Although the basic mechanisms of DNA synthesis are conserved across species, there are differences between simple and complex eukaryotes. Here, Errico and Costanzo reflect on the known distinctions—of poorly understood causes and consequences—between yeast and metazoans and analyse possible reasons for these diverging strategies.
Keywords: DNA replication, checkpoint, homologous recombination, yeast, metazoans
Abstract
Although the basic mechanisms of DNA synthesis are conserved across species, there are differences between simple and complex organisms. In contrast to lower eukaryotes, replication origins in complex eukaryotes lack DNA sequence specificity, can be activated in response to stressful conditions and require poorly conserved factors for replication firing. The response to replication fork damage is monitored by conserved proteins, such as the TIPIN–TIM–CLASPIN complex. The absence of this complex induces severe effects on yeast replication, whereas in higher eukaryotes it is only crucial when the availability of replication origins is limiting. Finally, the dependence of DNA replication on homologous recombination proteins such as RAD51 and the MRE11–RAD50–NBS1 complex is also different; they are dispensable for yeast S-phase but essential for accurate DNA replication in metazoans under unchallenged conditions. The reasons for these differences are not yet understood. Here, we focus on some of these known unknowns of DNA replication.
See Glossary for abbreviations used in this article.
Glossary.
- ACS
autonomous consensus sequence
- AND1
acidic nucleoplasmic DNA-binding protein 1
- ATM
ataxia telangiectasia mutated
- ATR
ataxia telangiectasia and Rad3-related
- ATRIP
ATR-interacting protein
- BRCA
breast cancer
- BRCT
BRCA1 C–terminus
- Cdc
cell division control
- CDK
cyclin-dependent kinase
- CDT1
chromatin licensing and DNA replication factor 1
- CHK1
checkpoint protein kinase 1
- Ctf4
chromosome transmission fidelity 4
- DDK
DBF4-dependent kinase
- Dpb11
DNA polymerase B (II)
- DSB
DNA double-strand break
- DT40
B-lymphocyte cell line
- ExoI
exonuclease 1
- FANCM
Fanconi anaemia, complementation group M
- GINS
Go, Ichi, Ni and San complex
- HeLa
cervical cancer cell line
- MCM
minichromosome maintenance protein
- Mrc1
mediator of replication checkpoint 1
- MRE11
meiotic recombination 11
- NBS1
Nijmegen breakage syndrome 1
- ORC
origin recognition complex
- Plx1
Xenopus Polo-like kinase 1
- POLα
polymerase-α
- Rad
radiation arrest deficient
- RECQL4
RecQ protein-like 4
- RPA
replication protein A
- Sgs1
slow growth suppressor 1
- Sld
synthetic lethal with Dpb11-1
- TIM1
Timeless
- TIPIN
Tim1-interacting protein
- TOPBP1
DNA topoisomerase 2 binding protein 1
Introduction
All eukaryotes use similar machinery and regulatory mechanisms for DNA duplication and cell division. Indeed, the main players have been conserved throughout evolution from unicellular organisms to mammals (Table 1). However, despite this high level of conservation, important differences can be observed between higher and lower eukaryotes. Many of these variations have been studied in the main DNA replication model systems, which are the Xenopus laevis egg extract and mammalian cell cultures for complex eukaryotes and metazoans, and yeast cells for unicellular eukaryotes. Here, we analyse the basic processes of DNA replication in which lower and higher eukaryotes differ, although the reasons for these differences remain obscure.
Table 1. Yeast and vertebrate homologues of proteins involved in different aspects of DNA replication.
| Vertebrates | Saccharomyces cerevisiae | Schizosaccharomyces pombe |
|---|---|---|
| Pre-replicative complex components | ||
| ORC1–6 | Orc1–6 | Orc1–6 |
| CDC6 | Cdc6 | Cdc6 |
| MCM2 | Mcm2 | Mcm2/Cdc19/Nda1 |
| MCM3 | Mcm3 | Mcm3 |
| MCM4 | Mcm4/Cdc54 | Cdc21 |
| MCM5 | Mcm5/Cdc46 | Mcm5/Nda4 |
| MCM6 | Mcm6 | Mcm6/Mis5 |
| MCM7 | Mcm7/Cdc47 | Mcm7 |
| Pre-initiation complex | ||
| MCM10 | Mcm10/Dna43 | Cdc23 |
| CDC45 | Cdc45/ Sld4 | Cdc45/Sna41 |
| TOPBP1 | Dbp11 | Cut5/Rad4 |
| — | Sld2 | Drc1 |
| — | Sld3 | Sld3 |
| SLD5 | Sld5 | Sld5 |
| PSF1 | Psf1 | Psf1 |
| PSF2 | Psf2 | Psf2 |
| PSF3 | Psf3 | Psf3 |
| Replication pausing complex | ||
| TIPIN | Csm3 | Swi3 |
| TIM1 | Tof1 | Swi1 |
| CLASPIN | Mrc1 | Mrc1 |
| AND1 | Ctf4 | Mcl1 |
| S-phase and DNA damage checkpoint | ||
| ATR | Mec1 | Rad3 |
| ATRIP | Ddc2 | Rad26 |
| ATM | Tel1 | Tel1 |
| CHK1 | Chk1 | Chl1/Rad27 |
| CHK2 | Rad53 | Cds1 |
| DNA repair (HR) | ||
| MRE11 | Mre11 | Rad32 |
| RAD50 | Rad50 | Rad50 |
| NBS1 | Xrs2 | Nbs1 |
| RAD51 | Rad51 | Rad51 |
| BRCA1 | Rad9 | Crb2 |
| BRCA2 | — | — |
| RAD54 | Rad54 | Rad54 |
| RAD52 | Rad52 | Rad52 |
| DNA repair (NHEJ) | ||
| LIGIV | LigIV | LigIV |
| XRCC4 | Lif1 | — |
| XLF | Nej1 | Xlf1/Nej1 |
| KU70 | Ku70 | Ku70 |
| DNA-PK | — | — |
HR, homologous recombination; NHEJ, non-homologous end-joining.
Establishment and selection of replication origins
DNA replication is a tightly controlled process. Several mechanisms have evolved to ensure that no regions of DNA are left unreplicated or are replicated more than once in every cell cycle. Prokaryotes use mostly a single origin to replicate their small genome, whereas eukaryotes—which have a high DNA content—have multiple replication origins distributed throughout the DNA. The first step in DNA replication is the assembly of a pre-replicative complex (pre-RC) at each origin (Diffley et al, 1995; Rowles et al, 1999), which consists in the binding of ORC1–6 (Bell & Stillman, 1992; Rabitsch et al, 2001), Cdc6 and Cdt1 (Gillespie et al, 2001) to chromatin, followed by the loading of the essential helicase activity, MCM2–7. In early S-phase, the pre-RC complex is converted into an initiation complex—which promotes DNA unwinding and polymerase loading (Diffley et al, 1995)—through the activity of S-phase kinases, the CDKs and DDK.
