Abstract
Following our previous study (Piquemal et al., New J. Chem., 2003, 27, 909), we present here a DFT study of the inhibition of the Tyrosinase enzyme. Broken-symmetry DFT computations are supplemented with Spin-Flip TD-DFT calculations, which, for the first time, are applied to such a dicopper enzyme. The chosen biomimetic model encompasses a dioxygen molecule, two Cu(II) cations, and six imidazole rings. The docking energy of a natural substrate, namely phenolate, together with those of several inhibitor and non-inhibitor compounds, are reported and show the ability of the model to rank the most potent inhibitors in agreement with experimental data. With respect to broken-symmetry calculations, the Spin-Flip TD-DFT approach reinforces the possibility for theory to point out potent inhibitors: the need for the deprotonation of the substrates, natural or inhibitors, is now clearly established. Moreover, Electron Localization Function (ELF) topological analysis computations are used to deeply track the particular electronic distribution of the Cu-O-Cu three-center bonds involved in the enzymatic Cu2O2 metallic core (Piquemal and Pilmé, J. Mol. Struct.: Theochem, 2006, 77, 764). It is shown that such bonds exhibit very resilient out-of-plane density expansions that play a key role in docking interactions: their 3D-orientation could be the topological electronic signature of oxygen activation within such systems.
Keywords: Density Functional Theory, Spin-Flip TD-DFT, copper, oxygenase, Tyrosinase, competitive inhibition
Introduction
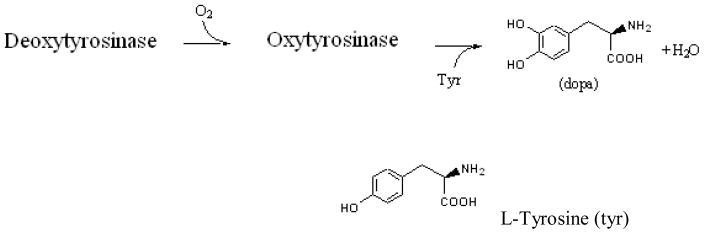
Tyrosinase is a monophenol oxygenase (IUBMB enzyme Nomenclature: EC 1.14.18.1) found widely within the living realm (Lerch, 1987; Dugas, 1989; Solomon et al., 2001). This enzyme is involved, for instance, in the formation of the melanin shield protecting plants from external stress, and is also responsible for the brown color of fruits upon contact with air (O2). Human Tyrosinase can be found in skin melanocytes or in the iris of eyes; it is responsible for the dark color of the cerebellum. Several mutations are associated with known pathologies, such as albinism which is linked to a thermal instability of the enzyme at physiological temperature. [Giebel et al., 1991 and references therein]. The chemical role of Tyrosinase is to catalyze the ortho-hydroxylation of monophenols into ortho-diphenols or, alternatively, the oxidation of ortho-diphenols into the corresponding ortho-quinones. In mammals, it performs for instance the hydroxylation of L-tyrosine into 3-(3,4-dihydroxyphenyl)-L-alanine (DOPA) which is oxidized into DOPA quinone (catechol oxidase activity), as depicted in Figure 1 (Decker and Tuczek, 2000; Kim and Uyama, 2005; and references therein).
Fig. 1.
Enzymatic activity of Tyrosinase
From a structural viewpoint, tyrosinase is classified as a type III copper enzyme because of the presence of two coupled copper cations in its active site, whose role is to activate dioxygen to initiate catalytic activity.(Ross et al., 1991). Two forms of the enzyme have thus to be considered: a deoxy state for the native protein, and an oxy state when dioxygen binds. The deoxy form is EPR-silent, and its UV-visible spectra does not exhibit any charge transfer bands. These findings are consistent with a singlet spin state and formal Cu(I)/3d10 redox states of the copper cations. The oxy form also is EPR-silent, but exhibits a much more complex electronic structure: it encompasses a peroxide ligand (O22−) bridging two formal Cu(II)/(3d9) cations exhibiting a strong antiferromagnetic coupling (Gherman and Cramer, 2009; Piquemal et al., 2006; and references therein).
