Abstract
Tuberous sclerosis complex (TSC) is an autosomal dominant neurocutaneous syndrome caused by mutations in TSC1 and TSC2. However, 10 to 15% TSC patients have no mutation identified with conventional molecular diagnostic studies. We used the ultra-deep pyrosequencing technique of 454 Sequencing to search for mosaicism in 38 TSC patients who had no TSC1 or TSC2 mutation identified by conventional methods. Two TSC2 mutations were identified, each at 5.3% read frequency in different patients, consistent with mosaicism. Both mosaic mutations were confirmed by several methods. Five of 38 samples were found to have heterozygous non-mosaic mutations, which had been missed in earlier analyses. Several other possible low frequency mosaic mutations were identified by deep sequencing, but were discarded as artifacts by secondary studies. The low frequency of detection of mosaic mutations, 2 (6%) of 33, suggests that the majority of TSC patients who have no mutation identified are not due to mosaicism, but rather other causes, which remain to be determined. These findings indicate the ability of deep sequencing, coupled with secondary confirmatory analyses, to detect low frequency mosaic mutations.
Keywords: TSC, mosaicism, 454 Sequencing, TSC1, TSC2
Introduction
Tuberous sclerosis complex (TSC) is an autosomal dominant neurocutaneous syndrome of high penetrance, characterized by a highly variable phenotype and the development of multiple hamartomas at various sites throughout the body (Crino et al. 2006; Gomez et al. 1999). Approximately 60 to 70% of TSC cases are sporadic, reflecting a high spontaneous mutation rate in the two genes, TSC1 and TSC2 (European Tuberous Sclerosis Consortium 1993; Sampson et al. 1989; van Slegtenhorst et al. 1997).
Comprehensive mutation detection studies have led to identification of mutations in 70 - 90% of TSC patients (Au et al. 2007; Dabora et al. 2001; Jones et al. 1999; Niida et al. 1999; Sancak et al. 2005) (http://chromium.liacs.nl/LOVD2/TSC/home.php). More than 1,500 mutations and more than 800 unique mutations have been identified in TSC1 and TSC2 combined. However, 10 to 15% of TSC patients have no mutation identified (NMI), despite a thorough molecular diagnostic assessment, including analysis for large genomic deletions. These NMI TSC subjects generally have milder clinical features of TSC than patients with identified TSC1 or TSC2 mutations (Dabora et al. 2001; Sancak et al. 2005).
Although a third gene for TSC is a possibility, there is no discrete evidence for this at this time. On the other hand, both somatic (generalized) and germline (confined gonadal) mosaicism for TSC1 and TSC2 mutations have been described in many TSC patients and their parents, respectively (Kozlowski et al. 2007; Kwiatkowska et al. 1999; Rose, et al. 1999; Sampson et al. 1997; Verhoef et al. 1999; Jones et al. 2001; Yates et al. 1997). In addition, mosaicism is known to occur at a high rate in several other tumor suppressor gene and other syndromes (Aretz et al. 2007; Kluwe and Mautner 1998; Leuer et al. 2001; Lietman et al. 2005; Maertens et al. 2006a; Vandenbroucke et al. 2004). Thus, mosaicism is a credible explanation for the failure to detect mutations in NMI patients.
Many different methods have been developed to identify mosaic mutations (Aretz et al. 2007; Emmerson et al. 2003; Janne et al. 2006; Lietman et al. 2005; Maertens et al. 2006b; Newton et al. 1989). A newer approach to the identification of mosaic mutations in DNA samples is to perform sequencing at the single molecule level, analyzing a large enough sample of individual amplicons/molecules that mosaicism can be readily detected (Mardis 2008; Margulies et al. 2005; Shendure et al. 2005; Smith et al. 2008; Thomas et al. 2006). Here we report the results of this approach in the analysis of TSC NMI patients, searching for mosaicism. We used the ultra-deep pyrosequencing technique of 454 Sequencing on the Genome Sequencer FLX system (Roche) to identify mosaicism mutations in two of 33 TSC patients. This method proved to be robust and sensitive. However, this low rate of mosaic mutation detection suggests that most TSC NMI patients are not explained by mosaicism.
Materials and Methods
TSC patients
All TSC patients who participated in this study provided informed consent for this research, and the study was approved by the Partners Human Research Committee, the Institutional Review Board for the Partners Hospitals.
Thirty-eight TSC patients, all of whom were sporadic cases without evidence of parental TSC, provided blood samples for DNA extraction, which was performed by standard means. All of the patients met standard diagnostic criteria for definite TSC (Roach et al. 1998), and had been previously studied by preceding DHPLC or direct exon sequencing in different diagnostic labs to identify mutations in TSC1 and TSC2, with no mutation identified.
Clinical information on the major manifestations of TSC was collected for these patients. This consisted of information on CNS involvement (seizure history, developmental history, subependymal nodules, subependymal giant cell astrocytomas, tubers, retina); skin involvement (white spots, facial angiofibroma, forehead plaque, shagreen patch, ungula fibroma, confetti macules); renal angiomyolipoma and cysts, and lymphangioleiomyomatosis; and cardiac rhabdomyoma. Each of these four was considered a different organ system, to assess the number of organ systems involved in an individual patient. Renal angiomyolipoma and lymphangioleiomyomatosis were considered one organ system because of evidence that they are closely related, and that the abnormal cells travel via the lymphatics and bloodstream from one organ to the other (Crino et al. 2006). Six of these 40 patients, all females, had lymphangioleiomyomatosis.
Examination for large genomic deletions
All DNA samples were examined for genomic deletions in TSC1 and TSC2 using multiplex ligation-dependent probe amplification including probe sets for each of the exons of TSC1 and TSC2, as described previously (Kozlowski et al. 2007).