In Saccharomyces cerevisiae, replication origins are specified by a particular DNA sequence known as autonomously replicating sequence (ARS), which recruits the ORC. The ARS consists of an essential 11 bp consensus sequence (known as ACS) and several elements that contribute to promote initiation. Replication origins in the fission yeast Schizosaccharomyces pombe are much larger—500–1,000 bp compared with the 150 bp in S. cerevisiae—and do not display a clear consensus other than being extremely rich in A+T (Dai et al, 2005; Segurado et al, 2003). In contrast to yeast, replication origins in higher eukaryotes are defined less rigidly and apparently do not have a specific sequence requirement. An extreme case of a lack of ACS for origin specification can be found in embryonic systems—such as X. laevis and Drosophila melanogaster—in which DNA replication initiates at seemingly random sites spaced 10–15 kb apart (Blow et al, 2001; Shinomiya & Ina, 1991). A main difference between embryonic and somatic cells is the absence of transcription in embryos, which initiates at the midblastula transition—a stage with more defined initiation zones (Hyrien et al, 1995; Sasaki et al, 1999). In somatic cells replication origins are less frequent, being present approximately once every 150 kb. Until recently, only about 40 origins had been characterized in 10 metazoan organisms, from fruit flies to humans, with little evidence of a common consensus (Dimitrova et al, 1996). Two genome-wide approaches have led to the mapping of a greater number of initiation sites in the HeLa cell line genome. The first study characterized the presence of 283 origins using the HeLa S3 suspension cell line (Cadoret et al, 2008) and the second, more recent study identified 150 new origins in adherent HeLa cells (Karnani et al, 2009). Although the two studies agree only partly about individual origins (Karnani et al, 2009), they both define specific features for the metazoan initiation sites, revealing a correlation between origins and transcription start sites (Cadoret et al, 2008; Karnani et al, 2009). These studies have started to clarify the connection between origin selection, gene regulation and chromatin structure. Future studies will probably lead to a clearer consensus that defines the metazoan origins of replication, although it seems unlikely to be related to the yeast origins.
The fact that metazoan origins occur at many sites in large initiation zones and tend to be organized in clusters is a further difference with respect to yeast. It has been proposed that these features confer a selective evolutionary advantage for complex organisms with large genomes, enabling them to easily replicate newly acquired DNA sequences (Hyrien & Mechali, 1993). These differences might also reflect the requirement for integrating the control of DNA replication with cell differentiation and organism development. In this case, origin specification could be dictated by the high-order structure of chromatin, which changes during cell differentiation.
Another intriguing feature of replication origin organization is that the number of MCM2–7 complexes loaded on DNA exceeds the number of ORC1–6 complexes. These extra MCM2–7 complexes have been proposed to be additional sites from which replication can start (Lei et al, 1996; Walter & Newport, 1997) but are clearly redundant, as reducing their amount on chromatin does not impair unchallenged DNA replication (Ibarra et al, 2008; Lei et al, 1996). Interestingly, these supplementary origins remain ‘dormant' during S phase in X. laevis and mammals and only fire when replication forks are stalled or slowed. This mechanism is potentially relevant to ensure the complete replication of the genome in the presence of obstacles to replication forks (Fig 1; Ge et al, 2007; Ibarra et al, 2008; Woodward et al, 2006). Whether dormant origins exist in yeast and what their role would be is not clear. A recent study has unexpectedly shown that the presence of double-strand breaks (DSBs) in yeast can trigger the firing of nearby dormant origins (Doksani et al, 2009), suggesting that this feature is conserved across species.
Figure 1.
The organization of replication origins. The number of MCM2–7 complexes loaded onto chromatin is in excess compared with ORC1–6. When a replication fork stalls (a), a nearby dormant origin (green circle) can be activated to resume DNA replication. If DNA replication continues unperturbed (b), dormant origins are replicated passively by the active replicon. CDC, cell division control; CDK, cyclin-dependent kinase; MCM, minichromosome maintenance protein; ORC, origin recognition complex.
The mechanism that leads to the firing of dormant origins is unclear. One hypothesis is that their firing is not due to an active mechanism, but to a kinetic and probabilistic process whereby, when forks stall, dormant origins have more time and a greater chance of being used before the region they occupy is replicated and inactivated by a fork coming from an adjacent active origin (Ge et al, 2007). There could also be an active process regulating dormant origins in response to replicative stress. The ATM/ATR-dependent intra-S-phase checkpoint regulates origin firing, thereby limiting the number of origins that actually fire in the presence of replication stress (Shechter et al, 2004). The intra-S-phase checkpoint needs to be downregulated transiently for the activation of dormant origins (Ge et al, 2007; Woodward et al, 2006) and Plx1 has been recently shown to have an important role in its suppression (Trenz et al, 2008). The ATM/ATR-dependent phosphorylation of MCM2 is essential for this Plx1-mediated function (Cortez et al, 2004; Yoo et al, 2004), as it promotes Plx1 binding to the MCM2–7 complex through its Polo box domain (Trenz et al, 2008). When this Plx1/MCM2–7 complex is in the proximity of stalled replication forks, it seems to be involved in the release of CHK1-mediated suppression of nearby dormant origins (Trenz et al, 2008). However, how Plx1 suppresses CHK1 activity in this process remains unclear. The phosphorylation of adaptors required for CHK1 activation might be involved in this pathway. Overall, these data suggest that MCM2-recruited Plx1 promotes the progression of replication in the presence of replication stress. Consistent with this, DNA replication in the absence of Plx1 leads to the accumulation of DSBs, further supporting the role of Plx1 in promoting genome stability during S phase (Trenz et al, 2008). These findings have been recently confirmed in other vertebrates by showing that PLK1—a Plx1 orthologue—is required to promote DNA replication recovery after fork stalling by releasing the inhibition on origin firing in DT40 cells that lack FANCM (Schwab et al, 2010). This pathway was also shown to be dependent on MCM2 phosphorylation by ATR, demonstrating that this mechanism is highly conserved in metazoans. Neither the phosphorylation of MCM2 by ATM/ATR, nor a role for Plx1 orthologues in DNA replication have been described in yeast cells. Further work is required to understand in detail how dormant origins are regulated and their role in promoting genome replication in different organisms.
