Abstract
Arrays of transparent, releasable micron-scale structures termed “microcups” were created for the purpose of patterning and isolating viable cells from small cell samples. Cells were captured by the microcups without the need for barriers or walls on the intervening substrate. Furthermore, in contrast to prior methods for creating cell arrays with releasable elements, no chemical modification of the substrate was required. Individual microcups were released from the array using a pulsed laser at very low energy. Improvements in microcup design enabled cells in suspension to be loaded into the microcups with greater than 90% efficiency. Cells cultured within the microcups displayed 100% viability and were cultured over 4 days yielding colonies that remained sequestered within the microcups to generate pure clonal populations. Standard microscopic imaging was used to identify cells or colonies of interest, and the microcups containing these cells were then released and collected. Individual target cells isolated in this manner remained viable as demonstrated by clonal expansion of 100% of collected cells. Direct comparisons with cell isolation by fluorescence-activated cell sorting and magnetic-bead-based isolation systems demonstrated that the microcup cell isolation procedure yielded higher purity, yield and viability than these standard technologies when separating samples with small numbers of cells. The power of this technique was demonstrated by the isolation of hematopoietic stem cells from a human bone marrow aspirate possessing only 4,000 total cells.
Microfabricated arrays for patterning cells have been extensively described in the literature as they are ideal tools for performing fundamental studies of subcellular, intracellular and cell surface interactions. Applications range from patterning of neuronal networks,1, 2 control of spindle formation,3 stem cell studies,4–9 gene expression monitoring,10–12 design and manufacture of implant biomaterial,13 drug screening,14–16 and cell-based biosensors.17, 18 One general scheme for cell arraying employs modifying the array surface to create regions permissive to cell attachment and growth with non-permissive areas surrounding these regions. Such patterned surfaces can be created by microcontact printing,19 photolithography,20 injection bioprinting,9, 21 optical tweezers,22 or magnetic forces.8 These surfaces have been used to create arrays of single or groups of cells, and have been used to control cell and colony morphology. This approach is used for adherent cell types that naturally grow attached to a surface.19, 23 A drawback to these arrays is that under most circumstances individual cells displaying a unique attribute (cell surface marker, morphology, etc.) cannot be isolated for further study due to their attachment to the culture surface. Recently, Revzin’s group successfully demonstrated the isolation of groups of cultured hepatocytes for genetic analysis from a micropatterned glass surface using a commercial laser microdissection system.24 Unfortunately, this cell retrieval process is inherently damaging to the collected cells, and isolation of viable cells from a micropatterned substrate has yet to be demonstrated.25
Another array format involves the creation of microwells whose walls serve as a physical barrier to capture and maintain cells in highly localized regions. These structures may be created by etching, molding, or embossing wells into a substrate, or by creating three dimensional structures on the substrate’s surface.4, 5, 26, 27 Since the wells act to keep cells in place with a physical barrier, both non-adherent and adherent cell types have been arrayed successfully. While in general more effort may be required to create these features, the structures are stable over time and chemical modification of their surface properties by deposition of biological molecules does not interfere with their barrier function. Recent technological innovation has made it possible to isolate nonadherent cells cultured in microwells. Toner’s group has utilized laser microdissection to retrieve nonviable lymphocytes from individual wells of a microwell array.27 Voldman’s group combined optical tweezers and microfluidic techniques to enable targeted lymphocytes to be manipulated into a defined region of a microfabricated device for collection by pipet.26 Prior literature on optical manipulation suggested cell viability would be retained, although a viability assay or expansion of isolated cells was not accomplished. To date, a technique to sort and collect viable adherent cells using a microwell format has yet to be demonstrated.
The Allbritton Group has pursued a third approach for isolating adherent cells by creating an array of transparent micron-scale posts (termed micropallets) formed on a glass substrate.28–30 The individual micropallets are removed using a pulsed laser and collected with their cells attached.31 To position cells on the pallets, the substrate between the micropallets must be chemically modified so that a continuous region of air (a “virtual wall”) is trapped between the array elements.32 Cells can then only attach to the micropallet upper surface. Isolation of cells cultured on the array relies on the virtual wall to maintain segregation of the cells on individual micropallets. This approach of creating a barrier of air presents a number of drawbacks. Silane-based surface modification with a hydrophobic moiety is required to create a hydrophobic region between the micropallets to entrap air. The Si-O bond formed with the glass surface is subject to hydrolysis, which necessitates stringent storage conditions prior to use and limits the length of time that air will remain trapped under cell culture conditions. Extracellular matrix proteins secreted by cells can also destabilize air entrapment. Furthermore, the geometry of the array is limited by the need to maintain the entrapped air between the array elements.32 Physical walls composed of the hydrogel poly(ethylene glycol) (PEG) have been used to replace the air walls.33 This material enables greater flexibility in the spacing of the arrayed elements, but also requires chemical modification of the glass substrate to promote PEG adhesion and significantly increases the effort needed to fabricate the array. In addition, swelling of the hydrogel upon immersion of the array in culture media results in friction between the PEG wall and the micropallets. This friction necessitates higher laser energies to release the micropallets which may influence the viability of some cell types. Finally, only adherent cell types can be isolated using the micropallet format.
The current work describes a new and unique array of releasable elements that combines the capabilities and advantages of the microwell and micropallet array formats. A two-step process is used to produce arrays of releasable micron-scale cups on a glass substrate. The efficiency of capturing cells as a function of percent total surface area occupied by the cups is determined and designs to enhance capture efficiency are studied. Culture of viable cells within the cups is demonstrated by viability assay and by generating and maintaining clonal colonies in culture. Isolation of individual viable cells is demonstrated by highly efficient single-cell cloning. Direct comparisons with cell isolation by fluorescence-activated cell sorting and magnetic-bead-based isolation systems demonstrated that the microcup cell isolation procedure produced higher purity, yield and viability for small sample sizes such as those composed of only one-thousand cells. The use of the technique for isolation of stem cells from a sample composed of only 4,000 cells obtained from a bone marrow aspirate show the potential of this new cell isolation technology.
