Abstract
The neurofibromatosis type 1 tumor suppressor protein neurofibromin, is a GTPase activating protein for H-, N-, K-, R-Ras and TC21/R-Ras2 proteins. We demonstrate that Schwann cells derived from Nf1-null mice have enhanced chemokinetic and chemotactic migration in comparison to wild-type controls. Surprisingly, this migratory phenotype is not inhibited by a farnesyltransferase inhibitor or dominant-negative (dn) (N17)H-Ras (which inhibits H-, N-, and K-Ras activation). We postulated that increased activity of R-Ras and/or TC21/R-Ras2, due to loss of Nf1, contributes to increased migration. Mouse Schwann cells (MSCs) express R-Ras and TC21/R-Ras2 and their specific guanine exchange factors, C3G and AND-34. Infection of Nf1-null MSCs with a dn(43N)R-Ras adenovirus (to inhibit both R-Ras and TC21/R-Ras2 activation) decreases migration by approximately 50%. Conversely, expression of activated (72L)TC21/R-Ras2, but not activated (38V)R-Ras, increases migration, suggesting a role of TC21/R-Ras2 activation in the migration of neurofibromin-deficient Schwann cells. TC21/R-Ras2 preferentially couples to the phosphatidylinositol 3-kinase (PI3-kinase) and MAP kinase pathways. Treatment with a PI3-kinase or MAP kinase inhibitor reduces Nf1-null Schwann cell migration, implicating these TC21 effectors in Schwann cell migration. These data reveal a key role for neurofibromin regulation of TC21/R-Ras2 in Schwann cells, a cell type critical to NF1 tumor pathogenesis.
Keywords: Schwann cell, TC21, neurofibromin, chemotaxis, R-Ras, Ras
Introduction
The Nf1 gene product neurofibromin is a GTPase activating protein (GAP) for Ras proteins. Neurofibromin negatively regulates Ras activity by accelerating the conversion of Ras-GTP to Ras-GDP (Ballester et al., 1990; Xu et al., 1990; Donovan et al., 2002). Neurofibromatosis type 1 (NF1) patients possess germline mutations in the NF1 gene and develop benign peripheral nerve tumors called neurofibromas and malignant peripheral nerve sheath tumors (MPNSTs) (Huson, 1994). Neurofibroma lysates (Guha et al., 1996) and MPNST cells (Basu et al., 1992; DeClue et al., 1992), both with reduced expression of neurofibromin, demonstrate increased levels of Ras-GTP, thus confirming neurofibromin’s role as a Ras-GAP. Neurofibromin’s GAP-related domain (GRD) is believed to be crucial to the development of NF1 disease, as missense mutations are detected in the GRD of NF1 patients. (Klose et al., 1998; Fahsold et al., 2000).
Biallelic NF1 mutations occur in MPNSTs (Legius et al., 1993) and Schwann cells from neurofibromas (Kluwe et al., 1999; Serra et al., 2001), indicating that Schwann cells are the primary pathogenic cell type in NF1 peripheral nerve tumors. Ras-GTP levels are elevated in Schwann cells purified from human neurofibromas and Nf1−/− mice (Kim et al., 1995; Sherman et al., 2000). Furthermore, Nf1 mutant mouse Schwann cells (MSCs) exhibit altered proliferation in comparison to wild-type cells. These proliferation defects are reversed by a farnesyltransferase inhibitor (FTI) (Kim et al., 1997) (which inhibits H-Ras processing and activation) and are mimicked by activated H-Ras in wild-type cells (Kim et al., 1995).
While the proliferation defects of Nf1-deficient Schwann cells can be ascribed to H-, N-, and/or K-Ras- GTP excess, inhibition of Ras family proteins does not result in reversal of all mutant phenotypes. Schwann cells isolated from human neurofibromas and Nf1 mutant mice are increasingly invasive in comparison to normal controls (Sheela et al., 1990; Muir, 1995; Kim et al., 1997;). This phenotype is not inhibited by FTI treatment (Kim et al., 1997), nor is it mimicked by H-Ras activation (unpublished observations).
We postulated that loss of Nf1 results in increased activity of the nonclassical Ras proteins R-Ras and/or TC21/R-Ras2, contributing to the migratory phenotype of neurofibromin-deficient Schwann cells. In vitro, neurofibromin stimulates the GTPase activity of R-Ras and TC21/R-Ras2, as well as of H-, N-, and K-Ras proteins (Rey et al., 1994; Ohba et al., 2000). The roles of R-Ras and TC21/R-Ras2, two closely related members of the R-Ras subfamily, are not well characterized. R-Ras has distinct biological functions from those of the classical Ras proteins. R-Ras binds some of the same effectors as H-, N-, and K-Ras, but most effectively activates the phosphatidylinositol 3-kinase (PI3-kinase)-Akt pathway, and possesses weak transforming activity (Chan et al., 1994; Cox et al., 1994; Herrmann et al., 1996; Marte et al., 1997). In contrast, constitutively active mutants of TC21/R-Ras2, the closest relative of R-Ras, strongly transform fibroblast and epithelial cell lines, suggesting that TC21/R-Ras2 shares more functional similarity with H-Ras than with R-Ras (Graham et al., 1994; Clark et al., 1996). TC21/R-Ras2 binds and activates p110, the catalytic subunit of PI3-kinase, in a GTP-dependent manner, and PI3- kinase activation is essential for TC21/R-Ras2-mediated transformation (Rosario et al., 2001; Murphy et al., 2002; Rong et al., 2002). TC21/R-Ras2 activation of the Raf/MAPK and RalGDS pathways remains controversial (Graham et al., 1999; Movilla et al., 1999; Rosario et al., 2001).
R-Ras and TC21/R-Ras2 have been implicated in cell adhesion and motility. The expression of constitutively active R-Ras enhances adhesion to matrix proteins via increased integrin ligand binding activity (Zhang et al., 1996). Furthermore, transfection of constitutively active R-Ras or TC21/R-Ras2 into breast epithelial cells promotes cell migration and invasion, while dominant-negative (dn) R-Ras and dnTC21/R-Ras2 inhibit cell migration (Keely et al., 1999).
Our data demonstrate that TC21 mediates a migratory phenotype of Nf1-deficient Schwann cells; this signaling pathway may contribute to tumor growth in human patients with NF1 disease.
Results
Nf1-deficient MSCs exhibit increased migration in vitro
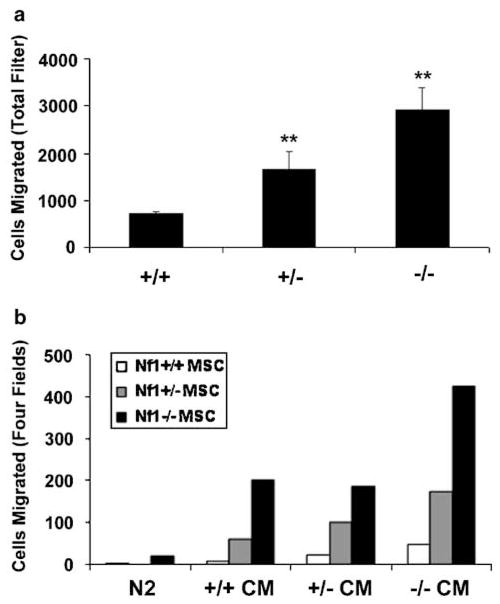
To test if neurofibromin loss alters cell motility, Schwann cells derived from wild-type (+/+), heterozygous (+/−), or homozygous (−/−) Nf1 mutant embryos were tested in a Boyden chamber assay using transwells coated on both sides with poly-L-lysine. Nf1−/− Schwann cells exhibited a fourfold increase in migration in comparison to wild-type cells; Nf1+/− cells demonstrated a twofold increase (Figure 1a). This trend was highly consistent in three separate experiments using cells derived from different embryos. In some experiments, we saw up to an eightfold increase in Nf1−/− Schwann cell migration. On laminin-coated transwells, Nf1-deficient MSCs demonstrated a fivefold increase in migration in comparison to wild-type controls; heterozygous cells had an intermediate migratory phenotype (data not shown). To evaluate whether Schwann cell-conditioned medium further potentiates cell migration, we performed chemotaxis assays. Nf1+/+, Nf1+/− and Nf1−/− MSCs were plated on transwells, and conditioned medium collected from Nf1+/+, +/−, −/− Schwann cells or N2 was added to the lower chamber. As illustrated in Figure 1b, Nf1−/− cell conditioned medium most potently stimulated the migration of Schwann cells of all three genotypes. Conditioned medium collected from wild-type and heterozygous Schwann cells less potently stimulated migration but was a more effective chemotaxis signal than N2 alone. Under all conditions, Nf1- null Schwann cells migrated to a greater extent than wild-type cells. Conditioned medium placed only in the upper chamber or in both upper and lower chambers was significantly less effective in stimulating Nf1−/− cell migration, thus confirming a chemotactic as opposed to chemokinetic mechanism of migration (data not shown). These results indicate that Nf1 loss in Schwann cells results in increased migratory potential in a cell autonomous fashion. Migration is further stimulated by unidentified autocrine/paracrine factor(s) in Schwann cell conditioned medium.