Initiation of DNA replication
Two replication factors—Sld2 and Sld3—have recently emerged as crucial in the cell-cycle-dependent control of DNA replication initiation in yeast (Tanaka et al, 2007; Zegerman & Diffley, 2007). Sld2 and Sld3 represent the minimal set of substrates that need to be phosphorylated by CDKs to initiate DNA replication (Tanaka et al, 2007; Zegerman & Diffley, 2007). Their phosphorylation allows them to interact with a Dpb11—a BRCT-containing protein—which seems to facilitate the loading of Cdc45 and, therefore, origin firing. Orthologues of Sld2, Sld3 and Dpb11 can be found in fungi—although sequence conservation is low even among related species—but their presence in other organisms is uncertain. Most importantly, although TOPBP1/CUT5/MUS101 is the recognized orthologue of Dpb11 (Table 1; Garcia et al, 2005), no clear orthologues of Sld2 and Sld3 have been identified so far in animal cells. RECQL4, which interacts with TOPBP1 and is required for DNA replication, has been recently suggested to be the putative orthologue of Sld2. However, RECQL4 has a limited homology to Sld2 and its function does not seem to be regulated by CDK-dependent phosphorylation (Sangrithi et al, 2005). Therefore, although RECQL4 is necessary for origin firing, it is possibly not a crucial CDK target. An additional difference between yeast and mammals emerged from a recent study on the formation of the CMG complex, which is an association between CDC45, MCM2–7 and the GINS complex that requires the presence of RECQL4, Ctf4/AND1 and MCM10 to be assembled. The CMG complex seems to have a crucial role in the formation and progression of replication forks and, surprisingly, the depletion of TOPBP1—which has an essential role in the chromatin loading of CDC45 and GINS in yeast cells—does not significantly affect CMG complex formation in mammals (Im et al, 2009).
Sld3 seems to be even more divergent, as no putative orthologue has been identified according to primary sequence. The CDK-dependent regulation of the initiation of DNA replication is a conserved process and therefore it is likely that functional orthologues of both Sld2 and Sld3 will be found. It is tempting to speculate that owing to the variety of different cells in multicellular organisms, replication initiation requires a more complex regulation, probably through the phosphorylation of many substrates that fulfil the roles Sld2 and Sld3 have in yeast.
Another interesting distinction between higher and lower eukaryotes is that several proteins—such as geminin, which is a CDT1-regulatory protein (McGarry & Kirschner, 1998), and MCM9—have been identified as the main regulators of DNA replication factors only in higher eukaryotes. In all eukaryotic organisms, the assembly of a new origin is suppressed by a high concentration of CDKs. In addition to CDK activity, multicellular eukaryotes use geminin to regulate the assembly of replication origins and prevent re-replication. Geminin interacts tightly with CDT1, thereby preventing the binding of the MCM2–7 complex to origins (Wohlschlegel et al, 2000). The binding of geminin to CDT1 blocks licensing, whereas it is enabled by MCM9 binding to CDT1—which prevents the loading of geminin onto chromatin during licensing (Lutzmann & Mechali, 2008). Furthermore, yeast Cdt1 binds directly to the Mcm2–7 complex, whereas in multicellular organisms, a direct interaction between these proteins in the absence of chromatin has not been verified in vivo (Seo et al, 2005). The requirement of complex organisms for a more sophisticated regulation to integrate DNA licensing and replication within a development programme might explain the existence of additional factors in the metazoan pre-RC.
MCM8, which is an additional member of the MCM2–7 family, has also been described only in higher eukaryotes (Maiorano et al, 2005). Studies performed in X. laevis egg extracts showed that MCM8 binds to chromatin after DNA synthesis is initiated and is required for the efficient progression of replication forks. These data suggest that MCM8 is not involved in origin licensing but functions specifically as a DNA helicase in vivo, perhaps contributing to DNA unwinding during the elongation process of DNA replication (Maiorano et al, 2005). The requirement for MCM8 in higher eukaryotes might be related to the size and the complexity of the genome, associated with the need to ensure efficient processivity in replicating large genomes.
Together, these data suggest that the proteins and mechanisms involved in the initiation of DNA replication in higher eukaryotes differ from those in yeast systems (Fig 2).
Figure 2.
Initial steps of DNA replication in yeast and metazoans. Orc1–6 defines the origins of replication. (A) In yeast, the loading of the Mcm2–7 helicase is regulated through the action of Cdc6 and Cdt1. Sld2 and Sld3 are required for origin firing after phosphorylation by CDKs. (B) In higher eukaryotes, there is an additional level of regulation of MCM2–7 loading due to the presence of MCM9 and geminin. MCM8 is only present in higher eukaryotes and seems to facilitate fork progression. PLK1/Plx1 regulates DNA replication under stressful conditions, a role that has only been shown for higher eukaryotes. Cdc, cell division control; CDKs, cyclin-dependent kinases; Cdt1, chromatin licensing and DNA replication factor 1; Dpb11, DNA polymerase B (II); Mcm, minichromosome maintenance protein; ORC, origin recognition complex; Plx1, Xenopus Polo-like kinase 1; Sld, synthetic lethal with Dpb11–1; TOPBP1, DNA topoisomerase 2 binding protein 1.
Stalled forks: a task for the replication pausing complex
Once two adjacent origins have fired, the two converging forks progress until they meet, ensuring the complete replication of the DNA segment. However, forks can stall if they encounter DNA damage. To ensure that replication will resume after the obstacle is removed, it is important to stabilize the replication fork so that the replisome components do not dissociate. To this end, several proteins that are not essential for DNA synthesis are present at the replication fork through their interaction with members of the replisome (Branzei & Foiani, 2005; Gambus et al, 2006). Among these, TIM1, TIPIN and CLASPIN have been identified—both in yeast and higher eukaryotes—as members of the ‘replication pausing complex' that contributes both to fork stabilization and to checkpoint activation. A central role in this S-phase checkpoint response is carried out by the DNA-damage-sensing kinase ATR, its functional homologues in budding and fission yeast—which are Mec1 and Rad3, respectively—and the ATR downstream kinase CHK1. These kinases are required to promote fork stability both in the absence and in the presence of DNA damage (Branzei & Foiani, 2005). The mechanism that senses fork lesions has been studied in many systems, including the X. laevis egg extract. Work with this model suggested that when the polymerase encounters a lesion its progression is blocked, whereas the helicase keeps unwinding the DNA (Byun et al, 2005). The uncoupling between the stalled polymerase and the helicase generates a segment of single-strand (ss) DNA that constitutes the signal for recruiting the ATR–ATRIP complex through RPA binding. In budding yeast, Tof1 (TIM1), Csm3 (TIPIN) and Mrc1 (CLASPIN) proteins are required for the Mec1/Rad53 (ATR/CHK2) checkpoint response that prevents the collapse of stalled replication forks and enables DNA replication to restart after recovery (Branzei & Foiani, 2005). In mammals TIM1, TIPIN and CLASPIN seem to mediate the ATR–CHK1 signalling cascade (Chou & Elledge, 2006; Errico et al, 2007; Unsal-Kacmaz et al, 2007). Intriguingly, these proteins are also part of the replisome in the absence of DNA damage and travel with the replication fork (Errico et al, 2007; Katou et al, 2003; Tanaka et al, 2009). Consistent with this, yeast proteins Mrc1 and Tof1 are important for the regulation of the normal progression of DNA replication (Hodgson et al, 2007; Katou et al, 2003; Tourriere et al, 2005), and a reduction in the expression levels of mammalian TIM1 results in a decreased rate of DNA synthesis (Chou & Elledge, 2006; Unsal-Kacmaz et al, 2007).
