Abstract
In mammals, sex differences are evident in many aspects of brain development, brain function and behaviour. Ultimately, such differences must arise from the differential sex chromosome complements in males and females: males inherit a single X chromosome and a Y chromosome, whilst females inherit two X chromosomes. One possible mechanism for sexual differentiation of the brain is via male-limited expression of genes on the small Y chromosome. Many Y-linked genes have been implicated in the development of the testes, and therefore could theoretically contribute to sexual differentiation of the brain indirectly, through influencing gonadal hormone production. Alternatively, Y-linked genes that are expressed in the brain could directly influence neural masculinisation. The present paper reviews evidence from human genetic studies and animal models for Y-linked effects (both direct and indirect) on neurodevelopment, brain function and behaviour. Besides enhancing our knowledge of the mechanisms underlying mammalian neural sexual differentiation, studies geared towards understanding the role of the Y chromosome in brain function will help to elucidate the molecular basis of sex-biased neuropsychiatric disorders, allowing for more selective sex-specific therapies.
INTRODUCTION
There is a substantial body of evidence showing that female and male mammals (including humans) differ with respect to many aspects of their physiology and behaviour. Neuropsychological studies in man (to use the term in its loosest sense) have demonstrated that males tend to outperform females on behavioural tasks tapping visuospatial and navigational skills, whereas females tend to score more highly on tasks assaying verbal and social proficiency [1]. Females also tend to exhibit superior performance in assays of object location memory [2,3], emotion recognition [4,5] and empathy [6], whilst males are more likely to exhibit higher levels of aggression and behavioural disinhibition [1,7]. A combination of early post mortem studies and more recent in vivo neuroimaging work has identified a number of brain structures whose anatomy is sex-specific, and whose different function in males and females may underlie the behavioural effects described above [8]; these include the amygdala (larger in males), the hippocampus (larger in females), the corpus callosum (larger in females, but see [9]) and regions of the cerebral cortex. With respect to the latter structure, women tend to have larger paralimbic and fronto-orbital areas, whereas men generally have a larger fronto-medial cortex [10]. Sexual dimorphisms have also been reported in the hypothalamus, an area implicated in sexual preference and numerous sex-related behaviours. Simon Le Vay’s work in particular has demonstrated that the interstitial nuclei of the anterior hypothalamus region 3 is approximately three times larger in heterosexual men than in homosexual men and heterosexual women [11]. Whilst many brain and behavioural findings regarding sex differences in humans are robust, and have been recapitulated in animal models [12-14], the magnitude, functional relevance, and even the veracity of others has been a matter for some debate [15]. Inconsistent findings between studies may be due to i) small effect size, ii) failure to take into account stage of the female menstrual cycle, iii) poorly specified phenotypes of interest, and iv) more general phenomena such as differential matching for IQ and social status between the sexes. The advent of more refined behavioural/neuropsychological testing procedures, and more sophisticated imaging techniques will improve the identification and characterisation of sexually dimorphic brain and behavioural substrates.
Sexual differentiation, the process by which females and males acquire their distinctive physiologies, is a downstream consequence of the fact that the sexes possess different chromosomal complements [16]. In mammals, males possess a single X chromosome (invariably inherited from their mother) and a single Y chromosome (inherited from their father), and, as such, have the karyotype 46,XY. Females, in contrast, inherit two X chromosomes, one from either parent (karyotype 46,XX). There are therefore three possible mechanisms through which sex-linked gene expression could contribute to sexual dimorphism in brain and behaviour (reviewed in [17]). Firstly, as females possess two X chromosomes, as opposed to the male’s one, genes which escape the process of X-inactivation will be expressed approximately twice as highly in female than male tissues; the most contemporary estimates suggest that ~20% of all X-linked genes on the human X chromosome may escape X-inactivation to some extent [18,19]. Secondly, as the two sexes differ with respect to the parental origins of their X chromosomes, any so-called imprinted genes on this chromosome may exhibit sexually dimorphic expression; the degree to which an X-linked imprinted gene exhibits sexually dimorphic expression will depend upon whether it is preferentially expressed from the paternally or maternally inherited allele and whether or not it is subject to X-inactivation [20]. The third possible genetic mechanism through which sexual differentiation of the brain may occur, and the one upon which the remainder of this review will focus, is via the male-limited expression of genes in the non-recombining region (NRY) of the Y chromosome (i.e. those genes that are Y-unique) [17]. Our discussion shall consider: i) the structure and genetic complement of the Y chromosome, ii) how the genes upon it may shape male brain function and behaviour through indirect or direct means and how we may dissociate between the two possibilities, and iii) the role of the Y chromosome in engendering vulnerability to male-biased neuropsychiatric disorders.
THE Y CHROMOSOME
Despite its profound effects on sexual differentiation (see later), the brain effects mediated by the Y chromosome have not attracted the same amount of interest as those mediated by its larger and cognition-gene rich counterpart, the X; this may be because of the former’s small size and low gene content [21]. In humans the X chromosome is ~155Mb in size and houses ~1500 genes, whereas the Y chromosome is just ~60Mb in size and houses ~350 genes, many of which are pseudogenes. Alternatively, it may be because, unlike the X chromosome, it is not subject to the intrinsically interesting epigenetic process of silencing, or because its repetitive structure means that it is not readily amenable to genetic and genomic studies.
THE HUMAN Y CHROMOSOME
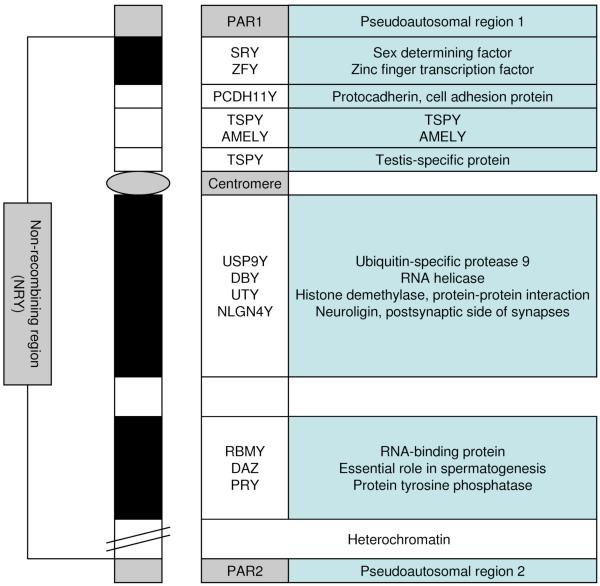
The human Y chromosome, like the autosomes and the X chromosome, consists of a short (Yp) and a long (Yq) arm (~11.5Mb and ~48.5Mb respectively) separated by a centromere (Fig. 1). At either end of the chromosome are regions which can recombine during meiosis with their equivalents on the X chromosome; as this recombinatory behaviour is reminiscent of that of the autosomes, these terminal domains are designated pseudoautosomal regions (or PARs). PAR1 is located on Yp, and contains ~10 genes. PAR2 is located on Yq and contains ~15 genes. Together, the PARs comprise ~5% of the basepair content of the chromosome. The remaining 95% of the chromosome constitutes the non-recombining region (NRY) referred to earlier (also known as the male-specific region or MSY). Currently, 156 transcription units (distinct regions of DNA which are transcribed into RNA) have been found on human NRY, 78 of which are protein-coding (27 distinct proteins or protein families) [22]. Genes on NRY fall into two categories: those that are expressed throughout the body (including the brain) and those that are expressed mainly, or exclusively, in testes and are likely to affect testis development and/or spermatogenesis. Microarray analysis comparing gene expression in post mortem male and female brains has suggested that ~20% of NRY-linked genes are expressed in this tissue, though this figure is probably an underestimate given sensitivity issues and the possibility of cross-hybridisation with closely related X-linked sequences [23]. The human NRY comprises three different types of euchromatic sequences (i.e. those which are likely to be transcribed)[22]: i) X-transposed sequences which have ~99% homology to equivalent regions on the X chromosome (although importantly they do not recombine) and are characterised by low gene and high repeat density ii) ampliconic sequences which exhibit a high degree of similarity (99.9%) to other NRY sequences and include genes mainly expressed in testes; these regions are usually large allowing for gene conversion, a phenomenon of non-reciprocal recombination between Y chromosome sequences that has resulted in eight large palindromes in the ampliconic sequences iii) X-degenerate sequences, which include single-copy genes and pseudogenes with an X chromosome homologue. X-degenerate genes are generally ubiquitously expressed - indeed, it is noteworthy that no ubiquitously expressed gene has yet been found amongst the other two types of sequence.
Fig. (1).
Schematic of the human Y chromosome showing key structural features and genes.
As the NRY does not recombine, it is generally passed unchanged from father to son with both sharing the same Y-linked polymorphism profile (or ‘haplotype’). As Y-linked haplotypes tend to vary across geographical regions, and even between neighbouring countries due to the differential fixation of de novo mutations, haplotype analyses have been important in addressing questions regarding human origins and patterns of migration [24]. The utility of haplotype analyses to neurobiological studies, and the caveats associated with their use are discussed later.
