Abstract
Chemotherapy resistance is a major obstacle in cancer treatment, yet the mechanisms of response to specific therapies have been largely unexplored in vivo. Employing genetic, genomic, and imaging approaches, we examined the dynamics of response to a mainstay chemotherapeutic, cisplatin, in multiple mouse models of human non-small-cell lung cancer (NSCLC). We show that lung tumors initially respond to cisplatin by sensing DNA damage, undergoing cell cycle arrest, and inducing apoptosis—leading to a significant reduction in tumor burden. Importantly, we demonstrate that this response does not depend on the tumor suppressor p53 or its transcriptional target, p21. Prolonged cisplatin treatment promotes the emergence of resistant tumors with enhanced repair capacity that are cross-resistant to platinum analogs, exhibit advanced histopathology, and possess an increased frequency of genomic alterations. Cisplatin-resistant tumors express elevated levels of multiple DNA damage repair and cell cycle arrest-related genes, including p53-inducible protein with a death domain (Pidd). We demonstrate a novel role for PIDD as a regulator of chemotherapy response in human lung tumor cells.
Keywords: Mouse models, cisplatin, Kras, p53, chemotherapy resistance, lung cancer
Lung cancer is the leading cause of cancer death in the United States, with a 5-year survival rate of only ∼15% (American Cancer Society 2007). The majority of patients with advanced non-small-cell lung cancer (NSCLC) are treated with combination therapy that includes a platinum-based compound. However, only ∼30% of patients with advanced NSCLC respond to this treatment (Socinski 2004). The remaining ∼70% of patients suffer negative side effects associated with drug toxicity without the therapeutic benefits of treatment. Among the ∼30% of patients that initially respond, most patients eventually develop resistant disease. Therefore, both inherent and acquired drug resistance are major barriers to successful platinum-based therapy. Cisplatin [cis-diamminedichloroplatinum(II)] is one of the most widely employed drugs in cancer therapy. Its activity as an anti-cancer agent was discovered >40 years ago (Rosenberg et al. 1969), and it became the first FDA-approved platinum compound for cancer treatment in 1978 (Kelland 2007). Cisplatin and platinum-based analogs like carboplatin are currently used to treat many malignancies, including lung, ovarian, head and neck, bladder, and testicular cancer (Socinski 2004). While the major barriers limiting the use and efficacy of platinum-based compounds are toxicity and resistance (Kelland 2007), there are currently no established approaches to identify patients who are likely to respond to cisplatin-based therapy.
Cisplatin and carboplatin bind DNA to form intrastrand and interstrand cross-links between purine bases. Platinated adducts distort the DNA helix in a manner that is recognized by high-mobility group (HMG) proteins and other proteins involved in the DNA damage response (Wang and Lippard 2005). These adducts impair replication and transcription, which can lead to stalled replication forks and the formation of double-strand breaks. A number of DNA repair pathways, including mismatch repair (MMR) and nucleotide excision repair (NER), have been implicated in platinum adduct repair, and, correspondingly, alterations in these pathways have been implicated in resistance (Wang and Lippard 2005; Helleday et al. 2008). Other signaling pathways—such as those involving NF-κB, c-ABL, JNK, and p73—have also been implicated in cisplatin response in vitro (Kharbanda et al. 1995; Gong et al. 1999; Hayakawa et al. 2004; Mabuchi et al. 2004; Leong et al. 2007).
Multiple mechanisms that mediate intrinsic or acquired resistance to cisplatin in vitro have been identified (Kelland 2007). Mechanisms that preclude the formation of platinum-DNA (Pt-DNA) adducts include decreased import, increased detoxification, and increased efflux (Hall et al. 2008). For example, impaired uptake of cisplatin due to down-regulation of the copper-transporter 1 (CTR1) protein has been demonstrated in ovarian cancer (Ishida et al. 2002; Holzer et al. 2006). Increased detoxification by conjugation of cisplatin to glutathione, coupled with increased export, has also been documented in ovarian cancer cell lines derived from the same patient before and after drug resistance (Lewis et al. 1988). However, numerous gene expression studies have failed to identify a single transporter that is altered universally in cisplatin-resistant cell lines. It is therefore likely that multiple genes involved in import, detoxification, and efflux can be involved in clinically relevant resistance. Tissue specificity of transporter expression may also impact the mechanisms of resistance in different tumor types (Bando et al. 1998).
Cisplatin resistance can also occur through enhanced DNA damage repair. NER is thought to be the predominant repair pathway for Pt-DNA adducts. The marked sensitivity of testicular cancer to cisplatin may be due to intrinsically lower levels of the NER pathway proteins ERCC1 and XPA (Welsh et al. 2004). Additionally, increased expression of ERCC1 in ovarian tumors and cancer cell lines has been associated with cisplatin resistance (Dabholkar et al. 1994; Selvakumaran et al. 2003). Recent clinical trials suggest that patients with tumors with low ERCC1 levels benefit preferentially from cisplatin-based chemotherapy (Olaussen et al. 2006). However, very few DNA repair genes have been functionally validated in vivo.
Finally, the role of the tumor suppressor p53 in mediating cisplatin response remains controversial and appears to be cell type-dependent. In some cell lines, p53 mutation is associated with cisplatin resistance (Perego et al. 1996). However, in other cell lines, loss of p53 increases cisplatin sensitivity (Pestell et al. 2000). Since p53 is mutated in ∼50% of human NSCLCs (Ahrendt et al. 2000; Skaug et al. 2000), elucidating its role in chemoresistance has important implications for treatment strategies.
Although much has been learned from studying resistance mechanisms in isolated cell lines, tumors in vivo encounter drugs in very different conditions. The tumor microenvironment may provide signals and physical barriers that alter signaling networks and the context in which cells respond to therapy (Olive et al. 2009). The immune system can also act as a barrier or promoter of tumor behavior. Finally, drug pharmacodynamics differ in vitro compared with in vivo. Therefore, a systematic attempt to model cisplatin response and resistance in vivo may provide insights that cannot be ascertained from in vitro studies. Observations in xenograft models first demonstrated that in vivo chemotherapy resistance mechanisms were distinct from those in vitro (Teicher et al. 1990). Few studies have examined the response of autochthonous tumors to platinum-based therapy in vivo. For example, responses to several chemotherapy agents, including cisplatin, were analyzed in mice bearing Brca1−/−;p53−/− mammary tumors (Rottenberg et al. 2007). Interestingly, these tumors developed resistance to doxorubicin and docetaxel but not to cisplatin, even after repeated doses. Thus, there is still a need for in vivo models of inherent and acquired resistance to platinum agents.
We described previously the development of mouse models for human lung cancer in which expression of oncogenic Kras (mutated in ∼30% of NSCLCs) is the initiating event (Jackson et al. 2001; Johnson et al. 2001). In the LSL-KrasG12D/+ model, Cre-mediated loss of a stop cassette permits expression of the oncogenic KrasG12D allele from its endogenous promoter. Mice develop lung adenomas with 100% penetrance that eventually progress to high-grade adenocarcinomas. LSL-KrasG12D/+ mice that possess conditional mutant or null alleles of p53 develop lung tumors with a shorter latency and advanced histopathology compared with mice with wild-type p53 (Jackson et al. 2005). We demonstrated previously a strong similarity between KrasG12D-initiated lung tumor models and human NSCLC at the level of gene expression (Sweet-Cordero et al. 2005). Since early-stage and advanced NSCLC are frequently treated with platinum compounds, we investigated the effects of cisplatin treatment on oncogenic KrasG12D-initiated lung tumors.
Results
Short-term response to cisplatin
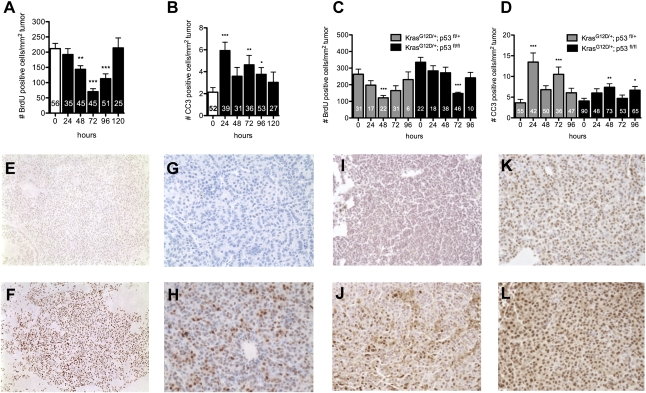
LSL-KrasG12D/+ mice were treated intraperitoneally (ip) with a single dose of cisplatin (7 mg/kg) 12–16 wk after tumor initiation by intranasal Adeno-Cre (AdCre) infection (higher doses led to death and excessive weight loss in pilot studies) (Supplemental Fig. S1). Mice were sacrificed at different time points following cisplatin treatment to analyze the effects on cell cycle and cell survival in tumors. As indicated by BrdU (5-bromo-2′-deoxyuridine) incorporation, cisplatin led to a reduction in the number of cells entering the cell cycle that was maximal 72 h after a single dose, with full recovery by 120 h (Fig. 1A). In contrast, the kinetics of the apoptotic response as measured by cleaved caspase 3 (CC3) staining had two waves of activity that peaked at 24 and 72 h, and returned to control levels by 120 h after treatment (Fig. 1B). A maximal decrease in mitotic index was observed 24 h after cisplatin treatment and persisted through 72 h (Supplemental Fig. S2).
Figure 1.