EXPERIMENTAL SECTION
Supplemental Experimental Materials and Methods
Details are presented in the Supporting Information for the following: materials, electron microscopy, cell culture, cell capture, viability assay, fabrication of collection plate, microcup collection, conjugation of Dynabeads with antibody, and processing of human bone marrow mononuclear cells.
Fabrication of the Microcup Arrays
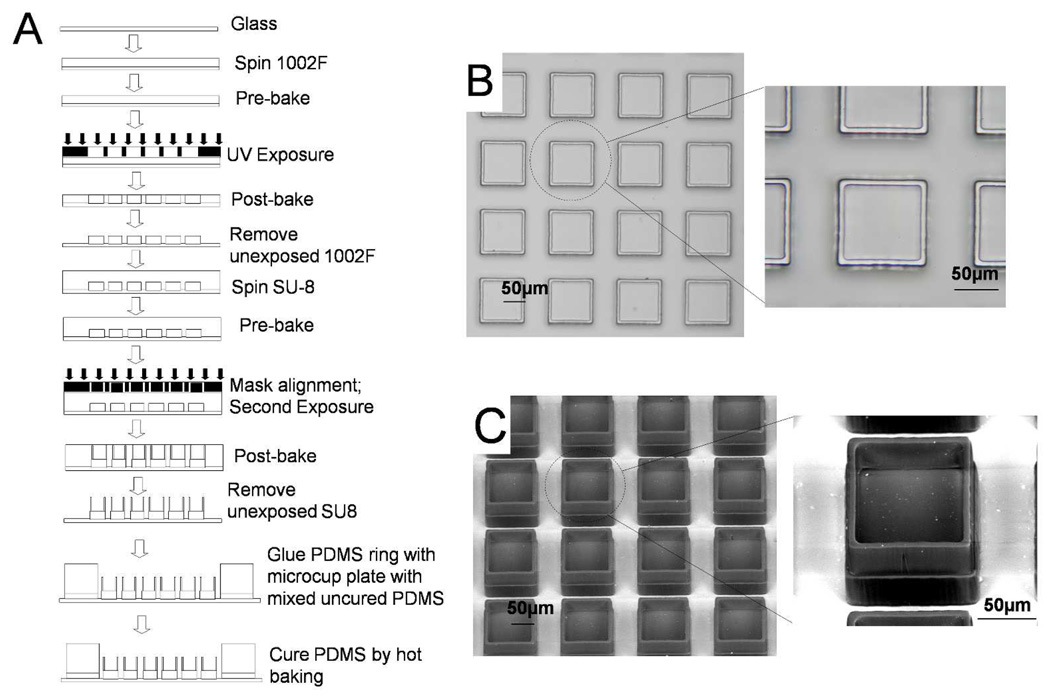
The fabrication of the microcup used a standard multilayer microfabrication process as shown in Fig. 1A. The photoresist 1002F-50 was prepared as previously described, and was then spun on glass slides using a spin coater (WS-400B-6NPP/LITE, Laurell Technologies Corporation, North Wales, PA) to create a 50 µm thick film by a two-step spin: an initial spin of 10 s at 500 rpm followed by a spin of 30 s at 2200 rpm.33 The coated slides were then soft baked in a convection oven (Isotemp Oven, Fisher Scientific, Pittsburgh, PA) at 95 °C for 45 min to remove organic solvent. After baking, the slides were allowed to cool to room temperature (~40 min). The first layer of the microcup structure, i.e. the cup base, was prepared by exposing the coated slides through a chrome mask using a collimated UV source (76 mW/cm2, Oriel Model #97435, Newport Inc., Stratford, CT). The post-exposure bake was performed by baking exposed slides in the convection oven for 10 min. The slides were then cooled to room temperature. After cooling, the slides were developed in a bath of SU-8 developer in a 100 mm glass Petri dish for 6 min, rinsed with 2-propanol, and blown dry in a stream of nitrogen. The dried slides were placed on a hot plate (825-HP, VWR, West Chester, PA) for 2 hrs at 95 °C to perform the hard bake. After cooling, the slides were inspected under a microscope to confirm the 1002F base fabrication was achieved before fabrication of the microcup wall. In some experiments, row numbers and column numbers were fabricated on the edges of the array to ease identification and tracking of the individual microcups during cell culture.
Figure 1.
Fabrication of microcup arrays. (A) Schematic of the process flow for fabrication of the microcup array. (B) Brightfield image of a section of a completed microcup array. (C) ESEM photo of the same array.
Prior to fabricating the microcup walls, the arrays were treated for 15 min in a plasma cleaner (Harrick Plasma, Ithaca, NY) to generate a hydrophilic surface, thus preventing air bubbles from being trapped between the base structures during the second photoresist spin cycle. After surface treatment, SU-8–100 was spun on the slides in a similar manner as the 1002F except that a speed of 1800 rpm was used in the second spin step. The slides were then baked in the convection oven at 95 °C for 25 min to remove the organic solvent. After cooling, the slides were aligned with the second chrome mask and UV exposed (MA6, SUSS Microtec, Germany) to create the wall. The arrays were then subjected to a post-exposure bake of 10 min at 95 °C, and allowed to cool to room temperature over a 45 min period. The developing process after exposure was a two step process. The initial step was carried out in a bath of SU-8 developer for 5 min followed by a 7 min SU-8 developer spray using a squeeze bottle. The slides were then rinsed with isopropyl alcohol followed by deionized water and dried on a hotplate at 95 °C for 1 hr. A reservoir was created for each array using molded PDMS glued to the glass substrate with a thin layer of PDMS.
Laser-based Microcup Release
Release of microfabricated elements similar to the microcups using a laser-based method has been previously described.30 Briefly, a laser pulse (5 ns, 532 nm) from a Q-switch Nd:YAG laser (Minilite I, Continuum Electro-Optics Inc., Santa Clara, CA) was focused by a microscope objective at the interface of the microcup base and glass substrate. The focused pulse formed a plasma and cavitation bubble. The expansion of the cavitation bubble between the base of the microcup and glass substrate dislodged the microcup.31 To minimize the laser energies used to release an individual microcup, a series of pulses directed at different areas of the microcup base was used (energy per pulse = 3 µJ, avg. pulse number = 10).
