Abstract
CPU time and memory usage are two vital issues that any numerical solvers for the Poisson-Boltzmann equation have to face in biomolecular applications. In this study we systematically analyzed the CPU time and memory usage of five commonly used finite-difference solvers with a large and diversified set of biomolecular structures. Our comparative analysis shows that modified incomplete Cholesky conjugate gradient and geometric multigrid are the most efficient in the diversified test set. For the two efficient solvers, our test shows that their CPU times increase approximately linearly with the numbers of grids. Their CPU times also increase almost linearly with the negative logarithm of the convergence criterion at very similar rate. Our comparison further shows that geometric multigrid performs better in the large set of tested biomolecules. However, modified incomplete Cholesky conjugate gradient is superior to geometric multigrid in molecular dynamics simulations of tested molecules. We also investigated other significant components in numerical solutions of the Poisson-Boltzmann equation. It turns out that the time-limiting step is the free boundary condition setup for the linear systems for the selected proteins if the electrostatic focusing is not used. Thus, development of future numerical solvers for the Poisson-Boltzmann equation should balance all aspects of the numerical procedures in realistic biomolecular applications.
Introduction
The electrostatic interaction is crucial for the structural, functional, and dynamic properties of biomolecules. Accurate and efficient modeling of the electrostatic interaction has been an active topic in computational chemistry.1–14 Since biomolecules are surrounded by solvent molecules, the solvent effect must be taken into account in modeling the electrostatics interaction. The solvent effect can be treated explicitly – explicit solvent molecules are placed around the studied biomolecules, or treated implicitly – solvent molecules are considered as a continuum. In the implicit treatment, the Poisson-Boltzmann equation (PBE) has been established as a fundamental equation to model the electrostatic interaction.1–14
When the PBE is applied to model biomolecular electrostatics, a significant challenge is how to obtain accurate solution of the PBE efficiently given the limited memory and speed of today’s computer servers because a typical biomolecule consists of tens of thousands to millions of atoms. This challenge becomes more difficult when incorporating the PBE in a typical dynamics simulation involving millions of time steps. In practice solving the PBE and interpolating the electrostatic energies and forces have to be accomplished within a few tenths second to finish the dynamics simulation within a reasonable wall-clock time.
Since analytic solution of the PBE can be achieved only in a few specific cases with simple solute geometry, only numerical solution of the PBE is possible for biomolecular applications. Many efforts have been invested to develop methods to obtain numerical solution of the PBE accurately and efficiently. The finite difference method (FDM),15–25 as the most widely-adopted method, commonly uses uniform grids to discretize the space and builds up a set of linear equations directly from the PBE. It has been implemented in several programs, such as DelPhi, 15,17,23 UHBD,16,18 APBS,19,25 and in related modules of Amber24,26 and CHARMM.17,22 To set up the dielectric constant map, classical FDMs only classify grid points as outside or inside the biomolecular surface. This simplification leads to a symmetric and positive-definite coefficient matrix of the linear system and some highly efficient solvers, such as preconditioned conjugate gradient, have been developed to solve the PBE.16,24,27–30 However, the treatment blurs the exact biomolecular boundary, leading to discretization error. Furthermore, the boundary jump conditions can not be enforced in the FDMs. Thus the solution is only first-order accurate. To achieve second-order accurate solution, researchers have used interface conditions with sub-grid resolution, as in the immersed interface method31,32 and the matched interface and boundary method.33,34 However, these methods lead to asymmetric and positive-indefinite coefficient matrices. Thus preconditioned conjugate gradient can no longer be applied to solve the linear systems.
Boundary element method (BEM)35–48 integrates the PBE at the biomolecular boundary to obtain a linear system whose unknowns are either the induced surface charges35–38,42,43,45,46 or the normal components of the electric displacement39–41,44,47,48 on the boundary. Compared with the FDM, the BEM reduces the number of unknowns by an order and describes the biomolecular boundary more accurately. However, the BEM involves an extra step of triangulization of the boundary. Furthermore, its efficiency is not directly related to the number of unknowns because integration operator along the macromolecular boundary is the most time limiting step.
The finite-element method (FEM)49–54 is based on the weak variational formulation. The electrostatic potential to be solved is approximated by a superposition of a set of basis functions. A linear or nonlinear system for the coefficients produced by the weak formulation has to be solved. A nice property of the FEM is that the mesh is irregular so that adaptivity can be achieved. Also body fitted mesh can be generated to fit nicely to the boundary or the interface, such as the molecular surface. However, this strategy may cause additional cost, i.e., constant remeshing, for dynamics simulations.
Even with the extensive development of numerical PBE solvers as reviewed above, only limited comparisons among a few FDM solvers were reported in the literature.16,19 No systematic analysis of their performance on biomolecules has been reported. Apparently a thorough analysis is important to guide users to choose appropriate PBE solvers for their applications at hand. Such an analysis is also helpful for further development of more robust PBE solvers. In this first attempt of systematic analysis of numerical PBE solvers, we focused on the widely-adopted FDM solvers and compared their performance in a large and diversified test set of biomolecules, including proteins, nucleic acids, and short peptides. We systematically analyzed factors related to the solver efficiency and memory usage in the context of the typical biomolecular systems.
Methods
In this work, we implemented and carefully optimized five different FDM solvers, successive over-relaxation,55 conjugate gradient,55 (modified) incomplete Cholesky conjugate gradient,24 geometric multigrid,56 and algebraic multigrid.57 In the following we review the fundamental concepts and algorithms that are used in our implementation of these numerical FDM solvers.