Cisplatin induces cell cycle arrest and cell death in KrasG12D-initiated lung tumors, independently of p53 activity. (A) Number of BrdU-positive cells per lung tumor area from LSL-KrasG12D/+ mice treated with a single dose (7 mg/kg body weight) of cisplatin and analyzed 0–120 h later. (B) Number of CC3-positive cells per lung tumor area, as in A. (C) Number of BrdU-positive cells per lung tumor area from LSL-KrasG12D/+ mice either heterozygous or homozygous for the p53fl/fl allele, treated as in A. (D) Number of CC3-positive cells per lung tumor area from LSL-KrasG12D/+ mice either heterozygous or homozygous for the p53flfl allele, treated as in A. In A–D, number of tumors analyzed is shown for each bar. Error bars represent standard error of the mean (SEM). Significant changes compared with control are indicated by (*) P < 0.04, (**) P < 0.006, or (***) P < 0.0001. (E–L) PBS-treated lung tumors (E,G,I,K) or cisplatin-treated lung tumors (F,H,J,L) stained with Pt-1,2-d(GpG) antibody (8 h) (E,F), γ-H2AX antibody (24 h) (G,H), anti-phospho Chk1 (Ser345) antibody (12 h) (I,J), or anti-phospho Chk2 (Thr68) antibody (12 h) (K,L).
To investigate whether p53 activation mediates apoptosis and cell cycle arrest in response to cisplatin in this model, we crossed LSL-KrasG12D/+ mice with conditional Trp53F2-10/F2-10 mice (Jonkers et al. 2001), hereafter referred to as p53fl/fl. Delivery of AdCre to the lungs of LSL-KrasG12D/+;p53fl/fl mice leads to simultaneous activation of oncogenic Kras and loss of p53 function (Jackson et al. 2005). KrasG12D/+ lung tumors null for p53 had significantly higher basal proliferation indices than tumors with wild-type p53 (P < 0.003), while p53 heterozygous lung tumors had intermediate levels of proliferation (Supplemental Fig. S3). However, in response to cisplatin, both p53 heterozygous and p53-null lung tumors exhibited cell cycle arrest similar to that seen in KrasG12D/+ tumors with wild-type p53 (Fig. 1C). The majority of tumors had significant decreases in BrdU incorporation 72 h after cisplatin, regardless of p53 status (Supplemental Fig. S4). While the maximum number of apoptotic cells observed in KrasG12D/+;p53fl/fl tumors in response to cisplatin was decreased compared with KrasG12D/+, we detected a statistically significant increase in both cases (Fig. 1B,D). Thus, cell cycle arrest induced by cisplatin is not dependent on p53 in this model, and apoptosis is at least partially p53-independent as well. We confirmed the lack of dependence on p53 for cell cycle arrest in this model by crossing LSL-KrasG12D/+ mice to mice lacking a functional allele of p21 (Brugarolas et al. 1995). KrasG12D/+; p21−/− lung tumors had similar cell cycle arrest and apoptosis profiles in response to cisplatin compared with controls (Supplemental Fig. S5). Taken together, these data suggest that cisplatin response in vivo is not dependent on the p53–p21 pathway.
To investigate the kinetics of cisplatin adduct formation and DNA damage signaling at a cellular level, we analyzed cisplatin-treated tumors for the presence of Pt-DNA adducts using a Pt-1,2-d(GpG) intrastrand cross-link-specific monoclonal antibody (Liedert et al. 2006). This antibody recognizes the most frequently occurring adduct formed by cisplatin, which is associated with its cytotoxicity and anti-cancer activity (Liedert et al. 2006; Dzagnidze et al. 2007). Pt-DNA adducts were detected in the lung as early as 3 h after a single dose of cisplatin (Fig. 1E,F; data not shown). Platinum adduct formation can cause stalling of replication forks, which leads to collapse and the generation of DNA double-strand breaks (Henry-Mowatt et al. 2003). This leads to activation of checkpoint kinases ATM and ATR, and their downstream substrates, Chk2 and Chk1, which recruit other repair proteins to sites of damaged DNA (Pabla et al. 2008). The phosphorylated form of the histone variant H2AX (γ-H2AX) is a critical component of this repair complex, and thus can be used as a marker of DNA damage signaling. In cisplatin-treated KrasG12D/+ tumors, we detected γ-H2AX 4 h (the earliest time point examined) after cisplatin treatment, with maximal staining 12–24 h following treatment (Fig. 1G,H; Supplemental Fig. S6; data not shown). Basal phosphorylation of the checkpoint kinase Chk2 (Thr68) was detected in untreated tumors, and increased phosphorylation of both Chk1 (Ser345) and Chk2 (Thr68) was clearly evident after cisplatin treatment (Fig. 1I–L). Taken together, these data demonstrate that tumors sense DNA damage in response to cisplatin within 4 h, and respond by cell cycle arrest and cell death associated with activation of both Chk1 and Chk2. In KrasG12D/+;p53fl/fl lung tumors analyzed 4–24 h after a single dose of cisplatin, we did not detect obvious differences in DNA damage signaling compared with p53 wild-type tumors (Supplemental Fig. S7). We observed very few tumors with patterns of BrdU or γ-H2AX staining that deviated significantly from the mean at the indicated time points, suggesting that most tumors initially respond to cisplatin-induced DNA damage in this model (Supplemental Fig. S4; data not shown).
Long-term response to cisplatin
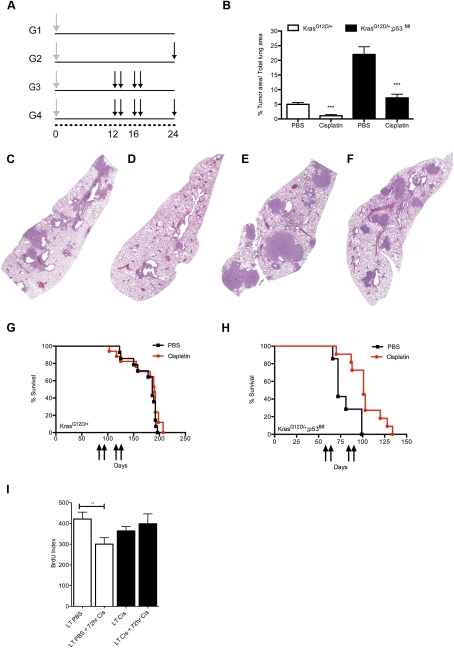
To analyze the long-term effects of cisplatin therapy on KrasG12D-initiated lung tumors, we treated mice 12 wk following AdCre infection with cisplatin once a week for 2 wk, followed by a 2-wk rest period to allow recovery from toxicity, and repeated this regimen for a total of four doses of cisplatin (Fig. 2A, Group 3). Tumor response was measured by determining the ratio of tumor area to total lung area (TA/LA) in histological sections. Treatment with cisplatin significantly reduced tumor burden in the treated G3 group (n = 8) compared with the control G1 mice (n = 7) (P < 0.0002) (Fig. 2B–D).
Figure 2.
Cisplatin treatment significantly reduces lung tumor burden in KrasG12D-initiated lung tumors regardless of p53 activity. (A) Treatment regimens for groups 1–4 (G1–G4). Mice were infected with AdCre to permit expression of KrasG12D at time 0 (gray arrow). Cisplatin was given at indicated time points in weeks (black arrows) for each group. (B) Tumor area/total lung area in control (G1) versus treated (G3) LSL-KrasG12D/+ mice (white bars; [***] P < 0.002) and in LSL-KrasG12D/+;p53fl/fl mice (black bars; [***] P < 0.0001). (C–F). Representative H&E stains at 2× magnification of PBS-treated (C,E) or cisplatin-treated (D,F) lungs from LSL-KrasG12D/+ mice (C,D) or LSL-KrasG12D/+;p53fl/fl mice (E,F). (G,H). Kaplan-Meier survival curves of LSL-KrasG12D/+ mice (G) and LSL-KrasG12D/+;p53fl/fl mice (H) treated with four doses of cisplatin (red) or PBS (black). Black arrows indicate cisplatin treatments at X number of days post-AdCre infection. For H, cisplatin significantly prolongs survival (P < 0.002). (I) Number of BrdU-positive cells per lung tumor area in LSL-KrasG12D/+ mice with or without a final 72-h dose of cisplatin ([**] P < 0.009). Error bars represent SEM.
To determine whether this response was dependent on intact p53, we treated LSL-KrasG12D/+;p53fl/fl mice with a similar treatment regimen. Upon sacrifice, the basal tumor volume in untreated LSL-KrasG12D/+;p53fl/fl mice was much greater than those with wild-type p53. However, despite this increase in volume, LSL-KrasG12D/+;p53fl/fl mice treated with cisplatin (n = 11) also had a significant reduction in tumor burden compared with controls (n = 10) (P < 0.0001), again demonstrating that wild-type p53 is not required for response to cisplatin (Fig. 2B,E,F).
Using another cohort of LSL-KrasG12D/+ and LSL-KrasG12D/+;p53fl/fl mice, we asked whether the impact of the four-dose regimen of cisplatin could prolong survival of tumor-bearing mice. Unexpectedly, despite the significant reduction in tumor burden in LSL-KrasG12D/+ mice observed after the treatment regimen (Fig. 2B), there was no improvement in survival (Fig. 2G). In contrast, LSL-KrasG12D/+;p53fl/fl mice treated with four doses of cisplatin survived significantly longer (n = 11) than mice treated with PBS (n = 8) (P < 0.002) (Fig. 2H). Tumors in LSL-KrasG12D/+ mice develop much more slowly than tumors that lack p53, and untreated LSL-KrasG12D/+ mice do not die from their lung tumor burden until 7–13 wk after treated mice receive the fourth dose of cisplatin—a considerable time frame for residual treated tumors to regrow. Indeed, tumor burden at the time of death in LSL-KrasG12D/+ mice treated with four doses of cisplatin was not significantly different from control animals (data not shown). In contrast, KrasG12D/+;p53-null lung tumors develop extremely rapidly, and these tumors typically kill untreated animals near the time when treated mice are receiving their fourth and final dose of cisplatin. When we treated LSL-KrasG12D/+ mice with continuous dosing of cisplatin beyond four doses, mice experienced a significant survival benefit (Supplemental Fig. S8). The fact that treatment with cisplatin significantly prolongs survival of mice with p53-null lung tumors further demonstrates that p53 is not required for drug response and therapeutic benefit. This suggests that loss of p53, while a predictor of poor prognosis and more aggressive tumors in mice, still permits therapeutic benefits from cisplatin.