THE Y CHROMOSOME IN RODENT MODELS
The Y chromosome of the most genetically amenable mammal, the mouse, is around the same size as its human equivalent (50-60Mb). As in man, it comprises two arms separated by a centromeric region, a minute Xp, and a long Xq. However, unlike in man, there is just one PAR, located at the Xq telomere [25]. The majority of the mouse Y chromosome has recently been sequenced [26]. This analysis has shown that the X-degenerate portion of the mouse Y is even more degenerate than that of the human Y, whilst the ampliconic portion of the mouse Y has expanded massively to cover ~95% of the chromosome with highly repetitive sequence. Many of the genes on the mouse Y chromosome have human orthologues and a large proportion influence testis development (e.g. Sry) [27,28] and/or spermatogenesis (e.g. Eif2s3y) [29]; consistent with the latter, the murine Y chromosome is highly condensed in somatic cells, becoming transcriptionally active only in germ cells [30]. Systematic deletion of Y-linked genes or gene families in mouse mutants has begun to unravel which genes are responsible for particular aspects of germ cell development [31]. As the sequencing of the Y chromosome of a second rodent, the rat, is ongoing it would be premature to draw any general conclusions about its structure. However, comparative studies of the rat and mouse Y chromosomes have noted the presence of species-specific repetitive sequences [32]. Moreover, it has been shown that TSPY, whose product is involved in spermatogenesis, is a functional gene in both rats and man, but is pseudogenic in the mouse [33].
SRY: The Sex-Determining Gene
Early work by Jacobs and Strong (amongst others) showed that, in mammals, in contrast to nematode worms and fruit flies, it was the inheritance of the Y chromosome (even in the presence of multiple X chromosomes) that initiated testis development and conferred male-typical physiological and behavioural traits [34]. Consequently, substantial effort was invested in trying to identify the locus responsible for the so-called testis-determining factor (TDF) within the critical sex-determining region of the Y. In 1990, a seminal Nature paper by Sinclair and colleagues reported a novel gene candidate for the TDF in man; this gene, located at Yp11.3, encoded a protein with a DNA-binding motif present in high motility group (HMG) proteins, and was designated SRY (for Sex-determining Region on the Y) [35]. The fact that this gene was deleted in 40,XY mice who appeared female provided converging evidence for its role as the crucial determinant of maleness [36]. Interestingly, SRY is the sole X-degenerate gene which is not ubiquitously expressed (it is predominantly expressed in testis). The precise molecular mechanisms through which SRY confers a male phenotype are beyond the scope of this review, and have been considered in detail elsewhere [37]. Briefly, data from mice suggests that during an expression period from embryonic day 10.5-12.5, Sry interacts with SF1 synergistically to upregulate Sox9 gene expression, ultimately leading to Sertoli cell differentiation and subsequent testes development [38]. Once Sry expression declines, Sox9 levels are maintained at a high level, inducing, in combination with a collection of other proteins (notably WT1, SF-1 and GATA4), the expression of Anti-Mullerian hormone, the function of which is to prevent the development of the mullerian ducts into the uterus. From approximately embryonic day 15 in rodents, Leydig cells in the developing testes synthesize testosterone, which is essential for the development of male genitalia [39]. In mammals, testosterone can affect masculinisation of the brain substantially in critical pre- and peri-natal periods (organisational effects) and in later postnatal life (activational effects). Testosterone exerts its effects either through its metabolite, estradiol, acting at estrogen receptors or directly by acting on androgen receptors. Conversion of testosterone to estradiol takes place locally in the brain through aromatisation by the P450 enzyme [40]. In rodents, brain areas that are sensitive to the influence of testosterone/estradiol include the medial amygdala, the medial preoptic area, the paraventricular nucleus, and the anteroventral periventricular nucleus of the hypothalamus [41,42]. In humans, masculinisation of the brain is thought to be caused primarily by the direct effects of testosterone on androgen receptors, and not through its aromatisation to estradiol [43]. Testosterone has been shown to influence a wide range of cognitive functions, notably visuospatial ability, object recognition and working memory, in both humans and rodents. Administering high levels of testosterone to female rats in utero improved their subsequent visuospatial performance [44], whilst performance on various behavioural tasks assaying visuospatial and working memory and object recognition has been shown to be impaired in gonadectomised male rats [45-48]. In humans, males with a higher level of testosterone have been reported to have superior visuospatial abilities [49]. Importantly, therefore, in mammals, SRY (and indeed any other Y-linked gene which affects testis development and subsequent gonadal hormone secretion) could influence the development of male-specific neurobiology by indirect means. In marsupials like the tammar wallaby many somatic sex differences are evident prior to testicular formation, implying that in this group, there is a greater degree of hormonal independence in sexual differentiation than in mammals (and, therefore a greater reliance on direct sex-linked gene effects)[50].
SRY in the Brain
Although SRY is chiefly expressed in the testes, it is also expressed to some extent in other tissues including heart, liver and kidney, and certain brain regions [51,52]. Hence, besides influencing male-specific traits indirectly, theoretically it could influence neurodevelopment and brain function in a direct cell-autonomous manner. In humans, SRY expression has been described in the medial rostral hypothalamus, frontal and temporal cortex [52]. In adult rodents, the gene is expressed in the hypothalamus and midbrain (notably the substantia nigra and the ventral tegmental area) [53,54]. In mice at least, the gene is expressed in two different forms depending upon developmental stage: during embryogenesis, circular (and untranslated) transcripts are produced, whilst linear (and translatable) transcripts are produced thereafter [55]. The transition between expressed forms is presumably due to a developmental switch in promoter usage, but the evolutionary and functional reasons behind it remain to be investigated. In mice and humans, Sry is a single copy gene; in contrast, in rats and certain other rodent species multiple Y-linked copies of the Sry gene have been reported [56-58]. Six Sry copies have been identified in the NRY of Rattus norvegicus: Sry1, Sry2, Sry3, Sry3B, Sry3B1, and Sry3C; all six copies have a conserved opening reading frame and the difference between them is less than 2% [56]. Our initial gene expression analyses have shown that in rat brain Sry2 is the predominant transcript (>90% of transcripts), whilst Sry1 may be expressed at low levels in some strains (unpublished observations). These findings mirror the results from other rat tissues: in testis, Sry2 constitutes ~50% of all Sry transcripts, whilst in the adrenal glands the Sry transcript pool is almost exclusively of the Sry2 variety [56].
The expression of Sry in brain and adrenal tissue rich in catecholaminergic cells led Milsted and colleagues to test whether the protein might be influencing the expression of genes important in catecholamine biosynthesis. Their in vitro assays showed that Sry appeared to bind at the promoter region of the gene encoding tyrosine hydroxylase (the rate-limiting enzyme in dopamine biosythesis) to enhance its transcription [59]. These data suggest that one way in which Sry acts to confer maleness is through affecting development of the dopaminergic system; they further imply that SRY (dys)function may contribute towards the male bias in certain conditions with a known catecholaminergic basis e.g. ADHD, addiction and hypertension [60-62]. Future work might be directed towards identifying other key transcriptional targets for Sry in the brain by using chromatin immunoprecipitation for example.
DOWNSTREAM BRAIN AND BEHAVIOURAL FUNCTIONS OF SRY
As mentioned above, Sry may influence brain and behaviour either indirectly, via effects on testis development and subsequent hormone secretion, or directly via its expression in neural tissue. The downstream effects of Sry on brain function and behaviour may be most readily addressed using rodents, and in this section, we describe two model systems which have recently been used to some effect.
i) 4-Core Genotype Cross
One mouse model which may be used to identify Sry-dependent and Sry-independent sexually dimorphic phenotypes is the so-called ‘4-core genotype cross’ (comprehensively reviewed by Arnold and Chen [63]). Sry-dependent phenotypes may be due to indirect effects of the protein (via effects on testis differentiation and function) or to direct effects on brain function. Sry-independent phenotypes are due to the influence of sex-linked genes other than Sry. The model makes use of a variant of the Y129 chromosome deleted for Sry (Tdym1 mutation [64]), designated Y−, and an autosomally located ubiquitously expressed Sry transgene. A cross between a 40,XY−Sry male (i.e. a mouse with the deletion of the Y-linked Sry gene, and an autosomal transgene) and a 40,XX female generates ‘four core genotypes’: 40,XX (chromosomally and gonadally female), 40,XXSry (chromosomally female, but gonadally male due to the presence of the autosomal Sry transgene), 40,XY− (chromosomally male, and gonadally female due to loss of endogenous Sry expression), and finally, 40,XY−Sry (chromosomally and gonadally male). The key point about the model is that through the use of pre-planned comparisons, Sry-dependent and independent processes may be dissociated. Specifically, a difference in a particular brain or behavioural parameter between 40,XX and 40,XXSry mice (or between 40,XY− and 40,XY− mice) would imply an effect which was dependent upon the presence of the Sry transgene. In contrast, a difference between 40,XX and 40,XY− mice (or between 40,XXSry and 40,XY−Sry mice) who are matched for gonadal sex but not for sex chromosome complement, would imply an Sry-independent sex-linked gene effect; the precise mechanism underlying this effect could then be elucidated using alternative models such as the 39,XO mouse [65].
