Abstract
CMF1 protein is expressed in developing striated muscle before the expression of contractile proteins, and depletion of CMF1 in myoblasts results in inability to express muscle-specific proteins. Previous studies of CMF1 identify a functional Rb-binding domain, which is conserved in the murine and human homologues. Here, we show that CMF1 binds Rb family members, while a CMF1 protein with deletion of the Rb-binding domain (Rb-del CMF1) does not. Myogenic cell lines over-expressing Rb-del CMF1 proliferate normally, but exhibit markedly impaired differentiation, including dramatically reduced contractile proteins gene expression and failure to fuse into myotubes. Furthermore, by quantitative real-time polymerase chain reaction, MyoD and Myf5 mRNA levels are comparable to wild-type, while myogenin and contractile protein mRNA levels are significantly attenuated. These data demonstrate that CMF1 regulates myocyte differentiation by interaction with Rb family members to induce expression of myogenic regulatory factors.
Keywords: CMF1, LEK1, LEK proteins, Rb, myocyte differentiation
INTRODUCTION
Skeletal myocyte differentiation is a stepwise process, beginning with expression of myogenic regulators, followed by permanent withdrawal from the cell cycle, expression of contractile proteins, and ultimately cell fusion (Andres and Walsh, 1996; Walsh and Perlman, 1997). This is an important distinction from cardiac muscle, which simultaneously proliferates and differentiates during embryonic and early neonatal development (Gill and Hamel, 2000; Pasumarthi and Field, 2002). The interplay between proliferation and differentiation is less well understood in cardiac muscle. Still, permanent withdrawal from the cell cycle in cardiomyocytes is characteristic of terminal differentiation, and occurs soon after birth in most species (Soonpaa et al., 1996). There are few common regulators of early differentiation in both types of striated muscle. Avian CMF1 protein is one such regulator. CMF1 is highly expressed in chick heart and somites during embryonic development, and its expression precedes the expression of contractile proteins (Wei et al., 1996; Dees et al., 2000; Pabon-Pena et al., 2000). CMF1 RNA is detectable in the cardiac crescent (Hamburger and Hamilton stage 4), and protein in cardiogenic mesoderm (stage 5–6; Pabon-Pena et al., 2000). RNA antisense disruption of CMF1 blocks differentiation, both in precardiac mesodermal explants and in cultured skeletal myoblasts (Wei et al., 1996; Dees et al., 2000). The precise mechanism of this block in differentiation, and how it occurs in both muscle cell types, was undetermined.
Recently, we demonstrated defects in both cell cycle progression and differentiation in a skeletal myogenic cell line overexpressing a CMF1 protein with a deletion of the nuclear localization sequence (Dees et al., 2006). Importantly, cells expressing a full-length CMF1 expression construct as a control (FL-CMF1) exhibited normal cell cycle progression and differentiation, suggesting that specific alteration in nuclear localization of the CMF1 protein was responsible for the phenotype. These cells, termed NLS-del CMF1, exhibited an incomplete cell cycle withdrawal characterized by reversible G0 arrest in response to differentiating conditions. This finding was associated with failure to express contractile proteins and to form multinucleate myotubes. Given that permanent cell cycle withdrawal precedes expression of differentiation markers in normal skeletal myocytes, one explanation for the observed phenotype was that the inability to differentiate resulted directly from the proliferation defect. Alternately, the proliferation and differentiation defects might represent separable functions of CMF1. While less straightforward, this model would have better applicability to cardiomyocyte differentiation, in which proliferation and differentiation are not mutually exclusive.
An important clue to CMF1 function comes from the identification of an Rb binding domain. Presence of this and other functional domains place CMF1 within the LEK family of proteins. Other family members include murine LEK1 and human CENP-F (Goodwin et al., 1999). CMF1 expression is restricted to developing striated muscle, while the other family members are ubiquitously expressed (Goodwin et al., 1999; Pabon-Pena et al., 2000). Human CENP-F was characterized first as a kinetochore binding protein with the ability to bind Rb in a yeast two-hybrid screen (Rattner et al., 1993; Zhu et al., 1995b). Both CMF1 and LEK1 can also bind Rb (Redkar et al., 2002; Ashe et al., 2004). Furthermore, we have shown that a C-terminal fragment of LEK1 containing the predicted Rb-binding domain coprecipitates all three known Rb proteins (Rb, p107, and p130; Ashe et al., 2004). LEK1 binds a specific site known as the pocket domain of the Rb proteins, and the Rb family is also termed the “pocket proteins.” The pocket domain binds several other proteins, most notably the E2F proteins (Ashe et al., 2004). Rb/ E2F interactions are well described as major regulators of the cell cycle, through Rb phosphorylation by the cyclin-dependent kinases (Huang et al., 1988; Goodrich et al., 1991; Weintraub et al., 1992; Helin et al., 1993; Dyson, 1998).
However, in addition to its important role in cell cycle control, Rb has direct regulatory roles in other cellular processes, including differentiation and apoptosis (Zacksenhaus et al., 1996; Kaelin, 1999; Novitch et al., 1999; Classon and Harlow, 2002). For example, in differentiating myocytes, Rb cooperates directly with myogenic regulators to activate transcription of muscle specific genes (Novitch et al., 1996, 1999). Several lines of evidence from our laboratory and others’ suggest that LEK protein/Rb interaction may be important in this transcriptional activation. Papadimou et al. recently examined Rb interaction with LEK1 in ES cell differentiation into the cardiomyocyte lineage (Papadimou et al., 2005; Puceat, 2005). Rb homozygous deletion in embryonic stem (ES) cells caused specific delays in differentiation, associated with delayed expression of the cardiac-specific transcription factors Nkx2.5 and Mef2c. They further showed that blocking LEK1 expression in wild-type ES cells, using antisense methods, delayed progression into the cardiac lineage in an identical manner to Rb knockout (Papadimou et al., 2005). These data suggest overlapping roles for Rb and LEK proteins in promoting expression of cardiac-specific transcription factors.
In the current study, we show that CMF1 can bind endogenous Rb family members, by means of the Rb-binding domain that is conserved among the LEK proteins. A CMF1 mutant lacking this 90 basepair domain causes profound defects in skeletal myocyte differentiation when stably overexpressed in a myogenic cell line. Specifically, our CMF1 Rb-del cells do not activate myogenin expression or contractile protein expression, and do not differentiate into multinucleate myotubes. These defects parallel the defects shown by Papidimou et al. in Rb null/LEK1–disrupted ES cells stimulated to differentiate into the cardiomyocyte lineage. Our data show that CMF1 stimulates skeletal myocyte differentiation by means of an Rb-dependent induction of myogenic gene expression, and support a model in which tissue-specific factors coordinate with Rb proteins to promote differentiation into a specific cell lineage.
RESULTS
CMF1 and Rb Coimmunoprecipitate In Vitro and Colocalize in Mitotically Active Myocytes
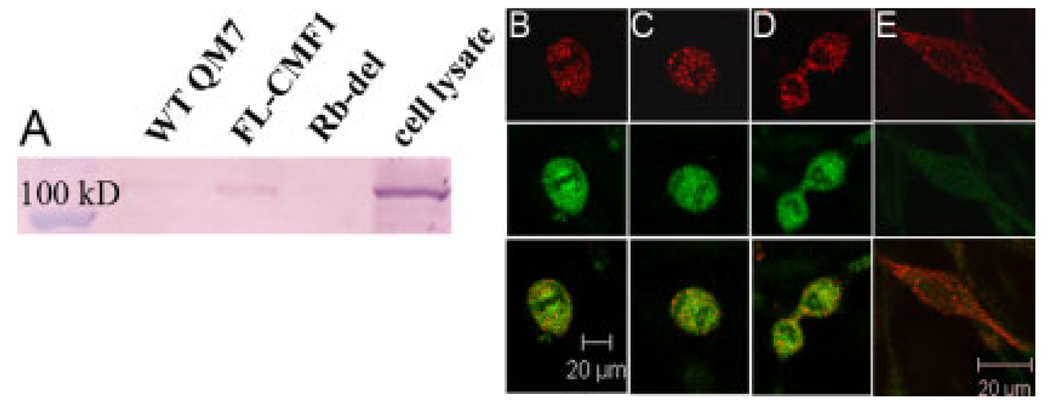
To determine whether CMF1 can interact with endogenous pocket proteins, we performed coimmunoprecipitation assays using transfected FLAG-labeled CMF1 and whole cell lysates as a source of endogenous pocket proteins. Transfected FLAG-tagged CMF1 protein was precipitated using protein G beads and anti-FLAG antibody. The precipitates were then incubated with total protein isolates from 3T3 fibroblasts, and the resulting complexes resolved by Western blotting. Anti-pocket protein antibodies were used for detection of coimmunprecipitated proteins. Results with anti-p107 antibody are shown in Figure 1A. Lysate from 3T3 cells is shown for reference, and a strong signal is seen migrating at 107 kDa. Full-length CMF1 (FL-CMF1) coimmunoprecipitates p107, while the Rb-deleted construct does not. The FL-CMF1 result is in agreement with previous reports. We have demonstrated coimmunoprecipitation of LEK1 and pocket proteins (Rb, p107 and p130) using GST-pulldown methods (Ashe et al., 2004). Redkar et al. also demonstrated CMF1/Rb interaction using an in vitro transcription and translation system, and coimmunoprecipitation using radiolabeled total protein isolated from embryonic tissues (Redkar et al., 2002).