Although the function of these proteins seems to be conserved across species, the phenotype observed in the yeast strains deficient for these proteins is more severe when compared with depletions of the same genes in higher eukaryotes. Recent work in X. laevis strengthened the idea that TIPIN and TIM1 are active components of the replisome but in contrast to yeast, the depletion of TIPIN and TIM1 only had a measurable effect on DNA replication when the dormant origins were suppressed (Errico et al, 2009). In the same study, TIPIN was shown to associate with Ctf4/AND1 and the TIPIN–TIM1–AND1 complex was shown to be required for the stable loading and association of POLα to the DNA (Errico et al, 2009). The AND1–POLα interaction is conserved in yeast, X. laevis and human cells (Tsutsui et al, 2005; Zhou & Wang, 2004; Zhu et al, 2007), whereas the TIPIN–AND1 interaction has only been reported in X. laevis (Errico et al, 2009). A conserved mechanism might exist whereby Ctf4/AND1—and probably other replisome components such as Csm3/TIPIN—couple the helicase to POLα on the lagging strand template (Errico et al, 2009; Gambus et al, 2009; Tanaka et al, 2009). Work in yeast has demonstrated that Mrc1 interacts with the catalytic subunit of DNA Polɛ, the leading strand polymerase, suggesting that Mrc1 is instead involved in coupling polymerization and unwinding on the leading strand at the replication fork (Lou et al, 2008).
Overall, these observations indicate that TIPIN, TIM1 and CLASPIN are structural components of the replication fork, representing a physical and functional link between the MCM2–7 helicase and other replication factors, such as DNA polymerases (Fig 3), and ensuring the stability of the replisome, which is a prerequisite for resuming DNA replication after stalling. The presence of orthologues of these proteins with similar functions in yeast indicates that the overall process of fork stabilization is conserved between complex and simple eukaryotes. However, these mechanisms seem to be partly redundant in higher eukaryotes to ensure that the disruption of the function of one gene is not detrimental to the whole process. This is consistent with a greater level of redundancy in relation to critical biological processes in higher eukaryotes.
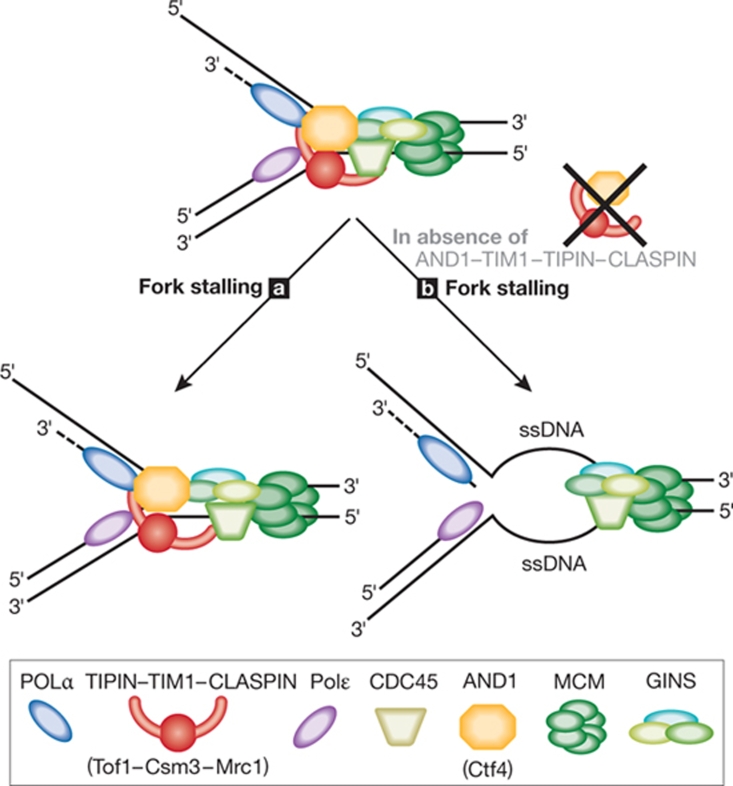
Figure 3.
The replication pausing complex. TIPIN–TIM1–AND1 might create a flexible bridge between replisome components such as CDC45, GINS, POLα and the MCM2–7 complex, which is required to stabilize POLα at replication forks. Mrc1 (CLASPIN) is also associated with TIPIN–TIM1 and is thought to couple Polɛ to the replisome. (a) When replication is halted, TIPIN–TIM1, AND1 and CLASPIN physically link the polymerase and helicase activities, preventing fork collapse. (b) The absence of these components could lead to excessive unwinding of DNA, thus destabilizing the replisome. AND1, acidic nucleoplasmic DNA-binding protein 1; CDC, cell division control; GINS, Go, Ichi, Ni and San complex; MCM, minichromosome maintenance protein; Mrc1, mediator of replication checkpoint 1; POLα/ɛ, polymerase-α/ɛ; TIM1, Timeless; TIPIN, Tim1-interacting protein.
Dealing with DSBs during DNA replication
DSBs are a significant threat to genome integrity and can be generated by genotoxic agents. However, the most common cause of these lesions in proliferating cells is aberrant DNA replication (Costanzo et al, 2001; Haber, 1998; Kuzminov, 2001). Eukaryotic cells repair DSBs through two main DNA repair pathways: homologous recombination (HR) and non-homologous end joining (NHEJ; Fig 4; Valerie & Povirk, 2003). HR uses an undamaged template—a sister chromatid or homologous chromosome—to restore chromosome integrity without any loss of genetic information. It occurs only during the S and G2 phases of the cell cycle, when sister chromatids are available, and relies on several proteins including Rad51/52/54/50, Mre11, Nbs1, RPA and Brca1/2 (Li & Heyer, 2008). By contrast, NHEJ is potentially mutagenic, as broken ends are processed and directly religated in the absence of homologous pairing (Lewis & Resnick, 2000; Lieber et al, 2003). The Mre11–Rad50–Nbs1 (MRN) complex has a crucial role in the DNA damage response and, together with ATM, is the primary sensor of DSBs. The MRN complex is also important in the initial steps of both HR and NHEJ (Mimitou & Symington, 2009).