Exon amplification and deep sequencing
The 62 TSC1 and TSC2 coding exons were amplified using 65 specially designed oligonucleotide primers (Simen et al. 2009). The composite primers each contained a 15–28 bp target-specific sequence at their 3′-end; and a common 19 bp region that is used in subsequent clonal amplification and sequencing reactions at their 5′-end. Amplicons ranged in size from 135bp to 393bp, with an average and median size of 254bp and 237bp, respectively. PCR primers were backed up from exon boundaries by a minimum of 10nt on the 5′ flanking side and a minimum of 6nt on the 3 ′ flanking side for all but a few exons, in the latter case due to primer design constraints.
For each patient sample, PCR was performed on 10–25 ng of genomic DNA using the FastStart High Fidelity PCR System (Roche) and standard thermocycling conditions on a PTC-200 thermocycler. PCR conditions were individualized for each amplicon, and the most common was: 5 min denaturation at 96°C, followed by 5 cycles of denaturation for 30 sec at 94°C, annealing for 30 sec at 55°C and extension for 45 sec at 72°C, 30 cycles of denaturation for 30 sec at 94°C, annealing for 30 sec at 60°C and extension for 45 sec at 72°C, and final extension for 10 min at 72°C. Amplicon products were assessed by agarose gel electrophoresis, purified using AMPure SPRI beads (Agencourt Bioscience Corporation, Beverly, US), quantified by measurement on a Nanodrop instrument (ThermoScientific), and then pooled at an equimolar ratio for each individual patient for sequencing.
Single PCR amplicon molecules were captured on individual 28 μm beads within an oil-water emulsion to enable clonal amplification in a second PCR process with universal primers that yields about 107 copies of the input DNA molecule. The emulsion was then disrupted, the beads were isolated, and loaded into picotiter plates containing wells of size 44 μm. Sequencing reactions are performed by synthesis using pyrosequencing (Margulies et al. 2005). This process, ultra-deep pyrosequencing (UDPS) technique of 454 Sequencing on the Genome Sequencer FLX system (Roche Applied Sciences, Indianapolis), was performed at the 454 facility in Branford, CT. To enhance sample throughput and reduce costs, individual patient samples were analyzed on picotiter plates in sets of 8, using a gasket device to provide separation among samples and wells.
The ultra-deep sequence data was analyzed using GS Amplicon Variant Analysis (AVA) Software to identify sequence variants in TSC1 and TSC2 (Simen et al. 2009). Amplicon nucleotide sequence reads were aligned to the Human Mar. 2006 (hg18) assembly genomic sequence of TSC1 and TSC2. The flowgram signals were used in concert with each read’s base-called nucleotides to facilitate alignment accuracy. Reads from both orientations were combined into a single alignment, and primer regions were automatically trimmed to avoid artifacts from the nucleotide content of the synthesized primers. The AVA software identifies all nucleotide variants, and provides read counts and frequencies. Individual flow grams were reviewed to examine and confirm all variant calls made by the software.
Allele-specific PCR
Allele-specific PCR was used to confirm low frequency variants. The allele-specific primer was designed to have its 3′ end nucleotide sit at the variant nucleotide, and to have an additional 3′ subterminal mismatch to enhance specificity of amplification. The primer sequences used can be supplied on request. Different annealing temperatures during PCR were tested for each variant to obtain maximum discrimination between wild type and variant sequences.
SNaPshot Analysis
SNaPshot analysis was used to both confirm and quantify the proportion of the mutation in patients with suspected mosaicism, following the manufacturer’s protocol (ABI Prisms SNaPshot TM Multiplex Kit; Applied Biosystems). SNaPshot is a single nucleotide extension sequencing method in which a single dye-labeled dideoxy nucleotide is added to primers localized adjacent to a site of suspected variation (Kaminsky et al. 2005). The products of the primer extension reaction were analyzed on an ABI 3100 sequencer (Applied Biosystems); and the proportion of normal and mutant DNA were quantified using GeneMapper version 3.0 (Applied Biosystems). In this analysis, small peaks are seen for variant nucleotides in many cases due to spontaneous base misincorporation. However, comparison with control samples permits discrimination of bona fide variant frequency down to 5% or less (van Oers et al. 2005 Lurkin et al. 2010). The degree of mosaicism, expressed as percentage of mutant to total DNA, was calculated as follows: the peak areas of the mutant (M) and wild (W) DNA were determined, and used in the formula: M/(M+W) × 100%. All experiments were performed in duplicate.
SURVEYOR digestion and fragment analysis by DHPLC
SURVEYOR nuclease recognizes mismatches present in heteroduplex DNA and cleaves both strands on the 3′ side of the mismatch distortion. This method was used to confirm deep sequencing findings as described previously (Janne et al. 2006). Briefly, DNA amplicons were treated with the SURVEYOR nuclease, purified, and then analyzed by high-performance liquid chromatography (HPLC) on the Transgenomic WAVE Nucleic Acid High Sensitivity Fragment Analysis System (WAVE HS system; Transgenomic, Omaha, NE).
Mass spectrometry analysis
Variant allele frequency was also determined using MALDI-TOF (Matrix-assisted laser desorption ionization - time of flight) mass spectrometry on the Sequenom (San Diego, CA) platform. Primers were designed using MassARRAY Assay Design version 3.1, and amplicons were subject to single base extension sequencing using the iPLEX chemistry (Sequenom), followed by mass spectrometry, and interpretation using Typer 4.0 software. Spectrometry profiles were imported into ImageJ v1.32j (W. Rasband, NIH) for quantification of variant allele frequency, using the same formula described above for SNaPshot.
Statistical analyses
Statistical comparisons were made using the Mann Whitney test for unpaired observations.
RESULTS
Initial results of deep sequencing analysis; identification of non-mosaic mutations
Thirty-eight sporadic TSC patient blood DNA samples were analyzed by the ultra-deep pyrosequencing technique of 454 Sequencing on the Genome Sequencer FLX system (Roche) (see Methods for details) to search for mosaic mutations. Sixty-five amplicons were used to cover the 62 coding exons of TSC1 and TSC2, with median and mean amplicon size of 237 and 254 bp, respectively. Median and mean read numbers, obtained using a gasket to enable analysis of 8 samples per plate, were 610 and 664, respectively. 95.1% of amplicons had read numbers > 200 while 98.5% had read numbers greater than 100. 73% of the nucleotides in TSC1 and TSC2, in total, were covered by bidirectional sequence reads.
