetal liver erythroid progenitors), but not in adult erythroid cells in which γ-globin genes are silenced (Sankaran et al. 2008, 2009). These short variants, however, do not correspond to reported BCL11A isoforms, and their identities remain unknown. In primary adult human erythroid cells, BCL11A is expressed as two major isoforms (designated XL and L) at both the protein and RNA levels (Supplemental Fig. 1; Sankaran et al. 2008, 2009). These isoforms differ only in usage of the 3′-terminal exon and function similarly in other settings (Liu et al. 2006). At the protein level, the XL and L isoforms are indistinguishable due to their similar size (Sankaran et al. 2008). At the mRNA level, the XL isoform is seven times more abundant than the L isoform in adult human erythroid progenitors and appears at a similar ratio in mouse erythroid cells (Supplemental Fig. 1B,C). To assess chromatin occupancy of both XL and L isoforms more comprehensively than before, we used three different antibodies directed to BCL11A in ChIP–chip experiments (Supplemental Fig. 1A). The ab1 and ab2 antibodies react with both XL and L isoforms, while the ab3 antibody reacts only with the XL isoform. The high-resolution mapping of chromatin occupancy using three antibodies reveals that BCL11A associates with the β-globin locus in vivo. As shown by the peaks, very similar patterns of BCL11A occupancy at the β-globin cluster were observed using all three antibodies (Fig. 1A). These data suggest that the XL isoform is the predominant form present in human erythroid progenitors and occupies discrete chromatin regions within the β-globin locus in vivo. Collectively, BCL11A binds strongly to the upstream HSs within the LCR, the embryonic ɛ-globin, and the intergenic region between Aγ-globin and δ-globin (Fig. 1A). The binding peaks within the δγ-intergenic region overlap previously reported deletion mutations in humans associated with elevated HbF expression (Bank 2006). Importantly, BCL11A does not detectably bind to the promoter regions of the γ-globin genes, suggesting that BCL11A may perform its function through long-distance interactions within the β-globin locus.
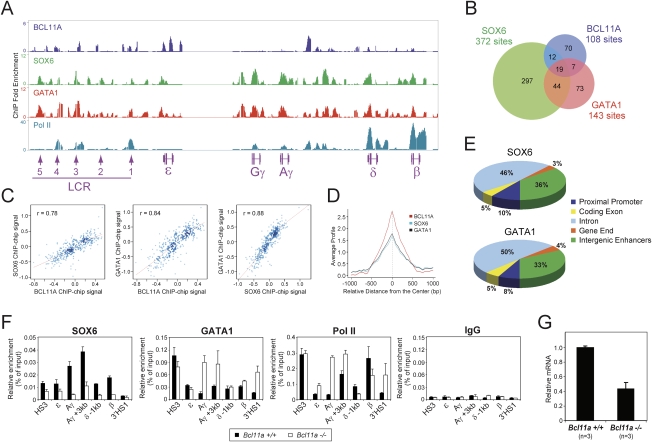
Figure 1.
Chromatin occupancy of BCL11A and common histone modifications within the human β-globin locus. (A) Genome browser representation of BCL11A.ab1, BCL11A.ab2, BCL11A.ab3, H3K9ac, H3K4me2, H3K4me3, H3K27me3, and RNA Pol II-binding patterns at the human β-globin cluster in primary adult human erythroid progenitors. The human β-globin locus is depicted at the bottom containing five β-like globin genes (ɛ, Gγ, Aγ, δ, and β) and the upstream DNase I HSs (1–5) within the LCR. (B) Venn diagram of genome-wide colocalization of BCL11A.ab1, BCL11A.ab2, and BCL11A.ab3 ChIP–chip sites. (C) Genome-wide analysis of BCL11A.ab1, BCL11A.ab2, BCL11A.ab3 co-occupancy. Scatter plots displays mutual enrichment between indicated ChIP–chip data sets relative to the input. Correlation values are shown. (D) Distribution of BCL11A enrichment peaks located in the promoter (−3 kb to +1 kb), coding exon, intron, gene end (5′UTR and 3′UTR), or intergenic enhancers (outside of these defined regions). The values were calculated using data sets combining BCL11A.ab1, BCL11A.ab2, and BCL11A.ab3. (E) Sitepro (Shin et al. 2009) analysis of the genome-wide correlation between the indicated ChIP–chip data sets. The average signal (from a WIG) is shown for the 2-kb region surrounding the center of the ChIP–chip sites from the BCL11A.ab1 data set.
Genome-wide analysis of BCL11A ChIP–chip data sets generated with the three antibodies reveals a significant number of shared sites (Fig. 1B). Out of 267 BCL11A sites, 204 (76.4%) were present in at least two data sets (Fig. 1B). Genomic-scale analysis of the ChIP–chip signal (from a WIG file) (Shin et al. 2009) also shows a strong correlation and mutual enrichment among three data sets (Fig. 1C,E). A motif search within sites enriched for BCL11A binding identified three de novo GC-rich motifs for BCL11A binding (Supplemental Fig. 2A). We next examined the location of BCL11A-binding sites (all three data sets combined) relative to annotated gene structures based on the University of California at Santa Cruz Genome Browser Database (Karolchik et al. 2008). Two percent of the peaks occur within proximal promoters (defined as −3 kb to +1 kb relative to transcription start site), 2% lie within coding exons, 45% are within introns, 2% are located within the 5′ untranslated region (UTR) and 3′UTR (gene end), and 49% are intergenic (Fig. 1D). These results suggest that BCL11A tends to associate with distal control regions, in contrast to other transcription factors (see below), in adult human erythroid cells.
BCL11A-binding sites are devoid of common histone modifications
To ascertain whether chromatin occupancy of BCL11A coincides with common histone modifications, ChIP–chip experiments were performed using antibodies directed to acetylated histone H3 Lys 9 (H3K9ac), dimethyl-H3 Lys 4 (H3K4me2), trimethyl-H3 Lys 4 (H3K4me3), and trimethyl-H3 Lys 27 (H3K27me3). H3 acetylation and H3K4 methylation are associated with transcriptionally active or poised genes, while H3K27 methylation is generally associated with transcriptional repression (Li et al. 2007). Peaks of acetylated H3 and di- and trimethylated H3K4 are observed at the upstream LCR, the actively transcribed δ-globin and β-globin genes, and an intergenic region between Aγ-globin and δ-globin (Fig. 1A). Weak peaks of active histone modifications are also observed at the largely inactive Gγ-globin and Aγ-globin genes. In contrast, peaks of trimethylated H3K27 are present only at the proximal promoters of the γ-globin genes, consistent with their very low-level expression in adult human erythroid cells. Importantly, high-resolution mapping of BCL11A binding and histone modifications reveals that BCL11A-binding sites are devoid of active histone modifications within the human β-globin locus (Fig. 1A). Particularly, the peaks of BCL11A at the ɛ-globin region bear no detectable enrichment of acetylated H3 and methylated H3K4 or H3K27. The other two strong peaks of BCL11A located at the HS3 site and the δγ-intergenic region are located in the “valley” of neighboring peaks for active histone modifications (H3K9ac, H3K4me2, and H3K4me3) (Fig. 1A). Global analysis of cooccupancy between BCL11A, H3K4, and H3K27 trimethylation also reveals a lack of shared binding sites between BCL11A and common histone modifications at a genome scale (Fig. 1E; Supplemental Fig. 2B,C).
BCL11A reconfigures the human β-globin locus
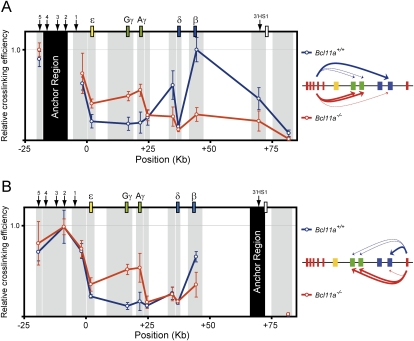
The lack of detectable association of BCL11A with the proximal promoter regions of the γ-globin genes suggests that BCL11A does not act in transcriptional repression locally at these genes. As an alternative mode of action, BCL11A might silence γ-globin expression through binding to distal control regions and mediate long-distance interaction by reconfiguration of the β-globin cluster at different development stages. To test this hypothesis, we employed chromatin conformation capture (3C) assay to examine the structure of the human β-globin locus in the presence or absence of BCL11A in fetal liver cells from human β-YAC mice. In this experiment, mouse fetal liver cells derived from β-YAC mice with either wild-type (Bcl11a+/+) or Bcl11a-null (Bcl11a−/−) alleles were examined (Fig. 2). Since these adult-type erythroid cells contain the human β-globin locus, we can assess chromatin structure in the presence or absence of BCL11A. In the 3C method, distances between a predefined “anchor region” and other positions in the locus are measured through a ligation assay. If the anchor region interacts physically with a downstream region, the relative cross-linking efficiency between those two regions will be higher than other regions lacking contact.
Figure 2.