Next, to investigate whether residual KrasG12D/+ tumors present at the end of the treatment regimen were resistant to cisplatin, we treated a cohort of LSL-KrasG12D/+ mice as described above with four total doses of cisplatin or PBS, waited 4 wk, and then treated them with a final 72-h dose of cisplatin before sacrifice (Fig. 2A, G2 vs. G4). When both sets of mice received a final dose of cisplatin, tumors from mice that had received previous cisplatin treatment no longer demonstrated a significant reduction in BrdU incorporation like the naïve tumors (Figs. 1A, 2I), suggesting that the pretreated tumors have acquired resistance to cisplatin treatment.
Dynamics of tumor response to cisplatin
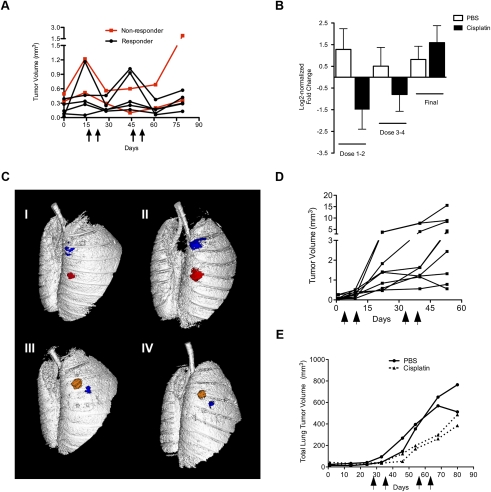
To gain further insight into the dynamics of cisplatin response in this model, we employed in vivo microcomputed tomography (microCT) imaging. LSL-KrasG12D/+ mice were treated with PBS or cisplatin according to the regimen described above and imaged prior to treatment, 5 d after the second dose of cisplatin, and 10 d after the fourth and final dose of cisplatin. We focused on tumors whose boundaries could be defined clearly in multiple scans over time. Untreated KrasG12D/+ lung tumors grew slowly (average tumor volume doubling time of ∼35 d), with highly variable growth rates. Following two doses of cisplatin, most tumors in the LSL-KrasG12D/+ model showed a reduction in tumor volume (Fig. 3A–C). During the dosing break (between doses 2 and 3), cisplatin-treated tumors resumed growth, but generally remained sensitive after the third and fourth doses (Fig. 3A,B). However, some tumors stopped responding to the third and fourth doses of cisplatin (Fig. 3A). Thus, while we cannot rule out that innate resistance occurs in individual clones within tumors, it does not appear to be a characteristic of bulk tumors. Importantly, in mice that received four doses of cisplatin and received a final dose of cisplatin ∼6 wk later, treated tumors no longer responded, again suggesting that tumors become resistant after four doses of cisplatin (Fig. 3B).
Figure 3.
In vivo microCT imaging reveals lung tumor regression and stasis in response to cisplatin in LSL-KrasG12D/+ mice, and decelerated growth in LSL-KrasG12D/+;p53fl/fl mice. (A) Tumor volume dynamics of individual cisplatin-treated tumors in LSL-KrasG12D/+ mice. Black arrows on the X-axis indicate cisplatin treatments. Red lines indicate tumors that stopped responding to treatment after three to four doses. The X-axis indicates days following the first pretreatment microCT scan, which occurred 14 wk post-AdCre infection. (B) Log2-normalized fold change in tumor volume of individual tumors in PBS-treated (white bars) and cisplatin-treated (black bars) mice. Tumor volumes were quantified before and after doses 1 and 2 (Dose 1–2), before and after doses 3 and 4 (Dose 3–4), and before and after one final dose (Final). (C) Representative microCT lung reconstructions before and after two doses of PBS (panels I,II) or cisplatin (panels III,IV) with individual lung tumors pseudocolored. (D) Tumor volume dynamics of individual cisplatin-treated tumors in response to cisplatin in LSL-KrasG12D/+;p53fl/fl mice. Black arrows on the X-axis indicate cisplatin treatments. (E) Total lung tumor volume in LSL-KrasG12D/+;p53fl/fl mice (n = 2 mice per group) treated with four doses of PBS (solid lines with circles) or cisplatin (dashed lines). Arrows on the X-axis (days following AdCre infection) indicate one dose of PBS or cisplatin.
KrasG12D/+;p53-null lung tumors grow much faster than those with wild-type p53 (doubling time of ∼7 d), and therefore it is more straightforward to observe a significant impact on tumor growth. Indeed, a single dose of cisplatin caused a significant reduction in tumor growth in this model, as observed by microCT (data not shown). Unlike KrasG12D/+ tumors with wild-type p53, p53-null tumors did not regress, but progressed despite therapy (Fig. 3D). In a smaller study, we quantified total tumor burden by microCT in LSL-KrasG12D/+;p53fl/fl mice treated with PBS or four doses of cisplatin. Cisplatin treatment clearly impeded tumor growth, but tumors continued to progress despite therapy (Fig. 3E).
Mechanism of cisplatin resistance in vivo
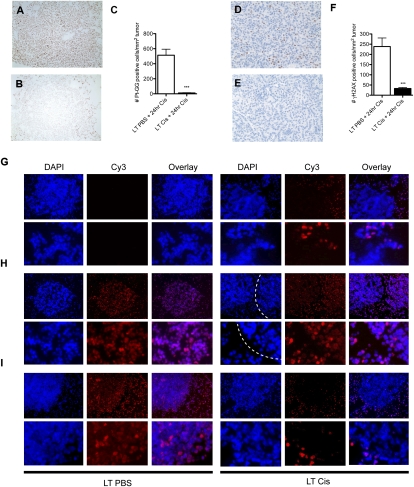
Previous studies suggest that cisplatin resistance in human cancer may be complex, as no single factor has been able to explain resistance in full. In vitro studies suggest that decreased uptake, increased detoxification, and increased efflux of platinum from cells may all be mechanisms of resistance. In addition, platinum adducts may be more rapidly repaired in resistant tumors. Finally, tumor cells may use error-prone translesion DNA polymerases in order to tolerate higher levels of adducts. To distinguish among these possibilities, we treated long-term PBS or cisplatin-treated mice with a final dose of cisplatin, and stained tumor sections with the antibody to Pt-1,2-intrastrand DNA cross-links to monitor the kinetics of adduct levels. Strikingly, 24 h after a final dose of cisplatin, long-term-treated tumors had significantly decreased levels of Pt-1,2-d(GpG) adducts compared with tumors from mice treated previously with PBS (G2 vs. G4) (Fig. 4A–C), whereas adduct levels in the normal surrounding lung cells were similar (Supplemental Fig. S9). Tumors that completely lacked adducts at this time point were found only in lungs from long-term cisplatin-treated animals. To support this observation, we quantified the levels of γ-H2AX in PBS and cisplatin-treated tumors that had received a final 24-h dose of cisplatin. We observed a significant reduction in γ-H2AX staining in resistant tumors (Fig. 4D–F), consistent with the lack of adducts at this time point. These data suggest that the mechanism of cisplatin resistance in this model is not mediated by tolerance of platinum adducts in vivo. However, these data do not discriminate between resistance mechanisms in which damage never occurs (i.e., import/detoxification/export), or in which damage occurs but is repaired rapidly. To discriminate between these possibilities, we used atomic absorption spectroscopy (AAS) to quantify platinum levels in lysates from individual lung tumors taken from animals treated with PBS or cisplatin (four total doses), plus a final dose of cisplatin given at 0, 2, 4, 12, 24, 48, or 72 h before sacrifice. Chronic cisplatin treatment did not cause a significant decrease in platinum levels within tumors at any time point examined (Supplemental Fig. S10), demonstrating that platinum is able to enter tumors similarly in naïve and pretreated tumors. This result suggests that decreased import and/or rapid efflux are not the driving forces behind cisplatin resistance in this model.
Figure 4.
Long-term cisplatin-treated lung tumors in LSL-KrasG12D/+ mice exhibit enhanced adduct repair in response to a final dose of cisplatin. (A) Representative sensitive lung tumor section (G2) stained with Pt-1,2-d(GpG) from long-term PBS-treated mice given a final 24-h dose of cisplatin. (B) Representative resistant tumor section (G4) from long-term cisplatin-treated mice, treated as in A. (C) Number of Pt-1,2-d(GpG)-positive cells per lung tumor area in long-term PBS-treated (white bar) or cisplatin-treated (black bar) mice given a final dose of cisplatin and sacrificed 24 h later ([***] P < 0.0001). (D) Representative sensitive tumor section (G2) stained with γ-H2AX from long-term PBS-treated mice given a final 24-h dose of cisplatin. (E) Representative resistant tumor (G4) from long-term cisplatin-treated mice, treated as in D. (F) Number of γ-H2AX-positive cells per lung tumor area in long-term PBS-treated (white bar) or cisplatin-treated (black bar) mice given a final dose of cisplatin and sacrificed 24 h later ([***] P < 0.0001). Error bars represent SEM. (G–I). Representative immunofluorescent images of lung tumor sections stained for nuclei (DAPI) or Pt-1,2-d(GpG) (Cy3), and an overlay of these images (Overlay) in mice treated with long-term PBS (LT PBS) or four doses of cisplatin (LT Cis) and given a final dose of cisplatin and analyzed after 0 h (G), 4 h (H), or 24 h (I). The top panels are 10× magnification, and the bottom panels are higher-magnification zooms. (G) Note that adducts persist in normal parts of the lung even after multiple weeks in long-term cisplatin (LT Cis, 0 h). In H (LT Cis, 4 h, DAPI), two tumors are separated by a dotted white line. Approximately 20% of tumors in long-term cisplatin mice had reduced adduct levels as early as 4 h after a final dose of cisplatin (left tumor) whereas the majority of tumors had similar levels of adducts (right tumor) at this time point.