Poisson-Boltzmann equation
In typical implicit solvation frameworks, both solute and solvent molecules are represented as continua. To compute the solute/solvent electrostatic interaction, the solute molecule is approximated as a low-dielectric continuum and the solvent molecules are collectively treated as a high-dielectric continuum. Solute atomic point charges in the molecular mechanics representation are assigned on atomic centers. The electrostatic field in the continuum representation of the solution system can be described by the Poisson’s equation in classical electrostatics
| (1) |
where ε is the dielectric constant, φ is the electrostatic potential, and ρ is the charge density. All three variables are functions of position vector r. If the solution is a dissolved electrolyte, a Boltzmann term describing the salt effect is added to the above equation to give the well-known Poisson-Boltzmann equation
| (2) |
where λ is the masking function for the Stern layer, ni is the number density of counterion of type i in the bulk solution, qi is the charge of the counterion of type i, k is the Boltzmann constant, and T is the temperature. Obviously the PBE is non-linear. When qiφ(r)/kT is small, the PBE can be linearized as
| (3) |
For biomolecules, only numerical solution of equation (2) or (3) can be obtained. In typical numerical procedures, the equation is discretized with the simplest finite-difference scheme. Under this discretization scheme, the PBE can be written as follows at each grid point
| (4) |
where h is the spacing in each dimension, i, j, and k are the grid indexes along x, y and z axes, respectively. εi(i, j, k) is the dielectric constant between grids (i, j, k) and (i+1, j, k). εj(i, j, k) and εk(i, j, k) are defined similarly. κ2 absorbs all the related coefficients in the Boltzmann term. q(i, j, k) is the total charge within the cubic volume centered at (i, j, k). The linear system can then be denoted as
| (5) |
where A is the coefficient matrix of dielectric constants and the Boltzmann term, and b is the constant vector of all charges on the grids.
To solve equation (5), a number of solvers have been developed, such as successive over-relaxation (SOR),55 conjugate gradient (CG),55 (modified) incomplete Cholesky conjugate gradient ((M)ICCG),27–30 geometric multigrid (GMG),56 and algebraic multigrid (AMG).57 All solvers proceed from an initial guess of φ(i, j, k) to approach the solution iteratively.
Successive Over-relaxation
SOR belongs to the classical iterative solvers for linear equations. It is a speed-up version of Gauss-Seidel.55 In this solver, the values of φ(i, j, k) are updated by
| (6) |
where w is called the over-relaxation factor. It can be proven that when w is between 0 and 2 SOR always converges.55 When w is equal to 1, SOR returns to Gauss-Seidel. It should be noted that φ(i, j, k) appears on the right hand side of equation (6) is the latest value, not the value updated in the previous iteration. Arelated update method is the Red-Black method. The grids in the space are colored by red or black in an interlaced manner, i.e., the neighbor grids of a red grid are all black and the neighbor grids of a black grid are all red. During iteration, φ(i, j, k) at black grids are first updated simultaneously and φ(i, j, k) at red grids are then updated simultaneously. The Red-Black method does not accelerate convergence, but leads to a parallel operation. A Red-Black SOR can be summarized as the following pseudo code:
set l = 0
calculate the norm of residues ||rl|| = ||b− Aφl|| If ||rl||/||b|| is less than a predefined convergence threshold δ, output φl as the solution. Otherwise go to the next step.
update the φl at black grids first and then update the φl at red grids according to Eq. (6)
set l = l+1 and go to step 2.
Conjugate Gradient
CG is suitable to solve linear systems with symmetric and positive-definite coefficient matrices. It approaches the exact solution along a series of conjugate directions. In theory, CG can obtain the exact solution after at most Ngrid iterations (Ngrid is the number of grids). However the exact solution can not be achieved in practice because of numerical errors. Thus, CG is always implemented in an iterative manner as SOR. CG can be summarized as follows:
set l = 0, p0 = r0
calculate the norm of residues ||rl||. If ||rl||/||b|| is less than a predefined convergence threshold δ, output φl as the solution. Otherwise go to the next step.
-
calculate
-
calculate
set l= l+1 and go to step 2
Modified Incomplete Cholesky Conjugate Gradient
The convergence speed of CG depends on the distribution of the eigenvalues of the coefficient matrix. It has been shown that its convergence behavior is optimal when the eigenvalues of the coefficient matrix distribute in a small range.27 Therefore, preconditioned conjugate gradient introduces a preconditioning matrix M into equation (5)
| (7) |
so that the new equivalent equation can be written as
| (8) |
An appropriate M makes à with all eigenvalues close to one to speed up the convergence. One widely-used class of preconditioning matrices comes from the incomplete LDLT factorizations
| (9) |
Where A= L+ D+ LT, L is the strictly lower triangular matrix of A, D is the positive diagonal matrix of A, and D̃ is an undetermined positive diagonal matrix. If the diagonal of M is equal to D, the corresponding preconditioned conjugate gradient is called ICCG. For the case in 3D, A is a symmetric and 7-banded matrix. If the dimension of A is denoted as nxnynz and the diagonal elements and the three off-diagonal elements at row i as ai,1, ai,2, ai,3, and ai,4, respectively, the diagonal elements of D̃ can be written as
| (10) |
In MICCG, an extension of ICCG, the diagonal elements of D̃ can be written as
| (11) |
where α is a relax coefficient. When α is equal to 0, MICCG goes back to ICCG.
Multigrid
Multigrid is highly efficient to solve linear equations or nonlinear equations. It builds up a hierarchy of discretization. Here we denote the finest discretization level as level 1 and larger level numbers denote coarser discretization levels. To construct a multigrid method, three operators are required. The first operator is the relaxation operator, which is used to obtain an improved solution at a certain level i. Gauss-Seidel and CG can both be used as relaxation operators. However, it should be noted that SOR is not suitable as a relaxation operator, because over-relaxation cannot maintain the small high-frequency error at the coarser levels. In this study, we have used Gauss-Seidel as the relaxation operator. The second operator is called the restriction operator, which restricts the residue terms at level i to construct the constant vector terms at level i+1. The third operator is the interpolation operator, which interpolates the solution at level i to correct the solution at level i−1. The restriction and interpolation operators transfer the information between neighboring levels. Their algorithms vary according to the problems at hand to pursue the highest computational efficiency. After these three operators are defined, one iteration between level 1 and level 2 can proceed as:
1. φ1 = S1(φ1,b1), r1 = b1 − A1φ1
2. b2 = R1 (r1)
3. φ2 = S2 (φ2, b2)
4. φ1 = φ1+ I2 (φ2)
5. φ1 = S1 (φ1,b1)
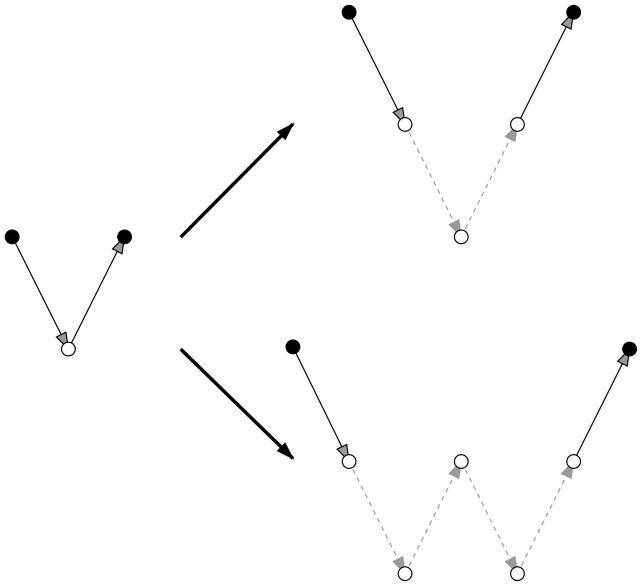
where S, R, and I represent the relaxation, restriction, and interpolation operator, respectively. The superscripts represent the level number. At step 3, we can relax the solution at level 2 by introducing level 3 and performing γ two-level iterations between level 2 and level 3. Recursively introducing coarser levels until the coarsest level builds up one cycle of multigrid. When γ =1, the cycle is called V-cycle and when γ =2, the cycle is called W-cycle, as shown in Figure 1.