Fluorescence Activated Cell Sorting (FACS)
Samples composed of a known number of RBL and GFP-expressing 3T3 cells were suspended in DMEM with 1% FBS (1 × 106 cells in 0.5 mL media and 1 × 103 cells in 200 µL media) and then transported to the flow cytometry core facility where an experienced technician performed the sort using a commercial flow sorter (MoFlo, Beckman-Coulter, Brea, CA). Cells were sorted based on forward scatter, side scatter, and GFP fluorescence using a singlet gate and a 100 µm tip. Before sorting, the FACS system required 6,000 – 11,000 cells from the sample in order to set the sort parameters. After set-up, the remaining cells in each sample were sorted. In one experiment, a single-cell sort was performed using the single-cell deposition accessory (Auto Clone, Beckman-Coulter, Brea). Individual cells were deposited into wells of a 96-well glass bottom plate preloaded with 100 µL DMEM with 10% FBS. The remainder of the sorted cells were deposited into a single well containing 100 µL DMEM with 10% FBS. These cells were used to determine yield and purity. The cells deposited on the 96-well plate were examined by microscopy and those wells containing cells were identified. The cells were then cultured in conditioned media under standard tissue culture conditions for 72 hrs. After that time, the cells were again examined and colony formation determined as the measure of viability.
Magnetic Cell Sorting
Magnetic cell sorting was performed using the Dynal Magnetic Cell Separation System (Dynabeads M-280, Invitrogen, Carlsbad, CA) in a negative selection mode. Dynabeads (2 × 108) were labeled with the capture antibody according to the manufacturer’s protocol (Supplementary Data). For each experiment, two samples (103 and 106 cells) containing a known number of RBL and GFP-expressing 3T3 cells were each suspended in a 1.5 mL centrifuge tube containing 1 mL cell separation buffer (PBS without Ca2+ and Mg2+, 0.1% (w/v) BSA and 2 mM EDTA, pH 7.4). Each sample was mixed with Dynabeads then incubated on a roller at 4 °C for 30 min. The suspension was placed in the magnet to pellet the Dynabeads and unwanted cells. The supernatant containing the suspension of target cells was removed by pipet and analyzed for yield, purity and viability. For the sample initially containing 103 cells, the supernatant was transferred under sterile conditions directly to a collection dish where fluorescence microscopy was used to determine yield and purity. The cells were placed in standard tissue culture conditions in conditioned media for 72 hr at which time cell viability was determined. For the sample initially containing 106 cells, the suspension obtained after the magnetic separation step was gently mixed, and cell number determined using a standard hemocytometer. A 100 µL aliquot was transferred to a collection dish where fluorescence microscopy was used to determine purity. The cells were then cultured as described above to determine viability at 72 hr.
RESULTS AND DISCUSSION
Cell Capture
The motivation for the current work was to fabricate a cell-based array with releasable elements that did not require chemical modification of the underlying glass substrate and that could be used for isolation of nonadherent and adherent cell types. The approach was aimed at overcoming the stability and manufacturing issues generated by the air or PEG barriers which require chemical modifications of the array substrate. An array of cup-shaped structures that could serve as a cell trap confining cells during culture and expansion was fabricated on a glass slide (Fig. 1A). Microcup dimensions were as follows unless otherwise noted. The side of each microcup was 100 ± 0.5 µm (n = 30) with a height of 50 µm. The height of the wall forming the microcups was 40 µm with a thickness of 10 µm. Evaluation of the structure of fabricated microcups by brightfield microscopy and ESEM confirmed their dimensions as well as the correct alignment of the base and wall (Fig. 1B & C).
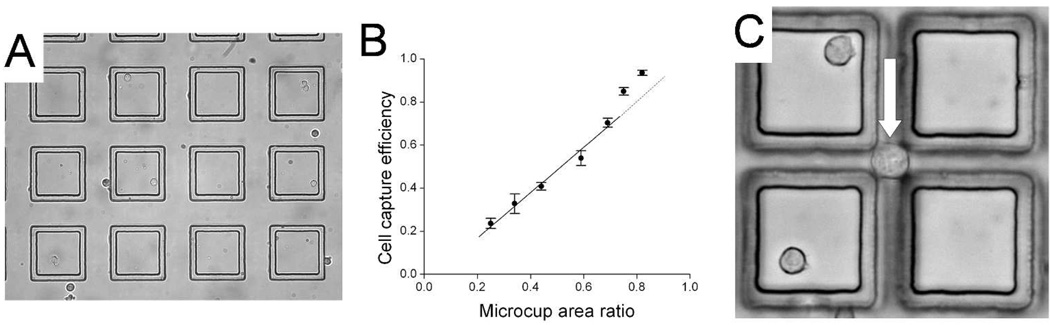
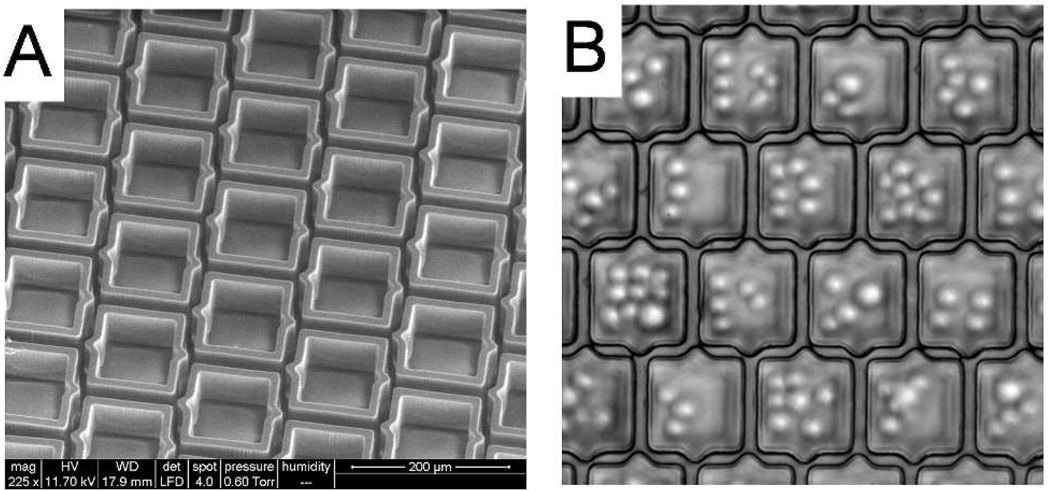