Figure 1.
Nf1-deficient MSCs exhibit increased migration in vitro. Passage-matched Nf1+/+, Nf1+/−, and Nf1−/− MSCs were plated onto poly-L-lysine-coated transwells and counted 16 h later. (a) N2 medium (N2) was added to the lower chamber of the transwell. (b) Conditioned medium (CM) derived from Nf1+/+, Nf1+/−, or Nf1−/− MSCs or N2 was added to the lower chamber of the transwell. Migration was quantified by counting the number of cells present on the entire transwell filter (a) or in four fields (b) using a fluorescent microscope. The average of duplicate samples is shown; duplicates varied by less than 10%. The number of cells that migrated was normalized to the total number of cells plated. **P<0.01 compared with wild-type control
Increased migration of Nf1−/− MSCs is not inhibited by dnH-Ras or by treatment with an FTI
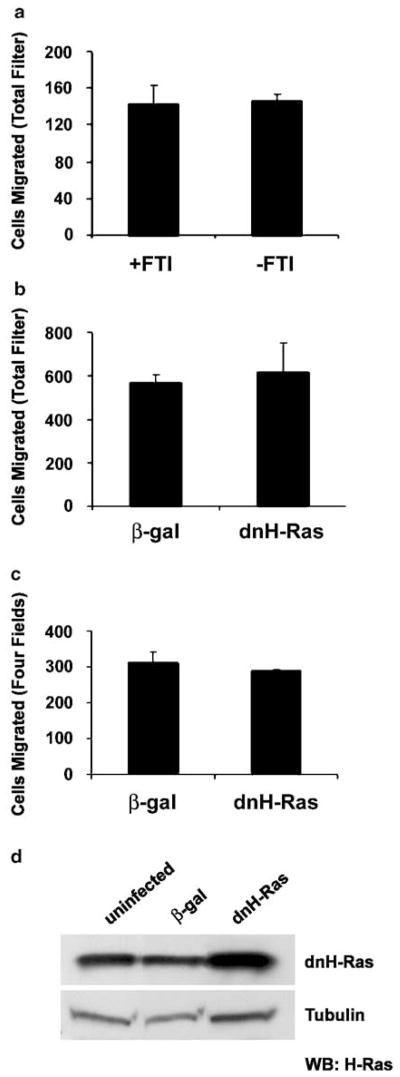
To clarify whether elevated Ras signaling contributes to the increased motility of Nf1 mutant Schwann cells, Nf1−/− MSCs were pretreated for 3 days with 1 μM of the FTI, L-744,832(Kohl et al., 1995). In our previous studies, FTI was shown to effectively inhibit processing of Ras proteins and to reverse the proliferation defect in Nf1−/− MSCs (Kim et al., 1997). FTI-treated cells migrated to the same extent as untreated controls (Figure 2a). Since FTI treatment most effectively inhibits H-Ras processing (Whyte et al., 1997; Omer et al., 2000), Nf1−/− cells were infected with a dnH-Ras adenovirus (which inhibits H-, N-, and K-Ras activation) or a β-galactosidase control. DnH-Ras-infected cells migrated to the same degree as β-galactosidase controls in the absence (Figure 2b) and presence (Figure 2c) of Nf1−/− conditioned medium. MSCs infected for 24 h with dnH-Ras demonstrate increased H-Ras expression in comparison to uninfected or β-galactosidase- infected cells, thus confirming expression of the dn form of the protein (Figure 2d). These results demonstrate that the migratory phenotype of Nf1- deficient MSCs is independent of their increased H-, N-, and/or K-Ras activation.
Figure 2.
Increased migration of Nf1−/− Schwann cells is not inhibited by FTI treatment or dnH-Ras. (a) Nf1−/− MSCs were pretreated with or without 1 μM L-744, 832(FTI) for 72 h and then tested in the cell migration assay in the absence of conditioned medium. (b, c) Nf1−/− MSCs were infected with dn(N17)H-Ras adenovirus (dnH-Ras) or a β-galactosidase control (β-gal) and tested for cell migration in the absence (b) and presence (c) of Nf1−/− mouse Schwann cell conditioned medium. Migration was quantified by counting the number of migrated cells present on the entire transwell filter or in four fields as indicated. Each condition was performed in triplicate and the number of cells that migrated was normalized to the total number of cells plated. The data shown are representative of three independent experiments. The values presented are the mean±s.d.; statistical significance was determined by t-test. (d) Nf1+/+ MSCs were infected for 24 h with β-galactosidase (β-gal) adenovirus, dnH-Ras adenovirus, or remained uninfected. The medium was then changed to normal growth medium and cells continued to grow for 24 h. Cell lysates were prepared and Western blots were probed for H-Ras. Blots were stripped and reprobed with antitubulin as a loading control
H-, N-, K-, R-Ras, and TC21/R-Ras2 are expressed by MSCs
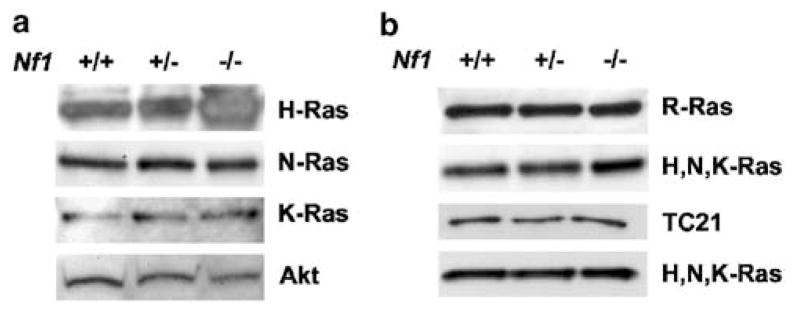
We postulated that loss of Nf1 results in the increased activity of the nonclassical Ras proteins R-Ras and/or TC21/R-Ras2, thus contributing to the migratory phenotype of neurofibromin-deficient Schwann cells. To confirm that Schwann cells are capable of signaling through R-Ras and TC21, we analysed the expression of the Ras and R-Ras family members in Nf1+/+, +/−, and −/− MSCs. H-, N-, and K-Ras were expressed in each of the lysates (Figure 3a). The R-Ras and TC21/R-Ras2 proteins were also detected in all three genotypes of Schwann cells (Figure 3b). The dosage of neurofibromin did not affect the expression level of any of the Ras proteins.
Figure 3.