Fig. 1.
CMF1 and Rb interactions. A: Coimmunoprecipitation: protein isolates from wild-type QM7 cells (WT QM7), QM7 cells transfected with full-length CMF1 (FL-CMF1), and QM7 cells transfected with Rb-deleted CMF1 (Rb-del), as labeled across the top, were precipitated by means of the FLAG epitope and incubated with whole cell 3T3 lysates. Resulting complexes were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis; 3T3 whole cell lysate (cell lysate) is included to show endogenous protein. Here, polyclonal anti-p107 antibody detects endogenous p107 protein (107 kDa) coimmunoprecipitated by FL-CMF1 protein but not by Rb-del protein, indicating that the Rb binding domain deletion abrogates binding. B–E: Colocalization: confocal microscopy, ×60 magnification, of wild-type QM7 myocytes immunostained with anti-CMF1 antibody (red) and anti-Rb antibody (green). The bottom row is an overlay of the red and green channels to show colocalization. Shown are examples of mitotic figures showing colocalization of CMF1 and Rb in metaphase (B), anaphase (C), and early in cytokinesis (D). E: An elongating cell, early in differentiation. Note that this cell is CMF1-positive but Rb-negative.
Next, we tested whether endogenous CMF1 and pocket proteins colocalize in wild-type cells before the onset of differentiation. To show this, we performed immunofluorescence studies using anti-CMF1 and anti-Rb antibodies in QM7 cells. These results are shown in Figure 1B. As we have shown previously, CMF1 protein (red) is nuclear in myoblasts, clusters around the chromatin in mitotic cells, and translocates to the cytoplasm in differentiating cells (Dees et al., 2006). Here both anti-CMF1 and anti-Rb (green) antibodies detect protein clustering around the chromatin during mitosis. Note there is some association of Rb protein with the chromatin that is not seen in CMF1, particularly in Figure 1B. Still the patterns in Figure 1B–D show considerable overlap, as seen in the bottom panel with dual exposures. Note, however, in Figure 1E that, while CMF1 is highly expressed in the cytoplasm in this differentiating cell, there is no expression of Rb protein. The coimmunoprecipitation and immunofluorescence data shown here indicate that CMF1 can bind pocket proteins, that the 90 base pair-binding domain is critical for this interaction, and that CMF1 and Rb proteins are coexpressed in cell nuclei before differentiation.
Generation of CMF1 Rb-Binding Domain Deleted Cell Lines (Rb-del CMF1)
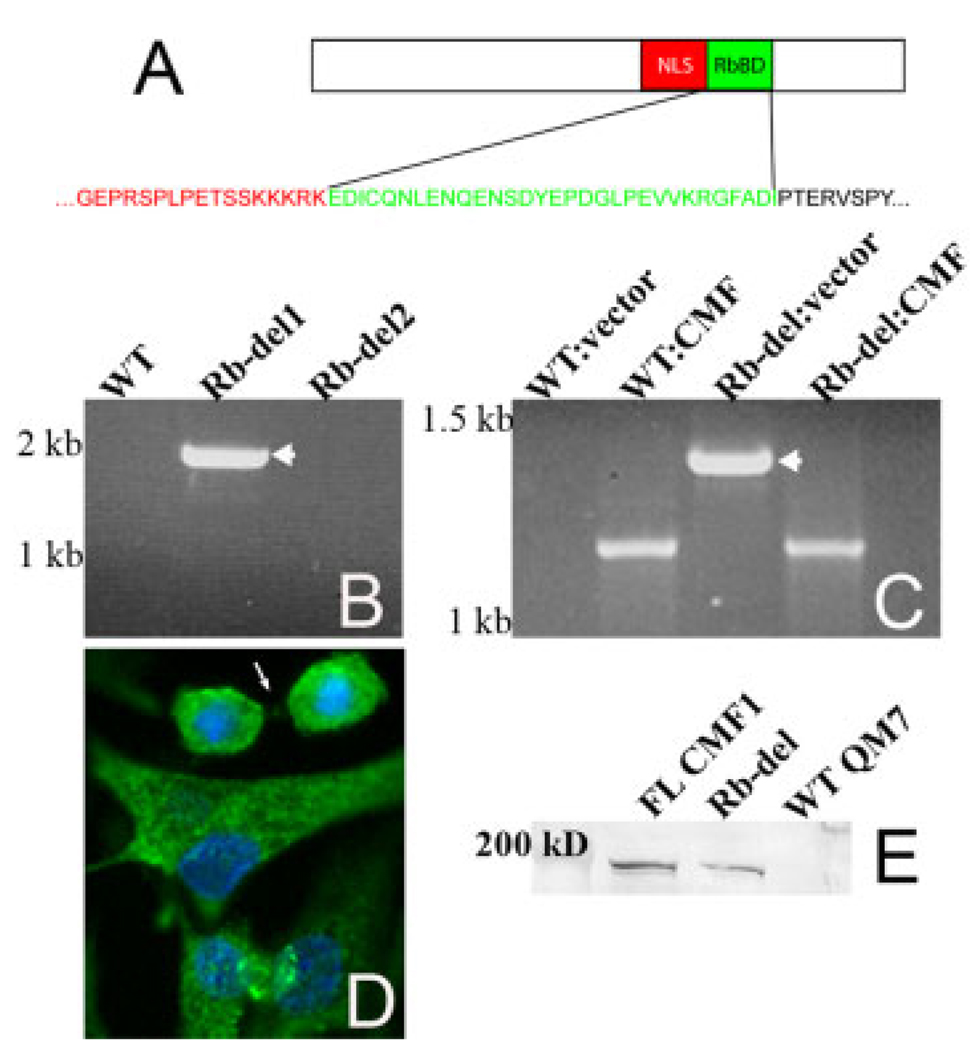
To determine whether CMF1/Rb interaction is necessary for skeletal myogenesis, we generated a stably transfected myocyte line overexpressing CMF1 protein with a specific deletion of the 90 base-pair Rb binding domain. The specific amino acids included in the deletion are shown in Figure 2A. The parental cell line was QM7, a quail myogenic line we have previously shown to express CMF1 with 94% nucleic acid homology to chick CMF1 (Dees et al., 2006). We verified stable incorporation of the construct (genomic polymerase chain reaction [PCR]), mRNA expression (reverse transcriptase-PCR [RT-PCR]) and production of altered CMF1 protein (immunocytochemistry), as shown in Figure 2B–D. A total of eight Rb-del clones were analyzed; two that did not pass genomic screening were not studied further. One of these clones (Rb-del2) is included in the genomic PCR data shown in Figure 2B. Total RNA was isolated from each cell line that passed genomic screening, and tested for CMF1 Rb-del mRNA by RT-PCR. Note that endogenous CMF1 is detected in addition to transfected message when CMF1-specific primers are used, and that these products can be distinguished by size (Fig. 2C). Finally, using the FLAG epitope included in the CMF1 Rb-del construct, CMF1 Rb-del protein is identified by immunofluorescence microscopy of cells labeled with anti-FLAG antibody (Fig. 2D). No signal is detected in wild-type QM7 (not shown). The Rb-del protein predominantly localizes to the cytoplasm, although it is also in the nucleus of some cells. Note how it clusters around the chromatin in the cell undergoing cytokinesis (arrow; compare with Fig. 1D). These resemble the endogenous CMF1 localization patterns previously reported (Dees et al., 2000, 2006) and suggest that CMF1 Rb-del protein of construct origin localizes properly within the cell. In Figure 2E, Western blotting of cell isolates demonstrates expression of the FLAG epitope in FL-CMF and in Rb-del CMF1 transfected cells, but not in wild-type QM7 cells, showing specificity of the FLAG marker for transfected CMF1 proteins.
Fig. 2.
