Hypothalamic circuits regulating energy balance are highly plastic and develop in response to nutrient and hormonal cues. To identify processes that could be susceptible to gestational influences in the mouse, we characterized the ontogeny of proopiomelanocortin (POMC) and neuropeptide Y (NPY) populations, which exert opposing influences on food intake and body weight. These analyses revealed that Pomc is broadly expressed in immature hypothalamic neurons and that half of embryonic Pomc-expressing precursors subsequently adopt a non-POMC fate in the adult. Moreover, nearly one quarter of the mature orexigenic NPY population shares a common progenitor with anorexigenic POMC neurons.
The rapid increase in the prevalence of childhood obesity and the concomitant rise in obesity-related medical morbidities and costs, lend urgency to the need for new insights into the causes and potential preventive measures for this disease 1. Mounting evidence supports the idea that the maternal environment can impart a lasting effect on susceptibility of offspring to obesity and type 2 diabetes2. The arcuate nucleus of the hypothalamus (ARH) is a critical component of the neuronal network regulating body weight, adiposity, and glucose homeostasis; and recent studies suggest that the development of arcuate neurons may be sensitive to maternal metabolic status 3. The discovery that ARH projections are influenced by leptin provided the first insight into potential mechanisms underlying “maternal programming” in the perinatal period 4. The gestational environment has also been shown to influence metabolic status of the offspring 5; however, little is known about the embryonic origins of arcuate lineages.
The two best-characterized arcuate populations – orexigenic neurons co-expressing neuropeptide Y (NPY) and agouti-related protein (AgRP) and anorexigenic neurons expressing proopiomelanocortin (POMC) – produce antagonistic effects on food intake in response to nutrient and hormonal signals of peripheral energy status (reviewed in 6). Signals of positive energy balance, such as leptin and glucose, stimulate subsets of POMC neurons leading to decreased food intake, while inhibiting the release of orexigenic peptides from neighboring NPY neurons 7,8. NPY neurons are active when the energy supply is not sufficient to meet system demands, releasing AgRP and γ-Aminobutyric acid (GABA) to inhibit melanocortin-mediated suppression of food intake 7,9. Together, NPY and POMC neurons integrate signals of energy homeostasis to direct physiological processes that regulate body weight 10. We focused our initial efforts on characterizing the ontogeny of NPY and POMC neuronal lineages during gestation because nutrient and hormonal cues influence the formation of NPY and POMC circuits, consistent with the idea that these developmental processes influence metabolic phenotypes.
