SUMMARY
The entry of human immunodeficiency virus (HIV-1) into cells is initiated by binding of the gp120 exterior envelope glycoprotein to the receptor, CD4. How does CD4 binding trigger conformational changes in gp120 that allow the gp41 transmembrane envelope glycoprotein to mediate viral-cell membrane fusion? The transition from the unliganded to the CD4-bound state is regulated by two potentially flexible topological layers (“Layers 1 and 2”) in the gp120 inner domain. Both layers apparently contribute to the non-covalent association of unliganded gp120 with gp41. After CD4 makes initial contact with the gp120 outer domain, Layer 1-Layer 2 interactions strengthen gp120-CD4 binding by reducing the off-rate. Layer 1-Layer 2 interactions also destabilize the activated state induced on HIV-1 by treatment with soluble CD4. Thus, despite lack of contact with CD4, the gp120 inner domain layers govern CD4 triggering by participating in conformational transitions within gp120 and regulating the interaction with gp41.
INTRODUCTION
Human immunodeficiency virus (HIV-1) entry into the host cell is mediated by the viral envelope glycoproteins (Env gps), which are derived by proteolytic cleavage of a trimeric, glycosylated gp160 Env gp precursor (Allan et al., 1985; Robey et al., 1985). The resulting mature Env gps, gp120 (SU) and gp41 (TM), constitute a trimeric complex that is anchored on the virion surface by the membrane-spanning segments of gp41 (Chan et al., 1997; Weissenhorn et al., 1997; Farzan et al., 1998; Zhu et al., 2003). The gp120 exterior Env gp is retained on the trimer via labile, noncovalent interactions with the gp41 ectodomain (Helseth et al., 1991). The gp120 glycoprotein binds the initial receptor, CD4 (Dalgleish et al., 1984; Klatzmann et al., 1984). CD4 binding triggers conformational changes in gp120 that promote its interaction with one of the chemokine receptors, CCR5 or CXCR4 (Alkhatib et al., 1996; Choe et al., 1996; Deng et al., 1996; Doranz et al., 1996; Dragic et al., 1996; Feng et al., 1996; Trkola et al., 1996; Wu et al., 1996). CD4 binding also induces conformational changes within the HIV-1 Env gp trimer that result in the exposure of a helical heptad repeat (HR1) segment of the gp41 ectodomain (Furuta et al., 1998; He et al., 2003; Koshiba and Chan, 2003; Si et al., 2004). Eventually, the conformational transition of the gp41 ectodomain from the CD4-induced “pre-hairpin intermediate” into a six-helix bundle composed of the HR1 and HR2 heptad repeat regions results in the fusion of the viral and target cell membranes (Lu et al., 1995; Chan et al., 1997; Weissenhorn et al., 1997).
The HIV-1 Env gps serve as a prototype for viral envelope proteins that are activated to mediate virus entry by engagement of the receptors. The energy required for viral-cell membrane fusion derives from the sequential transitions that the HIV-1 Env gps undergo, from the high-energy unliganded state to the low-energy six-helix bundle. The graded transitions down this energetic slope are initially triggered by CD4 binding. Thermodynamic, mutagenic and structural studies have provided insights into the HIV-1 gp120-CD4 interaction. The gp120 glycoprotein is composed of two domains: a gp41-interactive inner domain and a heavily glycosylated outer domain (Kwong et al., 1998). In the CD4-bound state, a bridging sheet minidomain connects the inner and outer domains (Kwong et al., 1998). CD4 initially binds a highly conserved region on the surface of the gp120 outer domain (Zhou et al., 2007). The interaction of gp120 and CD4 is accompanied by an unusually large, favorable enthalpic change, which offsets a large unfavorable change in entropy (Myszka et al., 2000). Certain antibodies and small molecules bind HIV-1 gp120 with similar thermodynamic signatures (Kwong et al., 2002; Schön et al., 2006). The binding of these ligands is thought to introduce order into the conformationally flexible unliganded gp120, accounting for the entropy decrease. The refolding of gp120 elements upon CD4 binding, which involves the formation of numerous internal bonds in gp120, contributes to the large change in enthalpy and to the slow off-rate of bound CD4 (Myszka et al., 2000; Kwong et al., 2002; Kassa et al., 2009).
Structural studies have revealed the contacts between HIV-1 gp120 and CD4 (Kwong et al., 1998). Several bridging sheet and outer domain gp120 residues that are highly conserved among HIV-1 strains contact CD4 residues phenylalanine 43 and arginine 59, contributing much of the binding energy of the gp120-CD4 interaction. Binding CD4 creates the “Phe 43 cavity”, an interfacial cavity bounded by all three gp120 domains and by phenylalanine 43 of CD4. The walls of the Phe 43 cavity are lined by well-conserved gp120 residues, rendering it suitable as a target for small-molecule inhibitors (Madani et al., 2008). Interventional approaches targeting the CD4-binding site, using either antibodies or small molecules, would benefit from a better understanding of the conformational transitions involved in receptor binding.
In previous structural studies (Kwong et al., 1998; Chen et al., 2005; Zhou et al., 2007), the N and C termini of gp120, which contribute to the non-covalent association with gp41 (Helseth et al., 1991; Binley et al., 2000), were truncated to facilitate crystallization. Recently, the structure of an HIV-1 gp120 variant with intact termini, bound to soluble CD4 and a neutralizing antibody fragment, has been solved (Pancera et al., 2010). This structure reveals the complete gp41-interactive region of gp120 in the CD4-bound conformation. The N- and C-terminal strands of gp120 project from the inner domain towards the viral membrane, presumably contacting the gp41 ectodomain. The inner domain, revealed in its entirety for the first time, can be subdivided into two parts: a) a β-sandwich, which mutagenesis studies suggest contributes to gp41 association (Helseth et al., 1991; Yang et al., 2003); and b) three loops that emanate from the strands of the β-sandwich and project towards the target cell. These loops form three topological layers that, like the leaves of a book, have been proposed to exhibit conformational flexibility; such flexibility would presumably contribute to the high entropy of the unliganded HIV-1 gp120 and allow transitions to the CD4-bound conformation. Here, we investigate the role of these inner domain layers in HIV-1 entry.
RESULTS
The HIV-1 gp120 inner domain mutants
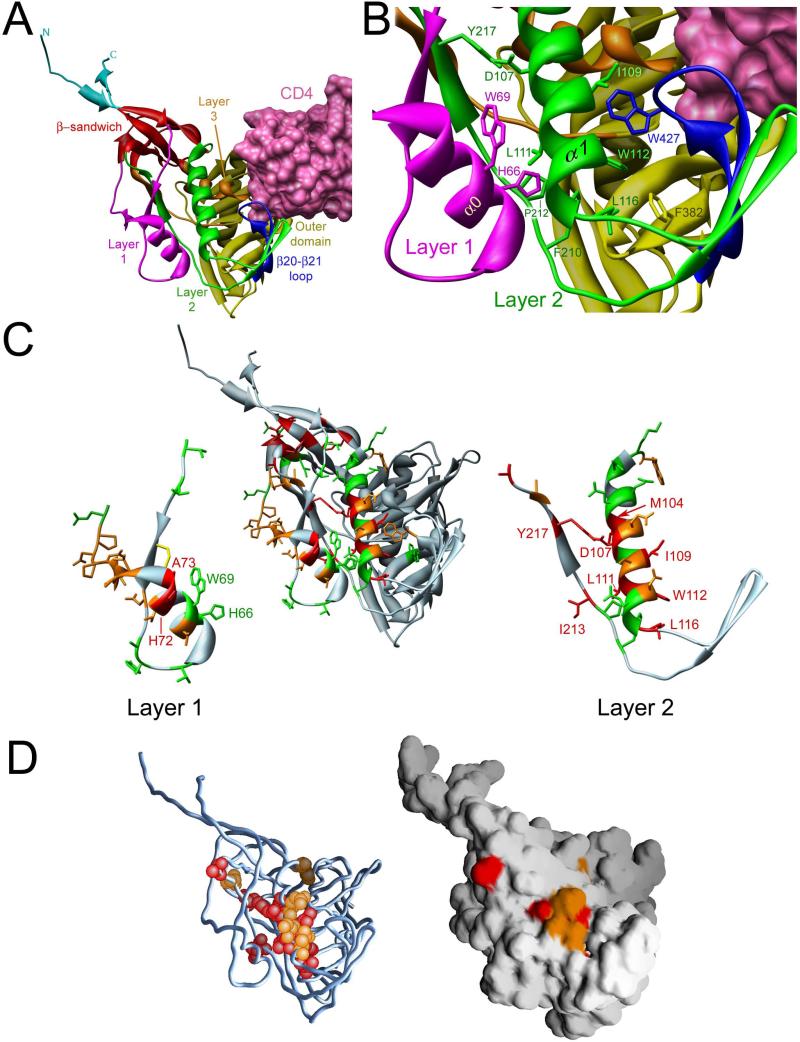
A recent x-ray crystallographic analysis of CD4-bound HIV-1 gp120 with intact N and C termini has revealed the structure of the complete gp120 inner domain (Pancera et al., 2010) (Figure 1A). A key component of the inner domain, a seven-stranded β-sandwich, serves as a point of departure for the gp120 N and C termini, which meander towards the viral membrane. The β-sandwich and the gp120 terminal strands have been implicated in the non-covalent association of gp120 with gp41 (Helseth et al., 1991; Binley et al., 2000; Yang et al., 2003; Sen et al., 2008; Wang et al., 2008). Projecting from the β-sandwich towards the target cell membrane are three excursions that compose the rest of the inner domain and, in two cases, transit into neighboring domains. These excursions form three topological layers (“Layers 1, 2 and 3”) that have been proposed to change conformation as gp120 undergoes the transition from the unliganded to the CD4-bound state (Pancera et al., 2010). To test this hypothesis, single amino acid changes were introduced into the Env gps of HIV-1YU2, a primary virus directly cloned from an HIV-1-infected individual (Li et al., 1992). The changes were focused on Layers 1 and 2, although a few alterations were introduced into adjacent gp120 structures.