Documented Sry-dependent brain and behavioural phenomena include progesterone receptor expression in the anteroventral periventricular nucleus, the medial preoptic nucleus, and the ventromedial nucleus [66], cortical thickness [67] and various sexual behaviours including degree of mounting, latency to mount and latency to thrust [68]. A dosage-dependent Sry effect on sexual behaviours is consistent with the observation that male mice with an additional Y chromosome (41,XYY) have shorter latencies to intromit, and to ejaculate than their 40,XY counterparts [69]. Our own ongoing work has shown that the expression of an Sry transgene may shape cognition, in that 40,XXSry and 40,XY−Sry mice made fewer errors in learning a maze-based task than animals without the transgene (unpublished results). Given that in mammals, the vast majority of known effects of the Y chromosome, are hormonally-mediated, most Sry-dependent effects, including those described above, are likely to be mediated via effects on testis differentiation and subsequent hormone (notably testosterone) release. There is a substantial literature describing the important and wide-ranging organisational and activational effects of testosterone on sexual differentiation of the brain [70].
Direct sex-linked gene effects (i.e. Sry-independent effects) have, to date, been described upon a multitude of measures including: autoimmune disease vulnerability [71], neural tube defects [72], nociception [73], habit formation [62], differentiation and development of mesencephalic dopaminergic neurons [74] and vasopressin immunoreactivity in the lateral septum [68,75].
ii) Knockdown of Sry Expression
To address the difficulty inherent in the 4-core genotype model of ascribing Sry-dependent effects to direct or indirect (hormonally-mediated) influences, one may turn to another model system. Specifically, the effects of artificially downregulating Sry expression at key brain sites in order to ascertain the effects on previously identified Sry-dependent phenotypes could be examined. If Sry downregulation has no effect on the phenotype of interest, it may be concluded that the phenotype is likely to be gonadal hormone-dependent; conversely, if the downregulation is effective, one might conclude that the phenotype is dependent upon the action of brain-expressed Sry. Recently, Dewing et al. used antisense oligonucleotides to knock down Sry expression in the rat substantia nigra [53]. This manipulation led to a decrease in tyrosine hydroxylase expression in this brain region (consistent with the previous data suggesting Sry as a transcriptional activator) and motor deficits. As the authors pointed out these results demonstrated ‘a direct male-specific effect on the brain by a gene encoded only in the male genome’. Furthermore, they speculated that the results may be of relevance to the molecular pathogenesis of Parkinson’s Disease which is more common in males [76], and which is characterised by motoric dysfunction and dopaminergic neuron loss in the substantia nigra. One exciting question which follows from this is whether, besides influencing motor function, Sry may directly influence behavioural (cognitive) endophenotypes, particularly those sensitive to dopaminergic manipulation; monitoring the brain and behavioural effects of knocking down Sry expression in the rat ventral tegmental area for example may help to address this point.
In addition to looking in animal models, we may also gain insights into the brain and behavioural functions of SRY by looking at humans with deletions of, or inactivating mutations in, the gene. However, as these mutations will abrogate SRY function in all tissues, as in the 4-core genotype cross, it will not be possible to dissociate between indirect and direct SRY effects. As these subjects are relatively rare, no comprehensive brain and behavioural analyses have been performed on them to date. Case studies of 46,XY females with SRY mutations and consequent gonadal dysgenesis [28,77-79] report that, in terms of psychosexual behaviour, such subjects tend to exhibit a female-typical profile in that they are attracted to men [79]. This observation indicates that SRY may play a fundamental role in sexual preference. Future work examining the behavioural/cognitive profiles of subjects with reduced or enhanced SRY dosage (notably 46,XY females or 46,XX males respectively) will help to clarify the role of SRY in brain function.
THE BRAIN AND BEHAVIOURAL EFFECTS OF THE SEX-LINKED STS GENE
A second sex-linked gene that has relatively well-defined effects on brain and behaviour is Sts, encoding the enzyme steroid sulfatase. Steroid sulfatase is responsible for the desulfation of various neuroactive steroids, notably of the GABAergic modulator dehydroepiandrosterone sulphate (DHEAS) to DHEA [80]. The enzyme is expressed most highly during embryogenesis in the placenta and the liver and in the cortex, thalamus and hindbrain [81]. In mice, Sts is the only known PAR gene [82,83], and is therefore expressed from both the X and Y chromosomes.
Aggression in mice is highly sexually dimorphic, with males exhibiting a far greater tendency to attack their conspecifics. A genetic study aiming to identify and characterise the Y chromosomal correlates of this sexual dimorphism localised the underlying region to the Y-PAR [84]. As the only known PAR gene, Sts immediately became an excellent genetic candidate for these effects on aggression. Follow-up pharmacological studies targeting the steroid sulfatase axis seem to confirm a role for the enzyme in the brain processes underlying aggression [85]. Besides influencing aggression, parallel genetic and pharmacological studies have demonstrated that steroid sulfatase may influence attention and impulsivity in mice [86,87]. As an aside, work by Maxson has shown that the NRY region of several pairs of inbred mouse strains can have differential effects on offense, suggesting Sry as a plausible positional candidate for effects on aggression [88].
Work in rats has shown that steroid sulfatase can influence learning and memory [89] and hippocampal acetylcholine release [90]. However, as Sts is non-pseudoautosomal in rats and has no detectable Y homologue [91], these neurobiological effects must be attributable to the X-linked Sts gene.
In man, subjects with deletions of the STS gene, or inactivating mutations within it, and thus presenting with the disorder X-linked ichthyosis, appear to display heightened vulnerability to autism and to predominantly-inattentive subtype ADHD [92]. Moreover, the STS gene has been associated with ADHD [93], suggesting that steroid sulfatase may underlie attentional processes in both rodents and humans. STS is located on the distal short arm of the X chromosome (Xp22.3), very close to PAR, and it escapes X-inactivation [94]. Unlike in the rat, in man, X-linked STS has a homologue located on the long arm of the Y chromosome, although a succession of base substitutions, and small insertions and deletions appear to have rendered this Y-linked version a non-expressed pseudogene [95]. The fact that STS escapes X-inactivation (and is therefore expressed from both X chromosomes), and that it has no functional Y homologue, could potentially explain the reported higher activity of the enzyme in female than male tissues [96,97]. Furthermore, this sex difference in steroid sulfatase activity could explain why males and females are differentially vulnerable to disorders of attention and impulse control such as ADHD and pathological gambling.
THE EFFECTS OF OTHER Y-LINKED GENES ON BRAIN AND BEHAVIOUR
SRY and STS are perhaps the best characterised genes resident on the Y chromosome in terms of their brain and behavioural functions, although it must be acknowledged that in many respects our knowledge about the role of these two genes is lacking. However, there are several other Y-linked genes in NRY, which, in that they are expressed in the brain, could also potentially contribute towards neural sexual differentiation. Xu and colleagues described six NRY genes (Dby (now Ddx3y) Ube1y, Smcy (now Kdm5d), Eif2s3y, Uty, and Usp9y) which were expressed at one or more developmental stages in male and 40,XY female mouse brain (the latter indicating a lack of requirement for testicular secretions)[98]. Of the genes analysed, all had an X-linked homologue, which was definitively known to escape X-inactivation in three cases (Smcx/Kdm5c, Utx and Eif2s3x). In several cases, the expression of the Y homologue in males was much lower than its X-linked homologue, and as such was not sufficient to ensure dosage compensation between the sexes. A further intriguing possibility when considering the genetic mechanisms underlying sexually dimorphic brain phenotypes, is that X and Y-linked homologues, in addition to being expressed at different levels, are expressed at different developmental stages and/or in different brain regions. Indeed, recent work by Xu et al. has shown that the paralogues Utx and Uty are differentially expressed in the paraventricular nucleus of the hypothalamus (high Uty expression) and in the amygdala (high Utx expression), possibly as a consequence of differential epigenetic marks [99]. To our knowledge, no comprehensive survey comparing the relative spatiotemporal expression dynamics of X and Y homologues has yet been performed, although it has been shown that that there is some consistency in the expression patterns of Eif2s3y and Eif2s3x, with highest expression of both in the thalamus, hypothalamus, hippocampus and cerebellum [100]. The expression patterns for many mouse NRY genes are now documented in resources such as the Allen Brain Atlas: of the Y-linked genes mentioned above, Ube1y and Eif2s3y are highly expressed throughout the hypothalamus. As the pituitary gland is pivotal in the secretion of hormones underlying sex-specific physiology, it would be worthwhile examining the expression of Y-linked genes in specific endocrine cell types of this tissue. In many cases, the brain and behavioural functions of NRY-linked genes are obscure, a fact probably attributable to the structural nature of the Y chromosome precluding the development of knockout models. Insights into the range of neural functions underpinned by NRY genes are likely to come from Y-chromosome mutant mice (in which Y-linked genes are spontaneously deleted or duplicated); alternatively, insights may come from a comparison of normal male mice and 39,XmO mice [65], the two groups only differing in the fact that the latter has no Y chromosome.