BCL11A reconfigures the human β-globin cluster. Locus-wide cross-linking frequencies show reconfiguration of the human β-globin cluster in the absence of Bcl11a. Relative cross-linking frequencies observed in wild-type (Bcl11a+/+) E14.5 fetal liver erythroid cells are shown in blue, and Bcl11a−/− cells are shown in red. Gray shading indicates position and size of the analyzed fragments, and black shading represents the “anchor region” HS2–HS4 (A) and 3′HS1 (B). Spatial organization of the human β-globin locus is indicated at the top. Schematic diagrams representing the three-dimensional interactions that occur within the human β-globin cluster are shown at the side. In each graph, the highest cross-linking frequency value was set to 1. The X-axis shows position (in kilobases) in the locus. All data are plotted as the mean ± SD of the measurement.
In mice wild type for BCL11A, the anchor region at the LCR has the highest cross-linking efficiency with the β-globin gene, indicating loop formation (Fig. 2A). The two γ-genes are cross-linked much less efficiently, indicating that the LCR regions interact preferentially with the adult genes in the presence of BCL11A. This observation is consistent with the predominant expression of β-gene at this stage (Supplemental Fig. 3; Sankaran et al. 2009). In contrast, in Bcl11a−/− cells, the locus is reconfigured such that the γ-genes are highly associated with the LCR, while the β-gene is cross-linked less efficiently (Fig. 2A). This finding, again, is consistent with the observation that γ-globins are the predominant subtypes expressed in these cells (Supplemental Fig. 3; Sankaran et al. 2009), indicating that the LCR regions strongly favor the interaction with the fetal genes in the absence of BCL11A. Similarly, use of the 3′HS1 as the anchor region reveals that the γ-genes are cross-linked at much higher efficiency in the absence of BCL11A (Fig. 2B). These results strongly support the hypothesis that BCL11A performs part of it function by reconfiguring the β-globin locus.
BCL11A and SOX6 are coexpressed during erythroid development
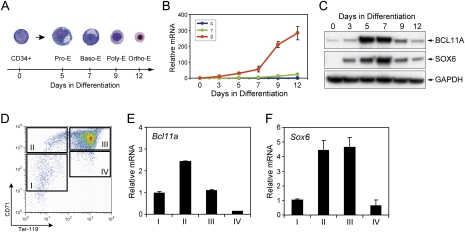
The study of human hemoglobin switching has relied largely on transgenic mouse models, such as human β-locus mice, or on in vitro culture systems that employ various cytokines to induce erythroid maturation of purified primary hematopoietic progenitors or pluripotent embryonic stem (ES)-like cells (Strouboulis et al. 1992; Gaensler et al. 1993; Peterson et al. 1993; Migliaccio et al. 2002). Although the transgenic mouse model provides a valid system for evaluating human developmental globin regulation, key limitations have been noted. Mice have no fetal globin equivalent and the human fetal (γ) globin genes behave largely as embryonic globin genes in these mice, demonstrating that critical differences in the trans-acting milieu have arisen during mammalian evolution (Sankaran et al. 2009). To circumvent these issues, we employed a serum-free two-phase liquid culture system to generate highly enriched populations of maturing, primary erythroblasts ex vivo from purified CD34+ human hematopoietic progenitors (Sankaran et al. 2008). In this experimental context, maturing erythroid precursors recapitulate the normal sequence of maturation from proerythroblasts to orthochromatic erythroblasts (Fig. 3A). By day 5 in culture medium that supports terminal erythroid maturation, the majority of cells acquire proerythroblast morphology. By day 7, cells exhibit basophilic erythroblast morphology. By days 9–12 of differentiation, the majority of cells are polychromatophilic and orthochromatophilic (Fig. 3A). Surface marker expression during differentiation includes loss of CD34 antigen, reduced expression of CD45, and acquisition of transferrin receptor (CD71), glycophorin A (CD235a), and antigen CD36 (Supplemental Fig. 4). This ex vivo culture system results in robust expression of adult hemoglobin (β-globin) and low-level HbF (γ-globin) production (Fig. 3B; Supplemental Fig. 5).
Figure 3.
BCL11A and SOX6 are coexpressed during erythroid development. (A) Human CD34+ cells were induced for erythroid differentiation in a serum-free two-phase liquid culture model. Representative cytospin images were shown for undifferentiated CD34+ cells, proerythroblasts (Pro-E), basophilic erythroblasts (Baso-E), polychromatic erythroblasts (Poly-E), and orthrochromatic erythroblasts (Ortho-E). (B) Globin mRNA levels were monitored by real-time RT–PCR in erythroid cells at various differentiation stages. All transcript levels were normalized against human GAPDH transcript levels. (C) The expression of human BCL11A and SOX6 proteins was measured by Western blot. GAPDH was analyzed as a loading control. (D) The CD71 and Ter119 expression pattern is shown for mouse fetal liver cells from E14.5 embryos. The cells were FACS-sorted into four populations (I, II, III, and IV) as described previously (Zhang et al. 2003). (E,F) Mouse Bcl11a and Sox6 mRNA levels were measured by real-time RT–PCR. Transcript levels were normalized against mouse Gapd transcript levels. All results are means ± SD of at least three independent experiments.
Because BCL11A does not reside at the promoters of the γ-genes (Fig. 1A), it is likely that other factors cooperate with BCL11A to achieve their proper silencing within the β-globin locus. SOX6 has been shown previously to silence the mouse embryonic ɛy-globin and βh1-globin expression, probably by direct association with the ɛy promoter (Yi et al. 2006). Given that the human γ-gene behaves as an embryonic gene in the mouse erythroid trans-acting environment (Sankaran et al. 2009), we hypothesized that SOX6 may also be involved in γ-gene control. As a first step in asking whether BCL11A and SOX6 might act together in globin gene regulation, we examined the expression of BCL11A and SOX6 in primary adult human erythroid cells. Both BCL11A (XL and L isoforms) and SOX6 are highly expressed during mid-maturation (days 5–7), corresponding to the proerythroblast and basophilic erythroblast stages (Fig. 3C; Supplemental Fig. 6). Expression of both proteins was much reduced in CD34+ hematopoietic progenitors and in more mature erythroblasts. Similarly, mouse Bcl11a and Sox6 mRNAs are both highly expressed in stage II (Ter119−CD71+) and stage III (Ter119+CD71hi) (Zhang et al. 2003) populations in mouse fetal liver (embryonic day 14.5 [E14.5]) erythroid cells (Fig. 3D–F). Thus, BCL11A and SOX6 are coexpressed during erythroid development in both humans and mice.
SOX6 comigrates and interacts physically with BCL11A
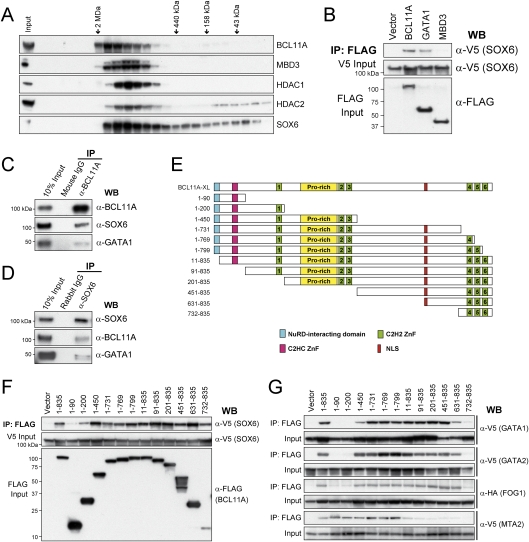
The coexpression of BCL11A and SOX6 during erythroid differentiation suggests that these factors, both of which have been described previously as mediators of hemoglobin switching and silencing of mouse embryonic globin gene expression (Yi et al. 2006; Sankaran et al. 2009), may function together in regulating globin switching in a protein complex. To explore this possibility, we first examined the migration of BCL11A and SOX6 in human erythroid nuclear extracts by gel filtration chromatography. As shown in Figure 4A, BCL11A migrates as a major peak centered between 440-kDa and 2-MDa markers. The BCL11A elution pattern overlaps substantially with the Mi-2/NuRD components MBD3, HDAC1, and HDAC2, suggesting that BCL11A strongly associates with NuRD complexes in human erythroid cells, similar to prior observations in mouse erythroleukemia (MEL) cells and B lymphoid cells (Cismasiu et al. 2005; Sankaran et al. 2008). Importantly, SOX6 also comigrates predominantly with BCL11A and NuRD components (Fig. 4A). A small proportion of SOX6 elutes in fractions corresponding to lower molecular mass (<440 kDa), indicating that SOX6 may also be present in association with other proteins. The chromatographic profile demonstrates that SOX6, together with BCL11A and NuRD complexes, comigrate in vitro and are likely part of a multiprotein complex.
Figure 4.