Figure 1.
V-cycle and W-cycle of multigrid. The relaxation at the coarse level (open circle on the left) is replaced by one two-level iteration in V-cycle or two two-level iterations in W-cycle (open circle and dash line on the right).
The advantage of multigrid rests with how the information spreads. In multigrid, the information propagates to the neighbor grids at level 1, just as SOR or CG. However, at other levels the information can be propagated to farther grids. In doing so, the high-frequency error is eliminated at finer levels, and low-frequency error is corrected at coarser levels.
A key issue in the all multigrid methods is how to construct the interpolation operator. In GMG, the space is divided with different spacing h1, h2, h3, …, at different levels. Usually, the grid spacing values between neighbor levels i and i+1 satisfy hi+1 = 2hi. The relationships of grid points at the neighbor levels are determined by their geometric position. When GMG is applied to solve the PBE for biomolecules in solution, attention should be paid to treatment of discontinuous dielectric constant. In general, the dielectric constant inside macromolecules is set as 1 to 4, and the dielectric constant outside biomolecules is set as 80. The discontinuity of dielectric constant leads to a large jump in the coefficient in the PBE, which then results in poor convergence of GMG. Sophisticated designs of interpolation have been the key in developing robust GMG methods. For example, Alcouffe et al. developed special harmonic averaging techniques to construct operator-induced interpolations.56 In the black-box multigrid method, Dendy simplified the interpolation procedure by considering the discretization stencils.58 In our implement, an interpolation suggested by Holst and Saied19 was adopted to retain a 7-banded coefficient matrix at each level.
AMG is an alternative multigrid method that bypasses the problem of discontinuous coefficient completely.59 Unlike GMG, AMG does not consider the geometric information in its operators, instead only the linear system is used. According to the original linear system, AMG automatically constructs a series of gradually smaller liner systems, whether the coefficients are continuous or discontinuous. The grids at the finer level Ch are split into two subsets: one set (CH) includes the grids kept at the coarser level, and the other set (FH) includes the rest. Then the interpolation operator can be defined as
| (12) |
The restriction operator is the reverse of interpolation operator, and the relaxation operator uses Gauss-Seidel. More grids in subset CH likely make the interpolation at grids in subset FH more accurate. On the other hand more grids in CH consume more computational resource. Therefore a good grid-splitting algorithm has to balance the interpolation accuracy and computational cost. The detail of the grid-splitting algorithm is reviewed in Ref 60. In this study, we investigated the performance of an AMG method, AMG1R5, as implemented by Ruge, Stuben and Hempel.57
Computational details
To insure a fair comparison, all tested FDM solvers were implemented within the PBSA module in the Amber 10 package.61 Thus the exactly same linear system was solved by all the solvers. Specifically, the following five FDM solvers were implemented: SOR, CG, MICCG, GMG, and AMG. Totally 558 biomolecular structures including proteins, nucleic acids, and short peptides in the Amber benchmark suite61 were used in the test. The atom numbers of these biomolecules range from 247 to 8,254 and their geometries are quite different. These molecules were assigned the charges of Cornell et al62 and the modified bondi radii. Unless specified otherwise in the Results and Discussion, the computations were conducted with the following conditions. The convergence criterion was set to be 10−3. The ratio of the grid dimension over the solute dimension (the fillratio keyword in Amber) was set as 1.5. The grid spacing was 0.5 Ǻ. No electrostatic focusing was used. The initial potential values on all grids were set to zero. The solvent dielectric constant was set to 80, while the solute dielectric constant was set to 1. The boundary dielectric constants were assigned as the harmonic average of the solvent and solute dielectric constants. Thus, the coefficient matrices are symmetric and positive-definite and all of the five solvers are able to solve the linear systems. Finally the free space boundary condition was used. In SOR, the over-relaxation factor was set to 1.95. In MICCG, the relax coefficient α was set to −0.3. All other parameters are set as default in the PBSA module of Amber 10 package.61
Results and Discussion
Once we can establish the numerical consistency among all linear solvers, we are mostly interested in their CPU usage or efficiency, which can be affected by many factors, such as system sizes, convergence criterion, grid spacing, initial guess of solution, and so on. The scaling of the implemented solvers with theses factors is presented in the following. The memory requirement of the solvers is also analyzed below. Finally, the computational cost of other significant components in the FDM solver is discussed.
Solver consistency
To validate the numerical consistency of the tested linear solvers, we first calculated the total electrostatic energies and forces with the P3M method26 given the computed grid potentials from the different solvers. With the typical convergence criterion set as 10−3 in dynamics simulations, the average relative difference in the total electrostatic energies among the different solvers is 5.2×10−6 and the largest relative difference is 3.9×10−5. We also analyzed the consistency in the atomic electrostatic forces computed with the same P3M strategy.26 Taking protein 1bci as an example, the average relative difference in the atomic electrostatic forces is 7.7×10−4 among the different solvers. These data indicate that the numerical precisions in electrostatic energies and forces from different linear solvers are quite consistent and the numerical uncertainties due to the use of different solvers are bounded by the convergence criterion.