Figure 1. Structure of the inner domain of HIV-1 gp120 in the CD4-bound conformation.
A. The structure (Pancera et al., 2010) of HIV-1HXBc2 gp120 (ribbon) complexed with two-domain CD4 (pink molecular surface) is shown, from the approximate perspective of the Env gp trimer axis. The outer domain of gp120 is colored yellow. The N and C termini are colored cyan. The components of the gp120 inner domain are the β-sandwich (red) and three loop-like extensions: Layer 1 (magenta), Layer 2 (green) and Layer 3 (orange). The β20-β21 strands of gp120 (blue) project from the outer domain and, in the CD4-bound conformation, compose two of the strands of the four-stranded bridging sheet. The other two strands of the bridging sheet are derived from the distal portion of Layer 2. B. A close-up view of the interactions of Layer 2 (green) with Layer 1 (magenta), and with the β20-β21 loop (blue), is shown from the same perspective as in A. The surface of CD4 (pink) is visible in the upper right-hand corner of the image. C. The center image is a ribbon diagram of the CD4-bound HIV-1 gp120 glycoprotein, shown in the same orientation as that in A. The ribbon and side chain residues that were altered in this and a previous study (Yang et al., 2003) are colored according to the gp120-gp41 association index (Red: association index < 0.5; Orange: 0.5 ≤ association index < 0.7; Green: association index ≥ 0.7). In the left- and right-hand images, Layer 1 and Layer 2 are separated from the rest of the gp120 glycoprotein. The disulfide bond in Layer 1 between cysteines 54 and 74 is colored yellow. D. The gp120 glycoprotein, in the CD4-bound conformation, is shown from the perspective in A. In the left image, on the Cα trace of gp120, the atoms of Layer 2 residues with association indices less than 0.5 are colored red; residues with association indices between 0.5 and 0.7 are colored orange. The image at the right shows the solvent-accessible surface of gp120, colored similarly.
The Env gp mutants were evaluated with respect to the following properties: proteolytic processing of the gp160 precursor, association of the gp120 and gp41 subunits, CD4 binding, functional ability to mediate cell-cell fusion and virus entry, and the effect of soluble CD4 (sCD4) on virus infectivity (Table 1). The conformation of gp120 in cell supernatants was assessed in two ways: a) analysis on non-reducing SDS-polyacrylamide gels allowed detection of aberrant, disulfide-linked dimers indicative of inefficient folding; and b) precipitation by a panel of monoclonal antibodies that recognize conformation-dependent epitopes (Moore and Sodroski, 1996; Supporting Table S1). The L52A, F53A, M100A and I215A mutants exhibited evidence of significant conformational disruption and were not considered further.
Table 1.
Characterization of HIV-1YU2 gp120 inner domain mutants
| Envelope glycoprotein | Residue Location | Processing indexa | Association indexa | CD4-Ig bindinga | Cell-cell fusiona | Relative Infectivitya | sCD4 IC90 (nM)a |
|---|---|---|---|---|---|---|---|
| Wild-type YU2 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 90 | |
| E47A | 0.49 | 0.64 | nd | nd | nd | nd | |
| T49A | Layer 1 | 0.77 | 1.07 | 1.14 | 0.96 | 1.06 | 90 |
| T51A | Layer 1 | 0.69 | 1.06 | 1.02 | 0.85 | 0.89 | nd |
| L52A | Layer 1 | 0.36 | 0.25 | 0.23 | 0.1 | 0.01 | nd |
| F53A | Layer 1 | 0.40 | 0.25 | 0.29 | 0.39 | 0.02 | nd |
| D57A | Layer 1 | 0.65 | 0.62 | 0.91 | 0.86 | 0.54 | nd |
| A60G | Layer 1 | 0.92 | 0.86 | 1.46 | 0.89 | 0.79 | nd |
| Y61A | Layer 1 | 0.95 | 0.89 | 1.33 | 0.81 | 0.74 | nd |
| T63A | Layer 1 | 0.98 | 0.99 | 1.79 | 0.43 | 1.11 | nd |
| H66A | Layer 1 | 1.08 | 0.97 | 0.50 | 0.82 | 0.88 | 200 |
| V68A | Layer 1 | 1.07 | 0.60 | 1.12 | 0.51 | 0.68 | nd |
| W69L | Layer 1 | 0.88 | 0.89 | 0.64 | 1.07 | 0.33 | >400 |
| T71A | Layer 1 | 0.79 | 0.58 | 0.85 | 0.92 | 0.03 | nd |
| H72A | Layer 1 | 1.02 | 0.33 | 1.11 | 0.56 | 0.04 | nd |
| A73S | Layer 1 | 1.13 | 0.46 | 1.19 | 0.32 | 0.86 | nd |
| V75A | Layer 1 | 0.75 | 0.55 | 0.72 | 0.40 | 0.19 | nd |
| P76A | Layer 1 | 0.85 | 0.64 | 1.02 | 0.37 | 0.11 | nd |
| T77A | Layer 1 | 1.66 | 0.56 | 1.52 | 0.95 | 0.65 | 90 |
| D78A | Layer 1 | 0.69 | 0.51 | 1.13 | 1.03 | 0.11 | 90 |
| P79A | Layer 1 | 1.09 | 0.64 | 1.29 | 0.90 | 0.54 | 88 |
| N80A | Layer 1 | 1.21 | 0.65 | 1.24 | 0.97 | 0.99 | 90 |
| P81A | Layer 1 | 1.18 | 0.61 | 1.10 | 0.89 | 0.81 | 90 |
| Q82A | Layer 1 | 1.17 | 1.03 | 1.12 | 0.92 | 0.98 | 100 |
| E83A | β-sandwich | 0.93 | 0.61 | 0.86 | 0.63 | 0.29 | 70 |
| L86A | β-sandwich | 0.65 | 0.36 | 0.48 | 0.58 | 0.07 | 20 |
| V89A | β-sandwich | 0.56 | 0.22 | 0.97 | 0.57 | 0.01 | nd |
| T90A | β-sandwich | 0.65 | 0.55 | 1.27 | 0.85 | 0.87 | 20 |
| E91A | β-sandwich | 0.53 | 0.41 | 1.45 | 0.26 | 0.07 | 80 |
| F93A | β-sandwich | 0.37 | <0.01 | nd | 0.01 | <0.01 | nd |
| W96A | Layer 2 | 1.35 | 0.65 | 0.66 | 2.79 | 0.96 | 110 |