Increased DNA repair has been proposed as a mechanism of platinum resistance (Martin et al. 2008). We reasoned that, if rapid repair was occurring, we should detect a difference in the kinetics of Pt-1,2-d(GpG) adduct formation and, potentially, markers of DNA damage signaling. To explore this possibility, we treated long-term PBS and cisplatin-treated mice with a final dose of cisplatin and examined the kinetics of platinum adducts at early time points (<24 h) following a final dose of cisplatin (Fig. 4G). Adduct levels were scored blindly as absent, low, or high (−, +, or ++) on at least 20 tumors per treatment group. As expected, adducts were not present in naïve tumors (long-term PBS), but, surprisingly, had persisted in nontumor lung areas of pretreated animals for at least 4 wk following their last dose of cisplatin (long-term cisplatin) (Fig. 4G). At 4 h after a final dose of cisplatin, 81% of cisplatin-pretreated tumors (21 of 26) had similar levels of adducts as naïve tumors treated with cisplatin (Fig. 4H). Thus, the majority of tumors showed similar levels of Pt-1,2-d(GpG) adducts regardless of whether or not they had previously received cisplatin. By 8 h after a final dose of cisplatin, 59% of cisplatin-pretreated tumors (13 of 22) had similar levels of adducts as naïve cisplatin-treated tumors (data not shown). By 24 h after a final dose of cisplatin, tumors that completely lacked adducts were found only in cisplatin-pretreated tumors (Fig. 4I). These data demonstrate that Pt-1,2-d(GpG) adducts are present in sensitive and most resistant tumors at early time points, but are cleared more rapidly in tumors pretreated with cisplatin.
Chk1 and Chk2 are checkpoint kinases that are activated by DNA damage signals mediated by ATM and ATR. Therefore, we analyzed Chk1 and Chk2 phosphorylation in long-term cisplatin-treated versus control mice with or without a final 12-h dose of cisplatin as another marker of whether cells were experiencing DNA damage. We observed a clear difference in the dynamics of phosphorylation of these two DNA damage signaling proteins. Chk1 phosphorylation occurred in response to cisplatin in tumors from both naïve and long-term-treated mice (G1 vs. G2, and G3 vs. G4) (Supplemental Fig. S11), demonstrating that resistant tumors activate the DNA damage response, and, thus, that cisplatin is entering these cells. In contrast, Chk2 phosphorylation was induced after an initial dose of cisplatin (G1 vs. G2), and then remained high even in tumors that had not been given cisplatin for several weeks (G3 in Supplemental Fig. S11). This finding suggests that these two signaling pathways may be responding to distinct DNA damage signals as a result of cisplatin treatment—one that is transient (Chk1), and another that is persistent (Chk2). Furthermore, it suggests there is a fundamental difference in the DNA damage response mechanism in naïve and long-term cisplatin-treated lung tumors. Taken together, our data strongly argue that increased DNA damage repair is the predominant mechanism of cisplatin resistance in vivo in this model.
Cross-resistance to platinum analogs
Cisplatin-resistant tumors in the clinical setting are often cross-resistant to other platinum analogs. To determine whether cisplatin-resistant tumors were cross-resistant to other platinum agents, we treated long-term PBS or cisplatin-treated tumors with a single dose of carboplatin (50 mg/kg in saline), and analyzed tumors 24 h later for the presence of Pt-1,2-d(GpG) adducts and DNA damage signaling (γ-H2AX). Carboplatin induces the same type of Pt-1,2-d(GpG) cross-links as cisplatin in cells, although at a slightly reduced frequency (Blommaert et al. 1995). Indeed, in our studies with carboplatin, staining for this adduct was less intense compared with a single dose of cisplatin (7 mg/kg) (data not shown). In long-term cisplatin-treated tumors, carboplatin produces fewer adducts (data not shown) and reduced DNA damage signaling, evident by γ-H2AX staining compared with naïve tumors (Supplemental Fig. S12). These data indicate that, just as encountered in clinical resistance (Wang and Lippard 2005), cisplatin-resistant tumors in this model are cross-resistant to other platinum analogs.
Comparative genomic analysis of naïve versus cisplatin-treated tumors
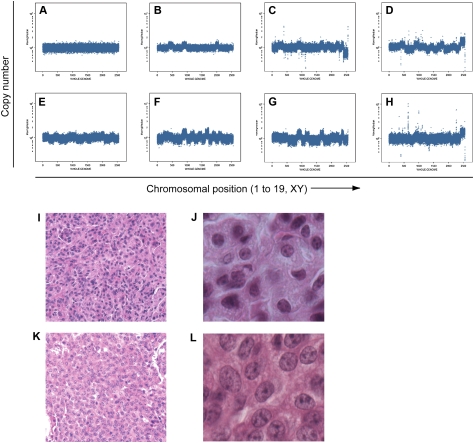
We performed DNA copy number analysis to identify potential genomic deletions or amplifications that might implicate particular genes involved in acquired resistance. LSL-KrasG12D/+ mice were treated with long-term PBS or cisplatin as described in G1 and G3 (Fig. 2), and then sacrificed ∼8 wk after their final dose of cisplatin—a total of 24–30 wk following tumor induction by AdCre. DNA was isolated from individually microdissected tumors and subjected to representational oligonucleotide microarray analysis (ROMA) (Lakshmi et al. 2006). Of 11 long-term PBS-treated lung tumors analyzed, only two (18%) had detectable whole-chromosomal aberrations (Fig. 5A). This observation is consistent with the low frequency of DNA copy number changes that we reported previously in this model using BAC arrays (Sweet-Cordero et al. 2006). In contrast, 19 of 23 long-term cisplatin-treated tumors (83%) harbored whole-chromosomal aberrations, including gains and losses of whole chromosomes (Fig. 5B–H). A subset of tumors was analyzed for copy number changes with independent methodologies, including Agilent Array CGH, Affymetrix SNP Arrays (Broad Institute, Cambridge, MA), and Solexa sequencing (Illumina); these techniques consistently validated the whole-chromosomal changes identified by ROMA (data not shown).
Figure 5.
DNA copy number profiling by ROMA reveals cisplatin treatment enhances the percentage of lung tumors from LSL-KrasG12D/+ mice with whole-chromosomal gains and losses. (A) Representative genomic profile of lung tumors from PBS-treated mice. Nine of 11 PBS-treated tumors did not exhibit genomic changes. (B–H) Representative genomic profiles of cisplatin-treated tumors with significant whole-chromosomal DNA copy number changes. Nineteen of 23 cisplatin-treated tumors harbored whole-chromosomal changes. The X-axis indicates chromosomal position from chromosome 1 to 19, and XY chromosomes. The Y-axis indicates copy number. (I,J) Representative H&E-stained tumor section from PBS-treated mice with low-grade tumor histology (20×, I), and a higher-magnification panel from the same tumor (40×, J). (K,L) Representative H&E-stained tumor section from cisplatin-treated mice with high-grade tumor histology (20×, K), and a higher-magnification panel from the same tumor (40×, L). Note the larger nuclei, more diffuse nuclear staining, and higher nuclear to cytoplasmic ratio in K and L compared with I and J.
Strikingly, histological analysis of a subset of these tumors revealed that the majority of cisplatin-treated tumors were higher grade (11 of 14 as Grade 2+, 79%) compared with untreated tumors (two of 10 as Grade 2+, 20%) (Fig. 5I–L). The only two PBS-treated tumors with whole-chromosomal changes were also scored blindly as Grade 2+ tumors, whereas eight PBS-treated tumors with undetectable copy number changes were scored as low-grade (Grade 2 or less). Therefore, high-grade tumors are associated consistently with chromosomal abnormalities, whereas low-grade tumors have apparently normal DNA copy numbers in this model. These data suggest that long-term cisplatin treatment selects for and/or promotes tumor progression accompanied by alterations in chromosome number.
Gene expression analysis of cisplatin response and resistance in vivo
The data presented above suggest that long-term cisplatin treatment creates tumors that are fundamentally different from naïve tumors. To characterize these potential differences, we performed gene expression analysis to examine cisplatin response and resistance. First, we examined the dynamics of gene expression changes in response to cisplatin using laser capture microdissection to isolate RNA from individual tumors at 24, 48, and 72 h after a single dose of cisplatin. We analyzed expression of p21, Mdm2, Bax, and Bcl2 using real-time PCR. Maximal differential expression of these genes occurred 72 h after cisplatin treatment despite the fact that DNA damage response and cell death occurred earlier (data not shown). We then performed a more global analysis of gene expression at the same time point after cisplatin therapy using microarrays. DNA from individually microdissected lung tumors (n = 49) was analyzed using Affymetrix 430A Genechips. Samples from mice treated in the four groups shown in Figure 2 were included: G1 (n = 13), G2 (n = 11), G3 (n = 9), and G4 (n = 7), as well as normal lung (n = 9) (Supplemental Table S1).