Solver scaling considerations
System sizes
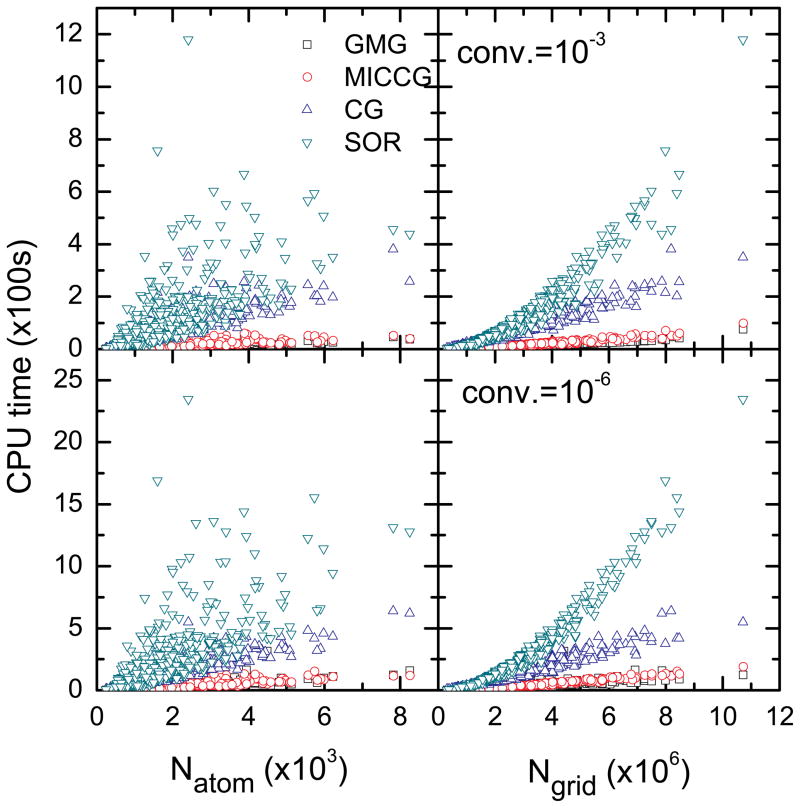
Efficiency scaling with respect to the system size is one of the crucial issues in the biomolecular applications of the numerical PBE solvers. The test set of 558 biomolecular structures61 was used to study the scaling behaviors of the implemented solvers. Due to the limitation of memory on our local computers, AMG was not included in this test (memory requirements of all the implemented solvers are discussed below). Figure 2 plots the CPU time (not including the setup time for the linear system) versus the number of atoms (Natom) and the number of grid points (Ngrid), respectively. Our analysis shows that the efficiency of CG is higher than that of SOR, but much lower than that of MICCG and GMG. However, it should be pointed out that we did not use “optimal” w values. Use of molecule-specific “optimal” w may improve the convergence rate of SOR.17,19 Finally the performances of MICCG and GMG are similar, and GMG is slightly better than MICCG under the testing conditions in this study: the CPU times are within 0.5% on average between the two solvers. Nevertheless, if we measure the slopes for the linear scaling of CPU times versus Ngrid, GMG is about 30% faster than MICCG.
Figure 2.
CPU time versus the number of atoms (Natom) or the number of grids (Ngrid) for GMG, MICCG, CG, and SOR, respectively. (Relative) convergence criterion is set to be 10−3 and 10−6, respectively.
It is interesting to note that the CPU times increase approximately linearly with Ngrid, when CG, MICCG, and GMG are used (Figure 2). The linear trend is less clear when the number of atoms is used. This is because the number of grid points depends on not only the numbers of atoms but also the solute geometries. The observed linear scaling for CG and MICCG is somewhat different from the observation in the literature.19 The primary difference between this work and the literature lies in the different scaling behaviors studied. In this study the solver scaling refers to its CPU time versus the numbers of grids for different solutes, while in the literature the solver scaling refers to its CPU time versus the numbers of grids from different grid spacing values for a single solute. Specifically the ratio of the grid dimension over the solute dimension (the fillratio keyword), was fixed for all solute biomolecules in this study. We also analyzed the scaling of CPU time versus the grid spacing (see below and Figure 4). It has to be pointed out that this scaling behavior is machine-dependent.
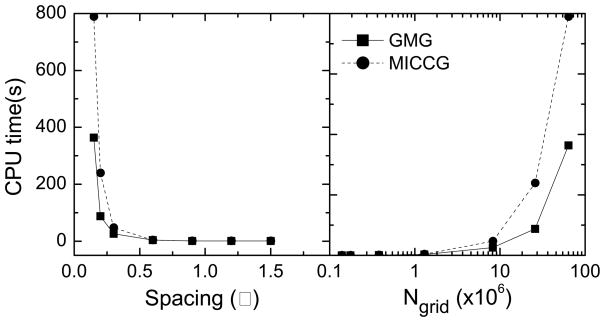
Figure 4.
CPU time versus grid spacing or Ngrid for GMG and MICCG, respectively. Here a small tested protein 1bci is used as an example due to the limited memory on the tested server.
Convergence criterion
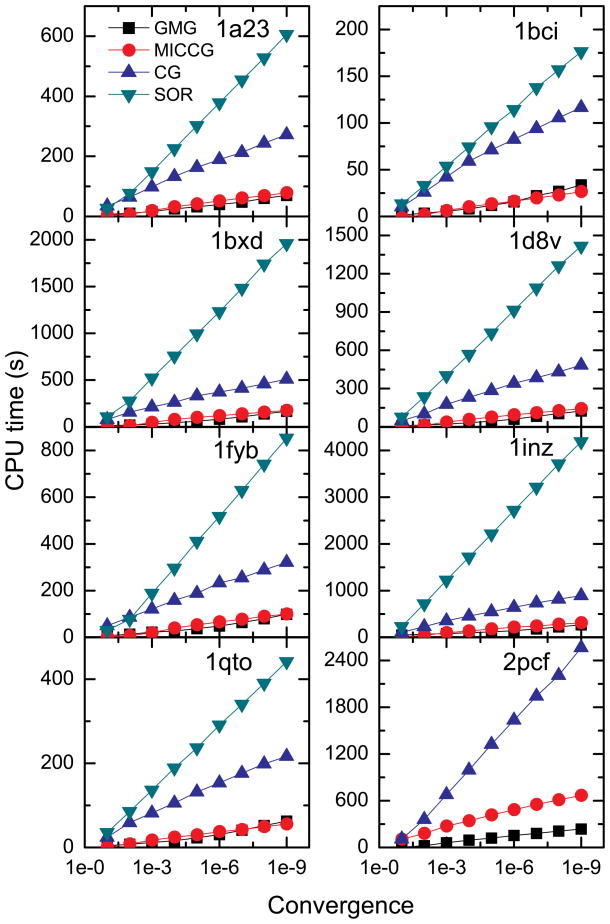
We also investigated the solver efficiency versus the convergence criterion. Eight representative proteins were selected in this analysis. Here, the atom numbers of the selected proteins vary from 1,619 to 4,211, and the numbers of grid points of these proteins vary from 2×106 to 107. The tested convergence criteria range from 10−1 to 10−9. As shown in Figure 3, again both GMG and MICCG scale better than the other two solvers. Furthermore the scaling behaviors of GMG and MICCG are quite consistent for all eight tested proteins. Their CPU times increase almost linearly with the negative logarithm of the convergence criterion.