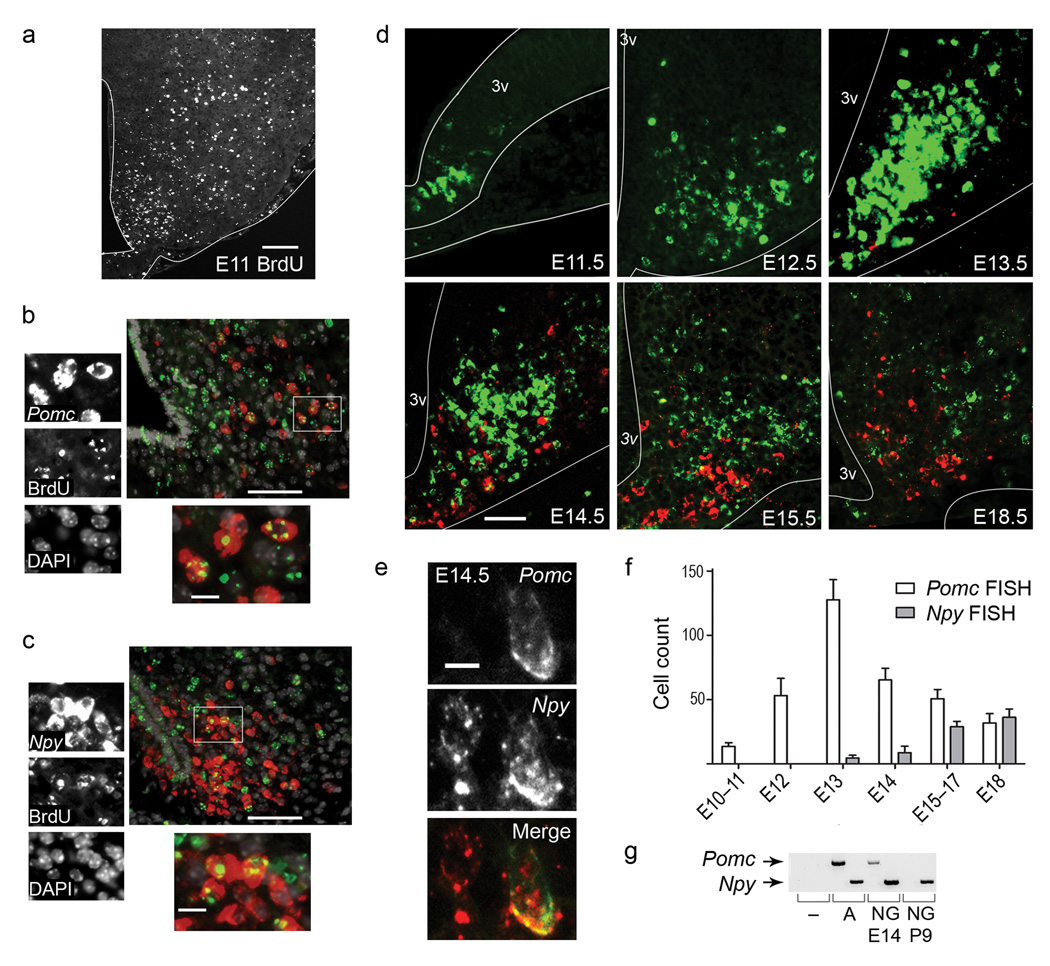
Using the GenePaint digital mouse atlas, we found that Pomc and Npy are expressed at embryonic day 14.5 (E14.5), whereas only Pomc is expressed at E10.5 (www.genepaint.org). Given the earlier onset of Pomc expression, together with the established lateromedial gradient of hypothalamic neurogenesis 11, we predicted that the lateral POMC neurons would be born before the medial NPY neurons. To determine the birthdates of POMC and NPY neurons in the ARH, we injected dams with a single pulse of bromodeoxyuridine (BrdU) between E11.5–E16.5 and assessed the retention of the BrdU label by immunohistochemistry (IHC) at postnatal day 9 (P9). Unexpectedly, analysis of BrdU label in conjunction with Pomc or Npy expression, as assessed by fluorescent in situ hybridization (FISH) demonstrated that both populations are born between E11.5–12.5 (Fig. 1a–c). The peak birthdate of ARH neurons is E11.5–12.5 (Fig. 1a and 12); however, we observed that BrdU injections at E13.5 labeled some non-POMC, non-NPY cells in the lateral ARH (Supplementary Figs. 1 and 2).
Figure 1.
Pomc is transiently expressed in a broad population of hypothalamic neurons during embryonic development. (a–c) We injected wild-type dams once with 200 mg kg−1 BrdU at E11.5 and sacrificed the offspring at P9 for analysis. (a) BrdU IHC. (b,c) Combined FISH with IHC using probes against Pomc (b) or Npy (c) in conjunction with an antibody against BrdU and counterstained for DAPI. Magnified view of boxed area to the left and below. (d) FISH for Pomc (green) and Npy (red) in ventral hypothalamus from E11.5–E18.5. (e) Confocal image of a cell containing both Npy and Pomc transcripts at E14.5. (f) We counted cells expressing Pomc and Npy as described in Supplementary Fig. 3. Each group represents the average counts of at least 5 coronal sections per animal spanning the rostrocaudal extent of the presumptive ARH with error bars representing mean ± SEM (n ≥ 3 animals for each group). (g) We dissociated cells from transgenic Npy-hrGFP hypothalami and FACS-purified GFP+ cells at E14.5 and P9. Pomc and Npy expression, as assessed by PCR on cDNA generated from sorted GFP+ cells. Abbreviation for sources of cDNA: no cDNA control (−); adult hypothalamus positive control (A); Npy-hrGFP (NG). 3V, third ventricle; Scale bar (a) 100 µm, (b,c) low mag 50 µm, high mag 10 µm (d) 50 µm, (e) 5 µm; all tissue 10 µm cryo-sections.