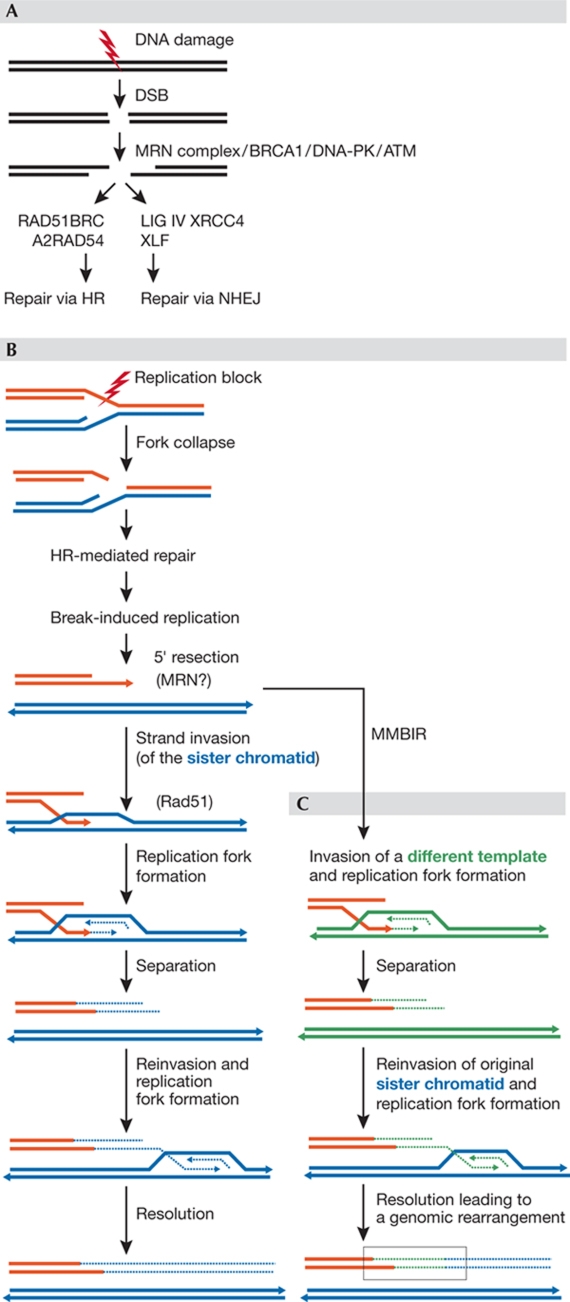
Figure 4.
Double-strand break repair pathways NHEJ, HR, BIR and MMBIR. (A) DSBs can be repaired by HR or NHEJ. (B) Replication forks collapse when they encounter a nick in the template, generating a DSB with only one end. The 5′ strand is then resected, which results in a 3′ overhang that can invade the sister molecule (blue) forming a D-loop, a process that is mediated by Rad51. The D-loop evolves in a replication fork with both leading and lagging strand synthesis. Whether BIR is resolved by cleavage of a Holliday junction or by helicase activity (separation step) is unknown. The separated end can dissociate and reinvade DNA templates, iterating the process until the replication of a chromosome segment is complete. (C) In MMBIR, microhomology-containing regions drive the strand invasion of non-sister templates, thereby leading to chromosomal rearrangements after a few rounds of invasion–replication–resolution. ATM, ataxia telangiectasia mutated; BIR, break-induced replication; DSB, double-strand break; HR, homologous recombination; MMBIR, microhomology-mediated BIR; NHEJ, non-homologous end-joining; Rad, radiation arrest deficient.
The relative contribution of NHEJ and HR to DSB repair varies substantially between budding yeast and mammalian cells. In S. cerevisiae, DSBs are repaired mainly through the HR pathway, whereas NHEJ has only a minor role (Aylon & Kupiec, 2004; Lee et al, 1999). By contrast, NHEJ seems to be of greater importance in mammalian cells (Critchlow & Jackson, 1998), as it is responsible for the repair of more than 60% of the exogenously induced DSBs in mouse embryonic stem cells (Liang et al, 1998). Surprisingly, mammalian cells that are deficient in NHEJ exhibit few spontaneous chromosome breaks and are viable, although this is the pathway that is preferentially used in vertebrates (Sonoda et al, 2006). This could be associated with the fact that NHEJ is dispensable and probably actively suppressed during S phase.
When sister chromatids are available, HR becomes the repair mechanism of choice for DSBs arising from collapsed replication forks. Intriguingly, despite the high degree of conservation of the single proteins, the requirement of each protein orthologue for cell survival and their contribution to the HR reaction differs profoundly across different species for many of the HR proteins. For example, yeast mutants lacking Rad51, Rad52 or Rad54 exhibit similar mild phenotypes unless they are challenged with DNA damaging agents. In marked contrast, the depletion of RAD51 results in cellular lethality in vertebrate cells, whereas mice carrying disrupted RAD52 or RAD54 genes do not show developmental abnormalities and are proficient in meiosis (Essers et al, 1997; Tan et al, 2003), suggesting that some HR proteins perform essential tasks during DNA replication in higher organisms. Similar to RAD51, the inactivation of other important HR genes such as MRE11, RAD50 and NBS1 is lethal in mice, indicating that the MRN complex also fulfils a task essential for cell survival (Luo et al, 1999; Xiao & Weaver, 1997; Zhu et al, 2001), probably owing to the role of MRN in preventing the accumulation of DSBs during normal DNA replication (Costanzo et al, 2001). Furthermore, the replacement of MRE11 with an allele that does not have nuclease activity induces the same phenotype as the complete knockout of MRE11 whereas mutations in the nuclease domain of Mre11 in S. cerevisiae have a limited effect, and Mre11-null cells are mostly viable (Bressan et al, 1998; Krogh et al, 2005; Lewis et al, 2004; Llorente & Symington, 2004). In summary, yeast mutants in many of the key HR proteins are viable, whereas the loss of the same proteins in higher eukaryotes results in cell or embryonic lethality.
The reasons behind this discrepancy are largely unclear. One possible explanation is the greater requirement for HR proteins—such as MRE11, RAD51 or BRCA2—for repairing and restarting stalled and collapsed forks in higher eukaryotes. The larger size of the genome might indeed lead to a higher percentage of stalled and collapsed forks in metazoans compared with yeast cells. For example, about 1 in 12 yeast cells lacking Rad52 gives rise to one dead and one living sister cell—as it would be expected if there were a DSB on one sister chromatid requiring repair (Jim Haber, personal communication). If this lesion frequency is scaled up to the vertebrate genome—which is 400 times larger—one would expect perhaps 30 lesions in the absence of RAD51, which would probably be sufficient to compromise the survival of a vertebrate cell. Therefore, these proteins are possibly just as necessary when measured in any defined region undergoing replication, and the same argument might apply to MRN proteins, which have so many different tasks. Other factors besides the genome size could contribute to this increased occurrence of corrupted forks, such as DNA sequence complexity, higher metabolic requirement, oxidative status or chromatin organization in higher organisms. Therefore, the replication machinery might rely more heavily on HR proteins to fix replication errors.
If HR is the main pathway to repair DSBs arising at replication forks, it should be noted that the collapse of a replication fork can generate a one-ended DSB (Fig 4), which is not the classical HR substrate. DSBs with only one free end are thought to be repaired by a sub-pathway of HR called break-induced replication (BIR; Poser et al, 2008; McEachern & Haber, 2006). The first step of this sub-pathway is similar to HR—the steps of which have been recently clarified (Mimitou & Symington, 2009)—in that the 5′ end of the broken arm is resected in a highly regulated fashion by a set of nucleases. In HR, this involves the sequential action of the Mre11 and Dna2 nucleases coupled to the Sgs1 helicase, which act redundantly with ExoI to produce a 3′ ssDNA filament (Mimitou & Symington, 2009) that is used to prime DNA synthesis on a new template (Fig 4). Once formed, this strand invades DNA templates in repeated attempts to find a suitable region of homology downstream or upstream from the point of fork collapse (Llorente et al, 2008; Smith et al, 2007). BIR could participate in replication fork recovery in yeast and, as such, it has been suggested to be the underlying mechanism of some chromosomal structural changes (Deem et al, 2008; Payen et al, 2008; Schmidt et al, 2006). However, the extent of BIR involvement in replication fork recovery in higher eukaryotes is unknown. It is tempting to speculate that BIR is a more important pathway to restart collapsed forks in higher eukaryotes, as it would be favoured by the presence of highly repetitive sequences that would facilitate homology-driven invasion. However, this remains to be established.
