Abstract
Group II introns are self-splicing RNAs found in eubacteria, archaea, and eukaryotic organelles. They are mechanistically similar to the metazoan nuclear spliceosomal introns; therefore, group II introns have been invoked as the progenitors of the eukaryotic pre-mRNA introns. However, the ability of group II introns to function outside of the bacteria-derived organelles is debatable, since they are not found in the nuclear genomes of eukaryotes. Here, we show that the Lactococcus lactis Ll.LtrB group II intron splices accurately and efficiently from different pre-mRNAs in a eukaryote, Saccharomyces cerevisiae. However, a pre-mRNA harboring a group II intron is spliced predominantly in the cytoplasm and is subject to nonsense-mediated mRNA decay (NMD), and the mature mRNA from which the group II intron is spliced is poorly translated. In contrast, a pre-mRNA bearing the Tetrahymena group I intron or the yeast spliceosomal ACT1 intron at the same location is not subject to NMD, and the mature mRNA is translated efficiently. Thus, a group II intron can splice from a nuclear transcript, but RNA instability and translation defects would have favored intron loss or evolution into protein-dependent spliceosomal introns, consistent with the bacterial group II intron ancestry hypothesis.
Keywords: Group II intron, yeast, ribozyme, RNA splicing, translation
Group II introns are self-splicing, mobile ribozymes that naturally inhabit diverse genes in the genomes of bacteria and eukaryotic organelles (Belfort et al. 2002; Dai et al. 2003; Pyle and Lambowitz 2006). They often encode a protein that, inter alia, helps fold the group II intron into its characteristic secondary and tertiary structure, required for its catalysis. These autocatalytic group II introns are mechanistically similar to the eukaryotic nuclear pre-mRNA introns, which require a dynamic spliceosomal complex consisting of five indispensable small nuclear RNAs (snRNAs) to catalyze splicing (Grabowski et al. 1985; Moore and Sharp 1993; Padgett et al. 1994; Sontheimer et al. 1999; Patel and Steitz 2003). Additionally, these snRNAs have structural and functional similarities to certain domains of group II introns, and boundary sequences between the group II and spliceosomal introns are similar (Cech 1986; Hetzer et al. 1997; Shukla and Padgett 2002; Toor et al. 2008; Keating et al. 2010).
Because of the parallels between bacterial group II and eukaryotic spliceosomal introns, the catalytic group II introns have been proposed to be the progenitors of the eukaryotic spliceosomal introns (Cech 1986; Cavalier-Smith 1991; Sharp 1991; Lynch and Kewalramani 2003; Martin and Koonin 2006; Roy and Gilbert 2006). It is widely speculated that group II introns entered the eukaryotic lineage with the mitochondrial endosymbiosis, invaded the nucleus, and evolved into more efficient spliceosome-dependent introns. However, unlike spliceosomal introns, group II introns are not found in the protein-coding genes of nuclear genomes. Also, there is no evidence for the functioning of group II introns outside of the bacteria-derived mitochondria and chloroplasts. It is therefore not known whether a group II intron can splice from a nuclear, RNA polymerase II (Pol II)-transcribed gene, and, if it does, what the destiny of the transcript might be.
Here, we probe the fate of bacterial group II intron-containing transcripts in Saccharomyces cerevisiae, a genetically tractable eukaryote that has many features of the pre-mRNA splicing and RNA surveillance machineries conserved from the ancestral eukaryotes (Lynch and Richardson 2002; Atkinson et al. 2008). We show that the Lactococcus lactis Ll.LtrB group II intron splices accurately and efficiently from a nuclear gene in yeast. However, the gene harboring the group II intron is poorly expressed because its RNA is subject to nonsense-mediated decay (NMD) and translational repression. In striking contrast, the gene bearing the Tetrahymena group I intron or the yeast spliceosomal ACT1 intron in the identical position is not subject to NMD or translational repression. The absence of group II introns from nuclear genes may therefore result from host restriction exerted on gene expression. Together, our findings provide the basis for understanding how and why group II introns might have evolved into spliceosomal introns.
Results
A nuclear gene bearing a group II intron is poorly expressed
To study the expression of group II intron-bearing protein-coding nuclear genes in yeast, we developed a CUP1-based intron-splicing reporter (GpII-CUP1), constitutively expressed from a 2-μ plasmid. The GpII-CUP1 fusion has the Ll.LtrB intron within its natural homing site fused to the 5′ end of the coding sequence of CUP1, which encodes a copper metallothionein that confers copper resistance (CuR). The CUP1 gene is read in-frame only upon precise Ll.LtrB splicing, leading to Cup1p expression (Fig. 1A). Splicing of the Ll.LtrB intron in bacteria requires its encoded protein, LtrA. Therefore, the ltrA ORF was expressed in trans from the GAL1 promoter on a separate plasmid. LtrA was tagged with the HA epitope and was expressed with or without the SV-40 T-antigen nuclear localization signal (NLS). Indirect immunofluorescence microscopy was used to show that NLS-tagged LtrA concentrates in the nucleus (Supplemental Fig. S1).
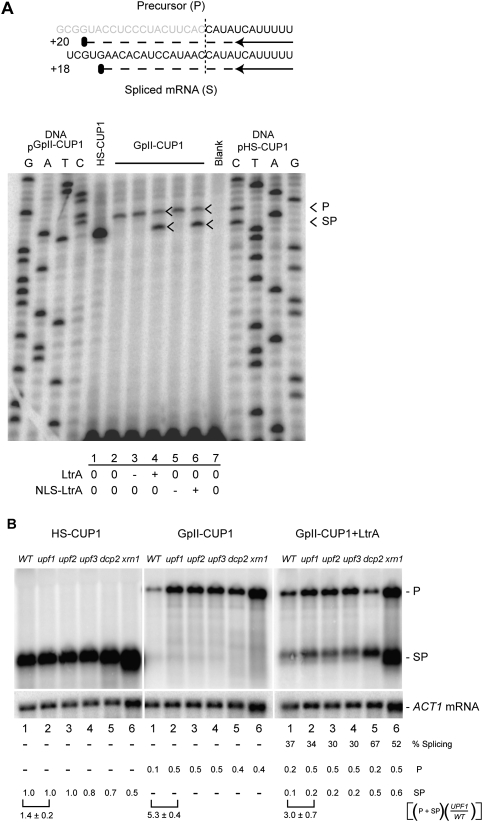
Figure 1.