When Ba/F3 cells (mean diameter of 15 µm ± 5) were plated on the array, cells settled both inside the microcups and in the gaps between the microcups (Fig. 2A). Under the conditions used (50 µm gap), the percentage of cells in the gaps between the microcups was 40 ± 2% (n = 3). A series of experiments was then undertaken to further assess and optimize the capture efficiency of the array. The capture efficiency was defined as the number of cells settling within the microcups vs. the total number of cells present in the field of evaluation. The microcup area ratio was defined as the area of the array covered by the microcups divided by the total area of the array. Not surprisingly, these experiments (see Supplemental Data, Fig. S1) indicated that gap size between the cups was the critical factor in designing the microcup arrays to maximize cell capture efficiency. Nevertheless, even in those experiments using a 10 µm gap which was less than the mean diameter of the cells being captured, the capture efficiency did not reach 100% (Fig. 2B). Further experiments identified the source of cell loss as primarily due to the settling of cells within intersections where the diagonal dimension was greater than the gap size (Fig. 2C, Supplemental Data and Figs. S2A & B). While further reduction of the gap could help to reduce the size of this region, the challenge of microfabricating such closely spaced elements became a limiting factor. To solve this problem, redesign of the shape and registration of the microcups was undertaken as shown in Fig. 3A. This interdigitated design created an array in which no inter-cup region would be greater in size than the gap, and which could be manufactured without increasing fabrication complexity. A microcup array using this design (cup size 70 µm, gap 15 µm, microcup area ratio 68%) was produced using the identical microfabrication process as that used for the square microcups. Cell capture efficiency was again assessed. With the newly designed arrays, no cells were observed in the gap intersection bounded by three microcups even at a cell density 8-fold greater than the total number of microcups (Fig. 3B). Furthermore, despite the decrease in the microcup area ratio compared with the earlier design (68% vs. 76% - see Supplemental Data), the cell capture efficiency using this new design increased from 77% to 90% ± 5% (n = 3, Fig. S2C). The 10% of cells not trapped in the microcups were seen to be straddling the upper aspect of adjacent microcup walls. The capture efficiency was enhanced to 98% ± 3% (n = 3) by gently agitating the media in the array during cell plating so that cells coming to rest on the walls were induced to settle into the microcups.
Figure 2.
Cell capture in square microcups. (A) Ba/F3 cells plated on a microcup array. Cells were present both within the microcup and in the inter-cup regions. (B) Plot of capture efficiency vs. microcup area ratio. The solid line is the best fit of the data points between the microcup area ratios of 0.25 to 0.7 to a straight line. (C) Image of a Ba/F3 cell with a diameter of ~20 µm settled on the glass substrate in the intersection of gaps formed by microcups with 70 µm sides and 15 µm gap.
Figure 3.
Interdigitated microcup design. (A) ESEM image of interdigitated microcups. (B) Image of Ba/F3 cells captured in the interdigitated microcups. No cells can be seen settled in the intersections of gaps between neighboring microcups. The focal plane is at the base of the microcup and glass substrate so that cells within the microcups are out of focus.
Culture of Cells on the Microcup Arrays
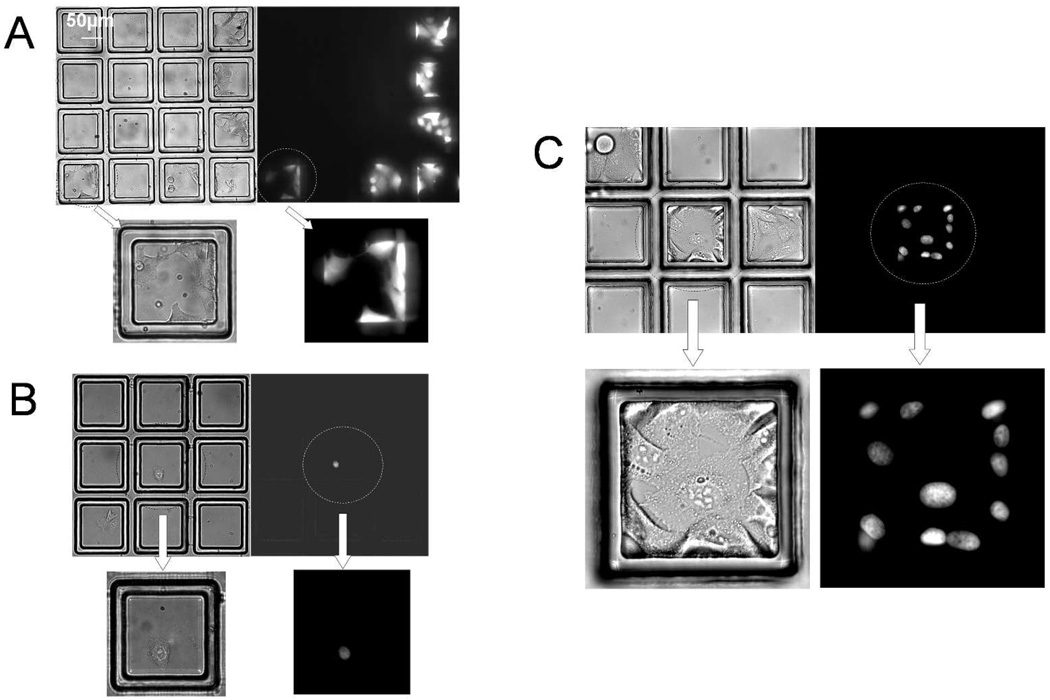
Although cells cultured in microcups appeared to be healthy based on their morphology, cell viability was formally tested with a viability dye and by following cell division on the arrays. HeLa cells in suspension (1,000 cells in total) were plated on a microcup array (gap 15 µm, nmicrocup = 1,200) and placed under standard tissue culture conditions for 72 hr. While still adherent within the microcups, the cells were labeled with the viability dye Calcein Red-Orange and observed by brightfield and fluorescence microscopy. The result demonstrated 100% of HeLa cells (≥100 cells analyzed per array) cultured within the microcups remained viable (n = 3, Fig. 4A).