| K97A | Layer 2 | 1.32 | 0.93 | 0.93 | 0.57 | 0.98 | 110 |
| N99A | Layer 2 | 1.18 | 0.70 | 0.97 | 1.28 | 0.82 | nd |
| M100A | Layer 2 | 0.75 | 0.29 | 0.50 | 0.75 | 0.39 | nd |
| E102A | Layer 2 | 1.36 | 0.79 | 0.81 | 0.83 | 1.03 | 50 |
| Q103A | Layer 2 | 0.77 | 1.07 | 1.15 | nd | 0.79 | 60 |
| Q103R | Layer 2 | 0.86 | 0.33 | 0.74 | 1.59 | 0.18 | 20 |
| M104A | Layer 2 | 1.38 | 0.47 | 1.25 | nd | 0.21 | 90 |
| E106A | Layer 2 | 0.86 | 0.61 | 0.90 | 0.59 | 0.58 | nd |
| D107A | Layer 2 | 0.24 | 0.32 | 0.66 | 0.39 | 0.09 | 20 |
| D107Q | Layer 2 | 0.78 | 0.61 | 0.69 | 0.79 | 0.01 | nd |
| D107Y | Layer 2 | 0.14 | 0.31 | nd | 0.62 | <0.01 | nd |
| I108A | Layer 2 | 0.68 | 0.79 | 1.48 | nd | 0.43 | 300 |
| I109A | Layer 2 | 0.95 | 0.44 | 0.49 | 1.95 | 0.41 | 50 |
| I109W | Layer 2 | 0.96 | 0.39 | 0.35 | 2.40 | 0.34 | 200 |
| S110A | Layer 2 | 0.69 | 0.62 | 0.83 | 1.04 | 0.09 | 90 |
| S110N | Layer 2 | 0.73 | 0.66 | nd | 1.09 | 0.21 | 90 |
| S110W | Layer 2 | 0.59 | 0.49 | nd | 0.98 | 0.03 | 20 |
| L111A | Layer 2 | 0.34 | 0.28 | 0.48 | 0.72 | 0.04 | >400 |
| L111F | Layer 2 | 0.32 | 0.10 | 0.75 | 0.46 | 0.02 | 20 |
| W112A | Layer 2 | 1.07 | 0.17 | 0.25 | 0.98 | 0.07 | 100 |
| D113A | Layer 2 | 1.08 | 0.55 | 1.79 | nd | 0.50 | 30 |
| Q114N | Layer 2 | 0.93 | 0.74 | 1.09 | 1.78 | 0.42 | 50 |
| S115A | Layer 2 | 0.98 | 1.12 | 1.36 | nd | 0.79 | 150 |
| L116A | Layer 2 | 1.04 | 0.25 | 0.67 | 0.25 | <0.01 | nd |
| F210A | Layer 2 | 0.51 | 1.33 | 0.56 | 0.85 | 0.06 | 40 |
| P212A | Layer 2 | 0.88 | 0.93 | 0.65 | 1.85 | 0.76 | 360 |
| I213A | Layer 2 | 0.37 | 0.48 | 0.62 | 0.92 | 0.04 | 160 |
| I215A | Layer 2 | 0.34 | 0.45 | 0.46 | 0.85 | 0.01 | 80 |
| Y217A | Layer 2 | 0.60 | 0.32 | 0.52 | 0.67 | 0.15 | nd |
| A221G | Layer 2 | 1.02 | 0.43 | 0.76 | 1.04 | 0.01 | nd |
| F223A | β-Sandwich | 0.90 | 0.23 | 0.85 | 0.63 | 0.19 | nd |
| S375W | Phe 43 cavity | 0.82 | 0.93 | 1.12 | 0.69 | 0.70 | 20 |
| W437R | β20-β21 loop | 1.07 | 0.64 | 0.09 | 0.46 | <0.01 | nd |
| K487A | β25 strand | 0.13 | 0.3 | nd | nd | nd | nd |
| K487E | β25 strand | 0.13 | 0.2 | nd | nd | nd | nd |
| W69L/L111W | Layer 1/Layer 2 | 0.62 | 0.28 | 0.72 | 0.30 | 0.02 | 140 |
| H66A/S375W | Layer 1/Phe 43 | 0.84 | 0.95 | 1.10 | 0.75 | 0.42 | 20 |
| W69L/S375W | Layer 1/Phe 43 | 0.88 | 0.81 | 1.11 | 1.09 | 0.28 | 20 |
| D107A/S375W | Layer 2/Phe 43 | 0.12 | 0.61 | 0.76 | 0.61 | nd | nd |
| L111A/S375W | Layer 2/Phe 43 | 0.62 | <0.01 | 1.32 | 0.34 | 0.03 | 20 |
The phenotypes of the wt and mutant Env gps were determined as described in Experimental Procedures. Results represent the mean values derived from at least three experiments. Less than 20% deviation from the mean value was typically observed. nd = not determined
The ability of mutant Env gps to bind CD4
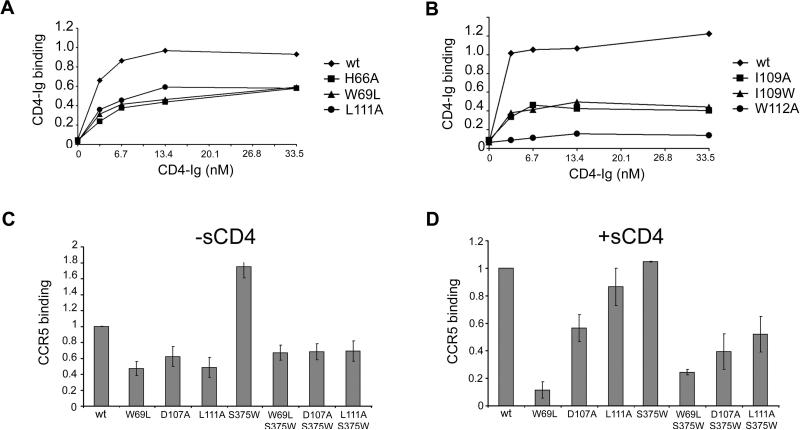
The effects of the changes introduced into the inner domain on the affinity of gp120 for CD4 were examined. Comparable amounts of radiolabeled gp120 in the supernatants of 293T cells expressing the Env gps were precipitated by CD4-Ig. CD4-Ig is a fusion protein in which the N-terminal two domains of CD4 are linked to the Fc component of immunoglobulin G (Chowdhury et al., 1991). Three sets of changes decreased the efficiency with which mutant gp120s were precipitated by CD4-Ig. The first set of changes involved residues (H66, W69, L111, F210 and P212) at the interface of Layer 1 and Layer 2 (Figure 1B and Table S2). These mutants exhibited moderate decreases in the efficiency with which they were precipitated by CD4-Ig (Figure 2A and Table 1). The observed decreases in CD4 binding were mainly due to the increased off-rates of the mutants compared with that of wild-type (wt) gp120 (Table 2). Thus, gp120 mutants with alterations in the Layer 1-Layer 2 interface fail to retain CD4 tightly upon initial binding.
Figure 2. Binding of gp120 receptors.
A, B. The effects of alterations in the Layer 1-Layer 2 interface (A) or in Layer 2 (B) on gp120 recognition by CD4-Ig were examined. Normalized amounts of radiolabeled wild-type mutant gp120 glycoproteins were incubated with increasing concentrations of CD4-Ig for 2 hours at 37°C. The precipitates were washed, run on SDS-polyacrylamide gels, and analyzed by densitometry. All values were normalized to a saturating value for the wt gp120. Representative results from at least two independent experiments are shown. C, D. Similar amounts of radiolabeled gp120 glycoproteins were incubated in the absence (C) or presence (D) of 200 nM sCD4 prior to addition to cells expressing CCR5. After two hours at 37°C, the amount of bound mutant gp120 was determined and normalized to the observed amount of bound wt gp120. Incubation with sCD4 increased the binding of wt gp120 8-fold. The data shown represent the means +/- SEM of two independent experiments.
Table 2.
Characterization of ligand binding to selected HIV-1YU2 gp120 mutants by surface plasmon resonance.