A group II intron-bearing nuclear gene is poorly expressed. (A) Schematic representation of the group II intron splicing reporter GpII-CUP1. (Bent arrow) Glyceraldehyde-3-phosphate dehydrogenase gene promoter (GPD); (inverted triangle) cytochrome-c gene terminator (CYC1); (E1 and E2) exon 1 and exon 2 of the Ll.LtrB homing site (Cousineau et al. 1998); (red line) ΔORF variant of the Ll.LtrB intron; (blue rectangle) CUP1 ORF; (c-Myc) c-Myc epitope. The presence of the Ll.LtrB intron renders the fusion out-of-frame (CuS), and Ll.LtrB splicing results in an in-frame fusion, HS-CUP1, which in theory confers increased copper resistance (CuR) in S. cerevisiae. (B) Growth of strains carrying pHS-CUP1 or pGpII-CUP1 on medium containing different concentrations of CuSO4. The expression plasmid was with or without LtrA or NLS-LtrA. Yeast cultures at OD600 ≈ 1.0 were applied in 5-μL spots. “Vector” indicates a plasmid without CUP1.
Expression of the constitutively transcribed GpII-CUP1 RNA in the presence and absence of LtrA was determined by measuring the CuR phenotype on selective plates containing increasing concentrations of Cu2+ (Fig. 1B). Although expression of an intron-less homing site-CUP1 fusion (HS-CUP1) conferred growth on agar with up to 0.5 mM Cu2+, no detectable CuR (resistance >0.01 mM Cu2+) was conferred by GpII-CUP1 expression, even when GpII-CUP1 was coexpressed with LtrA or NLS-LtrA (Fig. 1B; Supplemental Fig. S2). This phenotypic assay thus indicates that the presence of a group II intron imposes a block on expression of the CUP1 gene.
A bacterial group II intron can splice accurately from a nuclear gene
To determine whether the Ll.LtrB intron is spliced from the GpII-CUP1 RNA, primer extension termination assays were performed on GpII-CUP1 RNA expressed in the presence and absence of LtrA. We noted first that the level of GpII-CUP1 transcript was approximately fivefold lower than that of the HS-CUP1 transcript in the absence of LtrA expression, where, as expected, no spliced product was observed (Fig. 2A, cf. lanes 1 and 2,3,5). Second, we noted efficient LtrA-dependent Ll.LtrB splicing (53% ± 14%; spliced product/total RNA ± standard deviation, from three independent experiments) irrespective of the presence of the NLS fusion (Fig. 2A, lanes 4, 6). Accurate Ll.LtrB splicing was verified by sequencing across the splice junction of the PCR-amplified reverse transcript of the spliced product (Supplemental Fig. S3). Given the approximately fivefold lower level of GpII-CUP1 RNA relative to HS-CUP1 RNA, and the ∼50% efficiency of Ll.LtrB splicing, only a sevenfold to eightfold reduction in CuR was anticipated; however, a >50-fold reduction was observed (Fig. 1B; data not shown). These findings raise the possibilities that the precursor GpII-CUP1 RNA is unstable and/or that the spliced RNA is not translated efficiently.
Figure 2.
A group II intron can splice from eukaryotic Pol II transcripts, but is subject to NMD. (A) LtrA-dependent Ll.LtrB splicing. Schematic of the predicted primer extension termination products and their respective sizes is shown. (Gray) Intron; (black) exon; (vertical dashed line) intron–exon boundary or splice junction; (black arrow) primer; (horizontal dashed line) primer extension product; (black oval) extension termination. A primer extension termination assay with RNA from pHS-CUP1 or pGpII-CUP1, in the absence (0) or presence ([+] induced; [−] repressed) of LtrA- or NLS-LtrA-expressing plasmid. “Blank” indicates the absence of DNA or RNA template. Lanes labeled G, A, T, and C are sequencing ladders with pGpII-CUP1 and pHS-CUP1 DNA. Expression and localization of LtrA were confirmed by Western blotting (Fig. 3) and indirect immunofluorescence assay (Supplemental Fig. S1), respectively. (B) Northern blot analysis of HS-CUP1 and GpII-CUP1 RNA. RNA expressed in the presence or absence of LtrA was analyzed in the wild-type (WT) and mutant strains, with ACT1 mRNA as a loading control. The level of CUP1 RNA was normalized to ACT1 mRNA levels in each strain and quantitated relative to the HS-CUP1 level in the wild type. (P) GpII-CUP1 precursor RNA; (SP) HS-CUP1 mRNA or the spliced product; (% splicing) percentage of ligated exons relative to the total CUP1 RNA; [(P + SP)(upf1/WT)] the level of total CUP1 RNA in the upf1 mutant relative to that in the wild-type host is represented by a square bracket with standard deviation from at least two independent experiments. Calculations were made from GpII-CUP1 precursor RNA and HS-CUP1 mRNA or spliced product numbers to two decimal points from the independent experiments, leading to some apparent lack of correspondence in the ratios shown.
Unspliced group II intron transcripts are subject to NMD
Reduced expression of intron-bearing transcripts in eukaryotes can arise from NMD, which eliminates transcripts with premature translation termination codons (PTCs) (Muhlrad and Parker 1999b; Sayani et al. 2008). Group II introns, like many pre-mRNA introns, are rich in PTCs and are therefore potential substrates for NMD. To determine whether a group II intron-containing transcript is a substrate of NMD, the level of GpII-CUP1 RNA was measured in the NMD-null mutants upf1, upf2, and upf3 (Fig. 2B). Northern blot analysis revealed a threefold to fivefold increase in the GpII-CUP1 RNA in all of the NMD-null mutants, but little change in the level of HS-CUP1 RNA, suggesting that the group II intron-bearing transcript is a substrate of NMD (Fig. 2B, left and middle panels, cf. lanes 1 and 2–4). We also investigated the contribution of two 5′–3′ decay factors that act downstream from Upf1–3 in the NMD pathway: the decapping enzyme Dcp2p, and the exonuclease Xrn1p (Hilleren and Parker 1999). The GpII-CUP1 transcript level was again increased by approximately fourfold in dcp2 and xrn1 mutants. These results corroborate the reduction of GpII-CUP1 RNA relative to HS-CUP1 transcript observed by primer extension analysis (Fig. 2A), and ascribe the decrease to NMD (Fig. 2B).