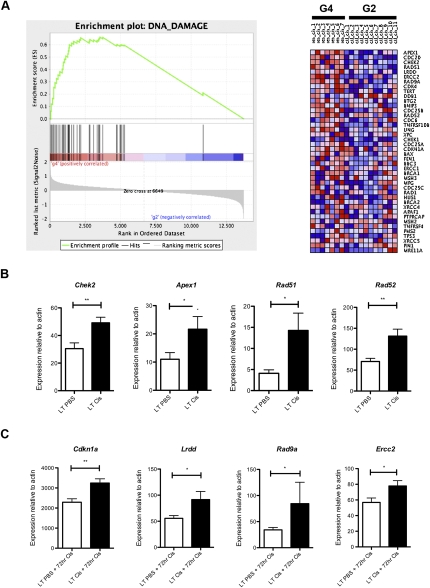
To identify cellular pathways altered in cisplatin-treated tumors, we used gene set enrichment analysis (GSEA) to identify gene sets representing molecular pathways with significant enrichment in control versus resistant tumors (Subramanian et al. 2005). GSEA provides an enrichment score (ES) that measures the degree of enrichment of a gene set at the top (highly correlated with class 1) or bottom (highly correlated with class 2) of a rank-ordered gene list derived from the data set. A nominal P-value is used to assess the significance of the individual ES score. We also used the pathway analysis tool MetaCore from GeneGO, Inc., to identify cellular processes significantly enriched between treatment groups. Pathways are defined in MetaCore as a set of curated consecutive signals or transformations that have been confirmed by experimental evidence or inferred relationships. We focused our analysis on the top-scoring 200 genes in each transition (G2 vs. G1: genes up-regulated in G2 compared with G1; G1 vs. G2: genes up-regulated in G1 compared with G2, etc.). Consistent with our data in Figure 2I, cell cycle and proliferation pathways were significantly enriched in naïve tumors compared with tumors treated with a single dose of cisplatin (G1 vs. G2; six of the top eight enriched pathways were associated with cell cycle and cell proliferation; false discovery rate [FDR] < 0.05, P < 0.003). However, cell cycle pathways were not similarly represented in cisplatin-pretreated tumors before and after treatment with the same dose (G3 vs. G4, and G4 vs. G3) (Supplemental Table S2). In addition to changes in cell cycle, pathways enriched in naïve tumors treated with a single dose of cisplatin (G2 vs. G1) included those related to adhesion, transport, and immune response (FDR < 0.25, P < 0.004). In tumors pretreated with four doses of cisplatin and treated with a final challenge of cisplatin, pathways enriched in pretreated tumors (G4 vs. G3) included those related to cell adhesion, G-protein-coupled receptor (GPCR) signaling, glutathione metabolism, and p53 signaling, whereas those depleted included pathways related to immune response and apoptosis/survival (FDR < 0.25, P < 0.05). Pathways enriched in cisplatin-pretreated tumors compared with naïve tumors (G3 vs. G1) were largely related to immune response (FDR < 0.25, P < 0.05). When comparing treatment of naïve tumors to cisplatin-pretreated tumors with a final dose of cisplatin (G4 vs. G2), pathways enriched in cisplatin-pretreated tumors included those related to cell cycle and DNA damage, glutathione and methionine metabolism, cell adhesion, and cytoskeletal remodeling, among others (FDR < 0.25, P < 0.05) (Supplemental Table S2).
Glutathione-mediated detoxification of cisplatin has been implicated previously in resistance, and we validated that a subset of glutathione-related genes (i.e., Mgst2 and GstT2) were up-regulated significantly in pretreated tumors (data not shown). Because our data suggest that the majority of cisplatin-resistant tumors repair adducts more quickly than naïve tumors, we decided to further pursue the cell cycle/DNA damage class of genes. In addition to the enrichment of cell cycle/DNA damage pathways using GeneGO, GSEA identified a DNA damage response gene set enriched in G2 versus G4 (Fig 6A). We validated the expression levels of a subset of these genes by real-time RT–PCR on an independent set of tumors. Cisplatin-resistant tumors expressed higher basal levels of some genes (Apex1, Chek2, Rad51, and Rad52) (Fig. 6B). Other genes were induced to a higher degree in cisplatin-resistant tumors compared with controls (Lrdd, Cdkn1a [p21], Ercc2, and Rad9) (Fig. 6C). Together, these data support our observation that cisplatin-resistant tumors have an enhanced ability to repair Pt-DNA adducts, and, additionally, they have the capacity to induce expression of genes known to play a role in multiple DNA repair pathways.
Figure 6.
Genes associated with DNA damage and repair are up-regulated in cisplatin-resistant lung tumors in vivo. (A) Enrichment plot of the DNA damage gene set identified by GSEA and corresponding heat map for G2 versus G4. Expression level is represented as a gradient from high (red) to low (blue). (B) Expression of indicated genes in long-term PBS (LT PBS) versus long-term cisplatin-treated (four doses, LT Cis) tumors. (C) Expression of indicated genes in long-term PBS (LT PBS) or long-term cisplatin (LT Cis) tumors treated with a final 72-h dose of cisplatin (LT PBS + 72 h Cis or LT Cis + 72 h Cis). All genes were analyzed in triplicate by real-time RT–PCR on six independent tumors per treatment group. Expression levels are normalized to β-actin. (**) P < 0.009; (*) P < 0.05. Error bars represent SEM.
p53-induced protein with a death domain (PIDD) expression induces cisplatin resistance in human cancer cell lines
Of these genes, Lrdd (Pidd) was notable because it had not been implicated previously in cisplatin resistance in vivo. PIDD was identified originally as a target gene of p53, whose expression promoted apoptosis in p53-null cell lines (Lin et al. 2000). Subsequently, it was shown that PIDD is an ∼90-kDa protein that is constitutively processed into two smaller C-terminal fragments, PIDD-C and PIDD-CC, by autocatalytic cleavage (Tinel et al. 2007). These fragments participate in different signaling complexes called PIDDosomes, which can act as prosurvival or prodeath signals in response to DNA damage, depending on the context (Tinel and Tschopp 2004; Janssens et al. 2005; Tinel et al. 2007; Shulga et al. 2009). More recently, PIDD has been implicated in cell cycle regulation in the context of DNA damage, particularly in nonhomologous end-joining (NHEJ) and the G2/M checkpoint (Shi et al. 2009).
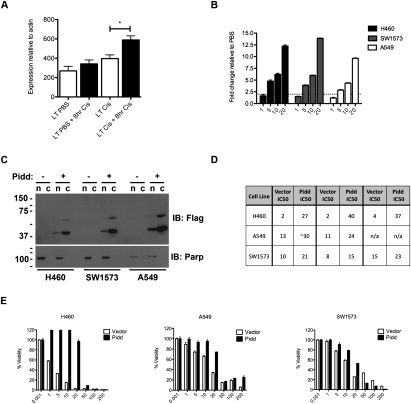
We reasoned that, if PIDD is playing a role in cell cycle arrest or repair in vivo, it should be induced early after a final dose of cisplatin in resistant tumors. We isolated an independent set of tumors from LSL-KrasG12D/+ mice (G1–G4) that were treated with or without a final 8-h dose of cisplatin and performed real-time RT–PCR for Pidd expression. Indeed, Pidd expression was significantly higher only in tumors pretreated with cisplatin (Fig. 7A). To examine the potential role of PIDD in cisplatin response in vitro, we treated three human NSCLC cell lines that have KRAS mutations and wild-type P53 with various doses of cisplatin and examined expression of PIDD 24 h following treatment. In all cell lines examined, cisplatin treatment led to increased levels of PIDD mRNA (Fig. 7B).
Figure 7.
Overexpression of PIDD confers resistance to cisplatin in human NSCLC cell lines. (A) Expression levels of Pidd mRNA in LSL-KrasG12D/+ lung tumors treated with PBS or four total doses of cisplatin, with or without a final 8-h dose of cisplatin (n = 6 tumors per group). P < 0.01. Error bars represent SEM. (B) Expression levels of PIDD mRNA in human NSCLC lines treated with increasing doses of cisplatin (micromolar) and harvested 24 h following treatment. The Y-axis is fold change relative to PBS-treated cells. Expression levels are normalized to ACTIN. (C) PIDD overexpression in human NSCLC lines by Western blot (IB) for Flag, and for Parp to confirm purity of nuclear/cytoplasmic fractions. Upon longer exposure, full-length PIDD is apparent in the cytoplasm, and both PIDD cleavage products are also present in the nucleus (data not shown). (D) IC50 values for cisplatin treatment in each cell line with MSCV Vector or MSCV-PIDD expression from three independent experiments performed in triplicate. (E) Representative survival plots for indicated cell lines expressing MSCV Vector or MSCV-Pidd treated with 0–200 μM cisplatin (X-axis) and analyzed 48 h later using CellTiter-Glo cell viability assay. The Y-axis represents percent of viable cells normalized to PBS-treated control. Error bars represent standard deviation.
To further investigate the role of PIDD in cisplatin response, we overexpressed PIDD by infecting cells with retroviruses carrying C-terminal Flag-tagged PIDD with a puromycin resistance cassette and selected cells with puromycin (Tinel et al. 2007). Overexpression was confirmed by Western blotting of nuclear and cytoplasmic cell lysates, with antibodies directed against Flag and PIDD (Fig. 7C; data not shown). These data demonstrate the presence of the autocatalytically cleaved forms of PIDD (∼51 kDa and ∼37 kDa), which were both present in the cytoplasm and also in the nucleus, although at lower levels (Fig. 7C; data not shown). Expression of PIDD led to reduced growth rate in each cell line (data not shown), with a corresponding increase in the percentage of cells in G1 of the cell cycle (Supplemental Fig S13). Importantly, in the presence of cisplatin, PIDD expression led to significantly enhanced cell viability (Fig. 7D,E). Strikingly, in H460 cells, overexpression of PIDD increased the IC50 by 13-fold to 20-fold (Fig. 7D,E). In addition, overexpression of PIDD contributed to increased resistance to other DNA-damaging agents, including gemcitabine and etoposide (Supplemental Fig. S14). Because PIDD has been implicated in NF-κB-mediated prosurvival signaling, we analyzed expression of the NF-κB subunit p65 by Western blot of nuclear and cytoplasmic cell fractions (Supplemental Fig. S15), but did not detect basal differences as a result of PIDD overexpression. This does not, however, rule out a role for PIDD in regulating NF-κB signaling, specifically in response to damage. Taken together, the mouse in vivo data and the human in vitro data support an important role for PIDD in cisplatin resistance in lung cancer.