There are human orthologues of Ddx3y, Kdm5d, Uty and Usp9y, therefore investigations in mouse models into the neurobiological functions of these genes are likely to shed light upon their role in male brain development in humans. There appear to be species differences between mice and humans with regard to some Y-linked brain-expressed genes, in that Ube1y and Eif2s3y have no human counterparts, whilst ZFY appears to be expressed in the hypothalamus and cortex of adult humans [52], but is not expressed at any developmental stage in mouse brain [98]. Hence, it is likely that the nature of the neural sexual differentiation process is, to a greater or lesser extent, species-specific. One X-Y homologous gene pair which has received a lot of interest regarding its role in neurodevelopment is PCDH11X/Y. The homologous genes are located within a hominid-specific region of the sex chromosomes (Xq21.3 and Xp11.2), and encode members of the protocadherin superfamily responsible for cell-cell interactions during development of the central nervous system [101]. Not only are PCDH11X and its Y counterpart structurally different (and therefore possibly functionally distinct) but they have been shown to exhibit differential expression patterns, most likely because the two genes possess different promoter regions [101]. In the brain, transcripts from both PCDH11X and PCDH11Y are present most highly in the cortex [102], and also in several subregions including the amygdala, caudate nucleus, hippocampus and thalamus [101]. Interestingly, PCDH11X seems to be the preferential transcript in the cerebellum; in the heart, transcripts are predominantly from PCDH11X, whereas in the kidney, liver, muscle and testis transcripts come mainly from PCDH11Y [101]. Together these data indicate that PCDH11X/Y genes may play key modulatory roles in the sexual differentiation of a wide variety of organs (including the brain) in hominid mammals. Exactly how PCDH11Y may act in the brain to modulate function remains to be resolved, but work in prostate cancer cell culture suggests that it may influence neuroendocrine tissue transdifferentiation via classical Wnt signalling pathways [103].
Y CHROMOSOME EFFECTS ON VULNERABILITY TO NEUROPSYCHIATRIC DISORDERS
For most multigenic human disorders, the underlying substrates are unknown or their functions are poorly understood. Neuropsychiatric disorders, in particular, are likely to have a complex and multifactorial basis, with their development and course being influenced by genetic and environmental factors, and by interactions between the two. Whilst there is some controversy regarding the extent and specificity with which behaviour differs between the sexes in healthy populations, it is clear that many, if not all, neuropsychiatric disorders show some sort of sex-bias in their presentation, even allowing for possible ascertainment biases; this observation implies that male and female brains are somehow differentially sensitive to such disorders. This sex-bias may be evident in terms of the incidence of the disorder, its age-at-onset, its progress, its response to conventional therapeutics, its underlying neurobiology, or a combination of some or all of these. In terms of specific conditions, we know that males are more commonly affected by developmental disorders such as ADHD [104] and that they may be more severely affected by some disorders: for example, a recent longitudinal study following patients with schizophrenia for 20 years, has shown that female schizophrenic patients have a significantly better course of illness and better global functioning than male patients even taking into the account the earlier age-at-onset for men (the mean age of onset is approximately 4 years earlier for men than for women) [105]. In contrast, females are more prone to affective disorders with a later onset, usually during adolescence, such as unipolar depression and anxiety [106]. The observation that males and females differ in their susceptibility to neurodevelopmen tal disorders such as autism and ADHD suggests that sex-linked genes may be particularly important in utero in directly modulating critical neurodevelopmental processes (either directly or via effects on prenatal gonadal hormone levels).
Case or small-scale studies have suggested a role for Y chromosome genes in some neuropsychiatric disorders. For example, individuals with 47,XYY syndrome are at elevated risk of developing antisocial behaviour [107] and may be at increased risk of developing schizoaffective disorder [108], probably as a consequence of NRY gene over-expression. A potential role for Y-linked genes in the pathogenesis of schizophrenia is also supported by the observation of an isodicentric Y chromosome in a schizophrenic patient [109]. A case study has implicated Y-linked genes in ADHD susceptibility: in this case, the affected boy possessed a rare deletion of Yq with duplication of Yp [110]. Interestingly, the duplicated region included the SRY gene, suggesting that overdosage of this specific gene may be responsible (either directly or indirectly) for the observed behavioural phenotype. One important caveat with studies such as these is that the co-occurrence of a particular cytogenetic mutation and a particular neuropsychiatric manifestation does not necessarily imply that the two are linked.
Although studies examining rare Y-linked mutations or rare polyploidies such as those described above may provide important proof-of-principle data about the involvement of sex-linked genes in neuropsychiatric phenotypes, ideally we would like to know the extent to which common Y-linked genetic variants may predispose to male-biased mental illnesses within the general population. In order to address this question, association studies have compared Y chromosome haplogroups (i.e. a group of haplotypes that share a common ancestor) in patient and control cohorts. This experiment represents an indirect way of examining whether Y chromosome polymorphisms are involved in a disorder and assumes that a variant conferring susceptibility arose against a specific Y chromosome haplogroup background - due to the lack of recombination within the NRY, the susceptibility variant would be co-inherited with the variants that define this haplogroup. In association studies of this type, ensuring control and patient groups are adequately matched is vital, given that Y chromosome haplogroups may vary substantially with geographical region [111]. This matching process has been facilitated by the work of the Y Chromosome Consortium, a collaborative body dedicated to studying variation within the human Y chromosome. To date, the consortium has resolved nomenclature problems associated with Y-linked variants and has produced a parsimonious phylogenetic tree including all known Y chromosome haplogroups, the latest revised version of which contains 311 haplogroups and approximately 600 single nucleotide polymorphisms (SNPs) [112].
Using the strategy outlined above, significant associations between particular Y chromosome haplogroups and male-biased behavioural traits (including alcohol dependence [113] and aggression [114]) have been reported, whilst a study examining the relationship between Y chromosome haplogroups and autism failed to find any association [115]. However, the results of studies like these must be interpreted with caution given that they often use small, heterogeneous samples with non-overlapping Y chromosome markers. In our ongoing work, we are testing the idea that Y chromosome variation may contribute to ADHD and schizophrenia vulnerability in men, by comparing the haplogroups of large samples of Caucasian patients and controls of U.K. origin. Preliminary analyses suggest that, whilst there is little evidence for association with either disorder, there is an apparent modifying effect of Y chromosome variants on cognitive performance (performance IQ) in the ADHD sample (unpublished results). An obvious, and important, question which arises from this finding, is whether the effect is generalisable to a non-clinical male sample - if it is, identifying and characterising the causal variant will be a priority. Such work would have important ramifications for explaining the evolutionary basis of cognitive sexual dimorphism.
With regard to specific candidate genes for the male-bias in neuropsychiatric disorders, numerous factors make the PCDH11Y gene an attractive proposition. Firstly, it appears to be expressed in males in a highly regulated and spatiotemporally dynamic manner and is involved in synapse formation and neuronal path finding in the brain, processes which go awry in a number of common male-biased mental conditions [116]. The fact that PCDH11Y is not expressed (or only weakly expressed) in the cerebellum, whereas it is expressed elsewhere in the brain, could potentially explain why males are especially vulnerable to disorders with known cerebellar pathology (e.g. ADHD and autism). Finally, as mentioned previously, PCDH11Y is specific to the hominid lineages and is absent in non-human primates such as chimpanzees and gorillas [117]; moreover, the gene has shown accelerated sequence change in the hominid lineage [118]. For this reason, it has been proposed, most vociferously by Timothy Crow and colleagues, that aberrant expression of the protein encoded by PCDH11Y could predispose males to disorders of human-specific functions such as language (thought to be a correlate of cerebral asymmetry), theory of mind and problem-solving flexibility [119]. There is convincing evidence that these types of human-specific function are impaired in a number of male-biased disorders, notably schizophrenia and autism [6,120]. However, initial, relatively small, candidate gene studies examining possible association between SNPs within PCDH11Y and a number of neurospsychiatric disorders (autism, ADHD, bipolar disorder, obsessive-compulsive disorder and schizophrenia/psychosis) have failed to find evidence for a link to date [102,119]. It is formally plausible that variation in PCDH11Y, or on the Y chromosome in general, may be responsible for vulnerability to these disorders in specific sub-groups. Alternatively, it may be that it is the expression pattern of the gene (which may be modulated epigenetically via environmental influences) which is more important in modulating its effect on male disease vulnerability than its sequence.