H-, N-, K-, R-Ras, and TC21/R-Ras2 are expressed by MSCs. Nf1+/+, +/−, and −/− Schwann cell lysates were analysed by Western blot. (a) Blots were probed with anti-H-Ras, anti-N-Ras or anti-K-Ras antibody. Blots were stripped and reprobed with an Akt antibody as a loading control. (b) Blots were probed with anti-R-Ras or anti-TC21/R-Ras2. Blots were stripped and reprobed with a pan-Ras antibody that detects H, N, and K-Ras as a loading control
Schwann cells express guanine exchange factors (GEFs) that specifically activate R-Ras family proteins
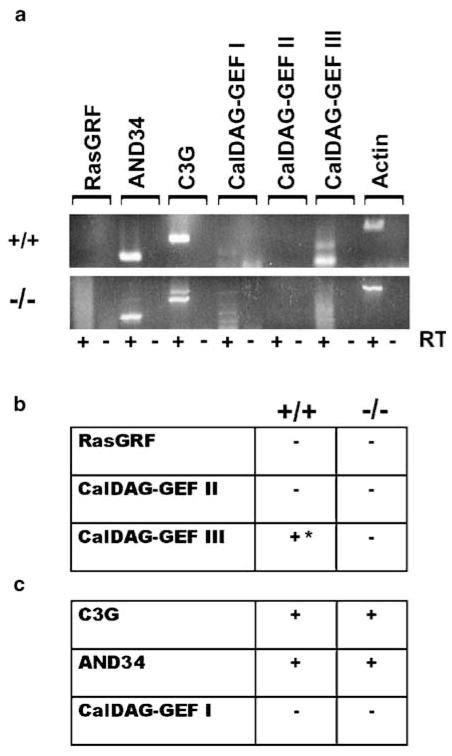
GEFs activate Ras proteins by mediating the cycling of Ras proteins from their GDP-bound state to their GTP-bound state. Therefore, activation of endogenous R-Ras and TC21/R-Ras2 in MSCs requires the expression of GEFs that promote the guanine nucleotide exchange of R-Ras and TC21/R-Ras2. The complement of GEFs expressed by wild-type and Nf1-null MSCs was ascertained by RT–PCR analysis (Figure 4a). We determined the expression of (1) GEFs that activate both the classical Ras and R-Ras family members and (2) GEFs that specifically activate R-Ras family proteins (Gotoh et al., 1997; Ebinu et al., 1998; Kawasaki et al., 1998; Gotoh et al., 2000; Ohba et al., 2000; Yamashita et al., 2000). Each of the primer pairs detected an appropriate size band in mouse brain cDNA (data not shown). Wild-type MSCs expressed one GEF that catalyses the activation of both Ras and R-Ras family members (CalDAG GEF III), while Nf1−/− MSCs expressed none (Figure 4b). Both Nf1+/+ and −/− MSCs express C3G and AND-34, two GEFs that specifically activate R-Ras family but not Ras family proteins (Figure 4c). Therefore, MSCs are capable of signaling through R-Ras and TC21/R-Ras2.
Figure 4.
Wild-type and Nf1-deficient Schwann cells express GEFs that specifically activate R-Ras and TC21/R-Ras2. cDNA was prepared from wild-type (+/+) and Nf1-null (−/−) MSCs. (a) RT–PCR was used to determine the expression of (b) GEFs that activate both Ras and R-Ras family members and (c) GEFs that specifically activate R-Ras family proteins. The + and − symbols denote the presence or absence of the message following a 40-cycle RT–PCR reaction. *Message was detectable only when using gene-specific cDNA. The RT symbol indicates presence or absence of the reverse transcriptase enzyme
DnR-Ras inhibits migration of Nf1−/− MSCs
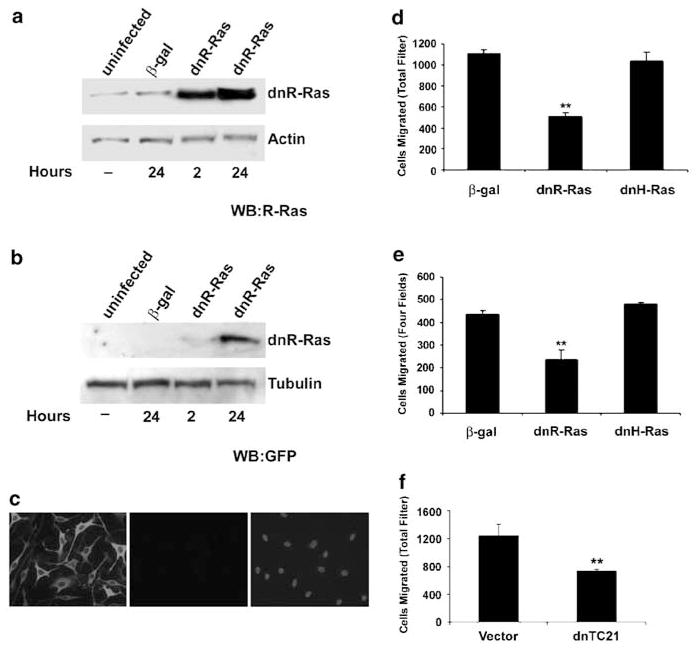
Dominant-negative Ras inhibits the activation of Ras family proteins by sequestering their guanine nucleotide exchange factors (reviewed in Feig, 1999). We constructed a bicistronic adenovirus that expresses a GFP marker and dn(43N)R-Ras. We confirmed expression of dnR-Ras protein in wild-type MSCs. Uninfected and β-gal- infected MSCs expressed a small, equal amount of endogenous R-Ras (Figure 5a). Infection with dnR-Ras for either 2 or 24 h resulted in a dramatic increase in RRas expression, reflecting expression of the dominant-negative form of the R-Ras protein. When these lysates were probed for GFP, a robust increase in GFP expression is apparent after 24 h of infection (Figure 5b). To visualize expression, we stained dnRRas- infected Schwann cells for GFP. In all, 90–100% of the cells exhibited a GFP signal, indicating that most cells express dnR-Ras protein (Figure 5c).
Figure 5.
DnR-Ras inhibits migration of Nf1−/− Schwann cells. Nf1−/− wild-type MSCs were infected with a bicistronic adenovirus that expresses dn(43N)R-Ras and GFP (dnR-Ras) or a β-galactosidase adenovirus (β-gal). Cells were incubated in the presence of adenovirus for 2 or 24 h. Medium was then changed to normal growth medium and cells continued to grow for 66 or 44 h. Cell lysates were prepared and analysed by Western blot. Blots were probed with (a) anti-R-Ras (WB: R-Ras) or (b) anti-GFP (WB: GFP). Blots were stripped and reprobed with anti-actin or anti-tubulin as a loading control. (c) +/+ MSCs were infected with dnR-Ras adenovirus and incubated in the presence of the virus for 24 h. Cells were stained for GFP (left). A corresponding sample that omitted incubation with primary antibody (middle panel) was stained with bisbenzimide to demonstrate presence of cells (right). (d, e) MSCs were infected with dn(43N)R-Ras (dnR-Ras), dn(N17)H-Ras (dnH-Ras), or a β-galactosidase adenovirus (β-gal). Infected cells were tested in the cell migration assay in the absence (d) and presence (e) of Nf1−/− mouse Schwann cell conditioned medium. Migration was quantified as in Figure 2. (f) Nf1−/− MSCs were cotransfected with β-galactosidase and dn(26A)TC21/R-Ras2-pcGN (dnTC21) or vector control. Cells were tested in the cell migration assay in the presence of Nf1−/− mouse Schwann cell conditioned medium and the migration was quantified by counting the number of β-galactosidase-positive cells that had migrated across the transwell filter. Each condition was performed in triplicate and the number of cells that migrated was normalized to the total number of β-galactosidase-positive cells plated. **P<0.01 compared with β-gal control
To determine the effect of R-Ras and TC21/R-Ras2 activity on the migration of neurofibromin-deficient Schwann cells, Nf1−/− MSCs were infected or transfected with dnR-Ras or dn(26A)TC21/R-Ras2 (each construct inhibits both R-Ras and TC21/R-Ras2 activation) and then tested in the Boyden chamber assay. While β-galactosidase- and dnH-Ras-infected cells migrated to the same extent, migration of the dnR-Ras-infected Nf1−/− MSCs was inhibited by approximately 50% in the absence (Figure 5d) and presence (Figure 5e) of Nf1−/− conditioned medium. Similarly, Nf1−/− cells expressing dnTC21/R-Ras2 had a 40% reduction in their migration in comparison to vector control in the presence of conditioned medium (Figure 5f). These data suggest that R-Ras and/or TC21/R-Ras2 activity contributes to the migration of Nf1-deficient Schwann cells.