Discussion
While molecularly targeted therapies hold promise for the future of cancer treatment, most patients are currently treated with cytotoxic agents. Cisplatin is an example of a widely employed anti-cancer drug about which we have very little understanding of whether a given patient will be responsive or resistant to treatment. An improved understanding of the molecular and genetic basis of cisplatin response and resistance could significantly impact clinical strategies. Previously, mouse models of hematopoietic malignancies were successfully used to study the genetics of chemotherapy response (Schmitt et al. 2000, 2002). However, few attempts have been made to model chemotherapy resistance in mouse models of epithelial cancers. Here we used genetically engineered mouse models of lung cancer to dissect the molecular and genetic mechanisms of response and resistance to cisplatin therapy in vivo.
We showed that KrasG12D-initiated lung tumors are responsive to cisplatin treatment regardless of loss of p53. Tumors initially respond to cisplatin by sensing damage and undergoing cell cycle arrest and death, leading to a significant decrease in tumor burden. We provide genetic evidence that cisplatin efficacy is independent of p53 loss and does not require the cdk inhibitor p21. Indeed, an intact p53–p21 pathway was not required for cell cycle arrest, apoptosis, inhibition of tumor growth, or survival benefit in this model. Thus, the KrasG12D/+;p53-null lung tumor model resembles human lung cancer in that P53 loss confers a poor prognosis, but it does not necessarily mean that therapy will not be beneficial (Tsao et al. 2007). However, even though p53-null tumors respond to cisplatin, our data suggest that there are fundamental differences in that response compared with tumors with wild-type p53. Specifically, p53-null tumors exhibit reduced apoptosis, and, instead of regressing in response to cisplatin like p53 wild-type tumors, their growth was simply impaired. Since most patients with p53 alterations have point mutations in p53, it will be important to compare the effects of cisplatin in these mouse models, which we are currently investigating.
Our studies differ from a recent report that investigated the response of Brca1−/−;p53−/− mouse mammary tumors to treatment with doxorubicin, docetaxel, and cisplatin. Tumors in this model acquired resistance to doxorubicin and docetaxel, which was in part mediated by overexpression of P-glycoprotein. Notably, cisplatin is not implicated as a substrate of P-glycoprotein, and Brca1−/−;p53−/− mammary tumors remained sensitive to cisplatin after multiple rounds of treatment (Rottenberg et al. 2007). We hypothesize that the discrepancy in these results could be a consequence of the genetic context of BRCA1 and P53 deficiency, since this combination of genetic alterations has been associated with cisplatin sensitivity (Bartz et al. 2006). Given that these mammary tumors are defective in homologous recombination (HR), and that tumors defective in HR are often sensitive to platinum-based compounds, these studies suggest that HR may be an important repair pathway contributing to cisplatin resistance. Indeed, restoration of wild-type BRCA2 in BRCA2-mutated tumors has been shown to be an important mechanism of therapeutic resistance to cisplatin (Edwards et al. 2008; Sakai et al. 2008). Other genes involved in HR are also up-regulated in resistant tumors in our model (i.e., Rad51, Rad52, and Rad9a). Thus, further studies to test the involvement of HR in resistance in this model may be warranted.
Importantly, we found that cisplatin treatment of LSL-KrasG12D/+ mice selected for tumors with increased genomic instability that were histologically more advanced. Two possibilities could explain these results. First, tumor cells with abnormal karyotypes could be present prior to chemotherapy, and are selected for by repeated doses of cisplatin. Alternatively, cisplatin treatment itself may induce DNA damage that is not accurately repaired, leading to chromosomal aberrations. Untreated KrasG12D/+ mice can develop higher-grade tumors with whole-chromosomal changes at low frequency (see the Results; Sweet-Cordero et al. 2006); thus, it is possible that cisplatin enhances the survival of these cells, which can eventually develop into more advanced tumors. In either case, our data suggest that, in some instances, treating with chemotherapy can have no survival benefit, and can actually lead to more advanced tumors—in this case, with increased chromosomal changes, more advanced histology, and increased drug resistance. Given that many human cancers have premalignant stages of tumor progression, it will be important to investigate whether treating low-grade tumors with DNA-damaging agents can facilitate tumor progression. This knowledge will become more important as the technology to detect earlier-stage disease advances. Whether treating high-grade genomically unstable tumors with DNA-damaging agents can promote further progression, such as metastasis, is not well understood. This model could be used to investigate this possibility. Notably, the observation that cisplatin treatment can promote genomic instability may not have been uncovered using tumor cell line models that have already acquired high levels of genomic instability.
We demonstrate that prolonged cisplatin treatment leads to resistance in KrasG12D-initiated lung tumors. Acquired cisplatin resistance appears to be mediated by mechanisms that inhibit the ability of cisplatin to sustain adducts on DNA. This result is in agreement with early work pointing to a critical role of 1,2-intrastrand d(GpG) cross-links in mediating the anti-cancer activity of cisplatin (Lippard 1982). Our data strongly suggest that the most predominant mechanism of resistance in this model is rapid repair of Pt-DNA adducts, based on the following observations: (1) Using AAS, both naïve and long-term cisplatin-treated tumors had similar levels of platinum following cisplatin treatment, ruling out resistance mechanisms based on platinum entry/export. (2) Analysis of adduct kinetics by immunofluorescence demonstrated that naïve and long-term cisplatin-treated tumors had similar levels of adducts early, but that long-term-treated tumors exhibited an enhanced ability to remove adducts within 24 h after a final dose of cisplatin. (3) Chk1 phosphorylation was induced in both naïve and cisplatin-pretreated tumors, suggesting that tumors were encountering DNA damage. Notably, Chk2 phosphorylation was persistent in lung tumors that had been treated multiple times with cisplatin, but had not received cisplatin for several weeks. This suggests that high basal phosphorylation of Chk2 is associated with, and could be causally involved in, cisplatin resistance, and that damage signaling between naïve and long-term-treated tumors is fundamentally different. (4) Cisplatin-pretreated tumors induced expression of genes that have been shown to facilitate DNA repair and resistance (including Apex1, Chek2, Rad51, and Rad52, which were basally higher, and Pidd, Cdkn1a [p21], Ercc2, and Rad9, which were induced to higher levels following treatment) (Furuta et al. 2002; Bartz et al. 2006; Wagner and Karnitz 2009; Wang et al. 2009). Together, these data suggest that the predominant mechanism of acquired resistance in this model is enhanced damage repair.
While our data suggest that import/export and translesional bypass are not frequent mechanisms of resistance, our data do not exclude the possibility that factors in addition to enhanced DNA damage repair may also contribute to resistance. For example, our gene expression analysis suggests that changes in glutathione metabolism and immune response may alter drug response. Furthermore, we observe heterogeneity in adduct formation in resistant tumors in response to a final challenge of cisplatin. In particular, a subset of resistant tumors (∼20%) have reduced adduct levels even at early time points (4 h) post-cisplatin (Fig. 4H). We hypothesize that detoxification of cisplatin by increased glutathione expression may be involved in reducing adduct formation in these tumors. This model will be useful for testing the role of other drug resistance mechanisms in vivo.
Our gene expression data suggested that Pidd induction correlated with and may play a role in chemotherapy resistance in vivo. We demonstrate for the first time that overexpression of PIDD in human lung tumor cells can facilitate cisplatin resistance. In the context of DNA damage, PIDD has been implicated previously in apoptosis, survival, NHEJ, and the G2/M checkpoint. Further studies will be necessary to determine whether PIDD-induced chemoresistance is related to its effects on prosurvival NF-κB signaling, the cell cycle, and/or DNA damage arrest and repair. Functional studies will be necessary to elucidate whether PIDD expression is sufficient to induce chemotherapy resistance in vivo, and whether inhibition of PIDD function could potentially have therapeutic applications by synergizing with chemotherapy treatment.
In summary, we established and characterized a model system for studying response and acquired resistance to cisplatin in lung cancer. In vivo treatment with cisplatin in this model recapitulates important features that are seen in the treatment of human lung cancer. Specifically, tumors acquire resistance to cisplatin after prolonged treatment, and this is associated with cross-resistance to other platinum analogs. This model will be useful for comparing the efficacy of novel platinum compounds and combination therapies, as well as their impact on the emergence of drug resistance.
Materials and methods
Mouse breeding and drug treatment
Mice were housed in an environmentally controlled room according to the Committee of Animal Care. All mice were bred onto a 129svJae background. Mice were infected with 3 × 107 plaque-forming units (PFU) of AdCre (University of Iowa) by nasal instillation as described previously (Jackson et al. 2001) and allowed to develop tumors for 12–16 wk prior to cisplatin treatment. Mice were given freshly prepared cisplatin in PBS at 7 mg/kg body weight ip as indicated (Sigma; prepared from K2PtCl4 supplied as a gift from Engelhard Corporation, now BASF) or carboplatin (50 mg/kg body weight in saline; Sigma). For BrdU labeling experiments, BrdU (Sigma) was injected ip (30 mg/kg) 24 h prior to sacrifice.
Immunohistochemistry
Antibodies and experimental conditions for immunohistochemistry are described in the Supplemental Material.
MicroCT
At indicated time points, mice were scanned for 15 min under isoflurane anesthesia using a small animal eXplore Locus microCT (GE Healthcare) at 45-μm resolution, 80 kV, with 450-μA current. Images were acquired and processed using GE eXplore software.