Figure 4.
Testing cell viability and growth on microcup arrays. (A) Test of cell viability: brightfield and fluorescence images of wild type Hela cells cultured on a microcup array for 72 hr and then labeled with Calcein Red-Orange. (B–C) Test of cell replication: brightfield and fluorescence images of HeLa cells captured in microcups 1 hr after plating (B) and after 96 hr in culture (C). (B) The cell in the center microcup possesses a fluorescent nucleus due to stable expression of a GFP/histone-H1 fusion protein. (C) Images of a region of the microcup array containing a clonal colony of GFP/histone-H1-expressing HeLa cells and colonies of non-fluorescent cells in adjacent microcups.
Assessment of the expansion of single cells into colonies on the array provided both an indication of cell health and a measure of the ability of the microcups to sequester captured cells over time. For these experiments, two cells lines that could be differentiated based on their fluorescence properties were used. To determine if single cells cultured in the microcups grew into colonies and remained localized to the microcup, a 1:10 mixture of HeLa cells stably transfected with a nuclear GFP fusion protein and wild-type HeLa cells were plated (103 total cells) on an array identical to that described in the previous paragraph. The number of cells plated was arrived at empirically to provide a majority of individual microcups possessing either 1 or 0 cells. Cell expansion and sequestration was tested by following individual microcups containing single fluorescent HeLa cells over a 4 day period in culture. The array was imaged by bright field and fluorescence microscopy 1 hr after plating the cells. Microcups (n = 5) were identified that contained a single fluorescent target cell and which were adjacent to one or more microcups containing non-fluorescent cells. The arrays were then placed under standard tissue culture conditions and followed at 24 hr intervals to track growth of the cells within the microcups. Each target cell was seen to grow into a colony composed of cells with fluorescent nuclei (Fig. 4B & C). Fluorescent cells remained sequestered for the duration of the experiment with an average of 16 ± 5 cells present in the microcups at the 96 hr time point. No non-fluorescent cells were seen within the colonies indicating that for these culture times non-target cells did not admix with the target cells. These results clearly show that cells remain viable and sequestered during culture to enable generation of clonal colonies on the array. Sequestration of a rapidly growing cell line such as HeLa for at least 4 days compares very favorably with other literature in which cell arrays based on patterned chemical surface modifications retained this cell type within the cell islands for less than 72 hr.19
Isolation of Single Cells with the Microcups
To determine if single cells could be isolated in a viable manner and clonally expanded, wild-type HeLa cells were plated on an array of square microcups. Microcups with single cells were identified and released (Fig. S3A & B). Following release, the detached microcups were collected using a pipet. All cells (n = 7) remained attached after release and transfer to a Petri dish for further expansion (Fig. S3C). In this single experiment, after 13 days in culture the single cells collected grew into clonal colonies (Fig. S3D). These data clearly demonstrated the practicality of viable cell isolation and single-cell cloning using the microcup array.
Cell Protection by the Microcups
It was reasoned that the 3-dimensional structure provided by the microcup might offer enhanced protection to cells during release and collection, thus enhancing post collection viability. Two different cell types, HeLa and 3T3 (103 cells per array) were plated on two microcup arrays (size 100 µm square, gap 50 µm, nmicrocup = 1,200) and two standard micropallet arrays (size 100 µm square, gap 50 µm, nmicropallet=1,200). Then the arrays were placed under standard tissue culture conditions for 1 hr to allow for cell attachment. Microcups and micropallets containing single cells were released (n = 3 independent experiments). The released microcups or micropallets were collected into a 35 mm Petri dish using a standard 5 mL pipette. Collected microcups or micropallets were observed under brightfield to determine whether the cell was retained during the collection process. In each experiment, ten microcups or micropallets were collected and evaluated. For standard micropallets, the cell loss was 40% ± 10% for the 3T3 cells and 53% ± 12% for the HeLa cells using this pipette collection technique. The cell loss for the microcups was much reduced with only 3% ± 5% for both the 3T3 cells and HeLa cells. To evaluate cell viability, the collected microcups and micropallets were placed under standard tissue culture conditions. After 48 hr, the micropallets and microcups were evaluated for expansion of the retained cells into colonies. For the micropallets plated with 3T3 cells 50% ± 15% possessed colonies, whereas those with HeLa cells showed expansion in only 40% ± 17% of collected micropallets. In contrast, microcups containing 3T3 cells possessed colonies in 80% ± 10% while 90% ± 10% of the HeLa-containing microcups possessed colonies. These results clearly show the 3-dimensional structure of the microcups improve the efficiency of collecting viable cells.
Comparison of the Microcup Array with FACS
The microcup array cell isolation procedure was compared with cell isolation by FACS. A standardized population of cells was prepared for these experiments by mixing the non-fluorescent wild type cell line rat basophil leukemia (RBL) with a fluorescent cell line of NIH 3T3 cells stably transfected with GFP. The non-fluorescent:fluorescent cells were mixed at ratio of 99:1. All comparison experiments were performed with samples composed of either one-million cells or one-thousand cells. On the day of the experiment, a single mixed population of cells in suspension was prepared and aliquoted in appropriate numbers for the comparisons to assure identical samples were being used with each technology. The arrays used for these experiments were composed of interdigitated microcups 100 µm in size with a gap of 15µm and nmicrocup = 10,800. All cell isolation experiments were repeated with 3 independent samples.