The structure-function analysis of the enzyme has been hampered for several years due to the lack of any crystallographic data: the first crystallographic structure has been reported only recently (Matoba et al., 2006). Nevertheless, several cross data including genetic sequence homology and X-ray spectroscopy (X absorption, EXAFS and XANES) had shown strong structural similarities between Tyrosinase and Hemocyanin (Decker and Tuczek, 2000 and references herein). The latter is also a class III dicopper protein and plays a role of oxygen carrier in mollusk and arthropod hemolymph. When the structure of Panulirus Interruptus hemocyamin was solved (Volbeda and Hol, 1989), it allowed a clear-cut picture of the bimetallic active site that appeared to involve six histidine residues. Based on these structures several bio-inspired models have been synthesized (Kitajima and Morooka, 1994; Karlin et al., 1998; Murthy et al., 2001; Palavinici et al., 2005; Mirica et al., 2006; Tolman, 2006; and references therein), some of them have been found to hydroxylate phenol derivatives to the corresponding quinones. Extensive theoretical studies were also performed focusing on the structure and reactivity of compounds exhibiting a Cu2O2 core. The computational approaches however have to face the complex electronic structures associated with such bimetallic cores and adequate quantum chemistry tools need to be employed (Gherman and Cramer, 2009). However, beyond the interest of theoreticians for such subtle electronic structures and as the crystal structure has been recently solved (Decker et al., 2006), little remains known about the effective mechanism of the enzyme. Indeed, understanding and inhibiting Tyrosinase would be important in medicine due to its clear involvement in Parkinson’s disease (Xu et al., 1997), melanoma (Prezioso et al., 1992), and hyperpigmentation phenomenon (Maeda and Fukuda, 1991). Moreover, inhibiting Tyrosinase can prevent the unwanted darkening of fruits and seafood, which has an important financial cost (Friedman, 1996). A computational approach would thus help to understand the details of such inhibition
Building on the immense amount of work available, we started to work on this problem in 2003 (Piquemal et al., 2003) and showed on the basis of Density Functional Theory (DFT) calculations, that it is possible to predict computationally the inhibition of a model of the enzyme by 2-aminophenol (2-APOH). It was first shown that both the substrate and the inhibitor should be deprotonated to form a stable complex with the active site. Second, it was shown that only phenolate binds to the oxy form. We finally suggested a competitive inhibition mechanism relying on the deprotonation of the substrates. Recently, such predictions have found a clear experimental confirmation (Mirica et al., 2006). In the present contribution, we report an extension of our previous study to other inhibitors, and compare the theoretical inhibition hierarchy to the experimental one. To improve our previous work, two levels of computations are employed here: a broken-symmetry approach (BS-DFT) and the recently introduced spin-flip TD-DFT technique (Shao et al., 2003; Wang and Ziegler, 2004; Neese, 2009), hereafter referred to as SF-DFT. To go deeper into the understanding of the inhibition mechanism, the analysis of the electronic structures is complemented by the Electron Localization Function (ELF) topological analysis (Becke and Edgecombe, 1990; Silvi and Savin, 1994). Such an analysis is expected to provide some insight into the particular role of the peculiar 3-center Cu-O-Cu bonds within the Cu2O2 core (Piquemal and Pilmé, 2006) in the regioselective approach of the substrate or inhibitors.
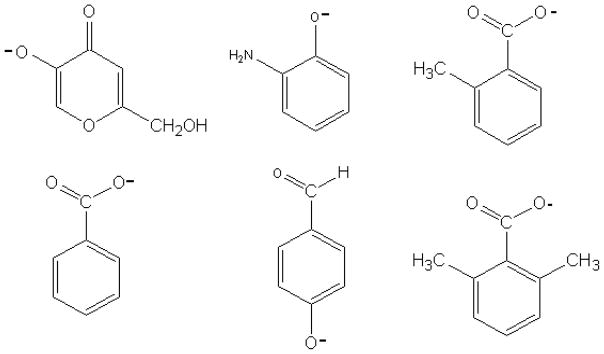
Several inhibitors having demonstrated experimental potency thanks to their structural similarity to phenol, a model of the tyrosine side-chain, will be tested under their deprotonated form, namely 2-aminophenol (2-APOH), benzoic and o-toluic acids; kojic acid (5-hydroxy-2-hydroxymethyl-4-pyrone) used in cosmetics, and 4-hydroxybenzaldehyde. A known non-inhibitor compound, 2,6-dimethylbenzoic acid, is also investigated to test the ability of the model to discriminate between inhibitor and non-inhibitor molecules. All compounds are depicted on Figure 2.
Fig. 2.
Semi-developed formulas of inhibitors (from left to right): deprotonated kojic acid, 2-APO−, o-toluate, benzoate, deprotonated 4-hydroxybenzaldehyde, 2, 4 dimethylbenzoate (non inhibitor)
The paper is organized as follows: we will first present the model of the tyrosinase active site, describe the used methodologies, and then discuss the results.
Choice of the model and methodologies
i) Model of the active site
The bio-inspired model of the oxy-tyrosinase active site, hereafter referred to as TYR-O2, encompasses two copper cations, six imidazole ligands, and dioxygen: the resulting complex [(Cu(ImH)3)2-O2]2+ recovers that of our previous study (Piquemal et al., 2003). Such a structure was initially retained by Lind et al. (1999) to model the six histidine residues of the active site. From a structural viewpoint, this model has been shown to be in fine agreement with experimental data and is consistent with the recent X-ray structure of tyrosinase (Decker et al. 2006). As previous studies showed that the (μ-O)2 arrangement is not catalytically relevant (Tolman, 2006), we only consider here the μ2:η2-η2 form. The substrate and the inhibitors, taken in their deprotonated forms, are presented on Figure 2. No solvent effects were considered in the previous investigations as the active site of tyrosinase is known to be encapsulated in a hydrophobic pocket of the whole protein (Conrad et al., 1994).
ii) Computational details
Complete geometry optimizations were carried out using Jaguar (Jaguar 4.1, 2000) at the DFT level of computation using the B3LYP functional (Becke, 1988; Lee et al., 1998) with the LACVP** basis set (McLean et al., 1980; Rasolov et al., 1998). Broken symmetry solutions were sought to account for the open-shell nature of the Cu2O2 singlet core. Indeed, despite its known spin delocalization problem, BS-DFT has been shown to be able to capture the main electronic features of type III copper active sites (Piquemal and Pilmé, 2006; Gherman et al., 2009; and references therein). In our BS-DFT computations, spin density values on the oxygen and copper atoms are consistent with those reported for comparable active site models (Cramer et al., 2006). Actually, recent studies even showed that DFT performs better than expected to reproduce accurate structures (Rode and Werner, 2005; Cramer et al., 2006) when compared to high-level multireferential post-Hartree-Fock approaches. It has moreover been shown by means of the ELF topological analysis that the essential electronic features of the central Cu2O2 core, especially the three-centre nature of the Cu-O-Cu bonds, were conserved when going from a CASSCF to a DFT formalism (Piquemal and Pilmé, 2006).