DNA copy number analysis
LSL-KrasG12D/+ mice were treated with long-term PBS or cisplatin (four total doses over 2 mo). After the fourth dose of cisplatin, mice were aged for ∼4–8 wk in order to allow residual tumors to increase in volume. Mice were sacrificed, and individual lung tumors were microdissected from the lung surface and snap-frozen. DNA was isolated from individual lung tumors and tail samples from the same animal using the Puregene DNA isolation kit (Gentra Systems). Genomic DNA was phenol-chloroform-extracted three times and submitted to Cold Spring Harbor Laboratories for ROMA. Briefly, DNA was digested using BglII enzyme, PCR-amplified using universal adaptors and primers, and labeled with fluorophores Cy3 or Cy5 (Lakshmi et al. 2006). Tumor and tail samples were hybridized onto NimbleGen chips containing 85,000 mouse probes. Lung tumor DNA was compared with tail DNA from the same animal. Raw array data were processed and normalized according to Lakshmi et al. (2006). A moving-median algorithm based on a window of five data points was used to smoothen the normalized data to visualize copy number gains and losses (Kendall et al. 2007).
Gene expression analysis
Mice were sacrificed by cervical dislocation. Lungs were inflated with RNAlater (Ambion), removed, and placed in the same solution. Visible tumors were microdissected and frozen immediately on dry ice. Frozen tumor samples were thawed in Trizol solution (Invitrogen), and then homogenized using first a Kontes pestle and then a polytron homogenizer. RNA and DNA were isolated from Trizol using the manufacturer's instructions. RNA was further purified using a Qiagen column. RNA was reverse-transcribed, linearly amplified, and labeled with biotin prior to hybridization to oligonucleotide using an Ovation amplification kit (Nugen). All samples were hybridized to Affymetrix 430A arrays. For the gene expression trial time course by laser capture, tumors were isolated from KrasLA2 mice (Johnson et al. 2001).
Microarray expression data were validated on at least six independent tumors per treatment group by real-time RT–PCR. RNA was isolated by Trizol as described, and 1 μg of total RNA was converted to cDNA using iScript cDNA synthesis kit (Bio-Rad). Real-time RT–PCR was performed using gene-specific primers and Sybr Green Supermix (Bio-rad) in triplicate on an iCycler real-time machine (Bio-Rad). Analysis was performed using iCycler software, and expression values were based on 10-fold serial dilutions of standards and normalized to Actin levels. Human and mouse primers are included in the Supplemental Material.
Statistical analysis
All statistical analyses were performed using Graphpad Prism 5.0. For column statistics to determine P-values, unpaired two-tailed Student's t-tests were performed. For survival curves, log-rank (Mantel-Cox) test was performed. For IC50 analysis, nonlinear fit-log(agonist) versus normalized response (variable slope) was performed.
Cell culture
Human NSCLC lines (H460, SW1573, and A549) were cultured according to the American Type Culture Collection. Cells were infected with retroviruses MSCV-Puro or MSCV-Puro-PIDD (Tinel and Tschopp 2004) and selected with puromycin. For viability assays, cells were seeded in triplicate (6 × 103 per well) in an opaque 96-well plate and treated the next day with increasing doses of cisplatin (0–200 μM). After 48 h of treatment, cell viability was measured using CellTiter-Glo (Promega) on a luminometer. PIDD overexpression was validated by Western blotting using antibodies to Flag (M2 clone, Sigma), PIDD (Anto-1 clone, Alexis), and PARP1 (46D11, Cell Signaling Technologies). For nuclear and cytoplasmic fractionations, lysates were prepared as described in the Supplemental Material.
Acknowledgments
We thank Etienne Meylan for helpful discussions and critical reading of the manuscript, and Antoine Tinel for kindly providing PIDD constructs. We thank the Koch Institute Core Facilities for technical support, including Eliza Vasile (microscopy) and Glenn Paradis (flow cytometry). This work was supported by the National Institutes of Health and the National Cancer Institute (5-UO1-CA84306 to T.J. and CA034992 to S.J.L.), and in part by Cancer Center Support (core) grant P30-CA14051 (T.J.) and a grant from the J. Manchot Foundation and Deutsche Forschungsgemeinschaft (J.T.). T.J. and T.G. are Investigators of the Howard Hughes Medical Institute, and T.J. is a Daniel K. Ludwig Scholar. A.S.C. was supported by grants from the Robert Woods Johnson Foundation (Harold Amos Medical Faculty Development Program), by a mentored clinical scientist grant from the National Cancer Institute, and by an American Cancer Society Research Scholar Award. T.O. is an ASPET-Merck post-doctoral fellow.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1897010.
Supplemental material is available at http://www.genesdev.org.
References
- Ahrendt SA, Chow JT, Yang SC, Wu L, Zhang MJ, Jen J, Sidransky D 2000. Alcohol consumption and cigarette smoking increase the frequency of p53 mutations in non-small cell lung cancer. Cancer Res 60: 3155–3159 [PubMed] [Google Scholar]
- American Cancer Society 2007. American Cancer Society Cancer Facts and Figures 2007. American Cancer Society, Atlanta, GA [Google Scholar]
- Bando T, Fujimura M, Kasahara K, Matsuda T 1998. Significance of Na+, K+-ATPase on intracellular accumulation of cis-diamminedichloroplatinum(II) in human non-small-cell but not in small-cell lung cancer cell lines. Anticancer Res 18: 1085–1089 [PubMed] [Google Scholar]
- Bartz SR, Zhang Z, Burchard J, Imakura M, Martin M, Palmieri A, Needham R, Guo J, Gordon M, Chung N, et al. 2006. Small interfering RNA screens reveal enhanced cisplatin cytotoxicity in tumor cells having both BRCA network and TP53 disruptions. Mol Cell Biol 26: 9377–9386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blommaert FA, van Dijk-Knijnenburg HC, Dijt FJ, den Engelse L, Baan RA, Berends F, Fichtinger-Schepman AM 1995. Formation of DNA adducts by the anticancer drug carboplatin: Different nucleotide sequence preferences in vitro and in cells. Biochemistry 34: 8474–8480 [DOI] [PubMed] [Google Scholar]
- Brugarolas J, Chandrasekaran C, Gordon JI, Beach D, Jacks T, Hannon GJ 1995. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature 377: 552–557 [DOI] [PubMed] [Google Scholar]
- Dabholkar M, Vionnet J, Bostick-Bruton F, Yu JJ, Reed E 1994. Messenger RNA levels of XPAC and ERCC1 in ovarian cancer tissue correlate with response to platinum-based chemotherapy. J Clin Invest 94: 703–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzagnidze A, Katsarava Z, Makhalova J, Liedert B, Yoon MS, Kaube H, Limmroth V, Thomale J 2007. Repair capacity for platinum-DNA adducts determines the severity of cisplatin-induced peripheral neuropathy. J Neurosci 27: 9451–9457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards SL, Brough R, Lord CJ, Natrajan R, Vatcheva R, Levine DA, Boyd J, Reis-Filho JS, Ashworth A 2008. Resistance to therapy caused by intragenic deletion in BRCA2. Nature 451: 1111–1115 [DOI] [PubMed] [Google Scholar]
- Furuta T, Ueda T, Aune G, Sarasin A, Kraemer KH, Pommier Y 2002. Transcription-coupled nucleotide excision repair as a determinant of cisplatin sensitivity of human cells. Cancer Res 62: 4899–4902 [PubMed] [Google Scholar]
- Gong JG, Costanzo A, Yang HQ, Melino G, Kaelin WG Jr, Levrero M, Wang JY 1999. The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature 399: 806–809 [DOI] [PubMed] [Google Scholar]
- Hall MD, Okabe M, Shen DW, Liang XJ, Gottesman MM 2008. The role of cellular accumulation in determining sensitivity to platinum-based chemotherapy. Annu Rev Pharmacol Toxicol 48: 495–535 [DOI] [PubMed] [Google Scholar]
- Hayakawa J, Mittal S, Wang Y, Korkmaz KS, Adamson E, English C, Ohmichi M, McClelland M, Mercola D 2004. Identification of promoters bound by c-Jun/ATF2 during rapid large-scale gene activation following genotoxic stress. Mol Cell 16: 521–535 [DOI] [PubMed] [Google Scholar]
- Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA 2008. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer 8: 193–204 [DOI] [PubMed] [Google Scholar]
- Henry-Mowatt J, Jackson D, Masson JY, Johnson PA, Clements PM, Benson FE, Thompson LH, Takeda S, West SC, Caldecott KW 2003. XRCC3 and Rad51 modulate replication fork progression on damaged vertebrate chromosomes. Mol Cell 11: 1109–1117 [DOI] [PubMed] [Google Scholar]
- Holzer AK, Manorek GH, Howell SB 2006. Contribution of the major copper influx transporter CTR1 to the cellular accumulation of cisplatin, carboplatin, and oxaliplatin. Mol Pharmacol 70: 1390–1394 [DOI] [PubMed] [Google Scholar]
- Ishida S, Lee J, Thiele DJ, Herskowitz I 2002. Uptake of the anticancer drug cisplatin mediated by the copper transporter Ctr1 in yeast and mammals. Proc Natl Acad Sci 99: 14298–14302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T, Tuveson DA 2001. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes & Dev 15: 3243–3248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson EL, Olive KP, Tuveson DA, Bronson R, Crowley D, Brown M, Jacks T 2005. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res 65: 10280–10288 [DOI] [PubMed] [Google Scholar]