Verification of stable transfection in Rb-del CMF1 cells. A: Schematic of the CMF1 Rb binding domain (RbBD), showing the amino acids included in the deletion. B: Genomic polymerase chain reaction of wild-type QM7 cells (WT) and two clones of Rb-del CMF1 cells (Rb-del 1 and 2) using vector-specific 3′ primer. Expected size for construct CMF1 is 1950 bp, seen only in Rb-del1 (arrow). The Rb-del2 clone was excluded from further analysis. C: Reverse transcriptase-polymerase chain reaction (RT-PCR) of CMF1 message from total RNA isolated from wild-type QM7 cells (WT) and Rb-del CMF1 cells (Rb-del). PCR amplification is performed in each sample using the same CMF1 5′ primer with both a vector-specific 3′ primer (vector) and a CMF1-specific 3′ primer (CMF). Only Rb-del CMF1 cells amplify message when the vector-specific 3′ primer is used (1350 bp, arrowhead). Endogenous message is present in both samples using CMF1-specific primers at 1200 bp. D: FLAG immunocytochemistry, green, and DAPI (4′,6-diamidine-2-phenylidole-dihydrochloride), blue, of Rb-del CMF1 cells showing expression of the FLAG epitope. Epifluorescence microscopy, ×40 magnification. E: FLAG epitope is detected by anti-FLAG antibody in cells transfected with FL-CMF1 and Rb-del constructs, but not in untransfected WT QM7 cells. FL-CMF1 migrates at 179 kDa and Rb-del at 176 kDa.
Rb-del CMF1 Cells Proliferate Normally
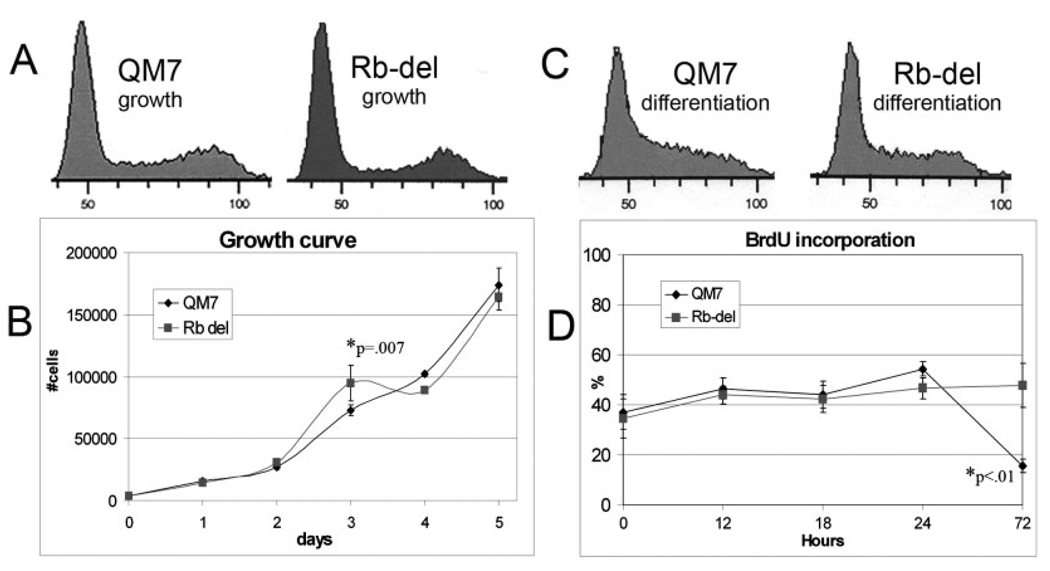
To determine whether deletion of the CMF1 Rb-binding domain disrupts proliferation, we performed fluorescence-activated cell sorting (FACS) analysis, immunocytochemistry, and growth curve analyses on proliferating Rb-del CMF1 cells. FACS analysis (Fig. 3A) shows a normal cell cycle profile for Rb-del CMF1 cells, as compared to wild-type, sampled at 70% confluence during rapid growth phase. Growth curves for wild-type CMF1 and Rb-del CMF1 from day 1 through 5 are shown in Figure 3B. A statistical difference is seen only at day 3, with higher numbers of Rb-del CMF1 cells than QM7. This trend is reversed in days 4 and 5, although not to statistical significance. For time points beyond day 5 (not shown), an increasing proportion of spontaneously differentiated myotubes in the QM7 population were observed; such multinucleate myotubes were excluded from cell counts. Still, counts for individual cells remained similar between QM7 and Rb-del CMF1 cells through day 10. We conclude from these analyses that overexpression of Rb del-CMF1 does not affect the progression of normal mitosis in growth-promoting culture conditions.
Fig. 3.
Normal proliferation in Rb-del CMF1 cells. A: A fluorescence-activated cell sorting (FACS) profile of wild-type QM7 and Rb-del CMF1 cells in growth phase, showing a normal distribution in all phases of the cell cycle. Propidium iodide was used to stain cell nuclei, and cells were sorted based on DNA content. B: Growth curves of wild-type QM7 and Rb-del CMF1 cells in growth medium showing similar patterns. The asterisk denotes the time point with statistically different values by Student’s t-test, n = 6 values for each time-point. C: FACS profiles of QM7 and Rb-del CMF1 cells at day 1 in differentiation medium. The patterns are similar and are consistent with cell cycle synchronization, but not block, in response to low serum medium. D: Bromodeoxyuridine (BrdU) labeling of cell lines as labeled placed in differentiation medium. The Rb-deleted cells incorporate BrdU similarly to wild-type, until the 72-hr time point when the majority of wild-type cells have differentiated (asterisk).
We next examined the cell cycle response to differentiating conditions. FACS analysis of cells assayed after 24 hours in differentiation medium is shown in Figure 3C and is similar between wild-type and Rb-del CMF1 cells. This remained true in multiple assays through 72 hours in differentiating conditions. It is important to note that differentiated cells (myotubes) are filtered out in this analysis, thus only the subset of undifferentiated cells that remain mononuclear is included. A more complete picture of these cultures is demonstrated by bromodeoxyuridine (BrdU) incorporation in Figure 3D. Here, all cells in the culture were analyzed, including differentiated myotubes. Rb-del CMF1 cells and wild-type QM7 show similar BrdU incorporation for 24 hours. By 72 hours, wild-type cells exhibit a sharp decline in BrdU incorporation, corresponding to the majority of cells in these cultures having differentiated into myotubes, which do not incorporate BrdU (Gill and Hamel, 2000; Dees et al., 2006). In contrast, Rb-del CMF1 cells exhibit the same percent of BrdU incorporation at 72 hours as at hour 0. These data indicate that the Rb-del mutation does not prevent continued proliferation in cells that are not responding to (or receiving) differentiation signals.
CMF1 Rb-del Cells Exhibit Impaired Differentiation
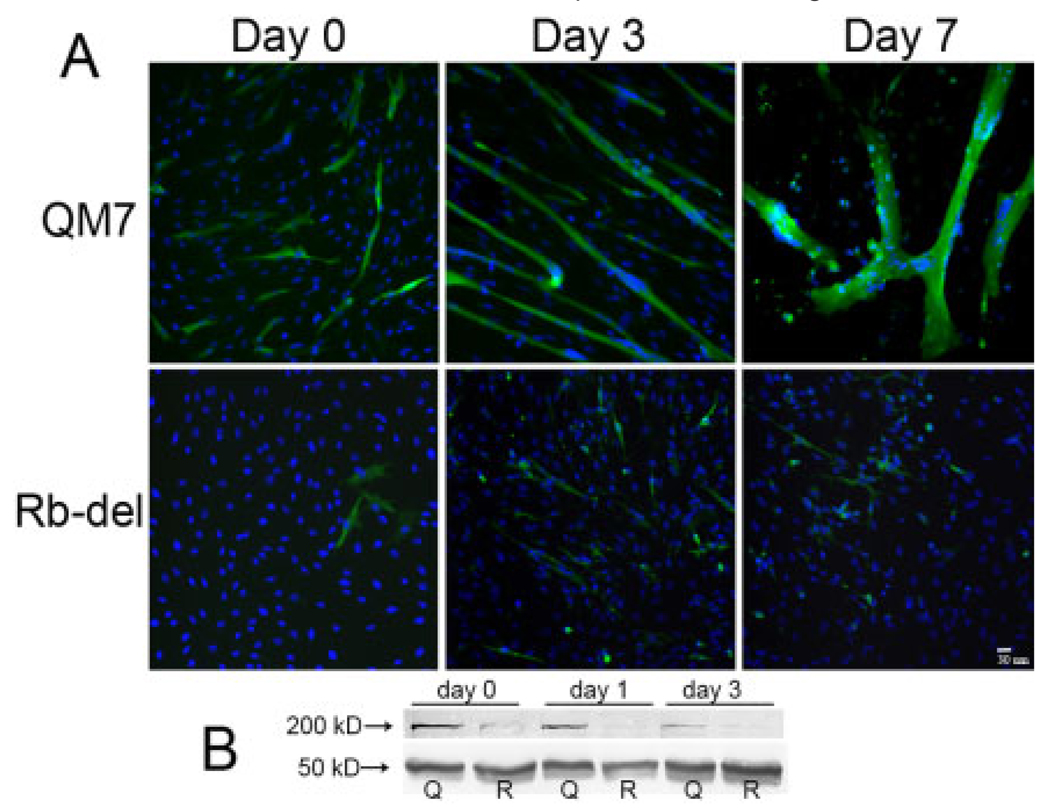
Rb-del CMF1 cells exhibited a striking impairment in their ability to differentiate in response to normal differentiation signals. This phenotype is similar, but more severe, than that observed in our previously reported NLS-del CMF1 cells and is shown in Figure 4. Rb-del CMF1 cells form very rare multinucleate myotubes, instead remaining as single cells (94%) for up to 7 days in differentiation (low serum) medium. In contrast, the majority of wild-type QM7 cells (74%) fuse into large multinucleate syncytia by 7 days in differentiation medium. Myosin is highly expressed in >75% of QM7 cells by day 7, compared to 15% in Rb-del cells by day 7. This differentiation abnormality is apparent even before switching to differentiation medium. Note at day 0, while the cells are still in growth medium, there are myosin-positive cells in wild-type QM7 cells (6%), some of which are beginning to fuse into myotubes. Rare Rb-del CMF1 cells are myosin-positive (1%), and no myotube fusion is seen. Western blotting (Fig. 4B) corroborates the morphologic and immunofluorescence finding of decreased myosin protein in Rb-del CMF1 cells compared with wild-type at confluence (day 0), and day 1 and day 3 in differentiation medium.