A second candidate gene that could potentially influence neuropsychiatric phenotypes is NLGN4Y, the Y homologue of NLGN4X. These genes encode cell adhesion molecules which interact with β-neurexins at the postsynaptic membrane during the process of synaptogenesis [121]. Early findings that mutations in NLGN4X were present in families with mental retardation and autism spectrum disorders suggested a possible causal link between the disorders and the gene mutation [122, 123]. The results of subsequent studies have suggested that, if mutations in NLGN4X are pathogenic, they are likely to be rare, and are not likely to explain the majority of cases of autism [124-127]. In a study explicitly focussing on sequence variants in NLGN4Y, Yan and co-workers identified one mis-sense variant present in one autistic patient and his learning-disabled father out of a population of 335 autistic or mentally retarded males [128]. This result is consistent with the idea that mutations in the NLGN4 genes may contribute to the etiology of autism, but only in a limited proportion of cases.
CONCLUSIONS
Despite its small size, and limited gene content, we have argued here that the Y chromosome may exert a considerable influence on brain function. As a consequence of its inheritance pattern, genes upon it may help to define male-specific brain phenotypes, and hence male-typical behaviours. An alternative perspective is that, in some cases, Y-linked genes may act to attenuate sex differences (e.g. where the Y homologue of an X-linked escaping inactivation performs a functionally equivalent role). In this context, Dewing and colleagues suggested that, in rats, ‘Sry could compensate for a factor that is only present in females and maintains tyrosine hydroxylase expression in substantia nigra neurons’, positing high levels of estrogens in females as such a factor [53]. A major goal for future work will be to describe the brain functions of Y-linked genes in terms of their relevance to selective evolutionary forces acting on the chromosome, such as sexual antagonism. Further studies on the Y chromosome will provide insights into the biological basis of neural sexual differentiation (or lack thereof), and will clarify the molecular basis of sex biases in common neuropsychiatric disorders.
ACKNOWLEDGEMENTS
EK is supported jointly by a Cardiff University Endowment Studentship and the Medical Research Council (MRC) UK. ES is supported by the Wellcome Trust and the MRC UK. PML is supported by the Biotechnology and Biological Sciences Research Council (BBSRC) UK. LSW is supported by a Cardiff University Link Award. WD is supported by a Research Councils UK (RCUK) Fellowship.
Footnotes
This is an open access article licensed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
REFERENCES
- [1].Craig IW, Harper E, Loat CS. The genetic basis for sex differences in human behaviour: role of sex chromosomes. Ann Hum Genet. 2004;68:269–84. doi: 10.1046/j.1529-8817.2004.00098.x. [DOI] [PubMed] [Google Scholar]
- [2].Lejbak L, Vrbancic M, Crossley M. The female advantage in object location memory is robust to verbalizability and mode of presentation of test stimuli. Brain Cogn. 2009;69:148–53. doi: 10.1016/j.bandc.2008.06.006. [DOI] [PubMed] [Google Scholar]
- [3].Eals M, Silverman I. The hunter-gatherer theory of spatial sex differences: proximate factors mediating the female advantage in recall of object arrays. Ethol Sociobiol. 1994;15:95–105. [Google Scholar]
- [4].Hall JA. Gender effects in decoding nonverbal cues. Psychol Bull. 1978;85:845–58. [Google Scholar]
- [5].McClure E. A meta-analytic review of sex differences in facial expression processing and their development in infants, childen and adolescents. Psychol Bull. 2000;126(3):424–53. doi: 10.1037/0033-2909.126.3.424. [DOI] [PubMed] [Google Scholar]
- [6].Baron-Cohen S. The extreme male brain theory of autism. Trends Cogn Sci. 2002;6(6):248–54. doi: 10.1016/s1364-6613(02)01904-6. [DOI] [PubMed] [Google Scholar]
- [7].Holden C. Sex and the suffering brain. Science. 2005;308:1574–7. doi: 10.1126/science.308.5728.1574. [DOI] [PubMed] [Google Scholar]
- [8].Cachill L. Why sex matters for neuroscience. Nat Rev Neurosci. 2006;7:477–84. doi: 10.1038/nrn1909. [DOI] [PubMed] [Google Scholar]
- [9].Sullivan EV, Rosenbloom MJ, Desmond JE, Pfefferbaum A. Sex differences in corpus callosum size: relationship to age and intracranial size. Neurobiol Aging. 2001;22:603–11. doi: 10.1016/s0197-4580(01)00232-9. [DOI] [PubMed] [Google Scholar]
- [10].Goldstein JM, Seidman LJ, Horton NJ, et al. Normal sexual dimorphism of the adult human brain assessed by in vivo magnetic resonance imaging. Cereb Cortex. 2001;11:490–7. doi: 10.1093/cercor/11.6.490. [DOI] [PubMed] [Google Scholar]
- [11].Le Vay S. A difference in hypothalamic structure between heterosexual and homosexual men. Science. 1991;253:1034–7. doi: 10.1126/science.1887219. [DOI] [PubMed] [Google Scholar]
- [12].Andreano JM, Cahill L. Sex influences on the neurobiology of learning and memory. Learn Mem. 2009;16(4):248–66. doi: 10.1101/lm.918309. [DOI] [PubMed] [Google Scholar]
- [13].Luine V, Dohanich G. Sex differences in cognitive function in rodents. In: Becker JB, Berkley KJ, Geary N, Hampson E, Herman JP, Young EA, editors. Sex differences in the brain, from genes to behavior. Oxford University Press Inc.; New York: 2008. pp. 227–51. [Google Scholar]
- [14].Fattore L, Altea S, Fratta W. Sex differences in drug addiction: a review of animal and human studies. Womens Health (Lond Engl) 2008;4:51–65. doi: 10.2217/17455057.4.1.51. [DOI] [PubMed] [Google Scholar]
- [15].Kaiser A, Haller S, Schmitz S, Nitsch C. On sex/gender related similarities and differences in fMRI language research. Brain Res. 2009 doi: 10.1016/j.brainres.2009.03.065. in press. [DOI] [PubMed] [Google Scholar]
- [16].Fleming A, Vilain E. The endless quest for sex determination genes. Dev Biol. 2004;67:15–25. doi: 10.1111/j.1399-0004.2004.00376.x. [DOI] [PubMed] [Google Scholar]
- [17].Davies W, Wilkinson LS. It is not all hormones: alternative explanations for sexual differentiation of the brain. Brain Res. 2006;1126:36–45. doi: 10.1016/j.brainres.2006.09.105. [DOI] [PubMed] [Google Scholar]
- [18].Carrel L, Cottle AA, Goglin KC, Willard HF. A first-generation X-inactivation profile of the human X chromosome. Proc Natl Acad Sci USA. 1999;96(25):14440–4. doi: 10.1073/pnas.96.25.14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–4. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- [20].Davies W, Isles AR, Burgoyne PS, Wilkinson LS. X-linked imprinting: effects on brain and behaviour. Bioessays. 2005;28:35–44. doi: 10.1002/bies.20341. [DOI] [PubMed] [Google Scholar]
- [21].Vallender EJ, Lahn BT. How mammalian sex chromosomes acquired their peculiar gene content. Bioessays. 2004;26:159–69. doi: 10.1002/bies.10393. [DOI] [PubMed] [Google Scholar]
- [22].Skaletsky H, Kuroda-Kawagushi T, Minx PJ, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–37. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- [23].Vawter MP, Evans S, Choudary P, et al. Gender-specific gene expression in post-mortem human brain: localization to sex chromosomes. Neuropsychopharmacology. 2004;29:373–84. doi: 10.1038/sj.npp.1300337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Jobling MA, Tyler-Smith C. The human Y chromosome: an evolutionary marker comes of age. Nat Rev Genet. 2003;4(8):598–612. doi: 10.1038/nrg1124. [DOI] [PubMed] [Google Scholar]
- [25].Bishop CE. Mouse Y chromosome. Mamm Genome. 1992;3:S289–S93. doi: 10.1007/BF00648439. [DOI] [PubMed] [Google Scholar]
- [26].Alfoldi JE. Sequence of the mouse Y chromosome. Massachusetts Institute of Technology; 2007. Ph.D thesis. [Google Scholar]
- [27].Goodfellow PN, Lovell-Badge R. SRY and sex determination in mammals. Annu Rev Genet. 1993;27:71–92. doi: 10.1146/annurev.ge.27.120193.000443. [DOI] [PubMed] [Google Scholar]