Zero-point vibrational energy (ZPE) and basis set superposition error (BSSE) are included in the reported energies. ZPE and entropies were evaluated within the harmonic approximation at T = 298 K.
To go a step further towards accuracy we have also considered the recent Spin-Flip Time-Dependent DFT formalism (SF-DFT) as an alternative to the BS-DFT techniques (de la Lande et al., 2007; de la Lande et al., 2008a; de la Lande et al., 2008b; de la Lande et al., 2009). This approach was shown to bevery promising for the treatment of mononuclear Cu/O2 adducts found in non-coupled copper monooxygenases. The present study thus offers the opportunity to test this formalism in the case of binuclear sites. All SF-DFT computations were performed at the BS-DFT minima using the Q-Chem package (Shao et al., 2006).
Topological analysis of the Electron Localization function
In order to follow the organization of the electron density at the Cu2O2 core by means of ELF topological analyses, we have used a modified TopMod package (Noury et al., 1999; Pilmé and Piquemal, 2008). We here briefly recall that the ELF function can be interpreted as the signature of the electronic-pair distribution. Its topological analysis allows the partition of space into domains, the so-called “basins”, which are related to the intuitive chemical picture of bonds. For a given atom A, core electrons generate core basins labelled C(A). Regions associated with lone pairs are labelled V(A) and bonding regions denoting chemical bonds between atoms A and B are noted V(A,B), or V(A,B,C) in the case of three-center bonds. These ELF regions match closely the domains of the VSEPR model. Details about ELF and its applications to bioinorganic chemistry can be found in a recent review paper (Piquemal et al., 2008).
Results
1) Energetics of oxy-tyrosinase model/inhibitor complexes
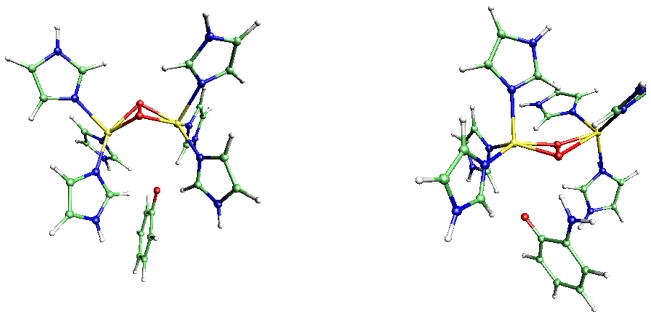
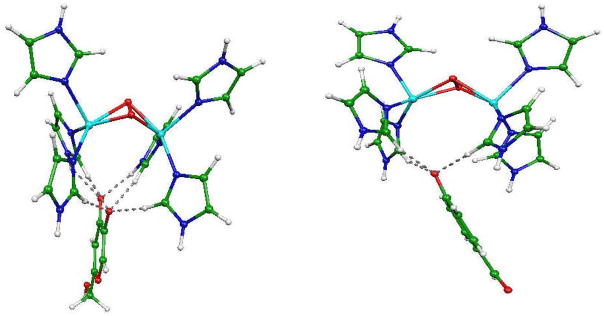
Representations of the optimized structures of the oxy-tyrosinase model/inhibitor complexes are displayed on Figures 3 to 5.
Fig. 3.
From left to right: TYR/O2/phenolate and TYR/O2/2-APO− complexes
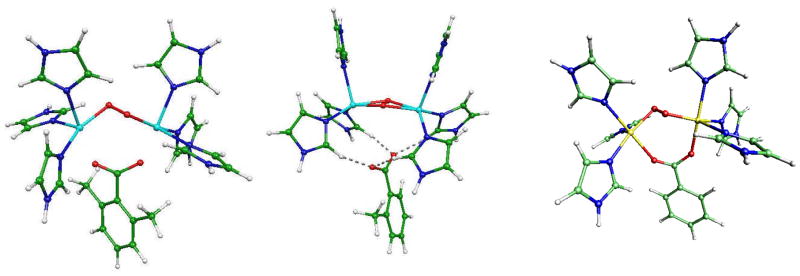
Fig. 5.
From left to right: TYR/O2/dimethylbenzoate, TYR/O2/o-toluate and TYR/O2/benzoate complexes
We computed the interaction energies (ΔE) between the tyrosinase model and its substrate/inhibitor:
To take thermodynamics (and therefore for entropic effects) into account, ΔE energies include several corrections: ΔΔG computations are then provided (at 298K). Final ΔΔG values include the ZPE and BSSE energies added to the inhibitor deprotonation energy denoted as Edeprot as the substrate and the inhibitor should be deprotonated to form a stable complex with the active site (Piquemal et al., 2003). Energetic results are gathered in Tables 1 and 2.
Table 1.
Total energies values (in a. u.) and ZPE energies (kcal.mol−1)
| Compound | E | ZPVE |
|---|---|---|
| Deprotonated 4-hydroxybenzaldehyde | −420.2423 | 68.9 |
| Deprotonated kojic acid | −532.5137 | 70.3 |
| o-toluate | −459.5796 | 87.8 |
| Benzoate | −420.2532 | 69.6 |
| 2,6-Dimethylbenzoate | −498,8870 | 105.2 |
Table 2.