Activated TC21/R-Ras2 enhances, whereas activated R-Ras inhibits, the migration of Nf1−/− MSCs
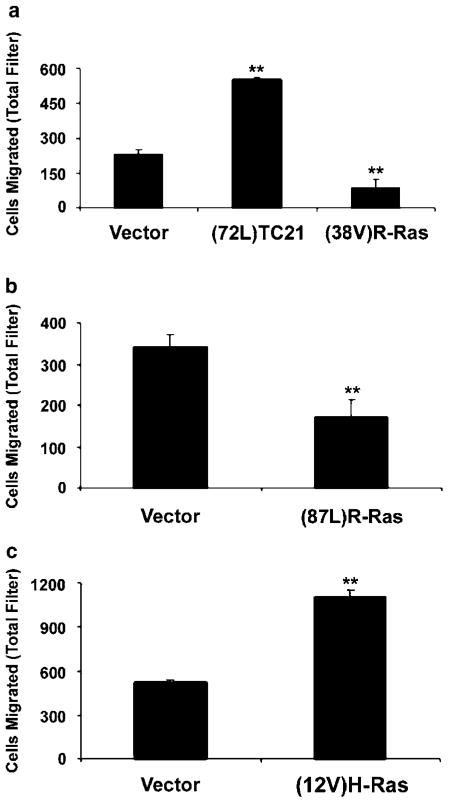
To differentiate the effects of R-Ras and TC21/R-Ras2 activation on Schwann cell migration, Nf1−/− MSCs were transfected with constitutively activated Ras alleles. Expression of either activated TC21/R-Ras2 or activated H-Ras significantly increased cell migration in comparison to vector control (Figure 6a and c). The expression of activated TC21/R-Ras2 in wild-type MSCs also increased their migration, although the full level of Nf1−/− Schwann cell migration was not achieved (data not shown). Surprisingly, expression of either of two activated R-Ras isoforms, (38V)R-Ras or (87L)R-Ras (analogous to mutations in Ras codons 12 and 61, respectively), inhibited the migration of Nf1-null Schwann cells by approximately 50–60% in comparison to controls (Figure 6a and b). These results are similar to those obtained with dnR-Ras. Thus, perturbing the balance of active to inactive R-Ras in either direction inhibits Nf1-null Schwann cell migration. These results suggest that the increased migratory potential of Nf1 mutant Schwann cells is the result of increased activity of TC21/R-Ras2 but not of R-Ras.
Figure 6.
Activated TC21/R-Ras2 and activated H-Ras, but not activated R-Ras, stimulate the migration of Nf1−/− Schwann cells. (a) Nf1−/− MSCs were cotransfected with β-galactosidase and (72L)TC21/R-Ras2-pZIP, (38V)R-Ras-pZIP, or vector control. (b) Nf1−/− MSCs were cotransfected with β-galactosidase and (87L)R-Ras-pCGN-Hyg or vector control. (c) Nf1−/− MSCs were cotransfected with β-galactosidase and (12V)H-Ras-pZIP, or vector control. Cells were tested in the cell migration assay in the presence of Nf1−/− mouse Schwann cell conditioned medium and the migration was quantified as in Figure 5. **P<0.01 compared with vector control
Inhibition of PI3-kinase and MAP-kinase attenuates the migration of Nf1−/− Schwann cells
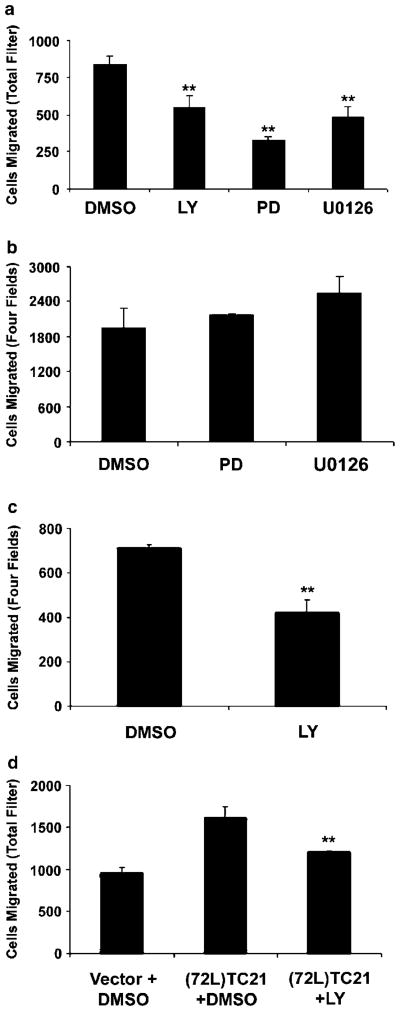
To begin to assess the role of downstream effectors of TC21/R-Ras2 in the migration phenotype, we treated Nf1-null MSCs with the PI3-kinase inhibitor LY294002 and two MAP-kinase inhibitors, PD98059 and U0126, over the course of the migration assay. Inhibition of PI3-kinase and MAP-kinase significantly decreased the migration of Nf1−/− cells in comparison to vehicle controls in the absence of conditioned medium (Figure 7a). Similar to dnR-Ras, the inhibition was only partial. Suprisingly, in the presence of Nf1−/− conditioned medium, both MAP-kinase inhibitors failed to inhibit Nf1-null Schwann cell migration (Figure 7b). In contrast, inhibition of PI3-kinase resulted in the decreased migration of Nf1−/− MSCs both in the absence and presence of conditioned medium (Figure 7a and c). To determine whether PI3-kinase inhibition could reverse TC21/R-Ras2-stimulated migration, Nf1−/− Schwann cells expressing activated TC21/R-Ras2 were treated with LY294002 in the presence of conditioned medium. Inhibition of PI3-kinase significantly decreased the migration induced by activated TC21/R-Ras2; migration was inhibited by 50%, suggesting that PI3-kinase activity contributes to TC21/R-Ras2-induced migration (Figure 7d). Treatment of Nf1−/− cells with an Akt inhibitor, a PKC inhibitor, or a ROCK inhibitor failed to inhibit migration (data not shown). Taken together, these results demonstrate that while PI3-kinase and MAP-kinase activity contributes to Nf1−/− mouse Schwann cell migration, other signaling pathways also play a role.
Figure 7.
Inhibition of PI3-kinase and MAP-kinase attenuates the increased migration of Nf1−/− Schwann cells. Nf1−/− MSCs were treated with 10 μM PI3-kinase inhibitor, LY294002 (LY), 25 μM MAP-kinase inhibitor PD98059 (PD), 2 μM MAP-kinase inhibitor U0126 (U0126), or DMSO during the course of the migration assay. The asssay was performed in the absence (a) or presence (b, c) of conditioned medium and quanitifed as in Figure 2. (d) Nf1−/− MSCs were cotransfected with β-galactosidase and (72L)TC21/R-Ras2- pZIP or vector control. Cells were treated with 10 μM LY294002 or DMSO during the migration assay. The asssay was performed in the presence of conditioned medium. Migration was quantified as in Figure 6. **P<0.01 compared with vehicle control or TC21(72L)
Discussion
We show that neurofibromin loss causes a dose-dependent increase in mouse Schwann cell migration. This increased migration is not inhibited by FTI treatment or dn(N17)H-Ras, but is inhibited by dn(43N)R-Ras and dn(26A)TC21-R-Ras/2. Furthermore, activated (72L)TC21/R-Ras2, but not activated (38V)R-Ras or (87L)R-Ras, potentiates the migratory phenotype of Nf1-null Schwann cells. We also implicate PI3-kinase and MAP-kinase activity in Nf1−/− mouse Schwann cell migration. These results suggest that Nf1 loss in Schwann cells increases the basal activity of TC21/R-Ras2 and its downstream effectors.