Fig. 4.
Rb-del CMF1 cells differentiate abnormally. A: Immunocytochemistry showing myosin staining (green, MF20 antibody) and DAPI stained nuclei in wild-type QM7 cells (top) and Rb-del CMF1 cells (bottom) at time points indicated. Epifluorescence microscopy, ×20 magnification. The majority of Rb-del CMF1 cells do not express myosin, and there is a complete lack of myocyte fusion into multinucleate myotubes. B: Western blot of total protein isolated from wild-type QM7 and Rb-del CMF1 cells at confluence (day 0), day 1, and day 3 in differentiation medium. Myosin protein is detected with MF20 antibody and anti-alpha tubulin is used as a loading control. Note lower myosin content in Rbdel CMF1 cells compared to wild-type.
CMF1 Rb-del Differentiation Is Disrupted at the Stage of Myogenin Activation
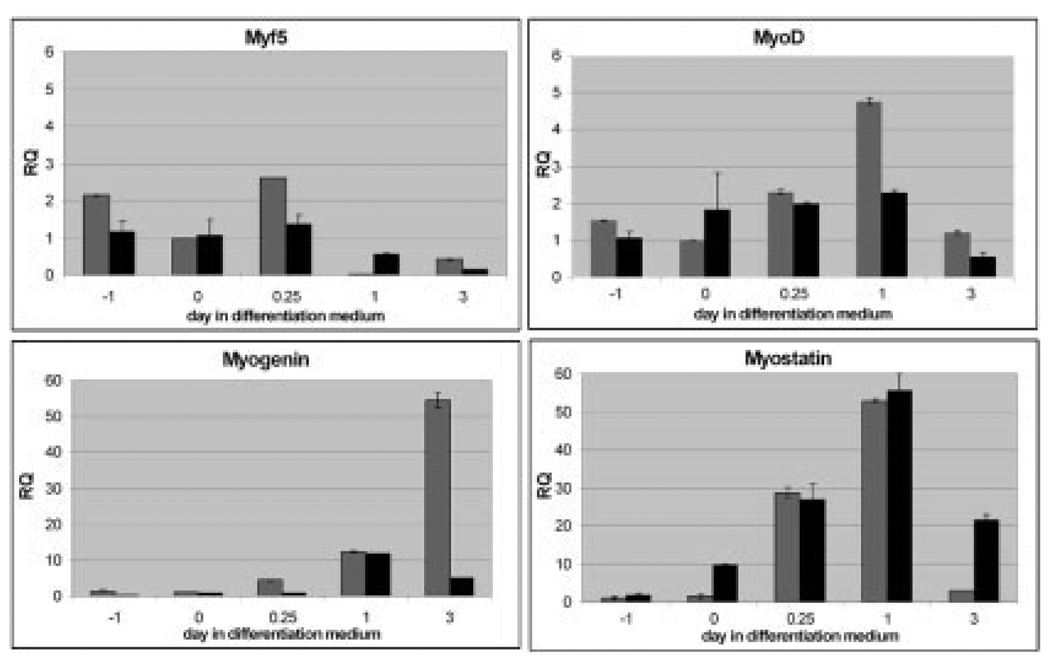
To determine the mechanism underlying abnormal differentiation in Rb-del CMF1 cells, we examined the message expression of known skeletal myogenic regulators in differentiating wild-type QM7 and Rb-del CMF1 cells by real-time PCR. The results are shown for wild-type QM7 (gray) and Rb-del CMF1 cells (black) in Figure 5, and are calibrated to message levels at day 0. Day 0 denotes cultures at confluence in high serum medium. Day −1 denotes subconfluent, proliferating cells 1 day before confluence, and each point after day 0 refers to the time after placement in low serum (differentiation) medium. MyoD and Myf5 transcript levels increased less than fivefold in both cell types, with peak of Myf5 at hour 6 and MyoD at hour 24 in both wild-type and Rb-del CMF1 cells. Myogenin expression, however, is nearly 60-fold greater in wild-type cells at day 3 in differentiation medium. This up-regulation of myogenin is completely blunted in the Rb-del CMF1 cells. We next tested the possibility that increased expression of the negative regulator, myostatin was suppressing myogenin expression. We saw significant up-regulation of myostatin in both wild-type QM7 and Rb-del CMF1 cells, with little difference between the two cell lines to implicate this to be the major mechanism for myogenin suppression. These data support a direct role for CMF1/Rb to activate myogenin expression.
Fig. 5.
Real-time quantitative reverse transcriptase-polymerase chain reaction (RT-PCR) of myogenic regulatory factors as labeled; wild-type QM7 cells are shown in gray and Rb-del CMF1 cells in black. Relative quantity (RQ) is calibrated to day 0 wild-type QM7 cells. Day −1 denotes subconfluent cells, day 0 confluent, and 0.25, 1, and 3 are days after switch to differentiation medium. Note the dramatic lack of activation of myogenin in Rb-del cells.
Failure of Myogenin Expression in Rb-del CMF1 Cells Results In Attenuated Contractile Protein Expression
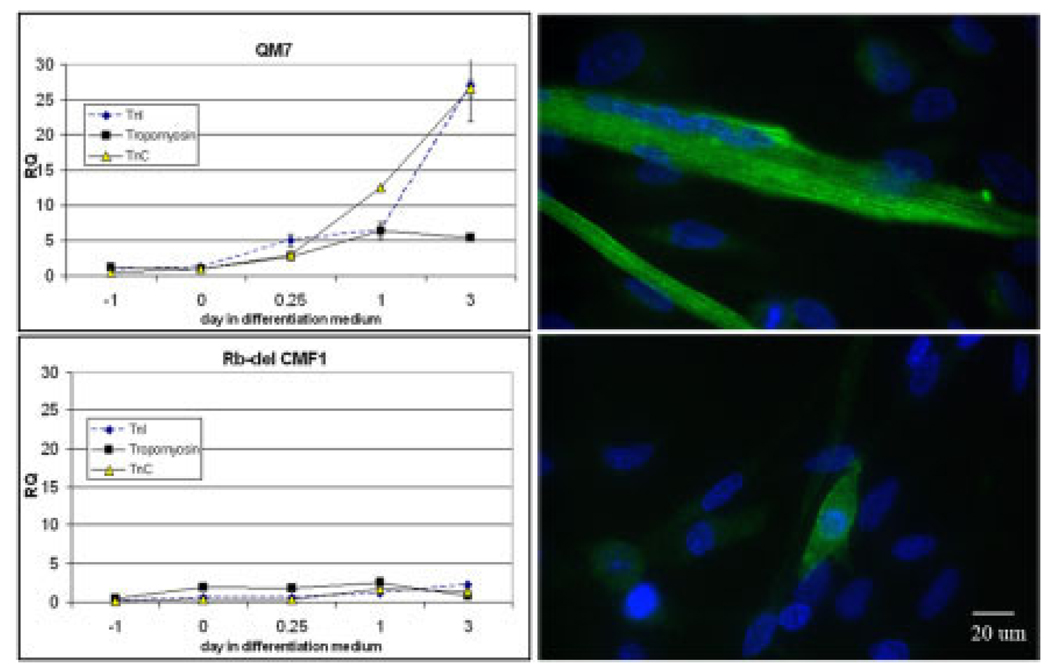
To verify that decreased myogenin expression in the Rb-del CMF1 cells is associated with decreased contractile protein gene expression, we examined message levels of troponin C (slow/cardiac isoform), troponin I (slow skeletal isoform), and α-tropomyosin message expression by real-time PCR. All of these isoforms are expressed in quail myoblasts as they differentiate (Hastings and Emerson, 1982; de la Brousse and Emerson, 1990; Antin and Ordahl, 1991). These data are shown in Figure 6. Note the increase in troponin C and I message levels in wild-type QM7 cells, beginning at hour 6, and increasing sharply to greater than 30-fold between day 1 and 3. Tropomyosin message levels also increased fivefold. The curves for all three messages are remarkably flat in the Rb-del CMF1 cells, indicating no significant activation of message transcription (P < 0.001 for troponin C and I; P = 0.03 for tropomyosin). Also shown is immunofluorescence staining of titin, a marker of sarcomere assembly (Dabiri et al., 1997; Pizon et al., 2002). Note that titin is extensively expressed in QM7 myotubes at day 5 of differentiation, while there is only scant staining along the periphery of Rb-del CMF1 cells. This, along with the data shown in Figure 4, confirms that contractile proteins are expressed at very low levels in the Rbdel CMF1 cell line. These data confirm that CMF1/Rb interaction is necessary for myocyte differentiation.