ΔE an ΔΔG binding energies (kcal/mol) of the tyrosinase substrate (phenolate) and some potential inhibitors on the [(ImH)6Cu2O2]2+ complex. ΔΔG computations include the ZPE and BSSE energies added to the inhibitor deprotonation energy denoted as Edeprot. ΔE= E TYR/O2/substrat or inhibitor − (E optimized TYR/O2 + E optimized substrate or inhibitor)
| TYR/O2 complexes | ΔE BS-DFT | ΔE SF-DFT | Edeprot | ΔΔG BS-DFT | ΔΔG SF-DFT |
|---|---|---|---|---|---|
| Phenolate | −147.2 | −148.4 | 359.1 | 0 | 0 |
| 2-APO− | −152.2 | −165.9 | 355.3 | −28.8 | −41.3 |
| Deprotonated kojic c acid | −166.3 | −158.9 | 359.1 | −17.9 | −9.3 |
| Benzoate c | −141.4 | −142.0 | 351.7 | −2.4 | −3.0 |
| o-toluate c | −136.4 | −138.4 | 351.5 | 0.3 | −0.5 |
| Deprotonated 4-hydroxybenzaldehydec | −131.5 | −135 | 339.1 | −12.6 | −14.9 |
| 2,6-dimethylbenzoatec | −127.1 | −146 | 349.8 | 5.5 | 12.2 |
2-APO− clearly appears as the best inhibitor. Its stabilization is 20 kcal/mol larger compared to that of the phenolate natural substrate. 2-APO− is followed by deprotonated kojic acid and 4-hydroxybenzaldehyde, the latter being favoured by its weak deprotonation energy. It is important to point out that SF-DFT discriminates more clearly 2-APO− from the other inhibitors: an additional −12.4 kcal/mol stabilization is found whereas deprotonated kojic acid is disfavoured by +9.4 kcal/mol.
Benzoate and o-toluate appear as the worst inhibitors. Toluate presents an energetic balance slightly superior to phenolate by 0.2 kcal/mol at the BS-DFT level and by −0.2 kcal/mol with SF-DFT. The inclusion of thermal corrections switches their relative stability: deprotonated o-toluic acid is now stabilized by −0.5 kcal/mol. However, considering the levels of calculations and their intrinsinc uncertainty coupled to thermal agitation under physiological conditions, we can consider that these two compounds have the same affinity for the active site.
Dimethylbenzoate does not appear to inhibit the active site model: it has a highly unfavorable energetic balance compared to phenolate.
2) Geometric parameters and ELF topological analysis
In all the structures, the Cu2O2 cores exhibit a strong butterfly effect oriented towards the inhibitors as can be seen on Figures 3 and 4. Detailed structural data are given in Table 3 for the optimized geometries. The more pronounced effect is observed for the most potent inhibitor, i.e. 2APO−: the Cu-Cu bond length (4.31Å) is increased when compared to that in the phenolate complex (3.33Å). It can be noted that the oxygen atoms of the deprotonated kojic acid appear in interaction with the hydrogen atoms of the imidazole residues.
Fig. 4.
From left to right: TYR/O2/deprotonated kojic acid and TYR/O2/deprotonated 4-hydroxybenzaldehyde complexes
Table 3.
Geometric parameters for the optimized geometries (distances in A, angles in degrees)
| Complexes | d(Cu-Cu) | d(Cu-O) | φ(CuOOCu) |
|---|---|---|---|
| TYR/O2/deprotonated kojic acid | 3.2 | 4.33/4.34 | 111.0 |
| TYR/O2/deprotonated 4-hydroxybenzaldehyde | 3.31 | 4.3/4.06 | 118.5 |
| TYR/O2/o-toluate | 3.77 | 3.7–4.2/4.1–3.9 | 164.6 |
| TYR/O2/benzoate | 3.68 | 2.05–2.24 | 125.2 |
| TYR/O2/2,6-dimethylbenzoate | 3.88 | 2.25–3.6/2.06–3.7 | 125.2 |
| TYR/O2/phenolate | 3.33 | 3.97/4.17 | 119.0 |
| TYR/O2/2-APO− | 4.31 | 3.54/5.31 | 163.0 |
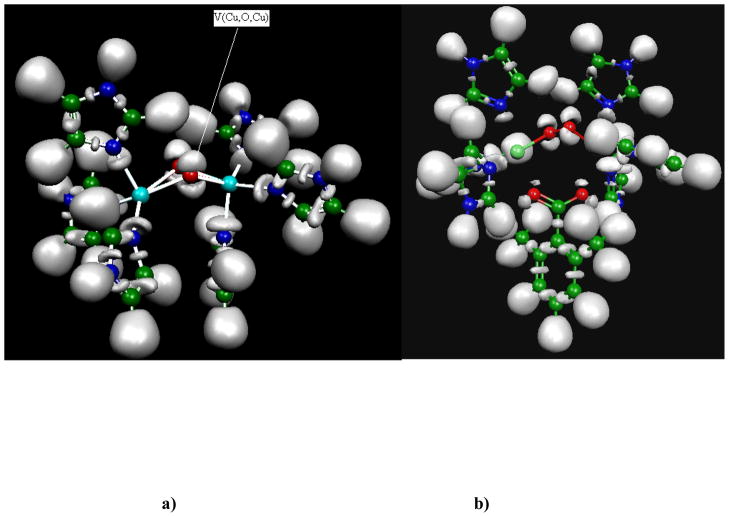
The optimized structure of the dimethylbenzoate complex shows a Cu2O2 metallic core presenting a deformation which is visually closer to a “zig-zag” form than to the usual μ–η2:η2 binding mode: the carboxylate oxygen atoms are oriented towards the copper cations. It is important to note that, despite the elongated O-O distance, an ELF analysis shows a conservation of the topological organization of the electronic structure of the μ–η2:η2 binding mode described previously (Piquemal and Pilmé. 2006). This topology involves two three-center Cu-O-Cu bonds being above and below the Cu2O2 central metallic core. This striking feature of the Cu2O2 core displayed in Figure 6 appears to be conserved in all the inhibitor/active site model calculations. In all cases, the Cu-O-Cu bond basin is oriented towards the anionic oxygen atoms of the inhibitor pointing out the importance of this unusual local organization of the electronic densities. For the non-inhibitor compound, (dimethylbenzoate complex), carboxylate oxygen atoms are oriented towards the copper atom and not toward the Cu-O-Cu bond. Nevertheless, the topology involving two three-center Cu-O-Cu bonds above and below the Cu2O2 central metallic core is conserved despite the apparent “zig-zag” configuration.