The use of dominant-negative Ras and R-Ras alleles allows us to discriminate between pathways mediated by classical Ras and R-Ras family proteins. Dominant-negative Ras proteins inhibit endogenous Ras activation by binding and sequestering GEFs, thereby preventing the efficient binding of Ras to GTP (reviewed in Feig, 1999). H-, N-, and K-Ras share common GEFs and have overlapping spatial localizations. Therefore, dnH-Ras inhibits the activation of all three family members (Matallanas et al., 2003). Nf1 also possesses GAP activity for M-Ras, a recently identified homologue of p21 Ras. Since M-Ras shares GEFs with the classical Ras proteins, M-Ras activity may also be inhibited by the dnH-Ras allele (Ehrhardt et al., 1999; Ohba et al., 2000). R-Ras and TC21/R-Ras2 are regulated by common GEFs and have similar subcellular localizations, so both proteins are likely inhibited by dnR-Ras and dnTC21/R-Ras2 (Ohba et al., 2000). RT–PCR analysis detected the expression of two GEFs, C3G and AND-34, in MSCs that specifically activate R-Ras and TC21/R-Ras2. This subset of exchange factors does not activate Ras proteins. Activation of the R-Ras family members is thus independent of dnH-Ras in Schwann cells.
FTI treatment failed to inhibit Nf1−/− mouse Schwann cell migration, consistent with the idea that this phenotype is mediated by TC21/R-Ras2 activity. FTIs inhibit Ras activity by blocking the addition of a farnesyl lipid to the C-terminus of the protein, preventing Ras localization to the plasma membrane (reviewed in Cox and Der, 2002). The clinical utility of FTIs is limited by the fact that they most effectively inhibit H-Ras activity. The activity of N-Ras and K-Ras is preserved, as they are alternatively gerangeranylated in the presence of FTI (Whyte et al., 1997). The C-terminal amino-acid motif specifying prenylation predicts addition of a gerangeranyl lipid rather than a farnesyl lipid to R-Ras and TC21/R-Ras2, thus rendering the R-Ras family of proteins insensitive to FTI treatment. This is supported by data demonstrating that FTI treatment fails to inhibit TC21/R-Ras2-mediated transformation (Carboni et al., 1995). FTIs inhibit the growth of neurofibromin-deficient cells (Yan et al., 1995; Kim et al., 1997), indicating that growth pathways in Schwann cells, unlike migratory pathways, may be mediated by FTI-sensitive Ras molecules including H-Ras.
We show that TC21/R-Ras2 activity is necessary and sufficient for the enhanced migration of Nf1-deficient Schwann cells. Activated H-Ras promotes Nf1−/− MSC migration to the same extent as activated TC21/R-Ras2, but dnH-Ras and FTI fail to inhibit the migratory phenotype. These data demonstrate that H-Ras activity is sufficient but not necessary for the migration of Nf1-null Schwann cells. Exogenous activation of H-Ras in Schwann cells may mimic TC21/R-Ras2-mediated migration by activating common effectors. While we cannot exclude the possibility that TC21 is downstream of H-Ras, there are no precedents for this signaling pathway. Surprisingly, both the activation and inhibition of R-Ras activity result in decreased Nf1−/− mouse Schwann cell migration. We hypothesize that perturbing the balance of active to inactive R-Ras in either direction inhibits the migration of Nf1-deficient Schwann cells. The precise level or duration of R-Ras signaling may be critical to Schwann cell migration. Sustained elevation of Ras activity has been shown to be required for PC12 cell differentiation (Qui and Green, 1992). In addition, R-Ras activates cell surface integrins (Zhang et al., 1996) and mediates cell migration that displays substrate specificity, suggesting that R-Ras signals to specific integrin receptors (Keely et al., 1999). Since the Schwann cell migration assays were performed on poly-L-lysine, a substrate that does not engage integrins, R-Ras signaling may be noncontributory in this context. We demonstrated that Nf1−/− Schwann cells have enhanced migration on laminin. Thus, R-Ras activity might play a role in the migration of Nf1−/− Schwann cells on laminin or fibronectin. Activated R-Ras (87L and 38V) and activated TC21/RRas2(72 L) stimulated breast epithelial cell migration across collagen but not fibronectin, suggesting that R-Ras and TC21 activity is modulated by different substrates (Keely et al., 1999).
Previous attempts to measure directly basal or stimulated activation of endogenous R-Ras and TC21 have been largely unsuccessful (Ohba et al., 2001; van Triest et al., 2001; Yu and Feig, 2002). We were unable to detect increased levels of R-Ras-GTP or TC21/R-Ras2-GTP in neurofibromin-deficient Schwann cells. Nf1+/+, Nf1+/−, and Nf1−/− mouse Schwann cell lysates were incubated with affinity probes (RafRBDGST, RalRBD-GST, and RlfRBD-GST) that bind the activated forms of R-Ras and TC21/R-Ras2 (Ohba et al., 2001; Rosario et al., 2001; L. Quilliam, personal communication). However, despite attempts to utilize a variety of neurofibromin-deficient cell systems and growth factor or cell-attachment stimuli, we failed to detect the GTP-bound form of either endogenous R-Ras or endogenous TC21/R-Ras2. Levels of activated, endogenous R-Ras and TC21/R-Ras2 in the Schwann cell may be below our threshold of detection. This idea is supported by detection of activated R-Ras in cells overexpressing a constitutively active form of R-Ras (unpublished observations). Furthermore, C3G-deficient mouse embryonic fibroblasts overexpressing wild-type R-Ras do not demonstrate a decrease in basal R-Ras-GTP, but do exhibit a small cell adhesion-dependent decrease in R-Ras-GTP in comparison to wild-type controls (Ohba et al., 2001). Endogenous R-Ras and TC21/R-Ras2 may also require an appropriate, but as yet unknown, stimulus for their robust, transient activation. Indeed, human blood platelets demonstrate no basal, endogenous R-Ras-GTP, but when stimulated with thrombin, a small increase in activated R-Ras is detected (van Triest et al., 2001).
Our results indicate that MAP-kinase and PI3-kinase activity contributes to the autonomous migration of Nf1−/− Schwann cells. MAP-kinase activity has been extensively implicated in cell migration in a variety of cell types (Klemke et al., 1997). In addition, it has been previously demonstrated that TC21/R-Ras2 interacts with and activates Raf-1 and B-Raf, and that activation of the Raf/MAPK pathway is required for TC21/RRas2- mediated transformation (Movilla et al., 1999; Rosario et al., 1999). Keely and colleagues, however, found that MEK inhibition did not affect migration induced by R-Ras or TC21. These discrepancies may be attributed to differences in cell type, substrates, or endogenous versus exogenous TC21 activation. MAP-kinase inhibition reduces the unstimulated migration of Nf1−/− Schwann cells, indicating that it acts downstream of neurofibromin. However, inhibition of MAP-kinase fails to attenuate the migratory phenotype in the presence of conditioned medium, thus indicating that MAP-kinase activity is not required for the conditioned medium effect.
PI3-kinase has been previously shown to mediate several effects of R-Ras and TC21/R-Ras2. The migration of breast epithelial cells expressing activated R-Ras or TC21/R-Ras2 is attenuated by inhibition of PI3- kinase (Keely et al., 1999). In addition, the ability of activated R-Ras to rescue fibroblast cell spreading is dependent upon PI3-kinase activity (Berrier et al., 2000). Similarly, inhibition of PI3-kinase activity partially decreased TC21/R-Ras2-stimulated migration, placing PI3-kinase downstream of TC21/R-Ras2 in MSCs. DnR-Ras, dnTC21/R-Ras2, and inhibition of PI3- kinase all diminish the migration of Nf1−/− MSCs in the presence of conditioned medium. These results further show that the TC21/R-Ras2 and PI3-kinase signaling pathway acts downstream of conditioned medium-stimulated migration. Nf1−/− mouse Schwann cell migration was only partially reduced by MAP-kinase and PI3-kinase inhibition. These two pathways may synergize or additional signaling pathways may contribute to the migratory phenotype. We could not demonstrate a role of Akt, PKC, or ROCK in the migration of Nf1-null Schwann cells. It remains to be determined whether other effectors, including Rac, Rho, p70 S6 kinase, and/or myosin light chain kinase, play a role (reviewed in Schmitz et al., 2000; Howe et al., 2002). Taken together, these data demonstrate that loss of Nf1 in MSCs results in the activation of two promigratory mechanisms. Loss of Nf1 may activate a Schwann cell migratory signaling cascade: TC21/R-Ras2 and its downstream effectors. In addition, neurofibromin deficiency results in the enhanced secretion of autocrine and/or paracrine factors that promote Schwann cell migration.