- [28].Harley VR, Clarkson MJ, Argentaro A. The molecular action and regulation of the testis-determining factors, SRY (sex-determining region of the Y chromosome) and SOX [SRY-related high-mobility group (HMG) box 9] Endocr Rev. 2003;24(4):466–87. doi: 10.1210/er.2002-0025. [DOI] [PubMed] [Google Scholar]
- [29].Mazeyrat S, Saut N, Grigoriev V, et al. A Y-encoded subunit of the translation initiation factor Eif2 is essential for mouse spermatogenesis. Nat Genet. 2001;29(1):49–53. doi: 10.1038/ng717. [DOI] [PubMed] [Google Scholar]
- [30].Singh L, Panicker SG, Nagaraj R, Majumdar KC. Banded krait minor-satellite (Bkm)- associated Y chromosome-specific repetitive DNA in mouse. Nucleic Acids Res. 1994;22(12):2289–95. doi: 10.1093/nar/22.12.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Burgoyne PS. The role of Y-encoded genes in mammalian spermatogenesis. Semin Cell Dev Biol. 1998;9:423–32. doi: 10.1006/scdb.1998.0228. [DOI] [PubMed] [Google Scholar]
- [32].Grutzner F, Himmelbauer H, Paulsen M, Ropers HH, Haaf T. Comparative mapping of mouse and rat chromosomes by fluorescence in situ hybridization. Genomics. 1999;55:306–13. doi: 10.1006/geno.1998.5658. [DOI] [PubMed] [Google Scholar]
- [33].Mazeyrat S, Mitchell MJ. Rodent Y chromosome TSPY gene is functional in rat and non-functional in mouse. Hum Mol Genet. 1998;7(3):557–62. doi: 10.1093/hmg/7.3.557. [DOI] [PubMed] [Google Scholar]
- [34].Jacobs PA, Strong JA. A case of human intersexuality having a possible XXY sex-determining mechanism. Nature. 1959;183:302–3. doi: 10.1038/183302a0. [DOI] [PubMed] [Google Scholar]
- [35].Sinclair AH, Berta P, Palmer MS, et al. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 1990;346:240–4. doi: 10.1038/346240a0. [DOI] [PubMed] [Google Scholar]
- [36].Gubbay J, Collignon J, Koopman P, et al. A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature. 1990;346:245–50. doi: 10.1038/346245a0. [DOI] [PubMed] [Google Scholar]
- [37].Wilhelm D, Koopman P. The making of maleness: towards an intergrated view of male sexual development. Nat Rev Genet. 2006;7:620–31. doi: 10.1038/nrg1903. [DOI] [PubMed] [Google Scholar]
- [38].Sekido R, Lovell-Badge R. Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer. Nature. 2008;453:930–4. doi: 10.1038/nature06944. [DOI] [PubMed] [Google Scholar]
- [39].Wilson CA, Davies DC. The control of sexual differentiation of the reproductive system and brain. Reproduction. 2007;133(2):331–59. doi: 10.1530/REP-06-0078. [DOI] [PubMed] [Google Scholar]
- [40].Hutchison JB. Gender-specific steroid metabolism in neural differentiation. Cell Mol Neurobiol. 1997;17(6):603–26. doi: 10.1023/A:1022581902880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Becker JB, Berkley KJ, Geary N, Hampson E, Herman JP, Young EA, editors. Sex differences in the brain, from genes to behavior. Oxford University Press Inc.; New York: 2008. [Google Scholar]
- [42].Morris JA, Jordan CL, Breedlove SM. Sexual differentiation of the vertebrate nervous system. Nat Neurosci. 2004;7(10):1034–9. doi: 10.1038/nn1325. [DOI] [PubMed] [Google Scholar]
- [43].Swaab DF. Sexual differentiation of the brain and behavior. Best Pract Res Clin Endocrinol Metab. 2007;21(3):431–44. doi: 10.1016/j.beem.2007.04.003. [DOI] [PubMed] [Google Scholar]
- [44].Roof RL. Neonatal exogenous testosterone modifies sex difference in radial arm and Morris water maze performance in prepubescent and adult rats. Behav Brain Res. 1993;53:1–10. doi: 10.1016/s0166-4328(05)80261-x. [DOI] [PubMed] [Google Scholar]
- [45].Aubele T, Kaufman R, Montalmant F, Kritzer MF. Effects of gonadectomy and hormone placement on a spontaneous novel object recognition task in adult male rats. Horm Behav. 2008;54(2):244–52. doi: 10.1016/j.yhbeh.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Joseph R, Hess S, Birecree E. Effects of hormone manipulations and exploration on sex differences in maze learning. Behav Neural Biol. 1998;24:364–77. doi: 10.1016/s0091-6773(79)90223-2. [DOI] [PubMed] [Google Scholar]
- [47].Spritzer MD, Gill M, Weinberg A, Galea LA. Castration differentially affects spatial working and reference memory in male rats. Arch Sex Behav. 2008;37(1):19–29. doi: 10.1007/s10508-007-9264-2. [DOI] [PubMed] [Google Scholar]
- [48].Williams CL, Bamett AM, Meck YM. Organizational effects of early gonadal secretions on sexual differentiation in spatial memory. Behav Neurosci. 1990;104:84–97. doi: 10.1037//0735-7044.104.1.84. [DOI] [PubMed] [Google Scholar]
- [49].Warren MF, Serby MJ, Roane DM. The effects of testosterone on cognition in elderly men: a review. CNS Spectrum. 2008;13(10):887–97. doi: 10.1017/s1092852900016990. [DOI] [PubMed] [Google Scholar]
- [50].Renfree MB, Short RV. Sex determination in marsupials: evidence for a marsupial-eutherian dichotomy. Philos Trans R Soc Lond B Biol Sci. 1988;322:41–53. doi: 10.1098/rstb.1988.0112. [DOI] [PubMed] [Google Scholar]
- [51].Clepet C, Schafer AJ, Sinclair AH, Palmer MS, Lovell-Badge R, Goodfellow PN. The human SRY transcript. Hum Mol Genet. 1993;2(12):2007–12. doi: 10.1093/hmg/2.12.2007. [DOI] [PubMed] [Google Scholar]
- [52].Mayer A, Lahr G, Swaab DF, Pilgrim C, Reisert I. The Y-chromosomal genes SRY and ZFY are transcribed in adult human brain. Neurogenetics. 1998;1(4):281–8. doi: 10.1007/s100480050042. [DOI] [PubMed] [Google Scholar]
- [53].Dewing P, Chiang CW, Sinchak K, et al. Direct regulation of adult brain function by the male-specific factor SRY. Curr Biol. 2006;16(4):415–20. doi: 10.1016/j.cub.2006.01.017. [DOI] [PubMed] [Google Scholar]
- [54].Lahr G, Maxson SC, Mayer A, Just W, Pilgrim C, Reisert I. Transcription of the Y chromosomal gene, Sry, in adult mouse brain. Brain Res Mol Brain Res. 1995;33(1):179–82. doi: 10.1016/0169-328x(95)00136-g. [DOI] [PubMed] [Google Scholar]
- [55].Mayer A, Mosler G, Just W, Pilgrim C, Reisert I. Developmental profile of Sry transcripts in mouse brain. Neurogenetics. 2000;3(1):25–30. doi: 10.1007/s100480000093. [DOI] [PubMed] [Google Scholar]
- [56].Turner ME, Martin C, Martins AS, et al. Genomic and expression analysis of multiple Sry loci from a single Rattus norvegicus Y chromosome. BMC Genet. 2007;8:11. doi: 10.1186/1471-2156-8-11. doi: 10.11 86/1471-2156-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Lundrigan BL, Tucker PK. Evidence for multiple functional copies of the male sex-determining locus, Sry, in African murine rodents. J Mol Evol. 1997;45:60–5. doi: 10.1007/pl00006202. [DOI] [PubMed] [Google Scholar]
- [58].Nagamine CM. The testis-determining gene, Sry, exists in multiple copies in old World rodents. Genet Res. 1994;64:151–9. doi: 10.1017/s001667230003281x. [DOI] [PubMed] [Google Scholar]
- [59].Milsted A, Serova L, Sabban EL, Dunphy G, Turner ME, Ely DL. Regulation of tyrosine hydroxylase gene transcription by Sry. Neurosci Lett. 2004;369(3):203–7. doi: 10.1016/j.neulet.2004.07.052. [DOI] [PubMed] [Google Scholar]
- [60].Andersen SL, Teicher MH. Sex differences in dopamine receptors and their relevance to ADHD. Neurosci Biobehav Rev. 2000;24:137–41. doi: 10.1016/s0149-7634(99)00044-5. [DOI] [PubMed] [Google Scholar]
- [61].Charchar FJ, Tomaszewski M, Strahorn P, Champagne B, Dominiczak AF. Y is there a risk to being male? Trends Endocrinol Metab. 2003;14(4):163–8. doi: 10.1016/s1043-2760(03)00032-8. [DOI] [PubMed] [Google Scholar]
- [62].Quinn JJ, Hitchcott PK, Umeda EA, Arnold AP, Taylor JR. Sex chromosome complement regulates habit formation. Nat Neurosci. 2007;10(11):1398–400. doi: 10.1038/nn1994. [DOI] [PubMed] [Google Scholar]