The shared birthdates of POMC and NPY neurons led us to consider whether these two antagonistic populations of neurons may be more closely related than expected. We characterized Pomc and Npy expression by two-color FISH across gestation (Fig. 1d–e). Pomc expression was first observed in the hypothalamic ventricular zone at E10.5–E11.5; from E12.5 expression was restricted to differentiated neurons, consistent with our birthdating studies. The number of Pomc-positive (Pomc+) cells reached a maximum at E13.5, after which its expression was extinguished in more than half of the population between E14.5 to 18.5 (Fig. 1f). Npy expression was not observed in the ventricular zone; it was first detected in laterally-situated cells in the rostralmost presumptive ARH at E13.5 and subsequently expanded to more medial and caudal regions. We did not detect appreciable levels of apoptotic cells by TUNEL stain, consistent with the idea that Pomc expression is turned off in a large percentage of immature hypothalamic neurons (data not shown and 13). These data argue that Pomc expression per se does not reflect the acquisition of a terminal cell fate; the gradual extinction of Pomc and progressive onset of Npy represent an ongoing maturation process that extends throughout gestation. Supporting this idea, POMC and NPY neurons do not acquire their terminal peptidergic phenotype, as reflected by Cart and Agrp expression, until the postnatal period in rodents 14,15.
Pomc and Npy are expressed in mutually exclusive cell populations in adults 16, yet we detected Pomc+ and Npy+ co-localization at mid-gestation (Fig. 1e). To substantiate the unprecedented finding that a subset of neurons co-expresses Pomc and Npy, we compared the expression profiles of NPY neurons isolated from embryonic versus postnatal stages. We used fluorescence activated cell sorting (FACS) to collect GFP-positive (GFP+) cells from Npy-hrGFP embryos, which express GFP under the control of Npy promoter and enhancer elements17. We detected Pomc transcripts by PCR on sorted cells from E14.5, and not from P9 (Fig. 1g and Supplementary Fig. 4a,b). These observations support the idea that during gestation, a subset of Pomc-expressing cells can differentiate into NPY neurons.
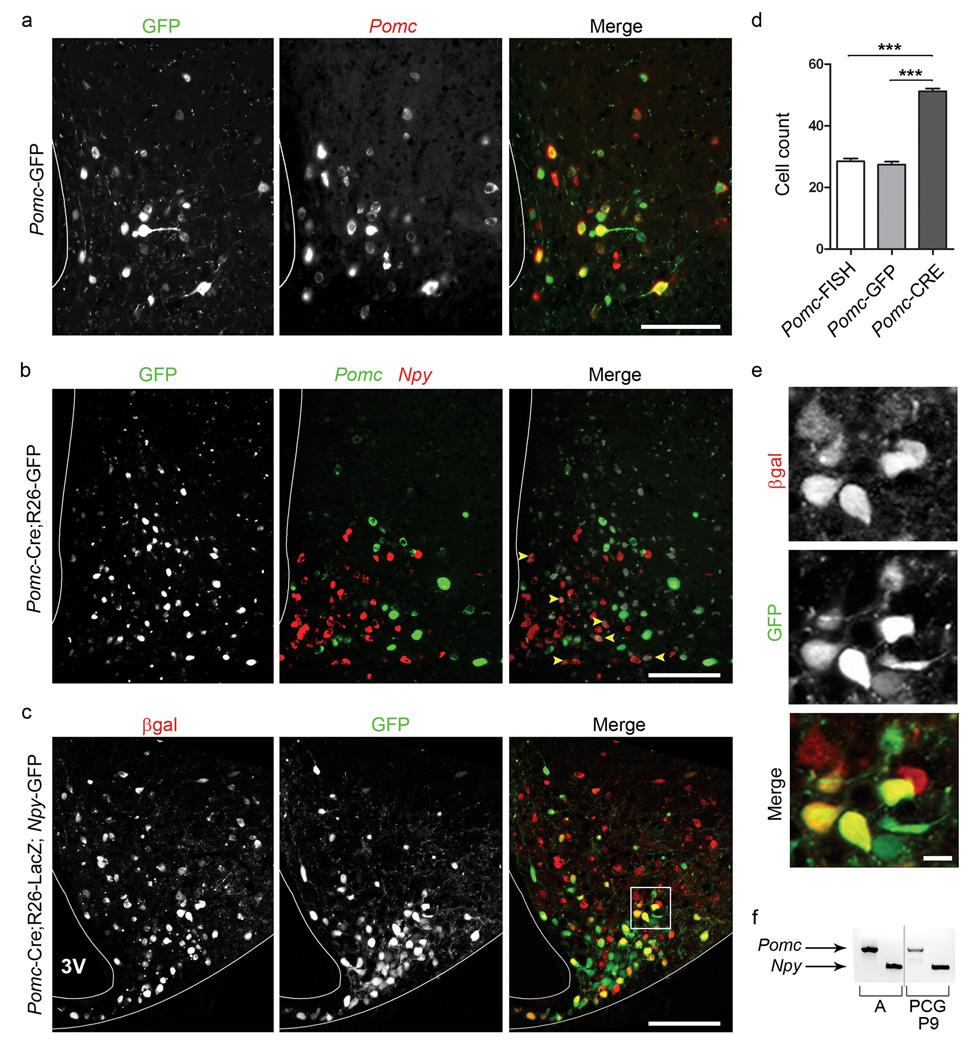