In the presence of LtrA, GpII-CUP1 splicing efficiency, defined as the percentage of the spliced product to total GpII-CUP1 RNA, was 30%–40% in the wild-type strain and upf mutants (Fig. 2B, right, lanes 1–4). The level of spliced product was elevated to 67% in the dcp2 mutant (Fig. 2B, lane 5) and to 52% in the xrn1 mutant (Fig. 2B, lane 6), likely reflecting the role of Dcp2p and Xrn1p in turnover of the spliced product. Regardless of the level of splicing, the observation that most of the GpII-CUP1 RNA is subject to NMD, which occurs exclusively in the cytoplasm (Kuperwasser et al. 2004), indicates that the majority of the GpII-CUP1 RNA is transported to the cytoplasm without being spliced. This result corroborates the finding that splicing levels were similar regardless of whether or not LtrA has an NLS and can enter the nucleus (Fig. 2A).
Spliced group II intron transcripts are poorly translated
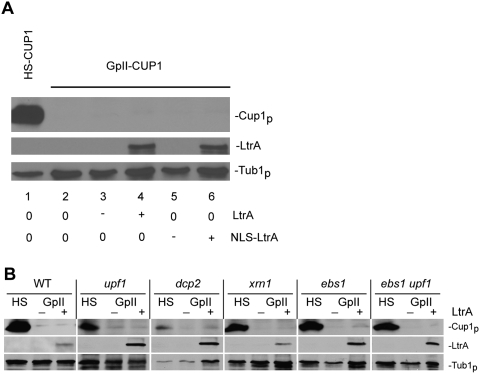
The aforementioned discrepancy between the CuR phenotype (Fig. 1B) and the levels of spliced GpII-CUP1 RNA (Fig. 2) suggested that the group II intron might also interfere with translation of the spliced message. To test whether the spliced GpII-CUP1 RNA was translationally repressed, we measured Cup1p expression by Western blotting with anti-c-Myc antibody (Fig. 3A). Indeed, Cup1p resulting from Ll.LtrB splicing from GpII-CUP1 RNA was undetectable, irrespective of whether LtrA or NLS-LtrA mediated splicing. In contrast, high levels of Cup1p were detected in the strain expressing HS-CUP1 (Fig. 3A).
Figure 3.
Spliced group II intron transcripts have an NMD-independent translational defect. (A) Western blot analysis of Cup1p made from pHS-CUP1 or pGpII-CUP1 in the wild-type strain. Expression was in the absence (0) or presence ([+] induced; [−] repressed) of LtrA or NLS-LtrA. LtrA and Tub1p expression was also determined. (B) Cup1p expression from pHS-CUP1 (HS) and pGpII-CUP1 (GpII) in the NMD mutant hosts. LtrA and Tub1p levels were determined in the presence (+) or absence (−) of LtrA in the wild-type (WT) and mutant strains, labeled above each set of three lanes.
We also measured Cup1p in the NMD mutants. Despite stabilization of the GpII-CUP1 RNA in the NMD-null mutants, Cup1p levels were barely detectable or were absent (Fig. 3B). Additionally, the translation defect persisted in the dcp2 or xrn1 mutant, despite the plethora of spliced GpII-CUP1 mRNA in these mutants. Furthermore, we asked whether Ebs1p, a translation repressor that is down-regulated by the NMD pathway (Ford et al. 2006), affects translation of the spliced GpII-CUP1 transcripts. Again, neither the ebs1 nor the ebs1 upf1 mutations suppressed the translation defect of the spliced GpII-CUP1 RNA (Fig. 3B). These results suggest that there is a discontinuity between spliced GpII-CUP1 mRNA levels and translation of the mRNA. Moreover, the defect in translation of spliced GpII-CUP1 appears to be NMD-independent.
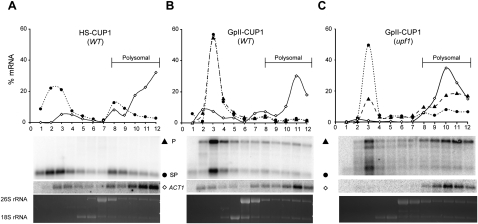
To establish the translational state of the GpII-CUP1 RNA, polysome analysis was performed with ACT1 mRNA as a control (Fig. 4). In the wild-type strain, HS-CUP1 mRNA was found in both the nonpolysomal (65%) and polysomal (35%) fractions (Fig. 4A). Intron-less genes in yeast and mammals are shown to be translated less efficiently than their intron-containing counterparts (Noé et al. 2003; Juneau et al. 2006), a likely explanation for the small fraction of HS-CUP1 relative to ACT1 mRNA in the polysomal fraction. In contrast to ACT1 and HS-CUP1 mRNA, in the presence of LtrA, both spliced and unspliced GpII-CUP1 RNA was found predominantly in the nonpolysomal fractions (83%) (Fig. 4B). Thus, neither spliced nor unspliced GpII-CUP1 RNA is translated as efficiently as HS-CUP1.
Figure 4.
Spliced group II intron transcripts are poorly translated. Polysome analysis of HS-CUP1 (A) and GpII-CUP1 RNA in the presence of LtrA in the wild type (B) and the upf1 mutant (C). ACT1 RNA served as a control for polysome accumulation of translatable RNA. Polysomes were analyzed by ethidium bromide staining of formaldehyde agarose gels (26S and 18S rRNA) prior to Northern blotting. The percentage of RNA from each fraction relative to the total RNA was calculated. (⋄) ACT1 mRNA; (●) HS-CUP1 mRNA or the group II intron spliced product (SP); (▲) GpII-CUP1 precursor RNA (P). The strain names are shown in parentheses.
To measure the contribution of NMD to the translation block, we analyzed polysome profiles in the upf1 mutant (Fig. 4C). Much of the unspliced GpII-CUP1 RNA was shifted to polysomal fractions in the upf1 mutant relative to the wild-type strain, suggesting that the Ll.LtrB-containing transcripts are relieved from sequestration by the NMD machinery, and thereby are available for translation in the upf1 mutant (Fig. 4C). In contrast, the spliced GpII-CUP1 RNA remained predominantly in the untranslated pool in the upf1 mutant (Fig. 4C). The paucity of spliced GpII-CUP1 RNA in polysomal fractions in the wild-type strain and the upf1 mutant indicates that the spliced group II intron transcripts are translationally repressed by an NMD-independent mechanism.