| sCD4 |
17b |
17b (+ sCD4) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| gp120 variant | on-rate constant (M−1s−1) | off-rate constant (s−1) | KD (M) (fold change) | on-rate constant (M−1s−1) | off-rate constant (s−1) | KD(M) (fold change) | on-rate constant (M−1s−1) | off-rate constant (s−1) | KD(M) (fold change) |
| wild-type | 1.19 × 104 | 7.83×10−5 | 6.58×10−9 (1.0) | 8.81×103 | 1.44×10−5 | 1.63×10−9 (1.0) | 3.55×104 | 1.64×10−4 | 4.62×10−9 (1.0) |
| H66A | 3.32×103 | 5.69×10−4 | 1.72×10−7 (26) | 4.71×102 | 2.69×10−4 | 5.72×10−7 (351) | 2.02×104 | 1.63×10−4 | 8.08×10−9 (1.7) |
| W69L | 9.69×103 | 5.26×10−4 | 5.42×10−8 (8.2) | 2.12×102 | 9.55×10−8 | 4.51×10−8 (28) | 2.08×104 | 1.80×10−4 | 8.63×10−9 (1.9) |
| D107A | 1.91×104 | 1.42×10−3 | 7.43×10−8 (11.3) | 2.85×102 | 2.98×10−5 | 1.04×10−7 (64) | 2.09×104 | 2.01×10−4 | 9.64×10−9 (2.1) |
| L111A | 1.32×104 | 4.26×10−4 | 3.23×10−8 (4.9) | 2.58×102 | 6.96×10−5 | 2.70×10−7 (166) | 5.51×104 | 1.77×10−4 | 2.84×10−9 (0.61) |
| L111F | 1.53×104 | 2.04×10−4 | 1.33×10−8 (2.0) | 2.67×102 | 1.51×10−5 | 5.65×10−8 (35) | 2.99×104 | 9.45×10−5 | 3.16×10−9 (0.68) |
| L111W | 2.09×103 | 1.30×10−3 | 6.21×10−7 (94) | 2.16×102 | 7.74×10−4 | 3.59×10−6 (2200) | 3.27×104 | 7.41×10−5 | 2.27×10−9 (0.49) |
| S375W | 1.38×104 | 6.49×10−7 | 4.69×10−11 (0.0071) | 2.43×104 | 1.21×10−5 | 4.98×10−10 (0.31) | 2.54×104 | 8.25×10−5 | 3.25×10−9 (0.70) |
| W69L/L111W | 5.54×102 | 4.83×10−4 | 8.71×10−7 (132) | 3.44×102 | 5.54×10−4 | 1.61×10−6 (988) | 2.42×104 | 6.25×10−5 | 2.58×10−9 (0.56) |
| H66A/S375W | 6.55×102 | 5.02×10−6 | 7.67×10−9 (1.2) | 1.69×103 | 9.31×10−5 | 5.51×10−8 (34) | 2.82×104 | 6.78×10−5 | 2.40×10−9 (0.52) |
| W69L/S375W | 1.18×104 | 9.98×10−6 | 8.46×10−10 (0.13) | 2.18×102 | 8.28×10−5 | 3.79×10−7 (233) | 4.65×104 | 1.28×10−4 | 2.76×10−9 (0.60) |
| L111A/S375W | 1.71×104 | 1.92×10−5 | 1.12×10−9 (0.17) | 5.19×102 | 2.38×10−5 | 4.59×10−8 (28) | 4.55×104 | 1.30×10−5 | 2.72×10−9 (0.59) |
The gp120-reactive ligand (sCD4 or 17b) was immobilized directly onto a CM5 sensor chip and the binding of the indicated gp120 gps evaluated as described in Experimental Procedures.
A second set of changes involving W112, I109 and L116 in Layer 2 resulted in reductions in CD4 binding (Figure 2B and Table 1). Although W112 does not contact CD4, it lines the Phe 43 cavity and, along with I109, interacts with W427, a cavity-lining residue that does contact CD4 (Lasky et al., 1987; Olshevsky et al., 1990; Kwong et al., 1998; Wyatt et al., 1998). Trp112 and L116 interact with F382, which also abuts the Phe 43 cavity. Thus, a network of interactions involving the distal end of the α1 helix in gp120 Layer 2 contributes to CD4 binding.
Finally, alteration of D107 or Y217, which interact with each other within Layer 2 (Figure 1B), decreased CD4-binding affinity (Tables 1 and 2).
Thus, multiple inner domain residues that do not contact CD4 nonetheless contribute to the affinity of the gp120-CD4 interaction.
Effect of filling the Phe 43 cavity on the phenotypes of inner domain mutants
Serine 375 flanks the Phe 43 cavity; substitution of a tryptophan residue for S375 fills the Phe 43 cavity with the indole ring (Zhou et al., 2007). As a result, the S375W mutant favors conformation(s) closer to that of the CD4-bound state (Xiang et al., 2002). To determine whether such a change in gp120 conformation would influence the phenotypes of the inner domain mutants, we introduced the S375W change into some of the gp120 variants described above. Remarkably, the presence of the S375W change completely restored the CD4-binding abilities of the gp120 mutants with alterations in H66, W69 and L111 (Table 2). In fact, the affinities of the W69L/S375W and L111A/S375W gp120 glycoproteins for sCD4 were greater than that of wt gp120. Thus, filling the Phe 43 cavity with the hydrophobic tryptophan side chain compensates for deficient Layer 1/Layer 2 contributions to the conformational transition of gp120 to the CD4-bound state.
Recognition of mutants by ligands that prefer the CD4-bound conformation
We examined the binding of some of the inner domain gp120 mutants by ligands, CCR5 and CD4-induced (CD4i) antibodies, that preferentially recognize the CD4-bound conformation (Trkola et al., 1996; Wu et al., 1996; Thali et al., 1993). These ligands bind overlapping, conserved regions on gp120 (Kwong et al., 1998; Wyatt and Sodroski, 1998).
In the absence of sCD4, the W69L, D107A and L111A changes in the inner domain reduced CCR5 binding approximately two-fold compared with that of the wt gp120 (Figure 2C). The S375W change that fills the Phe 43 cavity increased CCR5 binding of wt gp120 in the absence of sCD4, as expected (Xiang et al., 2002); however, the S375W change did not fully compensate for the diminished CCR5-binding ability of the W69L, D107A and L111A mutants. Addition of sCD4 partially restored the CCR5-binding ability of the L111A mutant, but not of the other mutants (Figure 2D). Thus, alteration of Layer 1 or Layer 2 residues in the gp120 inner domain moderately decreased CCR5 binding in a manner that was not completely compensated by the S375W change or by sCD4 binding.
The W69L, D107A and L111A mutants were precipitated by the 17b and 412d CD4i antibodies less efficiently than wt gp120 (Table 2 and Figure S1). Significant reductions in on-rates contributed to the decrease in 17b affinity (Table 2), indicating that these mutants do not spontaneously sample the conformation recognized by the 17b antibody as efficiently as wt gp120. Precipitation of the mutants by the 17b and 412d antibodies was enhanced by the S375W change, but was not restored to wt gp120 levels. However, incubation with sCD4 restored efficient 17b binding to all of the mutants (Table 2), indicating that these inner domain alterations do not directly disrupt the 17b epitope. Thus, Layer 1 and Layer 2 in the inner domain contribute to the ability of HIV-1 gp120 to assume spontaneously the conformation preferred by CD4i antibodies.
Proteolytic processing and subunit association of the Env glycoprotein mutants
Proteolytic processing of the gp160 precursor and association of the gp120 and gp41 subunits of each mutant Env gp were evaluated. All of the mutants were expressed efficiently, although a few mutants exhibited decreases in proteolytic processing of the gp160 Env gp precursor (Table 1). Several mutant Env gps exhibited a decrease in the non-covalent association of the gp120 and gp41 subunits (Table 1 and Figure 1C). These mutants contained alterations in three gp120 regions:
the inner domain β-sandwich – This structure has been previously implicated in gp120-gp41 interactions (Yang et al., 2003).
Layer 1 – Changes in A72 and H73 resulted in significant decreases in the association of gp120 and gp41. However, somewhat unexpectedly, given the predicted location of Layer 1 near the trimer axis, most changes in Layer 1 residues resulted in only modest reductions in gp120-gp41 association.
Layer 2 – Changes in several Layer 2 residues, particularly those located in the distal end of the α1 helix, significantly disrupted gp120-gp41 association. One of these Layer 2 residues (L111) is involved in interactions with Layer 1 in the CD4-bound conformation (Figure 1B); of note, changes in the cognate interacting residues (H66 and W69) in Layer 1 did not significantly decrease gp120-gp41 association. This suggests that the Layer 1-Layer 2 interactions that occur in the CD4-bound conformation of gp120 are not absolutely required for gp41 association in the unliganded Env glycoproteins. Most of the Layer 2 residues implicated in gp120-gp41 association are not solvent accessible in the CD4-bound conformation (Figure 1D). Thus, elements of gp120 Layer 2 that are relatively buried in the CD4-bound conformation apparently contribute, either directly or indirectly, to the non-covalent association with gp41 in the unliganded HIV-1 Env glycoproteins.
Function of the mutant HIV-1 Env gps
The mutant HIV-1 Env gps were assessed for the ability to mediate cell-cell fusion and to support virus entry into cells expressing CD4 and CCR5. In both assays, most of the mutant Env gps exhibited detectable activity (Table 1). The mutant Env gps with the lowest gp120-gp41 association indices also exhibited the lowest activities in these functional assays. A number of mutant Env gps with changes in Layer 1 (W69, T71, H72, D78, P79) and Layer 2 (Q103, D107, I109, S110, L111, W112, Q114, F210, I213, Y217 and A221) mediated cell-cell fusion much more efficiently than cell-free virus infectivity. This phenotype has previously been observed for alterations in the inner domain β-sandwich (Yang et al., 2003). Most of these mutants exhibited reductions in the efficiency of gp120-gp41 subunit association. Because the time between Env gp synthesis and engagement of the target cell is much longer in the virion infectivity assay than the cell-cell fusion assay, the former assay is more sensitive to decreases in gp120/gp41 stability.