To accomplish the FACS experiments with the two sample sizes, the sample containing 106 cells was performed initially utilizing a portion of the cells to adjust the gates for the sort. Then the remainder of the cells in that sample were sorted as described in the methods. The second sample containing 103 cells was then run using the gates defined with the 106 cell sample. The results for the 106 cell sample are shown in Table 1. The sample containing 103 cells yielded no cells in the output of the sorter in all 3 independent experiments. Conversely, the microcup array was not tested with a sample composed of 106 cells. At this stage, the platform remains a manual system for both analysis and cell isolation. It was, therefore, deemed unfeasible to attempt the analysis and isolation of target cells from this large a sample. On the other hand, the sample containing 103 cells was easily scanned and target cells isolated manually with near 100% purity, yield and cell viability (Table I).
TABLE I.
| FACS | Magnetic Cell Separation | |||||
|---|---|---|---|---|---|---|
| FACS 106 cells |
FACS 103 cells |
Microcup 103 cells |
Magnetic 106 cells |
Magnetic 103 cells |
Microcup 103 cells |
|
| Purity | 87 ± 4% | * | 100 ± 0% | 76% ± 6% | 14 ± 10% | 100 ± 0% |
| Yield | 30 ± 13% | * | 120 ± 20% | 98 ± 9% | 30 ± 10% | 130 ± 10% |
| Viability | 32 ± 7% | * | 97 ± 3% | 74 ± 6% | 61 ± 35% | 97 ± 3% |
| Time# | 60 mins | * | 100 mins | 60 mins | 60 mins | 110 mins |
Unsuccessful- see text
Total time required for cell preparation and isolation.
Comparison of the Microcup Array with Magnetic Cell Separation
The microcup array cell isolation procedure was compared with magnetic-bead-based cell isolation. A mixture of RBL and 3T3 cells was prepared in an identical fashion to that for the FACS comparison experiments. Cells were isolated by the commonly used negative selection protocol with the Dynabead magnetic cell sorting system. In this approach, the beads are conjugated with an antibody that recognizes a cell surface protein expressed by the non-target cells which is not expressed by the target cells.34 Unwanted cells bind to the beads which are retained by the magnet, and target cells are collected. Antibody to CD117 (aka c-kit receptor) was used to capture the non-target RBL cells.35 Target cells from both the 103 and 106 samples were collected with yield, purity and viability as shown in Table I. To compare the microcup array in an equivalent negative selection approach, a sample of 103 cells was prepared in which the non-target RBL cells were first immunofluorescently labeled with the red fluorophores Cy5 (Supplementary Data). Cells were plated on microarrays identical to the arrays in the FACS comparison, which were then analyzed under brightfield and epifluorescence (620ex/700em). To isolate cells by negative selection, cells not fluorescent at the Cy5 wavelength were identified and collected. Collected cells were then analyzed with both Cy5 and GFP filter sets to assess yield and purity (Table I).
Isolation of Stem Cells
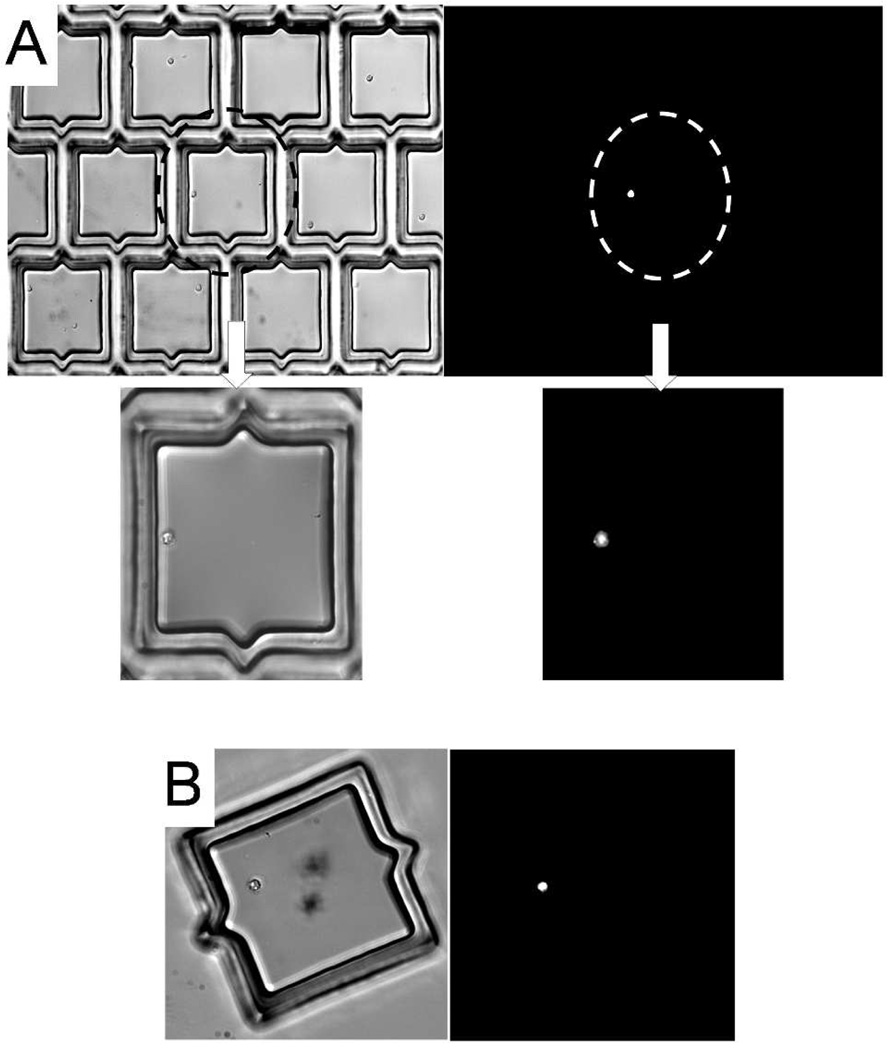
The data above suggested that the microcup array could be used to identify and isolate primary cells in samples composed of small numbers of cells. The feasibility of using the microcup arrays for isolating a clinically relevant subpopulation of cells from a small sample of primary cells was tested. These experiments used human bone marrow mononuclear cells (BMNCs), a non-adherent cell type obtained from a bone marrow aspirate. The cells were stained with antibody against the CD133 surface antigen and plated on microcup arrays. CD133 is a surface antigen that has been well validated as a marker of hematopoietic stem cells and cancer cells possessing stem-cell characteristics.36, 37 CD133+ cells typically make up less than 1–2% of cells obtained from bone marrow aspirates.38 After incubation with FITC-labeled anti-CD133, human BMNCs (4,000 cells) were plated on an array (interdigitated microcups 100 µm in size, gap 15 µm, nmicrocup = 20,000). In these experiments, the capture efficiency was 98% ± 2% (n = 3). CD133+ cells were identified by fluorescence microscopy. In these analyses, 1% ± 0.2% of cells in the microcups were found to be fluorescent (Fig. 5A). Positive cells (n = 5) were identified, released and collected. After collection, the collected microcups were re-examined under by fluorescence microscopy to confirm that all cells contained in the cups were CD133+ (Fig. 5B). This experiment demonstrates the feasibility of using the microcup array to isolate a targeted subpopulation of primary cells from a small sample.