- [63].Arnold AP, Chen X. What does the “four core genotypes” mouse model tell us about sex differences in the brain and other tissues? Front Neuroendocrinol. 2009;30:1–9. doi: 10.1016/j.yfrne.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Lovell-Badge R, Robertson E. XY female mice resulting from a heritable mutation in the primary testis-determining gene, Tdy. Development. 1990;109(3):635–46. doi: 10.1242/dev.109.3.635. [DOI] [PubMed] [Google Scholar]
- [65].Lynn PM, Davies W. The 39,XO mouse as a model for the neurobiology of Turner syndrome and sex-biased neuropsychiatric disorders. Behav Brain Res. 2007;179(2):173–82. doi: 10.1016/j.bbr.2007.02.013. [DOI] [PubMed] [Google Scholar]
- [66].Wagner CK, Xu J, Pfau JL, Quadros PS, De Vries GJ, Arnold AP. Neonatal mice possessing an Sry transgene show a masculinized pattern of progesterone receptor expression in the brain independent of sex chromosome status. Endocrinology. 2004;145(3):1046–9. doi: 10.1210/en.2003-1219. [DOI] [PubMed] [Google Scholar]
- [67].Markham JA, Jurgens HA, Auger CJ, DeVries GJ, Arnold AP, Juraska JM. Sex differences in mouse cortical thickness are independent of the complement of sex chromosomes. Neuroscience. 2003;116(1):71–5. doi: 10.1016/s0306-4522(02)00554-7. [DOI] [PubMed] [Google Scholar]
- [68].DeVries GJ, Rissman EF, Simerly RB, et al. A model system for study of sex chromosome effects on sexually dimorphic neural and behavioral traits. J Neurosci. 2002;22(20):9005–14. doi: 10.1523/JNEUROSCI.22-20-09005.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Park JH, Burns-Cusato M, Dominguez-Salazar E, et al. Effects of sex chromosome aneuploidy on male sexual behavior. Genes Brain Behav. 2008;7:609–17. doi: 10.1111/j.1601-183X.2008.00397.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Schwarz JM, McCarthy MM. Steroid-induced sexual differentiation of the developing brain: multiple pathways, one goal. J Neurochem. 2008;105:1561–72. doi: 10.1111/j.1471-4159.2008.05384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Smith-Bouvier DL, Divekar AA, Sasidhar M, et al. A role for sex chromosome complement in the female bias in autoimmune disease. J Exp Med. 2008;205(5):1099–108. doi: 10.1084/jem.20070850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Chen X, Watkins R, Delot E, et al. Sex difference in neural tube defects in p53-mull mice is caused by differences in the complement of X not Y genes. Dev Neurobiol. 2008;68(2):265–73. doi: 10.1002/dneu.20581. [DOI] [PubMed] [Google Scholar]
- [73].Gioiosa L, Chen X, Watkins R, et al. Sex chromosome complement affects nociception in tests of acute and chronic exposure to morphine in mice. Horm Behav. 2008;53(1):124–30. doi: 10.1016/j.yhbeh.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Carruth LL, Reisert I, Arnold AP. Sex chromosome genes directly affect brain sexual differentiation. Nat Neurosci. 2002;5:933–4. doi: 10.1038/nn922. [DOI] [PubMed] [Google Scholar]
- [75].Gatewood JD, Wills A, Shetty S, et al. Sex chromosome complement and gonadal sex influence aggressive and parental behaviors in mice. J Neurosci. 2006;26(8):2335–42. doi: 10.1523/JNEUROSCI.3743-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Van De Eeden SK, Tanner CM, Bernstein AL, et al. Incidence of Parkinson’s disease: variation by age, gender, and race/ethnicity. Am J Epidemiol. 2003;157(11):1015–22. doi: 10.1093/aje/kwg068. [DOI] [PubMed] [Google Scholar]
- [77].Cameron FJ, Sinclair AH. Mutations in SRY and SOX: Testis-determining genes. Hum Mutat. 1997;9:388–95. doi: 10.1002/(SICI)1098-1004(1997)9:5<388::AID-HUMU2>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- [78].Hawkins JR. Mutational analysis of SRY in XY females. Hum Mutat. 1993;2:347–50. doi: 10.1002/humu.1380020504. [DOI] [PubMed] [Google Scholar]
- [79].McCarthy BM, Migeon CJ, Meyer-Bahlburg HFL, Zacur H, Wisniewski AB. Medical and psychosexual outcome in women affected by complete gonadal dysgenesis. J Pediatr Endocrinol Metab. 2006;19:873–7. doi: 10.1515/jpem.2006.19.7.873. [DOI] [PubMed] [Google Scholar]
- [80].Reed MJ, Purohit A, Woo LWL, Newman SP, Potter BVL. Steroid sulphatase: molecular biology, regulation and inhibition. Endocr Rev. 2005;26(2):171–202. doi: 10.1210/er.2004-0003. [DOI] [PubMed] [Google Scholar]
- [81].Compagnone NA, Salido E, Shapiro LJ, Mellon SH. Expression of steroid sulfatase during embryogenesis. Endocrinology. 1997;138:4768–73. doi: 10.1210/endo.138.11.5504. [DOI] [PubMed] [Google Scholar]
- [82].Salido EC, Li XM, Yen PH, Martin N, Mohandas TK, Shapiro LJ. Cloning and expression of the mouse pseudoautosomal steroid sulphatase gene (Sts) Nat Genet. 1996;13:83–6. doi: 10.1038/ng0596-83. [DOI] [PubMed] [Google Scholar]
- [83].Perry J, Palmer S, Gabriel A, Ashworth A. A short pseudoautosomal region in laboratory mice. Genome Res. 2001;11(11):1826–32. doi: 10.1101/gr.203001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Roubertoux PL, Carlier M, Degrelle H, Haas-Dupertuis MC, Phillips J, Moutier R. Co-segretation of intermale aggression with the pseudoautosomal region of the Y chromosome in mice. Genetics. 1994;135:225–30. doi: 10.1093/genetics/136.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Nicolas LB, Pinoteau W, Papot S, Routier S, Guillaumet G, Mortaud S. Aggressive behavior induced by the steroid sulfatase inhibitor COUMATE and by DHEAS in CBA/H mice. Brain Res. 2001;922:216–22. doi: 10.1016/s0006-8993(01)03171-7. [DOI] [PubMed] [Google Scholar]
- [86].Davies W, Humby T, Isles AR, Burgoyne PS, Wilkinson LS. X-monosomy effects on visuospatial attention in mice: a candidate gene and implications for Turner syndrome and attention deficit hyperactivity disorder. Biol Psychiatry. 2007;61(12):1351–60. doi: 10.1016/j.biopsych.2006.08.011. [DOI] [PubMed] [Google Scholar]
- [87].Davies W, Humby T, Kong W, Otter T, Burgoyne PS, Wilkinson LS. Converging pharmacological and genetic evidence indicates a role for steroid sulfatase in attention. Biol Psychiatry. 2009;66(4):360–7. doi: 10.1016/j.biopsych.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Maxson SC. Searching for candidate genes with effects on an agonistic behavior, offense, in mice. Behav Genet. 1996;26:471–6. doi: 10.1007/BF02359751. [DOI] [PubMed] [Google Scholar]
- [89].Johnson DA, Wu TH, Li PK, Maher TJ. The effect of steroid sulfatase inhibition on learning and spatial memory. Brain Res. 2000;865:286–90. doi: 10.1016/s0006-8993(00)02372-6. [DOI] [PubMed] [Google Scholar]
- [90].Rhodes ME, Li PK, Burke AM, Johnson DA. Enhanced plasma DHEAS, brain acetylcholine and memory mediated by steroid sulfatase inhibition. Brain Res. 1997;773:28–32. doi: 10.1016/s0006-8993(97)00867-6. [DOI] [PubMed] [Google Scholar]
- [91].Li XM, Salido EC, Gong Y, et al. Cloning of the rat steroid sulfatase gene (Sts), a non-pseudoautosomal X-linked gene that undergoes X inactivation. Mamm Genome. 1996;7(6):420–4. doi: 10.1007/s003359900125. [DOI] [PubMed] [Google Scholar]
- [92].Kent L, Emerton J, Bhadravathi V, et al. X linked ichthyosis (steroid sulfatase deficiency) is associated with increased risk of attention deficit hyperactivity disorder, autism, and social communication deficits. J Med Genet. 2008;45(8):519–24. doi: 10.1136/jmg.2008.057729. [DOI] [PubMed] [Google Scholar]
- [93].Brookes KJ, Hawi Z, Kirley A, Barry E, Gill M, Kent L. Association of the steroid sulfatase (STS) gene with attention deficit hyperactivity disorder. Am J Med Genet B. 2008;147B:1531–5. doi: 10.1002/ajmg.b.30873. [DOI] [PubMed] [Google Scholar]
- [94].Li XM, Alperin ES, Salido E, Gong Y, yen P, Shapiro LJ. Characterization of the promoter region of human steroid sulphatase: a gene which escapes X-inactivation. Somat Cell Mol Genet. 1996;22:105–17. doi: 10.1007/BF02369901. [DOI] [PubMed] [Google Scholar]
- [95].Yen PH, Marsh B, Allen E, et al. The human X-linked steroid sulphatase gene and a Y-encoded pseudogene: evidence for an inversion of the Y chromosome during primate evolution. Cell. 1988;55:1123–35. doi: 10.1016/0092-8674(88)90257-7. [DOI] [PubMed] [Google Scholar]