Sensitivity of the mutant viruses to neutralization by sCD4
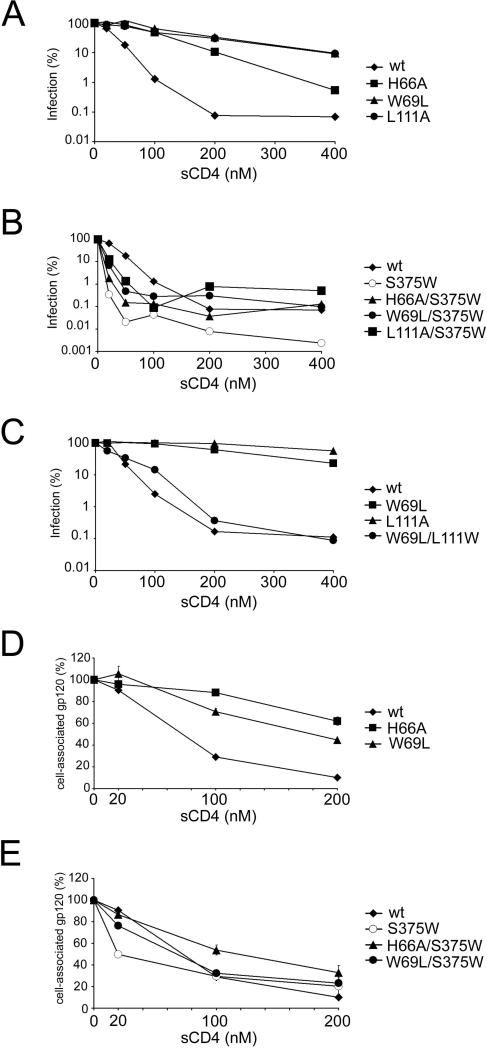
Changes in CD4-binding affinity can result in altered HIV-1 sensitivity to sCD4-mediated neutralization (Thali et al., 1991). Several viruses bearing Env gps with alterations in Layer 1 (H66A, W69L) or Layer 2 (I108A, I109W, L111A, S115A, P212A, I213A) were more resistant to sCD4 than viruses with wt Env gps (Figure 3A and Table 1). The introduction of the S375W change, which fills the Phe43 cavity (Xiang et al., 2002; Zhou et al., 2007), into the H66A, W69L and L111A mutant Env gps resulted in viruses that were neutralized by sCD4 as efficiently as a virus with wt Env gps (Figure 3B and Table 1).
Figure 3. Sensitivity of HIV-1 Env gp variants to soluble CD4 (sCD4).
A-C. Recombinant HIV-1 expressing luciferase and bearing wt or mutant HIV-1 Env gps were normalized by reverse transcriptase activity. Equal amounts of viruses were incubated with serial dilutions of sCD4 at 37°C for 1 hour prior to infection of Cf2Th-CD4/CCR5 cells. Infectivity at each dilution of sCD4 tested is shown as the percent of infection without sCD4 for each particular mutant. Duplicate samples were analyzed; data shown are representative of those obtained in at least three independent experiments. D, E. The sCD4-induced shedding of gp120 from HIV-1 Env gps expressed on the cell surface was measured. Transfected 293T cells were metabolically labeled with [35S]-methionine/cysteine for 16 hours in the presence of increasing concentrations of sCD4 (0 to 200 nM). Cell lysates were precipitated with a mixture of sera from HIV-1-infected individuals. Precipitates were analyzed by SDS-PAGE, autoradiography and densitometry. Data shown represent the means +/− SEM obtained from two independent experiments.
We also introduced a compensatory change (L111W) in Layer 2 that modeling suggested might restore interaction with the W69L mutant, which has an alteration of Layer 1. The sensitivity of this mutant to sCD4 increased considerably compared with that of the W69L mutant virus (Figure 3C), even though the W69L/L111W mutant did not completely regain the ability to bind CD4 (Table 1, Table 2 and data not shown). Other instances (e.g., I108A, S115A) were observed where altered sensitivity to sCD4 was not explained by CD4-Ig binding to gp120 monomers (Table 1). Thus, changes in the gp120 inner domain may specifically affect sCD4 binding to the Env gp trimer or may modulate the inhibitory consequences of sCD4 binding on HIV-1 infectivity (See below).
An extreme consequence of sCD4 binding is the shedding of gp120 from the Env gp trimer (Moore et al., 1990; Hart et al., 1991; Thali et al., 1992). The H66A and W69L mutants exhibited less sCD4-induced gp120 shedding than the wt Env gps (Figure 3D). The S375W change restored the efficiency of sCD4-induced gp120 shedding in the H66A and W69L mutant Env gps to levels near that of the wt Env gps (Figure 3E). Thus, for this group of mutants, sCD4 binding to gp120, sCD4 induction of gp120 shedding and sCD4 inhibition of HIV-1 infection correlate.
Infection of CD4-negative cells by viruses with mutant Env gps
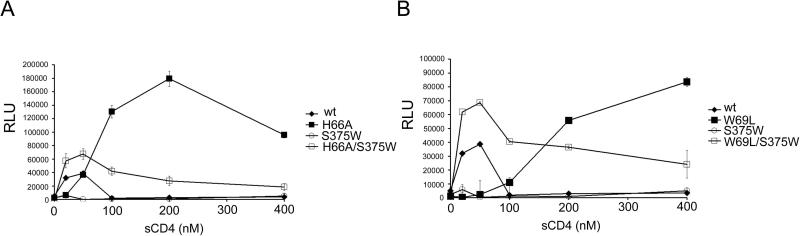
Soluble CD4 induces a transient activated state in the HIV-1 Env gps (Haim et al., 2009). To examine the effect of Layer 1 changes on this process, recombinant viruses bearing wt or mutant Env gps were incubated with CD4-negative, CCR5-expressing Cf2Th cells along with different concentrations of sCD4. In the absence of sCD4, no infection by any of the viruses was detected (Figure 4, A and B). The addition of a low concentration (20 nM) of sCD4 resulted in a slight increase in infectivity for viruses bearing the wt Env gps; this low level of infectivity was not observed at high concentrations (≥ 100 nM) of sCD4.
Figure 4. Effect of soluble CD4 on HIV-1 infection of CD4-negative, CCR5-expressing cells.
Recombinant luciferase-expressing HIV-1 with the indicated HIV-1 Env gps were normalized by reverse transcriptase activity and incubated with serial dilutions of sCD4 at 37°C for 1 hour prior to infection of Cf2Th-CCR5 cells. The level of infection is reported as Relative Light Units (RLU). For each data point, the infection was performed in duplicate; data shown represent the means +/- SEM derived from two independent experiments.
Interestingly, recombinant viruses bearing the H66A and W69L Env gps exhibited greater enhancement of infectivity by sCD4 than their wt counterpart (Figure 4, A and B). For these mutant viruses, the enhancement of infection was evident at high sCD4 concentrations (≥ 100 nM). Also of interest, the infectivity of viruses bearing the S375W Env glycoproteins was minimally enhanced, and only at a low concentration of sCD4; this is consistent with other studies suggesting that the CD4-bound conformation of HIV-1, which is favored by the S375W Env gps (Xiang et al., 2002; Zhou et al., 2007), is more prone to rapid inactivation (Haim et al., 2009; Kassa et al., 2009). Remarkably, introduction of the S375W change together with the W69L or H66A changes resulted in viruses that exhibited an enhancement of infectivity at low concentrations of sCD4 and retained significant infectivity at high concentrations of sCD4 (Figure 4, A and B). Similar results were observed when the T257S change was introduced along with the S375W change into the W69L and H66A mutants (Figure S2); the T257S change allows better accommodation of the tryptophan side chain in the Phe 43 cavity (Zhou et al., 2007). These double mutants apparently bind sCD4 comparably to viruses with the wild-type Env gps, because they are activated more efficiently at low sCD4 concentrations; moreover, for infection of cells expressing CD4 and CCR5, the double mutants were neutralized by sCD4 as efficiently as viruses with wild-type Env gps (Figure 3B and Figure S2). Thus, although viruses bearing the H66A/S375W and W69L/S375W Env gps interact efficiently with sCD4, they are less susceptible than wild-type viruses to inactivation at higher sCD4 doses.
DISCUSSION
Guided by a new crystal structure that includes the complete gp41-interactive region of the CD4-bound HIV-1 gp120 (Pancera et al., 2010), we identified a network of interactions involving the gp120 inner domain that modulate gp120-gp41 association, CD4 binding affinity, and susceptibility to inactivation by sCD4.