Figure 5.
Isolation of human stem cells.
(A) Brightfield and fluorescence images of a CD133+ cell segregated from multiple CD133− cells contained within microcups. (B) Brightfield and fluorescence images of a released and collected microcup containing a CD133+ BMNC.
CONCLUSION
A novel array composed of releasable microcup structures was fabricated by standard multilayer microfabrication processes. By matching the spacing of the structures to the cell size, cells loaded onto the array could be localized inside the microcups and excluded from the region between the structures to maximize capture efficiency. This design offers a number of advantages for cell-based arrays. First and foremost, a cell or cells of interest can be retrieved from the array while maintaining cell viability. Furthermore, the microcup arrays are compatible with both non-adherent and adherent cell types. The microcup wall acts as a physical barrier to simplify generation of the cell array by eradicating the chemical surface modification steps that are usually required for cell patterning. This physical barrier also remains constant over time without concern for the stability of the chemistries used to tailor the array surface. Importantly, high cell viability after release and collection was demonstrated by the efficient cloning of single cells using the microcup platform. The laser energy used to release the microcups (2 × 10−4 TJ/m2) is 50,000-fold less than the minimum energy required for detecting DNA damage after exposure to laser irradiation in other studies.39 This fact combined with the high cloning efficiency seen in the current and our previous studies strongly suggests that DNA damage is not an issue. In the current experiments, cells cultured in the microcups did not show evidence of growth arrest and continued to expand on the array, and after release and collection. It is conceivable that some cell types when cultured in a space limited region may undergo cell cycle arrest, thus restricting the expansion of the cell colony.40 This issue can be ameliorated by increasing the surface area of the microcup. For small sample sizes, purity, yield and cell viability were better than that seen with the standard technologies of FACS and Dynabeads. The microcup array was shown to be capable of performing efficient cell isolation with samples as small as 1,000 cells. The experiments to isolate CD133+ cells demonstrated a “real world” example of the ability to isolate stem cells from a very small sample, such as might be acquired from a biopsy or small animal model. This is in contrast to traditional sorting techniques, such as FACS or magnetic cell sorting where typical starting samples are composed of 105 – 106 cells.25, 41 The data presented in this paper validate the microcup array as a simple, inexpensive, and efficient means to identify and isolate viable cells from a mixed population for further analysis or expansion.
Supplementary Material
ACKNOWLEDGEMENT
This research was supported by the NIH (EB007612 and HG004843).
Footnotes
SUPPORTING INFORMATION AVAILABLE
Further details and data are provided in Supplemental Data. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Nam Y, Chang JC, Wheeler BC, Brewer GJ. IEEE Trans. Bio. Med. Eng. 2004;51:158–165. doi: 10.1109/TBME.2003.820336. [DOI] [PubMed] [Google Scholar]
- 2.Suzuki I, Sugio Y, Jimbo Y, Yasuda K. Lab Chip. 2005;5:241–247. doi: 10.1039/b406885h. [DOI] [PubMed] [Google Scholar]
- 3.Thery M, Racine V, Pepin A, Piel M, Chen Y, Sibarita JB, Bornens M. Nat. Cell Biol. 2005;7:947–953. doi: 10.1038/ncb1307. [DOI] [PubMed] [Google Scholar]
- 4.Chin VI, Taupin P, Sanga S, Scheel J, Gage FH, Bhatia SN. Biotechnol. Bioeng. 2004;88:399–415. doi: 10.1002/bit.20254. [DOI] [PubMed] [Google Scholar]
- 5.Parekkadan B, Berdichevsky Y, Irimia D, Leeder A, Yarmush G, Toner M, Levine JB, Yarmush ML. Neurosci Lett. 2008;438:190–195. doi: 10.1016/j.neulet.2008.03.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ong SM, Zhang C, Toh YC, Kim SH, Foo HL, Tan CH, van Noort D, Park S, Yu H. Biomaterials. 2008;29:3237–3244. doi: 10.1016/j.biomaterials.2008.04.022. [DOI] [PubMed] [Google Scholar]
- 7.Ungrin MD, Joshi C, Nica A, Bauwens C, Zandstra PW. PloS one. 2008;3:e1565. doi: 10.1371/journal.pone.0001565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ino K, Okochi M, Honda H. Biotechnol. Bioeng. 2009;102:882–890. doi: 10.1002/bit.22104. [DOI] [PubMed] [Google Scholar]
- 9.Phillippi JA, Miller E, Weiss L, Huard J, Waggoner A, Campbell P. Stem Cells. 2008;26:127–134. doi: 10.1634/stemcells.2007-0520. [DOI] [PubMed] [Google Scholar]