Next we used a genetic lineage tracing strategy 18 to visualize the mature POMC neuronal population, defined by Pomc expression in adults, in relation to the broad immature Pomc-expressing population in the embryo (Fig. 2). In Pomc-Cre;R26-GFP mice, Cre recombinase driven by Pomc regulatory elements directs the recombination of a floxed stop codon within the constitutively-active ROSA26 locus, permanently marking cells that expressed Pomc from gestation 19. To assess transcriptional activity in conjunction with a GFP reporter, we developed a technique to combine images of direct GFP fluorescence with FISH (Supplementary Figs. 3 and 5). When this assay was performed on adult tissue from Pomc-GFP transgenic animals, 95% of Pomc-GFP+ neurons co-express Pomc, validating the sensitivity of this technique (Fig. 2a) 7. In contrast, only half of the GFP+ cells in Pomc-Cre;R26-GFP mice express Pomc (Figs. 2b and Supplementary Fig. 6). Pomc-negative, GFP+ cells in Pomc-Cre;R26-GFP adults likely represent cells which turned off Pomc expression at some point after E13.5. GFP+ cell counts in Pomc-Cre;R26-GFP animals are consistently twice as high as those generated using Pomc FISH or direct fluorescence in Pomc-GFP animals (Fig. 2d).
Figure 2.
NPY neurons derived from a Pomc-expressing lineage persist to adulthood. (a,b) Images of direct GFP fluorescence obtained prior to FISH processing in Pomc-GFP (a left) or Pomc-Cre;R26-GFP (b left) adults, followed by FISH with Pomc alone (a center) or Pomc (green) plus Npy (red) probes (b center) or, composite images (a, b right) (technique described in Supplementary Fig. 5). (b) Cells expressing Npy and the Pomc-Cre lineage marker, GFP, are indicated with arrowheads. Because of its perinuclear localization, the FISH signal appears as a ring around the GFP signal. (a,b) All images acquired with a Nikon Eclipse 80i fluorescent microscope. (c) Confocal images of β-GAL IHC (red) in conjunction with direct GFP fluorescence (green) in Pomc-Cre;R26-LacZ;Npy-GFP adults. (d) Cell counts across the rostral-caudal axis of the ARH for Pomc transcript (Pomc-FISH), direct fluorescence of GFP from Pomc-GFP adults or from Pomc-Cre;R26-GFP (Pomc-CRE) adults are compared. (e) High magnification image of the region boxed in c. (f) We dissociated hypothalamic cells from transgenic Pomc-Cre;R26-GFP mice and FACS-purified GFP+ cells at P9. Pomc and Npy expression, as assessed by PCR on cDNA generated from sorted GFP+ cells. Adjacent lane doublets separated by the grey bar were run simultaneously on the same gel but not in neighboring lanes; they have been juxtaposed for the purposes of this figure. Abbreviation for sources of cDNA: adult hypothalamus positive control (A); Pomc-Cre;R26-GFP (PCG). 3V, third ventricle; Scale bar (a–c) 100 µm, (e) 10 µm. Error bars representing mean ± SEM (n ≥ 42 sections for each group, from n ≥ 6 animals), *** denotes p < 0.0001.