Prior to receptor engagement, gp120 must maintain its non-covalent association with gp41 and prevent gp41 from prematurely undergoing transitions to lower-energy conformations. In both unliganded and CD4-bound states, two HIV-1 gp120 elements, a 7-stranded β-sandwich and the N- and C-terminal extensions from this sandwich, play a major role in mediating gp120-gp41 association (Helseth et al., 1991; Binley et al., 2000; Yang et al., 2003; Sen et al., 2008; Wang et al., 2008). Two other gp120 regions apparently contribute to the interaction with gp41 in the unliganded Env gp trimer. The first region involves Layer 1, which is predicted to reside near the trimeric axis of the Env gp complex (Kwong et al., 2000; Pancera et al., 2010) and thus would be well-positioned to interact with gp41. Indeed, changes in two Layer 1 residues (H72 and A73) did decrease subunit association. Unexpectedly, however, most changes in Layer 1 residues (including those interacting with Layer 2) exerted modest or no effects on gp120-gp41 association. Thus, Layer 1 may play a modulatory rather than a primary role in the gp120-gp41 interaction. Such a role has been suggested for valine 65 in the Layer 1 α0 helix, based on the co-selection of gp120 and gp41 changes during virus adaptation to T-cell lines (Leavitt et al., 2003).
A second region important for gp120-gp41 association is Layer 2, particularly the distal end of the α1 helix. Most of these Layer 2 mutants mediate cell-cell fusion efficiently and are recognized by conformation-dependent antibodies, and thus are not globally misfolded. Many of the Layer 2 residues implicated in gp120-gp41 association are not surface-exposed in the CD4-bound conformation of gp120. Thus, any direct interaction between these Layer 2 residues and gp41 would require the existence of a different gp120 conformation in the unliganded state.
Other studies have suggested the possibility that the distal elements of Layer 2 and the adjacent β2-β3 strands, which compose part of the bridging sheet, undergo conformational changes upon CD4 binding (Kwong et al., 1998; Chen et al., 2005; Pan et al., 2005; Rits-Voloch et al., 2006). Indeed, in some structures of gp120 without CD4 (Chen et al., 2005; Zhou et al., 2007; Chen et al., unpublished data), the α1 helix is partially or completely unwound, potentially allowing direct contacts between some of the residues implicated in our study and gp41. In applying these observations to the unliganded Env gp complex, however, two caveats should be kept in mind. First, in all prior studies, gp120 inner domain components were deleted or modified to promote crystallization. Second, in light of the possibility that the inner domain interacts with gp41, structures of this region that are relevant to the unliganded Env gp trimer may require the inclusion of gp41 components.
Changes in the Layer 1-Layer 2 interface apparently decrease the spontaneous sampling of the CD4-bound conformation by HIV-1 gp120, lowering the on-rate, binding affinity and neutralization potency of CD4i antibodies (Table 2, Figure S1 and Kassa et al., 2009). CCR5 binding is also sensitive to these Layer 1-Layer 2 changes. By contrast, the initial contact of CD4 with gp120, reflected in its on-rate, is minimally affected by inner domain changes, and thus appears not to depend upon the sampling of the CD4-bound conformation by gp120 (Zhou et al., 2007; Rits-Voloch et al., 2006). Rather, changes in the integrity of the Layer 1-Layer 2 interface influence the off-rate of the CD4-gp120 complex; this effect cannot be explained by direct contacts of Layer 1 or Layer 2 residues with CD4 (Pancera et al., 2010). The S375W change, which fills the Phe 43 cavity, completely compensates for the inability of inner domain mutants to decrease the off-rate of bound CD4; however, in the absence of CD4, the S375W change is not sufficient to allow spontaneous sampling of the conformation required for efficient recognition by CCR5 and CD4i antibodies. Thus, Layers 1 and 2 in the inner domain contribute to the ability of gp120 to make the transition from the unliganded to the CD4-bound state; subtle differences exist in the requirements for binding CD4 on the one hand and CD4i antibodies/CCR5 on the other.
Alteration of gp120 residues in the Layer 1-Layer 2 interface (H66A, W69L, L111A and P212A) resulted in increased resistance to neutralization by sCD4. Differences in the affinity of monomeric gp120 for CD4 likely contribute to the sCD4 resistance of these mutants (Figure S3), as has been observed for other HIV-1 Env gp variants with decreases in CD4 binding (Thali et al., 1991). Consistent with this, the S375W change restored the CD4-binding affinity of the H66A, W69L and L111A mutants, and reverted the phenotypes of resistance to both sCD4 neutralization and sCD4 induction of gp120 shedding. Of interest, changes in the α0 helix of Layer 1 have been reported to segregate with sCD4 resistance during the passage of a primary HIV-1 isolate in a T-cell line (Orloff et al., 1995).
Soluble CD4 induces an activated state in the HIV-1 Env gps that rapidly decays into functionally inactive forms (Haim et al., 2009). Alterations in Layer 1 (e.g., H66A and W69L) apparently affect this decay process, even when sCD4 binding to the Env gps is restored by the S375W change in the Phe 43 cavity. During infection of CD4-negative, CCR5-positive target cells, viruses with the H66A/S375W and W69L/S375W mutant Env gps were activated at low sCD4 concentrations. During infection of CD4-expressing target cells, these mutant viruses were neutralized as efficiently by sCD4 as viruses with wild-type Env gps. Soluble CD4-induced shedding of gp120 was comparable for the wild-type, H66A/S375W and W69L/S375W Env gps. Therefore, the H66A/S375W and W69L/S375W Env gps apparently bind sCD4 efficiently; nonetheless, these mutants demonstrate activation at high sCD4 concentrations that, in parallel, do not activate wild-type HIV-1. In this respect, these HIV-1 mutants resemble SIV (Allan et al., 1992; Schenten et al., 1999). Thus, the H66A and W69L changes stabilize the sCD4-activated HIV-1 Env gp intermediate, slowing its decay into functionally inactive forms. The observed correlation between the longevity of the sCD4-activated state and the duration of sCD4-induced exposure of the gp41 HR1 region (Haim et al., 2009) is consistent with a role of Layer 1 in modulating conformational transitions in gp41. That H66A/S375W and W69L/S375W Env gps differ from wild-type Env gps in sCD4-induced activation-decay but not in gp120 shedding underscores previous suggestions that the functionally inactive products of these two processes are distinct (Si et al., 2004; Haim et al., 2009).
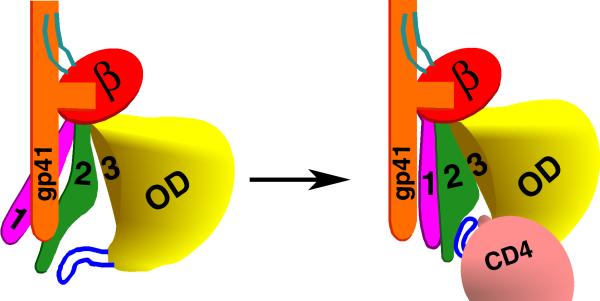
The involvement of Layer 1 and Layer 2 residues in gp41 association and in CD4 binding suggests a model for the unliganded gp120 in the assembled Env gp trimer, and for the conformational switch that occurs upon CD4 binding (Figure 5). According to this model, in the unliganded state, Layer 2 interacts with the gp41 ectodomain. By virtue of its own interaction with gp41, Layer 1 is shifted from its CD4-bound conformation, thus enhancing the opportunity for Layer 2 to contact gp41. The binding of CD4 to the wild-type HIV-1 Env gps results in the apposition of Layer 1 and Layer 2, as seen in the recent Pancera et al. crystal structure (Figure 1). The Layer 1-Layer 2 interaction reduces the off-rate of the bound CD4. The Layer 1-Layer 2 interaction also allows the gp41 ectodomain segment to undergo additional conformational changes involved in virus entry; this process may involve both the liberation of gp41 ectodomain segments through the loss of previous interactions with Layer 1 and Layer 2 residues, and the creation of new bonds between gp120 and gp41 that favor the formation of the pre-hairpin intermediate. This model helps to explain the relative lability of the sCD4-bound state (Haim et al., 2009; Kassa et al., 2009). Altered gp41 interactions with Layers 1 and 2 resulting from sCD4 binding might destabilize the gp41 ectodomain and either prematurely promote conformational changes related to those involved in virus entry or predispose to off-pathway inactivating events. Future studies will test the specific predictions of this model.
Figure 5. A model of the activation of the HIV-1 Env gps triggered by binding to the CD4 receptor.
One of the three subunits of the HIV-1 Env gp trimer is depicted, oriented so that the viral membrane is at the top of the figure. The gp41 ectodomain is colored orange, the gp120 N and C termini cyan, the β-sandwich red, Layer 1 magenta, Layer 2 green, Layer 3 yellow, outer domain (OD) yellow, and the β20-β21 loop blue. In the unliganded state (left figure), Layer 1 and Layer 2 assume conformations different from that observed in the CD4-bound state (Pancera et al., 2010), allowing both layers to interact with the gp41 ectodomain. CD4 binding results in the apposition of Layer 1 and Layer 2 (right figure). This rearrangement of the gp120 inner domain slows the off-rate of CD4 and allows the gp41 ectodomain to undergo additional conformational changes necessary for HIV-1 entry.