Exon context effects on host surveillance
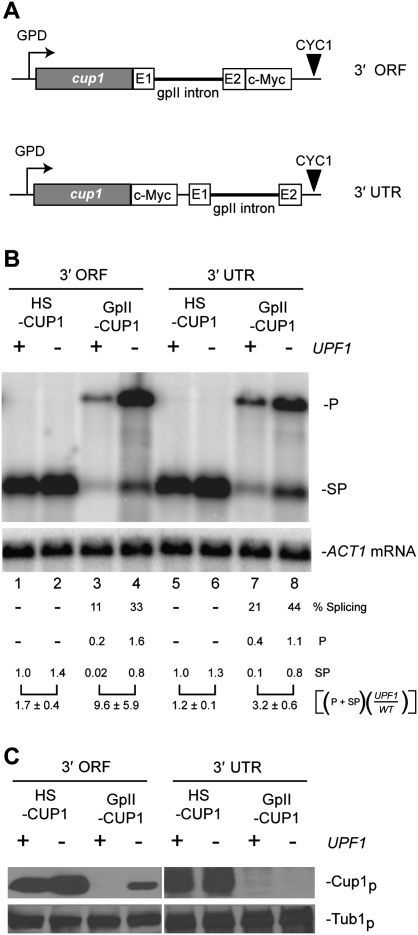
To determine whether the location of the group II intron within the transcript affects splicing, NMD, or translational repression, we measured GpII-CUP1 expression in constructs with the Ll.LtrB intron interrupting either the 3′ end of the ORF or the 3′ untranslated region (UTR) of the CUP1 mRNA (Fig. 5A), rather than the 5′ end of the ORF (Fig. 1A). Northern blot analysis revealed that the Ll.LtrB intron in the 3′ end of the ORF and in the 3′ UTR of the CUP1 mRNA spliced in the presence of LtrA (Fig. 5B). Furthermore, the presence of the group II intron subjected the transcripts to NMD, consistent with our observations of the intron in the 5′ end of the ORF (Figs. 2B, 5B). However, the efficiency of group II intron splicing and the severity of NMD varied depending on the position of the intron within the mRNA (Fig. 5B). The intron in the 3′ end of the ORF spliced relatively poorly (11%) and induced a more severe NMD effect (∼10-fold reduction in RNA) than the intron in the 5′ end of the ORF or in the 3′ UTR (threefold to fourfold reductions in RNA).
Figure 5.
Group II intron transcripts are poorly expressed regardless of exon context. (A) Schematic representation of the group II intron in the homing site (E1 and E2) fused to the 3′ end of the ORF or in the 3′ UTR of the CUP1 RNA (cf. 5′ ORF fusion) (Fig. 1A). (B) Northern blot analysis of RNA from pHS-CUP1 and pGpII-CUP1 in the 3′ ORF and 3′ UTR contexts. RNA was analyzed in the wild type (+) and upf1 mutant (−) with ACT1 RNA as a loading control. The level of CUP1 RNA was normalized to ACT1 mRNA levels in each strain and quantitated relative to the HS-CUP1 level in the wild-type strain. (P) GpII-CUP1 precursor RNA; (SP) HS-CUP1 mRNA or the spliced product; (% splicing) percentage of ligated exons relative to the total CUP1 RNA; [(P + SP)(upf1/WT)] the level of total CUP1 RNA in the upf1 mutant relative to that in the wild-type host is shown with standard deviation from at least two independent experiments, as in Figure 2B. (C) Western blot analysis. Cup1p expression from pHS-CUP1 and pGpII-CUP1 in the 3′ ORF and 3′ UTR contexts was determined in the wild type and upf1 mutant with Tub1p as the loading control. Cup1p expression in the upf1 mutant was verified by measuring the CuR phenotype (data not shown).
Western blot analysis once again revealed that Cup1p levels from the spliced GpII-CUP1 RNA were significantly lower than from the corresponding HS-CUP1 transcripts (Fig. 5C). Cup1p was not detected in wild-type cells, even when the Ll.LtrB intron was in the 3′ end of the ORF or in the 3′ UTR, indicating that the group II intron induces expression defects irrespective of position within the mRNA. However, with the Ll.LtrB intron in the 3′ end of the ORF, Cup1p was expressed in the upf1 mutant (Fig. 5C), unlike in the other exon contexts, suggesting that the translation defect can also result from NMD. Furthermore, a different reporter RNA that harbored a group II intron was also subject to NMD, as described below.
Group II intron splicing is predominantly cytoplasmic
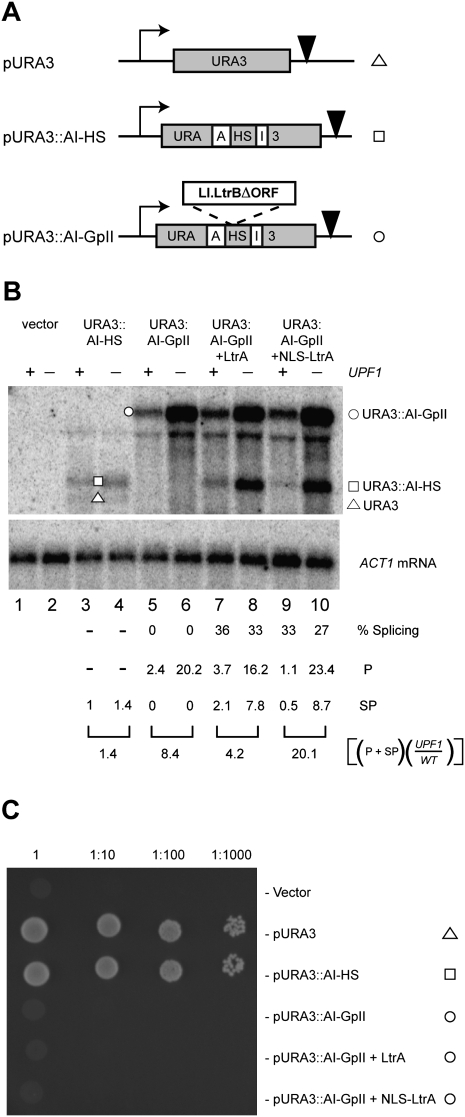
We used a different reporter system to verify that transcripts bearing group II introns are spliced predominantly in the cytoplasm and are therefore subject to NMD. In this URA3-based recursive splicing reporter (URA3∷AI-GpII), the URA3 gene is interrupted by an artificial spliceosomal intron (AI) (Yoshimatsu and Nagawa 1989), which is in turn interrupted by the Ll.LtrB intron (Fig. 6A). Spliceosomal introns splice only in the nucleus; therefore, URA3 will be expressed only if Ll.LtrB is spliced from URA3∷AI-GpII RNA in the nucleus.
Figure 6.