Physical interaction between BCL11A and SOX6. (A) Gel filtration fractions of nuclear extracts from human erythroid progenitors (day 5 in differentiation medium) were blotted for BCL11A, MBD3, HDAC1, HDAC2, and SOX6. There is significant overlap between BCL11A, SOX6, and components of the Mi-2/NuRD complexes. Elution positions of molecular mass standards (from left to right, 2 MDa, 440 kDa, 158 kDa, and 43 kDa) are indicated. (B) V5-tagged SOX6 cDNA was coexpressed in COS7 cells with a Flag-tagged empty vector, BCL11A, GATA1, and MBD3 cDNA. Nuclear extracts were immunoprecipitated using anti-Flag antibody, and copurified proteins were analyzed by Western blot with anti-V5 antibody. Inputs (5%) are shown. (C) Endogenous SOX6 and GATA1 were coimmunoprecipitated with BCL11A in human erythroid cells. Nuclear extracts from erythroid progenitors (day 5 in differentiation medium) were immunoprecipitated with anti-BCL11A (14B5) antibody or mouse IgG (negative control), and copurified proteins were analyzed by Western blot with antibodies against BCL11A, SOX6, and GATA1. (D) Endogenous BCL11A was coimmunoprecipitated with SOX6 in human erythroid cells. Nuclear extracts from human erythroid progenitors were immunoprecipitated with anti-SOX6 antibody or rabbit IgG (negative control). (E) Schematic diagram of BCL11A constructs used in F and G. (F) Co-IP of V5-tagged SOX6 coexpressed transiently with a Flag-tagged vector, BCL11A-XL (1–835 amino acids), or BCL11A truncation mutants in COS7 cells. Nuclear extracts were immunoprecipitated with an anti-Flag antibody, and copurified proteins were analyzed by Western blot with an anti-V5 antibody. Inputs (10%) are shown. (G) Co-IP of V5-tagged GATA1, GATA2, MTA2, or HA-tagged FOG1 coexpressed transiently with a Flag-tagged Vector, BCL11A-XL (1–835 amino acids), or BCL11A truncation mutants in COS7 cells. Co-IP and Western blot were performed as described in F. Inputs (10%) are shown.
To further explore the relationship of BCL11A and SOX6 proteins, we ectopically expressed V5-tagged SOX6 with Flag-tagged BCL11A (XL isoform), GATA1, or MBD3 in COS7 cells, followed by coimmunoprecipitation (co-IP). Both BCL11A and GATA1 efficiently coimmunoprecipitate SOX6, indicating that SOX6 interacts physically with BCL11A and GATA1 (Fig. 4B; Supplemental Fig. 7). NuRD components MBD3 and MTA2 are unable to interact with SOX6, suggesting that SOX6 makes independent contact with BCL11A (Fig. 4B; Supplemental Fig. 7). Furthermore, in human erythroid cell nuclear extracts, α-BCL11A antibody efficiently immunoprecipitates endogenous SOX6 and GATA1 proteins (Fig. 4C). Similarly, α-SOX6 antibody efficiently immunoprecipitates endogenous BCL11A and GATA1 proteins (Fig. 4D). These data demonstrate that SOX6 interacts directly with BCL11A and GATA1 under physiological conditions in human erythroid progenitors.
To further characterize the determinants mediating association between BCL11A, SOX6, GATA1, and other protein partners, we mapped the interaction domains by co-IP assays of epitope-tagged proteins expressed transiently in COS7 cells (Fig. 4E–G). These studies identify a broad region on BCL11A (amino acids 91–835) that is sufficient for BCL11A interaction with SOX6 (Fig. 4F). The C-terminal 104 amino acids (732–835 amino acids) of BCL11A protein encompass three zinc fingers and are sufficient for SOX6 binding. However, the first 90 amino acids at the N terminus are dispensable for SOX6 binding. These results indicate that SOX6 interacts with BCL11A through multiple domains, likely through multiple zinc fingers, similar to the interactions between GATA1 and FOG1 (Cantor and Orkin 2005). Interestingly, the three C-terminal zinc fingers (732–835 amino acids) are unable to interact with GATA1, GATA2, and FOG1 (Fig. 4G), suggesting that SOX6 makes independent contacts with BCL11A. The N-terminal 12 amino acids are highly homologous among BCL11A, BCL11B, FOG1, FOG2, and SALL1–4 proteins, and serve as a transcriptional repression motif that mediates interaction with the Mi-2/NuRD nucleosomal remodeling complexes (Lin et al. 2004). Nevertheless, our results demonstrate that the N-terminal 90 amino acids are necessary and sufficient for interaction of BCL11A with the NuRD component MTA2 (Fig. 4G).
BCL11A, SOX6, and GATA1 co-occupy the human β-globin cluster
We next examined the in vivo chromatin occupancy of BCL11A and SOX6 at the β-globin cluster in primary human erythroid progenitors by ChIP–chip. The alignment of BCL11A and SOX6 ChIP–chip peaks reveals partial overlap, particularly at the HS3 site of the upstream LCR and the embryonic ɛ-globin gene (Fig. 5A; Supplemental Fig. 8). Importantly, SOX6 strongly binds the Aγ and Gγ proximal promoters, reminiscent of the role of SOX6 in silencing mouse ɛy-globin expression by association with its proximal promoter (Yi et al. 2006). In contrast, BCL11A does not detectably bind to γ-promoters by ChIP–chip (Figs. 1A, 5A). Given the observation that these two proteins physically interact and likely associate within a protein complex (or complexes), these results suggest that SOX6 may act as a partner of BCL11A in silencing γ-globin expression. These data also suggest that SOX6, by interacting with γ-proximal promoters, may help recruit BCL11A to the proximal regions of the γ-genes during hemoglobin switching.
Figure 5.
Chromatin occupancy of BCL11A, SOX6, and GATA1 within the human β-globin locus in adult human erythroid progenitors. (A) In vivo chromatin occupancy of BCL11A, SOX6, GATA1, and RNA Pol II was examined by ChIP–chip in human erythroid progenitors. Precipitated DNA samples were amplified and hybridized to human ENCODE 2.0R array. A genome browser representation of binding patterns at the human β-globin cluster is shown. Relative positions of the human β-like globin genes and the upstream LCR consisting of five DNase I HSs (1–5) are shown at the bottom. (B) A Venn diagram of genome-wide colocalization of BCL11A, SOX6, and GATA1 ChIP–chip sites is shown. (C) Genome-wide analysis of BCL11A, SOX6, and GATA1 cooccupancy is shown. Scatter plots display mutual enrichment between indicated ChIP–chip data sets relative to the input. Correlation values are shown. (D) Sitepro (Shin et al. 2009) analysis of genome-wide correlation between the indicated ChIP–chip data sets. The average signal (from a WIG) is shown for a 2-kb region surrounding the center of ChIP–chip sites from the BCL11A.ab1 data set. (E) Distribution of SOX6 and GATA1 enrichment peaks located in the promoter (−3 kb to +1 kb), coding exon, intron, gene end (5′UTR and 3′UTR), or intergenic enhancers (outside of these defined regions) is shown. (F) In vivo chromatin occupancy of SOX6, GATA1, and RNA Pol II was examined by ChIP in E14.5 fetal liver cells from β-YAC control mice (Bcl11a+/+) and Bcl11a-null mice (Bcl11a−/−). Precipitated DNA samples were quantified using primers designed to amplify several control regions within the human β-globin locus as indicated below each graph. The ChIP signals are shown as a percentage of the input DNA signal. Results are mean ± SD of three independent experiments. (G) Relative mRNA expression of SOX6 was examined by real-time RT–PCR in FACS-sorted Ter119+CD71+ E14.5 fetal liver erythroid cells from Bcl11a+/+ (wild-type, n = 3) and Bcl11a−/− (n = 3) embryos. Transcript levels were normalized against mouse Gapd transcript levels. Results are means ± SD.
To obtain insight into whether other BCL11A-interacting partners are also recruited to the same chromatin regions in vivo, we mapped the binding of GATA1 across the β-globin cluster by ChIP–chip. The results indicate strong association of GATA1 with the upstream HS1, HS3, HS4, and HS5 within the LCR region, and the intergenic regions between the Aγ-genes and δ-genes (Fig. 5A; Supplemental Fig. 8). The binding peaks of GATA1 at the β-globin cluster overlap substantially with those of SOX6 and partially with peaks of BCL11A, suggesting that these proteins may function in a chromatin-associated multiprotein complex (or complexes) within the β-globin locus. The presence of BCL11A, SOX6, and GATA1 at the HS3 site within the LCR is consistent with a potential role of HS3 in mediating γ-globin transcription (Navas et al. 1998; Fang et al. 2005), likely through rearrangement of the configuration of the β-globin cluster by long-range interactions (Fig. 2A). In addition, RNA polymerase II (Pol II) is strongly associated with the LCR HS1, HS3, and HS4 sites, and the δ and β promoters. The strong binding of Pol II at the promoters of δ-globin and β-globin genes is consistent with their active transcription in adult erythroblasts (Figs. 3B, 5A).