The inner domain residues implicated in the CD4-induced conformational transition are well-conserved within the HIV-1 and SIVcpz lineage (Figure S4). Of interest, the corresponding residues are also conserved within the HIV-2/SIVsm lineage, although differences between the HIV-1/SIVcpz and HIV-2/SIVsm lineages are evident. For example, residue 375 is naturally a tryptophan in the HIV-2/SIVsm viruses (Kuiken et al., 2008); in HIV-1, substitution of tryptophan for serine 375 fills the Phe 43 cavity and diminishes the impact of alterations in Layers 1 and 2 on CD4 binding. Alanine substitutions in the distal part of Layer 2 (residues 206-214) in SIV gp120 have been previously reported to decrease gp41 association (Rits-Volloch et al., 2006); however, two equivalent HIV-1 gp120 mutants (F210A and P212A) studied herein exhibited wild-type levels of subunit association. Thus, while both immunodeficiency virus lineages preserve the potential to form a network of inner domain interactions, lineage-specific differences exist, probably driven by distinct biological necessities.
EXPERIMENTAL PROCEDURES
Cell lines
293T, Cf2Th and TZM-bl cells were propagated as described in the Supplemental Data.
Site-directed mutagenesis
Mutations were introduced into the pSVIIIenv plasmid expressing the full-length HIV-1YU2 Env gps using the QuikChange II XL site-directed mutagenesis protocol (Stratagene). The presence of the desired mutations was confirmed by DNA sequencing. All residues are numbered according to those of the prototypic HXBc2 sequence, as per current convention (Korber et al., 1998).
Ligand binding of gp120
The ability of mutant gp120 to bind CD4-Ig, monoclonal antibodies and CCR5 was assessed by immunoprecipitation or surface plasmon resonance biosensor analysis, as previously described (Johnsson et al., 1991). Detailed descriptions can be found in the Supplemental Data.
Infectivity and neutralization assays
The infectivity and neutralization sensitivity of single-round luciferase-expressing HIV-1 were assessed as described (Madani et al., 2008; Thali et al., 1991) and in the Supplemental Data.
Intramolecular interactions
LIGPLOT (Wallace et al., 1995) was used to predict the hydrophobic interactions and hydrogen bonds between gp120 residues.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ms. Yvette McLaughlin and Ms. Elizabeth Carpelan for manuscript preparation and Dr. Lavanya Krishnan for Biacore advice. This work was supported by grants from the National Institutes of Health (AI24755, GM56550 and AI67854), by the International AIDS Vaccine Initiative, and by the late William F. McCarty-Cooper. The authors have no conflicts of interest to report.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alkhatib G, Combadiere C, Broder CC, Feng Y, Kennedy PE, Murphy PM, Berger EA. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- Allan JS, Coligan JE, Barin F, McLane MF, Sodroski JG, Rosen CA, Haseltine WA, Lee TH, Essex M. Major glycoprotein antigens that induce antibodies in AIDS patients are encoded by HTLV-III. Science. 1985;228:1091–1094. doi: 10.1126/science.2986290. [DOI] [PubMed] [Google Scholar]
- Allan JS, Strauss J, Buck DW. Enhancement of SIV infection with soluble receptor molecules. Science. 1990;247:1084–8. doi: 10.1126/science.2309120. [DOI] [PubMed] [Google Scholar]
- Binley JM, Sanders RW, Clas B, Schuelke N, Master A, Guo Y, Kajumo F, Anselma DJ, Maddon PJ, Olson WC, et al. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 2000;74:627–43. doi: 10.1128/jvi.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DC, Fass D, Berger JM, Kim PS. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- Chen B, Vogan EM, Gong H, Skehel JJ, Wiley DC, Harrison SC. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature. 2005;433:834–841. doi: 10.1038/nature03327. [DOI] [PubMed] [Google Scholar]
- Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath PD, Wu L, Mackay CR, LaRosa G, Newman W, et al. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- Chowdhury IH, Koyanagi Y, Takamatsu K, Yoshida O, Kobayashi S, Yamamoto N. Evaluation of anti-human immunodeficiency virus effect of recombinant CD4-immunoglobulin in vitro: a good candidate for AIDS treatment. Med. Microbiol. Immunol. 1991;180:183–192. doi: 10.1007/BF00215247. [DOI] [PubMed] [Google Scholar]
- Dalgleish AG, Beverley PC, Clapham PR, Crawford DH, Greaves MF, Weiss RA. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312:763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton RE, Hill CM, et al. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- Doranz BJ, Rucker J, Yi Y, Smyth RJ, Samson M, Peiper SC, Parmentier M, Collman RG, Doms RW. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- Dragic T, Litwin V, Allaway GP, Martin SR, Huang Y, Nagashima KA, Cayanan C, Maddon PJ, Koup RA, Moore JP, et al. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- Farzan M, Choe H, Desjardins E, Sun Y, Kuhn J, Cao J, Archambault D, Kolchinsky P, Koch M, Wyatt R, et al. Stabilization of human immunodeficiency virus type 1 envelope glycoprotein trimers by disulfide bonds introduced into the gp41 glycoprotein ectodomain. J. Virol. 1998;72:7620–7625. doi: 10.1128/jvi.72.9.7620-7625.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Broder CC, Kennedy PE, Berger EA. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- Furuta RA, Wild CT, Weng Y, Weiss CD. Capture of an early fusion-active conformation of HIV-1 gp41. Nat. Struct. Biol. 1998;5:276–279. doi: 10.1038/nsb0498-276. [DOI] [PubMed] [Google Scholar]
- Haim H, Si Z, Madani N, Wang L, Courter JR, Princiotto A, Kassa A, DeGrace M, McGee-Estrada K, Mefford M, et al. Soluble CD4 and CD4-mimetic compounds inhibit HIV-1 infection by induction of a short-lived activated state. PLoS Pathogens. 2009;5:e1000360. doi: 10.1371/journal.ppat.1000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart TK, Kirsh R, Ellens H, Sweet RW, Lambert DM, Petteway SR, Jr., Leary J, Bugelski PJ. Binding of soluble CD4 proteins to human immunodeficiency virus type 1 and infected cells induces release of envelope glycoprotein gp120. Proc. Natl. Acad. Sci. U S A. 1991;88:2189–2193. doi: 10.1073/pnas.88.6.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Vassell R, Zaitseva M, Nguyen N, Yang Z, Weng Y, Weiss CD. Peptides trap the human immunodeficiency virus type 1 envelope glycoprotein fusion intermediate at two sites. J. Virol. 2003;77:1666–1671. doi: 10.1128/JVI.77.3.1666-1671.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helseth E, Olshevsky U, Furman C, Sodroski J. Human immunodeficiency virus type 1 gp120 envelope glycoprotein regions important for association with the gp41 transmembrane glycoprotein. J. Virol. 1991;65:2119–2123. doi: 10.1128/jvi.65.4.2119-2123.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsson B, Lofas S, Lindquist G. Immobilization of proteins to a carboxymethyldextran-modified gold surface for biospecific interaction analysis in surface plasmon resonance sensors. Anal. Biochem. 1991;198:268–277. doi: 10.1016/0003-2697(91)90424-r. [DOI] [PubMed] [Google Scholar]
- Kassa A, Madani N, Schön A, Haim H, Finzi A, Xiang S-H, Wang L, Princiotto A, Pancera M, Courter J, et al. Transitions to and from the CD4-bound conformation are modulated by a single-residue change in the HIV-1 gp120 inner domain. J. Virol. 2009;83:8364–8378. doi: 10.1128/JVI.00594-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klatzmann D, Champagne E, Chamaret S, Gruest J, Guetard D, Hercend T, Gluckman JC, Montagnier L. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature. 1984;312:767–768. doi: 10.1038/312767a0. [DOI] [PubMed] [Google Scholar]
- Korber B, Foley BT, Kuiken C, Pillai SK, Sodroski JG. Numbering Positions in HIV Relative to HXB2CG. Human Retroviruses and AIDS. 1998;III:102–111. [Google Scholar]
- Koshiba T, Chan DC. The prefusogenic intermediate of HIV-1 gp41 contains exposed C-peptide regions. J. Biol. Chem. 2003;278:7573–7579. doi: 10.1074/jbc.M211154200. [DOI] [PubMed] [Google Scholar]
- Kuiken C, Foley B, Marx P, Wolinsky S, Leitner T, Hahn B, McCutchan F, Korber B. HIV Sequence Compendium 2008. Los Alamos HIV Sequence Database. 2008 [Google Scholar]