Figure 3.
CPU time versus the (relative) convergence criterion for GMG, MICCG, CG, and SOR, respectively. The eight proteins listed in Table 1 are used as examples.
Grid spacing
Another interesting solver scaling issue is how efficient these solvers are when the grid spacing decreases, i.e. when a higher grid resolution is requested for a specific problem. We selected 1bci as the test system and changed the spacing from 0.15 Ǻ to 1.5 Ǻ. Here only GMG and MICCG were chosen in the test. Figure 4 plots the CPU time for both GMG and MICCG versus the grid spacing. When the grid spacing is large, the two methods need almost the same time to achieve the same convergence. However, when the grid spacing is small, for example 0.2 Ǻ, GMG is about 50% faster than MICCG. Note that there is nothing special about the grid spacing of 0.2 Ǻ. For other selected larger biomolecules, we can observe the same deteriorating performance even at grid spacing of 0.6 Ǻ in MICCG. Thus the deteriorating performance of MICCG might be due to the low memory utilization when the finite-difference grids become too large, i.e. there are too many cache misses when the potential update at (i, j, k) needs information on both grid plane k−1 and grid plane k+1 in the backward and forward substitution loops. Apparently, the aggressive memory usage of MICCG (to be discussed below) causes this to happen at relatively small problem sizes.
Solver performance in molecular dynamics
When the PBE is solved in molecular dynamics (MD), a very good initial guess for the solution is simply the solution at the previous time step due to the small atomic displacements per step at typical simulation temperatures. We should fully utilize this fact to accelerate the numerical solvers. Table 1 lists the average CPU times of GMG and MICCG for the eight selected proteins. The per-step CPU time is averaged over 100 MD steps. Interestingly, these data show that MICCG with the relax coefficient α of −0.3 can better take advantage of the very good initial guess than GMG for the tested proteins. Note too that the advantage of MICCG over GMG is quite consistent for the test proteins ranging from 1,600 to 4,200 atoms, with MICCG about 30% faster on average during dynamics simulations.
Table 1.
Average CPU times (in second) of GMG and MICCG for 100 MD step for eight selected proteins. No electrostatic focusing is used in the analysis.
| PROTEIN | Natom | Ngrid | Convergence 10−3 | Convergence 10−4 | ||
|---|---|---|---|---|---|---|
| GMG | MICCG | GMG | MICCG | |||
| 1a23 | 2955 | 175×159×143 | 7.58 | 2.90 | 12.61 | 8.50 |
| 1bci | 1969 | 159×111×111 | 3.56 | 1.35 | 5.64 | 3.68 |
| 1bxd | 2439 | 191×207×175 | 13.01 | 5.04 | 21.45 | 12.78 |
| 1d8v | 4211 | 175×191×175 | 10.83 | 4.15 | 17.27 | 11.26 |
| 1fyb | 1619 | 127×175×207 | 8.89 | 3.24 | 12.01 | 7.98 |
| 1inz | 2413 | 207×271×191 | 19.89 | 6.35 | 31.37 | 17.46 |
| 1qto | 1817 | 143×143×143 | 4.45 | 1.92 | 6.54 | 5.10 |
| 2pcf | 3884 | 239×223×159 | 16.66 | 7.23 | 28.47 | 19.22 |
It is possible that GMG can be further optimized to better take advantage of the update feature in applications to dynamics simulations. A feature that has to be exploited to achieve good performance in dynamics is the fast relaxation of local and rapidly changing errors since the long-range interactions change little between two adjacent dynamics steps. MICCG turns out to be able to address this challenge well by relaxing the local errors more rapidly. While in our current implementation of GMG, Gauss-Seidel is used as the relaxation operator, which is not as efficient as MICCG in relaxing local errors. An apparent way to improve the GMG in this regards is just to use MICCG or other preconditioned CG as the relaxation operator to better relax the local errors more efficiently in GMG.
Detailed analysis of the multigrid solvers
Our MICCG method has been extensively optimized. However, the multigrid methods implemented in this study may need further improvement to achieve higher efficiency. Our current implementation of GMG consists of four levels. A natural question to ask is how many levels are needed for typical biomolecular applications. Table 2 lists the CPU time at each level in the test case of 1inz, the test case with the largest number of grid points. Apparently, when the level becomes coarser, the time spent on that level drops rapidly. At the coarsest level (level 4), the time is only about 1 second. If more levels are used, the time saved at level 4 and lower is no more than 1 second, but the grid number at the finest level may increase. This is because the grid number at the finest level has to satisfy k *2N−1. Here k is an integer, and N is the number of levels. Thus more levels may actually lead to an increase of the CPU time. Table 2 thus shows that four levels are enough for GMG to achieve a high efficiency for the largest tested system. On contrast, this analysis also implies that use of four levels is probably too much for the smaller test cases in GMG. Thus a future direction to improve the GMG implementation is adaptively determine the number of levels needed for a given problem size at the finest grid level.
Table 2.
CPU Times (in second) for relaxation, interpolation, and restriction at each level in GMG. Here protein 1inz is used as an example.
| Operation | Level 1 | Level 2 | Level 3 | Level 4 |
|---|---|---|---|---|
| Relaxation | 6.84 | 0.86 | 0.09 | 0.75 |
| Interpolation | - | 22.73 | 2.71 | 0.31 |
| Restriction | 0.24 | 0.03 | 0.00 | - |
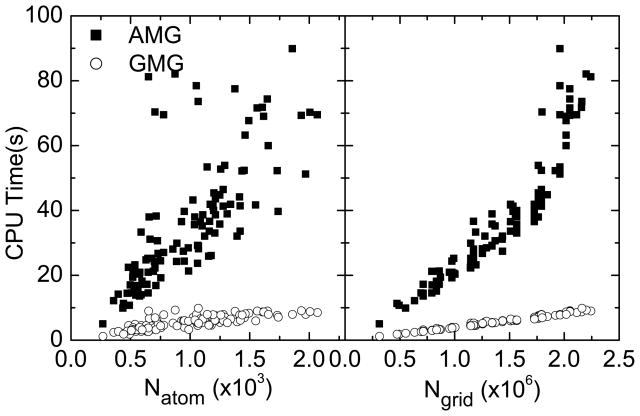
The large memory requirement of the general AMG method limits our testing of the method to smaller molecules in the test set. Nevertheless, Figure 5 shows that GMG is much faster than AMG for the 113 selected molecules that can be processed by AMG on our local computer servers.