Global analysis of BCL11A, SOX6, and GATA1 ChIP–chip data sets also reveals a significant number of shared sites (Fig. 5B) and mutual enrichment (Fig. 5C,D) at the genomic scale. Of the BCL11A sites, 28.7% (31 out of 108) and 24.1% (26 out of 108) are shared by SOX6 and GATA1, respectively, and 17.6% (19 out of 108) are co-occupied by all three factors (Fig. 5B). These include the HS3 site and ɛ-globin promoter at the β-globin locus (Fig. 5A). Genome-wide analysis of SOX6 and GATA1 distribution reveals a similar pattern (Fig. 5E). In contrast to BCL11A (Fig. 1D), a significant number of GATA1- and SOX6-binding sites are located within proximal promoters at a genomic scale.
If BCL11A, SOX6, and GATA1 bind the human β-globin cluster and silence γ-globin expression in a functional protein complex (or complexes), ablation of one critical component of the complex might lead to destabilization of the larger complex and/or its chromatin occupancy. We next determined whether the binding of SOX6 and GATA1 to the β-globin locus is dependent on the presence of BCL11A. ChIP was performed using mouse fetal liver cells derived from β-YAC mice with either wild-type or Bcl11a-null alleles. Since these adult-type erythroid cells contain the human β-globin locus, we can assess chromatin association of SOX6 in the presence or absence of BCL11A. In fetal liver cells wild-type for BCL11A, binding of SOX6 is enriched significantly at the Aγ promoter and the +3-kb downstream regions (Fig. 5F, Bcl11a+/+), similar to the pattern observed in the human erythroid cells (Fig. 5A; Supplemental Fig. 8). In Bcl11a-null cells, binding of SOX6 to the Aγ promoter region is decreased by 5.5-fold. The binding of SOX6 to other control regions within the β-globin locus is also decreased (Fig. 5F, Bcl11a−/−), indicating that loss of BCL11A leads to impaired chromatin occupancy of SOX6 within the human β-globin locus. In addition, the expression of SOX6 is reduced by 55% in the Bcl11a-null fetal liver erythroid cells as compared with the wild-type controls (Fig. 5G), suggesting that the reduced SOX6 chromatin occupancy may be due in part to the down-regulation of SOX6 expression in the absence of BCL11A. Interestingly, loss of BCL11A leads to increased GATA1 and RNA Pol II occupancy at the Aγ promoter, the γδ-intergenic regions (Aγ + 3 kb), and the 3′HS1 site. The enhanced Pol II occupancy at the Aγ promoter region is consistent with transcriptional reactivation of the γ-globin genes in the Bcl11a-null fetal liver cells (Supplemental Fig. 3; Sankaran et al. 2009). Taken together, these results demonstrate that loss of BCL11A in fetal liver definitive cells leads to diminished SOX6 occupancy within the β-globin locus, accompanied by elevated GATA1 and Pol II occupancy at the γ-globin promoter regions.
SOX6 cooperates with BCL11A in silencing mouse embryonic globins
The observation that repression of the endogenous embryonic ɛy-globin and βh1-globin genes is greatly impaired in definitive fetal liver erythrocytes of both Sox6-null mice and Bcl11a-null mice suggests that SOX6 and BCL11A may function collaboratively in their silencing (Yi et al. 2006; Sankaran et al. 2009). To test this hypothesis, we used lentivirus-mediated shRNAs to knock down the expression of BCL11A and/or SOX6 in MEL cells. In MEL cells, the mouse adult β-globins (βmajor and βminor) are expressed abundantly, and the embryonic ɛy-globin and βh1-globin are expressed at very low levels. MEL cells were transduced with control shRNA, BCL11A shRNA, or SOX6 shRNA lentiviruses, respectively. To knock down both BCL11A and SOX6 expression, MEL cells were transduced with SOX6 shRNA lentivirus (containing a puromycin-selectable cassette) and BCL11A shRNA lentivirus (containing a GFP cassette). Puromycin selection was imposed 1 d after transduction followed by GFP FACS sorting during the day of analysis (usually 4 d after transduction). As shown in Figure 6A, transduction of shRNA lentiviruses led to strong knockdown of BCL11A and/or SOX6 proteins. To determine the consequences of gene knockout, we examined the mRNA expression of ɛy-globin, βh1-globin, and βmajor/βminor-globins by quantitative RT–PCR (Fig. 6B). Knockdown of BCL11A alone led to a 116-fold increase of ɛy-globin mRNA transcripts. Knockdown of SOX6 alone led to a modest increase (15-fold) of ɛy mRNA. Similar effects on ɛy expression were observed with two other SOX6 shRNA lentiviruses (data not shown). Strikingly, combined knockdown of BCL11A and SOX6 led to a substantial increase of ɛy mRNA by 429-fold (Fig. 6B), indicating that SOX6 and BCL11A act together to maintain silencing of the ɛy-globin gene in MEL cells. Expression of βh1-globin was also derepressed to a lesser extent upon knockdown of BCL11A and SOX6, while expression of βmajor/βminor-globins remained largely unaffected (Fig. 6B). Taken together, these findings demonstrate that SOX6 cooperates with BCL11A to silence the expression of mouse embryonic β-like globin genes in MEL cells.
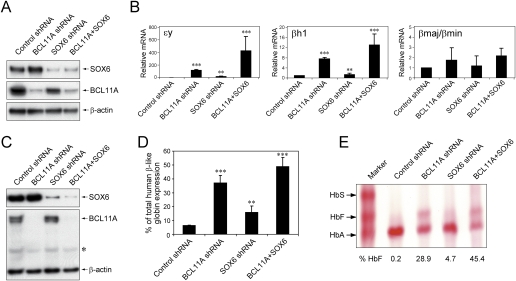
Figure 6.
BCL11A cooperates with SOX6 in silencing γ-globin gene expression. (A) Lentivirus-mediated shRNA delivery to MEL cells results in robust knockdown of both BCL11A and SOX6 proteins. Control samples were transduced with lentivirus prepared from the shRNA empty vector. Cells were harvested 4 d after transduction and analyzed by Western blot using antibodies for SOX6 and BCL11A. β-Actin was analyzed as a loading control. (B) shRNA-mediated knockdown of BCL11A and SOX6 results in elevations of ɛy-globin and βh1-globin mRNA levels. P < 0.001 (***) and P < 0.01 (**) in comparison with controls. (C) Lentiviral shRNA in human erythroid progenitors results in robust knockdown of both BCL11A and SOX6 proteins. Control samples were transduced with lentivirus prepared from the empty vector. Cells were harvested 7 d after transduction, which usually corresponded to day 5 in differentiation medium. Whole-cell lysates were prepared and analyzed by Western blot using antibodies for SOX6 and BCL11A. β-Actin was analyzed as a loading control. An asterisk (*) indicates nonspecific bands. (D) shRNA-mediated knockdown of BCL11A and SOX6 results in elevations of γ-globin mRNA levels (as a percentage of total β-like globin gene expression) on day 5 of differentiation. P < 0.001 (***) and P < 0.01 (**) in comparison with controls. (E) Hemolysates prepared from cells on day 14 of differentiation (terminal stage erythroblasts) show the presence of mature HbF. This was assessed using cellulose acetate hemoglobin electrophoresis with a size marker indicating the positions of HbA and HbF. The percentages indicating the proportion of hemoglobin species were determined by measurement from densitometry. All results are means ± SD from three independent experiments. Statistical significance was calculated using a Student t-test.
SOX6 contributes to the silencing of γ-globin expression in adult human erythroid cells
Because the human γ-globin genes are regulated largely as embryonic genes in the β-locus transgenic mouse, and SOX6 silences embryonic globin genes in mice, we hypothesized that SOX6 may be involved in γ-gene control. To assess a potential functional role for SOX6 in γ-globin expression, we employed shRNA knockdown in cultured primary adult human erythroid progenitors by lentivirus-mediated shRNAs as described previously (Moffat et al. 2006; Sankaran et al. 2008). As controls, we also infected the cells with either BCL11A shRNA alone, SOX6 shRNA alone, or control shRNA. Similar to the shRNA knockdown experiment in MEL cells, we employed puromycin selection and GFP FACS sorting to select for cells transduced with both SOX6 and BCL11A shRNA lentiviruses. When cells were transduced with the BCL11A and/or SOX6 shRNA lentiviruses and selection was imposed on the initiation of differentiation, we obtained an average of 99% and 95% knockdown of BCL11A and SOX6 proteins, respectively, by day 5 of erythroid maturation (Fig. 6C). No obvious morphological differences between the groups of cells were noted during differentiation, indicating that BCL11A knockdown and SOX6 knockdown do not impair differentiation of CD34+ cells into proerythroblasts (Supplemental Fig. 9; Sankaran et al. 2008). Remarkably, the level of γ-globin mRNA at day 5 of maturation was elevated by 4.8-fold, 2.1-fold, and 6.6-fold (from an average of 6.7% in the control to 31.8%, 14.2%, and 43.9% in cells transduced with either BCL11A shRNA alone, SOX6 shRNA alone, or both shRNAs, respectively) (Fig. 6D). Levels of mature HbF were also measured by hemoglobin electrophoresis (Fig. 6E). In control cells, only 0.2% of total hemoglobin produced is HbF. Upon knockdown of BCL11A alone, we obtained 28.9% HbF. Knockdown of SOX6 alone led to modest induction of HbF to 4.7%. Combined knockdown of BCL11A and SOX6 led to a substantial increase in HbF (to >45%). This level is greater than that observed upon knockdown of BCL11A, again suggesting that BCL11A and SOX6 act together to silence HbF expression. Quantification of HbF by high-performance liquid chromatography (HPLC) confirmed these observations (Supplemental Fig. 10). Taken together, these results argue persuasively that BCL11A cooperates with SOX6 to silence γ-globin expression in adult human erythroid cells.