Fig. 6.
a) Plot of the ELF function for the TYR/O2 optimized complex. The topology involves two sets of three-center Cu-O-Cu bonds above and below the Cu2O2 central metallic; b) Plot of the ELF function for the TYR/O2/dimethylbenzoate complex. The topology involving two three-center Cu-O-Cu bonds above and below the Cu2O2 central metallic core is conserved despite an apparent “zig-zag” configuration.
Such a “resilient” topological organization of the electron density (“out-of-plane” vs.” in-plane”) of dioxygen is likely to be linked to the “oxygen activation” and, then, may directly influence the regioselectivity of the enzyme by governing the orientation of the substrate/inhibitor approach.
COMPARISON WITH EXPERIMENT
Overall, the results are satisfactory and the model is able to qualitatively discriminate between inhibitor and non inhibitor compounds (Table 4). We now address the quantitative agreement of the computational results with the available experimental data (Maddaluno and Faull, 1988; Conrad et al., 1994; Bubacco et al., 2000; Jimenez et al., 2001).
Table 4.
Theoretical inhibition hierarchy
| Inhibitors |
|---|
| 2-APO−(a) |
| deprotonated kojic acid (b) |
| deprotonated 4-hydroxybenzaldehyde (c) |
| o-toluate (d) |
| benzoate(e) |
| 2,6-dimethylbenzoate (f) |
| Theoretical inhibition hierarchy : |
| BS-DFT: a > b > c > d > e > f. |
| SF-DFT: a > c > b > d > e > f. |
Several apparent inhibition constants (Ki) have been reported for Tyrosinase in the literature (Table 5). A note of caution has however to be brought as the genes encoding for Tyrosinase sometimes come from different living species. Here we consider the measurements reported for mushroom Tyrosinase (Maddaluno and Faull, 1988; Conrad et al., 1994; Jimenez et al., 2001) and for Streptomyces antibioticus Tyrosinase (Bubacco et al. 2000). This is a critical point: even if the active site is highly conserved between different species, the inhibition activity of a given drug may differ, probably due to the immediate proteic environment. For example, results for the benzoic acid indicate a good inhibiting power for the mushroom but a lesser one for Streptomyces Antibioticus. This observation is in agreement with a study which associated its inhibition capability to its ability to interact favourably with the hydrophobic enzymatic pocket (Conrad et al., 1994).
Table 5.
Experimental Ki inhibition constant (μM) of the different inhibitors and experimental inhibition hierarchy for mushroom and Streptomyces antibioticus tyrosinase (see references in the text). The error bar is indicated in parentheses (when given in the literature: Maddaluno et al., 1998; Conrad et al. 1994; Bubacco et al. 2000, Jimenez et al. 2001).
| Inhibiteur | Enzyme origin | |
|---|---|---|
| Mushroom (Ki) | Streptomyces antibioticus (Ki) | |
| 2-APO− (a) | 2 | |
| Deprotonated kojic acid (b) | 3.4 | |
| Deprotonated 4-hydroxybenzaldehyde (c) | 37 (+/−10) | |
| o-toluate (d) | 6.8 | |
| p-toluate (not calculated) | 80(+/−20) | 195(+/−0.9) |
| benzoate (e) | 18 (+/−1) | 800 +/−0.9 |
| 2,6-dimethylbenzoate (f) | Non inhibitor | |
| a >d> e > c > f | b >d > e | |
The role of entropy has also to be taken into account. However, our model encompasses only a limited number of atoms through an harmonic approximation. Indeed, as can be seen from Table 6 (experimental results for the mushroom), o-toluate appears to be a more efficient inhibitor compared to benzoate thanks to a more negative ΔS value and despite a less favourable ΔH value, which finally results in an overall more favorable free enthalpy (ΔG = ΔH − T. ΔS). This trend is also observed for Streptomyces Antibioticus: o-toluate is by far a better inhibitor than benzoate. From these results, and considering that our calculations do not take into account the stabilization (or destabilization) contribution that could be brought to the energy by the environment, we have to expect an inversion of benzoate and o-toluate.
Table 6.