Figure 5.
CPU time versus the number of atoms or the number of grids for AMG and GMG, respectively. Here 113 selected proteins that can be processed with AMG are used as examples.
Memory requirement
Besides computational efficiency, memory requirement is also crucial and needs to be taken into account in biomolecular applications. In this study, SOR turns out to use the smallest amount of memory. It only needs to allocate arrays to store the coefficient matrix, the residue, and the solution. Since the coefficient matrix is symmetric and 7-banded, its storage requirement is 4Ngrid. Thus the total storage of SOR is 6Ngrid. To implement CG, one additional working array of Ngrid is needed, so that its total storage need is 7Ngrid. MICCG is more aggressive in memory usage. It needs more working arrays (5Ngrid) than CG, so that its total storage is 11Ngrid. GMG needs to store the linear system at every level, but the storage need reduces by a factor of eight when going down one level, so that the total storage of GMG is predominantly determined by the finest level, which is about 6Ngrid. If we approximate the storage need of the next finest level as Ngrid, the total storage of GMG is about 7Ngrid. However, it should be pointed out that the 7-banded implementation of the coefficient matrices at coarser levels is not robust, resulting in its failure to converge in some test cases. A robust GMG would require the use of 27-banded coefficient matrices at all levels with a total storage requirement of about 20Ngrid. Finally AMG is a general-purpose algorithm and needs much more storage. In our experience, it needs to store 46Ngrid real numbers and 49Ngrid integer numbers.
Other significant components
Our analyses above only focus on the computational efficiency and memory usage of the linear solvers. However, the linear system setup and post-processing to obtain energies and forces from grid potentials cannot be neglected when these linear solvers are used in realistic biomolecular applications. A typical linear system setup includes distribution of atomic charges onto grid points by the trilinear method, calculation of the molecular surface to set up the dielectric map, and calculation of the boundary grid potential by the Debye-Huckle theory when the free space boundary condition is used. The charge distribution can be realized with an O(Natom) procedure. The molecule surface can be computed with ONatomneighbor × N) operations, where Nneighbor is the maximum number of neighbor atoms of a given atom with a distance cutoff (~9 Å is quite sufficient for molecular surface calculation). The setup of the boundary grid potential needs operations. Table 3 lists the time to set up the dielectric map, the boundary condition, and the CPU time to solve the PBE for selected proteins. Note that the time to set up the boundary condition is much longer than that to solve the linear system.
Table 3.
CPU times (in second) for linear system setup and solution in the GMG solver for eight selected proteins listed in Table 1. MS/EPS: time for calculation of molecular surface and setup of dielectric map. BC: time for setup of boundary condition. Solver: time for solving the linear system by the GMG solver.
| PROTEIN | MS/EPS | BC | Solver |
|---|---|---|---|
| 1a23 | 0.46 | 89.38 | 13.59 |
| 1bci | 0.25 | 37.56 | 6.56 |
| 1bxd | 0.63 | 112.54 | 23.51 |
| 1d8v | 0.66 | 167.67 | 19.91 |
| 1fyb | 0.42 | 57.39 | 17.84 |
| 1inz | 0.89 | 195.71 | 36.85 |
| 1qto | 0.31 | 44.74 | 9.84 |
| 2pcf | 0.82 | 222.96 | 29.24 |
To reduce the time to set up the boundary condition, the focusing method is often used. The focusing method first uses coarse finite-difference grids to discretize the space large enough to ensure good boundary condition and then uses fine finite-difference grids to discretize the space of interest. Such a treatment reduces the number of boundary grids leading to reduced setup time. The CPU time reduction by focusing can be estimated as follows. Denoting the fillratio keyword as Rfill and the ratio of the coarse grid spacing over the fine grid spacing as Rfocus, the setup time for the boundary condition can be reduced to , and linear solver time, including both the coarse and fine grids, can be reduced to . Usually Rfill is set to two to balance the accuracy of Debye-Huckle theory and the computational cost. Only when Rfocus is as large as 8 can the setup time for boundary condition be reduced to negligible portion of the overall CPU time (i.e as low as 1/64 of the setup cost for the non-focusing run). Another promising strategy is to introduce the periodical boundary condition into the PBE. In doing so, the boundary potentials can be obtained self-consistently by the periodical boundary condition. However, the CPU time might increase due to the use of more complex solvers to accommodate the periodic boundary condition.
So far we have left out the discussion of post-processing of electrostatic energies and forces from FDM grid potentials. This is not a trivial issue, though it is completely orthogonal to the solution of a FDM linear system. That is to say that a finer grid spacing always leads to more accurate energies and forces no matter what FDM solver is used. A detailed analysis of the accuracy of energies and forces was presented in one of our recent analyses.32 However, the scaling of these calculations is often not as bad as that of the linear system solvers, which scale at best as O(Ngrid). For example, the FDM electrostatic energy/forces scales as O(Natom) with the trilinear interpolation procedure, though the use of the more accurate P3M method would slow it down to O(NatomlogNatom). In addition the computation of electrostatic boundary forces scales as O(Nsurface), where Nsurface is the surface dielectric boundary grid points. Thus the computation of electrostatic energies and forces overall can be made more efficient than the linear system solution.