While neurofibromas are benign tumors that do not metastasize, Schwann cells within the tumor do lose contact with axons, migrating away from neurons and invading the abundant collagen matrix as the tumors grow. Nf1-null MSCs (Kim et al., 1997) and Schwann cells derived from human neurofibromas (Sheela et al., 1990) have an increased invasive capacity in comparison to normal controls. Cell invasion may reflect alterations in cellular motility, as documented here, and/or an altered balance of proteolytic enzymes and their inhibitors. Cutaneous neurofibroma Schwann cells secrete matrix metalloproteinase 1, 3, and 9 (Muir, 1995). Therefore, abnormal protease secretion coupled with the increased migratory potential demonstrated here may contribute to the invasive phenotype of Nf1 mutant Schwann cells.
Cell motility defects may be a common feature of neurofibromin deficiency. Nf1+/− astrocytes demonstrate decreased cell attachment and increased motility in comparison to wild-type controls (Gutmann et al., 2001). Further studies are required to assess the role of R-Ras and TC21/R-Ras2 signaling in the cell migratory defects of other neurofibromin-deficient cell types. In addition, our results suggest the importance of identifying other biological consequences of inappropriate R-Ras and TC21/R-Ras activity as a result of Nf1 loss.
Materials and methods
Mouse Schwann cell culture
Nf1−/− embryos obtained from timed matings of Nf1+/− C57Bl/6 mice at embryonic day 12.5 were identified by PCR genotyping (Brannan et al., 1994). MSCs were isolated from embryonic day 12.5 dorsal root ganglia as previously described (Kim et al., 1995). Wild-type, heterozygous, and Nf1-null Schwann cells were cultured on poly-L-lysine-coated plates in DMEM with 10% fetal bovine serum, 10 ng/ml rhGGF2(gift of Cambridge Neuroscience), and 2 μM forskolin (Calbiochem). Cells were used between passages 1 and 3. For conditioned medium, Schwann cell cultures at 90% confluency were switched to N2 medium (DMEM/F12+N2 supplements, GibcoBRL) and incubated for 48 h. Conditioned medium was harvested, centrifuged, and stored at −70°C.
Construction of the dnR-Ras adenovirus
The dn(43N)R-Ras adenovirus was created as described with minor modifications (He et al., 1998). A dn(43N)R-Ras plasmid was provided by Channing Der (UNC; Huff et al., 1997). The dn(43N)R-Ras cDNA was excised from the pZIP-NeoSV(x)1 shuttle vector, and cloned into the BamH1 site of the pIRES2-EGFP vector. Following XhoI/SspI digestion, the insert ((43N)R-Ras-e-GFP) was subcloned into KpnI/XhoI sites of the pAdCMV-MCS shuttle vector (Adeno-Easy system). The resultant plasmid was linearized by PmeI digestion, and subsequently cotransformed into Escherichia coli BJ5183 cells with an adenoviral backbone plasmid, pAdEasy-1. Recombinants were selected, confirmed by restriction endonuclease analysis, and retransformed into E. coli DH10B cells to generate plasmid DNA.
To produce virus, the PacI-linearized plasmid was transfected into the HEK293 packaging cell line. Viral production was monitored by GFP expression. At 7–12 days post-transfection, cells were collected, centrifuged, and resuspended in PBS. Following three freeze/thaw cycles, the supernatant was collected. Two semiconfluent flasks of 293 cells were infected with half of the viral supernatant and then incubated at 37°C. Cytopathic effect was evident after 2–3 days. Adenovirus was collected and purified by CsCl gradient centrifugation. Viral titer was estimated by OD 260nm measurements.
Adenovirus infection and transfection
For adenovirus experiments, MSCs were grown to 70–80% confluency and infected with a multiplicity of infection of 300. Dn(N17)H-Ras adenovirus was provided by Joe Nevins (Duke). Cells were incubated in the presence of the adenovirus for 24 h, and then changed to normal growth medium for 24 h. Transfections were performed using Fugene 6 (Roche Molecular Biochemicals) according to the manufacturer’s specifications. MSCs were grown to 90% confluency and then cotransfected with CMV-β-galactosidase and the construct of interest. Mutant constructs have been previously described (Cepko et al., 1984; Buss et al., 1989; Fiordalisi et al., 2001; Graham et al., 2001). (12V)H-Ras-pZIP was provided by Channing Der (UNC) and (26A)TC21-pCGN was provided by Patricia Keely (University of Wisconsin). Cells were plated for the migration assay 24 h after transfection.
Migration assay
The migratory response of MSCs was measured using the Boyden chamber assay (Boyden, 1962). Both sides of the transwell polycarbonate membranes (8 μm pore, Costar) were precoated with 50 μg/ml poly-L-lysine (Fisher). MSCs were detached with a cell scraper (Costar) and resuspended at 2.0×105 or 5.0×105 cells/ml of N2 medium with 0.1% BSA. For adenovirus and inhibitor experiments, 4×104 cells were plated per transwell. For transfection experiments, 1×105 cells were plated per transwell. Cell suspension (200 μl) was plated on the upper chamber of the transwell. The lower chamber contained 800 μl N2 medium with 0.1% BSA or 400 μl conditioned medium+400 μl N2 medium with 0.1% BSA. FTI experiments: cells were pretreated with 1 μM FTI, L- 744,832(Merck), for 72h prior to plating. Cells were not treated with FTI during the assay. Inhibitor experiments: cells were treated with 10 μM PI3-kinase inhibitor LY294002 (Calbiochem), 25 μM MAP-kinase inhibitor PD98059 (Calbiochem), 2 μM MAP-kinase inhibitor U0126 (Calbiochem), 10 μM Akt inhibitor, 1L-6-hydroxymethyl-chiro-inositol 2- (R)-2-O-methyl-3-O-octadecylcarbonate (Calbiochem), 50 μM PKC inhibitor PKCI19-27 (Calbiochem), 15 μM ROCK inhibitor Y27632 (Calbiochem), or DMSO control during the migration assay. Cells were incubated for 16 h at 37°C in 10% CO2. Nonmigrating cells were removed from the upper surface of the membrane with cotton swabs. The upper surface of one membrane from each experimental group remained unscraped to determine the total number of cells plated. Membranes were stained with bisbenzimide or processed for β-galactosidase staining and mounted onto glass slides. Migration was quantified by counting cells in four fields or on the entire membrane/filter as noted. Each condition was performed in duplicate or triplicate and the number of migrated cells was normalized to the total number of cells on the unscraped filter. The data shown are representative of three independent experiments and unless otherwise noted, values presented are the mean±s.d. Statistical significance was determined by t-test using Microsoft Excel software.
Western analysis
MSCs were grown to 90% confluency and lysed in RIPA buffer (150mM NaCl, 0.5% sodium deoxycholate, 1% NP-40, 0.1% SDS, 50mM Tris-HCl, pH 8.0) supplemented with protease inhibitors: 1mM PMSF, 1 μg/ml pepstatin, 100 μg/ml benzamidine, 1 μg/ml leupeptin, and 1 μg/ml aprotinin. Protein (20–50 μg) was separated on 4–20% gels (ISC BioExpress), followed by transfer to PVDF membrane (BioRad). Membranes were probed with anti-H-Ras, anti N-Ras, anti-K-Ras, anti-R-Ras, anti-TC21, or anti-GFP (1: 200, Santa Cruz). Blots were stripped and reprobed with a 1: 1000 dilution of anti-Ras C10 (Upstate Biotechnology), anti-Akt (Cell Signaling), antiactin (Santa Cruz), or antitubulin (Santa Cruz). Signals were detected using horseradish peroxidase-conjugated secondary antibodies (BioRad) in combination with ECL Plus developing system (Amersham Pharmacia).