Fig. 6.
Left, real-time quantitative reverse transcriptase-polymerase chain reaction (RT-PCR) of Troponin I (slow skeletal isoform), Troponin C (slow/cardiac isoform), and skeletal #-tropomyosin in wild-type QM7 and Rb-del CMF1 cell lines during the same differentiation time course as in Figure 5. Note an abrupt increase in message levels in wild-type cells between day 1 and 3 that is completely absent in Rbdel CMF1 cells. Right, Titin immunostaining (green) is shown with DAPI (blue, 4′,6-diamidine-2-phenylidole-dihydrochloride) in QM7 cells, top, and Rb-del CMF1 cells, bottom, after 5 days in differentiation medium. Note the lack of organized myofibrils in the Rb-del CMF1 cells. Epifluorescence microscopy, ×60 magnification.
DISCUSSION
Previously we showed that overexpression of full-length CMF1 in QM7 cells did not alter proliferation or differentiation, while overexpression of a CMF1 protein lacking the nuclear localization domain altered both (Dees et al., 2006). Altered nuclear localization likely has multiple effects on cells, including but not exclusive to interaction with Rb and other pocket proteins. Thus, the Rb-del CMF1 cell line reported in the current study is our most specific assay to date for determining CMF1 function. We demonstrate a coordinated role for CMF1 and pocket proteins in activating skeletal myogenic factors critical for differentiation. Our current data demonstrate that CMF1 binds endogenous pocket proteins, and that abrogation of this binding in Rb-del CMF1 cells blocks differentiation. We identify an inability of Rb-del CMF1 cells to activate myogenin mRNA expression, leading to an inability to activate contractile protein expression, as a mechanism by which this occurs. Cell cycle profiles suggest that CMF1/pocket protein interaction does not directly affect cell cycle progression. This suggests separable functions of both CMF1 and the pocket proteins affecting cell cycle regulation and stimulating differentiation.
Parallels to Cardiomyocyte Differentiation
Our findings compliment and extend those of Papadimou et al. who demonstrated specific LEK/Rb interaction in ES cells differentiated into cardiomyocytes (Papadimou et al., 2005). These investigators reported delayed differentiation in Rb homozygous null ES cells that was recapitulated by blocking LEK1 expression in wild-type ES cells. The authors used LEK1 antisense strategies, as well as a peptide blocker of the LEK1 Rb-binding domain. In each case, there was delayed expression of cardiac differentiation markers Nkx2.5 and Mef2c by quantitative RT-PCR and of contractile proteins by immunohistochemistry. The resulting phenotype was a delay, not complete block, of differentiation. The authors did not assay for increased p107 or p130 expression as a possible compensatory factor. However, they demonstrated that the phenotype could be rescued by overexpressing Nkx2.5, or by treating with soluble BMP2 or TGFβ. The authors noted a correlation of their findings to those of Pabon et al. showing preserved CMF1 expression in avian mesodermal explants cultured in the presence of the BMP antagonist, noggin (Pabon-Pena et al., 2000). We included BMP receptor 1A in our panel of real-time PCR assay to see if differences would be apparent in the Rb-del CMF1 cells (data not shown) but did not detect significant differences. Most studies demonstrate negative regulation of the skeletal myocyte lineage by BMPs (Reshef et al., 1998; Nakamura et al., 2005; Frank et al., 2006). However, such effects are likely to be complex, as different BMP family members can have opposing effects on differentiation, (Reshef et al., 1998; Frank et al., 2006) and timing of tissue exposure to BMPs may be critical to their effect (Yuasa et al., 2005).
Rb Null Cell Models of Skeletal Myocyte Differentiation
There are other precedents for a role for Rb protein in skeletal myocyte differentiation. Transfection of fibroblasts with the myogenic regulator MyoD, or with related factors Myf-5 or Mrf4, has been shown in multiple studies to recapitulate skeletal myocyte differentiation (Davis et al., 1990; Edmondson and Olson, 1990; Miner and Wold, 1990). However, when Rb null fibroblasts are transfected with MyoD, late muscle-specific genes such as myosin fail to activate (Novitch et al., 1996). Induction of earlier myogenic factors, including myogenin, occurs at levels comparable to wild-type. Using reporter activation assays, the transcription factor Mef2 has been implicated as the differentiation limiting step in Rb null cells. Specifically, without Rb protein Mef2 accumulates in the nucleus, binds DNA, but does not activate transcription (Novitch et al., 1999). More recent studies have shown that Mef2D cooperates with myogenin to recruit SWI/SNF ATP-dependent chromatin remodeling complexes to the promoters of late myogenic genes (de la Serna et al., 2005; Ohkawa et al., 2006). A possible role for CMF1/Rb complexes in directly inducing myogenin expression, or in promoting or stabilizing the myogenin/ Mef2-mediated recruitment of chromatin remodeling enzymes to myogenic gene promoters is intriguing. This question will be explored in future studies.
Partial Compensation of Rb Null Phenotype by p107
Our data indicate a block in myogenin expression, while Rb null fibroblasts have normal myogenin levels with a block further downstream in differentiation. A possible explanation for this discrepancy is that our CMF1 Rb-del protein is unable to bind any of the pocket proteins, and is not limited to Rb itself. Thus, any compensatory ability of alternate pocket proteins for Rb disruption would be blocked in our assay as well. A study by Schneider et al. looked at differentiation in Rb null myocytes, cloned from an Rb null murine teratoma (Schneider et al., 1994). Similar to the Rb null fibroblast studies, these investigators found normal myogenin levels but attenuated late differentiation markers/contractile proteins. Importantly, they also demonstrated abnormal up-regulation of p107 during myogenic differentiation in Rb null myocytes, and hypothesized that p107 played a compensatory role partially activating the myogenic pathway (Schneider et al., 1994). To support this, they showed that either Rb or p107 proteins can coimmunoprecipitate myogenin from cell extracts and that transfection of p107 into undifferentiated wild-type myoblasts accelerated normal differentiation (Schneider et al., 1994). Our data fit with such a model. Our Rb-del cells express CMF1 protein that lacks the capacity to bind any of the pocket proteins, thus compensatory effects of the other pocket proteins for normal CMF1/Rb functions would also be abrogated. If so, our findings would more truly reflect the in vivo function of Rb/CMF. Alternate factors may also contribute to the differences in results, including different species (avian vs. murine), different cell types, and the effects of transient overexpression of MyoD in the Rb null background on one hand and stable overexpression of CMF1 with wild-type Rb on the other. We are currently developing other models to test these possibilities.
Cell Cycle and Differentiation Effects of CMF/Rb Are Separable
Our cell cycle profiles of wild-type and Rb-del QM7 cells suggest that CMF1/Rb interaction does not directly affect cell cycle progression. However, several studies from our laboratory and others have shown direct effects of both CMF1 and the related proteins murine LEK1 and human CENP-F on proliferation. Treatment with a morpholino (synthetic antisense oligonucleotide) targeted against the 5′ untranslated region of murine LEK1 in 3T3 fibroblasts resulted in decreased cell numbers by greater than 50% after 3 days of treatment (Ashe et al., 2004; Dees et al., 2005). Our NLS-del CMF1 cells, a stably transfected QM7 line overexpressing a nuclear localization domain deleted CMF1 protein, exhibited defects both in the ability to permanently arrest in G0 and to differentiate when placed in low serum medium (Dees et al., 2006). Human CENP-F has been shown to bind the kinetochore during mitosis, participating directly in the mechanics of cell division (Rattner et al., 1993; Liao et al., 1995; Zhu et al., 1995a,b). The kinetochore binding domain has been determined, and is distinct from the Rb-binding domain (Zhu et al., 1995a). Recently, Evans et al. showed that overexpressing a C-terminal fragment of LEK1 (termed in this manuscript murine [m] CENP-F) caused delay in progression through the G2/M phase of mitosis in transiently transfected NIH 3T3 cells (Evans et al., 2007). These investigators also showed colocalization of endogenous pocket proteins and mCENP-F, with dynamic localization during mitosis, similar to our results for CMF1 and Rb shown in Figure 1. Evans et al. hypothesize that mCENP-F represents a link between pocket protein-mediated cell cycle regulation and mitosis regulation by means of kinetochore assembly (Evans et al., 2007). Our data fit with such a model, but underscore that these functions are separable. We show that functions related to pocket protein interactions, mediated by the CMF1 Rb-binding domain are not specific to cell cycle control. Functions related to cell cycle mechanics are distinct, and likely related to the kinetochore binding domains and possibly other mediators.