Group II intron expression from the URA3-based splicing reporter. (A) Schematic of the URA3-based splicing reporter. The wild-type URA3 (pURA3) and the URA3 with an artificial spliceosomal intron (AI) carrying the homing site of Ll.LtrB (pURA3∷AI-HS) are shown, along with the group II intron splicing reporter (pURA3∷AI-GpII), which has the Ll.LtrB ΔORF intron within its homing site in AI. (Bent black arrow) URA3 promoter; (inverted triangle) URA3 terminator. (B) Steady-state levels of URA3∷AI-HS and URA3∷AI-GpII RNA were measured by Northern blotting of total RNA from wild-type (+) and upf1 mutant (−) strains. (ACT1 mRNA) Loading control; (Vector) a plasmid without URA3. The square brackets represent numbers from one experiment, although duplicate experiments showed the same trend. (C) URA3 phenotypes. The ura3Δ0 strain carrying the vector, pURA3, pURA3∷AI-HS, or pURA3∷AI-GpII in the absence and presence of LtrA was assayed for growth on synthetic media lacking uracil. The undiluted yeast cultures at OD600 ≈ 1.0 and 10-fold serial dilutions were applied in 5-μL spots.
When URA3∷AI-GpII was transcribed from its native promoter on a CEN plasmid, Ll.LtrB was spliced, verifying the functionality of a group II intron within a different pre-mRNA (Fig. 6B, lanes 7–10, URA3∷AI-HS band). Despite Ll.LtrB splicing from URA3∷AI-GpII, the AI intron did not splice, resulting in an Ura− phenotype (Fig. 6C). The absence of AI splicing is likely due to cytoplasmic export of the URA3∷AI-GpII RNA prior to Ll.LtrB splicing, leading to the inability of the spliceosome to access the AI intron. The lack of URA3 expression from the URA3∷AI-GpII reporter underscores the inefficiency of Ll.LtrB group II intron splicing in the nucleus. Furthermore, the group II intron-containing URA3∷AI-GpII RNA accumulates to approximately eightfold higher levels in the upf1 mutant than in the wild-type strain (Fig. 6B, cf. lanes 5 and 6). Together, these experiments confirm that transcripts bearing the group II intron are subject to NMD, and argue in favor of cytoplasmic splicing of the group II intron.
Induction of NMD and translational repression is group II intron-specific
Besides spliceosomal pre-mRNA introns, group I introns are the only other class of introns that are known to function in eukaryotic Pol II transcripts (Haugen et al. 2005). The group I introns and the spliceosomal introns, like the group II introns, contain numerous PTCs and are potential substrates for cellular surveillance. Therefore, we introduced the self-splicing Tetrahymena pre-rRNA group I intron (tGpI) and the spliceosomal ACT1 pre-mRNA intron into the CUP1 reporter to measure splicing, NMD, and translational phenotypes. To facilitate Tetrahymena group I intron splicing, the flanking exons were modified to allow critical intron–exon interactions, P1 and P10 (Hagen and Cech 1999). Likewise, the ACT1 intron was embedded in exons conducive to splicing (Lesser and Guthrie 1993).
Not only did the Tetrahymena group I intron and the ACT1 intron splice efficiently from the CUP1 RNA, as measured by Northern blotting (>85%) (Fig. 7A), the precursor and spliced tGpI-CUP1 transcript levels were equivalent in the wild-type strain and NMD-null mutant (Fig. 7A). Even a construct containing suboptimal group I intron splicing signals (ptGpI*-CUP1) was insensitive to NMD. Additionally, the spliced ACT1-CUP1 and tGpI-CUP1 transcripts were translated efficiently, as measured by Western blotting (Fig. 7B). Furthermore, expression of tGpI-CUP1 and ACT1-CUP1 resulted in a CuR phenotype comparable with that conferred by HS-CUP1 (Fig. 7C).
Figure 7.
Spliceosomal and group I intron-containing transcripts are expressed efficiently in yeast. (A) Northern blot analysis with RNAs from pACT1-CUP1, ptGpI-CUP1, and ptGpI*-CUP1 (splicing-defective). RNA levels were analyzed in the wild type (+) and upf1 mutant (−) by Northern blotting, with ACT1 RNA as a loading control. The level of CUP1 RNA was normalized to ACT1 mRNA levels in each strain and was quantitated relative to the HS-CUP1 level in the wild-type strain. (P) ACT1-CUP1, tGpI-CUP1, or tGpI*-CUP1 precursor RNA; (SP) HS-CUP1 mRNA or the spliced product; (% splicing) percentage of ligated exons relative to the total CUP1 RNA; [(P + SP)(upf1/WT)] the level of total CUP1 RNA in the upf1 mutant relative to that in the wild-type host is shown with standard deviation from at least two independent experiments, as in Figure 2B. (B) Cup1p expression from pACT1-CUP1, ptGpI-CUP1, and ptGpI*-CUP1. Western blotting was used to determine protein levels in the wild type and upf1 mutant, with Tub1p as the loading control. (C) Strains carrying pHS-CUP1, pACT1-CUP1, ptGpI-CUP1, or ptGpI*-CUP1 were assayed for CuR as in Figure 1B.
Using a luciferase reporter system, we verified that the Tetrahymena group I intron spliced efficiently in yeast (>50%) from different positions in the luciferase ORF (Supplemental Fig. S4A,B). Furthermore, the spliced luciferase mRNA was translated as efficiently as the intron-less luciferase mRNA (Supplemental Fig. S4). Thus, in the yeast model system, the group II intron was distinctive among the three intron types in eliciting NMD and in blocking translation of nuclear transcripts.
Discussion
One of the longest abiding mysteries of molecular evolution
Group II introns have long been the focus of opposing theories in eukaryogenesis, including the origin of spliceosomal introns. How spliceosomal introns arose has been the subject of much debate (Sharp 1991; Koonin 2006; Roy and Gilbert 2006) and “one of the longest-abiding mysteries of molecular evolution” (Roy and Gilbert 2006). Moreover, the lack of evidence for the functioning of group II introns outside the endosymbiont-derived organelles in eukaryotes and the total absence of group II introns from the nuclear genomes present a conundrum. This study demonstrates for the first time that a bacterial group II intron residing in a nuclear protein-coding gene can splice in eukaryotes (Figs. 2, 5, 6). As in bacteria, Ll.LtrB splicing in S. cerevisiae is dependent on LtrA, the intron-encoded protein. Our observations pose an obvious question: If group II introns can splice from nuclear genes, what led to the absence of group II introns from nuclear genomes (Dai et al. 2003)? This study shows that accurate and efficient splicing of the L. lactis Ll.LtrB group II intron in S. cerevisiae is followed by roadblocks to gene expression (Figs. 2–6), providing the basis for understanding how and why group II introns might have evolved into spliceosomal introns.