- 10.Baghdoyan S, Roupioz Y, Pitaval A, Castel D, Khomyakova E, Papine A, Soussaline F, Gidrol X. Nucleic Acids Res. 2004;32:e77. doi: 10.1093/nar/gnh074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng X, Guerasimova A, Manke T, Rosenstiel P, Haas S, Warnatz HJ, Querfurth R, Nietfeld W, Vanhecke D, Lehrach H, Yaspo ML, Janitz M. Gene. 2009 doi: 10.1016/j.gene.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 12.Erfle H, Neumann B, Liebel U, Rogers P, Held M, Walter T, Ellenberg J, Pepperkok R. Nat. Protoc. 2007;2:392–399. doi: 10.1038/nprot.2006.483. [DOI] [PubMed] [Google Scholar]
- 13.Ito Y. Biomaterials. 1999;20:2333–2342. doi: 10.1016/s0142-9612(99)00162-3. [DOI] [PubMed] [Google Scholar]
- 14.Bhadriraju K, Chen CS. Drug Discovery Today. 2002;7:612–620. doi: 10.1016/s1359-6446(02)02273-0. [DOI] [PubMed] [Google Scholar]
- 15.Stett A, Egert U, Guenther E, Hofmann F, Meyer T, Nisch W, Haemmerle H. Anal. Bioanal.Chem. 2003;377:486–495. doi: 10.1007/s00216-003-2149-x. [DOI] [PubMed] [Google Scholar]
- 16.Wu MH, Huang SB, Cui ZF, Cui Z, Lee GB. Sens. Actuators, B. 2008;129:231–240. [Google Scholar]
- 17.Gu MB, Mitchell RJ, Kim BC. Adv. Biochem. Eng./Biotechnol. 2004;87:269–305. doi: 10.1007/b13533. [DOI] [PubMed] [Google Scholar]
- 18.Pancrazio JJ, Whelan JP, Borkholder DA, Ma W, Stenger DA. Ann. Biomed. Eng. 1999;27:697–711. doi: 10.1114/1.225. [DOI] [PubMed] [Google Scholar]
- 19.Rozkiewicz DI, Kraan Y, Werten MW, de Wolf FA, Subramaniam V, Ravoo BJ, Reinhoudt DN. Chem. Eur. J. 2006;12:6290–6297. doi: 10.1002/chem.200501554. [DOI] [PubMed] [Google Scholar]
- 20.Hahn MS, Taite LJ, Moon JJ, Rowland MC, Ruffino KA, West JL. Biomaterials. 2006;27:2519–2524. doi: 10.1016/j.biomaterials.2005.11.045. [DOI] [PubMed] [Google Scholar]
- 21.Roth EA, Xu T, Das M, Gregory C, Hickman JJ, Boland T. Biomaterials. 2004;25:3707–3715. doi: 10.1016/j.biomaterials.2003.10.052. [DOI] [PubMed] [Google Scholar]
- 22.Birkbeck AL, Flynn RA, Ozkan M, Song DQ, Gross M, Esener SC. Biomed. Microdevices. 2003;5:47–54. [Google Scholar]
- 23.Lussi JW, Falconnet D, Hubbell JA, Textor M, Csucs G. Biomaterials. 2006;27:2534–2541. doi: 10.1016/j.biomaterials.2005.11.027. [DOI] [PubMed] [Google Scholar]
- 24.Lee JY, Jones C, Zern MA, Revzin A. Anal. Chem. 2006;78:8305–8312. doi: 10.1021/ac0613333. [DOI] [PubMed] [Google Scholar]
- 25.Sims CE, Bachman M, Li GP, Allbritton NL. Anal. Bioanal.Chem. 2007;387:5–8. doi: 10.1007/s00216-006-0612-1. [DOI] [PubMed] [Google Scholar]
- 26.Kovac JR, Voldman J. Anal. Chem. 2007;79:9321–9330. doi: 10.1021/ac071366y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Revzin A, Sekine K, Sin A, Tompkins RG, Toner M. Lab Chip. 2005;5:30–37. doi: 10.1039/b405557h. [DOI] [PubMed] [Google Scholar]
- 28.Shadpour H, Sims CE, Thresher RJ, Allbritton NL. Cytometry Part A. 2009;75:121–129. doi: 10.1002/cyto.a.20672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sims CE, Allbritton NL. Biological Applications of Microfluidics. New York: John Wiley & Sons; 2008. pp. 29–64. [Google Scholar]
- 30.Wang Y, Young G, Aoto PC, Pai JH, Bachman M, Li GP, Sims CE, Allbritton NL. Cytometry Part A. 2007;71:866–874. doi: 10.1002/cyto.a.20424. [DOI] [PubMed] [Google Scholar]
- 31.Quinto-Su PA, To'a Salazar G, Sims CE, Allbritton NL, Venugopalan V. Anal. Chem. 2008;80:4675–4679. doi: 10.1021/ac800129a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y, Sims CE, Marc P, Bachman M, Li GP, Allbritton NL. Langmuir. 2006;22:8257–8262. doi: 10.1021/la061602k. [DOI] [PubMed] [Google Scholar]
- 33.Pai JH, Wang Y, Salazar GT, Sims CE, Bachman M, Li GP, Allbritton NL. Anal. Chem. 2007;79:8774–8780. doi: 10.1021/ac071528q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matheu MP, Cahalan MD. J. Vis. Exp. 2007;9:1. doi: 10.3791/409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ott VL, Moffitt LA, Cambier JC. Signal Transduction. 2005;5:11. [Google Scholar]
- 36.Mehra N, Penning M, Maas J, Beerepoot LV, van Daal N, van Gils CH, Giles RH, Voest EE. Clin. Cancer Res. 2006;12:4859–4866. doi: 10.1158/1078-0432.CCR-06-0422. [DOI] [PubMed] [Google Scholar]
- 37.Warmuth M, Kim S, Gu XJ, Xia G, Adrian F. Curr. Opin. Oncol. 2007;19:55–60. doi: 10.1097/CCO.0b013e328011a25f. [DOI] [PubMed] [Google Scholar]
- 38.Timmermans F, Plum J, Yoder MC, Ingram DA, Vandekerckhove B, Case J. J. Cell. Mol. Med. 2009;13:87–102. doi: 10.1111/j.1582-4934.2008.00598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mohanty SK, Rapp A, Monajembashi S, Gupta PK, Greulich KO. Radiat. Res. 2002;157:378–385. doi: 10.1667/0033-7587(2002)157[0378:camodd]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 40.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Science. 1997;276:1425–1428. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 41.Watson JV. Introduction to Flow Cytometry. New York: Cambridge University Press; 1991. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.