Certainly the Rb proteins have a well defined and critical role in cell cycle control. This is primarily by repressing the E2F family of transcription factors critical to cell cycle progression (Friend et al., 1986; Huang et al., 1988; Goodrich et al., 1991; Weintraub et al., 1995). Evans et al. tested the possibility that mCENP-F is involved in the aspect of Rb function. By reporter activation studies, transfection of mCENP-F constructs did not transactivate cell cycle regulatory protein expression, including pRb, E2F, p53, and c-myc, suggesting that mCENP-F/pocket protein interactions do not mediate E2F transcriptional regulation (Evans et al., 2007). Cell cycle abnormalities have been shown to occur in Rb null fibroblasts and Rb null myocytes, and serum stimulation induces Rb null partially differentiated myocytes to re-enter the cell cycle (Schneider et al., 1994; Novitch et al., 1996, 1999). Schneider et al. found that this aspect of Rb function was not compensated by p107; indeed p107 levels were decreased in serum stimulated Rb null cells compared with wild-type (Schneider et al., 1994). Novitch et al. also found that the function of Rb restricting mitosis in differentiating cells was separable from its effects on differentiation markers (Novitch et al., 1996). Interestingly, the mechanism by which Rb promotes cell cycle withdrawal may be by means of p21 up-regulation (Kang et al., 2004) by the same SWI/SNF chromatin remodeling complexes implicated in myogenin activation (Dunaief et al., 1994; Kang et al., 2004; de la Serna et al., 2005; Ohkawa et al., 2006). Thus, it is possible that CMF1 and other LEK proteins are cofactors for the differentiation aspect but not for the cell cycle withdrawal aspect of Rb/chromatin remodeling complex function.
EXPERIMENTAL PROCEDURES
Generation of CMF1 Constructs
Full-length CMF1 was tagged with a 5′ FLAG epitope and subcloned into the pCI-neo vector (Promega) using the polylinker restriction sites SalI and MluI. The pCI-neo vector contains CMV promoter/enhancer sequence and an SV40 late polyadenylation sequence (Dees et al., 2006). From this construct, the coding sequence for the Rb domain was deleted using pcr mutagenesis using techniques previously described (Knight et al., 2003). Primers are as shown in Table 1.
TABLE 1.
Primers Used to Generate CMF1 Constructsa
| Primer | Direction | Purpose | Sequence 5′ to 3′ |
|---|---|---|---|
| Rb F | Sense | Deletion construct generation |
caacgtccggatatcctgaatattctaaaagtggcc |
| Rb-F1 short | Sense | actagctcacccactgaaagagttagcccttatatt | |
| Rb-F1 long | Sense | cctttgccagagactagctcacccactgaaagagttagcccttatatt | |
| Rb-R1 short | Antisense | ttcagtgggtgagctagtctctggcaaaggagatct | |
| Rb-R1 long | Antisense | agggctaactctttcagtgggtgagctagtctctggcaaaggagatct | |
| CMF1 4160 | Sense | Genomic PCR | cagttgacacgcatggaac |
| CMF1 4770AS | Antisense | tgacttcaaatgctcaatctcc | |
| CMF1 4440 | Sense | RT-PCR | caggaagaatttactatggagagg |
| CMF1 5730AS | Antisense | gctgttcttgcttgggactgc | |
| T3 (Promega) | Antisense | All of the above | attaaccctcactaaaggga |
PCR, polymerase chain reaction; RT-PCR, reverse transcriptase-PCR.
A BsiM1 restriction site included in the RbF1 primer sequence and a Not1 restriction site were used for directional cloning back into the pCi-neo vector. The resultant clone was sequenced, confirming in-frame deletion of the Rb-binding domain.
PCR
Genomic DNA and total RNA were extracted from QM7 lines as separate procedures, using Trizol (Gibco/BRL) per manufacturer’s protocols. Genomic DNA was used for PCR, using primers flanking the Rb deletion, such that Rb-deleted CMF1 could be distinguished from wild-type. All primers used in these reactions are as shown in Table 1. For RT-PCR, the Access RT-PCR One Step system (Promega) was used to amplify CMF1 Rb-deleted fragments, using random decamers for the reverse transcriptase reaction and primers as shown in Table 1.
For real-time PCR, the TaqMan Gene expression Array system from Applied Biosystems was used according to the manufacturer’s instructions. The primers used were custom designed by Applied Biosystems from published quail sequences, and are listed in Table 2. Each primer set spans an intron/exon boundary. Primers to amplify the 18S ribosomal internal control were also obtained from Applied Biosystems (Applied Biosystems). Total RNA was extracted using Trizol, with an additional ethanol precipitation step. Samples were DNAse (Ambion) treated to remove contaminating genomic DNA. Reverse transcription was performed in 50-µl reactions using the Applied Biosystems TaqMan multiscript RT kit, using one microgram of total RNA for each reaction. Before beginning the experimental real-time PCR, the relative efficiencies for the target and reference primer pairs were estimated by serial dilution of cDNA. A standard curve was obtained from RNA quantities ranging from 5 to 50 ng, plotting ΔCt vs. log ng of RNA. The slopes of the curves ranged from 0 to 0.16; a slope less than 0.1, indicating sufficiently comparable efficiencies, according to the Applied Biosystems protocol. Experimental real-time PCR amplification was then performed in 10-µl total reaction volumes, using the primer/probes as listed in Table 2 and 10 ng of each template. Reactions were performed in 384-well plates using the ABI Prism 7900 Sequence Detection System (Applied Biosystems). PCR conditions were 50°C for 2 min, 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec, and 60°C for 1 min. Each sample was assayed in duplicate, and each reaction was performed two to three times. The Applied Biosystems RQ Manager software used to generate a melting curve and calculate Ct values. The relative quantity of mRNA was calculated by first taking the difference between Ct values for target message and 18S (=ΔCt), then between ΔCt and a calibrator ΔCt value (ΔΔCt). Wild-type QM7 cells at confluence were used as the calibrator for each primer set. The relative quantity of mRNA was then calculated by the formula 2 −ΔΔCt.
TABLE 2.
Primers Designed From Published Quail Sequences
| Quail gene | NCBI accession no. |
Forward primer | Reverse primer | FAM labeled probe | Figure |
|---|---|---|---|---|---|
| Myostatin | AF407340 | acaaccgaaacgattatcacaatgc | gcaacattttggttttccctccatt | acggagtctgattttc | 5 |
| QMF3 (Myf5) | L15472 | ccctttgtcccctctgctt | ggtcgcgcctccct | acgcacacagcccc | 5 |
| QMF1 (MyoD) | L16686 | ccgatggcatgatggagtacag | gtgtagtagctgctgtcataactgt | ccgccttgcagctct | 5 |
| QMF2 (myogenin) |
L15473 | ccccacagatcatctcctgagt | aggagagcgagtggaggtt | cctctgctgcatcatc | 5 |
| BMPR1A | AF189777 | agggaagatatggtgaagtgtggat | tgaaaaacactttgactgcgacttt | tcacccctccatttcc | - |
| Tropomyosin | M15043 | ctgagcttgaagaggagttgaaaac | gtcttctttctgcgagtacttctca | cctccagcgacttcag | 6 |
| Troponin I | U37118 | caacctgaagtctgtcaagaagga | tgcgccaatcaccaacct | acgggacgctccttc | 6 |
| Troponin C | M29722 | gagatgatcgatgaggtggatgag | ccgaaccatcataacaaggaactga | acggtgccgctgccat | 6 |
Immunocytochemistry/Immunoprecipitations
Cells grown on glass slides were fixed with methanol and processed for immunocytochemistry as previously described (Dees et al., 2000). Primary antibodies, dilutions or concentrations, and sources were as follows: anti-CMF 1:200 (Dees et al., 2000, 2006); MF20 1:2 (Bader et al., 1982); polyclonal anti-Flag, 5 µg/ml (Sigma); anti-titin 1:2,000 (Sigma); and polyclonal anti-Rb 1:100 (Santa Cruz Biotechnology, Inc.). Secondary antibodies used included Cy2 1:1,000 and Cy3 1:1,000 conjugated to donkey anti-mouse or anti-rabbit IgG (Jackson laboratories). In cases where only polyclonal antibodies were available, cross-reactivity was avoided by direct conjugation of primary antibodies to Alexa 488, using the Zenon Rabbit IgG labeling kit from Molecular Probes. DAPI 1:1,000 (4′,6-diamidine-2-phenylidoledihydrochloride; Boehringer-Mannheim) was used for nuclear visualization with epifluorescence. BrdU labeling experiments were performed according to the manufacturer’s protocol (Roche), with addition of DAPI. Cells were visualized using epifluorescence or confocal microscopy. For confocal microscopy, samples were imaged using the ×40, 1.3-numerical aperture F-Fluar objective lens of a Zeiss LSM 510 confocal microscope and software. Multiline scans (488- and 543-nm laser lines) were used to eliminate detection bleed-through for the green (bandpass 505–550 filter) and red (longpass 560 filter) fluorophores. Control cells incubated in the absence of primary antibody were also examined, and these cells showed negligible fluorescence. For widefield epifluorescence, a Nikon Eclipse microscope was used with ×20, ×40 or ×60 objectives. Nikon NIS Elements imaging software was used for measurements.