- [96].Cuevas-Covarrubias SA, Juarez-Oropeza MA, Miranda-Zamora R, Diaz-Zagoya JC. Comparative analysis of human steroid sulfatase activity in prepubertal and postpubertal males and females. Biochem Mol Biol Int. 1993;30:691–5. [PubMed] [Google Scholar]
- [97].Migeon BR, Shapiro LJ, Norum RA, Mohandas T, Axelman J, Dabora RL. Differential expression of steroid sulphatase locus on active and inactive human X chromosome. Nature. 1982;299:838–40. doi: 10.1038/299838a0. [DOI] [PubMed] [Google Scholar]
- [98].Xu J, Burgoyne PS, Arnold AP. Sex differences in sex chromosome gene expression in mouse brain. Hum Mol Genet. 2002;11(12):1409–19. doi: 10.1093/hmg/11.12.1409. [DOI] [PubMed] [Google Scholar]
- [99].Xu J, Deng X, Watkins R, Disteche CM. Sex-specific differences in expression of histone demethylases Utx and Uty in mouse brain and neurons. J Neurosci. 2008;28:4521–7. doi: 10.1523/JNEUROSCI.5382-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Xu J, Watkins R, Arnold AP. Sexually dimorphic expression of the X-linked gene Eif2s3x mRNA but not protein in mouse brain. Gene Expr Patterns. 2006;6(2):146–55. doi: 10.1016/j.modgep.2005.06.011. [DOI] [PubMed] [Google Scholar]
- [101].Blanco P, Sargent CA, Boucher CA, Mitchell M, Affara NA. Conservation of PCDHX in mammals; expression of human X/Y genes predominantly in brain. Mamm Genome. 2000;11:906–14. doi: 10.1007/s003350010177. [DOI] [PubMed] [Google Scholar]
- [102].Durand CM. Expression and genetic variability of PCDH11Y, a gene specific to Homo sapiens and candidate for susceptibility to psychiatric disorders. Am J Med Genet B. 2006;141B(1):67–70. doi: 10.1002/ajmg.b.30229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Yang X, Chen MW, Terry S, et al. Complex regulation of human androgen receptor expression by Wnt signaling in prostate cancer cells. Oncogene. 2006;25:3436–44. doi: 10.1038/sj.onc.1209366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Biederman J. Attention-deficit/hyperactivity disorder: a selective overview. Biol Psychiatry. 2005;57(11):1215–20. doi: 10.1016/j.biopsych.2004.10.020. [DOI] [PubMed] [Google Scholar]
- [105].Grossman LS, Harrow M, Rosen C, Faull R, Strauss GP. Sex differences in schizophrenia and other psychotic disorders: a 20-year longitudinal study of psychosis and recovery. Compr Psychiatry. 2008;49(6):523–9. doi: 10.1016/j.comppsych.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Steiner M, Dunn E, Born L. Hormones and mood: from menarche to menopause and beyond. J Affect Disord. 2003;74(1):67–83. doi: 10.1016/s0165-0327(02)00432-9. [DOI] [PubMed] [Google Scholar]
- [107].Götz MJ, Johnstone EC, Ratcliffe SG. Criminality and antisocial behaviour in unselected men with sex chromosome abnormalities. Psychol Med. 1999;29(4):953–62. doi: 10.1017/s0033291799008594. [DOI] [PubMed] [Google Scholar]
- [108].Mors O, Mortensen PB, Ewald H. No evidence of increased risk for schizophrenia or bipolar affective disorders in persons with aneuploidies of the sex chromosomes. Psychol Med. 2001;31:425–30. doi: 10.1017/s0033291701003476. [DOI] [PubMed] [Google Scholar]
- [109].Yoshitsugu K, Meerabux JMA, Asai K, Yoshikawa T. Fine mapping of an isodicentric Y chromosomal breakpoint from a schizophrenic patient. Am J Med Genet B. 2003;116B:27–31. doi: 10.1002/ajmg.b.10794. [DOI] [PubMed] [Google Scholar]
- [110].Mulligan A, Gill M, Fitzgerald M. A case of ADHD and a major Y chromosome abnormality. J Atten Disord. 2008;12(1):103–5. doi: 10.1177/1087054707311220. [DOI] [PubMed] [Google Scholar]
- [111].Rosser ZH, Zerjal T, Hurles ME, et al. Y-chromosomal diversity in Europe is clinal and influenced primarily by geography, rather than by language. Am J Hum Genet. 2000;67(6):1526–43. doi: 10.1086/316890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Karafet TM, Mendez FL, Meilerman MB, Underhill PA, Zegura SL, Hammer MF. New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res. 2008;18(5):830–8. doi: 10.1101/gr.7172008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Kittles RA, Long JC, Bergen AW, et al. Cladistic association analysis of Y chromosome effects on alcohol dependence and related personality traits. Proc Natl Acad Sci USA. 1999;96(7):4204–9. doi: 10.1073/pnas.96.7.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Shah SS, Ayub Q, Firasat S, Kaiser F, Mehdi SQ. Y haplogroups and aggressive behavior in a Pakistani ethnic group. Aggress Behav. 2009;35(1):68–74. doi: 10.1002/ab.20281. [DOI] [PubMed] [Google Scholar]
- [115].Jamain S, Quach H, Quintana-Murci L, et al. Y chromosome haplogroups in autistic subjects. Mol Psychiatry. 2002;7(2):217–9. doi: 10.1038/sj.mp.4000968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Grant SG. Synapse signalling complexes and networks: machines underlying cognition. Bioessays. 2003;25(12):1229–35. doi: 10.1002/bies.10381. [DOI] [PubMed] [Google Scholar]
- [117].Wilson ND, Ross LJN, Crow TJ, Volpi EV. PCDH11 is X/Y homologous in Homo sapiens but not in Gorilla gorilla and Pan troglodytes. Cytogenet Genome Res. 2006;114(2):137–9. doi: 10.1159/000093329. [DOI] [PubMed] [Google Scholar]
- [118].Williams NA, Close JP, Giouzeli M, Crow TJ. Accelerated evolution of Protocadherin11X/Y: a candidate gene pair for cerebral asymmetry and language. Am J Med Genet B. 2006;141B:623–33. doi: 10.1002/ajmg.b.30357. [DOI] [PubMed] [Google Scholar]
- [119].Giouzeli M, Williams NA, Lonie LJ, DeLisi LE, Crow TJ. ProtocadherinX/Y, a candidate gene-pair for schizophrenia and schizoaffective disorder: a DHPLC investigation of genomic sequence. Am J Med Genet B. 2004;129B:1–9. doi: 10.1002/ajmg.b.30036. [DOI] [PubMed] [Google Scholar]
- [120].Hill SK, Harris MS, Herbener ES, Pavuluri M, Sweeney JA. Neurocognitive allied phenotypes for schizophrenia and bipolar disorder. Schizophr Bull. 2008;34(4):743–59. doi: 10.1093/schbul/sbn027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Ylisaukko-oja T, Rehnstrom K, Auranen M, et al. Analysis of four neuroligin genes as candidates for autism. Eur J Hum Genet. 2005;13:1285–92. doi: 10.1038/sj.ejhg.5201474. [DOI] [PubMed] [Google Scholar]
- [122].Jamain S, Quach H, Betancur C, et al. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet. 2003;34:27–9. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Laumonnier F, Bonnet-Brilhault F, Gomot M, et al. X-linked mental retardation and autism are associated with a mutation in the NLGN4 gene, a member of the neuroligin family. Am J Hum Genet. 2004;74:552–7. doi: 10.1086/382137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Gauthier J, Bonnel A, St-Onge J, et al. NLGN3/NLGN4 gene mutations are not responsible for autism in the Quebec population. Am J Med Genet B. 2005;132B:74–5. doi: 10.1002/ajmg.b.30066. [DOI] [PubMed] [Google Scholar]
- [125].Vincent JB, Kolozsvari D, Roberts WS, Bolton PF, Gurling HMD, Scherer SW. mutation screening of X-chromosomal neuroligin genes: no mutations in 196 autism probands. Am J Med Genet B. 2004;129B:82–4. doi: 10.1002/ajmg.b.30069. [DOI] [PubMed] [Google Scholar]
- [126].Talebizadeh Z, Lam DY, Theodoro MF, Bittel DC, Lushington GH, Butler MG. Novel splice isoforms for NLGN3 and NLGN4 with possible implications in autism. J Med Genet. 2006;43:e21. doi: 10.1136/jmg.2005.036897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Wermter AK, Kamp-Becker I, Strauch K, Schulte-Korne G, Remschmidt H. No evidence for involvement of genetic variants in the X-linked neuroligin genes NLGN3 and NLGN4X in probands with autism spectrum disorder on high functioning level. Am J Med Genet B. 2008;147B:535–7. doi: 10.1002/ajmg.b.30618. [DOI] [PubMed] [Google Scholar]
- [128].Yan J, Feng J, Schroer R, et al. Analysis of the neuroligin 4Y gene in patients with autism. Psychiatr Genet. 2008;18(4):204–7. doi: 10.1097/YPG.0b013e3282fb7fe6. [DOI] [PubMed] [Google Scholar]