The 38 samples had 0 – 5 (median 0) heterozygous sequence variants detected in TSC1 and 0 – 7 (median 1) heterozygous sequence variants detected in TSC2. This included all of the common SNPs previously detected in each of these genes (http://chromium.liacs.nl/LOVD2/TSC/home.php).
Five of the 38 samples analyzed had readily observable mutations at read frequencies from 41 to 53% (Table 1). These mutations were verified by standard dideoxy sequencing analysis of fresh PCR amplicons. None of these mutations appeared to be mosaic based on review of the standard sequencing traces, consistent with the observed variant read frequencies.
Table 1.
Heterozygous (non-mosaic) mutations identified by ultra-deep 454 pyro-sequencing.
| Exon | Mutation | Protein | R (reported)or NR (not reported) | Frequency of reads (%) | Confirmed by standard sequencing |
|---|---|---|---|---|---|
| TSC1-E23 | 3184C>T | R1062W | NR | 45 | Yes |
| TSC2-E4 | 451A>T | R151X | NR | 45 | Yes |
| TSC2-E19 | 2210T>C | L737P | R | 43 | Yes |
| TSC2-E29 | 3520delC | Frameshift | NR | 51 | Yes |
| TSC2-E 37 | 4911_4913delGAA | K1638del | R | 53 | Yes |
Four of these 5 mutations were clearly pathogenic, having been identified previously in TSC patients (two cases), or having a chain-terminating effect (two cases). One variant caused a missense change (R1062W) in TSC1 which has not been reported previously and may not be pathogenic, but is a non-conservative amino acid change. One additional patient analyzed was homozygous for the rare allele of 7 different TSC2 SNPs, including a SNP for which the rare allele frequency is less than 1%, suggesting the possibility of an unusual family structure (inbreeding) or a gene conversion event in TSC2 that could not be detected by MLPA. All DNA samples were analyzed by MLPA for each coding exon of TSC1 and TSC2 (Kozlowski, et al., 2007), and had no evidence for a deletion.
Low frequency sequence variants and detection of mosaic mutations
Many sequence variants were detected in this analysis at a frequency of < 10% using the AVA software. We established the following criteria for selecting variants with high likelihood of being true variants: 1) variants detected in more than one sample at low frequency were excluded under the assumption that they arose as an artifact of the PCR or other process step; 2) variants detected in <5 sequencing reads were excluded; 3) manual review of variants was performed and any reads determined to be of poor quality were excluded. Thirty-one variants remained, which were seen at a frequency of 0.5 to 6% (Table 2). No variants were detected at a read frequency of 6 – 30%. Subsequent analysis focused on the 11 sequence variants that were seen at a read frequency of > 2%.
Table 2.
Read frequency of potential mosaic mutations.
| Read frequency | 0–1% | 1–2% | 2–3% | 3–4% | 4–5% | 5–6% |
|---|---|---|---|---|---|---|
| Number of variants | 3 | 17 | 7 | 2 | 0 | 2 |
Confirmation of Potential Mosaic Mutations
Two mosaic mutations, each at a read frequency of 5.3% (Table 3), were initially examined by allele-specific PCR. However, the distinction between the mosaic samples and control samples was slight, and we performed additional studies.
Table 3.
TSC2 mosaic mutations detected by 454 ultra-deep pyrosequencing
| Exon | Mutation | Protein | Number of reads over total reads | Frequency of reads % | |
|---|---|---|---|---|---|
| Forward | Reverse | ||||
| 14 | 1444-1G>A | splice | 36/584 | 20/464 | 5.34 |
| 40 | 5228G>A | R1743Q | 9/158 | 12/235 | 5.34 |
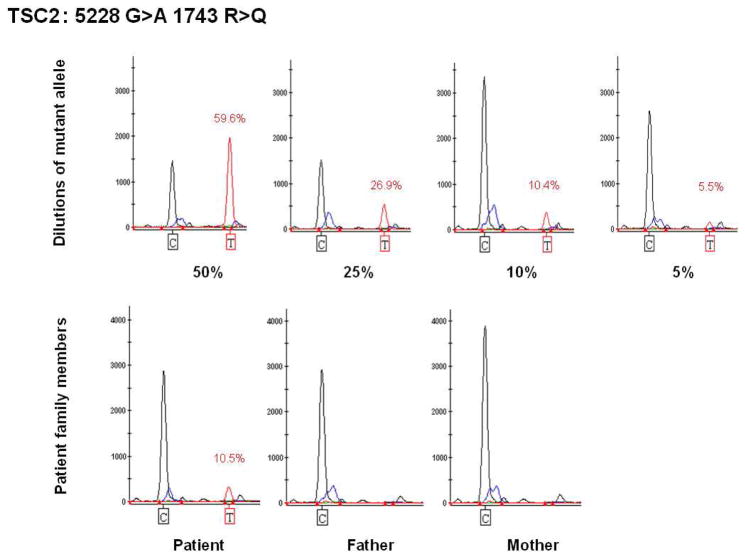
Both mutations were confirmed by SNaPshot single base extension sequencing (Figures 1 and 2). For the 5228G>A mutation, we performed a dilution experiment using serial mixtures of heterozygote DNA (provided by Drs. Au and Northrup) and a control DNA sample. The primer giving the clearest result was in the reverse direction. The observed values of the decreasing T-signal were close to expected values in the mixing experiment (Figure 1). Multiple replicate analyses of the patient’s DNA sample gave a T signal value of 10.5%. In contrast, parental leukocyte DNA samples showed no T signal when analyzed in parallel (Figure 1). For the 1444-1G>A mutation, we could not perform a mixing study due to lack of availability of a heterozygote patient sample. However, when analyzed by SNaPshot, an A (mutant) signal was detected in both patient and control DNA (due to base misincorporation), but was significantly higher in the patient (Figure 2).
Figure 1.