For coimmunoprecipitation assays of CMF1 with Rb protein, QM7 cells were transfected with full-length or Rb-deleted CMF1. Precleared protein isolates from these cells were incubated with anti-FLAG antibody and complexes were immunoprecipitated using horse anti-mouse secondary antibody conjugated to Protein G beads (Dynabeads, Invitrogen). The resulting complexes were washed and incubated with 3T3 whole cell lysates (two plates, each containing approximately 1 × 106 cells) overnight. The complexes were again isolated, washed, boiled in sample buffer, and resolved using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Western blotting was performed using standard methods (Harlow and Lane, 1988). Primary antibodies for Western blotting were polyclonal anti-Rb, p107, and p130 polyclonal antibodies at 1:200 (Santa Cruz), and anti-FLAG polyclonal antibody at 3.5 mg/ml (Sigma). MF20 1:2 (Bader et al., 1982) and anti–α tubulin 1:1,000 (Sigma) were also used in Western blotting, each with 1 hr incubation. Colorimetric detection with NBT/BCIP (Roche) was performed.
ACKNOWLEDGMENTS
The VUMC Institutional Flow Cytometry Core was supported by the Vanderbilt Ingram Cancer Center (P30 CA68485), and we thank Dr. James Higginbotham for his assistance. We also thank Dr. Yan Ru Su for her assistance with real-time RT-PCR, and members of the Bader laboratory, for helpful suggestions and comments. E.D. and D.B. were funded by the NIH.
Grant sponsor: Vanderbilt Ingram Cancer Center; Grant number: P30 CA68485; Grant sponsor: NIH; Grant number: K08 HL67049; Grant number: R01 HL37675.
REFERENCES
- Andres V, Walsh K. Myogenin expression, cell cycle withdrawal, and phenotypic differentiation are temporally separable events that precede cell fusion upon myogenesis. J Cell Biol. 1996;132:657–666. doi: 10.1083/jcb.132.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antin PB, Ordahl CP. Isolation and characterization of an avian myogenic cell line. Dev Biol. 1991;143:111–121. doi: 10.1016/0012-1606(91)90058-b. [DOI] [PubMed] [Google Scholar]
- Ashe M, Pabon-Pena L, Dees E, Price KL, Bader D. LEK1 is a potential inhibitor of pocket protein-mediated cellular processes. J Biol Chem. 2004;279:664–676. doi: 10.1074/jbc.M308810200. [DOI] [PubMed] [Google Scholar]
- Bader D, Masaki T, Fischman DA. Immunochemical analysis of myosin heavy chain during avian myogenesis in vivo and in vitro. J Cell Biol. 1982;95:763–770. doi: 10.1083/jcb.95.3.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Classon M, Harlow E. The retinoblastoma tumour suppressor in development and cancer. Nat Rev Cancer. 2002;2:910–917. doi: 10.1038/nrc950. [DOI] [PubMed] [Google Scholar]
- Dabiri GA, Turnacioglu KK, Sanger JM, Sanger JW. Myofibrillogenesis visualized in living embryonic cardiomyocytes. Proc Natl Acad Sci U S A. 1997;94:9493–9498. doi: 10.1073/pnas.94.17.9493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RL, Cheng PF, Lassar AB, Weintraub H. The MyoD DNA binding domain contains a recognition code for muscle-specific gene activation. Cell. 1990;60:733–746. doi: 10.1016/0092-8674(90)90088-v. [DOI] [PubMed] [Google Scholar]
- de la Brousse FC, Emerson CP., Jr Localized expression of a myogenic regulatory gene, qmf1, in the somite dermatome of avian embryos. Genes Dev. 1990;4:567–581. doi: 10.1101/gad.4.4.567. [DOI] [PubMed] [Google Scholar]
- de la Serna IL, Ohkawa Y, Berkes CA, Bergstrom DA, Dacwag CS, Tapscott SJ, Imbalzano AN. MyoD targets chromatin remodeling complexes to the myogenin locus prior to forming a stable DNA-bound complex. Mol Cell Biol. 2005;25:3997–4009. doi: 10.1128/MCB.25.10.3997-4009.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dees E, Pabon-Pena LM, Goodwin R, Bader D. Characterization of CMF1 protein in avian skeletal muscle. Dev Dyn. 2000;219:169–181. doi: 10.1002/1097-0177(2000)9999:9999<::aid-dvdy1055>3.3.co;2-2. [DOI] [PubMed] [Google Scholar]
- Dees E, Robertson JB, Ashe M, Pabon-Pena LM, Bader D, Goodwin RL. LEK1 protein expression in normal and dysregulated cardiomyocyte mitosis. Anat Rec A Discov Mol Cell Evol Biol. 2005;286A:823–832. doi: 10.1002/ar.a.20221. [DOI] [PubMed] [Google Scholar]
- Dees E, Robertson JB, Zhu T, Bader D. Specific deletion of CMF1 nuclear localization domain causes incomplete cell cycle withdrawal and impaired differentiation in avian skeletal myoblasts. Exp Cell Res. 2006;312:3000–3014. doi: 10.1016/j.yexcr.2006.06.027. [DOI] [PubMed] [Google Scholar]
- Dunaief JL, Strober BE, Guha S, Khavari PA, Alin K, Luban J, Begemann M, Crabtree GR, Goff SP. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell. 1994;79:119–130. doi: 10.1016/0092-8674(94)90405-7. [DOI] [PubMed] [Google Scholar]
- Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- Edmondson DG, Olson EN. A gene with homology to the myc similarity region of MyoD1 is expressed during myogenesis and is sufficient to activate the muscle differentiation program. Genes Dev. 1990;4:1450. doi: 10.1101/gad.4.8.1450. [DOI] [PubMed] [Google Scholar]
- Evans HJ, Edwards L, Goodwin RL. Conserved C-terminal domains of mCenp-F (LEK1) regulate subcellular localization and mitotic checkpoint delay. Exp Cell Res. 2007;313:2427–2437. doi: 10.1016/j.yexcr.2007.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank NY, Kho AT, Schatton T, Murphy GF, Molloy MJ, Zhan Q, Ramoni MF, Frank MH, Kohane IS, Gussoni E. Regulation of myogenic progenitor proliferation in human fetal skeletal muscle by BMP4 and its antagonist Gremlin. J Cell Biol. 2006;175:99–110. doi: 10.1083/jcb.200511036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friend SH, Bernards R, Rogelj S, Weinberg RA, Rapaport JM, Albert DM, Dryja TP. A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature. 1986;323:643–646. doi: 10.1038/323643a0. [DOI] [PubMed] [Google Scholar]
- Gill RM, Hamel PA. Subcellular compartmentalization of E2F family members is required for maintenance of the postmitotic state in terminally differentiated muscle. J Cell Biol. 2000;148:1187–1201. doi: 10.1083/jcb.148.6.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich DW, Wang NP, Qian YW, Lee EY, Lee WH. The retinoblastoma gene product regulates progression through the G1 phase of the cell cycle. Cell. 1991;67:293–302. doi: 10.1016/0092-8674(91)90181-w. [DOI] [PubMed] [Google Scholar]
- Goodwin RL, Pabon-Pena LM, Foster GC, Bader D. The cloning and analysis of LEK1 identifies variations in the LEK/centromere protein F/mitosin gene family. J Biol Chem. 1999;274:18597–18604. doi: 10.1074/jbc.274.26.18597. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. pp. 479–504. [Google Scholar]