Experimental thermodynamic data for the inhibitor/tyrosinase interaction (Conrad et al., 1994)
| ΔG (kcal/mol) | ΔH (kcal/mol) | ΔS (cal/mol/K) | |
|---|---|---|---|
| benzoate | −6.5 | −32 | −85 |
| o-toluate | −7.1 | −25 | −60 |
The inhibition ranking obtained from our calculations (Table 4) then appears in relatively good agreement with experiment. First, the two inhibitors known to be really potent, namely 2-APO− and deprotonated kojic acid, are ranked in the two first places, which is satisfactory. The increased theoretical stabilization of 2-APO− obtained at the SF-DFT level compared to the BS-DFT approach is more in line with the extremely small experimental Ki constant. Second, the non-inhibitor compound dimethylbenzoate appears last in the ranking: this shows the capability of the model to discriminate between an inhibitor and a compound without action on the enzyme.
Finally, deprotonated 4-hydroxybenzaldehyde appears to be slightly more stabilized than benzoate probably because of the absence of lateral chains in our model. Indeed, these latter have been shown to stabilize the benzoate adduct (Conrad et al., 1994).
Conclusion
Our study of the tyrosinase shows that it is possible to use quantum chemistry to perform in silico predictive docking computation using a relatively small active site model compared to reality. SF-DFT computations were for the first time performed on such an enzyme model. Results on enzyme/substrate interactions confirm that BS-DFT is able to reasonably describe such systems as a first theoretical inhibition hierarchy has been computed in reasonable agreement with experimental inhibition constants. SF-DFT computations were shown to be able to more clearly discriminate the best inhibitor.
The present computations tend to confirm that reactivity occurs when the active site is in oxygenated form and that the substrate and inhibitors are active in their deprotonated form, which confirms our previous hypothesis. This study also illustrates the importance of taking into account environment and entropy effects, and more generally the need to consider a greater number of residues to refine the model if aiming for a finer virtual screening of the inhibitors.
The ELF topological analysis has shown that the metallic core conserves a non conventional three-center bond organization along the docking process even in the case where a zig-zag configuration seems to be present. This very resilient topology clearly indicates that this peculiar density organization plays a direct role in the interaction as the Cu2O2 core acts as a single entity. As dioxygen is known to be activated in these complexes, such Cu2O2 three-center bonds could be the topological signature of dioxygen activation. Another aspect that has to be investigated is the efficiency of alternative spin states in the oxidative reaction of the substrate. This work is currently in progress and will be reported in due time. Thanks to the availability of a crystal structure, future work will focus on the inclusion of the whole protein in the simulations using QM/MM techniques and free energy perturbation to include environment dynamics and entropy. Such an approach should enable the screening of dozens of compounds, and maybe even more as the available computational resources increase.
These results will also be important for the design of advanced molecular modelling techniques able to handle the difficult electronic structure of the enzyme (Piquemal, J.-P., Williams-Hubbard, 2003; Gresh et al., 2007; Diedrich et al. 2008).
Acknowledgments
The computations were performed at CRIHAN (Saint-Etienne-du-Rouvray, France) on project 2008011, IDRIS (Orsay, France) and NIEHS (RTP, NC, USA). This research was supported by the Intramural Research program of the NIH and NIEHS (USA). The authors are deeply indebted to Dr Claude Giessner-Prettre who initiated the project.
References
- Becke AD. Correlation energy of an inhomogeneous electron gas: A coordinate-space model. J Chem Phys. 1988;88:1053–1062. [Google Scholar]
- Becke AD, Edgecombe KE. A simple measure of electron localization in atomic and molecular systems. J Chem Phys. 1990;92:5397–5403. [Google Scholar]
- Bubacco L, Vijgenboom E, Gobin C, Tepper AWJW, Salgado J, Canters G. Kinetic and paramagnetic NMR investigations of the inhibition of Streptomyces Antibioticus tyrosinase. J Mol Cat B: Enzymatic. 2000;8:27–35. [Google Scholar]
- Conrad JS, Dawso SR, Hubbard ER, Meyers TE, Strothkamp KG. Inhibitor binding to the binuclear active site of tyrosinase: Temperature, pH, and solvent deuterium isotope effects. Biochem. 1994;33:5739–5744. doi: 10.1021/bi00185a010. [DOI] [PubMed] [Google Scholar]
- Cramer CJ, Woch M, Piecuch P, Puzzarini C, Gagliardi L. Theoretical models on the Cu2O2 torture track: Mechanistic implications for oxytyrosinase and small-molecule analogues. J Phys Chem A. 2006;110:1991–2004. doi: 10.1021/jp056791e. [DOI] [PubMed] [Google Scholar]