- Kwong PD, Doyle ML, Casper DJ, Cicala C, Leavitt SA, Majeed S, Steenbeke TD, Venturi M, Chaiken I, Fung M, et al. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature. 2002;420:678–682. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong PD, Wyatt R, Sattentau QJ, Sodroski J, Hendrickson WA. Oligomeric modeling and electrostatic analysis of the gp120 envelope glycoprotein of human immunodeficiency virus. J. Virol. 2000;74:1961–1972. doi: 10.1128/jvi.74.4.1961-1972.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasky LA, Nakamura G, Smith DH, Fennie C, Shimasaki C, Patzer E, Berman P, Gregory T, Capon DJ. Delineation of a region of the human immunodeficiency virus type 1 gp120 glycoprotein critical for interaction with the CD4 receptor. Cell. 1987;50:975–985. doi: 10.1016/0092-8674(87)90524-1. [DOI] [PubMed] [Google Scholar]
- Leavitt M, Park EJ, Sidorov IA, Dimitrov DS, Quinnan GV., Jr. Concordant modulation of neutralization resistance and high infectivity of the primary human immunodeficiency virus type 1 MN strain and definition of a potential gp41 binding site in gp120. J. Virol. 2003;77:560–570. doi: 10.1128/JVI.77.1.560-570.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Hui H, Burgess CJ, Price RW, Sharp PM, Hahn BH, Shaw GM. Complete nucleotide sequence, genome organization, and biological properties of human immunodeficiency virus type 1 in vivo: evidence for limited defectiveness and complementation. J. Virol. 1992;66:6587–6600. doi: 10.1128/jvi.66.11.6587-6600.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Blacklow SC, Kim PS. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat. Struct. Biol. 1995;2:1075–1082. doi: 10.1038/nsb1295-1075. [DOI] [PubMed] [Google Scholar]
- Madani N, Schon A, Princiotto AM, Lalonde JM, Courter JR, Soeta T, Ng D, Wang L, Brower ET, Xiang SH, et al. Small-molecule CD4 mimics interact with a highly conserved pocket on HIV-1 gp120. Structure. 2008;16:1689–1701. doi: 10.1016/j.str.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JP, McKeating JA, Weiss RA, Sattentau QJ. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science. 1990;250:1139–1142. doi: 10.1126/science.2251501. [DOI] [PubMed] [Google Scholar]
- Moore JP, Sodroski J. Antibody cross-competition analysis of the human immunodeficiency virus type 1 gp120 exterior envelope glycoprotein. J. Virol. 1996;70:1863–1872. doi: 10.1128/jvi.70.3.1863-1872.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myszka DG, Sweet RW, Hensley P, Brigham-Burke M, Kwong PD, Hendrickson WA, Wyatt R, Sodroski J, Doyle ML. Energetics of the HIV gp120-CD4 binding reaction. Proc. Natl. Acad. Sci. U S A. 2000;97:9026–9031. doi: 10.1073/pnas.97.16.9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olshevsky U, Helseth E, Furman C, Li J, Haseltine W, Sodroski J. Identification of individual human immunodeficiency virus type 1 gp120 amino acids important for CD4 receptor binding. J. Virol. 1990;64:5701–5707. doi: 10.1128/jvi.64.12.5701-5707.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orloff SL, Bandea CI, Kennedy MS, Allaway GP, Maddon PJ, McDougal JS. Increase in sensitivity to soluble CD4 by primary HIV type 1 isolates after passage through C8166 cells: association with sequence differences in the first constant (C1) region of glycoprotein 120. AIDS Res. Hum. Retroviruses. 1995;11:335–342. doi: 10.1089/aid.1995.11.335. [DOI] [PubMed] [Google Scholar]
- Pan Y, Ma B, Russinov R. CD4 binding partially locks the bridging sheet in gp120 but leaves the β2/3 strands flexible. J. Mol. Biol. 2005;350:514–527. doi: 10.1016/j.jmb.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Pancera M, Majeed S, Ban Y-EA, Chen L, Huang C-C, Kong L, Kwon YD, Stuckey J, Zhou T, Robinson JE, Schief WR, Sodroski J, Wyatt R, Kwong PD. Structure of HIV-1 gp120 with gp41-interactive region reveals layered architecture and basis of conformational mobility. Proc. Natl. Acad. Sci. U S A. 2010 doi: 10.1073/pnas.0911004107. in the press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rits-Volloch S, Frey G, Harrison SC, Chen B. Restraining the conformation of HIV-1 gp120 by removing a flexible loop. EMBO J. 2006;25:5026–5035. doi: 10.1038/sj.emboj.7601358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robey WG, Safai B, Oroszlan S, Arthur LO, Gonda MA, Gallo RC, Fischinger PJ. Characterization of envelope and core structural gene products of HTLV-III with sera from AIDS patients. Science. 1985;228:593–595. doi: 10.1126/science.2984774. [DOI] [PubMed] [Google Scholar]
- Schenten D, Marcon L, Karlsson GB, Parolin C, Kodama T, Gerard N, Sodroski J. Effects of soluble CD4 on simian immunodeficiency virus infection of CD4-positive and CD4-negative cells. J. Virol. 1999;73:5373–5380. doi: 10.1128/jvi.73.7.5373-5380.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schön A, Madani N, Klein JC, Hubicki A, Ng D, Yang X, Smith AB, III, Sodroski J, Freire E. Thermodynamics of binding of a low-molecular-weight CD4 mimetic to HIV-1 gp120. Biochemistry. 2006;45:10973–10980. doi: 10.1021/bi061193r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen J, Jacobs A, Caffrey M. Role of the HIV gp120 conserved domain 5 in processing and viral entry. Biochemistry. 2008;47:7788–7795. doi: 10.1021/bi800227z. [DOI] [PubMed] [Google Scholar]
- Si Z, Madani N, Cox JM, Chruma JJ, Klein JC, Schon A, Phan N, Wang L, Biorn AC, Cocklin S, et al. Small-molecule inhibitors of HIV-1 entry block receptor-induced conformational changes in the viral envelope glycoproteins. Proc. Natl. Acad. Sci. U S A. 2004;101:5036–5041. doi: 10.1073/pnas.0307953101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thali M, Furman C, Helseth E, Repke H, Sodroski J. Lack of correlation between soluble CD4-induced shedding of the human immunodeficiency virus type 1 exterior envelope glycoprotein and subsequent membrane fusion events. J. Virol. 1992;66:5516–5524. doi: 10.1128/jvi.66.9.5516-5524.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thali M, Moore JP, Furman C, Charles M, Ho DD, Robinson J, Sodroski J. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J. Virol. 1993;67:3978–3988. doi: 10.1128/jvi.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thali M, Olshevsky U, Furman C, Gabuzda D, Li J, Sodroski J. Effects of changes in gp120-CD4 binding affinity on human immunodeficiency virus type 1 envelope glycoprotein function and soluble CD4 sensitivity. J. Virol. 1991;65:5007–5012. doi: 10.1128/jvi.65.9.5007-5012.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trkola A, Dragic T, Arthos J, Binley JM, Olson WC, Allaway GP, Cheng-Mayer C, Robinson J, Maddon PJ, et al. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature. 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- Wallace AC, Laskowski R, Thornton JM. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Prot. Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- Wang J, Sen J, Rong L, Caffrey M. Role of the HIV gp120 conserved domain 1 in processing and viral entry. J. Biol. Chem. 2008;283:32644–32649. doi: 10.1074/jbc.M806099200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- Wu L, Gerard NP, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso AA, Desjardins E, Newman W, et al. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- Wyatt R, Kwong PD, Desjardins E, Sweet RW, Robinson J, Hendrickson WA, Sodroski JG. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- Xiang SH, Kwong PD, Gupta R, Rizzuto CD, Casper DJ, Wyatt R, Wang L, Hendrickson WA, Doyle ML, Sodroski J. Mutagenic stabilization and/or disruption of a CD4-bound state reveals distinct conformations of the human immunodeficiency virus type 1 gp120 envelope glycoprotein. J. Virol. 2002;76:9888–9899. doi: 10.1128/JVI.76.19.9888-9899.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Mahony E, Holm GH, Kassa A, Sodroski J. Role of the gp120 inner domain beta-sandwich in the interaction between the human immunodeficiency virus envelope glycoprotein subunits. Virology. 2003;313:117–125. doi: 10.1016/s0042-6822(03)00273-3. [DOI] [PubMed] [Google Scholar]
- Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, Yang X, Zhang MY, Zwick MB, Arthos J, et al. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445:732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P, Chertova E, Bess J, Jr., Lifson JD, Arthur LO, Liu J, Taylor KA, Roux KH. Electron tomography analysis of envelope glycoprotein trimers on HIV and simian immunodeficiency virus virions. Proc. Natl. Acad. Sci. U S A. 2003;100:15812–15817. doi: 10.1073/pnas.2634931100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.