Splicing of a group II intron is predominantly cytoplasmic
Although we did not eliminate the possibility that some group II intron splicing can occur in the nucleus, it appears that, with both the CUP1 and URA3 reporter systems, Ll.LtrB splicing is predominantly cytoplasmic (Fig. 8). Extranuclear processing of group II intron-containing transcripts is suggested by their sensitivity to NMD (Figs. 2, 4–6), a cytoplasmic phenomenon, and by the inability of an NLS fusion to LtrA to improve the efficiency of splicing (Fig. 2). Moreover, a splicing-proficient group II intron within a spliceosomal intron blocked spliceosomal splicing (Fig. 6), consistent with group II intron splicing occurring outside the nucleus.
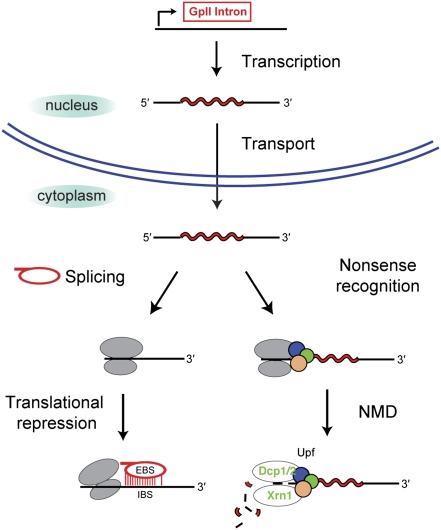
Figure 8.
Model for the fate of a group II intron-bearing nuclear transcript. After transcription and transport to the cytoplasm, the transcript can be silenced by NMD and/or translational repression. The EBS–IBS interaction between the intron lariat and the mRNA is shown for the hypothetical translational repression model.
Confinement of group II intron splicing to the cytoplasm may be one mechanism by which cells block invasion of the nuclear genome by new group II introns. It is hypothesized that the mutational decay of group II introns into spliceosomal introns, which are processed slowly relative to translation of the mRNA, provided the selective pressure for nucleus–cytoplasm compartmentalization (Martin and Koonin 2006). It is worth considering that the nuclear membrane evolved not only to partition ribosomes from nascent transcripts, but also to separate excised group II introns from the host genome.
A group II intron subjects RNA to NMD
We showed that precursor transcripts bearing the Ll.LtrB group II intron are substrates for NMD in yeast (Figs. 2, 5, 6, 8). The unspliced GpII-CUP1 RNA was subject to NMD irrespective of the position of the group II intron within the mRNA (Figs. 2, 5, 6). However, the effect of NMD was much more modest when the intron was in the 5′ coding region of the mRNA than when it was in the 3′ coding region (Figs. 2, 5). Interestingly, spliceosomal introns in S. cerevisiae are found predominantly in the 5′ region of the protein-coding sequences, leading to speculation that position-dependent mRNA surveillance against group II intron-bearing Pol II transcripts might have contributed to the locational bias (Sakurai et al. 2002).
The decay of group II intron-bearing nuclear transcripts by NMD affects gene expression by lowering the amount of RNA available for translation. It was proposed that NMD originated after the origin of spliceosomal introns to ensure translation of only the properly spliced nuclear transcripts (Martin and Koonin 2006). We suggest that the poor gene expression that results from NMD could have led to the loss of group II introns from nuclear genomes or to their evolution into spliceosomal introns, which have been proposed to be stimulated to proliferate by NMD (Lynch and Kewalramani 2003).
A group II intron imposes a translation block
Splicing of the group II intron, unlike splicing of a spliceosomal and a group I intron, gave rise to mature mRNAs that are not readily available for translation (Figs. 3–5, 7; Supplemental Fig. S4). Irrespective of the exon context, the group II intron imposed a translation block on the nuclear gene. While the block is partially NMD-dependent with the group II intron in the 3′ end of the ORF, repression with the intron at the 5′ end of the ORF or in the 3′ UTR is NMD-independent. The paucity of spliced GpII-CUP1 RNA on polysomes in the wild-type strain and upf1 mutant validates the NMD-independent nature of the translational block (Fig. 4). A barrier to translation could result from either premature export of the group II intron transcript into the cytoplasm or RNA sequestration by another host surveillance mechanism, such as confinement to stress granules (Buchan and Parker 2009).
Group II intron exclusivity in restricting gene expression
In contrast to the Ll.LtrB group II intron, the spliceosomal pre-mRNA ACT1 intron and the tGpI intron are not sensitive to NMD (Fig. 7). Although there is no unifying model to explain NMD transcript selection in S. cerevisiae, it has been shown that intron identity and splicing kinetics are key determinants in targeting unspliced intron-containing genes to NMD (Sayani et al. 2008). It remains to be determined whether specific NMD surveillance of group II introns in our model system results from intron type, splicing kinetics, or cytoplasmic compartmentalization of intron splicing. Interestingly, bacteria and eukaryotic organelles are free of such surveillance mechanisms (Gagliardi et al. 2004), likely accounting for the maintenance and proliferation of group II introns in these hosts.
Whereas the group II intron subjects the RNA to an NMD-independent translation block, the spliceosomal ACTI and Tetrahymena group I introns do not inhibit translation (Fig. 7; Supplemental Fig. S4). Curiously, the Tetrahymena group I intron-bearing transcripts were reported in mammalian cells to have a translation defect despite efficient intron splicing (Hagen and Cech 1999). These different translational outcomes suggest host-specific differences in the way messages are presented to the protein synthesis machinery after splicing.
The mechanism of the group II intron-specific translation block is the subject of speculation. One hypothesis is based on the ability of the intron to pair with its exon sequences via 14-base-pair (bp) exon-binding site (EBS)/intron-binding site (IBS) interactions (Fig. 8; Mohr et al. 2000). We postulate that the intron RNA, either by itself or in complex with the intron-encoded protein, interacts with the spliced product, blocking translation in any number of ways, such as forming a complex that stalls ribosomes or is transported to a storage compartment. Although group I introns also have an internal guide sequence that interacts with exon residues, over 12 bp for the Tetrahymena intron, group I introns are spliced and likely retained in the nucleus (Brehm and Cech 1983), and therefore not in the cytoplasm to “mark” the mRNA for repression.
Regardless of mechanism, the barriers imposed by a nuclear group II intron to gene expression are consistent with the proposals that the nucleus–cytoplasm compartmentalization and NMD arose as consequences of group II intron invasion during eukaryotic evolution (Lynch and Kewalramani 2003; Martin and Koonin 2006). The cytoplasmic compartmentalization of group II intron splicing might have restricted the proliferation of group II introns, whereas the poor expression of genes bearing group II introns might have threatened group II intron maintenance in nuclear genomes. Irrespectively, targeting of unspliced group II intron-bearing transcripts by NMD and translational repression of the spliced mRNAs are phenomena consistent with the idea that the nuclear gene encoding a group II intron was under selective pressure to expunge the intron and/or evolve properties that gave rise to spliceosomal introns.