Conclusion
Efficiency is one of the central issues in applying the Poisson-Boltzmann equation to molecular modeling of biomolecules. In this work, we implemented five numerical solvers for the linearized Poisson-Boltzmann equation, including successive over-relaxation, conjugate gradient, modified incomplete Cholesky conjugate gradient, geometric multigrid, and algebraic multigrid, and compared their efficiency with a large set of diversified biomolecules, ranging from 247 to 8,254 atoms. Our analysis shows modified incomplete Cholesky conjugate gradient and geometric multigrid are the most efficient among the tested solvers. Their CPU times increase approximately linearly with the numbers of grids. In addition the CPU times of the two most efficient solvers increase almost linearly with the negative logarithm of the convergence criterion for the selected test cases. The two most efficient solvers were also analyzed in the context of molecular dynamics simulations. In these applications a good initial guess of the electrostatic potential exists. Our comparison shows that modified incomplete Cholesky conjugate gradient uses ~30% less time as geometric multigrid under the testing conditions.
Memory usage is another important issue in numerical solutions of the Poisson-Boltzmann equation for biomolecular applications. In this work the 7-banded geometric multigrid is less demanding of memory than modified incomplete Cholesky conjugate gradient. However the 7-banded geometric multigrid may fail to converge in some cases. To improve its robustness, the 27-banded geometric multigrid is often used, leading to much higher memory usage.
Though not the focus of this analysis, we also investigated other significant components in numerical solutions of the Poisson-Boltzmann equation. It turns out that the time-limiting step is the boundary condition setup for the linear systems of selected proteins. It should be pointed out that the electrostatic focusing method is often used to reduce the boundary condition setup time. An alternative way is to apply the periodic boundary condition in the solution of the Poisson-Boltzmann equation, where the boundary potential is computed self-consistently with rest of the grid potentials. Of course modest revisions of the numerical solvers are needed to incorporate the new boundary condition.
Finally, it is worth discussing future directions in numerical solution of the linear Poisson-Boltzmann equation. It is well known that key sources of error in numerical solution of the Poisson-Boltzmann equation is the singular charge sources and discretization of the dielectric boundary as reviewed in our recent studies.32,63 Harmonic average was proposed to alleviate the discretization error, though it does not use any jump conditions at the sub-grid resolution. To obtain more accurate solution, the analytical molecular surface must be taken into account. New procedures, such as the immersed interface method31,32 and the matched interface and boundary method33,34, can be used to achieve second-order accuracy in the finite-difference methods. However, these new finite-difference discretization schemes produce asymmetric and positive-indefinite coefficient matrices, which can no longer be handled by conjugate gradient-based solvers, such as modified incomplete Cholesky conjugate gradient discussed here. Even the 7-banded geometric multigrid method is inadequate. This leaves only the 27-banded geometric multigrid method to solve such linear systems. Thus how to achieve a balance of efficiency and memory usage in biomolecular applications is clearly a direction to pursue when these higher-accuracy numerical procedures are used to solve the linearized Poisson-Boltzmann equation.
Acknowledgments
We thank Drs. Hong-Kai Zhao (UC, Irvine) and Zhi-Lin Li (NC State) for exciting discussions. This work is supported in part by NIH (GM069620 & GM079383 to RL).
Footnotes
Software Packages
All numerical finite-difference solvers documented here, except AMG, have been implemented in the Amber 11 package and will be released in early 2010.
References
- 1.Davis ME, McCammon JA. Chemical Reviews. 1990;90(3):509–521. [Google Scholar]
- 2.Sharp KA. Current Opinion in Structural Biology. 1994;4(2):234–239. [Google Scholar]
- 3.Gilson MK. Current Opinion in Structural Biology. 1995;5(2):216–223. doi: 10.1016/0959-440x(95)80079-4. [DOI] [PubMed] [Google Scholar]
- 4.Honig B, Nicholls A. Science. 1995;268(5214):1144–1149. doi: 10.1126/science.7761829. [DOI] [PubMed] [Google Scholar]
- 5.Roux B, Simonson T. Biophysical Chemistry. 1999;78(1–2):1–20. doi: 10.1016/s0301-4622(98)00226-9. [DOI] [PubMed] [Google Scholar]
- 6.Cramer CJ, Truhlar DG. Chemical Reviews. 1999;99(8):2161–2200. doi: 10.1021/cr960149m. [DOI] [PubMed] [Google Scholar]
- 7.Bashford D, Case DA. Annual Review Of Physical Chemistry. 2000;51:129–152. doi: 10.1146/annurev.physchem.51.1.129. [DOI] [PubMed] [Google Scholar]
- 8.Baker NA. Current Opinion in Structural Biology. 2005;15(2):137–143. doi: 10.1016/j.sbi.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 9.Chen JH, Im WP, Brooks CL. Journal of the American Chemical Society. 2006;128(11):3728–3736. doi: 10.1021/ja057216r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feig M, Chocholousova J, Tanizaki S. Theoretical Chemistry Accounts. 2006;116(1–3):194–205. [Google Scholar]
- 11.Im W, Chen JH, Brooks CL. In Peptide Solvation and H-Bonds. Elsevier Academic Press Inc; San Diego: 2006. pp. 173–198. [Google Scholar]
- 12.Koehl P. Current Opinion in Structural Biology. 2006;16(2):142–151. doi: 10.1016/j.sbi.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 13.Lu BZ, Zhou YC, Holst MJ, McCammon JA. Communications in Computational Physics. 2008;3(5):973–1009. [Google Scholar]
- 14.Wang J, Tan CH, Tan YH, Lu Q, Luo R. Communications in Computational Physics. 2008;3(5):1010–1031. [Google Scholar]