Discussion
Here we determined and analyzed the genome-wide occupancy of BCL11A in primary adult human erythroid cells using ChIP followed by hybridization to the human ENCODE array. The exceedingly high 7-bp resolution of the ENCODE array allows fine mapping of BCL11A-binding sites within the human β-globin locus, as well as other genomic regions. The high-resolution ChIP–chip analysis enables careful comparison between the patterns of BCL11A occupancy and other transcriptional marks, such as common histone modifications. These comparisons reveal that BCL11A-binding sites are generally excluded from activating histone marks, such as acetylated histone H3 and di- and trimethylated H3K4 (Fig. 1A). Specifically, the BCL11A-binding peaks at the upstream HS3 site within the LCR and the intergenic regions between Aγ-globin and δ-globin genes are located in the “valley” of peaks for active histone modifications (Fig. 1A). No detectable peaks of trimethylated H3K27, a mark generally associated with transcription repression, are observed at the above binding sites for BCL11A (Fig. 1A). The three strong BCL11A peaks at the ɛ-globin are also devoid of any histone modifications. Genome-wide analyses also reveal a similar lack of co-occupancy between BCL11A and common histone modifications (Fig. 1E; Supplemental Fig. 2). The pattern of chromatin occupancy for BCL11A is distinct from that of other transcription factors such as GATA1 and SOX6, as we and others have shown (Fig. 5; Cheng et al. 2009; Yu et al. 2009). Several possibilities may account for the lack of common histone modifications at BCL11A-binding sites. BCL11A and/or associated protein partners, once bound to their target regions, may protect the nucleosomes at their binding sites from histone-modifying enzymes. BCL11A-binding sites may bear specific chromatin features that make them inaccessible to histone-modifying enzymes. Alternatively, BCL11A may tend to bind to nucleosome-free regions. Nevertheless, this special feature of BCL11A chromatin occupancy may be essential for its action in repressing gene expression by modulating long-range interactions (Fig. 2).
Over the past years, various models for the molecular control of globin switching have been put forward. These include autonomous gene silencing, gene competition, chromosomal looping, and tracking (Orkin 1990; Stamatoyannopoulos 2005; Bank 2006). The “chromosomal looping” model assumes flexibility of the chromatin fiber and predicts that distal control elements and promoters communicate through direct interactions mediated by proteins bound to these DNA elements (Ptashne 1986). Recent observations that active β-globin genes interact physically with multiple distal cis-regulatory elements, with inactive genes looping out, has provided a mechanistic framework for this model (Tolhuis et al. 2002; Palstra et al. 2003; Simonis et al. 2006; Schoenfelder et al. 2010). It is important to note that, although literature exists to support different models, the various models are not necessarily mutually exclusive. Moreover, it is likely that each model oversimplifies what occurs in an actual erythroid progenitor during globin gene expression. For example, while strong evidence supports intrachromosomal looping of the upstream LCR to the downstream genes within the β-globin locus, looping by itself does not preclude transient tracking within the locus or autonomous gene silencing of individual globin genes (Tolhuis et al. 2002; Palstra et al. 2003; Vakoc et al. 2005; Sankaran et al. 2010). The findings we describe here argue strongly that multiple modes of action may be employed by BCL11A in controlling hemoglobin switching, as well as silencing of γ-globin genes. BCL11A binds to distal control regions (the upstream LCR and the γδ-intergenic sites) and reconfigures the architecture of the β-globin cluster by chromosomal looping (Fig. 2). In the presence of BCL11A, the upstream LCR and downstream 3′HS1 interact preferentially with the transcriptionally active adult (β-globin) gene in adult-type erythroid cells. In the absence of BCL11A, the intrachromosomal looping within the β-globin cluster is reconfigured such that the LCR and 3′HS1 strongly favor interaction with the fetal (γ-globin) genes (Fig. 2). Importantly, BCL11A may also silence γ-globin expression by local interactions with the chromatin-associated SOX6 proteins at the proximal promoters of the γ-globin genes, as discussed below. Therefore, the function of BCL11A in controlling hemoglobin switching and γ-globin silencing not only reflects the complexity of transcriptional regulation occurring at the human β-globin locus, but provides new insight into further dissection of the globin switching process for targeted reactivation of HbF in patients with β-hemoglobin diseases.
In the present study, we also show that SOX6 is coexpressed with BCL11A during erythroid development, interacts physically with BCL11A and GATA1, and co-occupies the human β-globin cluster in vivo. Moreover, depleting the expression of both SOX6 and BCL11A leads to substantial derepression of mouse embryonic β-like globins in MEL cells and reactivation of γ-globins in adult human erythroid precursors. These results suggest that involvement of both BCL11A and SOX6 is preserved during mammalian evolution; however, their exact roles in cellular function, such as silencing of the embryonic or HbF expression, can be divergent. Our findings also illustrate that some evolutionarily conserved trans-acting factors may perform divergent regulatory functions among different species.
While the precise role of SOX6 in repressing mouse embryonic β-like globins and human γ-globins merits further study, SOX6 can act as either an activator or a repressor, depending on its interacting proteins and promoter context (Lefebvre et al. 1998; Murakami et al. 2001). SOX6 contains an evolutionarily conserved HMG domain that is essential for its DNA-binding activity (Grosschedl et al. 1994). HMG domain factors are capable of binding to the minor groove of the DNA and four-way junctions, and induce drastic DNA bending on binding (Ferrari et al. 1992; Connor et al. 1994). Modulation of DNA structure by the HMG proteins can mediate long-range enhancer functions on both DNA and chromatin-assembled genes by bridging other distant regions of DNA and associated factors and facilitating the assembly of high-order complexes (Thomas 2001). Specifically, HMG proteins have been suggested to modulate the β-globin genes (Dyer et al. 1996, 1998; Bagga et al. 2000; Drew et al. 2000). Using a proteomic approach, we demonstrated previously that BCL11A interacts with the NuRD chromatin remodeling and repressor complex, the erythroid transcription factors GATA1 and FOG1, and the nuclear matrix component Matrin-3 in erythroid progenitors (Sankaran et al. 2008). In this study, we show that SOX6 interacts with BCL11A, GATA1, and FOG1, but not the NuRD components MTA2 and MBD3 (Fig. 4B–G; Supplemental Fig. 7). The physical interaction between BCL11A and SOX6 may help recruit BCL11A and NuRD repressor complexes to the proximity of γ-globin genes. Thus, SOX6 may act as an important cofactor of BCL11A in mediating globin switching and silencing of γ-globin transcription. Our results suggest a model in which BCL11A mediates silencing of γ-globin genes through both long-range interaction within the human β-globin cluster and local interactions with the chromatin-associated SOX6 proteins at the proximal promoters of the γ-globin genes (Fig. 7). In summary, these findings provide critical insight into the mechanism of BCL11A action, and identify a second factor required for effective γ-globin silencing. An in-depth understanding of the role for BCL11A and its interacting proteins in globin switching offers the prospect for the design of target-based activation of HbF in adult erythroid cells of patients with hemoglobin disorders.
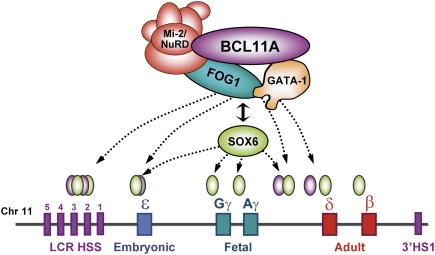
Figure 7.
Model of BCL11A-mediated silencing of γ-globin genes. The diagram illustrates the physical interaction between BCL11A and the Mi-2/NuRD complexes, erythroid transcription factors GATA1 and FOG1, and the HMG-box protein SOX6. Rather than binding to the promoters of the γ-globin or β-globin genes as these latter factors do, BCL11A protein occupies the upstream LCR and γδ-intergenic regions of the β-globin cluster in adult human erythroid progenitors. Our study suggests that transcriptional silencing of γ-globin expression by BCL11A involves long-range interaction within the β-globin cluster and local interactions with the chromatin-associated SOX6 proteins at the proximal promoters of the γ-globin genes.