- Hastings KE, Emerson CP., Jr cDNA clone analysis of six co-regulated mRNAs encoding skeletal muscle contractile proteins. Proc Natl Acad Sci U S A. 1982;79:1553–1557. doi: 10.1073/pnas.79.5.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helin K, Harlow E, Fattaey A. Inhibition of E2F-1 transactivation by direct binding of the retinoblastoma protein. Mol Cell Biol. 1993;13:6501–6508. doi: 10.1128/mcb.13.10.6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HJ, Yee JK, Shew JY, Chen PL, Bookstein R, Friedmann T, Lee EY, Lee WH. Suppression of the neoplastic phenotype by replacement of the RB gene in human cancer cells. Science. 1988;242:1563–1566. doi: 10.1126/science.3201247. [DOI] [PubMed] [Google Scholar]
- Kaelin WG., Jr Functions of the retinoblastoma protein. Bioessays. 1999;21:950–958. doi: 10.1002/(SICI)1521-1878(199911)21:11<950::AID-BIES7>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Kang H, Cui K, Zhao K. BRG1 controls the activity of the retinoblastoma protein via regulation of p21CIP1/WAF1/SDI. Mol Cell Biol. 2004;24:1188–1199. doi: 10.1128/MCB.24.3.1188-1199.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight RF, Bader DM, Backstrom JR. Membrane topology of Bves/Pop1A, a cell adhesion molecule that displays dynamic changes in cellular distribution during development. J Biol Chem. 2003;278:32872–32879. doi: 10.1074/jbc.M301961200. [DOI] [PubMed] [Google Scholar]
- Liao H, Winkfein RJ, Mack G, Rattner JB, Yen TJ. CENP-F is a protein of the nuclear matrix that assembles onto kinetochores at late G2 and is rapidly degraded after mitosis. J Cell Biol. 1995;130:507–518. doi: 10.1083/jcb.130.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miner JH, Wold B. Herculin, a fourth member of the MyoD family of myogenic regulatory genes. Proc Natl Acad Sci U S A. 1990;87:1089–1093. doi: 10.1073/pnas.87.3.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Wakitani S, Saito N, Takaoka K. Expression profiles of BMP-related molecules induced by BMP-2 or -4 in muscle-derived primary culture cells. J Bone Miner Metab. 2005;23:426–434. doi: 10.1007/s00774-005-0624-5. [DOI] [PubMed] [Google Scholar]
- Novitch BG, Mulligan GJ, Jacks T, Lassar AB. Skeletal muscle cells lacking the retinoblastoma protein display defects in muscle gene expression and accumulate in S and G2 phases of the cell cycle. J Cell Biol. 1996;135:441–456. doi: 10.1083/jcb.135.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitch BG, Spicer DB, Kim PS, Cheung WL, Lassar AB. pRb is required for MEF2-dependent gene expression as well as cell-cycle arrest during skeletal muscle differentiation. Curr Biol. 1999;9:449–459. doi: 10.1016/s0960-9822(99)80210-3. [DOI] [PubMed] [Google Scholar]
- Ohkawa Y, Marfella CG, Imbalzano AN. Skeletal muscle specification by myogenin and Mef2D via the SWI/SNF ATPase Brg1. EMBO J. 2006;25:490–501. doi: 10.1038/sj.emboj.7600943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabon-Pena LM, Goodwin RL, Cise LJ, Bader D. Analysis of CMF1 reveals a bone morphogenetic protein-independent component of the cardiomyogenic pathway. J Biol Chem. 2000;275:21453–21459. doi: 10.1074/jbc.M000518200. [DOI] [PubMed] [Google Scholar]
- Papadimou E, Menard C, Grey C, Puceat M. Interplay between the retinoblastoma protein and LEK1 specifies stem cells toward the cardiac lineage. EMBO J. 2005;24:1750–1761. doi: 10.1038/sj.emboj.7600652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasumarthi KB, Field LJ. Cardiomyocyte cell cycle regulation. Circ Res. 2002;90:1044–1054. doi: 10.1161/01.res.0000020201.44772.67. [DOI] [PubMed] [Google Scholar]
- Pizon V, Iakovenko A, Van Der Ven PF, Kelly R, Fatu C, Furst DO, Karsenti E, Gautel M. Transient association of titin and myosin with microtubules in nascent myofibrils directed by the MURF2 RING-finger protein. J Cell Sci. 2002;115:4469–4482. doi: 10.1242/jcs.00131. [DOI] [PubMed] [Google Scholar]
- Puceat M. Rb and LEK1: a “pas de deux”in cardiogenesis. Cell Cycle. 2005;4:1030–1032. doi: 10.4161/cc.4.8.1905. [DOI] [PubMed] [Google Scholar]
- Rattner JB, Rao A, Fritzler MJ, Valencia DW, Yen TJ. CENP-F is a.ca 400 kDa kinetochore protein that exhibits a cell-cycle dependent localization. Cell Motil Cytoskeleton. 1993;26:214–226. doi: 10.1002/cm.970260305. [DOI] [PubMed] [Google Scholar]
- Redkar A, deRiel JK, Xu YS, Montgomery M, Patwardhan V, Litvin J. Characterization of cardiac muscle factor 1 sequence motifs: retinoblastoma protein binding and nuclear localization. Gene. 2002;282:53–64. doi: 10.1016/s0378-1119(01)00789-2. [DOI] [PubMed] [Google Scholar]
- Reshef R, Maroto M, Lassar AB. Regulation of dorsal somitic cell fates: BMPs and Noggin control the timing and pattern of myogenic regulator expression. Genes Dev. 1998;12:290–303. doi: 10.1101/gad.12.3.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider JW, Gu W, Zhu L, Mahdavi V, Nadal-Ginard B. Reversal of terminal differentiation mediated by p107 in Rb−/− muscle cells. Science. 1994;264:1467–1471. doi: 10.1126/science.8197461. [DOI] [PubMed] [Google Scholar]
- Soonpaa MH, Kim KK, Pajak L, Franklin M, Field LJ. Cardiomyocyte DNA synthesis and binucleation during murine development. Am J Physiol. 1996;271:H2183–H2189. doi: 10.1152/ajpheart.1996.271.5.H2183. [DOI] [PubMed] [Google Scholar]
- Walsh K, Perlman H. Cell cycle exit upon myogenic differentiation. Curr Opin Genet Dev. 1997;7:597–602. doi: 10.1016/s0959-437x(97)80005-6. [DOI] [PubMed] [Google Scholar]
- Wei Y, Bader D, Litvin J. Identification of a novel cardiac-specific transcript critical for cardiac myocyte differentiation. Development. 1996;122:2779–2789. doi: 10.1242/dev.122.9.2779. [DOI] [PubMed] [Google Scholar]
- Weintraub SJ, Prater CA, Dean DC. Retinoblastoma protein switches the E2F site from positive to negative element. Nature. 1992;358:259–261. doi: 10.1038/358259a0. [DOI] [PubMed] [Google Scholar]
- Weintraub SJ, Chow KN, Luo RX, Zhang SH, He S, Dean DC. Mechanism of active transcriptional repression by the retinoblastoma protein. Nature. 1995;375:812–815. doi: 10.1038/375812a0. [DOI] [PubMed] [Google Scholar]
- Yuasa S, Itabashi Y, Koshimizu U, Tanaka T, Sugimura K, Kinoshita M, Hattori F, Fukami S, Shimazaki T, Ogawa S, Okano H, Fukuda K. Transient inhibition of BMP signaling by Noggin induces cardiomyocyte differentiation of mouse embryonic stem cells. Nat Biotechnol. 2005;23:607–611. doi: 10.1038/nbt1093. [DOI] [PubMed] [Google Scholar]
- Zacksenhaus E, Jiang Z, Chung D, Marth JD, Phillips RA, Gallie BL. pRb controls proliferation, differentiation, and death of skeletal muscle cells and other lineages during embryogenesis. Genes Dev. 1996;10:3051–3064. doi: 10.1101/gad.10.23.3051. [DOI] [PubMed] [Google Scholar]
- Zhu X, Chang K, He D, Mancini MA, Brinkley WR, Lee W. The C terminus of mitosin is essential for its nuclear localization, centromere/kinetochore targeting, and dimerization. J Biol Chem. 1995a;270:19545–19550. doi: 10.1074/jbc.270.33.19545. [DOI] [PubMed] [Google Scholar]
- Zhu X, Mancini MA, Chang K, Liu C, Chen C, Shan B, Jones D, Yang-Feng TL, Lee W. Characterization of a novel 350-kilodalton nuclear phosphoprotein that is specifically involved in mitoticphase progression. Mol Cell Biol. 1995b;15:5017–5029. doi: 10.1128/mcb.15.9.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]