Based on our finding that Npy and Pomc are co-localized in a subset of embryonic neurons, we considered whether some of the Pomc-negative, GFP+ neurons in Pomc-Cre;R26-GFP adults are NPY neurons. Npy expression was detected in 17±2% of GFP+ neurons in adult Pomc-Cre;R26-GFP mice (herafter referred to as NPYP) (Fig. 2b). We used two strategies to independently verify this observation. First, confocal images of IHC on Npy-GFP;Pomc-Cre;R26-LacZ mice confirmed that 25% of NPY (GFP+) neurons co-express the Pomc-Cre lineage trace (β-Gal IHC) (Figs. 2c,e). Second, RT-PCR on FACS-purified GFP+ cells from Pomc-Cre;R26-GFP demonstrated that some cells marked by the lineage trace express Npy (Fig. 2f and Supplimentary Fig. 4c).
These data provide evidence that NPYP neurons are derived from progenitors that are distinct from other ARH NPY neurons (NPYX), raising the possibility that they serve different functions within the hypothalamic feeding circuit, and thus may underlie the heterogeneous electrophysiological properties of NPY neurons 20. While the origins of NPY subpopulations may differ, their subsequent differentiation converges on an orexigenic, GABAergic phenotype, as we found that both NPYP and NPYX neurons express Agrp and Gad67 7. Our studies provide a framework to uncover the molecular mechanism underlying these differences within NPY neurons.
In this study, we report that Pomc is expressed in the vast majority of neurons in the presumptive ARH (Fig. 1d,f). During gestation, Pomc transcription is extinguished in more than half of these cells, some of which subsequently differentiate into NPY neurons and some of which adopt alternative terminal fates. Consistent with our FISH analyses, when Pomc-Cre;R26-GFP mice were used to trace Pomc-derived lineages in the adult hypothalamus, we found that half of the GFP-labeled neurons are non-POMC neurons. Therefore, use of this Pomc-Cre driver to investigate the roles played by POMC neurons in circuits that regulate energy homeostasis would also affect a subset of NPY/AgRP neurons and others whose functions have yet to be determined. Unanticipated effects on antagonistic populations that also express the Cre transgene (i.e. NPY/AgRP) could ameliorate phenotypes resulting from genetic manipulations intended for POMC neurons. Moreover, some functions ascribed to POMC neurons could be mediated by non-POMC neurons that also express the Cre transgene. Classification of functionally distinct subsets of neurons derived from a Pomc+ lineage is critical to elucidate how hormonal and nutrient signals are sensed by ARH neurons and relayed to downtream targets that regulate body weight and energy homeostasis.
Supplementary Material
Acknowledgements
We thank L. Sussel, H. Wichterle and D. Accili for critical reading of our manuscript and helpful comments; R. Leibel for support and critical funding for this project (RO1 DK52431-16); J. Overton for help with confocal imaging; L. Yang of the DERC Pathology Core for cryosectioning and C. Liu of the Irving Institute CTSA FACS core (NCRR UL1 RR024156); M. Low (University of Michigan Health Center), J. Elmquist (UT Southwestern Medical Center), and B. Lowell (Beth Israel Deaconess Medical Center) for generously providing mouse reagents. This work was supported by F31DK079372 (SLP), Institute of Human Nutrition Training Grant 2T32DK007647-21 (JSC), ADA 7-07RA-195 (LMZ), and Columbia DERC Pilot and Feasibility Award P30 DK63608-07 (LMZ), NY Obesity Research Center Pilot and Feasibility P30 DK26687-26 (LMZ).