- Janssens S, Tinel A, Lippens S, Tschopp J 2005. PIDD mediates NF-κB activation in response to DNA damage. Cell 123: 1079–1092 [DOI] [PubMed] [Google Scholar]
- Johnson L, Mercer K, Greenbaum D, Bronson RT, Crowley D, Tuveson DA, Jacks T 2001. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature 410: 1111–1116 [DOI] [PubMed] [Google Scholar]
- Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, Berns A 2001. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet 29: 418–425 [DOI] [PubMed] [Google Scholar]
- Kelland L 2007. The resurgence of platinum-based cancer chemotherapy. Nat Rev Cancer 7: 573–584 [DOI] [PubMed] [Google Scholar]
- Kendall J, Liu Q, Bakleh A, Krasnitz A, Nguyen KC, Lakshmi B, Gerald WL, Powers S, Mu D 2007. Oncogenic cooperation and coamplification of developmental transcription factor genes in lung cancer. Proc Natl Acad Sci 104: 16663–16668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharbanda S, Ren R, Pandey P, Shafman TD, Feller SM, Weichselbaum RR, Kufe DW 1995. Activation of the c-Abl tyrosine kinase in the stress response to DNA-damaging agents. Nature 376: 785–788 [DOI] [PubMed] [Google Scholar]
- Lakshmi B, Hall IM, Egan C, Alexander J, Leotta A, Healy J, Zender L, Spector MS, Xue W, Lowe SW, et al. 2006. Mouse genomic representational oligonucleotide microarray analysis: Detection of copy number variations in normal and tumor specimens. Proc Natl Acad Sci 103: 11234–11239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong CO, Vidnovic N, DeYoung MP, Sgroi D, Ellisen LW 2007. The p63/p73 network mediates chemosensitivity to cisplatin in a biologically defined subset of primary breast cancers. J Clin Invest 117: 1370–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis AD, Hayes JD, Wolf CR 1988. Glutathione and glutathione-dependent enzymes in ovarian adenocarcinoma cell lines derived from a patient before and after the onset of drug resistance: Intrinsic differences and cell cycle effects. Carcinogenesis 9: 1283–1287 [DOI] [PubMed] [Google Scholar]
- Liedert B, Pluim D, Schellens J, Thomale J 2006. Adduct-specific monoclonal antibodies for the measurement of cisplatin-induced DNA lesions in individual cell nuclei. Nucleic Acids Res 34: e47 doi: 10.1093/nar/gkl051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Ma W, Benchimol S 2000. Pidd, a new death-domain-containing protein, is induced by p53 and promotes apoptosis. Nat Genet 26: 122–127 [DOI] [PubMed] [Google Scholar]
- Lippard SJ 1982. New chemistry of an old molecule: Cis-[Pt(NH3)2Cl2]. Science 218: 1075–1082 [DOI] [PubMed] [Google Scholar]
- Mabuchi S, Ohmichi M, Nishio Y, Hayasaka T, Kimura A, Ohta T, Saito M, Kawagoe J, Takahashi K, Yada-Hashimoto N, et al. 2004. Inhibition of NFκB increases the efficacy of cisplatin in in vitro and in vivo ovarian cancer models. J Biol Chem 279: 23477–23485 [DOI] [PubMed] [Google Scholar]
- Martin LP, Hamilton TC, Schilder RJ 2008. Platinum resistance: The role of DNA repair pathways. Clin Cancer Res 14: 1291–1295 [DOI] [PubMed] [Google Scholar]
- Olaussen KA, Dunant A, Fouret P, Brambilla E, Andre F, Haddad V, Taranchon E, Filipits M, Pirker R, Popper HH, et al. 2006. DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N Engl J Med 355: 983–991 [DOI] [PubMed] [Google Scholar]
- Olive KP, Jacobetz MA, Davidson CJ, Gopinathan A, McIntyre D, Honess D, Madhu B, Goldgraben MA, Caldwell ME, Allard D, et al. 2009. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science 324: 1457–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabla N, Huang S, Mi QS, Daniel R, Dong Z 2008. ATR–Chk2 signaling in p53 activation and DNA damage response during cisplatin-induced apoptosis. J Biol Chem 283: 6572–6583 [DOI] [PubMed] [Google Scholar]
- Perego P, Giarola M, Righetti SC, Supino R, Caserini C, Delia D, Pierotti MA, Miyashita T, Reed JC, Zunino F 1996. Association between cisplatin resistance and mutation of p53 gene and reduced bax expression in ovarian carcinoma cell systems. Cancer Res 56: 556–562 [PubMed] [Google Scholar]
- Pestell KE, Hobbs SM, Titley JC, Kelland LR, Walton MI 2000. Effect of p53 status on sensitivity to platinum complexes in a human ovarian cancer cell line. Mol Pharmacol 57: 503–511 [DOI] [PubMed] [Google Scholar]
- Rosenberg B, VanCamp L, Trosko JE, Mansour VH 1969. Platinum compounds: A new class of potent antitumour agents. Nature 222: 385–386 [DOI] [PubMed] [Google Scholar]
- Rottenberg S, Nygren AO, Pajic M, van Leeuwen FW, van der Heijden I, van de Wetering K, Liu X, de Visser KE, Gilhuijs KG, van Tellingen O, et al. 2007. Selective induction of chemotherapy resistance of mammary tumors in a conditional mouse model for hereditary breast cancer. Proc Natl Acad Sci 104: 12117–12122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai W, Swisher EM, Karlan BY, Agarwal MK, Higgins J, Friedman C, Villegas E, Jacquemont C, Farrugia DJ, Couch FJ, et al. 2008. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature 451: 1116–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt CA, Rosenthal CT, Lowe SW 2000. Genetic analysis of chemoresistance in primary murine lymphomas. Nat Med 6: 1029–1035 [DOI] [PubMed] [Google Scholar]
- Schmitt CA, Fridman JS, Yang M, Lee S, Baranov E, Hoffman RM, Lowe SW 2002. A senescence program controlled by p53 and p16INK4a contributes to the outcome of cancer therapy. Cell 109: 335–346 [DOI] [PubMed] [Google Scholar]
- Selvakumaran M, Pisarcik DA, Bao R, Yeung AT, Hamilton TC 2003. Enhanced cisplatin cytotoxicity by disturbing the nucleotide excision repair pathway in ovarian cancer cell lines. Cancer Res 63: 1311–1316 [PubMed] [Google Scholar]
- Shi M, Vivian CJ, Lee KJ, Ge C, Morotomi-Yano K, Manzl C, Bock F, Sato S, Tomomori-Sato C, Zhu R, et al. 2009. DNA-PKcs-PIDDosome: A nuclear caspase-2-activating complex with role in G2/M checkpoint maintenance. Cell 136: 508–520 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Shulga N, Wilson-Smith R, Pastorino JG 2009. Hexokinase II detachment from the mitochondria potentiates cisplatin induced cytotoxicity through a caspase-2 dependent mechanism. Cell Cycle 8: 3355–3364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaug V, Ryberg D, Kure EH, Arab MO, Stangeland L, Myking AO, Haugen A 2000. p53 mutations in defined structural and functional domains are related to poor clinical outcome in non-small cell lung cancer patients. Clin Cancer Res 6: 1031–1037 [PubMed] [Google Scholar]
- Socinski MA 2004. Cytotoxic chemotherapy in advanced non-small cell lung cancer: A review of standard treatment paradigms. Clin Cancer Res 10: 4210s–4214s [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. 2005. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102: 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet-Cordero A, Mukherjee S, Subramanian A, You H, Roix JJ, Ladd-Acosta C, Mesirov J, Golub TR, Jacks T 2005. An oncogenic KRAS2 expression signature identified by cross-species gene-expression analysis. Nat Genet 37: 48–55 [DOI] [PubMed] [Google Scholar]
- Sweet-Cordero A, Tseng GC, You H, Douglass M, Huey B, Albertson D, Jacks T 2006. Comparison of gene expression and DNA copy number changes in a murine model of lung cancer. Genes Chromosomes Cancer 45: 338–348 [DOI] [PubMed] [Google Scholar]
- Teicher BA, Herman TS, Holden SA, Wang YY, Pfeffer MR, Crawford JW, Frei E 3rd, 1990. Tumor resistance to alkylating agents conferred by mechanisms operative only in vivo. Science 247: 1457–1461 [DOI] [PubMed] [Google Scholar]
- Tinel A, Tschopp J 2004. The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science 304: 843–846 [DOI] [PubMed] [Google Scholar]
- Tinel A, Janssens S, Lippens S, Cuenin S, Logette E, Jaccard B, Quadroni M, Tschopp J 2007. Autoproteolysis of PIDD marks the bifurcation between pro-death caspase-2 and pro-survival NF-κB pathway. EMBO J 26: 197–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao MS, Aviel-Ronen S, Ding K, Lau D, Liu N, Sakurada A, Whitehead M, Zhu CQ, Livingston R, Johnson DH, et al. 2007. Prognostic and predictive importance of p53 and RAS for adjuvant chemotherapy in non small-cell lung cancer. J Clin Oncol 25: 5240–5247 [DOI] [PubMed] [Google Scholar]
- Wagner JM, Karnitz LM 2009. Cisplatin-induced DNA damage activates replication checkpoint signaling components that differentially affect tumor cell survival. Mol Pharmacol 76: 208–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Lippard SJ 2005. Cellular processing of platinum anticancer drugs. Nat Rev Drug Discov 4: 307–320 [DOI] [PubMed] [Google Scholar]
- Wang D, Xiang DB, Yang XQ, Chen LS, Li MX, Zhong ZY, Zhang YS 2009. APE1 overexpression is associated with cisplatin resistance in non-small cell lung cancer and targeted inhibition of APE1 enhances the activity of cisplatin in A549 cells. Lung Cancer 66: 298–304 [DOI] [PubMed] [Google Scholar]
- Welsh C, Day R, McGurk C, Masters JR, Wood RD, Koberle B 2004. Reduced levels of XPA, ERCC1 and XPF DNA repair proteins in testis tumor cell lines. Int J Cancer 110: 352–361 [DOI] [PubMed] [Google Scholar]