Quantitative analysis of TSC2 5228G>A mutation using SNaPshot. Genemapper images are shown. Top row, analysis of a dilution series using a heterozygote patient with the 5228G>A mutation and a control sample. The expected percent of the mutant allele obtained from mixing of heterozygote and control samples is shown beneath each image, while the percent quantified by analysis of peak ratios of the mutant G and wild type A signals is shown in red. Bottom row, analysis of the mosaic patient and his parents. Note that the mutant T allele is detected only in the patient, but not in his parents. Other peaks seen are non-specific products, which do not vary consistently according to the DNA sample analyzed.
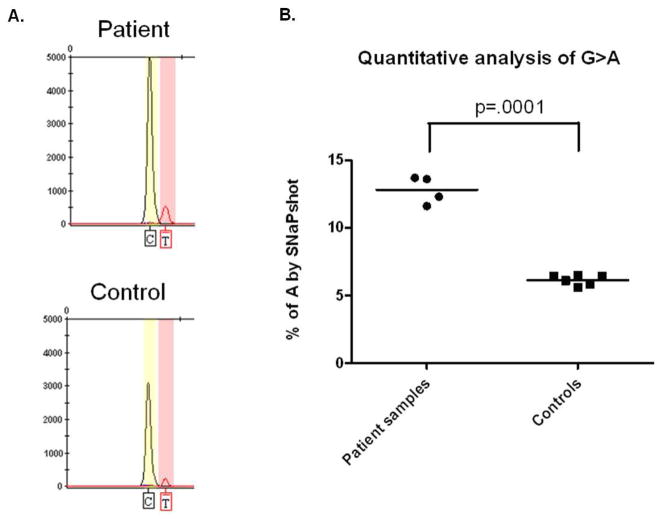
Figure 2.
Quantitative analysis of TSC2 1444-1G>A mutation using SNaPshot. A. Genemapper image shows that extension products corresponding to the mutant A allele are detected in both patient and control blood DNA (reverse primers being used here). B. The frequency of the mutant allele in the patient was significantly higher than in controls in replicate analyses (unpaired Mann Whitney test).
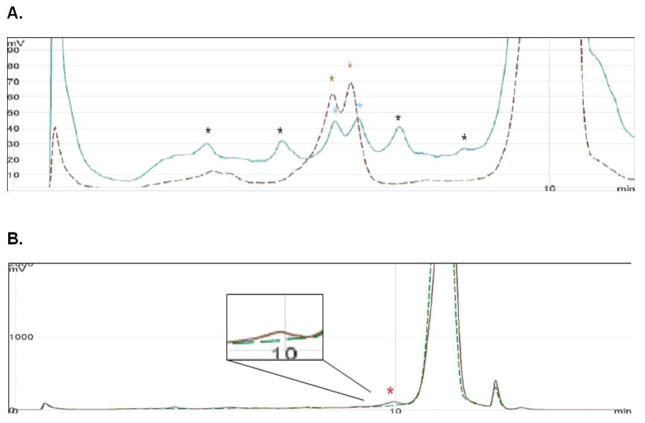
DHPLC following SURVEYOR digestion of fragments was also used to confirm the presence of a mutation in these samples. DHPLC elution curves for two exon 40 amplicons from two different DNA samples are shown in Figure 3A. The brown dashed line is derived from the 5228G>A heterozygote sample. Two brown stars indicate the fragments generated by SURVEYOR digestion of this sample at the site of mismatch. The blue line is derived from the patient who appears to be mosaic for the same 5228G>A mutation. Two blue stars indicate the presence of fragments whose size is similar to that from the heterozygote sample, but peak heights are much lower than that from the heterozygote sample. However, other peaks are also seen which are due to the occurrence of a polymorphism in this exon in this patient. For the 1444-1G>A mosaic sample, there was a weak signal at a mismatch point in the patient’s amplicon, but no signal was detected in control (Fig. 3B).
Figure 3.
SURVEYOR – HPLC analysis to confirm TSC2 mosaic mutations.
A. HPLC elution profile for TSC2 exon 40 amplicons from two DNA samples. The brown dashed line is derived from the 5228G>A heterozygote sample. Two brown stars indicate the fragments generated by digestion. The blue line is derived from the mosaic individual. Two blue stars indicate the presence of fragments whose size is similar to that from the heterozygote sample. Other peaks are due to a polymorphism in the mosaic individual.
B. DHPLC elution curve for TSC2 exon 14 amplicons from two DNA samples. The brown line is derived from the individual with the 1444-1G>A mosaic mutation, and the green dashed line is from a control sample. The red star indicates the fragment generated by SURVEYOR digestion of the mosaic sample, which is not seen in the control sample. This is shown at higher magnification in the inset.
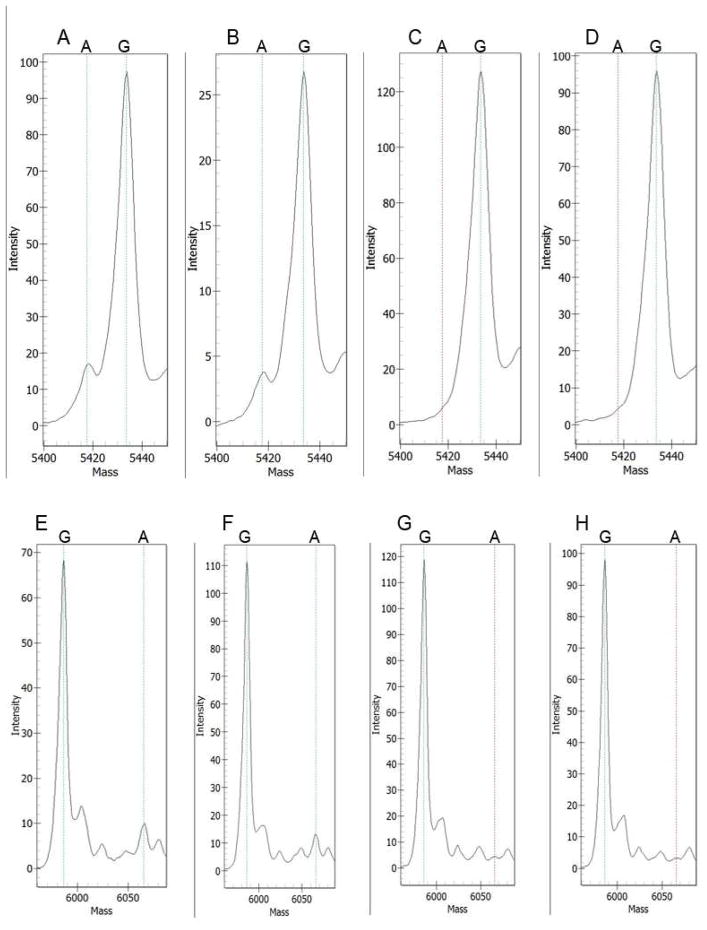
Mass spectrometry on the Sequenom platform was also used to confirm the presence of mosaicism in these two samples. Following single base extension sequencing, variant extension products determined by mass spectrometry represented 8.7% (range 7.7 – 9.8%, n = 3) of the total extension product for the 5228G>A variant; and 5.6% (range 4.6 – 6.8%, n = 3) of the total extension product for the 1444-1G>A variant (Figure 4). No (< 1%) extension product was seen for any of the control samples (n = 3 for each) by this method.