- 15.Klapper I, Hagstrom R, Fine R, Sharp K, Honig B. Proteins Structure Function and Genetics. 1986;1(1):47–59. doi: 10.1002/prot.340010109. [DOI] [PubMed] [Google Scholar]
- 16.Davis ME, McCammon JA. Journal of Computational Chemistry. 1989;10(3):386–391. [Google Scholar]
- 17.Nicholls A, Honig B. Journal of Computational Chemistry. 1991;12(4):435–445. [Google Scholar]
- 18.Luty BA, Davis ME, McCammon JA. Journal of Computational Chemistry. 1992;13(9):1114–1118. [Google Scholar]
- 19.Holst M, Saied F. Journal of Computational Chemistry. 1993;14(1):105–113. [Google Scholar]
- 20.Forsten KE, Kozack RE, Lauffenburger DA, Subramaniam S. Journal of Physical Chemistry. 1994;98(21):5580–5586. [Google Scholar]
- 21.Bashford D. Lecture Notes in Computer Science. 1997;1343:233–240. [Google Scholar]
- 22.Im W, Beglov D, Roux B. Computer Physics Communications. 1998;111(1–3):59–75. [Google Scholar]
- 23.Rocchia W, Alexov E, Honig B. Journal of Physical Chemistry B. 2001;105(28):6507–6514. [Google Scholar]
- 24.Luo R, David L, Gilson MK. Journal of Computational Chemistry. 2002;23(13):1244–1253. doi: 10.1002/jcc.10120. [DOI] [PubMed] [Google Scholar]
- 25.Holst MJ, Saied F. Journal of Computational Chemistry. 1995;16(3):337–364. [Google Scholar]
- 26.Lu Q, Luo R. Journal of Chemical Physics. 2003;119(21):11035–11047. [Google Scholar]
- 27.Meijerink JA, Vandervorst HA. Mathematics of Computation. 1977;31(137):148–162. [Google Scholar]
- 28.Gustafsson I. BIT Numerical Mathematics. 1978;18(1):142–156. [Google Scholar]
- 29.Eisenstat SC. Siam Journal on Scientific and Statistical Computing. 1981;2(1):1–4. [Google Scholar]
- 30.Meijerink JA, Vandervorst HA. Journal of Computational Physics. 1981;44(1):134–155. [Google Scholar]
- 31.Qiao ZH, Li ZL, Tang T. Journal of Computational Mathematics. 2006;24(3):252–264. [Google Scholar]
- 32.Wang J, Cai Q, Li ZL, Zhao HK, Luo R. Chemical Physics Letters. 2009;468(4–6):112–118. doi: 10.1016/j.cplett.2008.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou YC, Wei GW. Journal of Computational Physics. 2006;219(1):228–246. [Google Scholar]
- 34.Zhou YC, Zhao S, Feig M, Wei GW. Journal of Computational Physics. 2006;213(1):1–30. [Google Scholar]
- 35.Miertus S, Scrocco E, Tomasi J. Chemical Physics. 1981;55(1):117–129. [Google Scholar]
- 36.Hoshi H, Sakurai M, Inoue Y, Chujo R. Journal of Chemical Physics. 1987;87(2):1107–1115. [Google Scholar]
- 37.Zauhar RJ, Morgan RS. Journal of Computational Chemistry. 1988;9(2):171–187. [Google Scholar]
- 38.Rashin AA. Journal of Physical Chemistry. 1990;94(5):1725–1733. [Google Scholar]
- 39.Yoon BJ, Lenhoff AM. Journal of Computational Chemistry. 1990;11(9):1080–1086. [Google Scholar]
- 40.Juffer AH, Botta EFF, Vankeulen BAM, Vanderploeg A, Berendsen HJC. Journal of Computational Physics. 1991;97(1):144–171. [Google Scholar]
- 41.Zhou HX. Biophysical Journal. 1993;65(2):955–963. doi: 10.1016/S0006-3495(93)81094-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bharadwaj R, Windemuth A, Sridharan S, Honig B, Nicholls A. Journal of Computational Chemistry. 1995;16(7):898–913. [Google Scholar]
- 43.Purisima EO, Nilar SH. Journal of Computational Chemistry. 1995;16(6):681–689. [Google Scholar]
- 44.Liang J, Subramaniam S. Biophysical Journal. 1997;73(4):1830–1841. doi: 10.1016/S0006-3495(97)78213-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vorobjev YN, Scheraga HA. Journal of Computational Chemistry. 1997;18(4):569–583. [Google Scholar]
- 46.Totrov M, Abagyan R. Biopolymers. 2001;60(2):124–133. doi: 10.1002/1097-0282(2001)60:2<124::AID-BIP1008>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 47.Boschitsch AH, Fenley MO, Zhou HX. Journal of Physical Chemistry B. 2002;106(10):2741–2754. [Google Scholar]
- 48.Lu BZ, Cheng XL, Huang JF, McCammon JA. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(51):19314–19319. doi: 10.1073/pnas.0605166103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cortis CM, Friesner RA. Journal of Computational Chemistry. 1997;18(13):1591–1608. [Google Scholar]
- 50.Holst M, Baker N, Wang F. Journal of Computational Chemistry. 2000;21(15):1319–1342. [Google Scholar]
- 51.Baker N, Holst M, Wang F. Journal of Computational Chemistry. 2000;21(15):1343–1352. [Google Scholar]
- 52.Shestakov AI, Milovich JL, Noy A. Journal of Colloid and Interface Science. 2002;247(1):62–79. doi: 10.1006/jcis.2001.8033. [DOI] [PubMed] [Google Scholar]
- 53.Chen L, Holst MJ, Xu JC. Siam Journal on Numerical Analysis. 2007;45:2298–2320. [Google Scholar]
- 54.Xie D, Zhou S. BIT Numerical Mathematics. 2007;47(4):853–871. [Google Scholar]
- 55.Press WH, Teukolsky SA, Vetterling WT, Flannery BP. Numerical recipe: the art of scientific computing. Cambridge University Press; Cambridge [Cambridgeshire]; New York: 1986. [Google Scholar]
- 56.Alcouffe RE, Brandt A, Dendy JE, Painter JW. Siam Journal on Scientific and Statistical Computing. 1981;2(4):430–454. [Google Scholar]
- 57.Ruge JWKS. In: In Multigrid Methods. McCormick SF, editor. SIAM; Philadelphia: 1987. pp. 73–130. [Google Scholar]
- 58.Dendy JE. Siam Journal on Scientific and Statistical Computing. 1987;8(5):673–685. [Google Scholar]
- 59.Stuben K. Applied Mathematics and Computation. 1983;13(3–4):419–451. [Google Scholar]
- 60.Stuben K. Journal of Computational and Applied Mathematics. 2001;128(1–2):281–309. [Google Scholar]
- 61.Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Crowley M, Walker RC, Zhang W, Merz KM, Wang B, Hayik A, Roitberg A, Seabra G, Kolossvary I, Wong KF, Paesani F, Vanicek JXW, Brozell SR, Steinbrecher T, Gohlke H, Yang L, Tan C, Mongan J, Hornak V, Cui G, Mathews DH, Seetin MG, Sagui C, Babin V, Kollman PA. Amber. Vol. 10. University of California; San Francisco: 2008. [Google Scholar]
- 62.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. Journal of the American Chemical Society. 1995;117(19):5179–5197. [Google Scholar]
- 63.Cai Q, Wang J, Zhao HK, Luo R. Journal of Chemical Physics. 2009;130(14) doi: 10.1063/1.3099708. [DOI] [PubMed] [Google Scholar]