Immunocytochemistry
Cells were fixed with 4% paraformaldehyde, blocked with 10% goat serum, and permeabilized with 0.1% Triton X-100 (Sigma). Cells were stained using anti-GFP antibody (1: 50, Santa Cruz) followed by biotinylated goat anti-rabbit secondary antibody (1: 1000, Vector Laboratories), and visualized with avidin–rhodamine (1: 200, Jackson ImmunoResearch).
RT–PCR analysis
Messenger RNA isolated from wild-type and Nf1−/− MSCs (Micro FastTrack 2.0 kit, Invitrogen) was used to create cDNA (Superscript Preamplification System, GibcoBRL). OligodT primers and random hexamers were used in the reverse transcriptase reaction. Duplicate samples omitting reverse transcriptase were prepared to control for genomic DNA contamination. Gene-specific cDNA was prepared by substituting the sense and antisense primers for the gene of interest for the oligo-dT primers and random hexamers in the cDNA synthesis reaction. Primer sequences for the guanine exchange factors and β-actin can be requested. Each primer pair was tested in mouse brain cDNA as a positive control. RT–PCR reactions were amplified for 40 cycles and each reaction contained 1 μl of cDNA, 0.7 μM primers, 1.0–2.5mM MgCl2, 0.2mM dNTPs, 1×PCR buffer and 1.0U Taq (GibcoBRL). Absence of a message was judged by two criteria: (1) presence of an appropriate size band in mouse brain cDNA and (2) absence of an appropriate size band in the Schwann cell cDNA.
Acknowledgments
We thank the Ratner lab for helpful discussion and advice. We thank Merck Research Laboratories for the FTI. This work was supported by NIH-NS28840 to NR. FR was supported by the Medical Scientist Training Program’s John H Wulsin Fellowship.
References
- Ballester R, Marchuk D, Boguski M, Saulino A, Letcher R, Wigler M, Collins F. Cell. 1990;63:851–859. doi: 10.1016/0092-8674(90)90151-4. [DOI] [PubMed] [Google Scholar]
- Basu TN, Gutmann DH, Fletcher JA, Glover TW, Collins FS, Downward J. Nature. 1992;356:713–715. doi: 10.1038/356713a0. [DOI] [PubMed] [Google Scholar]
- Berrier AL, Mastrangelo AM, Downward J, Ginsberg M, LaFlamme SE. J Cell Biol. 2000;151:1549–1560. doi: 10.1083/jcb.151.7.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyden S. J Exp Med. 1962;115:453–466. doi: 10.1084/jem.115.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannan CI, Perkins AS, Vogel KS, Ratner N, Nordlund ML, Reid SW, Buchberg AM, Jenkins NA, Parada LF, Copeland NG. Genes Dev. 1994;8:1019–1029. doi: 10.1101/gad.8.9.1019. [DOI] [PubMed] [Google Scholar]
- Buss JE, Solski PA, Schaeffer JP, MacDonald MJ, Der CJ. Science. 1989;243:1600–1603. doi: 10.1126/science.2648572. [DOI] [PubMed] [Google Scholar]
- Carboni JM, Yan N, Cox AD, Bustelo X, Graham SM, Lynch MJ, Weinmann R, Seizinger BR, Der CJ, Barbacid M, Veeraswamy M. Oncogene. 1995;10:1905–1913. [PubMed] [Google Scholar]
- Cepko CL, Roberts BE, Mulligan RC. Cell. 1984;37:1053–1062. doi: 10.1016/0092-8674(84)90440-9. [DOI] [PubMed] [Google Scholar]
- Chan AM, Miki T, Meyers KA, Aaronson SA. Proc Natl Acad Sci USA. 1994;91:7558–7562. doi: 10.1073/pnas.91.16.7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark GJ, Kinch MS, Gilmer TM, Burridge K, Der CJ. Oncogene. 1996;12:169–176. [PubMed] [Google Scholar]
- Cox AD, Brtva TR, Lowe DG, Der CJ. Oncogene. 1994;9:3281–3288. [PubMed] [Google Scholar]
- Cox AD, Der CJ. Cancer Biol Ther. 2002;1:599–606. doi: 10.4161/cbt.306. [DOI] [PubMed] [Google Scholar]
- DeClue JE, Papageorge AG, Fletcher JA, Diehl SR, Ratner N, Vass WC, Lowy DR. Cell. 1992;69:265–273. doi: 10.1016/0092-8674(92)90407-4. [DOI] [PubMed] [Google Scholar]
- Donovan S, Shannon KM, Bollag G. Biochim Biophys Acta. 2002;1602:23–45. doi: 10.1016/s0304-419x(01)00041-5. [DOI] [PubMed] [Google Scholar]
- Ebinu JO, Bottorff DA, Chan EY, Stang SL, Dunn RJ, Stone JC. Science. 1998;280:1082–1086. doi: 10.1126/science.280.5366.1082. [DOI] [PubMed] [Google Scholar]
- Ehrhardt GR, Leslie KB, Lee F, Wieler JS, Schrader JW. Blood. 1999;94:2433–2444. [PubMed] [Google Scholar]
- Fahsold R, Hoffmeyer S, Mischung C, Gille C, Ehlers C, Kucukceylan N, Abdel-Nour M, Gewies A, Peters H, Kaufmann D, Buske A, Tinschert S, Nurnberg P. Am J Hum Genet. 2000;66:790–818. doi: 10.1086/302809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feig LA. Nat Cell Biol. 1999;1:E25–27. doi: 10.1038/10018. [DOI] [PubMed] [Google Scholar]
- Fiordalisi JJ, Johnson RL, II, Ulku AS, Der CJ, Cox AD. Methods Enzymol. 2001;332:3–36. doi: 10.1016/s0076-6879(01)32189-4. [DOI] [PubMed] [Google Scholar]
- Gotoh T, Cai D, Tian X, Feig LA, Lerner A. J Biol Chem. 2000;275:30118–30123. doi: 10.1074/jbc.M003074200. [DOI] [PubMed] [Google Scholar]
- Gotoh T, Niino Y, Tokuda M, Hatase O, Nakamura S, Matsuda M, Hattori S. J Biol Chem. 1997;272:18602–18607. doi: 10.1074/jbc.272.30.18602. [DOI] [PubMed] [Google Scholar]
- Graham SM, Cox AD, Drivas G, Rush MG, D’Eustachio P, Der CJ. Mol Cell Biol. 1994;14:4108–4115. doi: 10.1128/mcb.14.6.4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham SM, Oldham SM, Martin CB, Drugan JK, Zohn IE, Campbell S, Der CJ. Oncogene. 1999;18:2107–2116. doi: 10.1038/sj.onc.1202517. [DOI] [PubMed] [Google Scholar]
- Graham SM, Rogers-Graham K, Figueroa C, Der CJ, Vojtek AB. Methods Enzymol. 2001;333:203–216. doi: 10.1016/s0076-6879(01)33057-4. [DOI] [PubMed] [Google Scholar]
- Guha A, Lau N, Huvar I, Gutmann D, Provias J, Pawson T, Boss G. Oncogene. 1996;12:507–513. [PubMed] [Google Scholar]
- Gutmann DH, Wu YL, Hedrick NM, Zhu Y, Guha A, Parada LF. Hum Mol Genet. 2001;10:3009–3016. doi: 10.1093/hmg/10.26.3009. [DOI] [PubMed] [Google Scholar]
- He TC, Zhou S, da Costa LT, Yu J, Kinzler KW, Vogelstein B. Proc Natl Acad Sci USA. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann C, Horn G, Spaargaren M, Wittinghofer A. J Biol Chem. 1996;271:6794–6800. doi: 10.1074/jbc.271.12.6794. [DOI] [PubMed] [Google Scholar]