Figure 4.
Mass spectrometry analysis of mosaic mutations in TSC2. Mass spectrometry spectra are shown for two sets of single nucleotide sequencing reactions. A-D. Analysis of the TSC2 5228G>A variant; A, B are duplicate samples from the mosaic DNA sample; C, D are control samples from the parents of the mosaic individual. E-H. Analysis of the TSC2 1444-1G>A variant; E, F are duplicate samples from the mosaic DNA sample; G, H are control samples. Note the presence of a distinct peak for the variant A nucleotide only in the mosaic samples, A, B, E, and F. The mass shifts are different in the two reactions because the extension oligonucleotides were in the opposite orientation for these reactions.
Nine other sequence variants were seen at a read frequency of 2 – 4% by deep sequencing, of which two were indel mutations and 7 were point mutations (Table 2). All nine variants were evaluated by SNaPshot sequencing, and there was no evidence of the presence of these mutations in the original DNA samples by this method. In each case, replicate samples gave variant base incorporation signals that differed from control samples by < 1% (data not shown). Therefore, they did not appear to be bona fide mutations, but rather artifacts of the deep sequencing process.
Genotype – phenotype comparisons
Clinical information on the TSC manifestations in these 38 patients was used for comparison between mutation detection status and clinical phenotype. Clinical data was available for brain involvement, skin involvement, renal/lung involvement, and cardiac rhabdomyoma. Each of these four was considered a different organ system, to assess the number of organ systems involved in an individual patient. (Renal and lung involvement were combined because of evidence that there is a common origin for these pathologic processes (Crino et al. 2006).) There was a wide range of severity of manifestations among these patients, as commonly seen in TSC, including 8 patients with 4 organ systems involved, and 2 with only a single organ system involved. The clinical features were overall milder than ordinary TSC, as noted previously in NMI patients. We compared the number of organ systems involved in the 5 patients with heterozygote mutations identified here, the two patients with mosaic mutations, and the 31 patients who were still NMI after this analysis. The 5 patients with heterozygote mutations had more organ systems involved (median 4, mean 3.8) than the 31 patients with persistent NMI status (median 2, mean 2.5) (p=0.01, Mann Whitney test). The two patients with mosaic mutations had 2 and 3 different organ systems involved, and formal statistical comparison to the other groups could not be performed due to the limited sample size.
Discussion
In this work, we performed deep sequencing on 38 sporadic TSC patient blood DNA samples to search for mosaic mutations. Five (4 definite and 1 probable) mutations were identified in patients at heterozygote frequency. These 5 mutations were easily confirmed by standard sequencing analysis, implicating some kind of laboratory error or sample mix-up as the reason for their lack of detection in analyses performed prior to this study.
The frequency of mosaicism detection in these NMI patients (2 of 33, 6.1%) is lower than we had hypothesized. However, this study has some limitations. The depth of coverage, or read number, was not uniform for all TSC1 and TSC2 exons. This is a general problem when pooling amplicons for deep sequencing, as noted by others (Rohlin, et al., 2009). We had > 200 reads for 95.1% of amplicons, > 400 reads for 78.7% of amplicons, and a median read number of 610. For a mosaic mutation occurring at 5% allele frequency, the probability of detection of ≥ 5 reads would be 97.4% for 200 reads, and ≥ 99.9% for > 400 reads. Thus, our overall power for detection of mosaicism at 5% allele frequency was > 95%. At 2.5% allele frequency however, the probability of detection of ≥ 5 reads would be 61.6% for 200 reads, and ≥ 97.3% for > 400 reads. Thus, our overall power for detection of mosaicism at 2.5% allele frequency was > 90%. Clearly the power of detection for even lower frequency mosaicism (≤ 2% allele frequency) is relatively low with this number of reads. Deeper coverage would be more sensitive.
Although we could have pursued confirmation of the sequence variants detected by repeat deep sequencing, we felt it important to confirm the findings by alternative methodology. SNaPshot sequencing was the simplest method for confirmation in our experience, though it has limitations, and cannot reliably detect mosaicism at the level of 2% or less in our experience. However, as shown here and previously reported (van Oers et al. 2005 Lurkin et al. 2010), SNaPshot is capable of detection of mosaicism as low as 2.5% mixture of the variant allele in some cases, and reliably detects mosaicism down to 5%. Single base extension sequencing with mass spectrometry analysis appeared to be more sensitive, though it is clearly more costly and difficult to implement in the routine diagnostic laboratory. Confirmation of mosaicism at 2% or less is difficult by any common technique. As deep sequencing costs continue to fall, and equipment becomes more widely available, deep sequencing at very high read depths, or used in replicate manner for confirmation of initial findings may well become the best approach for both detection and confirmation of low level mosaicism.