Materials and methods
Cell culture and transfection
Primary human CD34+ cells were obtained from Yale Center of Excellence in Molecular Hematology (YCEMH). CD34+ cells were prepared from magnetically sorted mononuclear samples of G-CSF mobilized peripheral blood from donors. Cells were cultured initially for expansion in StemSpan SFEM Medium (StemCell Technologies, Inc.) supplied with 1× CC100 cytokine mix (StemCell Technologies, Inc.) and 2% penicillin–streptomycin (P/S) for 6 d. Cells were maintained at a density of 0.1 × 106 to 1 × 106 cells per milliliter with media changes every other day. On day 6, cells were pelleted and transferred into StemSpan SFEM Medium with 2% P/S, erythropoietin (1 U/mL), IL-3 (5 mg/mL), SCF (20 ng/mL), dexamethasone (2 μM), and β-estradiol (1 μM). Cells were incubated at 37°C with 5% CO2 and maintained at a density of 0.1 × 106 to 1 × 106 cells per milliliter by supplementing cultures every other day with fresh media. COS-7 and MEL cells were maintained in DMEM with 10% FCS. COS7 cells were transfected with the FuGene 6 (Roche) reagent according to the manufacturer's instructions. Lin− (excluding the Ter119 antibody) CD71+Ter119+ cells were FACS-sorted from E14.5 mouse fetal liver cells as described previously (Sankaran et al. 2008).
Experimental animals
The wild-type β-globin locus YAC transgenic (β-YAC) mice (Porcu et al. 1997; Peterson et al. 1998) were crossed with mice containing a Bcl11a floxed allele (Sankaran et al. 2009). To obtain Bcl11a-null β-YAC mice, these mice were crossed with Gata1-Cre mice and screened for germline deletions (Jasinski et al. 2001). All experiments were performed with the approval of the Children's Hospital Boston Animal Ethics Committee.
Antibodies and reagents
Antibodies against GATA1 (sc-265), HDAC2 (sc-7899), MBD3 (sc-9402), Pol II (sc-899), GAPDH (sc-25778), normal rabbit IgG (sc-2027), and normal mouse IgG (sc-2025) were purchased from Santa Cruz Biotechnologies. BCL11A (ab1: 15E3AC11; ab2: 14B5), SOX6 (ab64946), and GATA1 (ab47490) antibodies were purchased from Abcam. SOX6 (NBP1-19537) and BCL11A (ab3: NP600-261) antibodies were purchased from Novus Biologicals, LLC. HDAC1 antibody (06-720) was purchased from Upstate Biotechnologies. β-Actin antibody (MAB1501R) was purchased from Chemicon. SOX6 antibodies (αSox6C and αSox6N) were kindly provided by Dr. Paolo Sassone-Corsi (University of California at Irvine).
RNA isolation and RT–PCR
RNA was isolated using the RNeasy Plus Mini Kit (Qiagen) according to the manufacturer's protocol. cDNA was synthesized with the iScript cDNA synthesis kit (Bio-Rad). Real-time quantitative RT–PCR was performed using the iQ SYBR Green Supermix (Bio-Rad). Relative expression was quantified using the ΔΔCt method as described previously (Xu et al. 2009). Sequences of primers are available on request.
Co-IP and Western blot
For endogenous co-IP experiments, 1–5 mg of nuclear extract protein were treated with DNaseI (1 μg/mL), RNase A (1 μg/mL), and ethidium bromide (50 μg/mL), and incubated with 25 μg of BCL11A or SOX6 antibody on a rotating wheel overnight at 4°C. The protein complexes were collected by addition of protein G/A-agarose beads (Roche), followed by incubation for 3 h at 4°C. The beads were washed four times in BC139K for 15 min and then boiled for 5 min in sample buffer (Bio-Rad). Eluted material was used for Western blot analysis. For interaction mapping studies, COS-7 cells were transiently cotransfected with 5 μg of plasmids expressing either Flag-tagged or V5-His-tagged protein constructs in 10-cm culture plates. Two days after transfection, nuclear extracts were prepared, immunoprecipitated with anti-Flag M2 agarose beads (Sigma), and processed for Western blot as described previously (Sankaran et al. 2008).
Gel filtration chromatography
Nuclear extracts were prepared from human proerythroblasts ex vivo differentiated from CD34+ cells. Nuclear extract preparations and gel filtration chromatography were performed as described previously (Woo et al. 2008). Fractions were resolved on SDS-PAGE and processed for Western blot analysis.
3C
3C assay was performed as described previously (Palstra et al. 2003; Miele et al. 2006), with some modifications. Briefly, mouse fetal liver cells from 14.5-d post-coitum embryos were harvested and cross-linked with 2% formaldehyde for 10 min at room temperature. Cross-linked cells were lysed for 10 min with ice-cold lysis buffer (10 mM Tris-HCl at pH 8.0, 10 mM NaCl, 0.2% NP-40, 1 mM dithiothreitol). The nuclei were harvested and resuspended in appropriate restriction enzyme buffer containing 0.3% SDS and incubated for 1 h at 37°C with vigorous shaking. Triton X-100 was added to 2% to sequester the SDS, and samples were incubated for another 1 h at 37°C. Samples were digested with EcoRI overnight at 37°C. DNA ligation was performed for 4.5 h at 16°C and 30 min at room temperature. Cross-links were reversed, and DNA was then purified by phenol extraction and ethanol precipitation. All PCR products were amplified, cloned, and verified by DNA sequencing. To compare the cross-linking frequency and ligation efficiency between two restriction fragments present on an unrelated locus, PCR reactions were normalized to Ercc3 locus (Palstra et al. 2003). To correct for the PCR amplification efficiency of different primer sets, a BAC clone spanning the complete human β-globin locus (CTD-3055E11) and a BAC clone containing the mouse Ercc3 locus (RP24-97P16) were used as control templates. Equimolar amounts of human β-globin BAC and the mouse Ercc3 BAC were mixed, digested, and ligated. Quantitation of the data was performed by quantitative real-time PCR using the iQ SYBR Green Supermix (Bio-Rad).
ChIP and ChIP–chip
ChIP was performed as described previously with slight modifications (Yu et al. 2009). Briefly, 0.5 × 107 to 1 × 107 cells per immunoprecipitation were cross-linked with 1% formaldehyde for 10 min at room temperature. Chromatin was sonicated in RIPA buffer with 0.3 M NaCl to ∼500 bp. Dynabeads Protein A or G (Invitrogen) was used for collection of chromatin. For GATA1 ChIP, beads were incubated with rabbit anti-rat IgG (H + L) (Jackson Research Laboratories, Inc.) before incubation with the primary antibody. Antibodies were incubated with beads for 3 h before incubating with sonicated chromatin overnight. ChIP DNA was quantified by real-time PCR using the iQ SYBR Green Supermix (Bio-Rad). ChIP–chip experiments were performed using Affymetrix human ENCODE 2.0R array comprised of 30 Mb of DNA, or ∼1% of the human genome (http://www.nhgri.nih.gov/10005107) as described previously (Lupien et al. 2008). For each ChIP–chip experiment, two to three independent assays were performed. Model-based analysis of tiling array (MAT) (Johnson et al. 2006) was used to predict and map significant peaks (P < 1e-3) to the genome based on the human hg17 genome assembly. Genomic distribution of binding sites identified by ChIP–chip was performed using cis-regulatory element annotation system (CEAS) (Ji et al. 2006; Shin et al. 2009). Two binding sites were considered to overlap if they had at least 1 bp in common. The average size of ChIP–chip regions is ∼1 kb, which means that the center of the two binding sites had to be, on average, within 1 kb of each other to be considered overlapping.
Lentiviral RNAi
Lentiviral shRNA constructs in the pLKO vector were obtained from a large collection of shRNAs as described previously (Moffat et al. 2006; Sankaran et al. 2008). shRNA targeting human BCL11A gene contains the sequence CCGGCGCACAGAACACTCATGGATTCTCGAGAATCCATGAGTGTTCTGTGCGTTTTTG, shRNA targeting human SOX6 gene contains the sequence CCGGCCAGTGAACTTCTTGGAGAAACTCGAGTTTCTCCAAGAAGTTCACTGGTTTTT, shRNA targeting mouse Bcl11a gene contains the sequence CCGGGCATAGACGATGGCACTGTTACTCGAGTAACAGTGCCATCGTCTATGCTTTTTG, and shRNA targeting mouse Sox6 gene contains the sequence CCGGCCAGCCCTGTAACTCAAGTTACTCGAGTAACTTGAGTTACAGGGCTGGTTTTTG. The empty vector pLKO.1ps was used to produce control lentiviruses. Lentiviruses were prepared and transduction of cells was carried out as described (Moffat et al. 2006). The cells were washed three times with PBS and seeded in fresh media 24 h post-transduction. Selection with puromycin was initiated at 48 h post-transduction, which generally corresponded to the time when the cells were seeded into differentiation medium. To knock down both BCL11A and SOX6, cells were transduced with lentiviruses containing SOX6 shRNA (in pLKO vector with puromycin-selectable marker) and BCL11A shRNA (in pLKO vector with GFP). Cells were selected with puromycin as described above. GFP-positive cells were FACS-sorted on the day of harvest.