- Howe AK, Aplin AE, Juliano RL. Curr Opin Genet Dev. 2002;12:30–35. doi: 10.1016/s0959-437x(01)00260-x. [DOI] [PubMed] [Google Scholar]
- Huff SY, Quilliam LA, Cox AD, Der CJ. Oncogene. 1997;14:133–143. doi: 10.1038/sj.onc.1200815. [DOI] [PubMed] [Google Scholar]
- Huson SM. The Neurofibromatoses. Chapman and Hall; London: 1994. [Google Scholar]
- Kawasaki H, Springett GM, Toki S, Canales JJ, Harlan P, Blumenstiel JP, Chen EJ, Bany IA, Mochizuki N, Ashbacher A, Matsuda M, Housman DE, Graybiel AM. Proc Natl Acad Sci USA. 1998;95:13278–13283. doi: 10.1073/pnas.95.22.13278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keely PJ, Rusyn EV, Cox AD, Parise LV. J Cell Biol. 1999;145:1077–1088. doi: 10.1083/jcb.145.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HA, Ling B, Ratner N. Mol Cell Biol. 1997;17:862–872. doi: 10.1128/mcb.17.2.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HA, Rosenbaum T, Marchionni MA, Ratner N, DeClue JE. Oncogene. 1995;11:325–335. [PubMed] [Google Scholar]
- Klemke RL, Cai S, Giannini AL, Gallagher PJ, de Lanerolle P, Cheresh DA. J Cell Biol. 1997;137:481–492. doi: 10.1083/jcb.137.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose A, Ahmadian MR, Schuelke M, Scheffzek K, Hoffmeyer S, Gewies A, Schmitz F, Kaufmann D, Peters H, Wittinghofer A, Nurnberg P. Hum Mol Genet. 1998;7:1261–1268. doi: 10.1093/hmg/7.8.1261. [DOI] [PubMed] [Google Scholar]
- Kluwe L, Friedrich R, Mautner VF. Genes Chromosomes Cancer. 1999;24:283–285. doi: 10.1002/(sici)1098-2264(199903)24:3<283::aid-gcc15>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- Kohl NE, Omer CA, Conner MW, Anthony NJ, Davide JP, deSolms SJ, Giuliani EA, Gomez RP, Graham SL, Hamilton K, Handt Laurence K, Hartman George D, Koblan Kenneth S, Kral Astrid M, Miller Patricia J, Mosser Scott D, O’Neill Timothy J, Ranols Elaine, Schaber Michael D, Gibbs Jackson B, Oliff, Allen Nat Med. 1995;1:792–797. doi: 10.1038/nm0895-792. [DOI] [PubMed] [Google Scholar]
- Legius E, Marchuk DA, Collins FS, Glover TW. Nat Genet. 1993;3:122–126. doi: 10.1038/ng0293-122. [DOI] [PubMed] [Google Scholar]
- Marte BM, Rodriguez-Viciana P, Wennstrom S, Warne PH, Downward J. Curr Biol. 1997;7:63–70. doi: 10.1016/s0960-9822(06)00028-5. [DOI] [PubMed] [Google Scholar]
- Matallanas D, Arozarena I, Berciano MT, Aaronson DS, Pellicer A, Lafarga M, Crespo P. J Biol Chem. 2003;278:4572–4581. doi: 10.1074/jbc.M209807200. [DOI] [PubMed] [Google Scholar]
- Movilla N, Crespo P, Bustelo XR. Oncogene. 1999;18:5860–5869. doi: 10.1038/sj.onc.1202968. [DOI] [PubMed] [Google Scholar]
- Muir D. Clin Exp Metast. 1995;13:303–314. doi: 10.1007/BF00133486. [DOI] [PubMed] [Google Scholar]
- Murphy GA, Graham SM, Morita S, Reks SE, Rogers- Graham K, Vojtek A, Kelley GG, Der CJ. J Biol Chem. 2002;277:9966–9975. doi: 10.1074/jbc.M109059200. [DOI] [PubMed] [Google Scholar]
- Ohba Y, Ikuta K, Ogura A, Matsuda J, Mochizuki N, Nagashima K, Kurokawa K, Mayer BJ, Maki K, Miyazaki J, Matsuda M. EMBO J. 2001;20:3333–3341. doi: 10.1093/emboj/20.13.3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba Y, Mochizuki N, Yamashita S, Chan AM, Schrader JW, Hattori S, Nagashima K, Matsuda M. J Biol Chem. 2000;275:20020–20026. doi: 10.1074/jbc.M000981200. [DOI] [PubMed] [Google Scholar]
- Omer CA, Chen Z, Diehl RE, Conner MW, Chen HY, Trumbauer ME, Gopal-Truter S, Seeburger G, Bhimnathwala H, Abrams MT, Davide JP, Ellis MS, Gibbs JB, Greenberg I, Koblan KS, Kral AM, Liu D, Lobell RB, Miller PJ, Mosser SD, O’Neill TJ, Rands E, Schaber MD, Senderak ET, Oliff A, Kohl NE. Cancer Res. 2000;60:2680–2688. [PubMed] [Google Scholar]
- Qui MS, Green SH. Neuron. 1992;9:705–717. doi: 10.1016/0896-6273(92)90033-a. [DOI] [PubMed] [Google Scholar]
- Rey I, Taylor-Harris P, van Erp H, Hall A. Oncogene. 1994;9:685–692. [PubMed] [Google Scholar]
- Rong R, He Q, Liu Y, Sheikh MS, Huang Y. Oncogene. 2002;21:1062–1070. doi: 10.1038/sj.onc.1205154. [DOI] [PubMed] [Google Scholar]
- Rosario M, Paterson HF, Marshall CJ. EMBO J. 1999;18:1270–1279. doi: 10.1093/emboj/18.5.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosario M, Paterson HF, Marshall CJ. Mol Cell Biol. 2001;21:3750–3762. doi: 10.1128/MCB.21.11.3750-3762.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz AA, Govek EE, Bottner B, Van Aelst L. EMBO Rep. 2000;261:1–12. doi: 10.1006/excr.2000.5049. [DOI] [PubMed] [Google Scholar]
- Serra E, Rosenbaum T, Nadal M, Winner U, Ars E, Estivill X, Lazaro C. Nat Genet. 2001;28:294–296. doi: 10.1038/90148. [DOI] [PubMed] [Google Scholar]
- Sheela S, Riccardi VM, Ratner N. J Cell Biol. 1990;111:645–653. doi: 10.1083/jcb.111.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman LS, Atit R, Rosenbaum T, Cox AD, Ratner N. J Biol Chem. 2000;275:30740–30745. doi: 10.1074/jbc.M001702200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Triest M, de Rooij J, Bos JL. Methods Enzymol. 2001;333:343–348. doi: 10.1016/s0076-6879(01)33068-9. [DOI] [PubMed] [Google Scholar]
- Whyte DB, Kirschmeier P, Hockenberry TN, Nunez-Oliva I, James L, Catino JJ, Bishop WR, Pai JK. J Biol Chem. 1997;272:14459–14464. doi: 10.1074/jbc.272.22.14459. [DOI] [PubMed] [Google Scholar]
- Xu GF, Lin B, Tanaka K, Dunn D, Wood D, Gesteland R, White R, Weiss R, Tamanoi F. Cell. 1990;63:835–841. doi: 10.1016/0092-8674(90)90149-9. [DOI] [PubMed] [Google Scholar]
- Yamashita S, Mochizuki N, Ohba Y, Tobiume M, Okada Y, Sawa H, Nagashima K, Matsuda M. J Biol Chem. 2000;275:25488–25493. doi: 10.1074/jbc.M003414200. [DOI] [PubMed] [Google Scholar]
- Yan N, Ricca C, Fletcher J, Glover T, Seizinger BR, Manne V. Cancer Res. 1995;55:3569–3575. [PubMed] [Google Scholar]
- Yu Y, Feig LA. Oncogene. 2002;21:7557–7568. doi: 10.1038/sj.onc.1205961. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Vuori K, Wang H, Reed JC, Ruoslahti E. Cell. 1996;85:61–69. doi: 10.1016/s0092-8674(00)81082-x. [DOI] [PubMed] [Google Scholar]