- Decker H, Tuczek F. Tyrosinase/catecholoxidase activity of hemocyanins: structural basis and molecular mechanism. Trends in Biochemical Sciences. 2000;25:392–397. doi: 10.1016/s0968-0004(00)01602-9. [DOI] [PubMed] [Google Scholar]
- de la Lande A, Moliner V, Parisel O. Singlet-triplet gaps in large multireference systems: Spin-flip-driven alternatives for bioinorganic modeling. J Chem Phys. 2007;126:035102. doi: 10.1063/1.2423010. [DOI] [PubMed] [Google Scholar]
- de la Lande A, Gérard H, Parisel O. How to optimize a C-H cleavage with a mononuclear copper-dioxygen adduct? Int J Quant Chem. 2008a;108:1898–1904. [Google Scholar]
- de la Lande A, Parisel O, Gérard H, Moliner V, Reinaud O. Theoretical exploration on the oxidative properties of a [(tren)CuO2]+ adduct relevant to copper monoxygenase enzymes: insights into the competitive dehydrogenation vs. hydroxylation reactive pathways. Chem Eur J. 2008b;14:6465–6473. doi: 10.1002/chem.200701595. [DOI] [PubMed] [Google Scholar]
- Decker H, Schweikardt T, Tuczek F. Angew Chemie Int Ed. 2006;45:4546. doi: 10.1002/anie.200601255. [DOI] [PubMed] [Google Scholar]
- de la Lande A, Salahub D, Moliner V, Gerard H, Piquemal J-P, Parisel O. Dioxygen activation by mononuclear copper enzymes: Insights from a tripodal ligand mimicking their CuM coordination sphere. Inorg Chem. 2009;48:7003–7005. doi: 10.1021/ic900567z. [DOI] [PubMed] [Google Scholar]
- Diedrich C, Deeth RJ. On the performance of Ligand Field Molecular Mechanics for model complexes containing the peroxido-bridged [Cu2O2]2+ center. Inorg Chem. 2008;47:2494–2506. doi: 10.1021/ic701803g. [DOI] [PubMed] [Google Scholar]
- Dugas H. A Chemical Approach to Enzyme Action. Springer-Verlag; New-York: 1989. Bioorganic Chemistry. [Google Scholar]
- Friedman M. Food browning and its prevention: An overview. J Agric Food Chem. 1996;44:31–653. [Google Scholar]
- Gherman BF, Cramer CJ. Quantum chemical studies of molecules incorporating a Cu2O22+ core. Coord Chem Rev. 2009;253:723–753. [Google Scholar]
- Giebel LB, Tripathi, King RA, Spritz RA. A tyrosinase gene missense mutation in temperature-sensitive type I oculocutaneous albinism. A human homologue to the Siamese cat and the Himalayan mouse. J Clin Invest. 1991;87:1119–1122. doi: 10.1172/JCI115075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresh N, Cisneros GA, Darden TA, Piquemal J-P. Anisotropic, polarizable molecular mechanics studies of inter-, intra-molecular interactions, and ligand-macromolecule complexes. A bottom-up strategy. J Chem Theory Comput. 2007;3:1960–1986. doi: 10.1021/ct700134r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaguar 4.1. Schrodinger Inc; Portland OR, USA: 2000. [Google Scholar]
- Jimenez M, Chazarra S, Escribano J, Cabanes J, Garcia-Carmona FJ. Competitive inhibition of mushroom tyrosinase by 4-substituted benzaldehydes. Agric Food Chem. 2001;49:4060–4063. doi: 10.1021/jf010194h. [DOI] [PubMed] [Google Scholar]
- Karlin KD, Lee D-H, Obias HV, Humphreys KJ. Copper-dioxygen complexes: functional models for proteins. Pure Appl Chem. 1998;70:855–862. [Google Scholar]
- Kim Y-J, Uyama H. Tyrosinase inhibitors from natural and synthetic sources: structure, inhibition mechanism and perspective for the future. CMLS. 2005;62:1707–1723. doi: 10.1007/s00018-005-5054-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima N, Morooka Y. Copper dioxygen complexes. Inorganic and bioinorganic perspectives. Chem Rev. 1994;94:737–757. [Google Scholar]
- Krishnan R, Binkley JS, Seeger R, Pople JA. Self-consistent molecular orbital methods. A basis set for correlated wave functions. J Chem Phys. 1980;72:650–654. [Google Scholar]
- Lee C, Yang W, Parr RG. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys Rev B. 1988;37:785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- Lerch K. Molecular and active site structure of tyrosinase. Life Chem Rep. 1987;5:221–234. [Google Scholar]
- Lind T, Siegbahn PEM, Crabtree RH. A quantum chemical study of the mechanism of tyrosinase. J Phys Chem B. 1999;103:1193–1202. [Google Scholar]
- Maddaluno J, Faull KF. Inhibition of mushroom tyrosinase by 3-amino-L-tyrosine: molecular probing of the active site of the enzyme. Experientia. 1988;44:885–887. doi: 10.1007/BF01941189. [DOI] [PubMed] [Google Scholar]
- Maeda K, Fukuda M. In vitro effectiveness of several whitening cosmetic components in human melanocytes. J Soc Cosmet Chem. 1991;42:361–368. [Google Scholar]
- Matoba Y, Kumagai T, Yamamoto A, Yoshitsu H, Sugiyama M. Crystallographic evidence that the dinuclear copper center of tyrosinase is flexible during catalysis. J Biol Chem. 2006;281:8981–8990. doi: 10.1074/jbc.M509785200. [DOI] [PubMed] [Google Scholar]