Materials and methods
Strains and plasmids
All yeast strains, plasmids, and oligonucleotides used in this study are listed in Supplemental Tables S1A–C, respectively. Strains yRP1209, yRP1211, yRP1212, yRP1305, yRP1306, and yRP1307 were provided by Roy Parker (Muhlrad and Parker 1999a). The strains yRP1209 ebs1 and yRP1212 ebs1 were constructed by PCR amplification of the ebs1Δ∷KanMX allele from the yeast ORF deletion collection (Winzeler et al. 1999) using oligonucleotides IDT1745 and IDT1746, and transformation of the PCR product into yRP1209 and yRP1212, respectively. Likewise, strain JC3379 upf1 was constructed from the upf1Δ∷KanMX strain (Winzeler et al. 1999) using oligonucleotides PJ527 and PJ528, and transformation into JC3379 was constructed from the BY4741 strain (Brachmann et al. 1998) with a trp1∷hisG mutation made as described elsewhere (Alani et al. 1987). The plasmid construction is described in the Supplemental Material.
RNA and protein extraction
Using the hot phenol-chloroform extraction method (Schmitt et al. 1990), total cellular RNA was extracted from half of a 25-mL yeast culture grown to mid-log phase. The other half of the culture was used for protein extraction and phenotypic analysis, performed as described elsewhere (Lesser and Guthrie 1993).
Primer extension termination assay
Reverse transcription reactions were performed with DNase-treated total cellular RNA and a 3′ exon-specific γ-32P-labeled oligonucleotide, IDT0631, using the MMLV-RT kit (Ambion), and dCTP was exchanged for 2,3-dideoxycytidine triphosphate (ddCTP). Reactions were analyzed on a denaturing 7 M urea–12% polyacrylamide gel.
Northern blotting
Total RNA (10 μg) was resolved on a formaldehyde-agarose gel, blotted onto a Hybond N membrane (Amersham Biosciences), and then hybridized with γ-32P-labeled oligonucleotide probes (CUP1-specific IDT1597, URA3-specific IDT1832, Luciferase-specific IDT1325, or ACT1-specific IDT1384). The membrane was then exposed to a PhosphorImager screen overnight and quantified using ImageQuant software.
Western blotting
Total cellular protein (30 μg) was resolved on a denaturing 15% acrylamide–SDS gel. The protein was transferred onto a 0.4-mm PVDF membrane (Perkin-Elmer) and probed with mouse anti-c-Myc antibody (1:5000 dilution; Sigma), mouse anti-HA antibody (1:200 dilution; Santa Cruz Biotechnologies), or rat anti-α-tubulin antibody (1:500 dilution; Chemicon). Horseradish peroxidase (HRP)-conjugated goat anti-mouse antibody (1:10,000 dilution; Sigma) or donkey anti-rat antibody (1:5000 dilution; Upstate Biotechnologies) was used as a secondary antibody.
Polysome analysis
Yeast strains grown to mid-log phase were harvested into cold polysome lysis buffer (20 mM Tris-HCl at pH 8.0, 140 mM KCl, 5 mM MgCl2, 0.5 mM DTT, 1% Triton X-100, 1 mg/mL heparin). Cell extract was prepared from 15 OD600 units by glass bead lysis in 500 μL of polysome lysis buffer with Complete EDTA-free protease inhibitor cocktail (Roche) and 100 U/mL RNasin (Promega). The extract (50 A260 units) was applied to 5%–45% (w/v) sucrose gradients in polysome lysis buffer without Triton X-100. Sucrose gradients were formed using a BioComp Gradient Station (model 153, BioComp Instruments) set at an angle of 81.5° and a speed of 21 rpm for 1 min 24 sec. The gradients were centrifuged at 39,000 rpm for 2.5 h at 4°C using a SW41 rotor in an Optima L-90K ultracentrifuge (Beckman-Coulter). Twenty-four 450-μL fractions were collected directly into 550 μL of 8 M guanidine-HCl, and RNA was ethanol-precipitated from the even-numbered fractions and resuspended in 15 μL of DEPC-treated water. Finally, 7 μL of the RNA sample from each fraction were used for Northern blotting.
Acknowledgments
We thank Maryellen Carl and John Dansereau for help with manuscript and figure preparation, respectively; Roy Parker, Christine Guthtrie, and Tom Cech for strains and plasmids; Mariano Garcia Blanco, Tao Huang, and Patrick Maxwell for useful discussion; and the Wadsworth Center Molecular Genetics Core for DNA sequencing service. This work was supported by the National Institutes of Health grants GM39422, GM52077, and GM44844. V.R.C., M.J.C., and M.B. designed the experiments; V.R.C. performed the experiments; V.R.C., M.J.C., and M.B. analyzed data; and V.R.C., M.J.C., and M.B. wrote the paper.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1905010.
Supplemental material is available at http://www.genesdev.org.