Nine sequence variants detected by deep sequencing, seen at 2 – 4% read frequency, could not be confirmed by SNapShot analysis. Despite our use of a high fidelity Taq Polymerase during the PCR reactions for ultradeep sequencing (FastStart High Fidelity Taq Polymerase (Roche)), we suspect that they were due to spontaneous base misincorporation events occurring early during PCR amplification.
Since our rate of detection of bona fide mosaicism was quite low (2 of 33, 6.1%), this suggests that mosaicism is not the only mechanism which explains lack of molecular findings in TSC NMI patients. Mosaicism for TSC2 mutations has been found in as many as 27% of index patients with combined TSC2-polycystic kidney disease syndrome, due to genomic deletion of parts of both TSC2 and PKD1 (Sampson et al. 1997). Other studies have also shown that mosaicism appears generally to be more common with genomic deletion mutations than with smaller indels and point mutations in TSC2 (Cheadle et al. 2000; Kozlowski et al. 2007; Verhoef et al. 1999; Jones et al. 1999). Since genomic deletions in TSC1 or TSC2 are recognized in about 5–10% of all TSC patients, it is very unlikely that low level mosaic genomic deletions account for the 10–15% of TSC patients that are NMI.
TSC patients with mosaicism have been found to have less severe disease than those with full mutations, consistent with a dosage effect (Sampson et al. 1997). However, it has also been noted that TSC NMI patients have a milder phenotype than those with TSC2 mutations, consistent with mosaicism as a potential explanation in the NMI patients (Dabora et al. 2001; Sancak et al. 2005). We have replicated these previous observations here, as the 5 patients with heterozygote frequency mutations had more organ systems involved than the patients with persistent NMI status after this deep sequencing.
There are several possible explanations for the patients with persistent NMI status after deep sequencing. First, there may be some TSC patients who have generalized mosaicism at a level less than 2%, as well as some in whom there is localized somatic mosaicism. Two of the patients studied here had only a single organ system of involvement by TSC (one skin only, one brain only features), and these are good candidates for localized somatic mosaicism. However, this seems very unlikely for patients with 3 or more organ systems involved (44% of the persistent NMI group). A second possibility is that mutations present in introns (and thus undetected) or found in exons but unrecognized as causing splicing effects, account for a significant fraction of the NMI group. However, six of our persistent NMI patients had no variation at all within the coding exons of TSC1 or TSC2, and most sequence variants identified were relatively common SNPs found in many unaffected individuals. Thus, exonic variation causing splicing defects are not a likely explanation. Intronic variation might also cause splicing defects, but this is generally quite rare in human genetic disorders. Third, there is always the possibility of a third TSC gene. Fourth, promoter and enhancer mutations in upstream regions of TSC1 and TSC2 may cause loss of expression, and these regions are not commonly examined.
Mosaic mutations are common in many tumor suppressor gene syndromes in the first affected member of the family (Hall 1988; Kluwe and Mautner 1998). To our knowledge, our work is the first to identify mosaic mutations from blood DNA by deep sequencing for any human genetic disease. Although DHPLC analysis of heteroduplexes can detect mosaicism at a level as low as 6.5% in some cases (Jones et al. 2001), it is not clear how often mutations would be detected by DHPLC when present at this frequency. Moreover, one of the samples in which a mosaic mutation was identified here had been extensively screened by DHPLC analysis in more than one lab prior to the identification of this mosaic mutation.
Thus, overall this work shows that deep sequencing is an effective strategy for mosaicism detection. In addition, it appears to identify heterozygote mutations missed in some cases by conventional diagnostic methods. As deep sequencing costs continue to fall, enabling greater read depth, one can anticipate that this method will become even more effective for this purpose. However, at present, use of deep sequencing as a method for routine clinical diagnostic evaluation of TSC1 and TSC2 in NMI TSC patients cannot be recommended without further development, including significant improvement in the throughput to cost ratio.
Acknowledgments
We thank the TSC patients and families who participated in this study. We thank Paul Au and Hope Northup for the gift of TSC patient DNA. We also thank Edward Szekeres for assistance with AVA software and Michael Egholm of 454 Life Sciences for his support. Supported by NIH NINDS R01 2R37NS031535, and the Tuberous Sclerosis Alliance. BET and PB are employees of 454 Life Sciences. The remaining authors have no conflict of interest with regard to this work.
References
- Aretz S, Stienen D, Friedrichs N, Stemmler S, Uhlhaas S, Rahner N, Propping P, Friedl W. Somatic APC mosaicism: a frequent cause of familial adenomatous polyposis (FAP) Hum Mutat. 2007;28:985–92. doi: 10.1002/humu.20549. [DOI] [PubMed] [Google Scholar]