In principle, BIR is an accurate process that depends on recombination proteins and requires extensive homology for strand invasion. Nevertheless, it can lead to loss of heterozygosity and chromosomal rearrangements if the invading strand is paired with homologous allelic and non-allelic sequences (Deem et al, 2008; Payen et al, 2008; Smith et al, 2007). Indeed, BIR-based mechanisms can explain the complexity of the chromosomal structural changes that occur in cancer cells (Hastings et al, 2009; Lydeard et al, 2007; Smith et al, 2007). This is particularly relevant for a BIR-related pathway, microhomology-mediated BIR (MMBIR; Fig 4), that has been recently elucidated; this seems to be involved in the repair of one-ended DSBs that pair with stretches of non-related ssDNA molecules, which share microhomology with the invading 3′ ssDNA. MMBIR probably accounts for only a small fraction of DSB repair in yeast, whereas in mammalian cells it seems to be more efficient (Bentley et al, 2004). Genome-wide DNA sequencing studies of different cancer cell lines and primary tumours indicate that many rearrangements might derive from BIR and MMBIR-mediated events (Pleasance et al, 2009; Stephens et al, 2009).
Conclusions
The molecules and mechanisms that ensure a faithful DNA replication have been highly conserved throughout evolution. However, there are important differences between simple and complex organisms, just a few of which we have highlighted here. Among the differences that we have not considered there is an important class of genes, known as Fanconi anaemia (FA) proteins, that are involved in HR and DNA replication control in mammalian cells; except for a few members, FA proteins do not have clear homologues in unicellular eukaryotes.
A clear conclusion from this type of analysis is that the extrapolation from one organism to another cannot be considered universally reliable, although it has been extremely useful in studying DNA replication and its regulatory mechanisms. The challenge for the future is to understand the differences by taking advantage of more sophisticated approaches and innovative methods such as direct visualization of replication intermediates with advanced microscopy-based techniques. In addition, the further development of existing model systems that are capable of recapitulating DNA replication and repair will be useful for these studies (Sidebar A).
Sidebar A | In need of answers.
What consensus defines the metazoan origins of replication?
Are there Sld2 and Sld3 orthologues in higher eukaryotes? Do they represent the minimal set of CDK targets necessary for origin firing?
How many proteins that are important for cell cycle processes are present only in higher vertebrates?
Which DNA replication mechanisms have diverged and why?
Is HR required for DNA replication in metazoans? If yes, at which stage?
What is the impact of BIR and MMBIR on mammalian DNA replication?

Alessia Errico

Vincenzo Costanzo
Acknowledgments
We thank members of Clare Hall Laboratories and of the Genome Stability Unit for their comments. V. Costanzo is supported by Cancer Research UK, the Lister Institute of Preventive Medicine, the European Research Council start-up grant (206281) and the EMBO Young Investigator Programme.
References
- Aylon Y, Kupiec M (2004) DSB repair: the yeast paradigm. DNA Repair (Amst) 3: 797–815 [DOI] [PubMed] [Google Scholar]
- Bell SP, Stillman B (1992) ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature 357: 128–134 [DOI] [PubMed] [Google Scholar]
- Bentley J, Diggle CP, Harnden P, Knowles MA, Kiltie AE (2004) DNA double strand break repair in human bladder cancer is error prone and involves microhomology-associated end-joining. Nucleic Acids Res 32: 5249–5259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow JJ, Gillespie PJ, Francis D, Jackson DA (2001) Replication origins in Xenopus egg extract are 5–15 kilobases apart and are activated in clusters that fire at different times. J Cell Biol 152: 15–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D, Foiani M (2005) The DNA damage response during DNA replication. Curr Opin Cell Biol 17: 568–575 [DOI] [PubMed] [Google Scholar]
- Bressan DA, Olivares HA, Nelms BE, Petrini JH (1998) Alteration of N-terminal phosphoesterase signature motifs inactivates Saccharomyces cerevisiae Mre11. Genetics 150: 591–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA (2005) Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev 19: 1040–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadoret JC, Meisch F, Hassan-Zadeh V, Luyten I, Guillet C, Duret L, Quesneville H, Prioleau MN (2008) Genome-wide studies highlight indirect links between human replication origins and gene regulation. Proc Natl Acad Sci USA 105: 15837–15842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou DM, Elledge SJ (2006) Tipin and Timeless form a mutually protective complex required for genotoxic stress resistance and checkpoint function. Proc Natl Acad Sci USA 103: 18143–18147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D, Glick G, Elledge SJ (2004) Minichromosome maintenance proteins are direct targets of the ATM and ATR checkpoint kinases. Proc Natl Acad Sci USA 101: 10078–10083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo V, Robertson K, Bibikova M, Kim E, Grieco D, Gottesman M, Carroll D, Gautier J (2001) Mre11 protein complex prevents double-strand break accumulation during chromosomal DNA replication. Mol Cell 8: 137–147 [DOI] [PubMed] [Google Scholar]
- Critchlow SE, Jackson SP (1998) DNA end-joining: from yeast to man. Trends Biochem Sci 23: 394–398 [DOI] [PubMed] [Google Scholar]
- Dai J, Chuang RY, Kelly TJ (2005) DNA replication origins in the Schizosaccharomyces pombe genome. Proc Natl Acad Sci USA 102: 337–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deem A, Barker K, Vanhulle K, Downing B, Vayl A, Malkova A (2008) Defective break-induced replication leads to half-crossovers in Saccharomyces cerevisiae. Genetics 179: 1845–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley JF, Cocker JH, Dowell SJ, Harwood J, Rowley A (1995) Stepwise assembly of initiation complexes at budding yeast replication origins during the cell cycle. J Cell Sci Suppl 19: 67–72 [DOI] [PubMed] [Google Scholar]
- Dimitrova DS, Giacca M, Demarchi F, Biamonti G, Riva S, Falaschi A (1996) In vivo protein–DNA interactions at human DNA replication origin. Proc Natl Acad Sci USA 93: 1498–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doksani Y, Bermejo R, Fiorani S, Haber JE, Foiani M (2009) Replicon dynamics, dormant origin firing, and terminal fork integrity after double-strand break formation. Cell 137: 247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errico A, Costanzo V, Hunt T (2007) Tipin is required for stalled replication forks to resume DNA replication after removal of aphidicolin in Xenopus egg extracts. Proc Natl Acad Sci USA 104: 14929–14934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errico A, Cosentino C, Rivera T, Schwob E, Losada A, Hunt T, Costanzo V (2009) Tipin/Tim1/And1 a new protein complex required for DNA Polα association on the chromatin and establishment of sister chromatids cohesion. EMBO J 28: 3681–3692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers J, Hendriks RW, Swagemakers SM, Troelstra C, de Wit J, Bootsma D, Hoeijmakers JH, Kanaar R (1997) Disruption of mouse RAD54 reduces ionizing radiation resistance and homologous recombination. Cell 89: 195–204 [DOI] [PubMed] [Google Scholar]
- Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, Edmondson RD, Labib K (2006) GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol 8: 358–366 [DOI] [PubMed] [Google Scholar]
- Gambus A, van Deursen F, Polychronopoulos D, Foltman M, Jones RC, Edmondson RD, Calzada A, Labib K (2009) A key role for Ctf4 in coupling the MCM2–7 helicase to DNA polymerase alpha within the eukaryotic replisome. EMBO J 28: 2992–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia V, Furuya K, Carr AM (2005) Identification and functional analysis of TopBP1 and its homologs. DNA Repair (Amst) 4: 1227–1239 [DOI] [PubMed] [Google Scholar]
- Ge XQ, Jackson DA, Blow JJ (2007) Dormant origins licensed by excess Mcm2–7 are required for human cells to survive replicative stress. Genes Dev 21: 3331–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie PJ, Li A, Blow JJ (2001) Reconstitution of licensed replication origins on Xenopus sperm nuclei using purified proteins. BMC Biochem 2: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber JE (1998) The many interfaces of Mre11. Cell 95: 583–586 [DOI] [PubMed] [Google Scholar]
- Hastings PJ, Ira G, Lupski JR (2009) A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet 5: e1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson B, Calzada A, Labib K (2007) Mrc1 and Tof1 regulate DNA replication forks in different ways during normal S phase. Mol Biol Cell 18: 3894–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrien O, Mechali M (1993) Chromosomal replication initiates and terminates at random sequences but at regular intervals in the ribosomal DNA of Xenopus early embryos. EMBO J 12: 4511–4520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrien O, Maric C, Mechali M (1995) Transition in specification of embryonic metazoan DNA replication origins. Science 270: 994–997 [DOI] [PubMed] [Google Scholar]
- Ibarra A, Schwob E, Mendez J (2008) Excess MCM proteins protect human cells from replicative stress by licensing backup origins of replication. Proc Natl Acad Sci USA 105: 8956–8961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im JS, Ki SH, Farina A, Jung DS, Hurwitz J, Lee JK (2009) Assembly of the Cdc45–Mcm2–7–GINS complex in human cells requires the Ctf4/And-1, RecQL4, and Mcm10 proteins. Proc Natl Acad Sci USA 106: 15628–15632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnani N, Taylor CM, Malhotra A, Dutta A (2009) Genomic study of replication initiation in human chromosomes reveals the influence of transcription regulation and chromatin structure on origin selection. Mol Biol Cell [Epub 2 Dec 2009] doi:10.1091/mbc.E09-08-0707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katou Y, Kanoh Y, Bando M, Noguchi H, Tanaka H, Ashikari T, Sugimoto K, Shirahige K (2003) S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424: 1078–1083 [DOI] [PubMed] [Google Scholar]
- Krogh BO, Llorente B, Lam A, Symington LS (2005) Mutations in Mre11 phosphoesterase motif I that impair Saccharomyces cerevisiae Mre11–Rad50–Xrs2 complex stability in addition to nuclease activity. Genetics 171: 1561–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzminov A (2001) DNA replication meets genetic exchange: chromosomal damage and its repair by homologous recombination. Proc Natl Acad Sci USA 98: 8461–8468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SE, Paques F, Sylvan J, Haber JE (1999) Role of yeast SIR genes and mating type in directing DNA double-strand breaks to homologous and non-homologous repair paths. Curr Biol 9: 767–770 [DOI] [PubMed] [Google Scholar]
- Lei M, Kawasaki Y, Tye BK (1996) Physical interactions among Mcm proteins and effects of Mcm dosage on DNA replication in Saccharomyces cerevisiae. Mol Cell Biol 16: 5081–5090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis LK, Resnick MA (2000) Tying up loose ends: nonhomologous end-joining in Saccharomyces cerevisiae. Mutat Res 451: 71–89 [DOI] [PubMed] [Google Scholar]
- Lewis LK, Storici F, Van Komen S, Calero S, Sung P, Resnick MA (2004) Role of the nuclease activity of Saccharomyces cerevisiae Mre11 in repair of DNA double-strand breaks in mitotic cells. Genetics 166: 1701–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Heyer WD (2008) Homologous recombination in DNA repair and DNA damage tolerance. Cell Res 18: 99–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang F, Han M, Romanienko PJ, Jasin M (1998) Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc Natl Acad Sci USA 95: 5172–5177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber MR, Ma Y, Pannicke U, Schwarz K (2003) Mechanism and regulation of human non-homologous DNA end-joining. Nat Rev Mol Cell Biol 4: 712–720 [DOI] [PubMed] [Google Scholar]
- Llorente B, Symington LS (2004) The Mre11 nuclease is not required for 5′ to 3′ resection at multiple HO-induced double-strand breaks. Mol Cell Biol 24: 9682–9694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente B, Smith CE, Symington LS (2008) Break-induced replication: what is it and what is it for? Cell Cycle 7: 859–864 [DOI] [PubMed] [Google Scholar]
- Lou H, Komata M, Katou Y, Guan Z, Reis CC, Budd M, Shirahige K, Campbell JL (2008) Mrc1 and DNA polymerase epsilon function together in linking DNA replication and the S phase checkpoint. Mol Cell 32: 106–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G, Yao MS, Bender CF, Mills M, Bladl AR, Bradley A, Petrini JH (1999) Disruption of mRad50 causes embryonic stem cell lethality, abnormal embryonic development, and sensitivity to ionizing radiation. Proc Natl Acad Sci USA 96: 7376–7381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutzmann M, Mechali M (2008) MCM9 binds Cdt1 and is required for the assembly of prereplication complexes. Mol Cell 31: 190–200 [DOI] [PubMed] [Google Scholar]
- Lydeard JR, Jain S, Yamaguchi M, Haber JE (2007) Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature 448: 820–823 [DOI] [PubMed] [Google Scholar]
- Maiorano D, Cuvier O, Danis E, Mechali M (2005) MCM8 is an MCM2–7-related protein that functions as a DNA helicase during replication elongation and not initiation. Cell 120: 315–328 [DOI] [PubMed] [Google Scholar]
- McEachern MJ, Haber JE (2006) Break-induced replication and recombinational telomere elongation in yeast. Annu Rev Biochem 75: 111–135 [DOI] [PubMed] [Google Scholar]