Hemoglobin analysis
Hemolysates were prepared from cells (2 × 107 to 3 × 107) on days 12–15 of the differentiation phase of erythroid culture. Cells were lysed in 100 μL of hemolysate reagent (Helena Laboratories), and refrigerated overnight before centrifuging at 14,000 rpm for 10 min at 4°C to remove cellular debris. The cleared supernatant was used for characterization of hemoglobin production by cellulose acetate hemoglobin electrophoresis or HPLC as described previously (Sankaran et al. 2008).
Acknowledgments
We are grateful to P. Sassone-Corsi for providing α-SOX6 antibodies. We thank members of the Orkin laboratory for technical assistance and comments. We thank M. Lupien, H. He, S. Baily, X.S. Liu, M.H. Brilliant, and N. Hagiwara for helpful advice and discussions. This work was supported by funding from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) and the National Heart, Lung, and Blood Institute (NHLBI) of the National Institutes of Health (NIH) (to S.H.O.). S.H.O. is an Investigator of the Howard Hughes Medical Institute (HHMI). T.F.M. is supported by the Kay Kendall Leukaemia Fund. V.G.S. is supported by a Medical Scientist Training Program Award from the NIH. J.X. is an HHMI fellow of the Helen Hay Whitney Foundation.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1897310.
Supplemental material is available at http://www.genesdev.org.
References
- Bagga R, Michalowski S, Sabnis R, Griffith JD, Emerson BM 2000. HMG I/Y regulates long-range enhancer-dependent transcription on DNA and chromatin by changes in DNA topology. Nucleic Acids Res 28: 2541–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bank A 2006. Regulation of human fetal hemoglobin: New players, new complexities. Blood 107: 435–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender MA, Bulger M, Close J, Groudine M 2000. β-Globin gene switching and DNase I sensitivity of the endogenous β-globin locus in mice do not require the locus control region. Mol Cell 5: 387–393 [DOI] [PubMed] [Google Scholar]
- Cantor AB, Orkin SH 2005. Coregulation of GATA factors by the Friend of GATA (FOG) family of multitype zinc finger proteins. Semin Cell Dev Biol 16: 117–128 [DOI] [PubMed] [Google Scholar]
- Cheng Y, Wu W, Kumar SA, Yu D, Deng W, Tripic T, King DC, Chen KB, Zhang Y, Drautz D, et al. 2009. Erythroid GATA1 function revealed by genome-wide analysis of transcription factor occupancy, histone modifications, and mRNA expression. Genome Res 19: 2172–2184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cismasiu VB, Adamo K, Gecewicz J, Duque J, Lin Q, Avram D 2005. BCL11B functionally associates with the NuRD complex in T lymphocytes to repress targeted promoter. Oncogene 24: 6753–6764 [DOI] [PubMed] [Google Scholar]
- Cohen-Barak O, Erickson DT, Badowski MS, Fuchs DA, Klassen CL, Harris DT, Brilliant MH 2007. Stem cell transplantation demonstrates that Sox6 represses ɛ y globin expression in definitive erythropoiesis of adult mice. Exp Hematol 35: 358–367 [DOI] [PubMed] [Google Scholar]
- Connor F, Cary PD, Read CM, Preston NS, Driscoll PC, Denny P, Crane-Robinson C, Ashworth A 1994. DNA binding and bending properties of the post-meiotically expressed Sry-related protein Sox-5. Nucleic Acids Res 22: 3339–3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew LR, Tang DC, Berg PE, Rodgers GP 2000. The role of trans-acting factors and DNA-bending in the silencing of human β-globin gene expression. Nucleic Acids Res 28: 2823–2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumitriu B, Dy P, Smits P, Lefebvre V 2006. Generation of mice harboring a Sox6 conditional null allele. Genesis 44: 219–224 [DOI] [PubMed] [Google Scholar]
- Dyer MA, Naidoo R, Hayes RJ, Larson CJ, Verdine GL, Baron MH 1996. A DNA-bending protein interacts with an essential upstream regulatory element of the human embryonic β-like globin gene. Mol Cell Biol 16: 829–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer MA, Hayes PJ, Baron MH 1998. The HMG domain protein SSRP1/PREIIBF is involved in activation of the human embryonic β-like globin gene. Mol Cell Biol 18: 2617–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium 2004. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science 306: 636–640 [DOI] [PubMed] [Google Scholar]
- Fang X, Sun J, Xiang P, Yu M, Navas PA, Peterson KR, Stamatoyannopoulos G, Li Q 2005. Synergistic and additive properties of the β-globin locus control region (LCR) revealed by 5′HS3 deletion mutations: Implication for LCR chromatin architecture. Mol Cell Biol 25: 7033–7041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S, Harley VR, Pontiggia A, Goodfellow PN, Lovell-Badge R, Bianchi ME 1992. SRY, like HMG1, recognizes sharp angles in DNA. EMBO J 11: 4497–4506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester WC, Thompson C, Elder JT, Groudine M 1986. A developmentally stable chromatin structure in the human β-globin gene cluster. Proc Natl Acad Sci 83: 1359–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaensler KM, Kitamura M, Kan YW 1993. Germ-line transmission and developmental regulation of a 150-kb yeast artificial chromosome containing the human β-globin locus in transgenic mice. Proc Natl Acad Sci 90: 11381–11385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosschedl R, Giese K, Pagel J 1994. HMG domain proteins: Architectural elements in the assembly of nucleoprotein structures. Trends Genet 10: 94–100 [DOI] [PubMed] [Google Scholar]
- Grosveld F, van Assendelft GB, Greaves DR, Kollias G 1987. Position-independent, high-level expression of the human β-globin gene in transgenic mice. Cell 51: 975–985 [DOI] [PubMed] [Google Scholar]
- Jasinski M, Keller P, Fujiwara Y, Orkin SH, Bessler M 2001. GATA1-Cre mediates Piga gene inactivation in the erythroid/megakaryocytic lineage and leads to circulating red cells with a partial deficiency in glycosyl phosphatidylinositol-linked proteins (paroxysmal nocturnal hemoglobinuria type II cells). Blood 98: 2248–2255 [DOI] [PubMed] [Google Scholar]
- Ji X, Li W, Song J, Wei L, Liu XS 2006. CEAS: Cis-regulatory element annotation system. Nucleic Acids Res 34: W551–W554 doi: 10.1093/nar/gkl322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Li W, Meyer CA, Gottardo R, Carroll JS, Brown M, Liu XS 2006. Model-based analysis of tiling-arrays for ChIP–chip. Proc Natl Acad Sci 103: 12457–12462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Kuhn RM, Baertsch R, Barber GP, Clawson H, Diekhans M, Giardine B, Harte RA, Hinrichs AS, Hsu F, et al. 2008. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res 36: D773–D779 doi: 10.1093/nar/gkm966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre V, Li P, de Crombrugghe B 1998. A new long form of Sox5 (L-Sox5), Sox6 and Sox9 are coexpressed in chondrogenesis and cooperatively activate the type II collagen gene. EMBO J 17: 5718–5733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettre G, Sankaran VG, Bezerra MA, Araujo AS, Uda M, Sanna S, Cao A, Schlessinger D, Costa FF, Hirschhorn JN, et al. 2008. DNA polymorphisms at the BCL11A, HBS1L-MYB, and β-globin loci associate with fetal hemoglobin levels and pain crises in sickle cell disease. Proc Natl Acad Sci 105: 11869–11874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL 2007. The role of chromatin during transcription. Cell 128: 707–719 [DOI] [PubMed] [Google Scholar]
- Lin AC, Roche AE, Wilk J, Svensson EC 2004. The N termini of Friend of GATA (FOG) proteins define a novel transcriptional repression motif and a superfamily of transcriptional repressors. J Biol Chem 279: 55017–55023 [DOI] [PubMed] [Google Scholar]
- Liu H, Ippolito GC, Wall JK, Niu T, Probst L, Lee BS, Pulford K, Banham AH, Stockwin L, Shaffer AL, et al. 2006. Functional studies of BCL11A: Characterization of the conserved BCL11A-XL splice variant and its interaction with BCL6 in nuclear paraspeckles of germinal center B cells. Mol Cancer 5: 18 doi: 10.1186/1476-4598-5-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M 2008. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 132: 958–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel S, Garner C, Gut I, Matsuda F, Yamaguchi M, Heath S, Foglio M, Zelenika D, Boland A, Rooks H, et al. 2007. A QTL influencing F cell production maps to a gene encoding a zinc-finger protein on chromosome 2p15. Nat Genet 39: 1197–1199 [DOI] [PubMed] [Google Scholar]
- Miele A, Gheldof N, Tabuchi TM, Dostie J, Dekker J 2006. Mapping chromatin interactions by chromosome conformation capture. Curr Protoc Mol Biol 74: 21.11.1–21.11.20 doi: 10.1002/0471142727.mb2111s74 [DOI] [PubMed] [Google Scholar]