Footnotes
Author Contributions
S.L.P performed experiments, analyzed data, and wrote the paper; J.S.C. generated data for confocal analysis and contributed to data analysis; L.M.Z designed the study, analyzed the data, and wrote the paper.
References
- 1.Ogden CL, et al. Jama. 2006;295:1549–1555. doi: 10.1001/jama.295.13.1549. [DOI] [PubMed] [Google Scholar]
- 2.Taylor PD, Poston L. Exp Physiol. 2007;92:287–298. doi: 10.1113/expphysiol.2005.032854. [DOI] [PubMed] [Google Scholar]
- 3.Levin BE. Philos Trans R Soc Lond B Biol Sci. 2006;361:1107–1121. doi: 10.1098/rstb.2006.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouret SG, Draper SJ, Simerly RB. Science. 2004;304:108–110. doi: 10.1126/science.1095004. [DOI] [PubMed] [Google Scholar]
- 5.Shankar K, et al. Am J Physiol Regul Integr Comp Physiol. 2008;294:R528–R538. doi: 10.1152/ajpregu.00316.2007. [DOI] [PubMed] [Google Scholar]
- 6.Saper CB, Chou TC, Elmquist JK. Neuron. 2002;36:199–211. doi: 10.1016/s0896-6273(02)00969-8. [DOI] [PubMed] [Google Scholar]
- 7.Cowley MA, et al. Nature. 2001;411:480–484. doi: 10.1038/35078085. [DOI] [PubMed] [Google Scholar]
- 8.Ibrahim N, et al. Endocrinology. 2003;144:1331–1340. doi: 10.1210/en.2002-221033. [DOI] [PubMed] [Google Scholar]
- 9.Ollmann MM, et al. Science. 1997;278:135–138. doi: 10.1126/science.278.5335.135. [DOI] [PubMed] [Google Scholar]
- 10.Schwartz MW, Porte D., Jr Science. 2005;307:375–379. doi: 10.1126/science.1104344. [DOI] [PubMed] [Google Scholar]
- 11.Altman J, Bayer SA. J Comp Neurol. 1978;182:945–971. doi: 10.1002/cne.901820511. [DOI] [PubMed] [Google Scholar]
- 12.Shimada M, Nakamura T. Exp Neurol. 1973;41:163–173. doi: 10.1016/0014-4886(73)90187-8. [DOI] [PubMed] [Google Scholar]
- 13.Broad KD, Curley JP, Keverne EB. Dev Neurobiol. 2009;69:314–325. doi: 10.1002/dneu.20702. [DOI] [PubMed] [Google Scholar]
- 14.Cottrell EC, et al. Am J Physiol Regul Integr Comp Physiol. 2009;296:R631–R639. doi: 10.1152/ajpregu.90690.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nilsson I, Johansen JE, Schalling M, Hokfelt T, Fetissov SO. Brain Res Dev Brain Res. 2005;155:147–154. doi: 10.1016/j.devbrainres.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 16.Pinto S, et al. Science. 2004;304:110–115. doi: 10.1126/science.1089459. [DOI] [PubMed] [Google Scholar]
- 17.van den Top M, et al. Neuroscience. 2007;144:815–824. doi: 10.1016/j.neuroscience.2006.09.059. [DOI] [PubMed] [Google Scholar]
- 18.Zinyk DL, Mercer EH, Harris E, Anderson DJ, Joyner AL. Curr Biol. 1998;8:665–668. doi: 10.1016/s0960-9822(98)70255-6. [DOI] [PubMed] [Google Scholar]
- 19.Balthasar N, et al. Neuron. 2004;42:983–991. doi: 10.1016/j.neuron.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 20.Fioramonti X, et al. Diabetes. 2007;56:1219–1227. doi: 10.2337/db06-0567. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.