- McGarry TJ, Kirschner MW (1998) Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell 93: 1043–1053 [DOI] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS (2009) DNA end resection: many nucleases make light work. DNA Repair (Amst) 8: 983–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payen C, Koszul R, Dujon B, Fischer G (2008) Segmental duplications arise from Pol32-dependent repair of broken forks through two alternative replication-based mechanisms. PLoS Genet 4: e1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance ED et al. (2010) A comprehensive catalogue of somatic mutations from a human cancer genome. Nature 463:191–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poser I et al. (2008) BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nat Methods 5: 409–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabitsch KP et al. (2001) A screen for genes required for meiosis and spore formation based on whole-genome expression. Curr Biol 11: 1001–1009 [DOI] [PubMed] [Google Scholar]
- Rowles A, Tada S, Blow JJ (1999) Changes in association of the Xenopus origin recognition complex with chromatin on licensing of replication origins. J Cell Sci 112: 2011–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangrithi MN, Bernal JA, Madine M, Philpott A, Lee J, Dunphy WG, Venkitaraman AR (2005) Initiation of DNA replication requires the RECQL4 protein mutated in Rothmund–Thomson syndrome. Cell 121: 887–898 [DOI] [PubMed] [Google Scholar]
- Sasaki T, Sawado T, Yamaguchi M, Shinomiya T (1999) Specification of regions of DNA replication initiation during embryogenesis in the 65-kilobase DNApolα-dE2F locus of Drosophila melanogaster. Mol Cell Biol 19: 547–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt KH, Wu J, Kolodner RD (2006) Control of translocations between highly diverged genes by Sgs1, the Saccharomyces cerevisiae homolog of the Bloom's syndrome protein. Mol Cell Biol 26: 5406–5420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab RA, Blackford AN, Niedzwiedz W (2010) ATR activation and replication fork restart are defective in FANCM-deficient cells. EMBO J [Epub 7 Jan 2010] doi:10.1038/emboj.2009.385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segurado M, de Luis A, Antequera F (2003) Genome-wide distribution of DNA replication origins at A+T-rich islands in Schizosaccharomyces pombe. EMBO Rep 4: 1048–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo J, Chung YS, Sharma GG, Moon E, Burack WR, Pandita TK, Choi K (2005) Cdt1 transgenic mice develop lymphoblastic lymphoma in the absence of p53. Oncogene 24: 8176–8186 [DOI] [PubMed] [Google Scholar]
- Shechter D, Costanzo V, Gautier J (2004) ATR and ATM regulate the timing of DNA replication origin firing. Nat Cell Biol 6: 648–655 [DOI] [PubMed] [Google Scholar]
- Shinomiya T, Ina S (1991) Analysis of chromosomal replicons in early embryos of Drosophila melanogaster by two-dimensional gel electrophoresis. Nucleic Acids Res 19: 3935–3941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CE, Llorente B, Symington LS (2007) Template switching during break-induced replication. Nature 447: 102–105 [DOI] [PubMed] [Google Scholar]
- Sonoda E, Hochegger H, Saberi A, Taniguchi Y, Takeda S (2006) Differential usage of non-homologous end-joining and homologous recombination in double strand break repair. DNA Repair (Amst) 5: 1021–1029 [DOI] [PubMed] [Google Scholar]
- Stephens PJ et al. (2009) Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature 462: 1005–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan TL, Kanaar R, Wyman C (2003) Rad54, a jack of all trades in homologous recombination. DNA Repair (Amst) 2: 787–794 [DOI] [PubMed] [Google Scholar]
- Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, Araki H (2007) CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature 445: 328–332 [DOI] [PubMed] [Google Scholar]
- Tanaka H, Kubota Y, Tsujimura T, Kumano M, Masai H, Takisawa H (2009) Replisome progression complex links DNA replication to sister chromatid cohesion in Xenopus egg extracts. Genes Cells 14: 949–963 [DOI] [PubMed] [Google Scholar]
- Tourriere H, Versini G, Cordon-Preciado V, Alabert C, Pasero P (2005) Mrc1 and Tof1 promote replication fork progression and recovery independently of Rad53. Mol Cell 19: 699–706 [DOI] [PubMed] [Google Scholar]
- Trenz K, Errico A, Costanzo V (2008) Plx1 is required for chromosomal DNA replication under stressful conditions. EMBO J 27: 876–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui Y, Morishita T, Natsume T, Yamashita K, Iwasaki H, Yamao F, Shinagawa H (2005) Genetic and physical interactions between Schizosaccharomyces pombe Mcl1 and Rad2, Dna2 and DNA polymerase alpha: evidence for a multifunctional role of Mcl1 in DNA replication and repair. Curr Genet 48: 34–43 [DOI] [PubMed] [Google Scholar]
- Unsal-Kacmaz K, Chastain PD, Qu PP, Minoo P, Cordeiro-Stone M, Sancar A, Kaufmann WK (2007) The human Tim/Tipin complex coordinates an intra-S checkpoint response to UV that slows replication fork displacement. Mol Cell Biol 27: 3131–3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valerie K, Povirk LF (2003) Regulation and mechanisms of mammalian double-strand break repair. Oncogene 22: 5792–5812 [DOI] [PubMed] [Google Scholar]
- Walter J, Newport JW (1997) Regulation of replicon size in Xenopus egg extracts. Science 275: 993–995 [DOI] [PubMed] [Google Scholar]
- Wohlschlegel JA, Dwyer BT, Dhar SK, Cvetic C, Walter JC, Dutta A (2000) Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science 290: 2309–2312 [DOI] [PubMed] [Google Scholar]
- Woodward AM, Gohler T, Luciani MG, Oehlmann M, Ge X, Gartner A, Jackson DA, Blow JJ (2006) Excess Mcm2–7 license dormant origins of replication that can be used under conditions of replicative stress. J Cell Biol 173: 673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Weaver DT (1997) Conditional gene targeted deletion by Cre recombinase demonstrates the requirement for the double-strand break repair Mre11 protein in murine embryonic stem cells. Nucleic Acids Res 25: 2985–2991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo HY, Kumagai A, Shevchenko A, Shevchenko A, Dunphy WG (2004) Adaptation of a DNA replication checkpoint response depends upon inactivation of Claspin by the Polo-like kinase. Cell 117: 575–588 [DOI] [PubMed] [Google Scholar]
- Zegerman P, Diffley JF (2007) Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature 445: 281–285 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Wang TS (2004) A coordinated temporal interplay of nucleosome reorganization factor, sister chromatin cohesion factor, and DNA polymerase alpha facilitates DNA replication. Mol Cell Biol 24: 9568–9579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Petersen S, Tessarollo L, Nussenzweig A (2001) Targeted disruption of the Nijmegen breakage syndrome gene NBS1 leads to early embryonic lethality in mice. Curr Biol 11: 105–109 [DOI] [PubMed] [Google Scholar]
- Zhu W, Ukomadu C, Jha S, Senga T, Dhar SK, Wohlschlegel JA, Nutt LK, Kornbluth S, Dutta A (2007) Mcm10 and And-1/CTF4 recruit DNA polymerase alpha to chromatin for initiation of DNA replication. Genes Dev 21: 2288–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]