- Migliaccio G, Di Pietro R, di Giacomo V, Di Baldassarre A, Migliaccio AR, Maccioni L, Galanello R, Papayannopoulou T 2002. In vitro mass production of human erythroid cells from the blood of normal donors and of thalassemic patients. Blood Cells Mol Dis 28: 169–180 [DOI] [PubMed] [Google Scholar]
- Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, et al. 2006. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124: 1283–1298 [DOI] [PubMed] [Google Scholar]
- Murakami A, Ishida S, Thurlow J, Revest JM, Dickson C 2001. SOX6 binds CtBP2 to repress transcription from the Fgf-3 promoter. Nucleic Acids Res 29: 3347–3355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navas PA, Peterson KR, Li Q, Skarpidi E, Rohde A, Shaw SE, Clegg CH, Asano H, Stamatoyannopoulos G 1998. Developmental specificity of the interaction between the locus control region and embryonic or fetal globin genes in transgenic mice with an HS3 core deletion. Mol Cell Biol 18: 4188–4196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH 1990. Globin gene regulation and switching: Circa 1990. Cell 63: 665–672 [DOI] [PubMed] [Google Scholar]
- Palstra RJ, Tolhuis B, Splinter E, Nijmeijer R, Grosveld F, de Laat W 2003. The β-globin nuclear compartment in development and erythroid differentiation. Nat Genet 35: 190–194 [DOI] [PubMed] [Google Scholar]
- Palstra RJ, de Laat W, Grosveld F 2008. β-Globin regulation and long-range interactions. Adv Genet 61: 107–142 [DOI] [PubMed] [Google Scholar]
- Peterson KR, Clegg CH, Huxley C, Josephson BM, Haugen HS, Furukawa T, Stamatoyannopoulos G 1993. Transgenic mice containing a 248-kilobase human β locus yeast artificial chromosome display proper developmental control of human globin genes. Trans Assoc Am Physicians 106: 101–109 [PubMed] [Google Scholar]
- Peterson KR, Navas PA, Li Q, Stamatoyannopoulos G 1998. LCR-dependent gene expression in β-globin YAC transgenics: Detailed structural studies validate functional analysis even in the presence of fragmented YACs. Hum Mol Genet 7: 2079–2088 [DOI] [PubMed] [Google Scholar]
- Porcu S, Kitamura M, Witkowska E, Zhang Z, Mutero A, Lin C, Chang J, Gaensler KM 1997. The human β globin locus introduced by YAC transfer exhibits a specific and reproducible pattern of developmental regulation in transgenic mice. Blood 90: 4602–4609 [PubMed] [Google Scholar]
- Ptashne M 1986. Gene regulation by proteins acting nearby and at a distance. Nature 322: 697–701 [DOI] [PubMed] [Google Scholar]
- Sankaran VG, Menne TF, Xu J, Akie TE, Lettre G, Van Handel B, Mikkola HK, Hirschhorn JN, Cantor AB, Orkin SH 2008. Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science 322: 1839–1842 [DOI] [PubMed] [Google Scholar]
- Sankaran VG, Xu J, Ragoczy T, Ippolito GC, Walkley CR, Maika SD, Fujiwara Y, Ito M, Groudine M, Bender MA, et al. 2009. Developmental and species-divergent globin switching are driven by BCL11A. Nature 460: 1093–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaran VG, Xu J, Orkin SH 2010. Advances in the understanding of haemoglobin switching. Br J Haematol 10.1111/j.1365-2141.2010.08105.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepers GE, Teasdale RD, Koopman P 2002. Twenty pairs of sox: Extent, homology, and nomenclature of the mouse and human sox transcription factor gene families. Dev Cell 3: 167–170 [DOI] [PubMed] [Google Scholar]
- Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, et al. 2010. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet 42: 53–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedgewick AE, Timofeev N, Sebastiani P, So JC, Ma ES, Chan LC, Fucharoen G, Fucharoen S, Barbosa CG, Vardarajan BN, et al. 2008. BCL11A is a major HbF quantitative trait locus in three different populations with β-hemoglobinopathies. Blood Cells Mol Dis 41: 255–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H, Liu T, Manrai AK, Liu XS 2009. CEAS: Cis-regulatory element annotation system. Bioinformatics 25: 2605–2606 [DOI] [PubMed] [Google Scholar]
- Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W 2006. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat Genet 38: 1348–1354 [DOI] [PubMed] [Google Scholar]
- So CC, Song YQ, Tsang ST, Tang LF, Chan AY, Ma ES, Chan LC 2008. The HBS1L-MYB intergenic region on chromosome 6q23 is a quantitative trait locus controlling fetal haemoglobin level in carriers of β-thalassaemia. J Med Genet 45: 745–751 [DOI] [PubMed] [Google Scholar]
- Sripichai O, Kiefer CM, Bhanu NV, Tanno T, Noh SJ, Goh SH, Russell JE, Rognerud CL, Ou CN, Oneal PA, et al. 2009. Cytokine-mediated increases in fetal hemoglobin are associated with globin gene histone modification and transcription factor reprogramming. Blood 114: 2299–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatoyannopoulos G 2005. Control of globin gene expression during development and erythroid differentiation. Exp Hematol 33: 259–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strouboulis J, Dillon N, Grosveld F 1992. Developmental regulation of a complete 70-kb human β-globin locus in transgenic mice. Genes & Dev 6: 1857–1864 [DOI] [PubMed] [Google Scholar]
- Thein SL, Menzel S 2009. Discovering the genetics underlying foetal haemoglobin production in adults. Br J Haematol 145: 455–467 [DOI] [PubMed] [Google Scholar]
- Thein SL, Menzel S, Peng X, Best S, Jiang J, Close J, Silver N, Gerovasilli A, Ping C, Yamaguchi M, et al. 2007. Intergenic variants of HBS1L-MYB are responsible for a major quantitative trait locus on chromosome 6q23 influencing fetal hemoglobin levels in adults. Proc Natl Acad Sci 104: 11346–11351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thein SL, Menzel S, Lathrop M, Garner C 2009. Control of fetal hemoglobin: New insights emerging from genomics and clinical implications. Hum Mol Genet 18: R216–R223 doi: 10.1093/hmg/ddp401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JO 2001. HMG1 and 2: Architectural DNA-binding proteins. Biochem Soc Trans 29: 395–401 [DOI] [PubMed] [Google Scholar]
- Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W 2002. Looping and interaction between hypersensitive sites in the active β-globin locus. Mol Cell 10: 1453–1465 [DOI] [PubMed] [Google Scholar]
- Tuan D, Solomon W, Li Q, London IM 1985. The “β-like-globin” gene domain in human erythroid cells. Proc Natl Acad Sci 82: 6384–6388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uda M, Galanello R, Sanna S, Lettre G, Sankaran VG, Chen W, Usala G, Busonero F, Maschio A, Albai G, et al. 2008. Genome-wide association study shows BCL11A associated with persistent fetal hemoglobin and amelioration of the phenotype of β-thalassemia. Proc Natl Acad Sci 105: 1620–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakoc CR, Letting DL, Gheldof N, Sawado T, Bender MA, Groudine M, Weiss MJ, Dekker J, Blobel GA 2005. Proximity among distant regulatory elements at the β-globin locus requires GATA-1 and FOG-1. Mol Cell 17: 453–462 [DOI] [PubMed] [Google Scholar]
- Wegner M 1999. From head to toes: The multiple facets of Sox proteins. Nucleic Acids Res 27: 1409–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo AJ, Moran TB, Schindler YL, Choe SK, Langer NB, Sullivan MR, Fujiwara Y, Paw BH, Cantor AB 2008. Identification of ZBP-89 as a novel GATA-1-associated transcription factor involved in megakaryocytic and erythroid development. Mol Cell Biol 28: 2675–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Watts JA, Pope SD, Gadue P, Kamps M, Plath K, Zaret KS, Smale ST 2009. Transcriptional competence and the active marking of tissue-specific enhancers by defined transcription factors in embryonic and induced pluripotent stem cells. Genes & Dev 23: 2824–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi Z, Cohen-Barak O, Hagiwara N, Kingsley PD, Fuchs DA, Erickson DT, Epner EM, Palis J, Brilliant MH 2006. Sox6 directly silences ɛ globin expression in definitive erythropoiesis. PLoS Genet 2: e14 doi: 10.1371/journal.pgen.0020014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Riva L, Xie H, Schindler Y, Moran TB, Cheng Y, Yu D, Hardison R, Weiss MJ, Orkin SH, et al. 2009. Insights into GATA-1-mediated gene activation versus repression via genome-wide chromatin occupancy analysis. Mol Cell 36: 682–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Socolovsky M, Gross AW, Lodish HF 2003. Role of Ras signaling in erythroid differentiation of mouse fetal liver cells: Functional analysis by a flow cytometry-based novel culture system. Blood 102: 3938–3946 [DOI] [PubMed] [Google Scholar]