References
- Alani E, Cao L, Kleckner N 1987. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics 116: 541–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson GC, Baldauf SL, Hauryliuk V 2008. Evolution of nonstop, no-go and nonsense-mediated mRNA decay and their termination factor-derived components. BMC Evol Biol 8: 290 doi: 10.1186/1471-2148-8-290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belfort M, Derbyshire V, Cousineau B, Lambowitz A 2002. Mobile introns: Pathways and proteins. In Mobile DNA II (ed. Craig N et al. ), pp. 761–783 ASM Press, Washington, DC [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14: 115–132 [DOI] [PubMed] [Google Scholar]
- Brehm SL, Cech TR 1983. Fate of an intervening sequence ribonucleic acid: Excision and cyclization of the Tetrahymena ribosomal ribonucleic acid intervening sequence in vivo. Biochemistry 22: 2390–2397 [DOI] [PubMed] [Google Scholar]
- Buchan JR, Parker R 2009. Eukaryotic stress granules: The ins and outs of translation. Mol Cell 36: 932–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T 1991. Intron phylogeny: A new hypothesis. Trends Genet 7: 145–148 [PubMed] [Google Scholar]
- Cech TR 1986. The generality of self-splicing RNA: Relationship to nuclear mRNA splicing. Cell 44: 207–210 [DOI] [PubMed] [Google Scholar]
- Cousineau B, Smith D, Lawrence-Cavanagh S, Mueller JE, Yang J, Mills D, Manias D, Dunny G, Lambowitz AM, Belfort M 1998. Retrohoming of a bacterial group II intron: Mobility via complete reverse splicing, independent of homologous DNA recombination. Cell 94: 451–462 [DOI] [PubMed] [Google Scholar]
- Dai L, Toor N, Olson R, Keeping A, Zimmerly S 2003. Database for mobile group II introns. Nucleic Acids Res 31: 424–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford AS, Guan Q, Neeno-Eckwall E, Culbertson MR 2006. Ebs1p, a negative regulator of gene expression controlled by the Upf proteins in the yeast Saccharomyces cerevisiae. Eukaryot Cell 5: 301–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagliardi D, Stepien PP, Temperley RJ, Lightowlers RN, Chrzanowska-Lightowlers ZM 2004. Messenger RNA stability in mitochondria: Different means to an end. Trends Genet 20: 260–267 [DOI] [PubMed] [Google Scholar]
- Grabowski PJ, Seiler SR, Sharp PA 1985. A multicomponent complex is involved in the splicing of messenger RNA precursors. Cell 42: 345–353 [DOI] [PubMed] [Google Scholar]
- Hagen M, Cech TR 1999. Self-splicing of the Tetrahymena intron from mRNA in mammalian cells. EMBO J 18: 6491–6500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugen P, Simon DM, Bhattacharya D 2005. The natural history of group I introns. Trends Genet 21: 111–119 [DOI] [PubMed] [Google Scholar]
- Hetzer M, Wurzer G, Schweyen RJ, Mueller MW 1997. Trans-activation of group II intron splicing by nuclear U5 snRNA. Nature 386: 417–420 [DOI] [PubMed] [Google Scholar]
- Hilleren P, Parker R 1999. Mechanisms of mRNA surveillance in eukaryotes. Annu Rev Genet 33: 229–260 [DOI] [PubMed] [Google Scholar]
- Juneau K, Miranda M, Hillenmeyer ME, Nislow C, Davis RW 2006. Introns regulate RNA and protein abundance in yeast. Genetics 174: 511–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating KS, Toor N, Perlman PS, Pyle AM 2010. A structural analysis of the group II intron active site and implications for the spliceosome. RNA 16: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV 2006. The origin of introns and their role in eukaryogenesis: A compromise solution to the introns-early versus introns-late debate? Biol Direct 1: 22 doi: 10.1186/1745-6150-1-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuperwasser N, Brogna S, Dower K, Rosbash M 2004. Nonsense-mediated decay does not occur within the yeast nucleus. RNA 10: 1907–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesser CF, Guthrie C 1993. Mutational analysis of pre-mRNA splicing in Saccharomyces cerevisiae using a sensitive new reporter gene, CUP1. Genetics 133: 851–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Kewalramani A 2003. Messenger RNA surveillance and the evolutionary proliferation of introns. Mol Biol Evol 20: 563–571 [DOI] [PubMed] [Google Scholar]
- Lynch M, Richardson AO 2002. The evolution of spliceosomal introns. Curr Opin Genet Dev 12: 701–710 [DOI] [PubMed] [Google Scholar]
- Martin W, Koonin EV 2006. Introns and the origin of nucleus-cytosol compartmentalization. Nature 440: 41–45 [DOI] [PubMed] [Google Scholar]
- Mohr G, Smith D, Belfort M, Lambowitz AM 2000. Rules for DNA target-site recognition by a lactococcal group II intron enable retargeting of the intron to specific DNA sequences. Genes & Dev 14: 559–573 [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Sharp PA 1993. Evidence for two active sites in the spliceosome provided by stereochemistry of pre-mRNA splicing. Nature 365: 364–368 [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Parker R 1999a. Aberrant mRNAs with extended 3′ UTRs are substrates for rapid degradation by mRNA surveillance. RNA 5: 1299–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D, Parker R 1999b. Recognition of yeast mRNAs as “nonsense containing” leads to both inhibition of mRNA translation and mRNA degradation: Implications for the control of mRNA decapping. Mol Biol Cell 10: 3971–3978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noé V, MacKenzie S, Ciudad CJ 2003. An intron is required for dihydrofolate reductase protein stability. J Biol Chem 278: 38292–38300 [DOI] [PubMed] [Google Scholar]
- Padgett RA, Podar M, Boulanger SC, Perlman PS 1994. The stereochemical course of group II intron self-splicing. Science 266: 1685–1688 [DOI] [PubMed] [Google Scholar]
- Patel AA, Steitz JA 2003. Splicing double: Insights from the second spliceosome. Nat Rev Mol Cell Biol 4: 960–970 [DOI] [PubMed] [Google Scholar]
- Pyle AM, Lambowitz AM 2006. Group II introns: Ribozymes that splice DNA and invade RNA. In The RNA world, 3rd ed (ed.Gesteland RF et al. ), pp. 469–505 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Roy SW, Gilbert W 2006. The evolution of spliceosomal introns: Patterns, puzzles and progress. Nat Rev Genet 7: 211–221 [DOI] [PubMed] [Google Scholar]
- Sakurai A, Fujimori S, Kochiwa H, Kitamura-Abe S, Washio T, Saito R, Carninci P, Hayashizaki Y, Tomita M 2002. On biased distribution of introns in various eukaryotes. Gene 300: 89–95 [DOI] [PubMed] [Google Scholar]
- Sayani S, Janis M, Lee CY, Toesca I, Chanfreau GF 2008. Widespread impact of nonsense-mediated mRNA decay on the yeast intronome. Mol Cell 31: 360–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt ME, Brown TA, Trumpower BL 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res 18: 3091–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PA 1991. Five easy pieces. Science 254: 663. [DOI] [PubMed] [Google Scholar]
- Shukla GC, Padgett RA 2002. A catalytically active group II intron domain 5 can function in the U12-dependent spliceosome. Mol Cell 9: 1145–1150 [DOI] [PubMed] [Google Scholar]
- Sontheimer EJ, Gordon PM, Piccirilli JA 1999. Metal ion catalysis during group II intron self-splicing: Parallels with the spliceosome. Genes & Dev 13: 1729–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toor N, Keating KS, Taylor SD, Pyle AM 2008. Crystal structure of a self-spliced group II intron. Science 320: 77–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906 [DOI] [PubMed] [Google Scholar]
- Yoshimatsu T, Nagawa F 1989. Control of gene expression by artificial introns in Saccharomyces cerevisiae. Science 244: 1346–1348 [DOI] [PubMed] [Google Scholar]