- Au KS, Williams AT, Roach ES, Batchelor L, Sparagana SP, Delgado MR, Wheless JW, Baumgartner JE, Roa BB, Wilson CM, Smith-Knuppel TK, Cheung MY, Whittemore VH, King TM, Northrup H. Genotype/phenotype correlation in 325 individuals referred for a diagnosis of tuberous sclerosis complex in the United States. Genet Med. 2007;9:88–100. doi: 10.1097/gim.0b013e31803068c7. [DOI] [PubMed] [Google Scholar]
- Cheadle JP, Reeve MP, Sampson JR, Kwiatkowski DJ. Molecular genetic advances in tuberous sclerosis. Hum Genet. 2000;107:97–114. doi: 10.1007/s004390000348. [DOI] [PubMed] [Google Scholar]
- Crino PB, Nathanson KL, Henske EP. The tuberous sclerosis complex. N Engl J Med. 2006;355:1345–56. doi: 10.1056/NEJMra055323. [DOI] [PubMed] [Google Scholar]
- Dabora SL, Jozwiak S, Franz DN, Roberts PS, Nieto A, Chung J, Choy YS, Reeve MP, Thiele E, Egelhoff JC, Kasprzyk-Obara J, Domanska-Pakiela D, Kwiatkowski DJ. Mutational Analysis in a Cohort of 224 Tuberous Sclerosis Patients Indicates Increased Severity of TSC2, Compared with TSC1, Disease in Multiple Organs. Am J Hum Genet. 2001;68:64–80. doi: 10.1086/316951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmerson P, Maynard J, Jones S, Butler R, Sampson JR, Cheadle JP. Characterizing mutations in samples with low-level mosaicism by collection and analysis of DHPLC fractionated heteroduplexes. Hum Mutat. 2003;21:112–5. doi: 10.1002/humu.10159. [DOI] [PubMed] [Google Scholar]
- European Tuberous Sclerosis Consortium. Identification and characterization of the tuberous sclerosis gene on chromosome 16. Cell. 1993;75:1305–1315. doi: 10.1016/0092-8674(93)90618-z. [DOI] [PubMed] [Google Scholar]
- Gomez M, Sampson J, Whittemore V. The tuberous sclerosis complex. 3. Oxford University Press; Oxford, England: 1999. [Google Scholar]
- Hall JG. Review and hypotheses: somatic mosaicism: observations related to clinical genetics. Am J Hum Genet. 1988;43:355–363. [PMC free article] [PubMed] [Google Scholar]
- Janne PA, Borras AM, Kuang Y, Rogers AM, Joshi VA, Liyanage H, Lindeman N, Lee JC, Halmos B, Maher EA, Distel RJ, Meyerson M, Johnson BE. A rapid and sensitive enzymatic method for epidermal growth factor receptor mutation screening. Clin Cancer Res. 2006;12:751–8. doi: 10.1158/1078-0432.CCR-05-2047. [DOI] [PubMed] [Google Scholar]
- Jones AC, Shyamsundar MM, Thomas MW, Maynard J, Idziaszczyk S, Tomkins S, Sampson JR, Cheadle JP. Comprehensive Mutation Analysis of TSC1 and TSC2-and Phenotypic Correlations in 150 Families with Tuberous Sclerosis. Am J Hum Genet. 1999;64:1305–1315. doi: 10.1086/302381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AC, Sampson JR, Cheadle JP. Low level mosaicism detectable by DHPLC but not by direct sequencing. Hum Mutat. 2001;17:233–4. doi: 10.1002/humu.8. [DOI] [PubMed] [Google Scholar]
- Kaminsky ZA, Assadzadeh A, Flanagan J, Petronis A. Single nucleotide extension technology for quantitative site-specific evaluation of metC/C in GC-rich regions. Nucleic Acids Res. 2005;33:e95. doi: 10.1093/nar/gni094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluwe L, Mautner VF. Mosaicism in sporadic neurofibromatosis 2 patients. Hum Mol Genet. 1998;7:2051–5. doi: 10.1093/hmg/7.13.2051. [DOI] [PubMed] [Google Scholar]
- Kozlowski P, Roberts P, Dabora S, Franz D, Bissler J, Northrup H, Au KS, Lazarus R, Domanska-Pakiela D, Kotulska K, Jozwiak S, Kwiatkowski DJ. Identification of 54 large deletions/duplications in TSC1 and TSC2 using MLPA, and genotype-phenotype correlations. Hum Genet. 2007;121:389–400. doi: 10.1007/s00439-006-0308-9. [DOI] [PubMed] [Google Scholar]
- Kwiatkowska J, Wigowska-Sowinska J, Napierala D, Slomski R, Kwiatkowski DJ. Mosaicism in tuberous sclerosis as a potential cause of the failure of molecular diagnosis. N Engl J Med. 1999;340:703–7. doi: 10.1056/NEJM199903043400905. [DOI] [PubMed] [Google Scholar]
- Leuer M, Oldenburg J, Lavergne JM, Ludwig M, Fregin A, Eigel A, Ljung R, Goodeve A, Peake I, Olek K. Somatic mosaicism in hemophilia A: a fairly common event. Am J Hum Genet. 2001;69:75–87. doi: 10.1086/321285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lietman SA, Ding C, Levine MA. A highly sensitive polymerase chain reaction method detects activating mutations of the GNAS gene in peripheral blood cells in McCune-Albright syndrome or isolated fibrous dysplasia. J Bone Joint Surg Am. 2005;87:2489–94. doi: 10.2106/JBJS.E.00160. [DOI] [PubMed] [Google Scholar]
- Lurkin I, Stoehr R, Hurst CD, van Tilborg AA, Knowles MA, Hartmann A, Zwarthoff EC. Two multiplex assays that simultaneously identify 22 possible mutation sites in the KRAS, BRAF, NRAS and PIK3CA genes. PLoS One. 2010;5:e8802. doi: 10.1371/journal.pone.0008802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertens O, Brems H, Vandesompele J, De Raedt T, Heyns I, Rosenbaum T, De Schepper S, De Paepe A, Mortier G, Janssens S, Speleman F, Legius E, Messiaen L. Comprehensive NF1 screening on cultured Schwann cells from neurofibromas. Hum Mutat. 2006;27:1030–40. doi: 10.1002/humu.20389. [DOI] [PubMed] [Google Scholar]
- Maertens O, Legius E, Speleman F, Messiaen L, Vandesompele J. Real-time quantitative allele discrimination assay using 3′ locked nucleic acid primers for detection of low-percentage mosaic mutations. Anal Biochem. 2006;359:144–6. doi: 10.1016/j.ab.2006.07.039. [DOI] [PubMed] [Google Scholar]
- Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24:133–41. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–80. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton CR, Graham A, Heptinstall LE, Powell SJ, Summers C, Kalsheker N, Smith JC, Markham AF. Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS) Nucleic Acids Res. 1989;17:2503–16. doi: 10.1093/nar/17.7.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niida Y, Lawrence-Smith N, Banwell A, Hammer E, Lewis J, Beauchamp RL, Sims K, Ramesh V, Ozelius L. Analysis of both TSC1 and TSC2 for germline mutations in 126 unrelated patients with tuberous sclerosis. Hum Mutat. 1999;14:412–22. doi: 10.1002/(SICI)1098-1004(199911)14:5<412::AID-HUMU7>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Roach ES, Gomez MR, Northrup H. Tuberous sclerosis complex consensus conference: revised clinical diagnostic criteria. J Child Neurol. 1998;13:624–8. doi: 10.1177/088307389801301206. [DOI] [PubMed] [Google Scholar]
- Rohlin A, Wernersson J, Engwall Y, Wiklund L, Bjork J, Nordling M. Parallel sequencing used in detection of mosaic mutations: comparison with four diagnostic DNA screening techniques. Hum Mutat. 2009;30:1012–20. doi: 10.1002/humu.20980. [DOI] [PubMed] [Google Scholar]
- Rose VM, Au KS, Pollom G, Roach ES, Prashner HR, Northrup H. Germ-Line Mosaicism in Tuberous Sclerosis: How Common? Am J Hum Genet. 1999;64:986–992. doi: 10.1086/302322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson JR, Maheshwar MM, Aspinwall R, Thompson P, Cheadle JP, Ravine D, Roy S, Haan E, Bernstein J, Harris PC. Renal cystic disease in tuberous sclerosis: role of the polycystic kidney disease 1 gene. Am J Hum Genet. 1997;61:843–51. doi: 10.1086/514888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson JR, Scahill SJ, Stephenson JB, Mann L, Connor JM. Genetic aspects of tuberous sclerosis in the west of Scotland. J Med Genet. 1989;26:28–31. doi: 10.1136/jmg.26.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak O, Nellist M, Goedbloed M, Elfferich P, Wouters C, Maat-Kievit A, Zonnenberg B, Verhoef S, Halley D, van den Ouweland A. Mutational analysis of the TSC1 and TSC2 genes in a diagnostic setting: genotype - phenotype correlations and comparison of diagnostic DNA techniques in Tuberous Sclerosis Complex. Eur J Hum Genet. 2005;13:731–41. doi: 10.1038/sj.ejhg.5201402. [DOI] [PubMed] [Google Scholar]
- Shendure J, Porreca GJ, Reppas NB, Lin X, McCutcheon JP, Rosenbaum AM, Wang MD, Zhang K, Mitra RD, Church GM. Accurate multiplex polony sequencing of an evolved bacterial genome. Science. 2005;309:1728–32. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- Shi R, Otomo K, Yamada H, Tatsumi T, Sugawara I. Temperature-mediated heteroduplex analysis for the detection of drug-resistant gene mutations in clinical isolates of Mycobacterium tuberculosis by denaturing HPLC, SURVEYOR nuclease. Microbes Infect. 2006;8:128–35. doi: 10.1016/j.micinf.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Simen BB, Simons JF, Hullsiek KH, Novak RM, Macarthur RD, Baxter JD, Huang C, Lubeski C, Turenchalk GS, Braverman MS, Desany B, Rothberg JM, Egholm M, Kozal MJ. Low-abundance drug-resistant viral variants in chronically HIV-infected, antiretroviral treatment-naive patients significantly impact treatment outcomes. J Infect Dis. 2009;199:693–701. doi: 10.1086/596736. [DOI] [PubMed] [Google Scholar]
- Smith AD, Xuan Z, Zhang MQ. Using quality scores and longer reads improves accuracy of Solexa read mapping. BMC Bioinformatics. 2008;9:128. doi: 10.1186/1471-2105-9-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas RK, Nickerson E, Simons JF, Janne PA, Tengs T, Yuza Y, Garraway LA, LaFramboise T, Lee JC, Shah K, O’Neill K, Sasaki H, Lindeman N, Wong KK, Borras AM, Gutmann EJ, Dragnev KH, DeBiasi R, Chen TH, Glatt KA, Greulich H, Desany B, Lubeski CK, Brockman W, Alvarez P, Hutchison SK, Leamon JH, Ronan MT, Turenchalk GS, Egholm M, Sellers WR, Rothberg JM, Meyerson M. Sensitive mutation detection in heterogeneous cancer specimens by massively parallel picoliter reactor sequencing. Nat Med. 2006;12:852–5. doi: 10.1038/nm1437. [DOI] [PubMed] [Google Scholar]
- van Slegtenhorst M, de Hoogt R, Hermans C, Nellist M, Janssen B, Verhoef S, Lindhout D, van den Ouweland A, Halley D, Young J, Burley M, Jeremiah S, Woodward K, Nahmias J, Fox M, Ekong R, Osborne J, Wolfe J, Povey S, Snell RG, Cheadle JP, Jones AC, Tachataki M, Ravine D, Kwiatkowski DJ. Identification of the tuberous sclerosis gene TSC1 on chromosome 9q34. Science. 1997;277:805–8. doi: 10.1126/science.277.5327.805. [DOI] [PubMed] [Google Scholar]
- van Oers JM, Lurkin I, van Exsel AJ, Nijsen Y, van Rhijn BW, van der Aa MN, Zwarthoff EC. A simple and fast method for the simultaneous detection of nine fibroblast growth factor receptor 3 mutations in bladder cancer and voided urine. Clin Cancer Res. 2005;11:7743–8. doi: 10.1158/1078-0432.CCR-05-1045. [DOI] [PubMed] [Google Scholar]
- Vandenbroucke I, van Doorn R, Callens T, Cobben JM, Starink TM, Messiaen L. Genetic and clinical mosaicism in a patient with neurofibromatosis type 1. Hum Genet. 2004;114:284–90. doi: 10.1007/s00439-003-1047-9. [DOI] [PubMed] [Google Scholar]
- Verhoef S, Bakker L, Tempelaars AM, Hesseling-Janssen AL, Mazurczak T, Jozwiak S, Fois A, Bartalini G, Zonnenberg BA, van Essen AJ, Lindhout D, Halley DJ, van den Ouweland AM. High rate of mosaicism in tuberous sclerosis complex. Am J Hum Genet. 1999;64:1632–7. doi: 10.1086/302412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates JR, van Bakel I, Sepp T, Payne SJ, Webb DW, Nevin NC, Green AJ. Female germline mosaicism in tuberous sclerosis confirmed by molecular genetic analysis. Hum Mol Genet. 1997;6:2265–9. doi: 10.1093/hmg/6.13.2265. [DOI] [PubMed] [Google Scholar]