To the editor
Forward genetic screens for mutants in which specific biological processes are disrupted are a key strength of model systems like C.elegans or Drosophila. However, the road from isolating a phenotype-causing mutant strain to identifying the molecular nature of the genetic change, most often as little as a single point mutation, is cumbersome, traditionally involving time-consuming genetic mapping strategies. We have recently shown in a proof-of-principle study that conventional mapping steps can be shortcut through the use of whole genome sequencing (WGS) with massively parallel, deep-sequencing technology 1,2 (Suppl. Table 1). A similar proof-of-principle approach has proven successful for Drosophila mutant identification as well 3.
The key challenge of the WGS approach is the mapping of millions of small (<100bp) reads, obtained from sequencing the mutant genome, to a wild-type reference genome. Various mapping tools are available for this purpose, including ELAND or MAQ (Mapping and Assembly with Quality) 4. A downside of many of these tools is that a non-computer savvy wet lab biologist who obtained primary WGS data from, for example, a sequencing service facility, may find implementation, usage and data output format of these programs non-intuitive and may require outside bioinformatic support that may not be readily available.
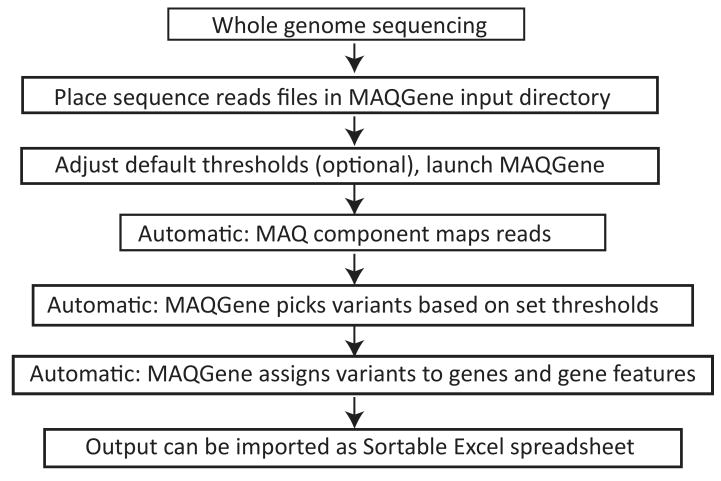
To circumvent this problem and thereby popularize the WGS approach, we developed a user-friendly, simple web browser interface, called MAQGene, that automatically launches the publicly available MAQ software and assembles a customized summary of the location and specific features of sequence variants of the mutant genome compared to a wild-type reference genome (Fig.1). The MAQGene submission form allows the user to select a specific set of parameters for aligning and interpreting WGS reads (see screenshot in Supplementary Information). Default parameters that we have used successfully for analyzing mutant C.elegans genomes are provided in the installation package and are easily reconfigured to suit individual preference. MAQGene may handle reads up to 127 bases long and map in both single read or paired-end modes. The output file (see Supplementary Information for sample output) is easily convertible to an Excel spreadsheet and allows easy browsing of sequence variants, as well as comparisons of different genomes (which is, for example, helpful to subtract background variants). Various measures are provided in the output file that allow the user to rapidly assess the degree of coverage for a given nucleotide position and the likelihood that a nucleotide variant is indeed real and of functional relevance. For example, provided the reference genome has all exons annotated, as is the case for the C.elegans genome, each variant is indicated as being intronic, intergenic, within a protein-coding gene (and if so whether the variant is silent, missense, splice site, or nonsense) or within an annotated non-coding RNA. These features are sortable in the output file, allowing for the generation of a “priority list” of variants which are to be chosen for validation by Sanger re-sequencing and for tests probing functional relevance. The output file can also be easily filtered so as to reveal variants present specifically in a genetically mapped interval.
Figure 1. Flowchart for using MAQGene.
Using data generated by an in-house Illumina Genome Analyzer II platform, we have used MAQGene for identifying sequence variants in more than half a dozen different C.elegans genomes compared to the wild-type C.elegans reference genome. In principle, MAQGene also provides the option to compare any input WGS reads (in fastq format) to any wild-type reference genome that is available in fasta format with GFF (general-feature format) coding exon annotations files, thereby easily allowing adaptation of MAQGene to analyze, for example, WGS data from Drosophila mutant strains.
MAQGene can be downloaded at http://maqweb.sourceforge.net. A detailed description of MAQGene, its installment and use can be found in the package itself and also in the Supplementary Information.
Supplementary Material
Acknowledgments
This work was supported by the HHMI, the NIH (R01NS039996-05; R01NS050266-03), a postdoctoral NIH training grant to H.B. (5T32HD055165-03) and a predoctoral NIH fellowship to S.S. (NS054540-01). We thank members of the Hobert lab for discussions.
References
- 1.Sarin S, Prabhu S, O'Meara MM, et al. Nat Methods. 2008;5(10):865. doi: 10.1038/nmeth.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shen Y, Sarin S, Liu Y, et al. PLoS ONE. 2008;3(12):e4012. doi: 10.1371/journal.pone.0004012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blumenstiel JP, Noll AC, Griffiths JA, et al. Genetics. 2009;182(1):25. doi: 10.1534/genetics.109.101998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li H, Ruan J, Durbin R. Genome Res. 2008;18(11):1851. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.