- McLean AD, Chandler GS. Contracted Gaussian basis sets for molecular calculations. I. Second row atoms, Z=11–18. J Chem Phys. 1980;72:5639–5648. [Google Scholar]
- Mirica LM, Rudd DJ, Vance MA, Solomon EI, Hodgson KO, Hedman B, Stack TDP. μ-η2:η2-Peroxodicopper(II) complex with a secondary diamine ligand: a functional model of tyrosinase. J Am Chem Soc. 2006;128:2654–2665. doi: 10.1021/ja056740v. [DOI] [PubMed] [Google Scholar]
- Murthy NN, Mahroof-Tahir M, Karlin KD. Dicopper(I) complexes of unsymmetrical binucleating ligands and their dioxygen reactivities. Inorg Chem. 2001;40:628–635. doi: 10.1021/ic000792y. [DOI] [PubMed] [Google Scholar]
- Neese F. Prediction of molecular spectra and molecular properties with Density Functional Theory: From fundamental theory to exchange coupling. Coord Chem Rev. 2009;253:526–563. [Google Scholar]
- Noury S, Krokidis X, Fuster F, Silvi B. Computational tools for the electron localization function topological analysis. Comput Chem. 1999;23:597–604. [Google Scholar]
- Palavicini S, Granata A, Monzani E, Casella L. Hydroxylation of phenolic compounds by a peroxodicopper(II) complex: Further insight into the mechanism of tyrosinase. J Am Chem Soc. 2005;127:18031–18036. doi: 10.1021/ja0544298. [DOI] [PubMed] [Google Scholar]
- Pilmé J, Piquemal J-P. Advancing beyond charge analysis using the Electronic Localization Function: Chemically intuitive distribution of electrostatic moments. J Comput Chem. 2008;29:1440–1449. doi: 10.1002/jcc.20904. [DOI] [PubMed] [Google Scholar]
- Piquemal J-P, Maddaluno J, Silvi B, Giessner-Prettre C. Theoretical study of phenol and 2-aminophenol docking at a model of tyrosinase active site. New J Chem. 2003;27:909–913. [Google Scholar]
- Piquemal J-P, Williams-Hubbard B, Fey N, Deeth RJ, Gresh N, Giessner-Prettre C. Inclusion of the ligand field contribution in a polarizable molecular mechanics: SIBFA LF. J Comput Chem. 2003;24:1963–1970. doi: 10.1002/jcc.10354. [DOI] [PubMed] [Google Scholar]
- Piquemal J-P, Pilmé J. Comments on the nature of the bonding in oxygenated dinuclear copper enzymes models. J Mol Struct: THEOCHEM. 2006;764:77–86. doi: 10.1016/j.theochem.2006.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piquemal J-P, Pilmé J, Parisel O, Gérard H, Fourré I, Bergès J, Gourlaouen C, de la Lande A, van Severen M-C, Silvi B. What can be learnt on biological or biomimetic systems with the topological analysis of the electron localization function? Int J Quant Chem. 2008;108:1951–1969. [Google Scholar]
- Prezioso JA, Epperly MW, Wang N, Bloomer WD. Effects of tyrosinase activity on the cytotoxicity of 4-S-cysteaminylphenol and N-acetyl-4-S-cysteaminylphenol in melanoma cells. Cancer Lett. 1992;63:73–79. doi: 10.1016/0304-3835(92)90092-a. [DOI] [PubMed] [Google Scholar]
- Rassolov V, Pople JA, Ratner M, Windus TL. 6–31G* basis set for atoms K through Zn. J Chem Phys. 1998;109:1223–1229. [Google Scholar]
- Rode MF, Werner HJ. Ab initio study of the O2 binding in dicopper complexes. Theor Chem Acc. 2005;114:309–317. [Google Scholar]
- Ross PK, Solomon EI. An electronic structural comparison of copper-peroxide complexes of relevance to hemocyanin and tyrosinase active sites. J Am Chem Soc. 1991;113:3246–3259. [Google Scholar]
- Shao Y, Head-Gordon M, Krylov AI. The spin-flip approach within time-dependent density functional theory: Theory and applications to diradicals. J Chem Phys. 2003;118:4807–818. [Google Scholar]
- Shao Y, et al. Advances in methods and algorithms in a modern quantum chemistry program package. Phys Chem Chem Phys. 2006;8:3172–3191. doi: 10.1039/b517914a. [DOI] [PubMed] [Google Scholar]
- Silvi B, Savin A. Classification of chemical bonds based on topological analysis of electron localization functions. Nature. 1994;371:683–686. [Google Scholar]
- Solomon EI, Chen P, Metz M, Lee SK, Palmer AE. Oxygen binding, activation, and reduction to water by copper proteins. Angew Chem Int Ed. 2001;40:4570–4590. doi: 10.1002/1521-3773(20011217)40:24<4570::aid-anie4570>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Tolman WB. Using synthetic chemistry to understand copper protein active sites: a personal perspective. J Biol Inorg Chem. 2006;11:261–271. doi: 10.1007/s00775-006-0078-9. [DOI] [PubMed] [Google Scholar]
- Volbeda A, Hol WGJ. Crystal structure of hexameric haemocyanin from Panulirus interruptus refined at 3.2 Å resolution. J Mol Biol. 1989;209:249–279. doi: 10.1016/0022-2836(89)90276-3. [DOI] [PubMed] [Google Scholar]
- Wang F, Ziegler T. Time-dependent Density Functional Theory based on a noncollinear formulation of the exchange-correlation potential. J Chem Phys. 2004;121:12191–12196. doi: 10.1063/1.1821494. [DOI] [PubMed] [Google Scholar]
- Xu Y, Stokes AH, Freeman WM, Kumer SC, Vogt BA, Vrana KE. Tyrosinase mRNA is expressed in human substantia nigra. Mol Brain Res. 1997;45:159–162. doi: 10.1016/s0169-328x(96)00308-7. [DOI] [PubMed] [Google Scholar]